Abstract
An opportunity has opened for research into primary prevention of psychotic disorders, based on progress in endophenotypes, genetics, and genomics. Primary prevention requires reliable prediction of susceptibility before any symptoms are present. We studied a battery of measures where published data supports abnormalities of these measurements prior to appearance of initial psychosis symptoms. These neurobiological and behavioral measurements included cognition, eye movement tracking, Event Related Potentials, and polygenic risk scores. They generated an acceptably precise separation of healthy controls from outpatients with a psychotic disorder.
Methods:
The Bipolar and Schizophrenia Network on Intermediate Phenotypes (B-SNIP) measured this battery in an ancestry-diverse series of consecutively recruited adult outpatients with a psychotic disorder and healthy controls. Participants include all genders, 16 to 50 years of age, 261 with psychotic disorders (Schizophrenia (SZ) 109, Bipolar with psychosis (BPP) 92, Schizoaffective disorder (SAD) 60), 110 healthy controls. Logistic Regression, and an extension of the Linear Mixed Model to include analysis of pairwise interactions between measures (Environmental kernel Relationship Matrices (ERM)) with multiple iterations, were performed to predict case-control status. Each regression analysis was validated with four-fold crossvalidation.
Results and Conclusions:
Sensitivity, specificity, and Area Under the Curve of Receiver Operating Characteristic of 85%, 62%, and 86%, respectively, were obtained for both analytic methods. These prediction metrics demonstrate a promising diagnostic distinction based on premorbid risk variables. There were also statistically significant pairwise interactions between measures in the ERM model. The strong prediction metrics of both types of analytic model provide proof-of-principle for biologically-based laboratory tests as a first step toward primary prevention studies. Prospective studies of adolescents at elevated risk, vs. healthy adolescent controls, would be a next step toward development of primary prevention strategies.
Keywords: Psychosis, prevention, diagnostic tests, prediction metrics, event-related potentials, cognition, eye movements, polygenic risk score
1. Introduction and hypothesis
1.1. Background.
Two decades ago the focus of prevention research in Schizophrenia converged onto prodromal patients who present with some features of the illness but not enough to yet meet diagnostic criteria (Cornblatt et al., 2002). Previously there were two separate research approaches pertinent to prevention, pre-prodrome research, focused largely on investigation of high risk persons based on family history, and prodromal studies focused on studying and treating early presentations of psychotic symptoms in persons at risk of progressing to chronic psychosis. The North American Prodrome Longitudinal Study (NAPLS) reflects the focus on prodrome, and has contributed significantly to understanding the early trajectory of psychotic disorders (Addington et al., 2020; Perkins et al., 2020).
In the decades before this shift, however, several salient aspects of the pre-prodromal state had been discovered. Particularly, eye movement tracking, and auditory Event Related Potential (ERP) abnormalities that are present in Schizophrenia were demonstrated to be present in persons who later developed Schizophrenia, or in persons at increased genetic risk of Schizophrenia (by family history). Furthermore, in the two decades since, the concept of Schizophrenia as a neurodevelopmental disorder beginning in utero has taken hold (Murray and Lewis, 1987; Weinberger, 1986), and genome-wide association has demonstrated specific sets of genotypes to be associated with Schizophrenia (including Schizoaffective disorder), in a polygenic pattern of inheritance (Ripke et al., 2014). The polygenic predisposition to Schizophrenia was found to overlap greatly with Bipolar disorder and Major Depressive Disorder (Purcell et al., 2009). Measurements of cognitive, eye movement, and Event Related Potentials (ERP), as well as other measurements, were shown to aggregate across three psychosis diagnoses (Schizophrenia, Schizoaffective disorder, and Bipolar disorder with psychosis), adding more weight to the concept of a psychosis diathesis (Clementz et al., 2016). Importantly these alterations are found in young relatives of psychotic patients, as well as in ill patients, consistent with the notion that they represent trait markers of risk for illness (Keshavan et al., 2005). The most convincing evidence on the pre-prodrome state in Schizophrenia would consist of prospective observations in patients prior to the onset of symptoms, including genotype observations that are made later but by their nature are prospective.
As an initial step toward primary prevention based on prospective prediction of a psychotic disorder, we develop here predictions of psychosis based on multivariate analysis of measures known to be abnormal prior to symptom onset. We apply prediction metrics, including specificity, sensitivity, and overall accuracy, to differentiate of patients with psychosis diagnoses from healthy controls. Prediction of case or control status is measured in subsets of the data, with cross-validation, where estimates based on part of the data are applied to the rest of it. We employ iterative multivariate analyses of cognition, Event Related Potentials (ERPs), and eye tracking movements, and satisfactorily separate a mixture of patients with psychotic disorders and healthy individuals. Further research would be needed, of course, to replicate these findings, and to demonstrate them prospectively in persons who later develop psychotic disorders. Other measurements, particularly static and functional brain imaging, and resting state EEG measures, and molecular phenotypes based on genetic information, might also prove useful, but we focus here on a particular set of measurements that are supported as prospective markers by earlier evidence.
1.2. Previous studies have used prediction metrics,
and machine learning, to distinguish psychotic disorder patients from controls. Sweeney et al. (Sweeney et al., 1994) studied eye tracking as a predictor of psychotic disorder, without overlapping the sample in the current study. They found that low pursuit gain has moderately high sensitivity but only modest specificity for schizophrenia. Johannsen et al. (Johannesen et al., 2013), examined the diagnostic efficiency of ERPs P50, P300, and N100 in schizophrenia (SZ) as compared with healthy (HN) and bipolar disorder samples. Cross-validation was used to demonstrate stability of results. N100 and spectral power measures improved classification accuracy of SZ vs HN, with sensitivity = .78, and specificity = .80.
Rozycki et al. (Rozycki et al., 2018), seeking a neuroanatomical signature of Schizophrenia, studied multiple measures including regional volumes, voxelwise measures, and complex distributed patterns. Analyses were performed by advanced machine learning methods, and the findings included cross-validated prediction accuracy of 0.76 and Area Under the Curve (AUC) of 0.84. These are impressive metrics, but there is no data on whether these variables are in the same state prior to onset of illness.
1.3. Evidence on presence of pre-prodromal associations with psychosis risk in
cognition, eye tacking movement, and ERP. Cognitive deficits are present in Schizophrenia, and there is a good deal of evidence that these deficits are present long before onset. Cannon et al (Cannon et al., 2000) reported on 72 patients with Schizophrenia or Schizoaffective disorder, 33 of their siblings without any mental health diagnoses or treatment, and 57 healthy controls without ill siblings, ascertained from a carefully followed US birth cohort of approximately 8000 individuals who had been evaluated with WISC and Stanford-Binet IQ, which are standardized tests of cognitive functioning, at 4 and 7 years of age. Follow-up diagnoses were based on these individuals at 30 to 35 years of age. The psychosis patients, and their siblings, had significantly lower IQ scores than the controls, after controlling for multiple demographic and environmental variables. Their conclusion was that “premorbid cognitive dysfunction in schizophrenia represents a relatively stable indicator of vulnerability deriving from primarily genetic (and/or shared environmental) etiologic influences.” These conclusions remain current (Woodberry et al., 2008) (Sheffield et al., 2018).
Abnormalities in auditory electrophysiological evoked responses (ERPs), which are currently thought to reflect a fundamental brain deficit in psychosis (Javitt and Sweet, 2015), have also been found in the children at increased genetic risk of psychosis. Reduction in Schizophrenia patients of the late-positive potential at ~300 msec after a particular auditory stimulus, one of the first and most replicated ERP findings in psychosis, was studied in children of patients with Schizophrenia by Friedman, Vaughan, and Erlenmeyer-Kimling (Friedman et al., 1982). They ascertained 41 children of patients admitted to a New York hospital with Schizophrenia, and compared them with 45 children of separately ascertained normal controls, and tested them in two samples, which had mean ages of 15 and 11. They found significant reductions in ERP amplitude in the children at risk under specific conditions, including stimulus relevance, which were consistent with the adult Schizophrenia findings.
Eye movement: Levy et al. (Levy et al., 2010) state, in an historical review, that the independent rediscovery of smooth pursuit eye movement impairment by Holzman and colleagues (Holzman et al., 1973; Holzman et al., 1974), otherwise known as eye tracking dysfunction (ETD), is one of the most widely replicated behavioral deficits in schizophrenia, and is found in unaffected relatives. Jacobsen et al. (Jacobsen et al., 1996) studied 17 schizophrenic children with onset of illness by age 12, 18 ADHD children, and 22 normal children. Eye tracking variables were compared across the three groups. Schizophrenic children exhibited significantly greater smooth pursuit impairments than either normal or ADHD subjects. Kukuchi et al (Kikuchi et al., 2018) did genome-wide association analysis of eye movements in schizophrenics and controls that included a horizontal position gain measurement of smooth pursuit (HPG). They found significant functional genomic associations with HPG at several chromosomal locations in patients, and one association that was significant in the combined patient and control sample. Together, these studies imply that the eye tracking abnormalities found in psychosis occur early in life and are genetically determined.
Most common diseases, including major mental disorders with psychosis, are largely polygenic, involving numerous small effect common variants, and in rare cases chromosomal rearrangements or small mutations. Based on genome-wide SNP analyses, there is considerable polygenic overlap of the psychotic disorders studied in this paper (Consortium and Smoller, 2013; Lee et al., 2013). In the psychotic disorders, Polygenic Risk Scores for Schizophrenia (PRS) were introduced in 2009, and included Schizoaffective disorder (Purcell et al., 2009). This score is applicable to other disorders with psychosis, including Bipolar Disorder, and we include PRS in the predictive tests studied here.
Laboratory tests that were predictors of the development of psychotic disorders, before any symptoms appeared, similar to serum LDL cholesterol in atherosclerotic disorders, might lead to very early detection of high risk, and strategies for primary prevention of these disorders. At this time, and despite the progress detailed above, we lack tests that would meet accepted standards for prediction metrics, which include accuracy, sensitivity, and specificity (Bzdok et al., 2020). Although there is very great statistical significance of PRS in comparing patients vs. controls (Khera et al., 2018), there is no current set of predictors, including PRS, that satisfy accepted prediction metrics to the extent that they are clinically useful (Vassos et al., 2017; Wald and Old, 2019). One plausible reason is that successful prediction requires incorporation of more complex models of disease into the prediction, including biology-based phenotypes, other nongenetic factors, and their interactions (Ni et al., 2019; Zhou et al., 2020b).
The hypothesis that endophenotypes (intermediate phenotypes) could serve to unravel the genetics and biology of psychosis is decades old (Gershon et al., 1971; Gottesman and Shields, 1973). The Bipolar and Schizophrenia Network on Intermediate Phenotypes (BSNIP) dataset. BSNIP was established to perform comprehensive endophenotype and genetic testing (deep phenotyping) on an unprecedented scale, to unravel the biology of psychosis disorders (Tamminga et al., 2013).
1.4. Hypothesis.
The B-SNIP collaboration has developed a unique data set appropriate for complex disease causation, including individual-level measurements of a large set of neurobiological and clinical intermediate phenotypes, genome-wide genotypes, and environmental variables, in a consecutive series of recruited psychosis patients and healthy controls. In this paper we study these patients and controls, with an eye toward future studies that attempt to predict psychosis in non-prodromal adolescents, using the results from this study.
We hypothesize that multivariate analysis of validated laboratory tests that distinguish individuals in the premorbid state from controls, in the domains of cognition, eye tracking movement, and Event Related Potentials, along with advances in genomic knowledge of the major psychotic disorders, will yield acceptable prediction metrics for presence or absence of a psychotic disorder, in a mixed sample of patients and controls, without diagnostic interview data. We also hypothesize that analysis of interactions among these tests will add to predictive efficacy of these measures. We test these hypotheses with data on psychosis patients and controls from the BSNIP collaboration.
2.0. Methods
2.1. Source of data.
The BSNIP collaboration recruited patients and controls at 5 sites in the US, over two periods, 2008-2013 and 2014 through the present.
All participants were given the Structured Clinical Interview for DSM-IV Axis I Disorders, Patient Edition (SCID-I/P) including the Global Assessment of Functioning (GAF) scale, the Positive and Negative Syndrome Scale (PANSS), the Young Mania Rating Scale, the Montgomery-Åsberg Depression Rating Scale (MADRS), the Schizo-Bipolar Scale, and the Birchwood Social Functioning Scale (Birchwood et al., 1990; Keshavan et al., 2011; Lançon et al., 2000; Montgomery and Asberg, 1979; Young et al., 1978). Diagnostic reliability was maintained by continued review and conferences across sites. Individuals were included as having a psychosis diagnosis (SZ, SAD, or BPP) or absence of Axis I disorder as healthy control. Additionally, no currently active substance abuse diagnosis was required of all participants.
The data described here are from BSNIP1, the first phase of the BSNIP study. We report on unrelated patients and healthy controls, verified for non-relatedness on genotypes by PREST-plus(Sun and Dimitromanolakis, 2014) and KING(Manichaikul et al., 2010) in order to remove individuals with 3rd degree or closer kinship. The participants are thus termed “unrelated.” Intermediate phenotypes were chosen that have evidence of predating illness (cognitive deficits, PRS), or of being found in well relatives of patients (Event Related Potentials and tracking eye movement abnormalities), as described above.
2.2. The BSNIP intermediate phenotypes
are described in several publications, including controlling for drug exposure (Ivleva et al., 2013; Lencer et al., 2017; Lencer et al., 2015; Mathew et al., 2014; Meda et al., 2015; Narayanan et al., 2015; Sheffield et al., 2017; Tamminga et al., 2013; Wang et al., 2015). The genetic and phenotypic overlap justification for pooling different psychotic diagnoses is described in those papers. Ethics: Consecutively recruited non-hospitalized patients with one of the psychotic disorders and healthy controls, studied under a uniform protocol approved by Institutional Review Boards (IRBs) at each site. The 5 BSNIP sites have concurrent measurement and diagnostic standardization across sites. All variables in the regression analyses are listed in Supplementary Table 1 (ST1).
2.3. Polygenic risk scores
(PRS) on these persons were calculated on the genotypic matrix of the current multi-ethnic dataset by Shafee et al.(Shafee et al., 2018), who provided scores to us on additional persons not in the referenced publication, using the PGC2 Schizophrenia data (Ripke et al., 2014) as a reference, and applied here with 7 Principal Components (PCs) as covariates.
2.4. Enrolled participants and ancestry.
883 unrelated individuals were enrolled in BSNIP1. The testing procedures required a great deal of time and effort of each volunteer (two non-consecutive full days after the diagnostic interview). Blood for genotyping was drawn at the diagnostic interview. Not all phenotypic tests were completed by all volunteers. Only 371 persons completed the phenotypes in this paper. Comparison of the volunteers who were enrolled and who were completed showed 66% psychosis patients in the enrolled group and 70% psychosis patients in the completers group, which was not a significant difference. It is not possible to compare the phenotypic values of the completing and non-completing participants, of course, because there is no data on the non-completers.
Ancestry classification of the participants (Table 1) was made by inspection of the first two PCs of the genotype matrix (Price et al., 2006).
Table 1.
Ancestry of completed participants
| Totals | African-American (AA) | Caucasian-European (CEU) | OTHER | |
|---|---|---|---|---|
| Healthy controls | 10 | 28 | 72 | 10 |
| Psychosis volunteers | 61 | 85 | 168 | 8 |
2.5. Statistical analysis: Multivariate analysis of risk and of interaction of the prediction variables.
The intermediate phenotype variables are listed in ST1.
2.5.1. Logistic regression of intermediate phenotypes on case-control status
(variables listed in ST1). We implemented a fitted logistic regression model using the Scikit statistical package (sklearn) described in Babcock (Babcock, 2016, Hackeling, 2014), with regularization and one round of Recursive Feature Elimination (RFE). The variables dropped after RFE are listed in ST2.
2.5.2. Extension of the Linear Mixed Model (LMM).
The LMM models are described in detail in Supplementary Information. The term Exposome (E) is applied to any non-genotype measure, and is equivalent in this context to Environment. The extension is to add the effects of interactions among environmental variables to the risk prediction. The term “EXE” (chosen to be analogous to Genetic by Environment interaction; GXE) refers to effects of interactions between all pairs of measured factors, including intermediate phenotypes and demographic covariates (listed in Table ST1), on the dependent variable (psychosis). We constructed Environmental kernel Relationship Matrices (ERM) based on all of these factors (Zhou et al., 2020a; Zhou et al., 2020b). We fit the kernel matrices jointly in the proposed models to estimate the variance components of neurobiological effects and their interactions, as E×E on psychosis. Given the multiple neurobiological measurements and other variables available in this project, this approach sheds some light on the latent biological architecture of psychosis, i.e. E×E interaction, undetected by existing methods (Bulik-Sullivan et al., 2015; Robinson et al., 2017; Yang et al., 2011). This ERM approach can estimate E×E interaction in which the Hadamard products of all possible pairs of measured variables are explicitly modelled to capture their interactions. The aggregated interactions can be parameterized as a single variance component, i.e., we use a powerful random-effects approach. Further discussion of the ERM model is presented in Supplementary Information.
2.6. Metrics of prediction.
The quality of any diagnostic test, or battery of tests, can be measured by how well they predict case or healthy control status (or risk) of individuals. It has been advocated for decades that diagnostic tests be accepted only after they have been validated by statistical prediction quality metrics (Cowley et al., 2019; Wasson et al., 1985). The quality of a binary classification test battery, which we study here, is not directly measured by the statistical significance of the case-control difference in a set of data (Bzdok et al., 2020). The standard metrics of prediction include having training and test datasets, and are concerned with the rates of True (T) vs. False (F), and Positive (P) vs. Negative (N) predictions in the test data (Hackeling, 2014). The accepted metrics for binary classification include Recall (sensitivity) = TP/(TP + FN), Specificity = TN/(TN+FP), Precision = TP/(TP + FP), and Accuracy (fraction of predictions that are correct. Additionally, the Area Under Curve (AUC) of the Receiver Operating Characteristic (ROC), quantifies the overall validity of a test. The ROC is a two-dimensional graph of TP vs. FP. AUC = 0.5 is a null test and AUC=1.0 is a perfect test.
AUC was calculated from actual diagnoses vs. p-values from Logistic regression, and from actual diagnoses vs. normally distributed regression values from the Linear Mixed Models (LMM) ERM analyses. For prediction metrics, the threshold for case status was determined as the quantile of the normally distributed regression values at a specific cumulative density of 0.71 that corresponded to the case proportion in the data (i.e., qnorm (1-0.71) in the R function). For Logistic Regression the threshold was regression-estimated probability of case status exceeding 0.5.
2.7. Four-fold cross-validation of predictions
was performed by random division of the data into four equal sized “folds”, and consecutively using each fold as a test subset and the remaining data as a training subset. Statistical analyses of cross-validation, including AUC and prediction metrics, were calculated on the test subsets in each analysis. In the Results, we report the mean and standard deviation of each of the metrics, from the results of 4-fold cross-validation.
3.0. Results
Our overall approach is to study psychosis risk prediction, and analysis of interactions among the predicting variables, in data collected by the B-SNIP consortium.
3.1. Logistic regression of intermediate phenotypes on case-control status.
As a proof-of-concept, we studied 51 phenotypic variables associated with patient-control differences in the literature and in the BSNIP1 dataset. The domains of these results are cognition, eye movement, and electrophysiological data, demographic covariates including years of education, and PRS, listed in Supplementary Table ST1.
Logistic regression was performed on 371 unrelated individuals. First, the Scikit-learn (sklearn) (Hackeling, 2014) Recursive Factor Eliminator (RFE) program was run, with scaled data, and 21 of the 51 variables could be dropped (Supplementary Table ST2). The pseudo-R-squared (variance for logistic regression) was 0.61 (Nagelkerke). We ran the Sckit-learn Logistic Regression program with four fold cross-validation on the non-dropped 26 variables, with regularization using L2 penalty, and sklearn.metrics for prediction metrics (Table 2). The AUC was 86%, sensitivity was 85% and specificity was 62%. These are robust prediction metrics, but the specificity value means that prediction of “no psychosis” is only correct in about 2/3 of instances.
Table 2.
Receiver Operating Characteristic Area Under Curve (AUC), accuracy, sensitivity, and specificity, by models averaged over 4 training-test (target) data divisions (cross-validation).
| Logistic regression | ERM Model 1: y = E + ε | ERM Model 2: y = E + EXE + ε (top 25 pairs of interaction for EXE) |
|
|---|---|---|---|
| AUC | 0.86±0.00 | 0.86±0.00 | 0.86±0.01 |
| Accuracy | 0.78±0.01 | 0.78±0.01 | 0.79±0.01 |
| Sensitivity | 0.85±0.01 | 0.84±0.02 | 0.85±0.02 |
| Specificity | 0.62±0.05 | 0.64±0.05 | 0.65±0.02 |
3.1.1. A note on PRS.
The most significant PRS patient-control difference on these 371 individuals was for PRS threshold p = 1E-8, p < 0.002. PRS as a predictor using logistic regression (Babcock, 2016), with age, sex, and 7 PCs as covariates showed pseudo-R-squared of 5.1% for the model. PRS had the highest Odds Ratio among the included factors (3.5), but only modest predictive power for diagnosis. AUC for the model was 0.49 after correction for covariates, very close to the null value of 0.5. We would attribute this performance to the multiancestry nature of the tested sample, and to the generally modest disease prediction effects of PRS scores (Wald and Old, 2019).
3.2. ERM approach to risk prediction.
Complementary to the genome (G), the concept of Exposome (E) has been proposed to capture the totality of human environmental exposures, encompassing external as well as internal environments over the lifetime of a given individual. We estimated the main and interaction effects of neurobiological and clinical variables on psychosis risk, where interaction effects from all pairs of the variables are simultaneously considered, termed as E×E interaction. E, in this context, includes the multiple neurobiological measurements and demographic covariates, and is chosen to contrast with genotype measurements. In the ERM approach, we constructed a kernel matrix, and a kernel matrix based on the Hadamard products of all possible pairs of measured variables (see Supplementary Information). We then fit these kernel matrices jointly to estimate environmental variance components and E×E interactions. The ERM models are described further in a section of Supplementary Information.
3.2.1. The proportion of phenotypic variance explained by additive effects of Environmental variables ().
From a linear mixed model fitting the additive effects of Environmental variables (model #1 is described in detail in Supplementary Information), we estimated (se = 0.06) on the observed scale. Using a population prevalence of psychotic disorders k=0.02 and the sample proportion of cases p=0.70, the estimated can be transformed to 0.24 (se = 0.05) on the liability scale (Lee et al., 2011).
It is remarkable that a substantial variance of the underlying liability is attributed to the 52 variables (Environmental +PRS), and the precision of the estimate is high (i.e. a low se) although the sample size is moderate. The high precision is because the variance of off-diagonal elements in the environmental kernel relationship matrix is high (Visscher et al., 2014). This was also confirmed with the GCTA power calculator (https://shiny.cnsgenomics.com/gctaPower/) (data not shown). Thus, the ERM approach with BSNIP data is a powerful approach.
Using an advanced Linear Mixed Model that fits an additional interaction term (model #2; see Supplementary Information), we also assessed how much additional variance was captured by the interaction term. As shown in Table ST4, around 10% of phenotypic variance on the liability scale was due to EXE interaction effects (p-value = 5.73E-06). Note that the main environmental variance () was slightly reduced on the liability scale as compared with observed variance (from 0.28 to 0.22 for top 50 EXE), which was probably due to collinearity between pairs of random effects (Zhou et al., 2020a).
The accuracy and AUC in model #2 were not significantly different from model #1 or the standard Logistic Regression (Table 2) (p-value = 0.22), possibly because of the relatively small sample size and small variance of off-diagonal elements in the ExE kernel matrix (Visscher et al., 2014). As in the preceding section, the prediction metrics are strong and very promising.
4.0. Discussion and Conclusions
4.1. The overall prediction metric,
AUC, is 86%, and the other prediction metrics, sensitivity, specificity, and accuracy, are in the range of 62% to 85%. This represents the first successful laboratory-test classification of psychotic disorders using the combined information of a dense phenotyping approach, although the metrics are not successful enough for clinical use at this point. If the specificity metric were in the range of the other metrics, we would consider them successful. Lower specificity than sensitivity was also reported by Sweeney et al. for eye tracking in SZ (Sweeney et al., 1994). Nonetheless, we submit that these findings are a first step toward statistically valid and precise prediction of psychosis risk, based on multiple features associated with the disorders. Prediction as tested here is on cross-validation of known cases and controls; to be proven effective, prediction would have to be applied prospectively to unaffected individuals at risk.
Prevention of the major psychotic disorders, before disease onset, has appeared to be an intractable challenge for many years. The known epidemiologic risk factors (Belbasis et al., 2018) were and remain too nonspecific (cannabis use) or non-addressable (obstetric complications, in utero viral exposures) for practical use. In an influential 2002 paper, Cornblatt et al (Cornblatt et al., 2002) described (and advocated) a shift away from research on premorbid tests: “High-risk researchers, who view the identification of accurate risk factors as necessarily preceding preventive programs, have begun to move from the premorbid to the prodromal phase as the most effective starting point.” This perspective dominates prevention research until the present time(McGorry and Nelson, 2020), even though considerable new knowledge on premorbid findings has accumulated (Keshavan et al., 2005).
4.2. Primary vs. secondary endophenotypes.
A crucial issue for primary prevention is which endophenotypic features of illness are secondary to disease, or to antipsychotic drug treatment. The evidence that cognitive deficits predate illness onset is strong, particularly in family studies (Kendler et al., 2016; van Oel et al., 2002), and higher polygenic risk score (PRS) for schizophrenia is associated with lower cognitive performance in healthy individuals(Shafee et al., 2018). As reviewed above, there are supportive data for premorbid presence of the major tests analyzed here. For other variables associated with psychosis, such as brain imaging measures (see Supplementary Tables ST6 and ST7), there are little data to determine if they are premorbid abnormalities.
4.3. Deep phenotyping of studied individuals.
Previous studies of mediation of the gene-disease association through intermediate phenotypes, with their own genetic and environmental associations, have focused on summary statistics of clinically reported variables such as age at onset (Bulik-Sullivan et al., 2015; Sekula et al., 2016). Direct measurement of multiple phenotypes in each person may serve to improve risk prediction, by enabling performance of intricate analyses of genetic and environmental interactions (Martin et al., 2019). The penalty of this approach, however, as evident in this paper, is that the number of persons to analyze becomes significantly limited by the arduousness of the phenotyping.
4.4. Potential additional phenotypes.
Among features of psychosis not included in the current analyses, derived molecular intermediate phenotypes are appealing as potential predictors. The Genotype-Tissue Expression (GTEx) collaboration has developed Normalized Effect Scores (NES) for genome-wide genotypes for specific gene expression in multiple tissues (Wang et al., 2016), and these are publicly available. The PsychEncode collaboration has found differential brain expression data in specific regions on a larger number of Schizophrenia and control brains (including the GTEx data) (Wang et al., 2018). One can develop expression scores for particular sets of genes, including the 320 genes with expression linked to Schizophrenia (Wang et al., 2018), and WGCNA gene coexpression modules from our own and others work(Chen et al., 2013; Radulescu et al., 2020). The HLA association with Schizophrenia has been one of the strongest molecular associations with psychosis for decades (Goldin and Gershon, 1983). These markers are contained in the large Human Histocompatibility Regions of chromosome 6. Within that region, the Complement protein C4 gene appears to be a major factor in disease association(Sekar et al., 2016). These represent additional molecular phenotypes to include in prediction studies, with attention to the population-specificity of these associations.
We would also expect incorporation of additional data such as connectivity derived from MRI data, and additional environmental variables such as cannabis use and childhood trauma history, might improve prediction metrics.
4.5. Limitations and next steps.
Not having prospective predictions is the major limitation we are aware of in this study. The specificities we report (62% to 65%) are too low for clinical applicability. Sample size is yet another limitation –we could include additional variables in a much larger sample, with less risk of overfitting. Another limitation, shared with other studies in the field, is that PRS scores do not apply well to persons of African ancestry. The attrition rate in the individuals studied, from 883 in the initial testing session to 371 who complete all sessions, may have reasons we are unaware of, that impact the results in this report.
Next steps: We are fortunate to have considerable ancestry diversity in the reported results, so that the results, if replicated and extended, would be widely applicable. Towards these ends we would do an independent replication of the current results, and add to the battery of tests to attempt improved prediction metrics so they are all above 90%, in a new sample. If that were achieved, an adequately powered prospective study would be in order, using the phenotypes and genetic measures developed from the earlier steps in this research, in children or adolescents. One might consider studying a population sample, but only 2% of a sample would actually develop illness. It would be more efficient to compare persons at increased genetic risk, such as relatives of known cases, to compare with controls. This would be a daunting study to propose and execute, but the results might lead to primary prevention strategies for psychotic disorders.
Supplementary Material
Figure 1.
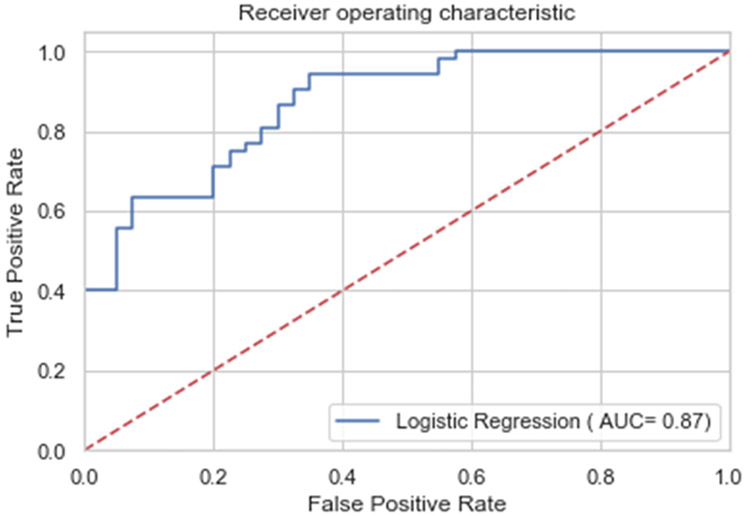
Area Under Curve (AUC) of a representative fold of Logistical Regression cross-validation. The axis titles are a legacy for AUC diagrams. In terms used in this paper, the vertical axis is sensitivity and the horizontal axis is (1-specificity). A perfect predictor would have AUC of 1.00 and a null predictor would have 0.5. For a summary of AUCs for all cross validation folds of Logistic Regression and of the two Linear Mixed Model ERM models, see Table 2 and Supplementary Table ST3.
Acknowledgements
Dr. Rebecca Shafee, of Harvard Medical School and the Broad Institute of MIT and Harvard, kindly supplied Polygenic Risk Scores for Schizophrenia for the data in this paper. John I. Nurnberger, Jr. of Indiana University offered comments and advice on a draft of this paper.
Grant support:
US National Institute of Mental Health: E.S.G., MH103368, B.A.C., MH103366, M.S.K. MH078113, G.D.P. MH077945, C.A.T. MH077851.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
Dr Sweeney consults to VeraSci.
References
- Addington J, Liu L, Brummitt K, Bearden CE, Cadenhead KS, Cornblatt BA, et al. , 2020. North American Prodrome Longitudinal Study (NAPLS 3): Methods and baseline description. Schizophr Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock J, 2016. Mastering Predictive Analytics with Python. Packt Publishing, Birmingham UK, Mumbai India. [Google Scholar]
- Belbasis L, Kohler CA, Stefanis N, Stubbs B, van Os J, Vieta E, et al. , 2018. Risk factors and peripheral biomarkers for schizophrenia spectrum disorders: an umbrella review of metaanalyses. Acta Psychiatr Scand 137(2), 88–97. [DOI] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics, C., et al. , 2015. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47(3), 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzdok D, Varoquaux G, Steyerberg EW, 2020. Prediction, Not Association, Paves the Road to PrecisionMedicine. JAMA psychiatry. [DOI] [PubMed] [Google Scholar]
- Cannon TD, Bearden CE, Hollister JM, Rosso IM, Sanchez LE, Hadley T, 2000. Childhood cognitive functioning in schizophrenia patients and their unaffected siblings: a prospective cohort study. Schizophr Bull 26(2), 379–393. [DOI] [PubMed] [Google Scholar]
- Chen C, Cheng L, Grennan K, Pibiri F, Zhang C, Badner JA, et al. , 2013. Two gene coexpression modules differentiate psychotics and controls. Mol Psychiatry 18(12), 1308–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementz BA, Sweeney JA, Hamm JP, Ivleva EI, Ethridge LE, Pearlson GD, et al. , 2016. Identification of Distinct Psychosis Biotypes Using Brain-Based Biomarkers. Am J Psychiatry 173(4), 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, C.-D.G.o.t.P.G., Smoller JW, 2013. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381(9875), 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornblatt B, Lencz T, Obuchowski M, 2002. The schizophrenia prodrome: treatment and high-risk perspectives. Schizophr Res 54(1–2), 177–186. [DOI] [PubMed] [Google Scholar]
- Cowley LE, Farewell DM, Maguire S, Kemp AM, 2019. Methodological standards for the development and evaluation of clinical prediction rules: a review of the literature. Diagnostic and Prognostic Research 3(1), 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D, Vaughan HG Jr., Erlenmeyer-Kimling L, 1982. Cognitive brain potentials in children at risk for schizophrenia: preliminary findings. Schizophr Bull 8(3), 514–531. [DOI] [PubMed] [Google Scholar]
- Gershon ES, Dunner DL, Goodwin FK, 1971. Toward a biology of affective disorders. Genetic contributions. Arch Gen Psychiatry 25(1), 1–15. [DOI] [PubMed] [Google Scholar]
- Goldin LR, Gershon ES, 1983. Association and linkage studies of genetic marker loci in major psychiatric disorders. Psychiatr Dev 1(4), 387–418. [PubMed] [Google Scholar]
- Gottesman II, Shields J, 1973. Genetic theorizing and schizophrenia. The British journal of psychiatry : the journal of mental science 122(566), 15–30. [DOI] [PubMed] [Google Scholar]
- Hackeling G, 2014. The Fundamentals of Machine learning, Mastering Machine Learning with scikit-learn. Pact Publishing Ltd., Birmingham, UK, pp. 7–20. [Google Scholar]
- Holzman PS, Proctor LR, Hughes DW, 1973. Eye-tracking patterns in schizophrenia. Science 181(4095), 179–181. [DOI] [PubMed] [Google Scholar]
- Holzman PS, Proctor LR, Levy DL, Yasillo NJ, Meltzer HY, Hurt SW, 1974. Eye-tracking dysfunctions in schizophrenic patients and their relatives. Arch Gen Psychiatry 31(2), 143–151. [DOI] [PubMed] [Google Scholar]
- Ivleva EI, Bidesi AS, Keshavan MS, Pearlson GD, Meda SA, Dodig D, et al. , 2013. Gray matter volume as an intermediate phenotype for psychosis: Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP). Am J Psychiatry 170(11), 1285–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen LK, Hong WL, Hommer DW, Hamburger SD, Castellanos FX, Frazier JA, et al. , 1996. Smooth pursuit eye movements in childhood-onset schizophrenia: comparison with attention-deficit hyperactivity disorder and normal controls. Biol Psychiatry 40(11), 1144–1154. [DOI] [PubMed] [Google Scholar]
- Javitt DC, Sweet RA, 2015. Auditory dysfunction in schizophrenia: integrating clinical and basic features. Nat Rev Neurosci 16(9), 535–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Ohlsson H, Mezuk B, Sundquist K, Sundquist J, 2016. A Swedish National Prospective and Co-relative Study of School Achievement at Age 16, and Risk for Schizophrenia, Other Nonaffective Psychosis, and Bipolar Illness. Schizophr Bull 42(1), 77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavan MS, Diwadkar VA, Montrose DM, Rajarethinam R, Sweeney JA, 2005. Premorbid indicators and risk for schizophrenia: a selective review and update. Schizophr Res 79(1), 45–57. [DOI] [PubMed] [Google Scholar]
- Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. , 2018. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 50(9), 1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi M, Miura K, Morita K, Yamamori H, Fujimoto M, Ikeda M, et al. , 2018. Genome-wide Association Analysis of Eye Movement Dysfunction in Schizophrenia. Scientific reports 8(1), 12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Wray N, Goddard M, Visscher P, 2011. Estimating Missing Heritability for Disease from Genome-wide Association Studies. Am J Hum Genet 88(3), 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, et al. , 2013. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 45(9), 984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencer R, Mills LJ, Alliey-Rodriguez N, Shafee R, Lee AM, Reilly JL, et al. , 2017. Genome-wide association studies of smooth pursuit and antisaccade eye movements in psychotic disorders: findings from the B-SNIP study. Transl Psychiatry 7(10), e1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencer R, Sprenger A, Reilly JL, McDowell JE, Rubin LH, Badner JA, et al. , 2015. Pursuit eye movements as an intermediate phenotype across psychotic disorders: Evidence from the B-SNIP study. Schizophr Res 169(1–3), 326–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DL, Sereno AB, Gooding DC, O’Driscoll GA, 2010. Eye tracking dysfunction in schizophrenia: characterization and pathophysiology. Current topics in behavioral neurosciences 4, 311–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM, 2010. Robust relationship inference in genome-wide association studies. Bioinformatics 26(22), 2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Daly MJ, Robinson EB, Hyman SE, Neale BM, 2019. Predicting Polygenic Risk of Psychiatric Disorders. Biol Psychiatry 86(2), 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew I, Gardin TM, Tandon N, Eack S, Francis AN, Seidman LJ, et al. , 2014. Medial temporal lobe structures and hippocampal subfields in psychotic disorders: findings from the Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP) study. JAMA psychiatry 71(7), 769–777. [DOI] [PubMed] [Google Scholar]
- McGorry PD, Nelson B, 2020. Clinical High Risk for Psychosis-Not Seeing the Trees for the Wood. JAMA psychiatry. [DOI] [PubMed] [Google Scholar]
- Meda SA, Wang Z, Ivleva EI, Poudyal G, Keshavan MS, Tamminga CA, et al. , 2015. Frequency-Specific Neural Signatures of Spontaneous Low-Frequency Resting State Fluctuations in Psychosis: Evidence From Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP) Consortium. Schizophr Bull 41(6), 1336–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray RM, Lewis SW, 1987. Is schizophrenia a neurodevelopmental disorder? 295(6600), 681–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan B, Ethridge LE, O’Neil K, Dunn S, Mathew I, Tandon N, et al. , 2015. Genetic Sources of Subcomponents of Event-Related Potential in the Dimension of Psychosis Analyzed From the B-SNIP Study. Am J Psychiatry 172(5), 466–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni G, van der Werf J, Zhou X, Hypponen E, Wray NR, Lee SH, 2019. Genotype-covariate correlation and interaction disentangled by a whole-genome multivariate reaction norm model. Nat Commun 10(1), 2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DO, Olde Loohuis L, Barbee J, Ford J, Jeffries CD, Addington J, et al. , 2020. Polygenic Risk Score Contribution to Psychosis Prediction in a Target Population of Persons at Clinical High Risk. Am J Psychiatry 177(2), 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D, 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38(8), 904–909. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, et al. , 2009. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460(7256), 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radulescu E, Jaffe AE, Straub RE, Chen Q, Shin JH, Hyde TM, et al. , 2020. Identification and prioritization of gene sets associated with schizophrenia risk by co-expression network analysis in human brain. Mol Psychiatry 25(4), 791–804. [DOI] [PubMed] [Google Scholar]
- Ripke S, Neale BM, Corvin A, Walters JT, Farh K, Holmans PA, et al. , 2014. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511(7510), 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MR, English G, Moser G, Lloyd-Jones LR, Triplett MA, Zhu Z, et al. , 2017. Genotype-covariate interaction effects and the heritability of adult body mass index. Nat Genet 49(8), 1174–1181. [DOI] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, et al. , 2016. Schizophrenia risk from complex variation of complement component 4. Nature 530(7589), 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekula P, Del Greco M F, Pattaro C, Kottgen A, 2016. Mendelian Randomization as an Approach to Assess Causality Using Observational Data. Journal of the American Society of Nephrology 27(11), 3253–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafee R, Nanda P, Padmanabhan JL, Tandon N, Alliey-Rodriguez N, Kalapurakkel S, et al. , 2018. Polygenic risk for schizophrenia and measured domains of cognition in individuals with psychosis and controls. Transl Psychiatry 8(1), 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield JM, Kandala S, Tamminga CA, Pearlson GD, Keshavan MS, Sweeney JA, et al. , 2017. Transdiagnostic Associations Between Functional Brain Network Integrity and Cognition. JAMA psychiatry 74(6), 605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield JM, Karcher NR, Barch DM, 2018. Cognitive Deficits in Psychotic Disorders: A Lifespan Perspective. Neuropsychology review 28(4), 509–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Dimitromanolakis A, 2014. PREST-plus identifies pedigree errors and cryptic relatedness in the GAW18 sample using genome-wide SNP data. BMC proceedings 8(Suppl 1 Genetic Analysis Workshop 18Vanessa Olmo), S23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney JA, Clementz BA, Haas GL, Escobar MD, Drake K, Frances AJ. Eye tracking dysfunction in schizophrenia: characterization of component eye movement abnormalities, diagnostic specificity, and the role of attention. J Abnorm Psychol. 1994. May;103(2):222–30. [DOI] [PubMed] [Google Scholar]
- Tamminga CA, Ivleva EI, Keshavan MS, Pearlson GD, Clementz BA, Witte B, et al. , 2013. Clinical phenotypes of psychosis in the Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP). Am J Psychiatry 170(11), 1263–1274. [DOI] [PubMed] [Google Scholar]
- van Oel CJ, Sitskoorn MM, Cremer MP, Kahn RS, 2002. School performance as a premorbid marker for schizophrenia: a twin study. Schizophr Bull 28(3), 401–414. [DOI] [PubMed] [Google Scholar]
- Vassos E, Di Forti M, Coleman J, Iyegbe C, Prata D, Euesden J, et al. , 2017. An Examination of Polygenic Score Risk Prediction in Individuals With First-Episode Psychosis. Biol Psychiatry 81(6), 470–477. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Hemani G, Vinkhuyzen AAE, Chen G-B, Lee SH, Wray NR, et al. , 2014. Statistical Power to Detect Genetic (Co)Variance of Complex Traits Using SNP Data in Unrelated Samples. PLoS Genet 10(4), e1004269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald NJ, Old R, 2019. The illusion of polygenic disease risk prediction. Genet Med 21(8), 1705–1707. [DOI] [PubMed] [Google Scholar]
- Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, et al. , 2018. Comprehensive functional genomic resource and integrative model for the human brain. Science 362(6420). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gamazon ER, Pierce BL, Stranger BE, Im HK, Gibbons RD, et al. , 2016. Imputing Gene Expression in Uncollected Tissues Within and Beyond GTEx. Am J Hum Genet 98(4), 697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Meda SA, Keshavan MS, Tamminga CA, Sweeney JA, Clementz BA, et al. , 2015. Large-Scale Fusion of Gray Matter and Resting-State Functional MRI Reveals Common and Distinct Biological Markers across the Psychosis Spectrum in the B-SNIP Cohort. Frontiers in psychiatry 6, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson JH, Sox HC, Neff RK, Goldman L, 1985. Clinical prediction rules. Applications and methodological standards. N Engl J Med 313(13), 793–799. [DOI] [PubMed] [Google Scholar]
- Weinberger DR, 1986. The pathogenesis of schizophrenia: a neurodevelopmental theory, in: Nasrallah HA, Weinberger DR (Eds.), The Neurology of Schizophrenia. Elseveier, Netherlands, pp. 397–406. [Google Scholar]
- Woodberry KA, Giuliano AJ, Seidman LJ, 2008. Premorbid IQ in schizophrenia: a metaanalytic review. Am J Psychiatry 165(5), 579–587. [DOI] [PubMed] [Google Scholar]
- Yang J, Lee S, Goddard M, Visscher P, 2011. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet 88(1), 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Im HK, Lee SH, 2020a. CORE GREML for estimating covariance between random effects in linear mixed models for complex trait analyses. Nat Comm In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, van der Werf J, Carson-Chahhoud K, Ni G, McGrath J, Hypponen E, et al. , 2020b. Whole-Genome Approach Discovers Novel Genetic and Nongenetic Variance Components Modulated by Lifestyle for Cardiovascular Health. J Am Heart Assoc 9(8), e015661. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


