Abstract
Clostridium perfringens type A isolates producing enterotoxin (CPE) are an important cause of food poisoning and non-food-borne human gastrointestinal (GI) diseases, including antibiotic-associated diarrhea (AAD). Recent studies suggest that C. perfringens type A food poisoning is caused by C. perfringens isolates carrying a chromosomal cpe gene, while CPE-associated non-food-borne GI diseases, such as AAD, are caused by plasmid cpe isolates. Those putative relationships, obtained predominantly with European isolates, were tested in the current study by examining 34 cpe-positive, C. perfringens fecal isolates from North American cases of food poisoning or AAD. These North American disease isolates were all classified as type A using a multiplex PCR assay. Furthermore, restriction fragment length polymorphism and pulsed-field gel electrophoresis genotyping analyses showed the North American AAD isolates included in this collection all have a plasmid cpe gene, but the North American food poisoning isolates all carry a chromosomal cpe gene. Western blotting demonstrated CPE expression by nearly all of these disease isolates, confirming their virulence potential. These findings with North American isolates provide important new evidence that, regardless of geographic origin or date of isolation, plasmid cpe isolates cause most CPE-associated AAD cases and chromosomal cpe isolates cause most C. perfringens type A food poisoning cases. These findings hold importance for the development of assays for distinguishing cases of CPE-associated food-borne and non-food-borne human GI illnesses and also identify potential epidemiologic tools for determining the reservoirs for these illnesses.
Clostridium perfringens is a gram-positive, spore-forming, anaerobic bacterium that produces at least 15 different protein toxins (13, 14, 20, 21, 22). However, each individual C. perfringens isolate expresses only a defined subset of this total toxin repertoire, providing the basis for a commonly used classification scheme that assigns C. perfringens isolates to one of five types (A through E), based upon their ability to produce alpha-, beta-, epsilon- and iota-toxin (20, 21).
About 2 to 5% of all C. perfringens isolates, mostly belonging to type A, produce C. perfringens enterotoxin (CPE), a 35-kDa single polypeptide (12, 17, 26). These CPE-producing C. perfringens type A isolates are an important cause of enteric disease in both humans and domestic animals (18, 19, 25). Traditionally, these bacteria are most recognized as the cause of C. perfringens type A food poisoning, which currently ranks as the third most commonly identified food-borne disease in the United States (18). Considerable epidemiologic evidence implicates CPE as the virulence factor responsible for most (if not all) diarrheal and cramping symptoms associated with C. perfringens type A food poisoning (18). Furthermore, recent studies fulfilling the molecular Koch's postulates have provided unambiguous proof that CPE expression is required for the gastrointestinal (GI) virulence of CPE-positive C. perfringens type A food poisoning isolates in animal models (23).
During the past 15 years, CPE-positive C. perfringens type A isolates have also become linked to several non-food-borne human GI diseases (2–4, 7, 8, 15). Some estimates suggest these bacteria account for 5 to 20% of all cases of antibiotic-associated diarrhea (AAD) and sporadic non-food-borne diarrhea (6). A direct role for CPE in the pathogenesis of CPE-associated non-food-borne human GI diseases receives support from both epidemiologic surveys (6) and recent studies fulfilling the molecular Koch's postulates, which demonstrated that CPE expression is required for the GI virulence of CPE-positive C. perfringens non-food-borne human GI disease isolates in animal models (23).
Recent studies have also established that the cpe gene can have either a chromosomal or plasmid location (7–9, 16, 24). Interestingly, initial studies have suggested that the cpe gene has a chromosomal location in food poisoning isolates (8, 9, 16) but is located on a plasmid in non-food-borne human GI disease isolates (8). If these initial cpe genotyping results are correct and particular cpe genotypes (chromosomal versus plasmid) do cause specific CPE-associated GI diseases (food poisoning versus non-food-borne GI diseases), then cpe genotype-based differential diagnostic assays might prove useful for distinguishing between cases of CPE-associated food-borne versus non-food-borne human GI diseases.
However, these relationships between cpe genotype and CPE-associated disease should still be considered tentative since they are based upon genotyping results using relatively few C. perfringens isolates, with limited diversity. For example, all 16 CPE-producing non-food-borne GI disease isolates (including 6 AAD isolates and 10 sporadic diarrhea isolates) genotyped to date had originated from patients sickened in England during the mid-1980s to early 1990s (7, 8). Furthermore, only a few North American food poisoning isolates, predominantly from a single C. perfringens type A food poisoning outbreak occurring in Vermont during the mid-1980s, were genotyped in previous studies (7, 8).
Therefore, the putative relationships noted between particular cpe genotypes and specific CPE-associated diseases clearly require verification by testing additional cpe-positive human GI disease isolates of different geographic and temporal origins from those isolates genotyped to date. In response, our current study has characterized a sizeable collection of C. perfringens fecal isolates associated with recent North American cases of CPE-associated human GI diseases. Notably, this study includes the first genotyping analysis of cpe-positive fecal isolates obtained from North American patients with CPE-associated non-food-borne GI diseases.
MATERIALS AND METHODS
Strains.
C. perfringens isolates used as controls in this study included F4969, a type A strain carrying the cpe gene on a plasmid (8); NCTC10239, a type A strain carrying a chromosomal cpe gene (8); ATCC 3624, a cpe-negative type A isolate (10); NCTC8533, a cpe-lacking, type B isolate (provided by Richard Titball); CN5383, a cpe-negative type C isolate (10); PS52, a cpe-negative type D isolate (provided by Ronald Labbe); and 853, a type E isolate carrying silent cpe sequences (1).
Among the North American human GI disease isolates examined in the present study were 16 cpe-positive C. perfringens fecal isolates associated with cases of CPE-associated AAD occuring in the state of Washington during 1998 and 1999, as well as 12 cpe-positive C. perfringens fecal isolates associated with cases of CPE-associated AAD occuring in British Columbia during 1999. All 28 of these North American AAD isolates came from the feces of different patients.
Also included in this study were six cpe-positive C. perfringens isolates obtained from feces of patients sickened with C. perfringens type A food poisoning. These six isolates originated from two different food poisoning outbreaks, which had occurred in Ohio and Virginia during the 1990s. These two outbreaks are distinct from the North American food poisoning outbreaks serving as the source for the C. perfringens food poisoning isolates genotyped in a previous study (8).
An initial screening using a cpe-specific PCR assay (17) had confirmed the presence of the cpe gene in all North American GI disease isolates included in this collection (data not shown).
Growth and sporulation conditions.
Starter vegetative cultures (6 ml) of each C. perfringens isolate were prepared by overnight growth at 37°C in fluid thioglycollate (FTG) medium (Difco). For DNA isolation, an aliquot (0.2 ml) of each FTG culture was inoculated into 10 ml of TGY broth (10), which was then incubated at 37°C overnight. Sporulating cultures of C. perfringens were obtained by inoculating an aliquot (0.2 ml) of each starter FTG culture into (i) 10 ml of Duncan-Strong (DS) sporulation medium (17), which was incubated at 37°C for 8 h; (ii) DS medium supplemented with 1.5% bile and 0.005% theophylline (DS-B), which was incubated at 37°C for 8 h (1); and (iii) 10 ml of raffinose-caffeine modified DS (RC), which was incubated for 5 h at 43°C (17). After the desired incubation, the presence of sporulating cells in each DS, DS-B, or RC culture was confirmed by phase-contrast microscopy.
Multiplex PCR toxin genotyping of C. perfringens isolates.
Total C. perfringens DNA was isolated from the overnight TGY cultures using a previously described protocol (10). That isolated DNA was then subjected, as described previously (1, 26), to multiplex PCR diagnostic screening for detection of gene sequences encoding C. perfringens alpha-toxin, beta-toxin, epsilon-toxin, iota-toxin, and CPE. After multiplex PCR, the presence of amplified toxin gene sequences was then analyzed by subjecting an aliquot of each PCR sample to electrophoresis at 100 V in 1.5% agarose gels, followed by ethidium bromide staining and visualization under UV illumination.
Preparation of DIG-labeled cpe probes for southern blot experiments.
A 639-bp digoxigenin (DIG)-labeled, double-stranded, cpe-specific DNA gene probe was prepared by a previously described (10), two-step PCR amplification method using the primer set 5′-GGTACCTTTAGCCAATCA-3′ (primer 2F) and 5′-TCCATCACCTAAGGACTG-3′ (primer 5R).
Restriction fragment length polymorphism (RFLP) Southern blot analyses.
Isolated C. perfringens DNA samples, prepared as described above, were digested to completion with NruI, separated by electrophoresis on 0.8% agarose gels, transferred to positively charged nylon membranes (Boehringer Mannheim), and UV fixed to those membranes, as described previously (23). The blots were then hybridized with the DIG-labeled cpe probe, as described in The Genius System Users Guide for Filter Hybridization (Roche). The hybridized cpe probe was then detected using a DIG-chemiluminescence detection system, using CSPD substrate (Roche).
PFGE Southern blot analyses.
Overnight TGY cultures were collected by centrifugation, with those pelleted cells then used to prepare agarose plugs containing genomic C. perfringens DNA, as previously described (5, 8, 9). An aliquot (100 μl) of each agarose plug was incubated overnight at 37°C in the presence or absence of 4 U of I-CeuI (New England Biolabs). These samples were then analyzed by pulsed-field gel electrophoresis (PFGE), using 1% agarose gels prepared with PFGE-grade agarose (Bio-Rad). PFGE was performed with a Bio-Rad CHEF-DR II apparatus, with pulse times ramped from 50 to 90 s over 20 h (1). These PFGE gels were then subjected to cpe Southern analysis, using the same procedure described above for RFLP Southern analysis.
CPE Western blot analysis.
When phase-contrast microscopy revealed the presence of spores, DS, DS-B, or RC cultures were lysed by six pulses (1 min each) with a Heat Systems Ultrasonics model W-375 sonicator set to 70% duty cycle and output level 5. This sonication procedure was sufficient to lyse >95% of all cells (lysis was monitored by phase-contrast microscopy). After sonication, each DS, DS-B, or RC sporulating culture lysate was analyzed for the presence of CPE using a previously described CPE Western immunoblot procedure (17).
RESULTS
Multiplex PCR toxin typing of cpe-positive C. perfringens isolates associated with North American human GI diseases.
Previous surveys (12, 17, 26) have determined that most cpe-positive C. perfringens isolates classify as type A; i.e., these enterotoxigenic isolates typically carry the alpha-toxin gene but not the genes encoding beta-, iota-, or epsilon-toxin (19, 20). However, some CPE-positive isolates belonging to other toxin types have also been identified (19, 20). Therefore, we first subjected our collection of cpe-positive fecal isolates associated with North American cases of GI diseases to multiplex PCR analysis in order to determine their toxin genotype (A through E).
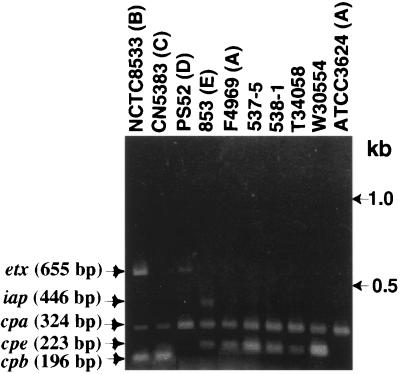
To ensure the reliability of multiplex PCR results obtained using template DNAs prepared from North American human GI disease isolates, control PCRs were run using template DNA prepared from known C. perfringens types A, B, C, D, or E isolates. As shown in Fig. 1, an ∼324-bp PCR product matching the expected product size that should be amplified from the alpha-toxin gene (cpa) by this multiplex assay (1, 26) was observed using template DNA prepared from all C. perfringens control strains, regardless of their toxin type. However, that product was absent if Escherichia coli template DNA was subjected to this multiplex PCR assay (data not shown). These results are consistent with the known presence of cpa in C. perfringens isolates of all toxin types.
FIG. 1.
Multiplex PCR analysis of North American human GI disease isolates. Representative results shown are for multiplex PCR using primers designed to amplify genes encoding each “typing” toxin and CPE. Migration of PCR products derived from each toxin gene are indicated on the left. As denoted on the figure by the letter shown in parentheses to their right, control typing strains used include NCTC8533 (type B), CN5383 (type C), PS52 (type D), 853 (type E), ATCC 3624 (cpe lacking, type A), and F4969 (cpe-positive, type A). Representative North American human GI disease isolates shown include 537-5 and 538-1 (food poisoning isolates) and T34058 and W30554 (AAD isolates). Molecular sizes of DNA markers are noted on the right of the figure.
An additional ∼223-bp product, which matches the expected product size that should be amplified from the enterotoxin gene (cpe), was also present when template DNA prepared from F4969, a known cpe-positive type A isolate, was subjected to this same multiplex PCR analysis. A similar ∼223-bp product was also amplified using DNA template prepared from the type E isolate 853, which is known to carry silent cpe sequences that hybridize the cpe primer set used in this multiplex PCR assay (1). However, no ∼223-bp PCR product was obtained using template DNA prepared from either (i) ATCC 3624, a cpe-negative type A isolate, or (ii) the type B, C, or D control strains. Since those four C. perfringens isolates all lack cpe sequences (reference 10 and data not shown), the absence of the ∼223-bp product using template DNA prepared from known cpe-negative control strains confirms the ∼223-bp PCR product as an amplification product of cpe gene sequences. Collectively, these control results confirm the ability of the multiplex PCR to reliably distinguish cpe-positive isolates from cpe-negative isolates.
Additional PCR products of ∼196, ∼655, and ∼446 bp were amplified when known type C, D, or E isolates, respectively, were used as the source of template DNA for the multiplex PCR (Fig. 1). Those three PCR products match the expected sizes of products that the multiplex PCR assay should amplify from genes encoding beta-toxin, epsilon-toxin, or the iota-toxin A component, respectively. Since the genes encoding beta-toxin, epsilon-toxin, and the iota-toxin A component are present in type C, D, and E isolates, respectively (20, 21), these PCR results confirm the ability of this multiplex PCR assay to specifically identify isolates belonging to any of the five C. perfringens toxin types (note: both ∼196- and ∼655-bp products were observed using template DNA prepared from the type B isolate NCTC8533, as would be expected since type B isolates carry both beta- and epsilon-toxin genes [20, 21]).
When template DNA isolated from each of our 16 AAD fecal isolates from Washington and 12 AAD fecal isolates from British Columbia was subjected to this same multiplex PCR analysis, PCR products of ∼223 and ∼324 bp were invariably obtained (see Fig. 1 for representative results). Therefore, these 28 AAD fecal isolates all clearly classify as cpe-positive, type A isolates. Similarly, the multiplex PCR also identified all six fecal isolates obtained from food poisoning victims as cpe-positive, type A isolates (see Fig. 1 for representative results).
RFLP genotyping of North American human GI disease isolates.
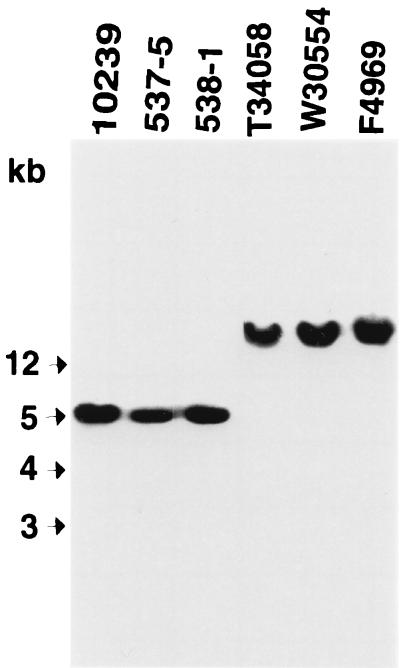
Our collection of cpe-positive type A fecal isolates associated with North American cases of human GI diseases was next subjected to cpe genotyping. Initially, these AAD and food poisoning isolates were subjected to NruI RFLP Southern blot analysis, which is now well-established as a reliable presumptive test for distinguishing between isolates carrying chromosomal versus plasmid cpe genes (8, 9, 16). Briefly, previous results with this cpe RFLP assay (8, 9, 16) have shown that the chromosomal cpe gene is invariably present on an ∼5-kb NruI DNA fragment, while the plasmid cpe gene is always present on NruI-digested DNA fragments of >20 kb.
To confirm the reliability of cpe RFLP results generated with our North American human GI disease isolates, we first digested DNA from control C. perfringens isolates F4969 or NCTC10239 with NruI and then Southern blotted that digested DNA with a cpe-specific probe. As shown in Fig. 2, this cpe-specific probe hybridized to a 5-kb fragment of NruI-digested DNA from isolate NCTC10239, consistent with previous studies identifying that isolate as a chromosomal cpe strain (8, 9, 16). In contrast, the same cpe-specific probe hybridized to NruI-digested DNA of >20 kb from isolate F4969, which was previously shown to carry a plasmid cpe gene (8).
FIG. 2.
RFLP Southern blot analysis of NruI-digested DNA from North American human GI disease isolates. Southern blots were probed with a 639-bp DIG-labeled cpe-specific probe. Control isolates shown include 10239 (NCTC10239; a chromosomal cpe, food poisoning isolate) and F4969 (a plasmid cpe, non-food-borne human GI disease isolate). Representative North American human GI disease isolates shown include food poisoning isolates 537-5 and 538-1 and AAD isolates T34058 and W30554. Molecular sizes of DNA markers are given to the left of the blot.
When the same NruI RFLP Southern blot assay was applied to our collection of 16 AAD Washington isolates and 12 AAD British Columbia isolates (see representative results in Fig. 2), the cpe probe always hybridized to DNA of >20 kb; i.e., these isolates all appeared to carry a plasmid cpe gene. However, that same cpe probe hybridized to a 5-kb NruI fragment of DNA isolated from all six food poisoning isolates, strongly suggesting those isolates carry a chromosomal cpe gene (see representative results in Fig. 2).
PFGE genotyping analysis of cpe-positive North American human GI disease isolates.
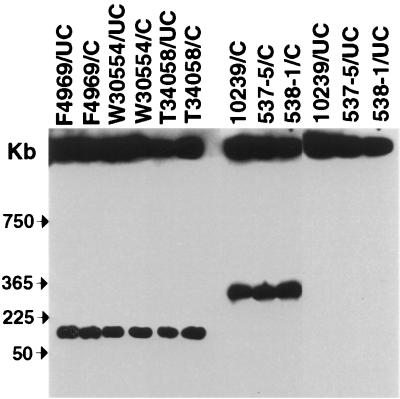
To verify the presumptive cpe genotyping results generated with the cpe RFLP assay, selected North American GI disease isolates in our collection were subjected to PFGE Southern blot analysis, which has been used in previous studies (8, 9, 16) to formally establish the chromosomal or plasmid localization of the cpe gene. Briefly, the principle of this method is that, without any restriction enzyme digestion, C. perfringens chromosomal DNA is too large to enter a pulsed-field gel. However, because of its smaller size, some plasmid DNA should enter a pulsed-field gel, even without any restriction enzyme treatment. Also, since I-CeuI sites are located exclusively on the C. perfringens chromosome, digestion of DNA samples with I-CeuI should produce chromosomal DNA fragments that can enter pulsed-field gels but should not affect the migration of plasmid DNA. Therefore, when DNA from chromosomal cpe isolates is subjected to this PFGE analysis and then cpe Southern blotted, all cpe-containing DNA should remain in the gel wells in samples without I-CeuI treatment, but some cpe-containing DNA should enter these gels (as an ∼360-kb fragment [8, 9, 16]) when samples are digested with I-CeuI prior to electrophoresis. However, when DNA from a plasmid cpe isolate is analyzed by this technique, some cpe-containing DNA should enter the pulsed-field gels even in the absence of restriction enzyme digestion, and the migration of this cpe-containing DNA should be unaffected by I-CeuI digestion.
To confirm the reliability of this PFGE Southern blot assay for our current studies, we first analyzed undigested DNA from control strain NCTC10239. As shown in Fig. 3, no cpe-containing DNA from that strain migrated into the pulsed-field gels without restriction enzyme digestion. However, an ∼360-kb, cpe-containing DNA fragment did enter the gel if NCTC10239 DNA was digested with I-CeuI prior to PFGE. These results are fully consistent with previous cpe PFGE genotyping analyses establishing NCTC10239 as a chromosomal cpe strain (8, 9, 16).
FIG. 3.
PFGE Southern blot analysis of selected North American human GI disease isolates. PFGE and Southern hybridization analysis of undigested (UC) and I-CeuI cut (C) DNA from selected isolates. Blots are probed with a cpe-specific probe. Control isolates shown include 10239 (NCTC10239; a chromosomal cpe, food poisoning isolate) and F4969 (a plasmid cpe, non-food-borne human GI disease isolate). Representative North American human GI disease isolates shown include food poisoning isolates 537-5 and 538-1 and AAD isolates T34058 and W30554. The pulsed-field gel was calibrated with Lambda DNA markers, whose migration is shown at the left of the blot.
In contrast, when DNA from control strain F4969 was subjected to the same cpe PFGE Southern blotting procedure, some cpe-containing DNA did migrate into pulsed-field gels, even in the absence of I-CeuI digestion (Fig. 3). Furthermore, the migration of this cpe-containing DNA was unaffected by I-CeuI treatment. These results are fully consistent with previous genotyping analyses (8) identifying F4969 as a plasmid cpe isolate.
Since the results for control strains NCTC10239 and F4969 confirmed the ability of the cpe PFGE Southern assay to distinguish between C. perfringens isolates carrying a chromosomal cpe gene and those carrying a plasmid cpe gene, that technique was employed to confirm the reliability of the RFLP-based presumptive genotype assignments using selected North American AAD and food poisoning isolates from our collection. As shown in Fig. 3, cpe-containing DNA of North American AAD strains T34058 and W30554 entered pulsed-field gels in the absence of restriction enzyme digestion. Furthermore, the migration of that cpe-containing DNA was unaffected by I-CeuI digestion. These PFGE results confirm the Fig. 2 RFLP results that had presumptively identified these two North American AAD isolates as plasmid cpe isolates.
In contrast, cpe-containing DNA of North American food poisoning strains 538-1 and 537-5 failed to enter pulsed-field gels in the absence of restriction enzyme treatment. However, an ∼360-kb restriction fragment was visible when DNA samples prepared from either of those two strains was digested with I-CeuI prior to cpe PFGE Southern blot analysis. Collectively, these PFGE results confirm the Fig. 2 RFLP results presumptively identifying these two North American food poisoning isolates as chromosomal cpe isolates.
Western Blot analysis of CPE expression.
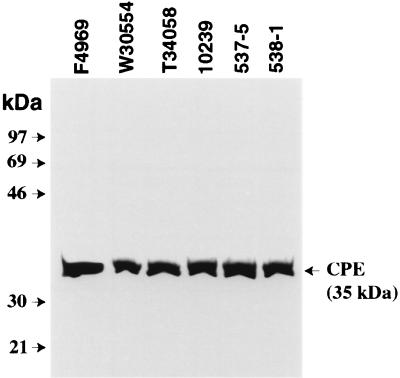
Finally, the CPE-expressing ability of the 34 North American AAD and food poisoning isolates in our current collection was also examined. Because CPE expression is strongly sporulation related (7, 10, 11), it was first necessary to obtain in vitro sporulation for these isolates. All but two of the 34 isolates showed significant sporulation (i.e., sporulating cells represented >25% of total cells present in a culture) in at least one of the three sporulation media (DS-, DS-B, or RC) used in this study. When lysates prepared from these sporulating cultures were analyzed by CPE Western blotting, 31 isolates were found to produce CPE; i.e., these culture lysates contained a 35-kDa protein that reacts with CPE antibodies and comigrates with purified CPE (see representative results in Fig. 4). Interestingly, one Washington AAD isolate (S10653) sporulated very well (80 to 90% sporulation) but failed to express any CPE detectable by Western blot analysis.
FIG. 4.
Western Blot analysis of CPE expression by selected North American human GI disease isolates. The expression of CPE by sporulating cultures of control and disease isolates of C. perfringens was evaluated using a CPE-specific Western immunoblot procedure. Isolates were grown in sporulation media as described in the Materials and Methods and then sonicated. An aliquot (40 μl) of each sonicated sporulating culture lysate was then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by immunoblotting with CPE antibodies. The blot was developed for chemiluminescence detection to identify immunoreactive species. Results for control isolates shown include 10239 (NCTC10239, a chromosomal cpe, food poisoning isolate) and F4969 (a plasmid cpe, non-food-borne human GI disease isolate). Results for representative North American human GI disease isolates shown include food poisoning isolates 537-5 and 538-1 and AAD isolates T34058 and W30554. Molecular mass markers are shown at left; the arrow at right indicates the migration of purified CPE.
DISCUSSION
The present study reports the first genotyping analyses of cpe-positive fecal isolates associated with North American cases of CPE-associated non-food-borne human GI diseases. All 28 of the cpe-positive North American AAD isolates examined in our current study were found to classify as type A isolates. Furthermore, every one of these North American AAD isolates appears to carry its cpe gene on a plasmid, as was also true of the previously examined English AAD isolates (8). It is also notable that the North American AAD isolates examined in our current study had been obtained in the late 1990s, while all English AAD isolates genotyped previously came from patients sickened during the mid-1980s. Collectively, these results now strongly suggest that, regardless of date of isolation or geographic origin, most (if not all) cpe-positive C. perfringens isolates causing CPE-associated AAD genotype as type A isolates carrying their cpe gene on a plasmid.
Six North American food poisoning isolates, originating from two separate outbreaks, were also genotyped in the present study. These food poisoning isolates were all identified as C. perfringens type A isolates carrying a chromosomal cpe gene, which is consistent with results from a previous study showing that eight of eight North American food poisoning isolates also genotyped as type A isolates carrying a chromosomal cpe gene. Furthermore, since 11 of 11 C. perfringens European food poisoning isolates (isolated as early as the 1950s) that were genotyped in previous studies also classified as type A isolates carrying a chromosomal cpe gene, it is now apparent that, regardless of geographic location or date of isolation, most (if not all) C. perfringens type A food poisoning outbreaks involve C. perfringens type A isolates carrying a chromosomal cpe gene.
These new data significantly strengthen the association between specific cpe genotypes (plasmid versus chromosomal) and particular CPE-associated GI diseases (food borne versus non-food borne), thereby offering important support for the possible use of cpe genotyping as a presumptive diagnostic assay for distinguishing between cases of C. perfringens type A food poisoning and cases of CPE-associated non-food-borne GI diseases. These new findings also support the use of cpe RFLP and PFGE assays as tools to investigate the reservoirs and transmission of cpe-positive food poisoning and AAD isolates.
It is possible that, with further sampling, exceptions will be discovered to the apparent relationships between chromosomal cpe isolates and food poisoning, or plasmid cpe isolates and non-food-borne GI diseases. However, emerging evidence indicates these genotype-disease relationships have a physiologic basis, suggesting that exceptions will be relatively uncommon. For example, both the vegetative cells and spores of C. perfringens type A food poisoning isolates carrying a chromosomal cpe gene have recently been shown (24) to exhibit significantly more heat resistance than the cells or spores of non-food-borne human GI disease isolates carrying a plasmid cpe gene. This greater heat resistance probably helps explain why the chromosomal cpe isolates are so strongly associated with C. perfringens type A food poisoning, since their survival will be favored in inadequately cooked or held foods, which are the two risk factors most commonly responsible for C. perfringens type A food poisoning outbreaks (18). Other studies (S. Brynestad, M. R. Sarkar, B. A. McClane, P. E. Granum, and J. I. Rood, submitted for publication) have recently shown that the cpe plasmid can be transferred, in vitro, to naturally cpe-negative isolates via conjugation. If similar conjugative transfer of the cpe plasmid can occur in vivo during the non-food-borne human GI diseases involving plasmid cpe strains, this might result in transfer of the cpe plasmid from a relatively few infecting cpe-positive strains to some of the many naturally cpe-negative C. perfringens isolates already present in the normal intestinal flora. Since C. perfringens normal intestinal flora strains have presumably been selected for their ability to persist and grow in the GI tract, genetic transfer of the cpe plasmid to normal flora strains could be a critical step for establishing CPE-associated non-food-borne human GI disease. It also could result in a prolonged presence of cpe-positive isolates in the GI tract, perhaps explaining why the symptoms of CPE-associated non-food-borne GI diseases last longer and are more severe than the symptoms of C. perfringens type A food poisoning.
Finally, demonstrating CPE expression by 31 of the 32 sporulating cpe-positive fecal isolates associated with North American human GI diseases supports the enteropathogenic potential of these isolates. It is possible that the one cpe-positive isolate which sporulated but did not express CPE lost the ability to produce CPE during subculture in the laboratory. This isolate does retain cpe sequences, so its loss of CPE expression is not simply due to loss of the cpe plasmid. In any event, this represents (to our knowledge) the first identification of a cpe-positive type A isolate that sporulates well but does not express the enterotoxin. Further studies are planned to determine why this isolate does not produce CPE when sporulating.
ACKNOWLEDGMENTS
This research was supported by U.S. Department of Agriculture grant 9802822 from the Ensuring Food Safety Research Program and by Public Health Service grant AI19844-17, from the National Institute of Allergy and Infectious Diseases.
We thank Ronald Labbe and Richard Titball for providing control C. perfringens strains.
REFERENCES
- 1.Billington S J, Wieckowski E U, Sarker M R, Bueschel D, Songer J G, McClane B A. Clostridium perfringens type E animal enteritis isolates with highly conserved, silent enterotoxin sequences. Infect Immun. 1998;66:4531–4536. doi: 10.1128/iai.66.9.4531-4536.1998. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Borriello S P. Newly described clostridial diseases of the gastrointestinal tract: Clostridium perfringens enterotoxin-associated diarrhea and neutropenic enterocolitis due to Clostridium septicum. In: Borriello S P, editor. Clostridia in gastrointestinal disease. Boca Raton, Fla: CRC Press, Inc.; 1985. pp. 223–228. [Google Scholar]
- 3.Borriello S P, Barclay F E, Welch A R, Stringer M F, Watson G N, Williams R K T, Seal D V, Sullens K. Epidemiology of diarrhea caused by enterotoxigenic Clostridium perfringens. J Med Microbiol. 1985;20:363–372. doi: 10.1099/00222615-20-3-363. [DOI] [PubMed] [Google Scholar]
- 4.Brett M M, Rodhouse J C, Donovan T J, Tebbut G M, Hutchinson D N. Detection of Clostridium perfringens and its enterotoxin in cases of sporadic diarrhea. J Clin Pathol. 1992;45:609–611. doi: 10.1136/jcp.45.7.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canard B, Saint-Joanis B, Cole S T. Genomic diversity and organization of virulence genes in the pathogenic anaerobe Clostridium perfringens. Mol Microbiol. 1992;6:1421–1429. doi: 10.1111/j.1365-2958.1992.tb00862.x. [DOI] [PubMed] [Google Scholar]
- 6.Carman R J. Clostridium perfringens in spontaneous and antibiotic-associated diarrhoea of man and other animals. Rev Med Microbiol. 1997;8Suppl.:S43–S45. [Google Scholar]
- 7.Collie R E, Kokai-Kun J F, McClane B A. Phenotypic characterization of enterotoxigenic Clostridium perfringens isolates from non-foodborne human gastrointestinal diseases. Anaerobe. 1998;4:69–79. doi: 10.1006/anae.1998.0152. [DOI] [PubMed] [Google Scholar]
- 8.Collie R E, McClane B A. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with nonfoodborne human gastrointestinal diseases. J Clin Microbiol. 1998;36:30–36. doi: 10.1128/jcm.36.1.30-36.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornillot E, Saint-Joanis B, Daube G, Katayama S, Granum P E, Carnard B, Cole S T. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Mol Microbiol. 1995;15:639–647. doi: 10.1111/j.1365-2958.1995.tb02373.x. [DOI] [PubMed] [Google Scholar]
- 10.Czeczulin J R, Collie R E, McClane B A. Regulated expression of Clostridium perfringens enterotoxin in naturally cpe-negative type A, B, and C isolates of C. perfringens. Infect Immun. 1996;64:3301–3309. doi: 10.1128/iai.64.8.3301-3309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czeczulin J R, Hanna P C, McClane B A. Cloning, nucleotide sequencing, and expression of the Clostridium perfringens enterotoxin gene in Escherichia coli. Infect Immun. 1993;61:3429–3439. doi: 10.1128/iai.61.8.3429-3439.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daube G, Simon P, Limbourg B, Manteca C, Mainil J, Kaeckenbeeck A. Hybridization of 2,659 Clostridium perfringens isolates with gene probes for seven toxins (α, β, ɛ, ι, θ, μ and enterotoxin) and for sialidase. Am J Vet Res. 1996;57:496–501. [PubMed] [Google Scholar]
- 13.Gibert M, Jolivet-Reynaud C, Popoff M R. Beta2 toxin, a novel toxin produced by Clostridium perfringens. Gene. 1997;203:65–73. doi: 10.1016/s0378-1119(97)00493-9. [DOI] [PubMed] [Google Scholar]
- 14.Hatheway C. Toxigenic clostridia. Clin Microbiol Rev. 1990;3:66–76. doi: 10.1128/cmr.3.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson S, Yip-Chuck D, Clark J, Brodsky M. Diagnostic importance of Clostridium perfringens enterotoxin analysis in recurring enteritis among the elderly, chronic care psychiatric patients. J Clin Microbiol. 1986;23:748–751. doi: 10.1128/jcm.23.4.748-751.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katayama S I, Dupuy B, Daube G, China B, Cole S T. Genome mapping of Clostridium perfringens strains with I-Ceu I shows many virulence genes to be plasmid-borne. Mol Gen Genet. 1996;251:720–726. doi: 10.1007/BF02174122. [DOI] [PubMed] [Google Scholar]
- 17.Kokai-Kun J F, Songer J G, Czeczulin J R, Chen F, McClane B A. Comparison of Western immunoblots and gene detection assays for identification of potentially enterotoxigenic isolates of Clostridium perfringens. J Clin Microbiol. 1994;32:2533–2539. doi: 10.1128/jcm.32.10.2533-2539.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McClane, B. A.Clostridium perfringens. In M. P. Doyle, L. R. Beuchat, and T. J. Montville (ed.), Food microbiology: fundamentals and frontiers, 2nd ed., in press. ASM Press, Washington, D.C.
- 19.McClane B A, Lyerly D M, Moncrief J S, Wilkins T D. Enterotoxic clostridia: Clostridium perfringens type A and Clostridium difficile. In: Fischetti V A, Novick R P, Ferretti J J, Portnoy D A, Rood J, editors. Gram-positive pathogens. Washington, D.C.: ASM Press; 2000. pp. 551–562. [Google Scholar]
- 20.McDonel J L. Toxins of Clostridium perfringens types A, B, C, D, and E. In: Dorner F, Drews H, editors. Pharmacology of bacterial toxins. Pergamon Press, Oxford, United Kingdom. 1986. pp. 477–517. [Google Scholar]
- 21.Rood J, Cole S T. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol Rev. 1991;55:621–648. doi: 10.1128/mr.55.4.621-648.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rood J I. Virulence genes of Clostridium perfringens. Annu Rev Microbiol. 1998;52:333–360. doi: 10.1146/annurev.micro.52.1.333. [DOI] [PubMed] [Google Scholar]
- 23.Sarker M R, Carman R J, McClane B A. Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe-positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops. Mol Microbiol. 1999;33:946–958. doi: 10.1046/j.1365-2958.1999.01534.x. [DOI] [PubMed] [Google Scholar]
- 24.Sarker M R, Shivers R P, Sparks S G, Juneja V K, McClane B A. Comparative experiments to examine the effects of heating on vegetative cells and spores of Clostridium perfringens isolates carrying plasmid versus chromosomal enterotoxin genes. Appl Environ Microbiol. 2000;66:3234–3240. doi: 10.1128/aem.66.8.3234-3240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Songer J G. Clostridial enteric diseases of domestic animals. Clin Microbiol Rev. 1996;9:216–234. doi: 10.1128/cmr.9.2.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Songer J G, Meer R M. Genotyping of Clostridium perfringens by polymerase chain reaction is a useful adjunct to diagnosis of clostridial enteric disease in animals. Anaerobe. 1996;2:197–203. [Google Scholar]