Abstract
Engineering synthetic minimal cells provides a controllable chassis for studying the biochemical principles of natural life: increasing our understanding of complex biological processes. Recently, synthetic cell engineering has enabled communication between both natural live cells and other synthetic cells.
System such as these enable studying interactions between populations of cells, both natural and artificial, and engineering small molecule cell communication protocols for a variety of basic research and practical applications. In this review, we summarize recent progress in engineering communication between synthetic and natural cells, and we speculate about the possible future directions of this work.
Introduction
While often referred to as the “building blocks of life,” living cells are complex, poorly understood entities carrying out millions of chemical reactions every second [1]. The traditional top-down studying of natural cells is complicated by a dense and nebulous network of signaling pathways: membrane-bound and membrane-less organelles, proteins, DNA, RNA, and all other cell constituents and processes that are necessary to sustain life: all contribute to background interference.
On the contrary, building cell mimetic vesicles from the bottom-up, termed synthetic or artificial cells, allows for the isolation of unique variables outside of the complexity of the full molecular and biochemical workings of a natural cell; these synthetic cells can be used to study the biology and biochemistries that allow for life to happen in a less background occluded system.
Biological and biochemical technologies have advanced substantially in recent years; however, the basic concept of artificial cells has remained since it was first proposed by Dr. Thomas Ming Swi Chang in 1957 [2]. Synthetic cells are compartmentalized cell-free systems that mimic the functions of natural cells [3]. The basic chassis of synthetic minimal cells are spherical liposomes, usually around 0.1 to 100 microns in diameter, and often constructed from phospholipids and cholesterol; however, hydrogel droplets and coacervate based systems also exist and provide alternative transduction kinetics and morphologies [4]. Membrane pores in liposome based synthetic cells allow for communication and are created by integrating proteins such as hemolysin into the lipid bilayer. Additionally, transcription and translation machinery including genes, tRNAs, small molecules, other proteins and polymerases necessary for life, such as the ribosome, are encapsulated within a lipid bilayer to create programmable, biomimetic vessels [5].
The engineering of a robust and autopoietic synthetic minimal cell system is the central pursuit of synthetic biology. This burgeoning reductionist methodology can be harnessed to probe the origins of modern life as well as to provide a tool to understand both age-old and emerging biological questions with applications in the biomedical, industrial, and basic science spaces. This review provides commentary on functional aspects of synthetic cells designed from the bottom-up and discusses the development of diverse technologies within recent years that has allowed for advancements in synthetic-synthetic and synthetic-natural communication.
Background
Formation of the compartment
The formation of a phospholipid bilayer is one of the principal methods of bio-compartmentalization and serves as a backbone into which additional chemistries can be combined to functionalize and increase the complexity of synthetic cells. The process of forming nano-scale liposome membranes has been investigated extensively as nano-scale liposomes are of interest in pharmacology; thin film rehydration and subsequent extrusion – which redistributes the upper tail of the size distribution – as well as ethanol injection and microfluidics flow-focusing are amongst the most prominent nano-scale formation methods [6-9].
For the formation of microscale liposomes, thin-film rehydration and extrusion can be used to achieve a size distribution of liposomes which includes micro-scale dimensions; though additionally, a method based on the same principals, where a thin-film is swelled in heated buffer, can be used to form GUVs [10]. Additionally, the same principals which facilitate the formation of liposomes with the reverse emulsion method also have led to the development of microfluidics methods such as droplet-shooting and size-filtration (DSSF), a microcapillary and centrifuge based microfluidics method, as well as octanol-assisted liposome assembly (OLA), an increasingly used PDMS based microfluidics system [11-13]. DLS has shown microfluidics system to be impressively uniform with regard to liposome size distribution which furthers the notion of uniform data associated with synthetic cells.
Membrane characteristic
Computational modeling and molecular dynamics has just begun to compile and elucidate the phospholipid bilayer of living cells and its proteins and cofactors [14]. Similarly, membrane pores like hemolysin are being used to tailor and elicit membrane fluidity and logic-controlled cross-membrane communication in synthetic cells [15,16]. Furthermore, there has been considerable success with regard to reconstituting functional membrane proteins such as ATP-synthase in synthetic cell systems [17,18].
Membrane fluidity and permeability can also be augmented through the presence of divalent ions such as Mg+2 as well as the presence of fatty acids or other amphiphiles within a heterologous phospholipid membrane [19,20]. Membrane permeability can also be affected by the chirality of the lipids, with homochiral membranes exhibiting enantioselective permeability. [21]
Live cells rarely conform to a circular geometry and therefore various efforts have investigated the deformation and shaping of synthetic cell membranes [22]. The confinement of synthetic cells in PDMS based microfluids traps has been used to study internal fluid kinematics of synthetic cells; likewise, cholesterol and the internal polymerization of proteins such as MreB have been used to form tetrahedral and rod-shaped vesicles respectively [23-25].
Growth and division
Living systems must grow and divide to perpetuate. The same is true for artificial cells, thus there is much interest in the development of an autopoietic cell-free system. Without self-production of membrane components, descendants of the initial artificial cell will undergo significant shrinkage [26].
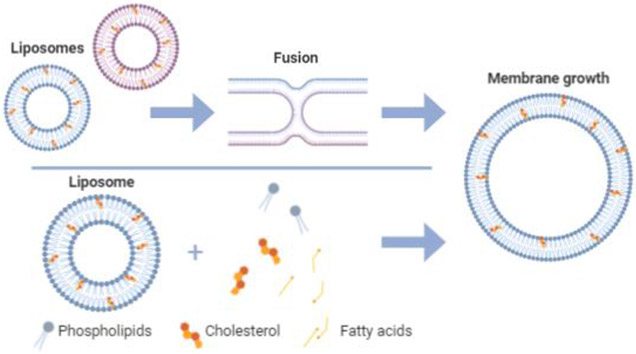
When a cell grows, the lipid bilayer increases in size to incorporate new lipid molecules into the membrane (Figure 1). Various bottom-up efforts have focused on synthesizing lipids within liposomes for incorporation into the membrane to stimulate growth [27-29]. Another approach to liposomal growth describes the fusion of smaller vesicles to create a larger vesicle as mediated by osmotic pressure [30]. It has been shown that the presence of fatty acids or other small molecules can catalyze membrane growth, division, or regulate membrane characteristics such as permeability [31-35] Recently, it was observed that when phospholipid vesicles are incubated in a bath of fatty acids, changes in the concentration of those fatty acids, vesicle size, and osmotic pressure differences across the membrane can lead to insertion of fatty acids into the membrane of vesicles that contain high levels of osmolytes, resulting in vesicle growth [33]. Also due to osmotic pressure, these vesicles became unstable and divided into daughter vesicles. This approach describes liposomal growth and division in one environment, a phenomenon crucial for the development of lifelike synthetic cells.
Figure 1:
Membrane growth of liposomes driven by liposomal fusion and fatty acid/lipid insertion.
Additional methods for catalyzing the division of synthetic cells or vesicles have been investigated, including chemical stimuli, temperature driven self-replication [36], microfluidic approaches [37], the internal organization of polymeric structures [38], osmolarity induced osmotic stress [39-41], and internal lipid or amphiphile synthesis [42,43]. However, a truly de novo system has yet to be described, as the findings reported here are limited by starting reagents or require intervention to induce non-persistent catalysis or reagent reuptake.
Quantitative analysis
The development of characterization methodologies applicable to synthetic cell systems has been of interest in the synthetic cell community for some time. Chiefly, FRET and various electron and light microscopy modalities are used in order to observe membrane growth and stability as well as to validate formation and examine morphology[44,45]. The imaging of liposomes and synthetic cells is confounded due to the fact that liposomes readily wet glass and are therefore effectively annihilated on contact with glass slides or cover slips; however the immobilization of the vesicle with PEG linkers, the treatment of glass with blocking agents such as BSA, or the formation of planar bilayers on cover slips or glass slides has shown to be effective at increasing membrane stability [46]. Similarly, sugars have been shown to increase membrane stability likely due to hydrogen bonding [47]. Parallel work in the field of NMR with regard to 31P spectroscopy and DOSY represent a promising area of study regarding possible synthetic cell related assays which might provide useful insights into vesicle morphology and shape, size, and laminarity as well as the relative amount of encapsulated material within the lumen as opposed to exogenously within solution [48-52].
Genetic robustness
Self-replication of DNA
Perhaps the most critical aspect of creating a functional and prolific synthetic cell is the ability to transcribe and translate DNA into proteins. As discussed, synthetic cells need to grow and divide without losing material to perpetuate; thus, creation of a de novo DNA self-replicating system with transcription and translation capabilities is a significant objective for synthetic biologists. Protein synthesis Using Recombinant Elements (PURE) uses RNA polymerases and machinery from E. coli to synthesize proteins [53,54].
A modified PURE system in conjunction with Phi29-transcription-translation-coupled DNA replication (TTcDR) has been used to self-replicate a 116 kB genome [55]. While this system provides a framework for gene replication, protein expression was at a minimal level. Similarly, this system requires multiple proteins for replication, and a simpler method would be preferred. A subsequent system utilizes rolling-circle replication of circular DNA by a self-synthesized phi29 DNA polymerase and Cre recombinase [56].
DNA logic gates/cascades
Parallel to engineering systems for replication of genomic information, controlling the expression of genes beyond replication machinery is a key objective. Encapsulation of TXTL constituents within a liposome enables a compartment to function as a synthetic minimal cell [57]. This general scheme has enabled the engineering of genetic circuits and cascades within a synthetic cell system, allowing for the control of communication by external signals, the combination of otherwise incompatible reactions, fusion of liposomes, and catalysis of enzymatic reactions [15,58,59].
Boolean logic gates provide a framework for controlling genetic circuits, but the non-binary nor discrete nature of biological signals means that quantum and multi-valued logics may be used in the design of biological circuits. NAND/NOR logic gates have been used to create a modular regulatory network by controlling binary inputs to effect binary outputs [60]. A recent study documents a membrane AND logic gate used to release membrane bound cargo in response to a stimulus, which could be harnessed to work with other genetic circuits to add another level of control within artificial cells [61]. However, differing sizes of liposomes, the effect of crosstalk between circuits, and undesirable interactions between chemical components leave room for further investigation and demonstrates the quasi-evolutionary necessity of further sub-compartmentalization: synthetic organelles.
Communication – between synthetic cells and with natural cells
With other microorganisms
Synthetic cells are an ideal vessel for studying communication from the bottom-up as they can be programmed to carry out select functions without the busy environment of a living cell. There has been much work to create mechanisms of communication among synthetic populations in consortium with natural cells (Figure 3). Harnessing the power of quorum sensing in synthetic cells in conjunction with natural cells has proven to be a useful approach to this challenge. A quorum sensing-based sender-receiver model using IPTG and AHL as inducers has been described [62]. Receiver cells composed of encapsulated bacteria express GFP under the inducible pLuxR promoter in response to AHL produced by sender cells. GFP fluorescence was visualized diffusing into both adjacent and distal receiver cells. Similarly, diffusion was observed from AHL synthesizing bacteria into neighboring receiver droplets. A similar study used bacterial quorum sensing molecules expressed in artificial cells to create a biological-Turing test applied to natural cells to determine the level of bio-mimesis achieved with these artificial cells [63]. Using fluorescence, luminescence, and RNA sequencing to compare the expression of sequences normally produced when V. fischeri bind a quorum molecule, it was found that while some overlap in expression was present, a natural cell’s response to an artificial cell is not identical, thus failing the Turing test. Studying artificial cells in the presence of natural cells provides a substantial opportunity to explore and expand the capabilities of synthetic cells using a well-characterized system.
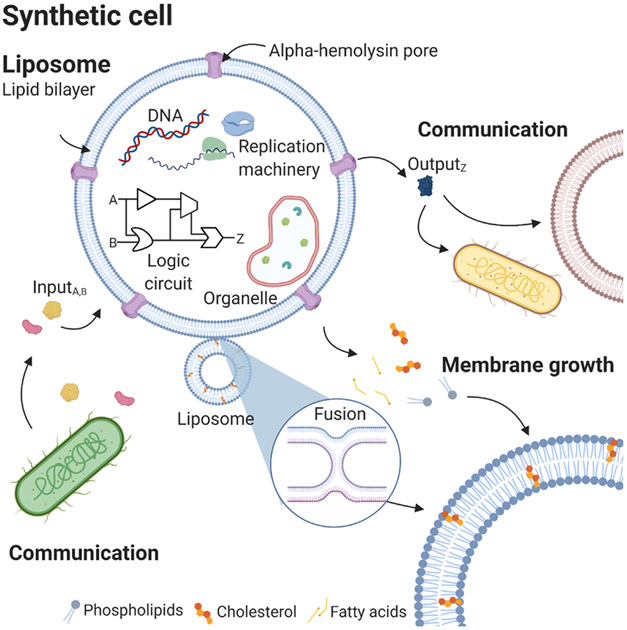
Figure 3: Schematic of synthetic cell.
Synthetic cell encapsulating various DNAs and proteins. Synthetic cells can receive chemical signals from natural cells as well as secrete signals through hemolysin pores to drive communication. Membrane growth can result from fusion with another vesicle or insertion of fatty acids into the existing membrane.
With other synthetic cells
Less is known about communication among entirely synthetic populations. A population of coacervate-based droplets containing proteases were able to electrostatically attach to proteinosomes, a type of protocell with a flexible, semi-permeable membrane constituted of amphiphilic enzymes and other protein-polymer building blocks [64]. After attaching, the coacervate-proteases degraded the proteinosome membrane and seized its internal contents. This work is an exciting display of an exclusively “protocell ecosystem”. While one of the populations was droplet-based, this displays the diversity of possible synthetic ecosystems. Moreover, this work highlights the predatory capabilities of synthetic populations. However, communication between populations of synthetic cells composed of a biomimetic lipid bilayer has yet to be described.
Organelles
The development of discrete intracellular compartments was a major evolutionary breakthrough in terms of eukaryogenesis and facilitated an increase in the complexity of intracellular biochemistry; moreover, it is widely accepted that mitochondria and plastids are the result of precursory endosymbiosis events [65-67]. Because of their ability to biorthogonally increase functionality and complexity, synthetic organelles could be used to build-up or increase the complexity of synthetic cell systems (Figure 3). Various efforts have investigated the engineering of synthetic internal compartmentalization structures based off of natural endosymbionts [68-71], oligolamellar vesicles [17,18], lipid sponge droplets [72], proteinaceous microcompartments [73-75], and coacervate-like structures [76-78]. The inherent complexity of organelle systems from the standpoint of biochemical communication should also be noted. For example, recent work to encapsulate living cells within a liposome epitomizes intramembrane communication between a synthetic cell chassis and a natural cell endosymbiont [68]. Similarly, these intramembrane communication logics associated with organelles can be reconstructed entirely from synthetic components and demonstrate Szostakian compartmentalization.
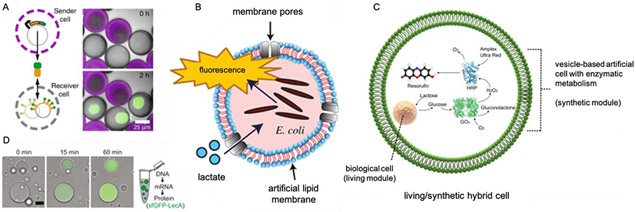
Recently, bottom-up synthetic oligolamellar organelles capable of anabolic catalysis mimicking photosynthesis were successfully engineered, though such systems remain non-autopoietic [17,18]. A clay-based DNA hydrogel was engineered to serve as a means of genetic compartmentalization in synthetic cells, and in addition, a proto-nucleus was shown to sequester transcriptional factors where transduction of such factors intracellularly represents an internal communication or passive logic [78]. Similarly, protein based microcompartments have served as the basis for organelles designed to sequester iron atoms [73], replicate and optimize the function of carboxisomes [75], and serve as a site of cell-free TXTL [74]. Moreover, sponge phase lipid droplets have been used to mimic the interconnected structure of the endoplasmic reticulum and Golgi apparatus from a biophysical or morphological standpoint [72]. Droplet systems were also used to facilitate synthetic cell – like bioreactors for evolution of active enzymes. From a top-down perspective, both prokaryotic, and mammalian cells have been used for the basis of synthetic endosymbionts [68-71]. Encapsulated E. coli can function as a sensing component within the top-down synthetic cell system and exhibit a communication logic whereby environmental lactate results in the fluorescence of the E. coli synthetic organelle [69].
In terms of fully synthetic organelle systems, communication has been observed within an artificial cell between two proto-organelles [79]. In this system, a glucose oxidase (GOx) containing proto-organelle was encapsulated within a GUV, followed by inward budding of the GUV to create a secondary proto-organelles encapsulating horseradish peroxidase and Amplex red. Glucose diffused into the GOx vesicle creating hydrogen peroxide, which then transduced the membrane of the HRP vesicle, oxidizing Amplex red to produce a fluorescent readout. This model describes successful uni-directional communication between fully synthetic organelles, although a reciprocal communication system has yet to be established.
Biosensing
There is a need for the development of synthetic biosensors able to detect small molecules for agricultural, biomedical, and environmental applications. For example, a paper-based, cell-free biosensor that expresses sfGFP in the presence of heavy metals as well as certain date rape drugs has been described [80]. However, artificial cells may allow for expanded capabilities beyond whole-cell and in vitro sensing.
Synthetic cells consisting of encapsulated bacteria have been useful for biosensing efforts. E. coli encapsulated within a liposome has been used to measure lactate levels in the external environment [69]. By using engineered bacteria employing the lldPRD operon that expresses GFP in the presence of lactate, fluorescence can be used to assess the amount of lactate present in a sample. Liposome-based approaches are particularly useful for pathogens that secrete pore-forming toxins, such as Listeriolysin O (LLO) secreted by Listeria monocytogenes (L. monocytogenes). A recent paper describes an assay for detecting the presence of LLO [81]. LLO forms pores on liposomes containing encapsulated cysteine, resulting in the release of the cysteine and the subsequent aggregation of gold nanoparticles. This aggregation results in a colorimetric change from red to purple/blue, denoting the presence of the bacterial toxin.
Encapsulating sensing pathways within the lipid membrane of an artificial cell allows for the compartmentalization of reactions from external environments where they may have been otherwise incompatible. This would represent the creation of simple and programmable bioreactors capable of sensory logics and may be better suited for instances where introducing bacteria or similar microorganisms is undesirable.
Conclusion
Within recent years, there has been sufficient progress in the engineering of biomimetic synthetic minimal cells. Emerging technologies in microfluidics, microscopy, NMR, biophysics, biochemistry, etc. have contributed to advances in synthetic cell formation, growth and division, organelle development, communication, biosensing, protein evolution [81] and the development of robust and self-sustaining genomic constructs. (Figure 3) Looking towards the future, one should expect rigorous research in the entirety of the field of synthetic cell engineering due to its burgeoning exposure. More specifically, it is likely that research will focus on the construction of autopoietic systems capable of more diverse communication with other synthetic cells and natural cells alike. In this way, self-sustaining synthetic cells capable of passing the equivalent of a biological Turing test – that is, the predation on and by natural cells – will be one of the next major evolutions of the field. As a result, the field of synthetic cell research will have to develop a more taxonomically useful characterization methodology for synthetic cell lines, possible arising out of the molecular and biological provenance of constituent parts.
Additionally, it is likely that the design and engineering of synthetic tissues, predicated on the development of synthetic extracellular matrixes, will develop into a robust field of its own withing the next few years.
Figure 2: Synthetic organelles and communication.
A) A porous synthetic cell with a nucleus-like DNA-hydrogel that expresses display proteins and communicates with neighboring synthetic cell through protein diffusion [78]. B) encapsulated E. coli functions as lactate sensor in synthetic cell [69]. C) Internal lipid droplet with sponge morphology enables expression regulation and mimics interconnected membrane morphologies of Golgi apparatus and endoplasmic reticulum [72]. D) internal synthetic cell chemistry communicating with encapsulated bacterial and eukaryotic cells to display logic gate encodable mutualism [68].
Acknowledgments
Figures created with BioRender.com.
A.O.R. is supported by the NIH Training for Future Biotechnology Development Training Grant 9277706 and the NSF Award 1844313. O.M.V. is supported by the NSF Award 1807461. K.P.A. is supported by the John Templeton Foundation award “Engineer synthetic cells of increased complexity and analyze properties of Boolean networks of metabolic reactions inside populations of synthetic cells.”, National Science Foundation grant 1844313 and by the National Aeronautics and Space Administration grant 80NSSC18K1139.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
Papers of particular interest, published within the period of review, have been highlighted as:
* * of outstanding interest
- 1.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P: Catalysis and the use of Energy by Cells. 2002. [Google Scholar]
- 2.Chang TMS: 50th anniversary of artificial cells: their role in biotechnology, nanomedicine, regenerative medicine, blood substitutes, bioencapsulation, cell/stem cell therapy and nanorobotics. Artif Cell Blood Subs Biotechnol 2007, 35:545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Y D, F W, C T: Synthetic biology: a bridge between artificial and natural cells. Basel, Switzerland: Life; 2014, 10.3390/life4041092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akbarzadeh A, Rezaei-Sadabady R, Davaran S, Joo SW, Zarghami N, Hanifehpour Y, Samiei M, Kouhi M, Nejati-Koshki K: Liposome: classification, preparation, and applications. Nanoscale Res Lett 2013, 8:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE: Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science 1996, 274:1859–1866. [DOI] [PubMed] [Google Scholar]
- 6.Daraee H, Etemadi A, Kouhi M, Alimirzalu S, Akbarzadeh A: Application of liposomes in medicine and drug delivery. Artif Cells Nanomed Biotechnol 2016, 44:381–391. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H: Thin-film hydration followed by extrusion method for liposome preparation BT - liposomes: methods and protocols. In Edited by D’Souza GGM. Springer; New York; 2017:17–22. [DOI] [PubMed] [Google Scholar]
- 8.Charcosset C, Juban A, Valour J-P, Urbaniak S, Fessi H: Preparation of liposomes at large scale using the ethanol injection method: effect of scale-up and injection devices. Chem Eng Res Des 2015, 94:508–515. [Google Scholar]
- 9.Aghaei H, Solaimany Nazar AR: Continuous production of the nanoscale liposome in a double flow-focusing microfluidic device. Ind Eng Chem Res 2019, 58:23032–23045. [Google Scholar]
- 10.Needham D, McIntosh TJ, Evans E: Thermomechanical and transition properties of dimyristoylphosphatidylcholine/cholesterol bilayers. Biochemistry 1988, 27:4668–4673. [DOI] [PubMed] [Google Scholar]
- 11.Pautot S, Frisken BJ, Weitz DA: Production of unilamellar vesicles using an inverted emulsion. Langmuir 2003, 19:2870–2879. [Google Scholar]
- 12.Morita M, Onoe H, Yanagisawa M, Ito H, Ichikawa M, Fujiwara K, Saito H, Takinoue M: Droplet-shooting and size-filtration (DSSF) method for synthesis of cell-sized liposomes with controlled lipid compositions. Chembiochem 2015, 16:2029–2035. [DOI] [PubMed] [Google Scholar]
- 13.Deshpande S, Dekker C: On-chip microfluidic production of cell-sized liposomes. Nat Protoc 2018, 13:856–874. [DOI] [PubMed] [Google Scholar]
- 14.Marrink SJ, Corradi V, Souza PCT, Ingólfsson HI, Tieleman DP, Sansom MSP: Computational modeling of realistic cell membranes. Chem Rev 2019, 119:6184–6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adamala KP, Martin-Alarcon DA, Guthrie-Honea KR, Boyden ES: Engineering genetic circuit interactions within and between synthetic minimal cells. Nat Chem 2017, 9:431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hilburger CE, Jacobs ML, Lewis KR, Peruzzi JA, Kamat NP: Controlling secretion in artificial cells with a membrane and gate. ACS Synth Biol 2019, 8:1224–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Berhanu S, Ueda T, Kuruma Y: Artificial photosynthetic cell producing energy for protein synthesis. Nat Commun 2019, 10. ** Here proteoliposomes containing ATP synthase and bacteriorhodopsin were encapsulated within a giant unilamellar vesicle. These synthetic organelles were then able to fuel protein synthesis inside the vesicle via a photosynthetic production of ATP. This demonstrates a total bottom-up synthesis of a biochemical energy flux and represents the synthetic engineering of a “simple” synthetic chloroplast.
- 18.Lee KY, Park S-J, Lee KA, Kim S-H, Kim H, Meroz Y, Mahadevan L, Jung K-H, Ahn TK, Parker KK, et al. : Photosynthetic artificial organelles sustain and control ATP-dependent reactions in a protocellular system. Nat Biotechnol 2018, 36:530. [DOI] [PubMed] [Google Scholar]
- 19.Jin L, Kamat NP, Jena S, Szostak JW: Fatty acid/phospholipid blended membranes: a potential intermediate state in protocellular evolution. Small 2018, 14:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brea RJ, Bhattacharya A, Bhattacharya R, Song JJ, Sinha SK, Devaraj NK: Highly stable Artificial cells from galactopyranose-derived single-chain amphiphiles. J Am Chem Soc 2018, 140:17356–17360. [DOI] [PubMed] [Google Scholar]
- 21.Hu J, Cochrane WG, Jones AX, Blackmond DG, Paegel BM: Chiral lipid bilayers are enantioselectively permeable. Nat Chem 2021, 13:786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhattacharya A, Devaraj NK: Tailoring the shape and size of artificial cells. ACS Nano 2019, 13:7396–7401. [DOI] [PubMed] [Google Scholar]
- 23.Fanalista F, Birnie A, Maan R, Burla F, Charles K, Pawlik G, Deshpande S, Koenderink GH, Dogterom M, Dekker C: Shape and size control of artificial cells for bottom-up biology. ACS Nano 2019, 13:5439–5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanasescu R, Lanz MA, Mueller D, Tassler S, Ishikawa T, Reiter R, Brezesinski G, Zumbuehl A: Vesicle origami and the influence of cholesterol on lipid packing. Langmuir 2016, 32:4896–4903. [DOI] [PubMed] [Google Scholar]
- 25.Garenne D, Libchaber A, Noireaux V: Membrane molecular crowding enhances MreB polymerization to shape synthetic cells from spheres to rods. Proc Natl Acad Sci 2020, 117:1902 LP–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buddingh’ BC, van Hest JCM: Artificial cells: synthetic compartments with life-like functionality and adaptivity. Acc Chem Res 2017, 50:769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Exterkate M, Caforio A, Stuart MCA, Driessen AJM: Growing membranes in vitro by continuous phospholipid biosynthesis from free fatty acids. ACS Synth Biol 2018, 7:153–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bhattacharya A, Brea RJ, Niederholtmeyer H, Devaraj NK: A minimal biochemical route towards de novo formation of synthetic phospholipid membranes. Nat Commun 2019, 10:1–8.30602773 ** De novo formation of phospholipid membranes was demonstrated by encapsulating a minimal transcription/translation system expressing FadD10, an enzyme that generates phospholipids from a provided supply of single-chain amphiphilic precursors, in liposomes.
- 29.Liu L, Zou Y, Bhattacharya A, Zhang D, Lang SQ, Houk KN, Devaraj NK: Enzyme-free synthesis of natural phospholipids in water. Nat Chem 2020, 12:1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deshpande S, Wunnava S, Hueting D, Dekker C: Membrane tension-mediated growth of liposomes. Small 2019, 15. e1902898. [DOI] [PubMed] [Google Scholar]
- 31.Castro JM, Sugiyama H, Toyota T: Budding and division of giant vesicles linked to phospholipid production. Sci Rep 2019, 9:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noireaux V, Libchaber A: A vesicle bioreactor as a step toward an artificial cell assembly. Proc Natl Acad Sci U S A 2004, 101:17669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dervaux J, Noireaux V, Libchaber AJ: Growth and instability of a phospholipid vesicle in a bath of fatty acids. Eur Phys J Plus 2017, 132:1–5. [Google Scholar]
- 34.Zhu TF, Adamala K, Zhang N, Szostak JW: Photochemically driven redox chemistry induces protocell membrane pearling and division. Proc Natl Acad Sci 2012, 109. 9828 LP–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adamala KP, Engelhart AE, Szostak JW: Collaboration between primitive cell membranes and soluble catalysts. Nat Commun 2016, 7:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walde P, Wick R, Fresta M, Mangone A, Luisi PL: Autopoietic self-reproduction of fatty acid vesicles. J Am Chem Soc 1994, 116:11649–11654. [Google Scholar]
- 37.Deshpande S, Spoelstra WK, van Doorn M, Kerssemakers J, Dekker C: Mechanical division of cell-sized liposomes. ACS Nano 2018, 12:2560–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noireaux V, Maeda YT, Libchaber A: Development of an artificial cell, from self-organization to computation and self-reproduction. Proc Natl Acad Sci U S A 2011, 108:3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andes-Koback M, Keating CD: Complete budding and asymmetric division of primitive model cells to produce daughter vesicles with different interior and membrane compositions. J Am Chem Soc 2011, 133:9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cans A-S, Andes-Koback M, Keating CD: Positioning lipid membrane domains in giant vesicles by micro-organization of aqueous cytoplasm mimic. J Am Chem Soc 2008, 130:7400–7406. [DOI] [PubMed] [Google Scholar]
- 41.Long MS, Cans A-S, Keating CD: Budding and asymmetric protein microcompartmentation in giant vesicles containing two aqueous phases. J Am Chem Soc 2008, 130:756–762. [DOI] [PubMed] [Google Scholar]
- 42.Murtas G: Internal lipid synthesis and vesicle growth as a step toward self-reproduction of the minimal cell. Syst Synth Biol 2010, 4:85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takakura K, Toyota T, Sugawara T: A novel system of self-reproducing giant vesicles. J Am Chem Soc 2003, 125:8134–8140. [DOI] [PubMed] [Google Scholar]
- 44.Kreye S, Malsam J, Söllner TH: In vitro assays to measure SNARE-mediated vesicle fusion BT - exocytosis and endocytosis: Ivanov AI. Humana Press; 2008:37–50. [DOI] [PubMed] [Google Scholar]
- 45.Robson A-L, Dastoor PC, Flynn J, Palmer W, Martin A, Smith DW, Woldu A, Hua S: Advantages and limitations of current imaging techniques for characterizing liposome morphology. Front Pharmacol 2018, 9:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morita M, Katoh K, Noda N: Direct observation of bacterial growth in giant unilamellar vesicles: a novel tool for bacterial cultures. ChemistryOpen 2018, 7:844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolkers WF, Oldenhof H, Tablin F, Crowe JH: Preservation of dried liposomes in the presence of sugar and phosphate. Biochim Biophys Acta Biomembr 2004, 1661:125–134. [DOI] [PubMed] [Google Scholar]
- 48.Miller MC, Klyosov A, Platt D, Mayo KH: Using pulse field gradient NMR diffusion measurements to define molecular size distributions in glycan preparations. Carbohydr Res 2009, 344:1205–1212. [DOI] [PubMed] [Google Scholar]
- 49.Pinheiro TJT, Vaz WLC, Geraldes CFGC, Prado A, da Costa MS: A 31P-NMR study on multilamellar liposomes formed from the lipids of a thermophilic bacterium. Biochem Biophys Res Commun 1987, 148:397–402. [DOI] [PubMed] [Google Scholar]
- 50.Dubinnyi MA, Lesovoy DM, Dubovskii PV, Chupin VV, Arseniev AS: Modeling of 31P-NMR spectra of magnetically oriented phospholipid liposomes: a new analytical solution. Solid State Nucl Magn Reson 2006, 29:305–311. [DOI] [PubMed] [Google Scholar]
- 51.Fröhlich M, Brecht V, Peschka-Süss R: Parameters influencing the determination of liposome lamellarity by 31P-NMR. Chem Phys Lipids 2001, 109:103–112. [DOI] [PubMed] [Google Scholar]
- 52.Elst Vander L, Piérart C, Fossheim SL, Raux J-C, Roch A, Muller RN: Enumeration of liposomes by multinuclear NMR and photon correlation spectroscopy. Supramol Chem 2002, 14:411–417. [Google Scholar]
- 53.Shimizu Y, Inoue a, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T: Cell-free translation reconstituted with purified components. Nat Biotechnol 2001, 19:751–5. [DOI] [PubMed] [Google Scholar]
- 54.van Nies P, Westerlaken I, Blanken D, Salas M, Mencía M, Danelon C: Self-replication of DNA by its encoded proteins in liposome-based synthetic cells. Nat Commun 2018, 9:1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Libicher K, Hornberger R, Heymann M, Mutschler H: In vitro self-replication and multicistronic expression of large synthetic genomes. Nat Commun 2020, 11:1–8.31911652 ** Authors engineered a PURE reaction for the self-replication of more than 116 kb genome, similar to the predicted genome size necessary to encode a minimal cell, that includes E. coli translation machinery, RNA and DNA polymerases, ribosomal RNAs, and an energy regeneration system.
- 56.Sakatani Y, Yomo T, Ichihashi N: Self-replication of circular DNA by a self-encoded DNA polymerase through rolling-circle replication and recombination. Sci Rep 2018, 8:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garamella J, Marshall R, Rustad M, Noireaux V: The all E. coli TX-TL toolbox 2.0: a platform for cell-free synthetic biology. ACS Synth Biol 2016, 5:344–355. [DOI] [PubMed] [Google Scholar]
- 58.Korman TP, Opgenorth PH, Bowie JU: A synthetic biochemistry platform for cell free production of monoterpenes from glucose. Nat Commun 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang W, Liu M, You C, Li Z, Zhang Y-HP: ATP-free biosynthesis of a high-energy phosphate metabolite fructose 1,6-diphosphate by in vitro metabolic engineering. Metab Eng 2017, 42:168–174. [DOI] [PubMed] [Google Scholar]
- 60.Goñi-Moreno A, Amos M: A reconfigurable NAND/NOR genetic logic gate. BMC Syst Biol 2012, 6:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weitz M, Mückl A, Kapsner K, Berg R, Meyer A, Simmel FC: Communication and computation by bacteria compartmentalized within microemulsion droplets. J Am Chem Soc 2014, 136:72–75. [DOI] [PubMed] [Google Scholar]
- 62.Lentini R, Martín NY, Forlin M, Belmonte L, Fontana J, Cornella M, Martini L, Tamburini S, Bentley WE, Jousson O, et al. : Two-way chemical communication between artificial and natural cells. ACS Cent Sci 2017, 3:117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ugrinic M, Zambrano A, Berger S, Mann S, Tang T-YD, deMello A: Microfluidic formation of proteinosomes. Chem Commun 2018, 54:287–290. [DOI] [PubMed] [Google Scholar]
- 64.Koonin EV: The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol 2010, 11:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gray MW, Burger G, Lang BF: Mitochondrial evolution. 80 Science 1999, 283. 1476 LP–1481. [DOI] [PubMed] [Google Scholar]
- 66.Howe CJ, Barbrook AC, Nisbet RER, Lockhart PJ, Larkum AWD: The origin of plastids. Philos Trans R Soc B Biol Sci 2008, 363:2675–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Elani Y, Trantidou T, Wylie D, Dekker L, Polizzi K, Law RV, Ces O: Constructing vesicle-based artificial cells with embedded living cells as organelle-like modules. Sci Rep 2018, 8:1–8.29311619 ** This work demonstrates a repurposing of living cells for use of synthetic organelles in synthetic systems. However, while this remains pertinent from an evolutionary standpoint, in terms of endosymbiosis, this more importantly demonstrates a communication between a synthetic cell and cytoplasm with a living cell endosymbiont.
- 68.Ces O, Elani Y, Trantidou T, Dekker L, Polizzi K: Functionalizing cell-mimetic giant vesicles with encapsulated bacterial biosensors. 2018, 10.6084/m9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Holler S, Hanczyc MM: Autoselective transport of mammalian cells with a chemotactic droplet. Sci Rep 2020, 10:5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Agapakis CM, Niederholtmeyer H, Noche RR, Lieberman TD, Megason SG, Way JC, Silver PA: Towards a synthetic chloroplast. Plos One 2011, 6:e18877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bhattacharya A, Niederholtmeyer H, Podolsky KA, Bhattacharya R, Song J-J, Brea RJ, Tsai C-H, Sinha SK, Devaraj NK: Lipid sponge droplets as programmable synthetic organelles. bioRxiv 2020, 10.1101/2020.01.20.913350. ** Lipid sponge droplets were encapsulated and functioned as synthetic organelles mimetic of interconnected organelle membranes such as the endoplasmic reticulum. This demonstrates further complexity with regard to morphological possibilities in terms of synthetic organelle engineering. Moreover, this shows that different membrane morphologies of synthetic organelles yield diverse diffusion and communication kinetics.
- 72.Giessen TW, Orlando BJ, Verdegaal AA, Chambers MG, Gardener J, Bell DC, Birrane G, Liao M, Silver PA: Large protein organelles form a new iron sequestration system with high storage capacity. Elife 2019, 8. e46070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jia H, Heymann M, Bernhard F, Schwille P, Kai L: Cell-free protein synthesis in micro compartments: building a minimal cell from biobricks. N Biotechnol 2017, 39:199–205. [DOI] [PubMed] [Google Scholar]
- 74.Bonacci W, Teng PK, Afonso B, Niederholtmeyer H, Grob P, Silver PA, Savage DF: Modularity of a carbon-fixing protein organelle. Proc Natl Acad Sci 2012, 109. 478 LP–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mason AF, van Hest JCM: Multifaceted cell mimicry in coacervate-based synthetic cells. Emerg Top Life Sci 2019, 3:567–571. [DOI] [PubMed] [Google Scholar]
- 76.Mason AF, Yewdall NA, Welzen PLW, Shao J, van Stevendaal M, van Hest JCM, Williams DS, Abdelmohsen LKEA: Mimicking cellular compartmentalization in a hierarchical protocell through spontaneous spatial organization. ACS Cent Sci 2019, 5:1360–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Niederholtmeyer H, Chaggan C, Devaraj NK: Communication and quorum sensing in non-living mimics of eukaryotic cells. Nat Commun 2018, 9:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li S, Wang X, Mu W, Han X: Chemical signal communication between two protoorganelles in a lipid-based artificial cell. Anal Chem 2019, 91:6859–6864.31020837 ** Authors engineered a chemical communication system between two lipid-based protoorganelles using a cascade of reactions to oxidize Amplex red into red resorufin as a fluorescent readout of communication.
- 79.Gräwe A, Dreyer A, Vornholt T, Barteczko U, Buchholz L, Drews G, Ho UL, Jackowski ME, Kracht M, Lüders J, et al. : A paper-based, cell-free biosensor system for the detection of heavy metals and date rape drugs. Plos One 2019, 14:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mazur F, Tran H, Kuchel RP, Chandrawati R: Rapid detection of Listeriolysin O toxin based on a nanoscale liposome-gold nanoparticle platform. ACS Appl Nano Mater 2020, 3:7270–7280. [Google Scholar]
- 81. Holstein JM, Gylstorff C, Hollfelder F: Cell-free directed evolution of a protease in microdroplets at ultrahigh throughput. ACS Synth Biol 2021, 10:252–257.33502841 ** A microfluidic approach to create water-in-oil emulsion droplets is used to carry out a three-step reaction that was previously incompatible. Employing this workflow allowed for in vitro evolution of the protease savinase, resulting in a higher-activity and faster enzyme.