Abstract
Background: The “cytokine storm” (CS) in COVID-19 leads to the worst stage of illness which can be controlled only with timely intervention. There is an urgent need to identify laboratory markers of disease progression for optimum allocation of resources in developing countries like India. Methods: A cross-sectional study was conducted on 100 COVID-19 positive patients over two months. The cases were sub-classified based on disease severity into mild to moderate (n=61), severe (n=26) and very severe (n=13) and into survivors (n=85) and non-survivors (n=15) based on survivor status. These patients were tested for hematological parameters (total blood lymphocyte counts, NLR, PLR, platelet indices etc.), coagulation markers (D-dimer, fibrin degradation products (FDP), fibrinogen etc.) and biochemical markers (LDH, ferritin, IL-6, procalcitonin, hs-CRP). Results: Statistically significant differences were observed in hematological variables (ANC, NLR and ESR), coagulation parameters (D-dimer, FDP, fibrinogen and thrombin time) and biochemical markers (LDH, ferritin, IL-6, procalcitonin and hs-CRP) with regard to subcategories based of disease severity as well as survivor status. There was strong correlation between NLR, D-dimer, IL-6, procalcitonin and ferritin. IL-6 emerged as the single best marker of disease severity (AUC: 0.997, P=0.00), however procalcitonin, LDH, D-dimer, FDP and NLR could also predict severe disease with a good sensitivity and specificity. Conclusion: To conclude, study demonstrates a plethora of biomarkers which could be utilized to accurately identify the hyperinflammation and tissue damage reminiscent of cytokine storm in COVID-19 patients so that timely, safe, and effective therapies can be administered to prevent progression and potentially reduce mortality.
Keywords: COVID-19, hematological indices, NLR, D-dimer, coagulation, biochemical marker, IL-6, acute phase reactant
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS CoV-2) implicated in COVID-19 disease was reported for the first time in Wuhan, China in December 2019 and has rapidly evolved from an epidemic outbreak into a global pandemic infecting around 150 million individuals across the world till date [1,2]. On 30th Jan 2020, the first case was reported in India from the state of Kerala. The patient had a positive travel history to Wuhan and ever since 20 million cases of COVID-19 have been documented.
India is currently witnessing world’s worst COVID-19 crisis with daily infection rates of 3-4 lakhs and death toll of 3000-4000 [1]. Healthcare infrastructure is crippling with an acute shortage of hospital beds, oxygen, ventilators etc. In the battle against COVID-19 disease, there is an urgent need for recognition of clinical and laboratory predictors of disease progression to serious and critical forms. These markers will aid in clinical management by focussing on patients at increased risk of developing a serious disease and optimizing allocation of resources in the ongoing pandemic especially in developing countries like India with limited human and technological resources.
The pathogenesis of COVID-19 is complex with multi-system involvement including hematopoietic system, hemostasis and immune system. There is formidable evidence suggesting features of hyper inflammation in critically ill patients, consisting of elevated serum C-reactive protein (CRP), procalcitonin, D-Dimer, and hyperferritinemia [3]. Moreover, lymphopenia is strongly correlated with the disease severity [4]. The neutrophil-lymphocyte ratio (NLR) and platelet-lymphocyte ratio (PLR) have both emerged as good indicators of subclinical systemic inflammation in many diseases like cardiovascular diseases, cancers, autoimmune diseases and also in COVID-19 [5]. The “cytokine storm” (CS) in COVID-19 leads to the worst stage of illness. Increased serum expression of interleukin (IL)-2R and IL-6 appears to be predicting the severity and prognosis of COVID-19 patients [6].
Taking into account the aforementioned, the present study was conducted with the aim to estimate serum levels of (1) acute phase reactants/biochemical markers (LDH, ferritin, IL-6, procalcitonin, hs-CRP), (2) hematological parameters (total blood lymphocyte counts, NLR, PLR, platelet indices etc.) as well as (3) coagulation markers (D-dimer, fibrin degradation products (FDP), fibrinogen etc.) in patients with COVID-19 disease. Further we aim to explore the correlation between the levels of the above parameters with disease severity and survivor status.
Material and methods
The present study was a prospective cross-sectional study conducted for two months (October and November 2020) on all RTPCR positive COVID-19 patients admitted to ESIC Medical College and Hospital, Faridabad. Institutional Ethics Committee provided approval for the study (No. 134/A/11/16/Academics/MC/2016/151) after written informed consent from the patients. Clinical and demographical details of the patients were recorded.
Sample collection
The venous blood samples were collected on day 1-3 of admission to the isolation ward, in three vacutainers (Plain, EDTA and citrate). The following tests were performed: 1) Biochemical-LDH, ferritin, IL-6, procalcitonin, hs-CRP. Among these, ferritin, hs-CRP and procalcitonin was performed on VITROS XT 7600 while LDH on Randox Rx Daytona. IL-6 was done by enzyme linked immunosorbent assay (ELISA). 2) Hematological parameters-Complete blood counts (CBC) including absolute neutrophil count (ANC), absolute lymphocyte count (ALC), absolute monocyte count (AMC), platelet count, platelet indices-MPV, platelet distribution width (PDW), plateletcrit (PCT), NLR, PLR, and erythrocyte sedimentation rate (ESR). A fully automated 6 Part hematology analyzer (Sysmex XN 1000) was used for CBC while VESMATIC CUBE 80 analyzer was used for ESR. NLR, PLR, MLR were calculated from hematological parameters. 3) Coagulation markers-D-dimer, fibrin degradation products (FDP), fibrinogen, prothrombin time, activated partial thromboplastin time, INR, thrombin time. Fully automated coagulation analyser (Stago STA compact max) was used for performing all the coagulation tests.
All requisite precautions as per the standard guidelines and protocols for COVID 19 sample handling were followed from collection of samples, processing to discarding.
Inclusion criteria
All patients above 18 years of age who were real time reverse transcriptase polymerase chain reaction (RT-PCR) positive were included in the COVID-19 positive cases. The cases were further categorized into three groups: mild to moderate, severe and very severe based on clinical severity.
Operational definitions [7]
Mild to moderate: a patient who presented with nonspecific symptoms like fever, sore throat, cough etc. not qualifying as severe/very severe disease according to criteria.
Severe COVID-19: Any patient meeting one of the three criteria: (1) Respiratory distress with respiratory rate more than 30 times/min; (2) Oxygen saturation ≤93% in resting state; (3) PaO2/FiO2≤300 mmHg (1 mmHg=0.133 kPa).
Very severe COVID-19: Any patient meeting one of the following criteria: (1) Respiratory failure in need of mechanical ventilation; (2) Shock; (3) Other organ dysfunction [7].
The patients were categorized into survivors and non-survivors as well.
Exclusion criteria
The patients with history of thromboembolic/cardiovascular disorders, inflammatory bowel disease, hematological disorders, surgeries or trauma in last six months or patients who are bedridden, pregnant females or drugs affecting hemostasis/platelets, liver or kidney disease, cancers, were excluded from the study.
Data and statistical analysis
The data collected for 100 COVID-19 cases was entered in Microsoft excel sheet and analyzed using SPSS version 21.0 (SPSS Inc., Chicago, IL, USA). Mean, median and box plots were used to present continuous data. Test of normality were performed on the continuous variables in the three groups viz., moderate, severe and very severe. Mann Whitney U test was used to compare the non-normally distributed variables in two groups (survivor and non-survivor) while Kruskal Walis test was used to compare three groups (Moderate, Severe and Very severe). Normally distributed variables were analysed using independent t test and ANOVA test respectively. The categorical data was presented as proportions and tested using chi square as test of significance between groups. To study correlation between laboratory parameters in COVID positive cases, Pearson’s correlation coefficient was calculated. To analyze the efficiency of parameters in predicting disease severity, receiver operation characteristic curve (ROC curve) was used. For calculating area under the curve (AUC) for parameters, the severe and very severe cases as defined above were taken as primary outcome. P value <0.05 was considered statistically significant.
Results
Out of a total of 100 COVID-19 positive patients which were enrolled for the study, 61 had mild to moderate disease, 26 had severe disease and 13 had very severe disease. Most of the patients (68%) belonged to 46-65 years age group while 67% non survivors were above 60 years of age. Males outnumbered females with a ratio of 1.78:1. Among the 100 COVID cases, 15 patients expired during the course of treatment, the mortality rate being 15% among hospital admissions. ALC<1000/cumm was observed in 33% with an NLR>3.3 in 60% of COVID 19 patients. D-dimer levels >1000 ng/ml were noted in 35% patients, moreover 87% of patients with severe and very severe disease had high D-dimer levels (>1000 ng/ml). Most of the patients (83%) had LDH levels above 300 U/l. The demographic and laboratory data are depicted in Table 1.
Table 1.
A Comparative analysis of demographic and laboratory parameters among the different subcategories of COVID-19 cases based on disease severity
| Variable | Subcategories | Very Severe (n=13) | Severe (n=26) | Mild to Moderate (n=61) | Total cases (n=100) | P value |
|---|---|---|---|---|---|---|
| Age (in years) | 18-45 | 2 | 2 | 25 | 29 | 0.002 |
| 46-65 | 9 | 23 | 36 | 68 | ||
| >66 | 2 | 1 | 0 | 3 | ||
| Gender | Male | 7 | 15 | 40 | 64 | 0.44 |
| Female | 6 | 11 | 21 | 36 | ||
| Survivor Status | Survivor | 1 | 23 | 61 | 85 | <0.001 |
| Non survivor | 12 | 3 | 0 | 15 | ||
| ALC (/cumm.) | <1000 | 8 | 11 | 14 | 33 | 0.01 |
| >1000 | 5 | 15 | 47 | 67 | ||
| NLR | <3.3 | 3 | 8 | 29 | 40 | 0.14 |
| >3.3 | 10 | 18 | 32 | 60 | ||
| PLR | <180 | 5 | 14 | 45 | 64 | 0.03 |
| >180 | 8 | 12 | 16 | 36 | ||
| D dimer (ng/ml) | <1000 | 0 | 5 | 60 | 65 | <0.001 |
| >1000 | 13 | 21 | 1 | 35 | ||
| LDH (U/L) | <300 | 0 | 3 | 14 | 17 | <0.001 |
| >300 | 13 | 23 | 47 | 83 | ||
| Ferritin (ng/ml) | <500 | 6 | 22 | 60 | 88 | 0.13 |
| >500 | 7 | 4 | 1 | 12 | ||
| Hs-CRP (mg/l) | <10 | 0 | 21 | 54 | 75 | <0.001 |
| >10 | 13 | 5 | 7 | 25 | ||
| IL-6 (pg/ml) | <7 | 0 | 2 | 49 | 51 | <0.001 |
| >7 | 13 | 24 | 12 | 49 | ||
| Procalcitonin (ng/ml) | <2 | 0 | 21 | 51 | 72 | <0.001 |
| >2 | 13 | 5 | 10 | 28 |
The characteristic hematological abnormalities observed in COVID-19 were leucocytosis, neutrophilia, lymphopenia and monocytosis. Statistically significant differences were observed in ANC, NLR, PLR, MLR, platelet count and ESR among the three categories based on disease severity. The ratios NLR, PLR and MLR were found to increase uniformly between mild to moderate vs. severe vs. very severe groups. Statistically significant differences were observed between the three subcategories of patients for D-dimer, FDP, fibrinogen, INR, thrombin time and LDH, ferritin, IL-6, procalcitonin, hs-CRP. Comparison of hematological, coagulation and biochemical parameters between the subcategories based on disease severity is shown in Table 2.
Table 2.
Comparative analysis of Hematological, Coagulation and Biochemical parameters in subcategories of COVID 19 patients based on disease severity
| Very severe (n=13) | Severe (n=26) | Moderate (n=61) | P value | |
|---|---|---|---|---|
| ALC | 1.61+0.91 | 1.65±1.82 | 2.19±2.57 | 0.49 |
| ANC | 14.72±7.48 | 10.42±9.01 | 7.82±7.57 | 0.016 |
| NLR | 22.25±5.68 | 13.29±4.92 | 5.99±4.52 | 0.00 |
| PLR | 339.65±391.22 | 227.74±188.95 | 162.64±209.43 | 0.043 |
| MLR | 1.52±2.25 | 0.68±0.492 | 0.49±0.32 | 0.001 |
| RDW SD | 55.48±8.16 | 50.05±6.17 | 52.1±7.65 | 0.099 |
| RDW CV | 16.18±1.21 | 15.74±42.14 | 16.29±2.29 | 0.557 |
| PLATELET | 227.3±95.53 | 200.15±88.99 | 168.95±81.95 | 0.05 |
| MPV | 12.71±1.02 | 12.26±1.17 | 11.99±1.25 | 0.139 |
| PDW | 18.32±4 | 16.23±3.26 | 15.72±3.7 | 0.070 |
| Plateletcrit | 0.19±0.07 | 0.23±0.09 | 0.25±0.09 | 0.062 |
| ESR | 30.38±5.32 | 26.54±7.99 | 21.33±6.79 | 0.00 |
| CRP | 17.33±5.99 | 7.01±4.66 | 4.72±2.73 | 0.00 |
| LDH | 1036±382.45 | 707.5±329.37 | 408.61±162.89 | 0.00 |
| Ferritin | 624.62±314.75 | 368.42±131.07 | 235.15±174.43 | 0.00 |
| Procalcitonin | 32.31±25.76 | 13.01±6.28 | 4.64±18.11 | 0.00 |
| IL-6 | 90.92±54.07 | 26.27±18.25 | 4.22±2.73 | 0.00 |
| D-Dimer | 4188.23±1637.56 | 2894.04±2172.35 | 449.3±712.93 | 0.00 |
| FDP | 15.73±3.07 | 8.07±7.24 | 0.83±0.40 | 0.00 |
| Fibrinogen | 952.46±235.55 | 748.85±344.26 | 397.03±131.91 | 0.00 |
| Thrombin Time | 27.31±4.84 | 22.09±4.98 | 17.75±2.28 | 0.00 |
| INR | 1.26±0.21 | 1.30±0.32 | 1.08±0.22 | 0.001 |
On classifying COVID-19 patients into survivors and non-survivors, statistically significant differences were observed in hematological variables (ANC, NLR and ESR), coagulation parameters (D-dimer, FDP, fibrinogen and thrombin time) and biochemical markers (LDH, ferritin, IL-6, procalcitonin and hs-CRP). Table 3 depicts the comparative analysis of various biomarkers among the COVID-19 survivors and non survivors.
Table 3.
Comparative analysis of Hematological, Coagulation and Biochemical parameters in COVID 19 patients based on survivor status
| Non survivor (n=15) | Survivor (n=85) | P value | |
|---|---|---|---|
| ALC | 1.41±1.26 | 2.15±2.38 | 0.40 |
| ANC | 13.18±8.01 | 8.73±8.12 | 0.05 |
| NLR | 19.30±6.32 | 8.36±6.31 | <0.001 |
| PLR | 299.53±373.88 | 185.48±206.9 | 0.27 |
| MLR | 1.37±2.12 | 0.55±0.38 | 0.16 |
| RDW SD | 54.09±8.41 | 51.64±7.28 | 0.24 |
| RDW CV | 15.89±1.52 | 16.18±2.23 | 0.64 |
| PLATELET | 216.2±92.05 | 179.08±85.86 | 0.13 |
| MPV | 12.48±1.16 | 12.09±1.22 | 0.26 |
| PDW | 17.29±3.5 | 15.99±3.72 | 0.21 |
| Plateletcrit | 0.20±0.07 | 0.24±0.09 | 0.09 |
| ESR | 27.87±7.57 | 23.15±7.15 | 0.03 |
| CRP | 14.87±7.42 | 5.56±6.28 | <0.001 |
| LDH | 897.6±365.88 | 509.71±292.18 | <0.001 |
| Ferritin | 569.13±320.87 | 276.54±176.51 | <0.001 |
| Procalcitonin | 28.43±25.48 | 7.23±16.28 | 0.01 |
| IL-6 | 87.67±50.76 | 9.5±11.61 | <0.001 |
| D-Dimer | 4647.13±2231.38 | 1028.14±1359.57 | <0.001 |
| FDP | 12.71±6.72 | 3.23±5.39 | <0.001 |
| Fibrinogen | 870.6±305.22 | 506.02±272.19 | <0.001 |
| Thrombin Time | 26.48±5.19 | 18.99±3.83 | <0.001 |
| INR | 1.25±0.20 | 1.15±0.27 | 0.16 |
A strong correlation of NLR with D-dimer, ferritin, IL-6, procalcitonin, LDH, PLR and MLR was observed. Similarly, D-dimer also showed a significant correlation with all biochemical markers (ferritin, IL-6, procalcitonin, LDH) and NLR. The biochemical markers including ferritin, IL-6, procalcitonin, LDH exhibited statistically significant correlation between each other as well as with NLR and D-dimer. The correlation of various parameters with each other is depicted in Table 4.
Table 4.
Correlation between various Biomarkers in COVID-19 patients
| FERRITIN | IL-6 | Procalcitonin | LDH | NLR | PLR | MLR | D Dimer | FDP | FIB | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FERRITIN (NG/DL) | Pearson Correlation | 1 | .478 | .237 | .409 | .397 | .077 | .112 | .428 | .496 | .425 |
| Sig. (2-tailed) | .000 | .017 | .000 | .000 | .444 | .269 | .000 | .000 | .000 | ||
| IL-6 | Pearson Correlation | .478 | 1 | .445 | .408 | .540 | .422 | .529 | .575 | .571 | .499 |
| Sig. (2-tailed) | .000 | .000 | .000 | .000 | .000 | .000 | .000 | .000 | .000 | ||
| Procalcitonin | Pearson Correlation | .237 | .445 | 1 | .261 | .296 | .192 | .478 | .299 | .396 | .315 |
| Sig. (2-tailed) | .017 | .000 | .009 | .003 | .056 | .000 | .003 | .000 | .001 | ||
| LDH (U/L) | Pearson Correlation | .409 | .408 | .261 | 1 | .504 | -.043 | .071 | .449 | .705 | .426 |
| Sig. (2-tailed) | .000 | .000 | .009 | .000 | .675 | .482 | .000 | .000 | .000 | ||
| NLR | Pearson Correlation | .397 | .540 | .296 | .504 | 1 | .349 | .316 | .568 | .637 | .522 |
| Sig. (2-tailed) | .000 | .000 | .003 | .000 | .000 | .001 | .000 | .000 | .000 | ||
| PLR | Pearson Correlation | .077 | .422 | .192 | -.043 | .349 | 1 | .456 | .133 | .129 | .150 |
| Sig. (2-tailed) | .444 | .000 | .056 | .675 | .000 | .000 | .189 | .202 | .137 | ||
| MLR | Pearson Correlation | .112 | .529 | .478 | .071 | .316 | .456 | 1 | .205 | .293 | .156 |
| Sig. (2-tailed) | .269 | .000 | .000 | .482 | .001 | .000 | .041 | .003 | .122 | ||
| D Dimer | Pearson Correlation | .428 | .575 | .299 | .449 | .568 | .133 | .205 | 1 | .542 | .483 |
| Sig. (2-tailed) | .000 | .000 | .003 | .000 | .000 | .189 | .041 | .000 | .000 | ||
| FDP | Pearson Correlation | .496 | .571 | .396 | .705 | .637 | .129 | .293 | .542 | 1 | .699 |
| Sig. (2-tailed) | .000 | .000 | .000 | .000 | .000 | .202 | .003 | .000 | .000 | ||
| Fibrinogen | Pearson Correlation | .425 | .499 | .315 | .426 | .522 | .150 | .156 | .483 | .699 | 1 |
| Sig. (2-tailed) | .000 | .000 | .001 | .000 | .000 | .137 | .122 | .000 | .000 | ||
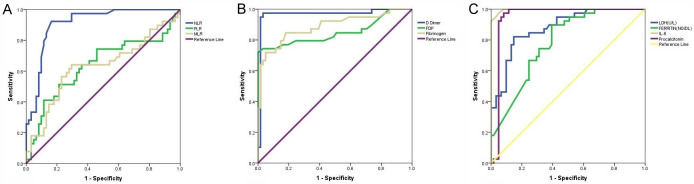
IL-6 (AUC: 0.997, P<0.001) emerged as the single best parameter as per area under curve of ROC in distinguishing mild to moderate cases from severe cases (severe and very severe), followed by D-dimer (AUC: 0.966, P<0.001), procalcitonin (AUC: 0.949, P<0.001), NLR (AUC: 0.910, P<0.001), fibrinogen (AUC: 0.886, P<0.001), LDH (AUC: 0.879, P<0.001) and so on. ROC analysis showed IL-6 at a cut-off value 10.5 pg/ml, predicting severe disease with a sensitivity of 94.9% and specificity of 96.7%, D-dimer at a cut-off value 890 ng/ml, predicting severe disease with a sensitivity of 94.9% and specificity of 98.4%, procalcitonin at a cut-off value 7.19 pg/ml, predicting severe disease with a sensitivity of 92.3% and specificity of 95.1%, LDH at a cut-off value 541 U/l, predicting severe disease with a sensitivity of 82.1% and specificity of 85.4%, FDP at a cut-off value 1.23/ml, predicting severe disease with a sensitivity of 74.4% and specificity of 96.7% and NLR at a cut-off value 10.17, predicting severe disease with a sensitivity of 84.6% and specificity of 86.9% (Table 5; Figure 1).
Table 5.
Area under the curve for various Biomarkers in severe and very severe versus moderate COVID 19 patients
| Test Result Variable | Cut off | Area | Std. Error | Sig. | 95% Confidence Interval | Sensitivity (%) | Specificity (%) | |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Lower Bound | Upper Bound | |||||||
| LDH (U/l) | 541 | .879 | .034 | .000 | .812 | .945 | 82.1% | 85.2% |
| FERRITIN (ng/dl) | 413 | .782 | .045 | .000 | .695 | .870 | 66.7% | 75.4% |
| IL-6 | 10.5 | .997 | .003 | .000 | .991 | 1.000 | 94.9 | 96.7 |
| Procalcitonin | 7.19 | .949 | .027 | .000 | .895 | 1.000 | 92.3 | 95.1 |
| CRP (MG/L) | 3.69 | .779 | .045 | .000 | .690 | .868 | 82.1 | 62.3 |
| D Dimer | 890 | .966 | .023 | .000 | .921 | 1.000 | 94.9 | 98.4 |
| FDP | 1.23 | .848 | .047 | .000 | .756 | .941 | 74.4 | 96.7 |
| Fibrinogen | 661.50 | .886 | .037 | .000 | .813 | .959 | 71.8 | 95.1 |
| NLR | 10.17 | .910 | .029 | .000 | .853 | .968 | 84.6 | 86.9 |
| PLR | 111.52 | .637 | .061 | .021 | .518 | .756 | 66.7 | 55.9 |
| MLR | 0.55 | .628 | .061 | .032 | .509 | .747 | 64.1 | 70.5 |
Figure 1.
ROC Analysis using laboratory parameters in the diagnosis of severe cases of COVID-19. The positive sample is the test results of severe and very severe cases while the negative sample is the results of mild to moderate cases. (A) ROC plot of biochemical markers (LDH, ferritin, IL-6, procalcitonin, hs-CRP), (B) ROC plot of NLR, PLR and MLR and (C) ROC plot of coagulation parameters (D-dimer, fibrin degradation products (FDP), fibrinogen).
Discussion
Coronaviruses are categorised into four genera: alpha, beta, delta, and gamma [8]. Among these, Severe Acute Respiratory Syndrome (SARS), Middle East Respiratory Syndrome (MERS), and the Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-Cov-2) are the notable beta genus viruses causing human diseases [9]. SARS was endemic in China in 2002-2003 with rapid spread to other countries and a mortality of around 10% [10], followed by MERS in 2012 in the Middle East with about 35% mortality [11]. Severe acute respiratory syndrome coronavirus 2 (SARS CoV-2) was reported for the first time in Wuhan, China in December 2019 and has been incriminated in the current, devastating global pandemic COVID-19 disease infecting more than 150 million people worldwide [1].
The symptoms of COVID-19 disease can range from mild (cough, sore throat, fever, malaise etc.) to severe (respiratory impairment, chest pain etc.) to critical. The most common demographic factors associated with adverse prognosis and mortality in COVID-19 have been reported as male gender, increasing age, and co-morbidities like obesity, diabetes, cardiovascular disease, hypertension, and chronic respiratory diseases [12-14].
The COVID-19 disease has a complex pathogenesis. The most serious complication in COVID-19 patients is the cytokine storm (CS) due to an exaggerated immune response to the virus triggered by inflammatory cell infiltration in the lungs, activation of T-helper 1 reactions, and excessive release of proinflammatory cytokines into the blood [15]. The CS leads to multi organ dysfunction and intravascular coagulopathy, documented by the presence of venous thromboembolism and microthrombi in arterioles and venules in COVID-19 patient autopsies [16]. Therefore, some authors suggest that the key to reducing the mortality among COVID-19 patients is the prompt and appropriate curtailment of this CS in its early stage, through immunomodulators, corticosteroids, and cytokine antagonists [17].
On account of the surge in the number of COVID-19 cases and its varied clinical presentation, there is a dire necessity to identify early signs of deterioration in order to initiate timely intensification of care in the patients whose illness shows a rapid worsening from mild to severe. The current study aimed at exploring the role of hematological, coagulation and biochemical markers of inflammation and tissue damage in predicting prognosis and disease severity.
Hematological parameters in COVID-19 patients
Inflammatory cytokines may significantly alter the actions of hematopoietic cells, predominantly neutrophils, lymphocytes and monocytes. In the present study, the difference between WBC and absolute neutrophil counts was statistically significant among the subcategories based on disease severity as well as survivor status akin to Chen et al. [2], Wang et al. [18], Wu et al. [19], Huang et al. [20] and Ding et al. [21]. Superimposed bacterial pneumonia reported by Li et al. [22] in some non-survivors with COVID-19 could be one of the reasons. Lymphopenia was a consistent feature reported by most of the studies [17-22], although it was observed in our study but it did not attain statistical significance. The difference in platelet counts between very severe, severe and mild to moderate COVID-19 disease was found to be statistically significant. On the contrary, Lippi et al. [23] and Liu et al. [24] observed that thrombocytopenia was associated with probability of severe COVID-19 disease and mortality.
The derived ratios like NLR, PLR and MLR are a better reflection of the inflammatory process compared to the absolute cell counts as documented by several authors [18,21,25-28] as they are relatively stable. Among different subgroups based on disease severity, significant differences were noted in NLR, PLR and MLR which is in concordance with several authors [18,21,25-29]. Wang et al. [18] studied a variety of combination parameters and concluded that NLR & RDW-SD was the best hematology index which can aid in predicting the severity of COVID-19 patients, followed by NLR & RDW-CV. In our prior study on COVID-19, the combined parameter which emerged as the marker with best diagnostic efficiency (AUC=0.871) in predicting severe disease was NLR and RDW-CV [30].
Coagulation parameters in COVID-19
Microorganisms and their components trigger tissue factor expression, in turn stimulating the immune system to release proinflammatory cytokines which activate the coagulation cascade thereby leading to disseminated intravascular coagulation (DIC) [31]. Among 184 COVID-19 patients, Klok et al. reported the incidence of thrombosis to be 31% [32].
Fibrinolysis leads to release of D-dimer and other Fibrin degradation products (FDPs) into the circulation, the blood levels of these in turn aid in the diagnosis of thrombosis [33]. Many authors have documented the role of elevated D-dimer and FDP levels in COVID-19 with regard to disease severity and survivor status [5,34,35]. Zhang et al. [34] and Zhou et al. [5] showed that D-dimer levels >2.0 µg/mL and >1 µg/mL respectively on admission is associated with mortality among COVID-19 patients. In the index study, according to ROC analysis, D-dimer at a cut-off value 890 ng/ml could predict severe disease with a high sensitivity and specificity. Therefore, D-Dimer could Serve as an early indicator of severity to improve the management of COVID-19 patients.
Fibrinogen is a serum glycoprotein produced by the liver. On activation of coagulation system in tissue and vascular damage, thrombin leads to conversion of fibrinogen to fibrin resulting in clot formation. Tang et al. found increased PT and D-dimer and decreased fibrinogen to be associated with DIC in severe COVID-19 patients [36]. On the contrary, Thachil et al. [37] demonstrated prothrombic diathesis in critically ill patients with extremely high fibrinogen levels similar to study by Zou et al. [38] and the current study. However in the advanced stages, thrombolysis results in decreased fibrinogen levels and increased fibrin-degradation products.
Thrombin time (TT) showed an increasing trend with disease intensity and mortality which was statistically significant. Our findings are similar to Long et al. [39], though, Hans et al. [40] could not establish any such trend.
Biochemical markers in COVID-19
In the current study, significant differences were noted in LDH, ferritin, IL-6, procalcitonin and hs-CRP among the three subgroups based on disease severity and between survivors and non-survivors. Chen et al. [2] also demonstrated that infection-related biomarkers like IL-6, procalcitonin, ESR, serum ferritin, and CRP were elevated by 6%, 52%, 85%, 63%, and 86% respectively among COVID-19 patients. Zeng et al. [41] in their meta-analysis of 16 studies showed that patients in the non-severe group had lower CRP, ESR, Procalcitonin, and serum ferritin levels as compared to those in the severe groups. In a meta-analysis by Zhang et al. [42], the most important observations were increased CRP (73.6%), decreased albumin (62.9%), increased ESR (61.2%), decreased Eosinophils (58.4%), increased IL-6 (53.1%), lymphopenia (47.9%), and increased LDH (46.2%).
Ferritin is an intracellular protein which is the major storage form of iron. A small amount of ferritin is present in the blood as an iron carrier. In infections or cancers, ferritin level increases significantly. In addition, ferritin is also an acute phase reactant [33]. Although the precise cause of increase in ferritin in COVID-19 is still unknown, the possible mechanism could be enhanced ferritin synthesis due to cytokines such as IL-6 or leakage of intracellular ferritin due to cellular damage [43]. Cytokine storm and raised ferritin on account of secondary hemophagocytic lymphohistiocytosis have been reported in severe COVID-19 disease. It has been postulated that COVID-19 could be part of the broader spectrum of hyperinflammatory syndromes such as the secondary hemophagocytic lymphohistiocytosis where increased serum ferritin is a cardinal feature [44].
The cytokine storm in COVID-19 patients is responsible for the worst form of disease with complications such as acute respiratory distress syndrome (ARDS). Interleukin-6 is the most commonly implicated cytokine in COVID-19 disease [33]. In our study, IL-6 emerged as the single best parameter as per area under curve of ROC in predicting disease severity. Coomes et al. [45] observed that interleukin-6 increases by about 2.9 fold in severe disease, quite similar to Liu et al. [46] and Ruan et al. [47]. Huang et al. [48] also found IL-6 to be increased in patients with SARS CoV2 infection and its association with viral RNA level in blood and disease progression.
LDH is an intracellular enzyme found throughout the body, therefore LDH levels cannot be considered a specific marker of a particular disease. LDH is released into the circulation when LDH-containing cells are damaged or diseased. Cancers, muscle injury, pancreatitis, liver and lung diseases such as COVID-19 stimulate the production of lactate dehydrogenase [33]. Many authors have shown that LDH levels correlate with disease severity in COVID-19 patients much alike our findings [35,49].
C-reactive protein (CRP) is a plasma protein produced by the liver in response to infammatory mediators such as IL-6. It serves as a nonspecifc, acute phase reactant [33]. Wang et al. [50] concluded that raised CRP is strongly associated with COVID-19 severity and critical state. Chen et al. [2] showed that CRP levels in COVID-19 patients who expired were much higher than survivors. Liu et al. [51] observed that CRP was elevated in 65% of COVID-19 patients on admission and elevated in 93.9% of severe COVID-19 patients. High sensitivity C reactive protein (hsCRP) levels have been acknowledged to be an independent risk factor for predicting clinical outcome of various diseases [52]. On extensive search of literature, there is single study on hsCRP in COVID-19 by Liu et al. [53] who concluded that variation in hsCRP is a major independent risk factor for ICU admission in young and middle-aged COVID-19 inpatients, but not in the elderly patients.
Procalcitonin (PCT) is a precursor of the hormone calcitonin released by the thyroid parafollicular cells. It is well recognized as a biomarker of bacterial infection. Recently, in the wake of COVID-19 pandemic, several authors have documented its positive association with the severity of COVID-19 [20,35,54]. A meta-analysis also observed that increased PCT is related to a 5-fold higher risk of severe SARS-CoV-2 infection [55]. Procalcitonin turned out to be a good biomarker for predicting severe disease in the present study at a cut-off value 7.19 pg/ml with good sensitivity and specificity.
Cappanera et al. [17] were the first to develop a new score for prompt identification of COVID-19 patients in a hyperinflammatory stage and to rapidly institute treatment in turn reducing the risk of intubation. Lymphopenia (<1000 (×103/mm3) was selected as the first major criterion for this CS score (CSs) alongwith at least two of the following three markers: D-dimer >1000 ng/mL, ferritin >500 ng/mL, or lactate dehydrogenase (LDH)>300 IU/L. The CSs was positive, and a hyperinflammatory stage of COVID-19 was suspected if the above criteria were met. In patients with lymphopenia and derangement of only one of D-Dimer, ferritin, or LDH, then if the C-reactive protein (CRP) was greater than 10 mg/dL, CSs was considered positive. According to authors, CSs could accurately identify COVID-19 patients in the early stages of a CS, so that timely, safe, and effective administration of immunomodulators, corticosteroids, and cytokine antagonists could be undertaken preventing progression of disease and hence reduce mortality.
The limitations of the present study were that the impact of certain probable confounding factors cannot be ruled out. Moreover, the associations observed between various biomarkers and disease severity or mortality cannot be considered as causal on account of the cross-sectional nature of the study. To overcome these limitations, large scale multicentre cohort studies are necessary.
Conclusion
To conclude, our study demonstrates a plethora of biomarkers which could be utilized to accurately identify the hyperinflammation and tissue damage reminiscent of cytokine storm in COVID-19 patients so that timely, safe, and effective therapies can be administered to prevent progression and potentially reduce mortality. Statistically significant differences were observed in hematological variables (ANC, NLR and ESR), coagulation parameters (D-dimer, FDP, fibrinogen and thrombin time) and biochemical markers (LDH, ferritin, IL-6, procalcitonin and hs-CRP) with regard to subcategories based of disease severity as well as survivor status. There was strong correlation between NLR, D-dimer, IL-6, procalcitonin and ferritin. IL-6 emerged as the single best marker of disease severity (AUC: 0.997, P=0), however procalcitonin, LDH, D-dimer, FDP and NLR could also predict severe disease with a good sensitivity and specificity. It is of utmost importance if the most standard, non-invasive and cost-effective tests like CBC, coagulation and biochemical tests could serve as a guide in determining disease severity for judicious allocation of medical resources in developing countries like India with limited funds and supplies for a massive population in the midst of such a pandemic.
Acknowledgements
We are extremely thankful to Dr. Asim Das, Dean and Dr. AK Pandey, Registrar Academics, ESIC Medical College, Faridabad for their exemplary leadership in handling the COVID-19 pandemic.
Disclosure of conflict of interest
None.
References
- 1. https://www.worldometers.info/coronavirus as of 5th May 2021.
- 2.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang I, Pranata R, Lim MA, Oehadian A, Alisjahbana B. C-reactive protein, procalcitonin, D-Dimer, and ferritin in severe coronavirus disease-2019: a meta-analysis. Ther Adv Respir Dis. 2020;14:1753466620937175. doi: 10.1177/1753466620937175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Velavan TP, Meyer CG. Mild versus severe COVID-19: laboratory markers. Int J Infect Dis. 2020;95:304–7. doi: 10.1016/j.ijid.2020.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. Clinical course and risk factors for mortality of adult in patients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao M. Cytokine storm and immunomodulatory therapy in COVID-19: role of chloroquine and anti-IL-6 monoclonal antibodies. Int J Antimicrob Agents. 2020;55:105982. doi: 10.1016/j.ijantimicag.2020.105982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin S, Huang M, Li D, Tang N. Difference of coagulation features between severe pneumonia induced by SARS-CoV2 and non-SARS-CoV2. J Thromb Thrombolysis. 2020;51:1107–1110. doi: 10.1007/s11239-020-02105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pormohammad A, Ghorbani S, Khatami A, Farzi R, Baradaran B, Turner DL, Turner RJ, Bahr NC, Idrovo JP. Comparison of confrmed COVID-19 with SARS and MERS cases-clinical characteristics, laboratory fndings, radiographic signs and outcomes: a systematic review and meta-analysis. Rev Med Virol. 2020;30:e2112. doi: 10.1002/rmv.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hui DS, Chan MC, Wu AK, Ng PC. Severe acute respiratory syndrome (SARS): epidemiology and clinical features. Postgrad Med J. 2004;80:373–381. doi: 10.1136/pgmj.2004.020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hui DS, Azhar EI, Kim YJ, Memish ZA, Oh MD, Zumla A. Middle east respiratory syndrome coronavirus: risk factors and determinants of primary, household, and nosocomial transmission. Lancet Infect Dis. 2018;18:e217–e227. doi: 10.1016/S1473-3099(18)30127-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Xia L. Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. Am J Roentgenol. 2020;214:1280–1286. doi: 10.2214/AJR.20.22954. [DOI] [PubMed] [Google Scholar]
- 13.Bansal M. Cardiovascular disease and COVID-19. Diabetes Metab Syndr. 2020;14:247–250. doi: 10.1016/j.dsx.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Xu S, Yu M, Wang K, Tao Y, Zhou Y, Shi J, Zhou M, Wu B, Yang Z, Zhang C, Yue J, Zhang Z, Renz H, Liu X, Xie J, Xie M, Zhao J. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J Allergy Clin Immunol. 2020;146:110–118. doi: 10.1016/j.jaci.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasparyan AY, Misra DP, Yessirkepov M, Zimba O. Perspectives of immune therapy in coronavirus disease 2019. J Korean Med Sci. 2020;35:e176. doi: 10.3346/jkms.2020.35.e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salerno M, Sessa F, Piscopo A, Montana A, Torrisi M, Patanè FG, Murabito P, Volti GL, Pomara C. No autopsies on COVID-19 deaths: a missed opportunity and the lockdown of science. J Clin Med. 2020;9:1472. doi: 10.3390/jcm9051472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cappanera S, Palumbo M, Kwan SH, Priante G, Martella LA, Saraca LM, Sicari F, Vernelli C, Di Giuli C, Andreani P, Mariottini A, Francucci M, Sensi E, Costantini M, Bruzzone P, D’Andrea V, Gioia S, Cirocchi R, Tiri B. When does the cytokine storm begin in COVID-19 patients? A quick score to recognize it. J Clin Med. 2021;10:297. doi: 10.3390/jcm10020297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C, Deng R, Gou L, Fu Z, Zhang X, Shao F, Wang G, Fu W, Xiao J, Ding X, Li T, Xiao X, Li C. Preliminary study to identify severe from moderate cases of COVID-19 using combined hematology parameters. Ann Transl Med. 2020;8:593. doi: 10.21037/atm-20-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, Huang H, Zhang H, Zhou X, Du C, Zhang Y, Song J, Wang S, Chao Y, Yang Z, Xu J, Zhou X, Chen D, Xiong W, Xu L, Zhou F, Jiang J, Bai C, Zheng J, Song Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding X, Yu Y, Lu B, Huo J, Chen M, Kang Y, Lou J, Liu Z. Dynamic profile and clinical implications of hematological parameters in hospitalized patients with coronavirus disease 2019. Clin Chem Lab Med. 2020;58:1365–1371. doi: 10.1515/cclm-2020-0411. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Wang L, Yan S, Yang F, Xiang L, Zhu J, Shen B, Gong Z. Clinical characteristics of 25 death cases with COVID-19: a retrospective review of medical records in a single medical center, Wuhan, China. Int J Infect Dis. 2020;94:128–132. doi: 10.1016/j.ijid.2020.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lippi G, Plebani M, Henry BM. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: a meta-analysis. Clin Chim Acta. 2020;506:145–8. doi: 10.1016/j.cca.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Sun W, Guo Y, Chen L, Zhang L, Zhao S, Long D, Yu L. Association between platelet parameters and mortality in coronavirus disease 2019: retrospective cohort study. Platelets. 2020;31:490–496. doi: 10.1080/09537104.2020.1754383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang AP, Liu J, Tao W, Li HM. The diagnostic and predictive role of NLR, d-NLR and PLR in COVID-19 patients. Int Immunopharmacol. 2020;84:106504. doi: 10.1016/j.intimp.2020.106504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Du X, Chen J, Jin Y, Peng L, Wang HHX, Luo M, Chen L, Zhao Y. Neutrophil-to lymphocyte ratio as an independent risk factor for mortality in hospitalized patients with COVID-19. J Infect. 2020;81:e6–e12. doi: 10.1016/j.jinf.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hengeveld PJ, Khader AO, de Bruin LHA, Geelen IGP, van Baalen EA, Jansen E, Bouwer NI, Balak O, Riedl JA, Langerak AW, Westerweel PE, Levin MD. Blood cell counts and lymphocyte subsets of patients admitted during the COVID-19 pandemic: a prospective cohort study. Br J Hematol. 2020;190:e201–e204. doi: 10.1111/bjh.16983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qu R, Ling Y, Zhang YH, Wei LY, Chen X, Li XM, Liu XY, Liu HM, Guo Z, Ren H, Wang Q. Platelet-to-lymphocyte ratio is associated with prognosis in patients with coronavirus disease 19. J Med Virol. 2020;92:1533–1541. doi: 10.1002/jmv.25767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lissoni P, Rovelli F, Monzon A, Privitera C, Messina G, Porro G, Fede GD, Lissoni A, Colciago M, Pelizzoni F. Evidence of abnormally low lymphocyte-to-monocyte ratio In COVID-19-induced severe acute respiratory syndrome. J ImmunoAllerg. 2020;1:1–6. [Google Scholar]
- 30.Pujani M, Raychaudhuri S, Verma N, Kaur H, Agarwal S, Singh M, Jain M, Chandoke RK, Singh K, Sidam D, Chauhan V, Singh A, Katarya K. Association of Hematologic biomarkers and their combinations with disease severity and mortality in COVID-19-an Indian perspective. Am J Blood Res. 2021;11:180–190. [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson SP, Darbousset R, Schoenwaelder SM. Thromboinfammation: challenges of therapeutically targeting coagulation and other host defense mechanisms. Blood. 2019;133:906–918. doi: 10.1182/blood-2018-11-882993. [DOI] [PubMed] [Google Scholar]
- 32.Klok FA, Kruip MJHA, van der Meer NJM, Arbous MS, Gommers DAMPJ, Kant KM, Kaptein FHJ, van Paassen J, Stals MAM, Huisman MV, Endeman H. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb Res. 2020;191:145–147. doi: 10.1016/j.thromres.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong LZ, Shou ZX, Zheng DM, Jin X. The most important biomarker associated with coagulation and infammation among COVID-19 patients. Mol Cell Biochem. 2021;476:2877–2885. doi: 10.1007/s11010-021-04122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Yan X, Fan Q, Liu H, Liu X, Liu Z, Zhang Z. D-dimer levels on admission to predict in-hospital mortality in patients with COVID-19. J Thromb Haemost. 2020;18:1324–1329. doi: 10.1111/jth.14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS China Medical Treatment Expert Group for Covid-19. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang N, Bai H, Chen X, Gong J, Li D, Sun Z. Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J Thromb Haemost. 2020;18:1094–1099. doi: 10.1111/jth.14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thachil J, Agarwal S. Understanding the COVID-19 coagulopathy spectrum. Anaesthesia. 2020;75:1432–1436. doi: 10.1111/anae.15141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zou Y, Guo H, Zhang Y, Zhang Z, Liu Y, Wang J, Lu H, Qian Z. Analysis of coagulation parameters in patients with COVID-19 in Shanghai China. Biosci Trends. 2020;14:285–289. doi: 10.5582/bst.2020.03086. [DOI] [PubMed] [Google Scholar]
- 39.Long H, Nie L, Xiang X, Li H, Zhang X, Fu X, Ren H, Liu W, Wang Q, Wu Q. D-Dimer and prothrombin time are the significant indicators of severe COVID-19 and poor prognosis. Biomed Res Int. 2020;2020:6159720. doi: 10.1155/2020/6159720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han H, Yang L, Liu R, Liu F, Wu KL, Li J, Liu XH, Zhu CL. Prominent changes in blood coagulation of patients with SARS-CoV-2 infection. Clin Chem Lab Med. 2020;58:1116–1120. doi: 10.1515/cclm-2020-0188. [DOI] [PubMed] [Google Scholar]
- 41.Zeng F, Huang Y, Guo Y, Yin M, Chen X, Xiao L, et al. Association of inflammatory markers with the severity of COVID-19: a meta-analysis. Int J Infect Dis. 2020;96:467–74. doi: 10.1016/j.ijid.2020.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang ZL, Hou YL, Li DT, Li FZ. Laboratory findings of COVID-19: a systematic review and meta-analysis. Scand J Clin Lab Invest. 2020;80:441–447. doi: 10.1080/00365513.2020.1768587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin Z, Long F, Yang Y, Chen X, Xu L, Yang M. Serum ferritin as an independent risk factor for severity in COVID-19 patients. J Infect. 2020;81:647–679. doi: 10.1016/j.jinf.2020.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ HLH Across Speciality Collaboration, UK. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coomes EA, Haghbayan H. Interleukin-6 in COVID-19: a systematic review and meta-analysis. Rev Med Virol. 2020;30:1–9. doi: 10.1002/rmv.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu B, Li M, Zhou Z, Guan X, Xiang Y. Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID19)-induced cytokine release syndrome (CRS)? J Autoimmun. 2020;111:102452. doi: 10.1016/j.jaut.2020.102452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan. Intensive Care Med. 2020;46:846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang L, Zhao X, Qi Y, Li H, Ye G, Liu Y, Zhang Y, Gou J. Sepsis-associated severe interleukin-6 storm in critical coronavirus disease 2019. Cell Mol Immunol. 2020;17:1092–1094. doi: 10.1038/s41423-020-00522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han Y, Zhang H, Mu S, Wei W, Jin C, Tong C, Song Z, Zha Y, Xue Y, Gu G. Lactate dehydrogenase, an independent risk factor of severe COVID-19 patients: a retrospective and observational study. Aging (Albany NY) 2020;12:11245–11258. doi: 10.18632/aging.103372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L. C-reactive protein levels in the early stage of COVID-19. Med Mal Infect. 2020;50:332–334. doi: 10.1016/j.medmal.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu F, Li L, Xu M, Wu J, Luo D, Zhu Y, Li B, Song X, Zhou X. Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J Clin Virol. 2020;127:104370. doi: 10.1016/j.jcv.2020.104370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y, Zhong X, Cheng G, Zhao C, Zhang L, Hong Y, Wan Q, He R, Wang Z. Hs-CRP and all-cause, cardiovascular, and cancer mortality risk: a meta-analysis. Atherosclerosis. 2017;259:75–82. doi: 10.1016/j.atherosclerosis.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 53.Liu Z, Wu D, Han X, Jiang W, Qiu L, Tang R, Yu X. Different characteristics of critical COVID-19 and thinking of treatment strategies in non-elderly and elderly severe adult patients. Int Immunopharmacol. 2021;92:107343. doi: 10.1016/j.intimp.2020.107343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang JJ, Dong X, Cao YY, Yuan YD, Yang YB, Yan YQ, Akdis CA, Gao YD. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020;75:1730–1741. doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- 55.Lippi G, Plebani M. Procalcitonin in patients with severe coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chim Acta. 2020;505:190–1. doi: 10.1016/j.cca.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]