Abstract
Activated macrophages undergo metabolic reprogramming, which not only supports their energetic demands but also allows for the production of specific metabolites that function as signaling molecules. Several Krebs cycles, or Krebs-cycle-derived metabolites, including succinate, α-ketoglutarate, and itaconate, have recently been shown to modulate macrophage function. The accumulation of 2-hydroxyglutarate (2HG) has also been well documented in transformed cells and more recently shown to play a role in T cell and dendritic cell function. Here we have found that the abundance of both enantiomers of 2HG is increased in LPS-activated macrophages. We show that L-2HG, but not D-2HG, can promote the expression of the proinflammatory cytokine IL-1β and the adoption of an inflammatory, highly glycolytic metabolic state. These changes are likely mediated through activation of the transcription factor hypoxia-inducible factor-1α (HIF-1α) by L-2HG, a known inhibitor of the HIF prolyl hydroxylases. Expression of the enzyme responsible for L-2HG degradation, L-2HG dehydrogenase (L-2HGDH), was also found to be decreased in LPS-stimulated macrophages and may therefore also contribute to L-2HG accumulation. Finally, overexpression of L-2HGDH in HEK293 TLR4/MD2/CD14 cells inhibited HIF-1α activation by LPS, while knockdown of L-2HGDH in macrophages boosted the induction of HIF-1α-dependent genes, as well as increasing LPS-induced HIF-1α activity. Taken together, this study therefore identifies L-2HG as a metabolite that can regulate HIF-1α in macrophages.
Keywords: L-2-hydroxyglutarate, macrophage, inflammation, immunometabolism, hypoxia-inducible factor (HIF), interleukin 1β (IL-1β), glycolysis
Abbreviations: 2HG, 2-hydroxyglutarate; 2HGDH, 2-hydroxyglutarate dehydrogenase; αKG, α-ketoglutarate; BMDM, bone-marrow-derived macrophages; HIF-1α, hypoxia-inducible factor 1α; IDH, isocitrate dehydrogenase; IL-1β, interleukin 1β; KDM, Jmjj-domain containing histone lysine demethylases; LDH, lactate dehydrogenase; LPS, lipopolysaccharide; MDH, malate dehydrogenase; mtROS, mitochondrial reactive oxygen species; OXPHOS, oxidative phosphorylation; PHD, HIF prolyl hydroxylase; SDH, succinate dehydrogenase; TET, ten-eleven translocation DNA hydroxylase
Recently, much attention has been focused on how the metabolism of immune cells changes upon activation and whether these changes can be linked to specific responses. For macrophages, the remodeling of their metabolism following activation supports the adoption of different effector functions, depending on the activating stimulus. Not only does this support increased energy demands, but different activation states require distinct metabolic processes (1). Macrophages activated with the Toll-like receptor-4 (TLR4) agonist, lipopolysaccharide (LPS) increase glycolysis to maintain production of ATP, and oxidative phosphorylation (OXPHOS) is impaired. Macrophages activated with IL-4, so-called alternatively activated macrophages, rely mainly on the Krebs cycle and OXPHOS (2). Metabolites themselves can also act as signaling molecules, for example, roles for the Krebs cycle metabolites succinate and α-ketoglutarate (αKG) in macrophage differentiation have been described (3). LPS promotes the oxidation of succinate by succinate dehydrogenase (SDH), which can drive the production of mitochondrial reactive oxygen species (mtROS), or following its mitochondrial export, stabilize HIF-1α (4, 5, 6, 7). However, the balance between succinate and αKG must be sufficiently skewed in favor of αKG in order for alternative macrophage activation to occur, as αKG is required for the Jmjd3-dependent epigenetic regulation of specific genes (7).
2HG is a metabolite derived from αKG, which has yet to be characterized in macrophages. Since 2HG is a chiral metabolite, two distinct isoforms of 2HG exist, L-2HG and D-2HG. Their accumulation has been linked to the metabolic disorders termed 2-hydroxyglutaric acidurias (2HGA) and certain cancers (8, 9). Increased D-2HG is associated with tumors possessing mutant isocitrate dehydrogenase 1 or 2 (IDH1 or IDH2), which are frequently found in low-grade glioma and acute myeloid leukaemia (10), while an increase in L-2HG has been seen in renal cell carcinoma (11). In healthy tissues, accumulation is controlled by enantiomer-specific dehydrogenases (D-2HGDH and L-2HGDH), which oxidize 2HG back in to αKG.
Both D-2HG and L-2HG have been shown to act as competitive inhibitors of multiple αKG-dependent dioxygenases, including the ten-eleven translocation (TET) family of DNA hydroxylases, the Jmjj-domain containing histone lysine demethylases (KDMs), and the HIF prolyl hydroxylases (PHDs), where L-2HG has been shown to be a much more effective inhibitor than D-2HG (12).
Here, we have examined a role for 2HG in macrophages. Our data suggests that the accumulation of L-2HG driven by LPS stimulation increases HIF-1α stability and therefore activity. The subsequent increase in HIF-target gene expression contributes to a proinflammatory activation state, including the adoption of a highly glycolytic metabolism and expression of the HIF-1α -dependent gene, notably that encoding interleukin 1β (IL-1β).
Results
2-HG accumulates in LPS-activated macrophages
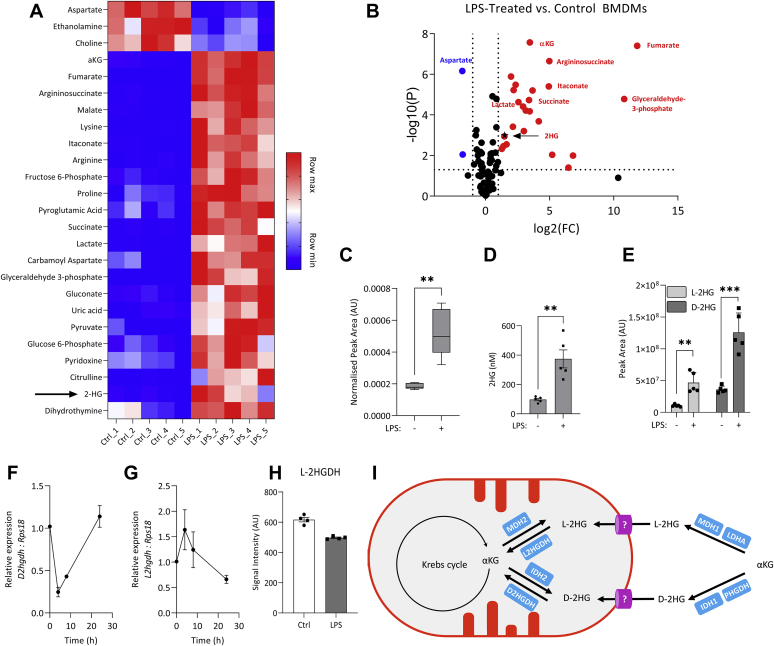
LC-MS was used to analyze changes in metabolite abundance in control and LPS-stimulated bone-marrow-derived macrophages (BMDMs) (Fig. 1, A and B). Consistent with previously published reports, aspartate was decreased in LPS-stimulated cells, while abundance of fumarate, itaconate, and succinate was increased (Fig. 1, A and B). 2HG also accumulated in LPS-stimulated cells (Fig. 1C), and the intracellular concentration reached 0.4 μM (Fig. 1D). Enantiomer-specific analysis combining chiral derivatization with LC-MS (13) revealed that abundance of both L-2HG and D-2HG was increased by LPS (Fig. 1E), with D-2HG making a greater contribution to the total pool of 2HG than L-2HG. Expression of the enzymes responsible for degradation of both D-2HG and L-2HG, D-2HGDH and L-2HGDH, respectively, both decreased following LPS stimulation, though the kinetics of this differed. Downregulation of D2hgdh was seen early after LPS stimulation, and by 24 h, poststimulation expression had returned to basal levels (Fig. 1F). Downregulation of L2hgdh was a later event, with expression reduced by half at 24 h (Fig. 1G). An unbiased quantitative proteomic screen confirmed decreased L-2HGDH in LPS-stimulated BMDMs (Fig. 1H), though D-2HGDH was not detected in this screen. The increase in D-2HG and L-2HG may therefore in part be due to a decrease in their respective dehydrogenases. A schematic of 2-HG synthesis and degradation is shown in Figure 1I. Figure S1 illustrates the strategy for derivatizing the 2-HG enantiomers and the chromatograph of labeled D-2HG and L-2HG.
Figure 1.
Abundance of 2-hydroxyglutarate is increased in LPS-stimulated macrophages.A, relative metabolite abundance in control and LPS-stimulated (100 ng/ml, 24 h) BMDMs. B, metabolites significantly upregulated (red) or downregulated (blue) following 24 h LPS stimulation, n = 5; FC, Fold change. C, 2HG levels in control and LPS stimulated (24 h) BMDMs, (D) intracellular 2HG concentration, and (E) relative abundance of each enantiomer of 2HG in control and LPS-stimulated (100 ng/ml, 24 h) BMDMs, n = 5. Expression of (F) D2hgdh and (G) L2hgdh in BMDMs stimulated with LPS, n = 9 from three independent experiments. H, relative changes in L-2HGDH protein level quantified with TMT coupled LC-MS, n = 4. I, possible routes of synthesis and degradation of L-2HG and D-2HG. Graphs represent median ± min/max (C); mean ± S.D. (D–E), mean ± S.E.M. (F–G); ∗∗ p < 0.01, ∗∗∗ p < 0.001, unpaired t test. 2HG, 2-hydroxyglutarate; BMDM, bone-marrow-derived macrophage; LPS, lipopolysaccharide.
Octyl-L-2HG increases HIF-1α activity in macrophages
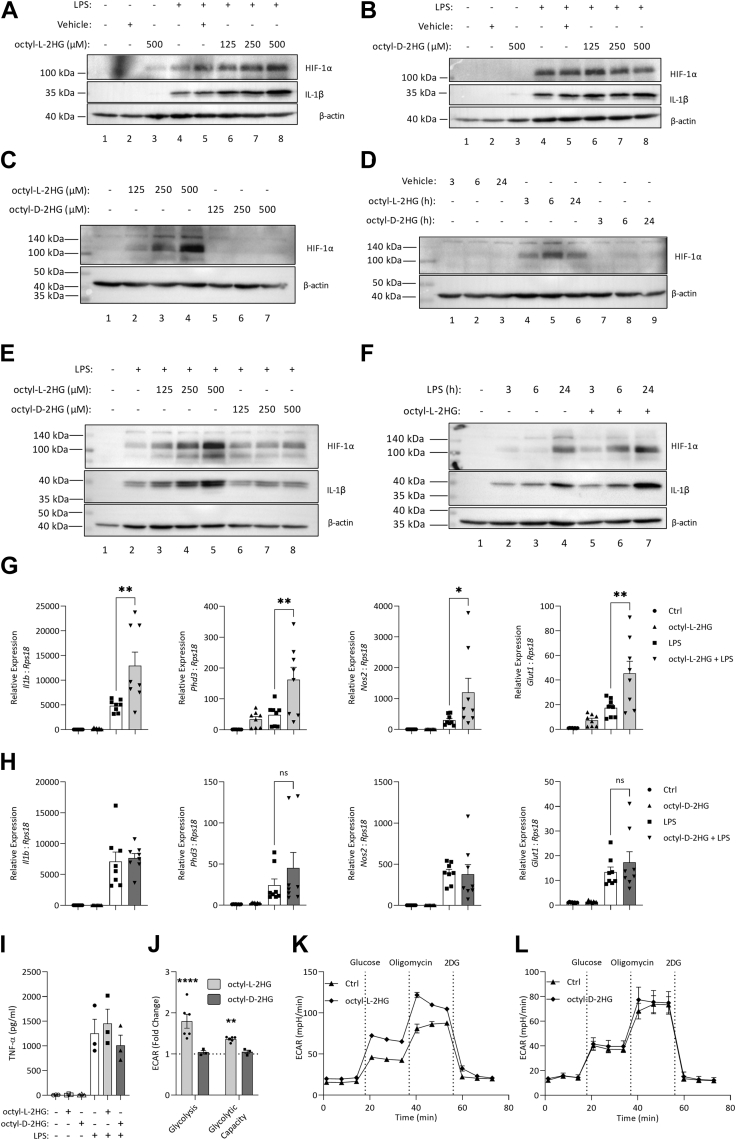
As it has been reported that L-2HG can inhibit the PHDs, we next used a cell-permeable analogue of each enantiomer to examine the effects on HIF-1α and several downstream gene products (12, 14). Octyl-L-2HG dose-dependently increased both HIF-1α and pro-IL-1β in LPS-stimulated BMDMs (Fig. 2A, compare lanes 6–8 with lane 5). Notably, even in the absence of LPS, HIF-1α could be detected in BMDMs treated with octyl-L-2HG (lane 3). In contrast, in cells treated with octyl-D-2HG, no change in either HIF-1α or pro-IL-1β was seen (Fig. 2B). These results demonstrated that the two enantiomers of 2HG might play distinct roles in macrophage function. The increase in HIF-1α in unstimulated cells treated with octyl-L-2HG was confirmed to be dose-dependent (Fig. 2C) and peaked at 6 h after treatment (Fig. 2D). Further dose (Fig. 2E) and time course (Fig. 2F) studies in LPS-stimulated BMDMs confirmed that octyl-L-2HG treatment increased both LPS-induced HIF-1α and pro-IL-1β, while octyl—D-2HG treatment had no effect on either protein. The expression of several HIF-target genes was then examined. In cells treated with octyl-L-2HG, expressions of Il1b, Phd3, Nos2, and Glut1 were increased, and in the case of Phd3 and Glut1 this occurred either with L-2HG alone or in combination with LPS (Fig. 2G). Octyl-D-2HG had no effect either on its own or in combination with LPS (Fig. 2H). LPS-driven TNF-α secretion was unaltered by treatment with either octyl-L-2HG or octyl-D-2HG (Fig. 2I), showing some specificity in the proinflammatory phenotype driven by octyl-L-2HG. Treatment with octyl-L-2HG increased both glycolysis and glycolytic capacity in BMDMs (Fig. 2, J and K), while treatment with octyl-D-2HG had no effect (Fig. 2, J and L). These data indicate that L-2HG, unlike D-2HG, can modulate HIF-1α activity in macrophages.
Figure 2.
Octyl-L-2HG increases HIF-1α activity in macrophages. LPS (24 h) induced HIF-1α and pro-IL-1-β in BMDMs pretreated for 3 h with octyl-L-2HG (A), or octyl-D-2HG (B). HIF-1α as detected in cells treated with octyl-2HG as indicated (C) or with 500 μM octyl-2HG (D). LPS (24 h) induced HIF-1α and pro-IL-1β in BMDMs pretreated for 3 h with octyl-2HG at the indicated concentration (E) or treated simultaneously with octyl-2HG (500 μM) and LPS (F). Expression of Il1b, Phd3, Nos2, and Glut1 in LPS stimulated (24 h) BMDMs pretreated with (G) octyl-L-2HG or (H) octyl-D-2HG (500 μM, 3 h), n = 8 from three independent experiments. TNF-α (I) in BMDMs treated with octyl-D-2HG or octyl-L-2HG (500 μM for 3 h), stimulated with LPS (3 h), n = 3. Glycolysis and glycolytic capacity (fold over unstimulated control) in octyl-D-2HG or octyl-L-2HG (500 μM) treated BMDMs (J), n = 3. ECAR in unstimulated and octyl-L-2HG (500 μM) (K), or octyl-D-2HG (L) treated BMDMs. Western blots are representative of n = 4 (A–B), or n = 3 (C–F), graphs represent the mean ± S.E.M.; ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗∗ p < 0.001; ns, not significant; one-way ANOVA. 2HG, 2-hydroxyglutarate; BMDM, bone-marrow-derived macrophage; LPS, lipopolysaccharide.
L-2HG increases HIF-1α activity in macrophages
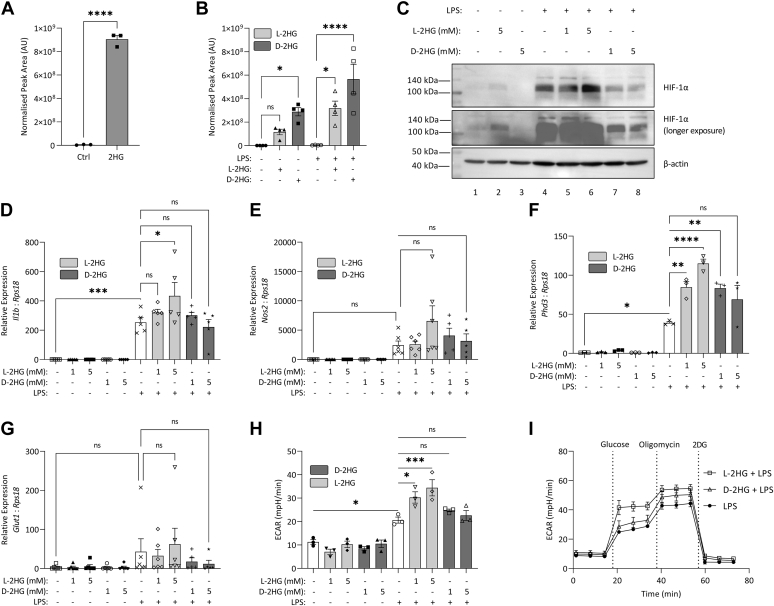
Other immune cells, such as both CD8+ and CD4+ T cells, dendritic cells (DCs), microglia, and B cell blasts have been reported to take up 2HG from their environment, though whether macrophages are capable of this is unknown (15, 16, 17). We therefore next tested 2-HG. Intracellular 2HG increased in macrophages treated with 2HG (Fig. 3A), and when treated with enantiomer-specific 2HG, a preference for D-2HG uptake over L-2HG was observed (Fig. 3B). In agreement with the increase in HIF-1α seen in cells treated with octyl-L-2HG, L-2HG treatment not only increased HIF-1α in LPS-stimulated macrophages (Fig. 3C, lanes 5 and 6 compared with lane 4) but also allowed for the detection of HIF-1α in unstimulated cells (Fig. 3C, middle panel (longer exposure), compare lanes 1 and 2). Expressions of the HIF-target genes, Il1b, Phd3, Nos2, and Glut1, were also increased in LPS-stimulated macrophages treated with L-2HG but not D-2HG (Fig. 3, D–G). Glycolysis was also boosted in LPS-stimulated macrophages treated with L-2HG (Fig. 3, H and I). Altogether, these results show that L-2HG, but not D-2HG, can increase HIF-1α activity in macrophages.
Figure 3.
L-2HG increases HIF-1α activity in macrophages.A, intracellular 2HG in BMDMs treated with 2HG (2 mM) for 24 h, n = 3. B, intracellular 2HG in BMDMs treated with L-2HG (5 mM), or D-2HG (5 mM) for 24 h, n = 3. C, BMDMs treated with L-2HG or D-2HG (5 mM) for 24 h, prior to stimulation with LPS (100 ng/ml) for 24 h. HIF-1α was measured by Western blot, n = 4. Expression of (D) Il1b, (E) Nos2, (F) Phd3, and (G) Glut1 in BMDMs pretreated with L-2HG or D-2HG (24 h) prior to stimulation with LPS (100 ng/ml, 24 h), n = 6 (D, E, G), n = 3 (F). E, glycolysis in BMDMs treated with L-2HG or D-2HG (5 mM) for 24 h, prior to stimulation with LPS (100 ng/ml) for 24 h, n = 3. Graphs represent the mean ± S.E.M.; ∗ p < 0.05, ∗∗p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001; ns, not significant; one-way ANOVA. 2HG, 2-hydroxyglutarate; BMDM, bone-marrow-derived macrophage; LPS, lipopolysaccharide.
L-2HG stabilizes HIF-1α via inhibition of PHDs
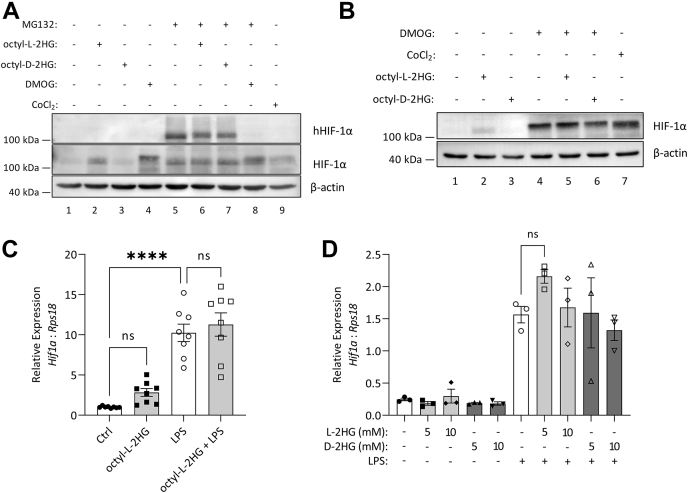
We next examined HIF-1α hydroxylation. BMDMs were treated with the proteasomal inhibitor, MG132, in order to prevent degradation of hydroxylated HIF-1α (hHIF-1α), and octyl-L-2HG and octyl-D-2HG treatment led to a slight reduction in the level of hHIF-1α (Fig. 4A, upper panel compare lanes 6 and 7 with lane 5). The known PHD inhibitors, DMOG and CoCl2, completely inhibited HIF-1α hydroxylation (upper panel lanes 8 and 9). Treatment with octyl-L-2HG did not cause an increase in total HIF-1α in cells where its degradation was blocked, indicating that the accumulation of HIF-1α in the presence of L-2HG is not as a result of increased synthesis (Fig. 4A, middle panel compare lane 6 with lane 5). Fitting with this when cells were treated with octyl-L-2HG in the presence of DMOG, no further increase in HIF-1α was seen (Fig. 4B, compare lane 5 with lane 4). The lack of an additive effect in cells treated with both DMOG and L-2HG would suggest that L-2HG can increase HIF-1α by blocking its degradation similar to DMOG, as if L-2HG boosted synthesis of HIF-1α, then treatment with both DMOG and octyl-L-2HG should cause a boost in HIF-1α when compared with DMOG alone. Neither treatment with octyl-L-2HG (Fig. 4C) nor L-2HG (Fig. 4D) increased Hif1a transcription. In macrophages, the two main pathways that control HIF-1α transcription, independent of oxygen tension, are NF-κB and mTOR signaling (1, 18). In BMDMs treated with either octyl-L-2HG or octyl-D-2HG, there was no effect on NF-κB or mTOR signaling (Fig. S2, A–C). Both octyl-D-2HG, and D-2HG treatment also had no effect on Hif1a transcription (Figs. 4D, S2D). Altogether, this indicated to us that increased HIF-1α activity driven by L-2HG was likely to be due to the inhibition of the PHD enzymes leading to the subsequent stabilization of HIF-1α.
Figure 4.
L-2HG stabilizes HIF-1α via inhibition of the PHDs.A, BMDMs treated with 10 μM MG132 for 1 h prior to octyl-L-2GH or octyl-D-2HG (500 μM), or DMOG (1 mM) or CoCl2 (100 μM) for 5 h. B, BMDMs treated with DMOG (1 mM) for 2 h prior to treatment with octyl-L-2HG or octyl-D-2HG (500 μM) for 3 h. C, Hif1a expression in BMDMs treated with octyl-L-2HG (500 μM) for 3 h prior to LPS (100 ng/ml) for 24 h. D, Hif1a expression in BMDMs treated with L-2HG or D-2HG at the indicated concentration for 24 h prior to LPS stimulation (100 ng/ml) for 24 h, n = 3. Western blots are representative of n = 3, graphs represent the mean ± S.E.M.; ∗∗∗∗ p < 0.0001; ns, not significant; one-way ANOVA. 2HG, 2-hydroxyglutarate; BMDM, bone-marrow-derived macrophage; LPS, lipopolysaccharide.
L-2HGDH expression negatively regulates HIF-1α activity
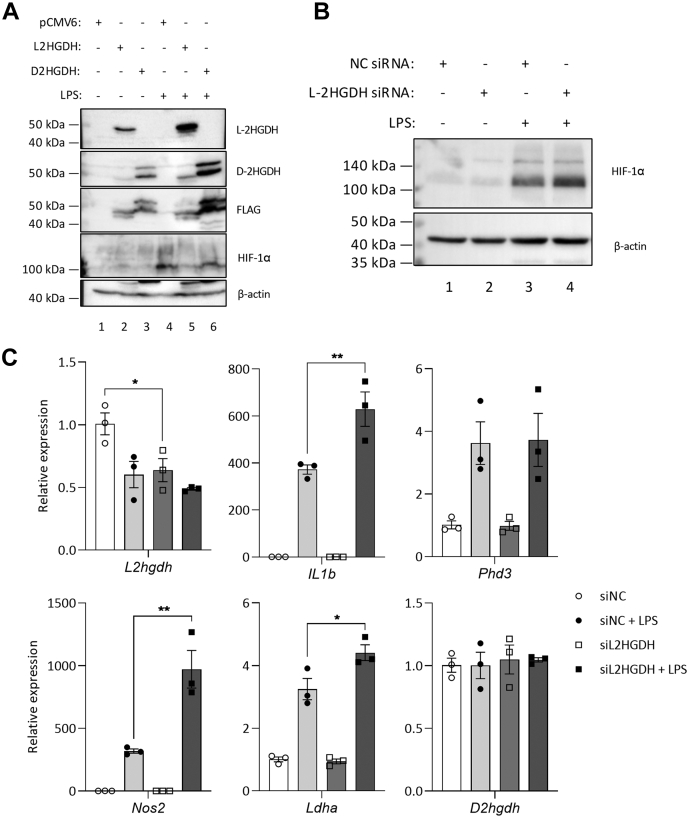
To examine a role for endogenous 2-HG in HIF-1α activation, we utilized HEK293 TLR4/MD2/CD14 (HEK293 MTC) cells in which we overexpressed L-2HGDH and D-2HGDH. HEK293 cells can produce both L-2HG and D-2HG (19), and so overexpression of the degrading enzymes for each enantiomer would be expected to reduce abundance of endogenous 2HG in an enantiomer-specific manner. LPS treatment resulted in increased HIF-1α, similar to LPS-activated BMDMs (Fig. 5A, fourth panel, compare lane 4 with lane 1). Where L-2HGDH was overexpressed, the LPS-induced increase in HIF-1α was reduced, while in cells where D-2HGDH was overexpressed, no change in HIF-1α was seen from the LPS-stimulated control (Fig. 5A, compare lanes 5 and 6 with lane 4). To determine whether the accumulation of endogenous L-2HG was sufficient to modulate HIF-1α, siRNA-mediated knockdown of L-2HGDH in BMDMs was also tested. In BMDMs treated with siRNA targeting L2hgdh, an increase in LPS-induced HIF-1α was seen (Fig. 5B). An increase in expression of Il1b, Nos2, and Ldha was also seen in LPS-stimulated BMDMs (Fig. 5C). These results demonstrated that, by altering L-2HGDH expression and therefore L-2HG abundance, HIF-1α activity and HIF-1α -dependent genes could also be modulated by endogenous L-2HG. This would suggest that changes in levels of endogenous L-2HG are sufficient to alter HIF-1α activity, implicating L-2HG in HIF-1α activation in macrophages treated with LPS.
Figure 5.
L-2HGDH expression is important for the modulation of HIF-1a in macrophages.A, HEK293 MTC cells were transfected with FLAG-tagged L-2HGDH or D-2HGDH for 24 h, prior to stimulation with LPS (100 ng/ml, 24 h). HIF-1α, IL-1β, L-2HGDH, D-2HGDH, and FLAG were measured by Western blot, n = 2. BMDMs were transfected with L-2HGDH or control siRNA (50 nM) for 144 h, prior to LPS (100 ng/ml) for 24 h, and HIF-1α and IL-1β measured by Western blot, n = 4 (B). C, expressions of L2hgdh, Il1b, Phd3, Nos2, Ldha, and D2hgdh are shown as fold over the unstimulated control, n = 3. Graphs represent the mean ± S.E.M.; ∗ p < 0.05, ∗∗ p < 0.01; one-way ANOVA. 2HGDH, 2-hydroxyglutarate dehydrogenase; BMDM, bone-marrow-derived macrophage; LPS, lipopolysaccharide.
Discussion
Metabolic reprogramming in macrophages is now well understood to be essential for an appropriate immune response (6). This includes the production of metabolites with nonmetabolic signaling roles, such as the Krebs cycle metabolites succinate, citrate, and αKG. Our work identifies a role for L-2HG in HIF-1α activation in LPS-activated macrophages. HIF-1α activation, and the resulting increase in glycolytic metabolism, has been shown to be essential for inflammatory macrophage activation (20, 21).
Several reports have shown L-2HG to be a more potent inhibitor of certain αKG-dependent enzymes than D-2HG (14, 22). One of these studies reported that the IC50 value for PHD2 was 419 μM in the case of L-2HG, but 7.3 mM for D-2HG (14). A similar effect was seen with factor inhibiting HIF (FIH), whereby L-2HG had a much lower IC50 value than D-2HG. However, IC50 values for two JMJD histone demethylases were comparable between the two enantiomers. This supports our findings that D-2HG does not have an effect on HIF-1α. Other metabolites have been reported to increase HIF-1α activity, the most notable in the case of macrophage function being succinate (4, 5). Succinate accumulation and subsequent export from the mitochondria can lead to PHD inhibition, while mtROS generated by reverse electron transport at SDH may also contribute to HIF-1α stabilization. The switch toward glycolytic metabolism in macrophages therefore allows the mitochondria to be repurposed from ATP production, and Krebs cycle intermediates to exert immunomodulatory effects. Our study indicates that L-2HG is another HIF-1α regulator in LPS-treated macrophages.
Our data also demonstrate the ability of macrophages to take up both D-2HG and L-2HG, as other immune cells have been shown to do (16). A transporter in T cells for D-2HG has been identified, SCL13A3, but this does not appear to be expressed on BMDMs (16). There is also a question of the subcellular localization of L-2HG. As L-2HGDH and D-2HGDH are both mitochondrial enzymes, this suggests that it is within this organelle that either enantiomer predominately accumulates. Both D-2HG and L-2HG are produced from αKG by promiscuous enzyme activity, whereby an enzyme is capable of catalyzing a side reaction in addition to its main activity (23). For L-2HG the enzymes responsible for this are both malate dehydrogenases (MDH1 and MDH2) and lactate dehydrogenase A (LDHA) (19, 24). Both MDH1 and LDHA are cytosolic enzymes, while MDH2 is located in the mitochondria. Depending on the enzyme responsible for L-2HG synthesis, it may be necessary for L-2HG to be transported out of the mitochondria to exert any effect on the PHD enzymes. Further work would be required in order to determine the enzyme responsible for L-2HG synthesis in macrophages and whether mitochondrial transporters exist. Our study shows that one mechanism for 2-HG accumulation in macrophages is downregulation of L-2HGDH and D-2HGDH.
Altogether, our results show that L-2HG is a HIF-1α activator. However, as the concentration of L-2HG seen in LPS-stimulated macrophages is relatively low, the question remains as to whether this increase in endogenous L-2HG is sufficient to modulate HIF-1α activity. The data presented here suggest that this is in fact the case, as altering L-2HGDH expression was sufficient to see altered HIF-1α expression and expression of downstream target genes. Metabolic reprogramming in macrophages coordinates several pathways that result in accumulation of metabolites that may activate HIF-1α. It is therefore likely that L-2HG accumulation acts in tandem with succinate, and possibly fumarate, to achieve this both through product inhibition of the PHDs and also by reducing αKG levels (5, 25).
While this study has focused on the role of L-2HG in modulating HIF-1α signaling, it is likely that the increase in both L-2HG and D-2HG seen in LPS-activated macrophages acts to inhibit other αKG-dependent enzymes. Loss of function of the electron transport chain complex III in hematopoietic stem cells has been shown to cause L-2HG accumulation with increases in DNA and histone methylation, which block their ability to differentiate (26). Similarly, in Treg cells, loss of complex III will inhibit their suppressive ability and is associated with increases in 2HG and succinate and a hypermethylated phenotype (27). D-2HG has been shown to promote TH17 cell differentiation from naïve CD4+ cells via an epigenetic mechanism resulting in the promoter methylation and suppression of Foxp3, the transcription factor known to drive iTreg differentiation (28). Further investigation will reveal whether either enantiomer of 2HG has an effect on macrophage function via epigenetic modifications.
In conclusion, our work provides evidence for the regulation of HIF-1α by L-2HG. These data suggest that L-2HG can, through inhibiting the PHDs, increase the stability of HIF-1α. The increased abundance of L-2HG in LPS-stimulated macrophages may be important for the stabilization of HIF-1α and the induction of proinflammatory events downstream of HIF-1α signaling. This includes the adoption of a glycolytic phenotype and the expression of IL-1β. While D-2HG abundance is also increased in LPS-stimulated macrophages, it did not affect HIF-1α signaling. L-2HG may therefore be designated as an immunometabolite that has a nonmetabolic signaling role in inflammatory macrophage activation.
Experimental procedures
Animals and cells
C57BL/6J mice used for the generation of BMDMs were purchased from Harlan UK and maintained under specific pathogen-free conditions in line with Irish and European Union regulations. HEK293 MTC cells were purchased from Invivogen (catalogue no. 293-htlr4md2cd14).
Generation of BMDMs
Mice were euthanized in a CO2 chamber, and death was confirmed by cervical dislocation. Bone marrow was extracted from the tibia, fibula, and hip using a 23-gague needle and DMEM. Cells were differentiated in DMEM (10% fetal calf serum (FCS), 1% penicillin/streptomycin, 20% L929 supernatant). After 6 days, cells were counted and plated for experiments.
Reagents
Octyl-L-2HG and octyl-D-2HG were purchased from Caymen Chemical. L-2HG sodium salt, D-2HG sodium salt, oligomycin, DMOG, CoCl2, and L-glutamine were purchased from Sigma. Ultrapure rough LPS from Escherichia coli (serotype EH100, catalogue number ALX-581-010) was purchased from Alexis, and used at 100 ng/ml. 2DG was purchased from Fisher-Scientific. MG132 was purchased from Merck.
Untargeted polar metabolomics and quantification of 2HG
Cells were washed three times with ice-cold PBS, with all the PBS being removed after the last wash. Extraction solution (methanol/acetonitrile/water, 50:30:20 v/v/v) was added (500 μl per 1 × 106 cells), and samples were incubated for 15 min on dry ice. The resulting suspension was transferred to ice-cold microcentrifuge tubes. Samples were agitated for 15 min at 4 °C in a thermomixer, incubated at −20 °C for 1 h, then centrifuged at maximum speed for 10 min at 4 °C. The supernatant was transferred into a new tube and centrifuged again at maximum speed for 10 min at 4 °C. For untargeted metabolite screens, 85 μl of each supernatant was transferred to autosampler vials and stored at −80 °C prior to analysis by LC-MS.
The remainder of the supernatant from each sample was used for chiral derivatization. Standards were made in metabolite extraction buffer. Samples and standards were dried using a Savant SC110 SpeedVac Concentrator and stored at −80 °C overnight. 5 mM N-(p-toluenesulfonyl)-L-phenylalanyl chloride (TSPC) (Santa Cruz) was made up in acetonitrile, and pyridine (Fluorochem) was added (2 μl/160 μl TSPC). In total, 160 μl TSPC was added to each sample and standard, vortexed, incubated for 10 min at room temperature, before being dried with a SpeedVac concentrator. Samples and standards were resuspended in the same volume as they were initially before chiral derivatization.
LC-MS analysis was performed using a Q Exactive mass spectrometer coupled to a Dionex U3000 UHPLC system (Thermo). The liquid chromatography system was fitted with a Sequant ZIC-pHILIC column (150 mm × 2.1 mm) and guard column (20 mm × 2.1 mm), both from Merck Millipore and temperature maintained at 45 °C. The mobile phase was composed of 20 mM ammonium carbonate with 0.1% ammonium hydroxide and acetonitrile. The flow rate was set at 200 μl/min with the gradient described by Mackay et al. (29). The mass spectrometer was operated in full MS and polarity switching mode. Samples were randomized to avoid bias due to machine drift and processed blindly. The acquired spectra were analyzed using XCalibur Qual Browser and XCalibur Quan Browser software (Thermo Scientific). LC-MS analysis of TSPC derivatized samples was carried out as described by Cheng et al. (13). Absolute quantification of selected metabolites was performed by interpolation of the corresponding standard curve obtained from serial dilutions of commercially available standards (Sigma Aldrich) running with the same batch of samples. Intracellular concentration of metabolites was calculated using cell volume measurements, as previously described (30). Further analysis of metabolite abundance was carried out with MetabolAnalyst 4.0 software.
TMT-couples LC/MS
Following treatment, cells were lysed in HEPES pH 7.5, EDTA, glycerol, and NP40. In total, 2 mM TCEP and 50 mM NEM were added in a buffer containing 50 mM HEPES, 2% SDS, 125 mM NaCl, pH 7.2, and samples were incubated for 60 min at 37 °C in the dark to reduce and alkylate all unmodified protein cysteine residues. In total, 20% (v/v) TCA was added to stabilize thiols and incubated overnight at 4 °C and then pelleted for 10 min at 4000g at 4 °C. The pellet was washed three times with cold methanol (2 ml) and then resuspended in 2 ml 8 M urea containing 50 mM HEPES (pH 8.5). Protein concentrations were measured by BCA assay prior to protease digestion. Protein lysates were diluted to 4 M urea and digested with LysC (Wako, Japan) in a 1/100 enzyme/protein ratio and trypsin at a final 1/200 enzyme/protein ratio for 4 h at 37 °C. Protein extracts were diluted further to a 2.0 M urea and LysC at 1/100 enzyme/protein ratio and trypsin at a final 1/200 enzyme/protein ratio were added again and incubated overnight at 37 °C. Protein extracts were diluted further to a 1.0 M urea concentration, and trypsin was added to a final 1/200 enzyme/protein ratio for 6 h at 37 °C. Digests were acidified with 250 μl of 25% acetic acid to a pH ∼2 and subjected to C18 solid-phase extraction (50 mg Sep-Pak, Waters). In total, 6–7 M excess TMT label was added to each digest for 30 min at room temperature (repeated twice). The reaction was quenched using 4 μL of 5% hydroxylamine. Samples were subjected to an additional C18 solid-phase extraction (50 mg Sep-Pak, Waters).
Data were collected using an Orbitrap Fusion Lumos mass spectrometer coupled with a Proxeon EASY-nLC 1200 LC pump. Peptides were separated on a 100 μm inner diameter microcapillary column packed with 45 cm of Accucore C18 resin (2.6 μm, 100 Å). Peptides were separated using a 3 h gradient of 6–22% acetonitrile in 0.125% formic acid with a flow rate of ∼400 nl/min. Each analysis used an MS3-based TMT method as described previously (31). The data were acquired using a mass range of m/z 400–14,000, resolution 120,000, AGC target 1 × 106, maximum injection time 100 ms, dynamic exclusion of 120 s for the peptide measurements in the Orbitrap. Data-dependent MS2 spectra were acquired in the ion trap with a normalized collision energy (NCE) set at 35%, AGC target was set to 1.8 × 104 and a maximum injection time of 120 ms. MS3 scans were acquired in the Orbitrap with a HCD collision energy set to 55%, AGC target was set to 1.5 × 105, maximum injection time of 150 ms, resolution at 50,000 and with a maximum synchronous precursor selection (SPS) precursors set to 10.
A compendium of in-house software was used to convert.raw files to mzXML format, as well as to adjust monoisotopic m/z measurements and correct erroneous peptide charge state assignments. Assignment of MS2 spectra was performed using the SEQUEST (v.28, rev.12) algorithm (32). All experiments utilized the Mouse UniProt database (downloaded 4/2016) where reversed protein sequences and known contaminants such as human keratins and bovine albumin were appended. SEQUEST searches were performed using a 20 ppm precursor ion tolerance, while requiring each peptide's N/C terminus to have trypsin protease specificity and allowing up to two missed cleavages. TMT tags on peptide N termini/lysine residues (+229.162932 Da) and cysteine (+125.047679 Da) were set as static modifications while methionine oxidation (+15.99492 Da) and cysteine alkylation (+4.978931 Da) were set as variable modifications.
For quantification, a 0.003 m/z window centered on the theoretical m/z value of each reporter ion was utilized for the nearest signal intensity. Reporter ion intensities were adjusted to correct for the isotopic impurities from the different TMT reagents as per manufacturer specifications. The signal-to-noise values for all peptides were summed within each TMT channel. For each peptide, a total minimum sum signal-to-noise value of 150 and an MS1 and MS2 isolation specificity of greater than 70% were required. A target decoy database strategy and a false discovery rate (FDR) of 1% were set for peptide-spectrum matches following filtering by linear discriminant analysis (LDA) (33, 34). The FDR for final collapsed proteins was 1%. The mass tolerance for fragment ions was 25 ppm. The peaklist generating software used was GFY Core (version 3.8). The database searched was M. musculus proteome (Uniprot 0/2014) with added common contaminant proteins.
qPCR
RNA was isolated using the PureLink RNA minikit (Ambion) and quantified using a NanoDrop 2000 spectrometer. cDNA was prepared by RT-PCR using a High Capacity cDNA reverse transcription kit (Applied Biosystems). Real-time quantitative PCR (qPCR) was performed on cDNA using a 7500 Fast Real-Time PCR System (Applied Biosystems) using PowerUp SYBR Green Master Mix (Applied Biosystems). Primer pair (Eurofins Genomics) sequences were as follows:
L2hgdh 5′- CCA AGA AGC AGG TGG CTC TAT-3′ (forward) and 5′- ACA CCG AAT CTC CTT TCC CTT G-3′ (reverse); D2hgdh 5′- AGC AAC TGC AGA CAT GCA AC-3′ (forward) and 5′- TAG CAG TGC CTA AGA ATC TGG G-3′ (reverse); Phd3 5′- TGC TGA AGA AAG GGC AGA AG-3′ (forward) and 5′- GCA CAC CAC AGT CAG TGT TTA-3′ (reverse); Il1b 5′- GGA AGC AGC CCT TCA TCT TT-3′ (forward) and 5′- TGG CAA CTG TTC CTG AAC TC-3′ (reverse); Nos2 5′- CCA AGC CCT CAC CTA CTT CC-3′ (forward) and 5′- CTC TGA GGG CTG ACA CAA GG-3′ (reverse); Glut1 5′- GAT CAC TGC AGT TCG GCT ATA A-3′ (forward) and 5′- GTA GCG GTG GTT CCA TGT T-3′ (reverse); Ldha 5′- ATC TTG ACC TAC GTG GCT TGG A-3′ (forward) and 5′- CCA TAC AGG CAC ACT GGA ATC TC-3′ (reverse); Hif1a 5′- GGG TAC AAG AAA CCA CCC AT-3′ (forward) and 5′- GAG GCT GTG TCG ACT GAG AA-3′ (reverse).
Western Blotting
Protein samples from cultured cells were prepared by direct lysis in 5× SDS sample buffer, followed by heating at 95 °C for 5 min. Samples were resolved on 8–12% SDS–polyacrylamide gels followed by transfer to PVDF membranes. Membranes were blocked in 5% (w/v) dried milk in Tris-buffered saline/Tween (TBST) for 1 h at room temperature, after which they were incubated with primary antibody, followed by the correct horseradish-peroxidase-conjugated secondary antibody. Blots were developed using Western Bright ECL substrate (Advansta) or Immobilon Western chemiluminescent substrate (Millipore) using a Bio-Rad Gel Doc. Antibodies used were anti-HIF-1α (catalogue no. 14179), anti-IκB-α (9242), anti-phospho-NF-κB-p65 (3033), anti-NF-κB-p65 (8242), anti-phospho-p38 MAPK (9211), anti-p38 MAPK (catalogue no. 9212), anti-phospho-ERK (9101), anti-ERK (4695), anti-phospho-p70-S6K (catalogue no. 9205), anti-p70-S6k (9202), all purchased from Cell Signaling Technology, anti-IL-1β (R&D Systems, AF401-NA), anti-β-actin (Sigma-Aldrich, AC-74), anti-FLAG (Sigma-Aldrich, F1804), L-2HGDH (Antibody Genie, CAB7996), and D-2HGDH (Antibody Genie, CAB16213). Anti-HIF-1α from Novus (catalogue no. NB100-449) was used for human blots. Secondary antibodies used were horseradish-peroxidase-conjugated anti-mouse IgG, anti-goat IgG and anti-rabbit IgG (Jackson ImmunoResearch).
Seahorse
Cells were plated at 1 × 105 cells/well in 100 μl DMEM (10% FCE, 1% P/S, 10% L929 supernatant) in a 96-well Seahorse plate (Agilent). The four corner wells were left without cells in order to perform background calibration measurements. Cells were treated and stimulated as normal. A utility plate was prepared by adding 200 μl calibrant fluid to each well and placing it with the injector cartridge into a CO2-free incubator overnight. One hour prior to the run, cells were washed with warm PBS, and 180 μl XF assay media was added to each well, and the plate was place in a CO2-free incubator. Glucose (10 mM), oligomycin (1 μ), and 2DG (50 mM) were diluted in XF assay media and added to the injector cartridge, concentrations indicated show final concentration when added to the cell plate. The injector cartridge and utility plate were loaded into a Seahorse Xfe96 (Agilent) for calibration. Following calibration, the utility plate was removed, the cell plate was loaded in its place, and measurements commenced as per the Glycolysis Stress Kit program.
ELISA
Cytokine concentrations in cell supernatants were measured using DuoSet ELISA kits (R&D Systems) for murine TNF-α (DY410) and TMB Substrate Reagent (BioLegend) according to the manufacturer's instructions. Optical densities were measured with a FLUOstar Optima plate reader (BMG Labtech), and concentrations were calculated using a four-parameter curve fit.
Overexpression
L-2HGDH (catalogue number MR207410), D-2HGDH (catalogue number MR208571), and pCMV6 (catalogue number PS100001) plasmids were purchased from Origene. Lipofectamine 2000 was purchased from Thermo Fisher Scientific. Cells were plated at 2 × 105 cells per well in 24-well plates overnight. Before transfection, the media was replaced with 500 μl of DMEM (without FCS or PS). Two sets of microcentrifuge tubes were prepared for each plasmid to be transfected. OptiMEM (100 μl/well) was added to each tube. Lipofectamine 2000 (4 μl/well) was added to one set of tubes, and plasmid (1.6 μg/well) was added to the other. The tubes were incubated for 5 min, then mixed and incubated for a further 20 min. The transfection mix (200 μl per well) was then added to the cell plate and left for 24 h, after which the cells were treated as desired.
siRNA
L-2HGDH siRNA (catalogue number 4390771, assay ID s104072), negative control siRNA (catalogue number 4390844) and Lipofectamine RNAiMAX were purchased from Thermo Fisher Scientific. Cells were plated at 1 × 106 cells per well in 12-well plates overnight. Before transfection, the medium was replaced with 500 μl of DMEM (without FCS or PS). Two sets of microcentrifuge tubes were prepared for each siRNA to be transfected. DMEM (250 μl/well) was added to each tube. RNAiMAX (5 μl/well) was added to one set of tubes, and siRNA (50 nM final well concentration) was added to the other. The contents of the RNAiMAX tubes were added to the siRNA tubes, mixed by pipetting, and then incubated for 15 min. The transfection mix (500 μl per well) was then added to the cell plate and left for 144 h.
Statistical analysis
Statistical analysis was performed using Prism 8 (GraphPad). Data were analyzed by unpaired Student's t test or one-way ANOVA, as indicated in the text. Differences were considered significant at the values of p < 0.05.
Data availability
All relevant data are contained within this article and in the supporting information. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD029155.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors would like to thank Aaron Douglas for helpful discussions relating to the manuscript.
Author contributions
N. C. W. and D. G. R. conceptualization; A. S. H. C. and M. P. J. formal analysis; S. M. C. and L. A. O. funding acquisition; N. C. W., D. G. R., A. S. H. C., E. L. M., and M. P. J. investigation; N. C. W. methodology; C. F. resources; L. A. O. supervision; N. C. W. writing—original draft; L. A. O. writing—review and editing.
Funding and additional information
This work was supported by funding from the European Research Council (ECFP7-ERC-MICROINNATE), Science Foundation Ireland (SFI12/IA/1531) and the Wellcome Trust (109443/Z/15/Z) obtained by L. A. O., as well as funding from Science Foundation Ireland (FRL 4862) obtained by S. M. C.
Edited by Dennis Voelker
Footnotes
Present address for Dylan G. Ryan: MRC Cancer Unit, University of Cambridge, Hutchison/MRC Research Centre, Cambridge Biomedical Campus, Cambridge, UK.
Present address for Ana S. H. Costa: Department of Environmental Medicine and Public Health, Icahn School of Medicine at Mount Sinai, New York, USA.
Present address for Christian Frezza: CECAD Research Center, Faculty of Medicine, University Hospital Cologne, Cologne, Germany.
Supporting information
References
- 1.Kelly B., O'Neill L.A.J. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 2015;25:771–784. doi: 10.1038/cr.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galván-Peña S., O'Neill L.A.J. Metabolic reprograming in macrophage polarization. Front. Immunol. 2014;5:420. doi: 10.3389/fimmu.2014.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryan D.G., Murphy M.P., Frezza C., Prag H.A., Chouchani E.T., O'Neill L.A., Mills E.L. Coupling Krebs cycle metabolites to signalling in immunity and cancer. Nat. Metab. 2019;1:16–33. doi: 10.1038/s42255-018-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mills E.L., Kelly B., Logan A., Costa A.S.H., Varma M., Bryant C.E., Tourlomousis P., Dabritz J.H.M., Gottlieb E., Latorre I., Corr S.C., McManus G., Ryan D., Jacobs H.T., Szibor M., et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell. 2016;167:457–470.e13. doi: 10.1016/j.cell.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tannahill G.M., Curtis A.M., Adamik J., Palsson-McDermott E.M., McGettrick A.F., Goel G., Frezza C., Bernard N.J., Kelly B., Foley N.H., Zheng L., Gardet A., Tong Z., Jany S.S., Corr S.C., et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryan D.G., O'Neill L.A.J. Krebs cycle reborn in macrophage immunometabolism. Annu. Rev. Immunol. 2020;38:289–313. doi: 10.1146/annurev-immunol-081619-104850. [DOI] [PubMed] [Google Scholar]
- 7.Liu P.S., Wang H., Li X., Chao T., Teav T., Christen S., Di Conza G., Cheng W.C., Chou C.H., Vavakova M., Muret C., Debackere K., Mazzone M., Huang H.D., Fendt S.M., et al. α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat. Immunol. 2017;18:985–994. doi: 10.1038/ni.3796. [DOI] [PubMed] [Google Scholar]
- 8.Ye D., Guan K.-L., Xiong Y. Metabolism, activity, and targeting of D- and L-2-hydroxyglutarates. Trends Cancer. 2018;4:151–165. doi: 10.1016/j.trecan.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Struys E.A. 2-Hydroxyglutarate is not a metabolite; d-2-hydroxyglutarate and l-2-hydroxyglutarate are! Proc. Natl. Acad. Sci. U. S. A. 2013;110 doi: 10.1073/pnas.1318777110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dang L., Su S.M. Isocitrate dehydrogenase mutation and (R)-2-hydroxyglutarate: From basic discovery to therapeutics development. Annu. Rev. Biochem. 2017;86:305–331. doi: 10.1146/annurev-biochem-061516-044732. [DOI] [PubMed] [Google Scholar]
- 11.Shim E.H., Livi C.B., Rakheja D., Tan J., Benson D., Parekh V., Kho E.Y., Ghosh A.P., Kirkman R., Velu S., Dutta S., Chenna B., Rea S.L., Mishur R.J., Li Q., et al. L-2-Hydroxyglutarate: An epigenetic modifier and putative oncometabolite in renal cancer. Cancer Discov. 2014;4:1290–1298. doi: 10.1158/2159-8290.CD-13-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koivunen P., Lee S., Duncan C.G., Lopez G., Lu G., Ramkissoon S., Losman J.A., Joensuu P., Bergmann U., Gross S., Travins J., Weiss S., Looper R., Ligon K.L., Verhaak R.G., et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng Q.Y., Xiong J., Huang W., Ma Q., Ci W., Feng Y.Q., Yuan B.F. Sensitive determination of onco-metabolites of D- and L-2-hydroxyglutarate enantiomers by chiral derivatization combined with liquid chromatography/mass spectrometry analysis. Sci. Rep. 2015;5:15217. doi: 10.1038/srep15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chowdhury R., Yeoh K.K., Tian Y.M., Hillringhaus L., Bagg E.A., Rose N.R., Leung I.K., Li X.S., Woon E.C., Yang M., McDonough M.A., King O.N., Clifton I.J., Klose R.J., Claridge T.D., et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Böttcher M., Renner K., Berger R., Mentz K., Thomas S., Cardenas-Conejo Z.E., Dettmer K., Oefner P.J., Mackensen A., Kreutz M., Mougiakakos D. D-2-hydroxyglutarate interferes with HIF-1α stability skewing T-cell metabolism towards oxidative phosphorylation and impairing Th17 polarization. OncoImmunology. 2018;7 doi: 10.1080/2162402X.2018.1445454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bunse L., Pusch S., Bunse T., Sahm F., Sanghvi K., Friedrich M., Alansary D., Sonner J.K., Green E., Deumelandt K., Kilian M., Neftel C., Uhlig S., Kessler T., von Landenberg A., et al. Suppression of antitumor T cell immunity by the oncometabolite (R)-2-hydroxyglutarate. Nat. Med. 2018;24:1192–1203. doi: 10.1038/s41591-018-0095-6. [DOI] [PubMed] [Google Scholar]
- 17.Ugele I., Cárdenas-Conejo Z.E., Hammon K., Wehrstein M., Bruss C., Peter K., Singer K., Gottfried E., Boesch J., Oefner P., Dettmer K., Renner K., Kreutz M. D-2-hydroxyglutarate and L-2-hydroxyglutarate inhibit IL-12 secretion by human monocyte-derived dendritic cells. Int. J. Mol. Sci. 2019;20:742. doi: 10.3390/ijms20030742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Uden P., Kenneth N.S., Rocha S. Regulation of hypoxia-inducible factor-1alpha by NF-kappaB. Biochem. J. 2008;412:477–484. doi: 10.1042/BJ20080476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oldham W.M., Clish C.B., Yang Y., Loscalzo J. Hypoxia-mediated increases in L-2-hydroxyglutarate coordinate the metabolic response to reductive stress. Cell Metab. 2015;22:291–303. doi: 10.1016/j.cmet.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cramer T., Yamanishi Y., Clausen B.E., Förster I., Pawlinski R., Mackman N., Haase V.H., Jaenisch R., Corr M., Nizet V., Firestein G.S., Gerber H.P., Ferrara N., Johnson R.S. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–657. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang T., Liu H., Lian G., Zhang S.-Y., Wang X., Jiang C. HIF1α-induced glycolysis metabolism is essential to the activation of inflammatory macrophages. Mediators Inflamm. 2017;2017:9029327. doi: 10.1155/2017/9029327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu W., Yang H., Liu Y., Yang Y., Wang P., Kim S.H., Ito S., Yang C., Wang P., Xiao M.T., Liu L.X., Jiang W.Q., Liu J., Zhang J.Y., Wang B., et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta R.D. Recent advances in enzyme promiscuity. Sustainable Chem. Process. 2016;4:2. [Google Scholar]
- 24.Intlekofer A.M., Dematteo R.G., Venneti S., Finley L.W., Lu C., Judkins A.R., Rustenburg A.S., Grinaway P.B., Chodera J.D., Cross J.R., Thompson C.B. Hypoxia induces production of L-2-hydroxyglutarate. Cell Metab. 2015;22:304–311. doi: 10.1016/j.cmet.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isaacs J.S., Jung Y.J., Mole D.R., Lee S., Torres-Cabala C., Chung Y.L., Merino M., Trepel J., Zbar B., Toro J., Ratcliffe P.J., Linehan W.M., Neckers L. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: Novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8:143–153. doi: 10.1016/j.ccr.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 26.Anso E., Weinberg S.E., Diebold L.P., Thompson B.J., Malinge S., Schumacker P.T., Liu X., Zhang Y., Shao Z., Steadman M., Marsh K.M., Xu J., Crispino J.D., Chandel N.S. The mitochondrial respiratory chain is essential for haematopoietic stem cell function. Nat. Cell Biol. 2017;19:614–625. doi: 10.1038/ncb3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinberg S.E., Singer B.D., Steinert E.M., Martinez C.A., Mehta M.M., Martinez-Reyes I., Gao P., Helmin K.A., Abdala-Valencia H., Sena L.A., Schumacker P.T., Turka L.A., Chandel N.S. Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature. 2019;565:495–499. doi: 10.1038/s41586-018-0846-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu T., Stewart K.M., Wang X., Liu K., Xie M., Ryu J.K., Li K., Ma T., Wang H., Ni L., Zhu S., Cao N., Zhu D., Zhang Y., Akassoglou K., et al. Metabolic control of TH17 and induced Treg cell balance by an epigenetic mechanism. Nature. 2017;548:228–233. doi: 10.1038/nature23475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackay G.M., Zheng L., van den Broek N.J., Gottlieb E. Analysis of cell metabolism using LC-MS and isotope tracers. Methods Enzymol. 2015;561:171–196. doi: 10.1016/bs.mie.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 30.Mills E.L., Ryan D.G., Prag H.A., Dikovskaya D., Menon D., Zaslona Z., Jedrychowski M.P., Costa A.S.H., Higgins M., Hams E., Szpyt J., Runtsch M.C., King M.S., McGouran J.F., Fischer R., et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. 2018;556:113–117. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McAlister G.C., Nusinow D.P., Jedrychowski M.P., Wuhr M., Huttlin E.L., Erickson B.K., Rad R., Haas W., Gygi S.P. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 2014;86:7150–7158. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eng J.K., McCormack A.L., Yates J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 33.Huttlin E.L., Jedrychowski M.P., Elias J.E., Goswami T., Rad R., Beausoleil S.A., Villén J., Haas W., Sowa M.E., Gygi S.P. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beausoleil S.A., Villén J., Gerber S.A., Rush J., Gygi S.P. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are contained within this article and in the supporting information. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD029155.