Take Home Message
We reviewed the literature to identify studies investigating circulating tumor DNA (ctDNA) in patients with renal cell carcinoma (RCC). We found that the ctDNA levels are low in RCC, but the use of tumor-guided analysis improves detection rates and cell-free methylated DNA immunoprecipitation and high-throughput sequencing is a sensitive method for ctDNA detection in RCC.
Keywords: Cell-free DNA, Circulating tumor DNA, Kidney neoplasms, Renal cell carcinoma, Liquid biopsy
Abstract
Context
Over the past decade there has been increasing interest in the potential of liquid biopsies and systematic biomarkers in the diagnosis and management of kidney cancer, as they may provide a tool for early detection of disease and monitoring of treatment response.
Objective
To identify and summarize relevant published data on circulating tumor DNA (ctDNA) in patients with renal cell carcinoma (RCC).
Evidence acquisition
We performed a systematic review according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement of studies identified in PubMed, MEDLINE, EMBASE, and Cochrane Library up to January 15, 2021. Two reviewers independently screened all articles and performed the data extraction.
Evidence synthesis
Nineteen studies investigating ctDNA in RCC (1237 patients) were included and analyzed in the final review. The study size and design varied widely, and the studies were divided into five groups according to the method used for ctDNA detection. The outcome data included (1) the sensitivity/specificity if available; (2) the method used for ctDNA detection; and (3) the main findings in the studies.
Conclusions
The studies highlight that the level of ctDNA in RCC appears to be low. Studies using multiple methods for ctDNA detection indicate that tumor-guided analysis improves the ctDNA detection rate and suggest that cell-free methylated DNA immunoprecipitation and high-throughput sequencing may be a very sensitive method for ctDNA detection in RCC.
Patient summary
We systematically reviewed the literature to identify all relevant studies investigating circulating tumor DNA in patients with kidney cancer to investigate its use and potential in this highly malignant disease. We found that the level of circulating tumor DNA is low in kidney cancer and that very sensitive methods have to be used for detection in this disease.
1. Introduction
Urological malignancies are an increasing health care problem worldwide, with more than an estimated 1 million patients newly diagnosed annually [1]. Renal cell carcinoma (RCC) is a highly malignant disease and represents 3% of all malignancies in Western countries, with its incidence increasing annually [2].
Diagnosis and treatment of patients with RCC present many challenges. This is mainly because RCC is characterized by asymptomatic manifestation in the early stages and a poor response to radiotherapy and chemotherapy in metastatic stages [3], [4]. Over the past several decades, there has been increasing interest in research on liquid biopsies and circulating cancer biomarkers because they might provide a tool for early detection of disease and for monitoring of treatment response. Ellinger et al [5] reported that the presence of cell-free DNA (cfDNA) in the bloodstream was first detected more than 50 yr ago. cfDNA can easily be obtained from peripheral blood, and it has been shown that it is present in patients with multiple solid malignancies [6]. The presence of cfDNA in the bloodstream of patients and changes in the levels of these circulating nucleic acids are associated with tumor load and tumor progression. However, studies differ as to whether they have been able to show correlation between cfDNA levels and tumor stage and grade in RCC [7], [8], [9]. Circulating tumor DNA (ctDNA) is cfDNA derived from apoptotic or necrotic tumor cells, secretions from macrophages, or circulating tumor cells (CTCs) [10]. Analysis of ctDNA may open the possibility of noninvasive detection of the mutational profile of a specific cancer during tumor progression, and studies have shown that the presence of ctDNA correlates with advanced disease and disease progression during treatment [7], [11], [12]. While several studies on ctDNA in RCC have been performed, there are still no clear recommendations or summaries of the literature.
We report here on a systematic review of all published data on ctDNA in patients with RCC to investigate the use and potential of ctDNA in RCC.
2. Evidence acquisition
2.1. Search strategy
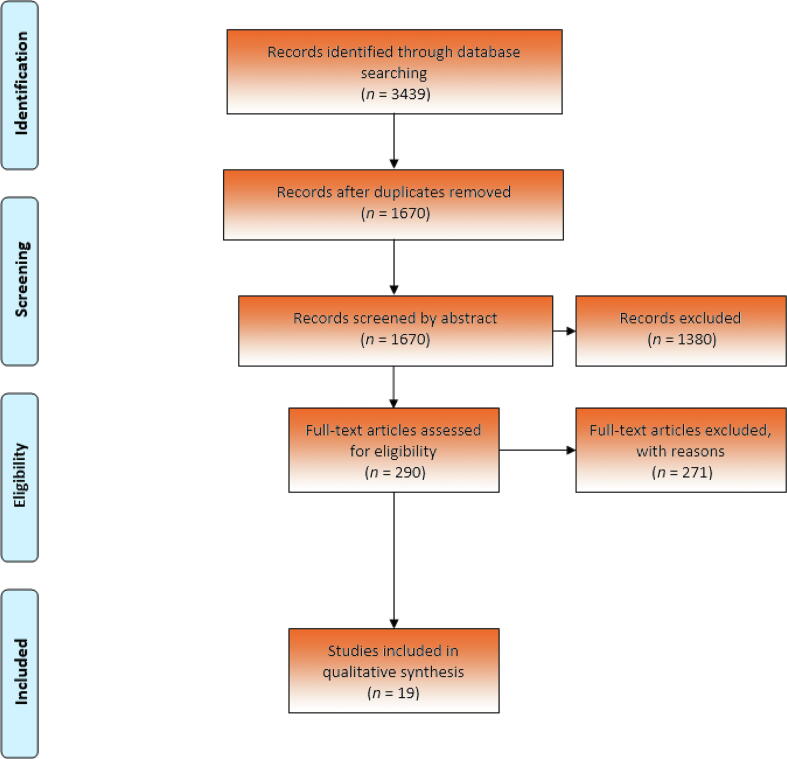
This systematic review was performed according to Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [13] (Fig. 1). The review was registered on the PROSPERO platform under identification number CRD42020208730. One author (L.G.) and an expert librarian conducted the search, for which the strategy is available in the Supplementary material.
Fig. 1.
Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flowchart of study selection.
The literature search was performed in the PubMed, MEDLINE, EMBASE, and Cochrane Library databases to identify relevant studies up to January 15 2021, with sorting and removal of duplicate articles performed in Covidence.
2.2. Screening and selection criteria
Study selection was performed in three sequential steps: articles were first assessed by title, second by abstract, and third by full text. Studies were included if they met the following inclusion criteria: (1) patients diagnosed with any stage of RCC; (2) analysis of DNA in plasma/serum; (3) analysis of mutations at a DNA level or epigenetic changes; and (4) article written in English. Articles regarding malignancies other than RCC, animal and cell-line studies, studies published before 2010, and small pilot studies containing fewer than five patients were excluded.
Two authors (L.G. and K.M.K.) independently screened the titles and abstracts and selected potentially eligible studies. The full text of potentially eligible studies was then screened. The authors discussed any disagreements until consensus was reached.
2.3. Data extraction
A data extraction sheet was developed and prespecified outcomes of interest were collected. Two reviewers (L.G. and K.M.K.) independently extracted data from the studies included. Disagreements were resolved via consensus.
The trial characteristics extracted included: (1) first author’s name and year of publication; (2) method applied for detection of ctDNA; (3) number of patients/cases; and (4) number of controls. The outcome data included: (1) sensitivity and specificity when available; (2) method used for ctDNA detection; and (3) key findings in the particular studies.
Quality assessment was not performed according to predefined criteria because of the variation in study design and methods in the studies included. The quality of the studies was assessed by one author (K.M.K.) with wide experience with methods used for ctDNA detection.
3. Evidence synthesis
3.1. Studies included
According to our search criteria, 3439 potentially relevant papers were identified and screened in the primary search. All papers were added into EndNote and duplicates were removed, leaving 1670 studies. The titles, abstracts, and full texts were screened manually, and 19 studies met the inclusion criteria [9], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31]. All studies included were published between 2010 and 2020. They were published in full, but the size and design of the studies varied widely, and therefore they were divided into five groups according to the method used for ctDNA detection: (1) tumor-guided analysis of plasma; (2) targeted sequencing of plasma; (3) global sequencing of plasma; (4) targeted methylation analysis of plasma; and (5) global methylation analysis of plasma.
To cover the use of ctDNA in RCC, we screened the studies in each of the ctDNA detection groups to find the ctDNA detection rate and the sensitivity and specificity if available. Furthermore, we identified studies investigating the concordance between ctDNA samples and tumor tissue from the same patient, and studies investigating the potential of ctDNA for prognosis and disease monitoring. Finally, we summarized the methods used for ctDNA detection and which method to choose on the basis of our findings.
3.2. ctDNA detection rates
3.2.1. Tumor-guided analysis of plasma
Five studies [14], [15], [16], [17], [18] applied tumor-guided analysis of plasma (Table 1). This type of analysis is based on identification of mutations in tumor tissue DNA and subsequent analysis of these mutations in cfDNA. The advantage of this method is that very high technical sensitivity can be achieved when the mutations of the tumor are known. The downside is that the analysis does not identify newly acquired mutations. The five studies included between five and 24 RCC patients and the ctDNA detection rate ranged from 17% to 54%.
Table 1.
Tumor-guided analysis of plasma.
| Study | Cases | Stage | Method | ctDNA detection rate (%) | Findings |
|---|---|---|---|---|---|
| Bettegowda 2014 [14] | 5 | mRCC | PCR or targeted amplicon sequencing of mutations identified in tumor tissue using sequencing. | 40 | ctDNA detected in only 2 of 5 patients with mRCC leading to the conclusion that RCC is a low-ctDNA cancer type. |
| Corrò 2017 [15] | 9 | nmRCC | Targeted sequencing of VHL (tumor and plasma samples), depth 7880× to 10 895× in plasma. Mutation-specific PCR in a single patient. | 17 | Mutation only identified in one patient where mutants-specific PCR was performed. No mutations identified by sequencing. |
| Smith 2020 [16] | 29 | Various | Customized sequencing targeting on average 297 variants per each patient identified by exome sequencing of tumor tissue; > 20 000 informative reads for a sample to pass quality control. | 54 | RCC has low levels of ctDNA compared to other types of cancer.Targeted, personalized approach improved ctDNA detection compared to untargeted approach. Heterogeneity analysis in two patients with multiple spatially distinct tumor tissue samples: ctDNA analysis may overcome tumor heterogeneity. |
| Wan 2020 [17] | 24a | Various | Exome sequencing of tumor tissue and subsequent custom panel sequencing of plasma targeting identified mutations. Approximate sensitivity of 10−5. | 42 (at specificity of 90%) | RCC has low levels of ctDNA compared to other types of cancer. ctDNA fraction of approximately 10-4 in RCC patients. |
| Lasseter 2020 [18] | 23 | mRCC | Tumor-guided amplicon sequencing of 47 variants identified in tumor tissue using panel sequencing of 27 genes. Depth of 122 035× for plasma samples. | 52 | Validation of tumor variants detected 16 variants in plasma that had already been detected by initial panel sequencing, and an additional 9 tumor-variants present at low frequency in plasma. Thus, tumor-guided analysis increased the sensitivity of ctDNA detection. VAF correlated with response to treatment in 5 patients with serial samples. |
ctDNA = circulating tumor DNA; mRCC = metastatic renal cell carcinoma; nmRCC = nonmetastatic RCC; PCR = polymerase chain reaction; VAF = variant allele frequency.
Subgroup of patients investigated in study by Smith et al [16].
In an early ctDNA landmark study, Bettegowda et al [14] investigated the presence of ctDNA in 640 patients with various cancer types, including five patients with metastatic RCC. Detection of ctDNA in plasma was via polymerase chain reaction (PCR)- or sequencing-based analysis of a mutation identified in the tumor. ctDNA was detected in 40% of RCC patients with metastatic disease, which led to classification of RCC as a low-ctDNA malignancy. Although the classification was based on a very low number of patients, it has been supported by multiple studies performed since then [15], [16], [17], [18].
The studies by Corrò et al [15] and Lasseter et al [18] both used targeted deep sequencing of one or a few mutations identified in tumor tissue and had a ctDNA detection rate of 17% and 52%, respectively. The patients in the study by Corrò et al had nonmetastatic disease, whereas the patients in the study by Lasseter et al had metastatic RCC. The difference in detection rate may result from the different patient cohorts; however, the relatively low detection rates in these studies are surprising, since the tumor-informed strategy has the potential to be one of the most technically sensitive methods. The study by Lasseter et al also showed that sequencing of cfDNA to an extremely high depth (122 035×) improved ctDNA detection because it increases the detection of variants present at very low frequencies.
Two very recent studies with mixed patient cohorts (nonmetastatic and metastatic RCC) [16], [17] applied ctDNA analysis of a large number of variants identified in tumor tissue. In the study by Smith et al [16], 91 patients were investigated using a variety of methods (Table 1, Table 2, Table 3). The authors found that a personalized approach targeting an average of 297 tumor-specific variants per patient using custom panel sequencing of mutations identified via exome sequencing of tumor tissue improved ctDNA detection compared to an untargeted approach. Tumor-guided analysis of plasma was performed for 29 patients and identified ctDNA in 54% of the patients for whom the analysis passed quality control. Wan et al [17] applied a similar methodological approach to 24 patients and found that the fraction of ctDNA in RCC patients was approximately 10−4. This finding highlights that extremely sensitive methods are required for ctDNA analysis in RCC.
Table 2.
Targeted sequencing of plasma.
| Study | Cases | Stage | Method | ctDNA detection rate (%) | Findings |
|---|---|---|---|---|---|
| Hahn 2017 [19] | 19 | mRCC | Panel sequencing, 72 genes; depth not stated. | 68 | Median mutation rate: 1 in ctDNA and 3 in tumor tissue. Concordance with matched tumor tissue 8.6%. |
| Maia 2017 [20] | 26 | mRCC | Panel sequencing, 73 genes; average depth 15 000×. | 53 | Median mutation rate in ctDNA: 2. Significantly higher tumor burden in patients with detectable ctDNA. Significant correlation between number of mutations and tumor burden. |
| Pal 2017 [21] | 220 | mRCC | Panel sequencing, 73 genes; average depth 15 000× | 79 | Median mutation rate in ctDNA: 1. Median VAF 20% in patients receiving first-line treatment and 24% in later-line treatment. Longitudinal samples from 13 patients: Increasing discordance between ctDNA profiles with increasing time between samplings, indicative of clonal evolution of the tumor. |
| Yamamoto 2019 [22] | 53 | Various | Panel sequencing, 48 genes; average depth 204× | 30 | Median mutation rate in ctDNA: 2. Median VAF 10%. ctDNA dynamics correlated with tumor burden in 6 patients with longitudinal samples. Mutant ctDNA fragments tends to be shorter than corresponding wild-type cfDNA fragments. |
| Bacon 2020 [23] | 55 | mRCC | Panel sequencing, 982 genes; median depth 938×, detection limit 1% VAF | 33 | Average ctDNA VAF 3.9% for ctDNA-positive patients. Concordance with matched tumor tissue 77%. ctDNA-positive patients had significantly shorter survival than ctDNA-negative patients. |
| Zhang 2020 [24] | 50 | mRCC | Panel sequencing, 120 genes; median depth 2980× | 100 | Median mutation rate in ctDNA: 2. Significant correlation between number of aberrations in plasma and number of lines of therapy. |
| Smith 2020 [16] | 43 | Mostly mRCC | Panel sequencing, 10 genes; median raw depth 9688× | 19 | Longitudinal samples from 37 patients: ctDNA levels fluctuate in accordance with clinical response to treatment. |
| Lasseter 2020 [18] | 40 | mRCC | Panel sequencing, 27 genes; median depth 989× | 28 | Analysis also identified 2 germline variants and 6 variants resulting from clonal hematopoiesis. Amplicon sequencing of an additional 31 variants identified in tumor tissue detected 9 of these at low frequency in plasma, ie, tumor-guided analysis increased the sensitivity of ctDNA detection. VAF correlated with response to treatment in 5 patients with serial samples. |
cfDNA = cell-free DNA; ctDNA = circulating tumor DNA; mRCC = metastatic renal cell carcinoma; VAF = variant allele frequency.
Table 3.
Global sequencing of plasma.
| Study | Cases | Controls | Stage | Method | ctDNA detection rate (%) | Findings |
|---|---|---|---|---|---|---|
| Mouliere 2018 [25] | 33 | 65 | Not stated | sWGS, depth 0.4×. | 65a (at specificity of 95%) | Large study of 344 plasma samples from 200 cancer patients. Enrichment of fragments of 90–150 bp improved ctDNA detection. RCC is a low-ctDNA cancer. |
| Smith 2020 [16] | 48 | 70 | Various | sWGS on average 16.4 million reads per sample. Predicted limit of detection: 0.3% ctDNA fraction | 23 (at specificity of 95%b) | RCC has low levels of ctDNA compared to other cancers. Targeted, personalized approach improved ctDNA detection compared to untargeted approach. |
| 43 | 42 | Mostly mRCC | Modified fast aneuploidy screening test-sequencing system and ichorCNA analysis of sWGS data (sensitivity 3% ctDNA fraction) | 33 (specificity not stated) | Longitudinal samples from 37 pts: ctDNA levels fluctuate in accordance with clinical response to treatment. |
ctDNA = circulating tumor DNA; mRCC = metastatic renal cell carcinoma; sWGS = shallow whole-genome sequencing.
Low-ctDNA cancers: renal, pancreatic, and glioma; not specified for RCC alone.
Determined by Mouliere et al [25].
3.2.2. Targeted sequencing of plasma
Eight studies [16], [18], [19], [20], [21], [22], [23], [24] (Table 2) applied targeted panel sequencing of DNA from plasma without prior analysis of tumor tissue DNA. The advantage of this strategy is that it does not require a tissue sample (ie, it is less invasive) and allows for identification of newly acquired mutations. However, the analysis may be less sensitive than tumor-guided analysis. In general, the studies included a larger number of patients, ranging from 19 to 220, and the detection rate was considerably better, ranging from 19% to 100%. Most of the studies were performed among patients with metastatic disease; only 14 patients in the study by Yamamoto et al [22] and two patients in the study by Smith et al [16] had nonmetastatic RCC (detection rates of 30% and 19%, respectively). The high detection rates may thus reflect a high tumor load in many of these patients. The findings in the study by Maia et al [20] support this hypothesis. The authors found that ctDNA detection correlated with tumor burden; tumor burden was significantly higher in patients with detectable ctDNA than in patients without ctDNA. Furthermore, the number of mutations in ctDNA was also significantly correlated with tumor burden.
Bacon et al [23] found a relatively low detection rate of 33% in comparison to similar studies. This may be because in 40 of 55 patients, plasma samples were collected after surgical removal of the primary tumor. However, in a study by Hahn et al [19] in which 15 of 19 patients also had their primary tumor surgically removed before sample collection, the ctDNA detection rate was 68%. The low detection rate could also be explained by the technical sensitivity of the assay, which did not allow for detection of variants present at a variant allele frequency (VAF) of <1% [23].
There seems to be a trend towards better detection in studies in which ctDNA analysis was based on a gene panel covering a high number of genes, very high sequencing depths, or a combination of these. For example, the studies by Maia et al [20] and Pal et al [21] both sequenced 73 genes with 15 000× coverage and had detection rates of 53% and 79%, respectively. By comparison, Smith et al [16] sequenced ten genes to a depth of 9688× and Lasseter et al [18] sequenced 27 genes to a depth of 989× and had ctDNA detection rates of 19% and 28%, respectively.
3.2.3. Global sequencing of plasma
Two studies [16], [25] that used global sequencing of plasma were identified (Table 3), both of which applied shallow whole-genome sequencing (sWGS). This untargeted method requires no prior knowledge of specific mutations in the tumor and has low sensitivity for single-base mutations but may identify large deletions and duplications. sWGS is relatively inexpensive and easy to perform. Mouliere et al [25] performed sWGS to a depth of 0.4× on 344 samples from 200 patients with various types of cancer including 33 with RCC (unknown stage of disease). ctDNA detection was based on analysis of cfDNA fragment lengths in cancer patients and healthy individuals. They found that in silico enrichment of fragments between 90 and 150 bp in size improved detection of ctDNA across cancer types. However, in RCC the proportion of fragments <150 bp was very similar to that in healthy individuals, indicating a low amount of ctDNA in RCC. The ctDNA detection rate was 65% for “low-ctDNA cancers” (RCC, pancreatic cancer, and glioma).
Smith et al [16] applied the same in silico size selection of cfDNA fragments as Mouliere et al [25] and found that this improved detection of ctDNA. In a cohort of patients with various stages of RCC and a cohort of mRCC patients they found that the ctDNA detection rate was 23% (11 of 48) and 33% (14 of 43), respectively, when the untargeted sWGS approach was combined with size selection.
Although there are some advantages of this untargeted and relatively less expensive method, the results suggest that it has relatively low sensitivity for RCC and that optimization is required.
3.2.4. Targeted methylation analysis of plasma
Targeted methylation analysis of plasma using a PCR-based method is very inexpensive and easy to perform. The disadvantage is that very few genes are analyzed and if the patients does not have a mutation in one of these genes, then the ctDNA test result will be negative. We identified five studies [9], [28], [29], [30], [31] that used this method (Table 4) and included between 27 and 157 patients with various stages of RCC. Four of the studies included a group of healthy controls.
Table 4.
Targeted methylation analysis of plasma.
| Gene | de Martino 2012 [9] | Hauser 2013 [28] | Skrypkina 2016 [29] | Lin 2017 [30] | Jung 2019 [31] | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| (157 cases, 43 controls) |
(35 cases, 54 controls) |
(27 cases, 15 controls) |
(142 cases, 34 controls) |
(100 cases, 0 controls) |
||||||
| SSY (%) | SPY (%) | SSY (%) | SPY (%) | SSY (%) | SPY (%) | SSY (%) | SPY (%) | SSY (%) | SPY (%) | |
| Single genes | ||||||||||
| RASSF1 | 45.9 | 93 | 22.9 | 98.2 | 62.9 | 93.3 | – | – | ||
| PTGS2 | 38.2 | 65.1 | 22.9 | 96.3 | – | – | – | |||
| VHL | 50.3 | 90.7 | – | 0 | 100 | – | – | |||
| P16 | 46.5 | 55.8 | – | – | – | – | ||||
| APC | – | 54.3 | 90.7 | 51.9 | 93.3 | – | – | |||
| GSTP1 | – | 17.1 | 98.1 | – | – | – | ||||
| p14ARF | – | 14.3 | 100 | – | – | – | ||||
| RAR-β | – | 40 | 85.2 | – | – | – | ||||
| LRRC3B | – | – | 74.1 | 66.7 | – | – | ||||
| FHIT | – | – | 55.6 | 100 | – | – | ||||
| PCDH17 | – | – | – | 57.7 | 100 | – | ||||
| SHOX2 | – | – | – | – | 12 | NA | ||||
| Gene combinations | ||||||||||
| APC or GSTP1 | 57.19 | 88.9 | ||||||||
| APC or PTGS2 | 60.0 | 87.0 | ||||||||
| APC or RAR-β | 74.3 | 77.8 | ||||||||
| PTGS2 or GSTP1 | 62.9 | 87.0 | ||||||||
| RASSF1, FHIT, APC | 92.3 | 86.7 | ||||||||
| RASSF1, FHIT | 77.8 | 93.3 | ||||||||
| RASSF1, APC | 77.8 | 93.3 | ||||||||
SSY = sensitivity; SPY = specificity; NA = not available.
In general, the authors concluded that methylation analysis provided high specificity but low sensitivity in distinguishing cancer patients from healthy controls. Hauser et al [28] and Skrypkina et al [29] performed combinatorial analysis of multiple genes, which increased the diagnostic sensitivity and specificity. Lin et al [30] and Jung et al [31] investigated the methylation status of PCDH17 and SHOX2, respectively. Lin et al found that the presence of PCDH17 correlated with advanced disease stage, and Jung et al found that patients with SHOX2-positive plasma had a significantly higher risk of death.
PCR-based analysis can be much less expensive to perform than sequencing-based analysis but it only allows for investigation of a limited number of mutations. Both sensitivity and specificity are often relatively low, as seen in the studies by Hauser et al [28] and de Martino et al [9]. Technically more sensitive methods such as digital droplet PCR might improve sensitivity for patients whose tumor has a mutation in the targeted genomic position [32]. However, it is likely that other methods targeting larger genomic regions are required for ctDNA detection in RCC.
3.2.5. Global methylation analysis
Three studies [18], [26], [27] investigated global methylation patterns (epigenetic aberrations) in cfDNA (Table 5). Epigenetic alterations are more abundant than genetic alterations in RCC, thereby providing more tumor-specific alterations. Global methylation analysis may thus achieve higher sensitivity. Cell-free methylated DNA immunoprecipitation and high-throughput sequencing (cfMeDIP-seq) is a bisulfite-free technique that can detect whole-genome methylation of cfDNA. The advantages of this technique is that it can enrich genome-wide CrG methylated cfDNA with a low DNA input (<10 ng) [33]. cfMeDIP-seq was applied in two studies: Nuzzo et al [26] had a ctDNA detection rate of 97% (at specificity of 100%) in a study of 69 patients with various stages of RCC, and Lasseter et al [18] had a ctDNA detection rate of 100% in a study of 34 patients with mRCC, although at lower specificity of 88%.
Table 5.
Global methylation analysis of plasma.
| Study | Cases | Controls | Stage | Method | ctDNA detection rate (%) | Findings |
|---|---|---|---|---|---|---|
| Nuzzo 2020 [26] | 69 | 13 | Various | cfMeDIP-seq | 97 (at specificity of 100%) | Classifier built on top 300 differentially methylated regions: 67 of 69 RCC plasma samples had higher methylation score than all of the healthy controls. |
| Lasseter 2020 [18] | 34 a | 38 | mRCC | cfMeDIP-seq; classifier trained on a separate cohort with 38 mRCC samples | 100 (at specificity of 88%) | cfMeDIP-seq increased ctDNA detection considerably compared to variant analysis, although at lower specificity. |
| Liu 2020 [27] | 56 (1531 cancer cases) | 1521 (training cohort) | Various | Whole-genome bisulfite sequencing; depth 30× | 21 (at specificity of 99.8%) | Large study with multiple cancer types. Sequencing data used to build a classifier to detect ctDNA and identify the cancer type (TOO). Sensitivity of ctDNA detection increased with disease stage. ctDNA detection in RCC was low compared to other cancers. TOO was classified correctly for all ctDNA-positive RCC samples. |
| 25 (654 cancer cases) | 610 (validation cohort) | Various | Methylation sequencing of 103 456 regions; median depth 139× | 12 (at specificity of 99.3%) |
cfMeDIP-seq = cell-free methylated DNA immunoprecipitation and high-throughput sequencing; ctDNA = circulating tumor DNA; mRCC = metastatic renal cell carcinoma; TOO = tissue of origin.
Total of 40 patients in the study.
In a large study including more than 2000 patients with different types of cancer (including 81 patients with various stages of RCC), Liu et al [27] found that the ctDNA detection rate in RCC was low compared to other types of cancer. The study was conducted in two steps using whole-genome bisulfite sequencing data from 1531 cancer cases and 1521 control samples to build a classifier for ctDNA detection. The authors applied the classifier to a validation cohort of 654 cancer cases and 610 control samples. The detection rate in this study was 21% (specificity of 99.8%) in the training cohort and 12% (specificity of 99.3%) in the validation cohort, and ctDNA detection had increasing sensitivity with increasing stage of disease.
These studies indicate that cfMeDIP-seq may be a promising strategy for ctDNA detection in RCC, but validation in larger studies is of utmost importance before any clear conclusions can be drawn.
3.3. Concordance with tumor tissue
One of the potential applications of ctDNA analysis is its ability to overcome genetic heterogeneity. Genetic heterogeneity occurs in space (spatial) and time (temporal), and poses a substantial challenge in the management of cancer. Spatial heterogeneity is the presence of genetic aberrations in regions/subclones of the primary tumor or in metastatic lesions. If targetable aberrations are not present in biopsy tissue from the primary tumor because of heterogeneity, they will remain unidentified and potentially lead to suboptimal treatment of the patient. Temporal heterogeneity arises over time as a result of clonal evolution of the tumor, which may be caused by selective pressure due to systemic treatment of the disease or genomic instability of the tumor.
Smith et al [16] performed heterogeneity analysis in one extensively characterized patient with ten spatially distinct tumor tissue samples and observed that region-specific mutations from nine of ten tumor regions were identified in plasma, thus indicating that ctDNA analysis may overcome tumor heterogeneity.
Another way to explore the potential ability of ctDNA to overcome genetic heterogeneity is to investigate the concordance between a plasma sample and a single tissue biopsy from the same patient. Bacon et al [23] used this approach and found concordance of 77% in four ctDNA-positive patients with available tumor tissue samples. Hahn et al [19] found concordance of only 8.6%. However, this is probably because the plasma samples in this patient cohort were collected several months after the corresponding tumor tissue samples (mean 22 mo, range 0–70) and sometimes after surgical resection of the primary tumor. Stratification of patients according to the time between tissue and plasma sample collection slightly increased the concordance rate to 11.4% for patients with ≤6 mo between tumor tissue and plasma collection. This indicates clonal evolution of the tumor over time, which would explain the low concordance between tumor tissue and plasma samples collected at different time points. This hypothesis is supported by findings in the study by Pal et al [21] in which 13 patients had ctDNA analysis performed on more than one sample, with the second sample obtained at a median of 157 d (range 21–360) after the first. The concordance between aberrations detected in first and second samples decreased with increasing time between sample collection, thus indicating clonal evolution of the tumor.
3.4. Prognosis and disease monitoring
One of the potential applications of ctDNA analysis is the ability to provide prognostic information, which was investigated in four studies. Jung et al, Lin et al, and Yamamoto et al all found that ctDNA detection in patients with various stages of RCC was significantly associated with either higher risk of death [31], or shorter progression-free survival [22], [30], cancer-specific survival [22], or overall survival [30]. Bacon et al [23] found that ctDNA-positive patients had shorter overall survival and progression-free survival on first-line therapy in comparison to ctDNA-negative patients. More studies are needed, but these findings indicate that ctDNA may have potential as a prognostic biomarker.
Another potential application of ctDNA analysis is in disease monitoring during systemic treatment and follow-up of patients. Four studies [16], [18], [21], [22] carried out longitudinal sampling of patients and in general found that the level of ctDNA fluctuated in accordance with the clinical course of the disease. Smith et al [16] analyzed 252 longitudinal samples from 37 patients. They observed that the level of ctDNA increased before initiation of treatment, decreased with response to treatment, and increased or remained elevated with disease progression or lack of response to treatment. The studies by Lasseter et al [18] and Yamamoto et al [22], who investigated ctDNA levels in longitudinal samples from five and six patients, respectively, support the findings reported by Smith et al. These studies thus support the notion that ctDNA may potentially be a biomarker suitable for monitoring of treatment response and disease progression.
3.5. Line of therapy and ctDNA
Two studies [21], [24] investigated the effect of first-line versus later-line therapy on genetic aberrations in ctDNA. Pal et al [21] performed a large study of 220 patients investigating the differences in genetic alterations found between patients receiving first-line and second- or later-line systemic treatment. They found that the median VAF was 20% for patients receiving first-line treatment but increased to 24% for patients receiving later-line treatment. Furthermore, they observed different mutation frequencies for several genes between patients receiving first-line therapy and those receiving later-line therapy. Zhang et al [24] found that the number of genetic aberrations in ctDNA was associated with the number of lines of therapy received. The two studies indicate that ctDNA may have a role in identifying genetic aberrations acquired during systemic treatment and could potentially provide a way to identify mechanisms of therapeutic resistance.
3.6. Methods for ctDNA detection: which to choose?
ctDNA is often present in minute amounts against a background of cfDNA released from healthy cells. Low ctDNA levels require highly sensitive methods for detection, especially in RCC, in which the levels of ctDNA generally appear to be low compared to other cancers. Comparison and optimization of methods are therefore of utmost importance.
Two of the studies compared different methods for detection of ctDNA. Smith et al [16] applied three methods to two different RCC patient cohorts; tumor-guided analysis, targeted panel sequencing, and global sequencing of plasma. They found that tumor-guided analysis had the highest ctDNA detection rate, whereas global sequencing of plasma had the lowest. This finding is supported by the study carried out by Lasseter et al [18], who also applied three methods for ctDNA detection in a single RCC patient cohort: global methylation analysis (cfMeDIP-seq), tumor-guided analysis, and targeted panel sequencing of plasma. They found that cfMeDIP-seq performed best and that tumor-guided analysis was more sensitive than targeted panel sequencing. A suggested reason for the superiority of cfMeDIP-seq was that epigenetic aberrations are more frequent than genetic variants in RCC [34].
Several studies indicate that the sensitivity of ctDNA analysis in RCC may be improved by enriching for shorter DNA fragments. Mouliere et al [25] performed WGS of plasma and found that the ctDNA detection rate in several cancer types was improved by selection of DNA fragments of between 90 and 150 bp, which is supported by results reported by Smith et al [16] for their study using sWGS and tumor-guided sequencing analysis. Yamamoto et al [22] analyzed cfDNA fragment length using data from targeted panel sequencing and found that DNA fragments with mutations tended to be shorter than corresponding nonmutated fragments and, similarly, that patients with positive ctDNA analysis tended to have a higher proportion of short versus long fragments. The findings in these studies together indicate that fragment size analysis may also be used to identify samples with certain levels of ctDNA to avoid expensive analysis on samples with very low ctDNA levels.
There are many possible explanations as to why the ctDNA detection rates in the studies included are below 100%; it may because of the sensitivity of the methods used for detection or other factors related to the analysis, for example, if the tumor mutations identified were subclonal and only present in a small part of the tumor [35]. Other possible reasons are the type of cancer, the anatomic location of the tumor causes ctDNA to be released to urine instead of blood, tumor DNA is cleared faster than in other tumors, or simply that renal tumors shed less DNA than other tumors [15]. A simple solution could be to analyze DNA from a larger amount of plasma. However, this would proportionally increase the cost of the analysis in most cases. We hope that future studies will cast light on some of these questions and improve the ability to detect ctDNA in RCC.
4. Conclusions
The number of studies investigating ctDNA in RCC patients is still low and the number of patients included in these studies is limited, although larger studies have appeared in the past few years. The studies highlight that the level of ctDNA in RCC appears to be low. However, patients with mRCC have a higher amount of ctDNA than patients with localized disease. Studies using multiple methods for ctDNA detection indicate that tumor-guided analysis improves the ctDNA detection rate compared to unguided methods and suggest that cfMeDIP-seq may potentially be a very sensitive method for ctDNA detection in RCC. More studies are needed to determine the potential applications of ctDNA analysis in RCC and to improve methods for these applications.
Author contributions: Lars Lund had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Geertsen, Magaard Koldby, Thomassen, Kruse, Lund.
Acquisition of data: Geertsen, Magaard Koldby.
Analysis and interpretation of data: Geertsen, Magaard Koldby.
Drafting of the manuscript: Geertsen, Magaard Koldby.
Critical revision of the manuscript for important intellectual content: Thomassen, Kruse, Lund.
Statistical analysis: Geertsen, Magaard Koldby.
Obtaining funding: None.
Administrative, technical, or material support: Geertsen, Magaard Koldby.
Supervision: Lund.
Other: None.
Financial disclosures: Lars Lund certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
Funding/Support and role of the sponsor: None.
Associate Editor: Guillaume Ploussard
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.euros.2021.12.006.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Parkin D.M., Bray F., Ferlay J., Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Ljungberg B, Albiges L, Bensalah K, et al. EAU guidelines: renal cell carcinoma 2020 V3-1. Arnhem, The Netherlands: European Association of Urology; 2020. https://uroweb.org/wp-content/uploads/EAU-Guidelines-on-Renal-Cell-Carcinoma-2020V3.pdf.
- 3.Danske Multidisciplinære Cancer Grupper. Renalcellecarcinomer – patologi. https://www.dmcg.dk/siteassets/forside/kliniske-retningslinjer/godkendte-kr/darenca/darenca_patologi_adm-godk_jbh100919.pdf.
- 4.Baldewijns M.M., van Vlodrop I.J.H., Schouten L.J., Soetekouw P.M.M.B., de Bruïne A.P., van Engeland M. Genetics and epigenetics of renal cell cancer. Biochim Biophys Acta. 2008;1785:133–155. doi: 10.1016/j.bbcan.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Ellinger J., von Rücker A., Bastian P.J., Müller S.C. Circulating cell-free serum DNA: significance as a new biomarker for urological malignancies. Urologe A. 2010;49 doi: 10.1007/s00120-010-2342-4. 1131-2, 1134. [DOI] [PubMed] [Google Scholar]
- 6.Schwarzenbach H., Hoon D.S., Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 7.Esposito A., Bardelli A., Criscitiello C., et al. Monitoring tumor-derived cell-free DNA in patients with solid tumors: clinical perspectives and research opportunities. Cancer Treat Rev. 2014;40:648–655. doi: 10.1016/j.ctrv.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Fleischhacker M., Schmidt B. Circulating nucleic acids (CNAs) and cancer–a survey. Biochim Biophys Acta. 2007;1775:181–232. doi: 10.1016/j.bbcan.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 9.de Martino M., Klatte T., Haitel A., Marberger M. Serum cell-free DNA in renal cell carcinoma: a diagnostic and prognostic marker. Cancer. 2012;118:82–90. doi: 10.1002/cncr.26254. [DOI] [PubMed] [Google Scholar]
- 10.Kidess E., Jeffrey S.S. Circulating tumor cells versus tumor-derived cell-free DNA: rivals or partners in cancer care in the era of single-cell analysis? Genome Med. 2013;5:70. doi: 10.1186/gm474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kukita Y., Uchida J., Oba S., et al. Quantitative identification of mutant alleles derived from lung cancer in plasma cell-free DNA via anomaly detection using deep sequencing data. PLoS One. 2013;8 doi: 10.1371/journal.pone.0081468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwarzenbach H., Stoehlmacher J., Pantel K., Goekkurt E. Detection and monitoring of cell-free DNA in blood of patients with colorectal cancer. Ann NY Acad Sci. 2008;1137:190–196. doi: 10.1196/annals.1448.025. [DOI] [PubMed] [Google Scholar]
- 13.Liberati A., Altman D.G., Tetzlaff J., et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009;6 doi: 10.1371/journal.pmed.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [DOI] [PMC free article] [PubMed]
- 15.Corrò C., Hejhal T., Poyet C., et al. Detecting circulating tumor DNA in renal cancer: an open challenge. Exp Mol Pathol. 2017;102:255–261. doi: 10.1016/j.yexmp.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Smith C.G., Moser T., Mouliere F., et al. Comprehensive characterization of cell-free tumor DNA in plasma and urine of patients with renal tumors. Genome Med. 2020;12:23. doi: 10.1186/s13073-020-00723-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan JCM, Heider K, Gale D, et al. ctDNA monitoring using patient-specific sequencing and integration of variant reads. Sci Transl Med 2020;12:eaaz8084. [DOI] [PubMed]
- 18.Lasseter K., Nassar A.H., Hamieh L., et al. Plasma cell-free DNA variant analysis compared with methylated DNA analysis in renal cell carcinoma. Genet Med. 2020;22:1366–1373. doi: 10.1038/s41436-020-0801-x. [DOI] [PubMed] [Google Scholar]
- 19.Hahn A.W., Gill D.M., Maughan B., et al. Correlation of genomic alterations assessed by next-generation sequencing (NGS) of tumor tissue DNA and circulating tumor DNA (ctDNA) in metastatic renal cell carcinoma (mRCC): potential clinical implications. Oncotarget. 2017;8:33614–33620. doi: 10.18632/oncotarget.16833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maia M.C., Bergerot P.G., Dizman N., et al. Association of circulating tumor DNA (ctDNA) detection in metastatic renal cell carcinoma (mRCC) with tumor burden. Kidney Cancer. 2017;1:65–70. doi: 10.3233/KCA-170007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pal S.K., Sonpavde G., Agarwal N., et al. Evolution of circulating tumor DNA profile from first-line to subsequent therapy in metastatic renal cell carcinoma. Eur Urol. 2017;72:557–564. doi: 10.1016/j.eururo.2017.03.046. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto Y., Uemura M., Fujita M., et al. Clinical significance of the mutational landscape and fragmentation of circulating tumor DNA in renal cell carcinoma. Cancer Sci. 2019;110:617–628. doi: 10.1111/cas.13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bacon JVW, Annala M, Soleimani M, et al. Plasma circulating tumor DNA and clonal hematopoiesis in metastatic renal cell carcinoma. Clin Genitourin Cancer 2020;18:322–31.e2. [DOI] [PubMed]
- 24.Zhang J., Liu Y., Xu B., et al. Circulating tumor DNA analysis of metastatic renal cell carcinoma. Mol Clin Oncol. 2021;14:16. doi: 10.3892/mco.2020.2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mouliere F, Chandrananda D, Piskorz AM, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med 2018;10:eaat4921. [DOI] [PMC free article] [PubMed]
- 26.Nuzzo P.V., Berchuck J.E., Korthauer K., et al. Detection of renal cell carcinoma using plasma and urine cell-free DNA methylomes. Nat Med. 2020;26:1041–1043. doi: 10.1038/s41591-020-0933-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu MC, Oxnard GR, Klein EA, Swanton C, Seiden MV, CCGA Consortium. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol 2020;31:745–59. [DOI] [PMC free article] [PubMed]
- 28.Hauser S., Zahalka T., Fechner G., Müller S.C., Ellinger J. Serum DNA hypermethylation in patients with kidney cancer: results of a prospective study. Anticancer Res. 2013;33:4651–4656. [PubMed] [Google Scholar]
- 29.Skrypkina I., Tsyba L., Onyshchenko K., et al. Concentration and methylation of cell-free DNA from blood plasma as diagnostic markers of renal cancer. Dis Markers. 2016;2016:3693096. doi: 10.1155/2016/3693096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin Y.L., Wang Y.P., Li H.Z., Zhang X. Aberrant promoter methylation of PCDH17 (protocadherin 17) in serum and its clinical significance in renal cell carcinoma. Med Sci Monit. 2017;23:3318–3323. doi: 10.12659/MSM.902077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung M., Ellinger J., Gevensleben H., et al. Cell-free SHOX2 DNA methylation in blood as a molecular staging parameter for risk stratification in renal cell carcinoma patients: a prospective observational cohort study. Clin Chem. 2019;65:559–568. doi: 10.1373/clinchem.2018.297549. [DOI] [PubMed] [Google Scholar]
- 32.Milbury C.A., Zhong Q., Lin J., et al. Determining lower limits of detection of digital PCR assays for cancer-related gene mutations. Biomol Detect Quantif. 2014;1:8–22. doi: 10.1016/j.bdq.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi J., Hong B., Tao R., et al. Prediction model for malignant pulmonary nodules based on cfMeDIP-seq and machine learning. Cancer Sci. 2021;112:3918–3923. doi: 10.1111/cas.15052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013;499:43–9. [DOI] [PMC free article] [PubMed]
- 35.Ball M.W., Gorin M.A., Guner G., et al. Circulating tumor DNA as a marker of therapeutic response in patients with renal cell carcinoma: a pilot study. Clin Genitourin Cancer. 2016;14:e515–e520. doi: 10.1016/j.clgc.2016.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.