Abstract
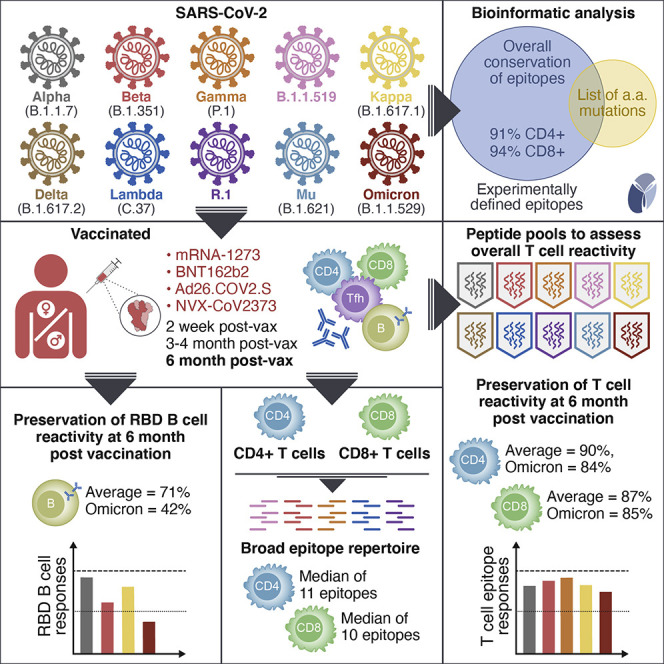
We address whether T cell responses induced by different vaccine platforms (mRNA-1273, BNT162b2, Ad26.COV2.S, and NVX-CoV2373) cross-recognize early SARS-CoV-2 variants. T cell responses to early variants were preserved across vaccine platforms. By contrast, significant overall decreases were observed for memory B cells and neutralizing antibodies. In subjects ∼6 months post-vaccination, 90% (CD4+) and 87% (CD8+) of memory T cell responses were preserved against variants on average by AIM assay, and 84% (CD4+) and 85% (CD8+) preserved against Omicron. Omicron RBD memory B cell recognition was substantially reduced to 42% compared with other variants. T cell epitope repertoire analysis revealed a median of 11 and 10 spike epitopes recognized by CD4+ and CD8+ T cells, with average preservation > 80% for Omicron. Functional preservation of the majority of T cell responses may play an important role as a second-level defense against diverse variants.
Keywords: T cells, B cells, SARS-COV-2, VOC, VOI, Delta, Omicron, epitopes, COVID-19 vaccines
Graphical abstract
Human memory T cells induced by SARS-CoV-2 vaccines maintain the ability to recognize viral variants, including the Omicron variant.
Introduction
The emergence of numerous SARS-CoV-2 variants of interest (VOI) and of concern (VOC) is one of the most important developments in the COVID-19 pandemic (Callaway, 2021). Our understanding of the virological and immunological features associated with the main VOCs is key to inform health policies, including boosting and vaccination schedules, and also inform the development of potential variant-specific or pan-coronavirus vaccines. Important aspects include whether the different variants are more infectious, more easily transmissible, linked to more severe disease, or escape immune responses induced by either vaccination or natural infection.
The Alpha (B.1.1.7), Beta (B.1.351), and Gamma (P.1) VOCs were reported in the late 2020 to May 2021 period (Harvey et al., 2021; Walensky et al., 2021). Several additional variants were described more recently (May to Oct 2021) (Chakraborty et al., 2021; Otto et al., 2021), including Mu (B.1.621) (Uriu et al., 2021) and Delta (B.1.617.2) (Mlcochova et al., 2021), with the latter quickly becoming the most dominant SARS-CoV-2 lineage worldwide. Omicron (B.1.1.529) is the latest VOC, reported in November 2021, and stands out with multiple attributes: a larger number of spike mutations compared with other VOCs (Karim and Karim, 2021), transmissibility even in the presence of Delta, and an ability to spread in populations with high levels of immunity. Omicron is expected to become dominant globally early in 2022.
A number of knowledge gaps remain in terms of our understanding of VOI/VOCs in relation to T and B cell immune reactivity. While the impact of variant-associated mutations has been established for most variants in terms of antibody reactivity (Garcia-Beltran et al., 2021; Stamatatos et al., 2021), including studies on Omicron (Liu et al., 2021; Planas et al., 2021; Schmidt et al., 2021), less is available for memory T cells and B cells. Memory T cell and B cell recognition of variants is an important issue. Several lines of evidence point to potential roles for T cells in reducing COVID-19 disease severity and contributing to disease protection (Gagne et al., 2021; Sette and Crotty, 2021; Tan et al., 2021). The continued maturation of B cell responses over time (Cho et al., 2021; Dan et al., 2021; Goel et al., 2021) may play an important role in adapting immunity to SARS-CoV-2 VOCs. Regarding memory T cells, we and others previously demonstrated that for the early variants—Alpha, Beta, Gamma, and Epsilon—the impact of mutations is limited and the majority of CD4+ and CD8+ T cell responses are preserved in both vaccinated and natural infection conditions (Collier et al., 2021; Geers et al., 2021; Keeton et al., 2021; Melo-González et al., 2021; Riou et al., 2021; Tarke et al., 2021b).
Studies on the impact of newer variants on T cells, including Mu and Omicron in particular, are limited or missing (Madelon et al., 2021). If the majority of T cell responses are maintained, memory T cells may play an important role as a second line of defense in light of the substantial escape of Omicron from antibodies. In this study, we focus on a large panel of variants to understand the impact of more recent variants on memory T cells and B cells compared with early variants, particularly in the context of COVID-19 vaccination and evaluation of the adaptive responses induced by different vaccine platforms.
Results
Cohort of COVID-19 vaccinees to assess T cell responses to a panel of SARS-CoV-2 variants
To assess the cross-recognition capability of T cell responses induced by different vaccine platforms, we enrolled a cohort of 96 adults vaccinated with different vaccines currently in use in the United States under FDA emergency use authorization (EUA) or approval: mRNA-based mRNA-1273, mRNA-based BNT162b2, and the adenoviral vector-based Ad26.COV2.S. Subjects immunized with a protein recombinant vaccine NVX-CoV2373, currently approved in the EU and in clinical assessment for the US, were also enrolled. To determine the longevity of T cell cross-recognition of SARS-CoV-2 variants, we studied samples from four different time points: 2 weeks after the first dose of vaccine, 2 weeks after the second dose of vaccine, and 3.5 months and 5–6 months after the last vaccine dose received. Based on sample availability, the study design was cross-sectional. A control donor cohort was also enrolled, comprising early convalescent donors who had mild disease (collected approximately 1 month post symptom onset, range 21–43 days).
Characteristics of the donor cohorts are summarized in Table S1. Sub-cohorts were approximately matched for gender and age across time points. For each time point, the days post-vaccination (dPV) and the SARS-CoV-2 S protein receptor binding domain (RBD) immunoglobulin G (IgG) ELISA titers are detailed as a function of the vaccine platform analyzed and the time point of sample collection. In addition, nucleocapsid (N) IgG was also run to assess previous infection, with the highest frequency of positive response in Ad26.COV2.S recipients (14% observed at time point 3, Table S1). HLA typing for the vaccinated cohort is presented in Table S2.
We previously reported T cell reactivity to the Alpha, Beta, and Gamma VOCs (Tarke et al., 2021b). Since then, by July 2021, an additional 5 SARS-CoV-2 VOI/VOCs had emerged, namely, B.1.1.519, Kappa (B.1.617.1), Delta (B.1.617.2), Lambda (C.37), and R.1. To estimate the impact of the different variants on T cell responses after vaccination, we mapped the specific spike protein mutations (amino acid replacements and deletions) as compared with the SARS-CoV-2 Wuhan ancestral sequence (Table S3). For the Wuhan ancestral sequence and each of the variants analyzed, we generated megapools (MPs) of 15-mer peptides, overlapping by 10 amino acids, spanning the entire spike protein.
Spike-specific CD4+ and CD8+ T cell reactivity to SARS-CoV-2 variants in vaccinated individuals
We evaluated T cells from the vaccine cohorts for their capacity to cross-recognize MPs spanning the entire spike sequences of different variants, compared with a control MP spanning the ancestral spike. First, T cell responses were determined from blood samples of fully immunized subjects 2 weeks after the second immunization with mRNA-based vaccines mRNA-1273 and BNT162b2, and 6 weeks post-immunization with the adenoviral vector-based Ad26.COV2.S. To measure the T cell responses, we combined activation-induced marker (AIM) assays (Tarke et al., 2021b) with cytokine intracellular staining (ICS) (Mateus et al., 2021). A comparison of the AIM and ICS protocols performed separately with the AIM+ICS combined protocol showed no significant differences in the markers analyzed (Figures S1 A–S1C).
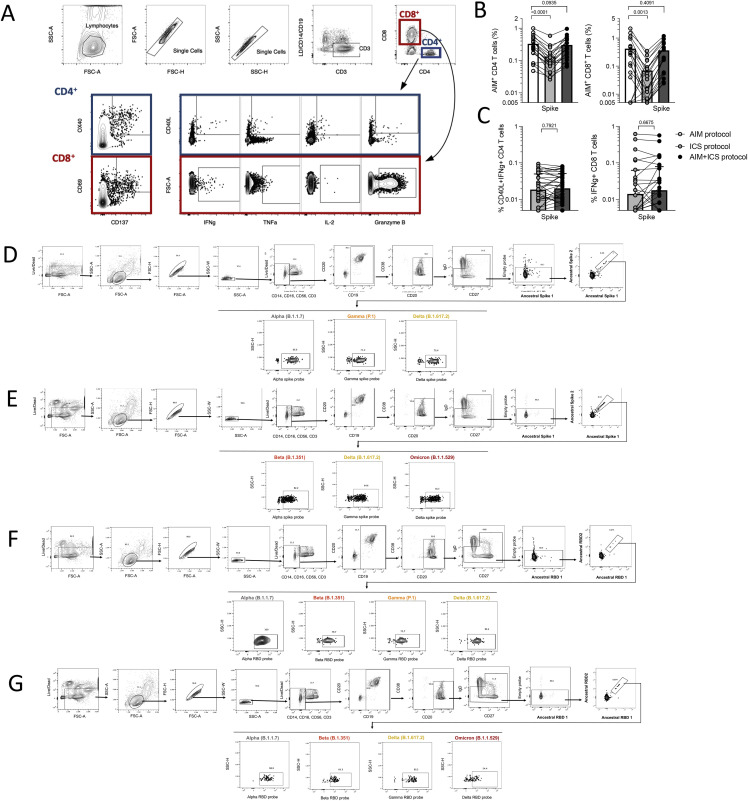
Figure S1.
Assessment of SARS-CoV-2-specific T and B cells by flow cytometry-based assays, related to Figures 1, 2, 3, 5, and 6
(A) Gating strategy for T cell AIM, ICS, and AIM+ICS assays included in this study. These gates and antibodies are the same for all time points. Spike-specific responses are measured for both CD4+ and CD8+ T cells within the same donors using the indicated AIM markers or cytokines.
(B and C) Validation of a combined AIM/ICS assay. The addition of a cocktail of brefeldin and Monesin in the ICS assay significantly decreases the detection of AIM markers, while the inclusion of the CD137 antibody in culture concomitantly, repristinates the response (B) and does not impact the IFNγ detection (C). Data are shown after background subtraction and stimulation index >2. Statistical analyses are performed using a paired Wilcoxon test.
(D–G) Representative gating strategy for the memory B cell assays using spike protein at time point 3 (D) or 4 (E), or RBD at time point 3 (F) or 4 (G).
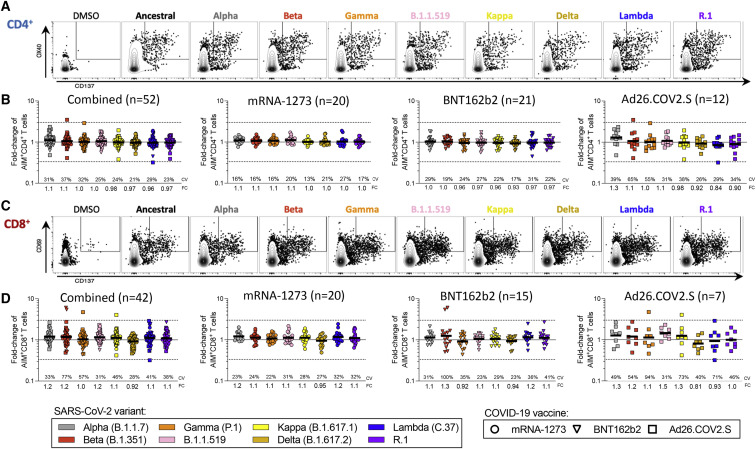
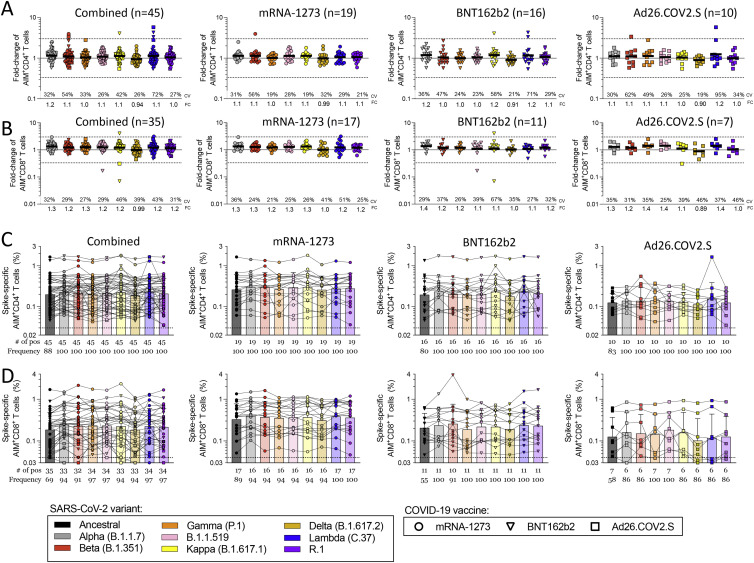
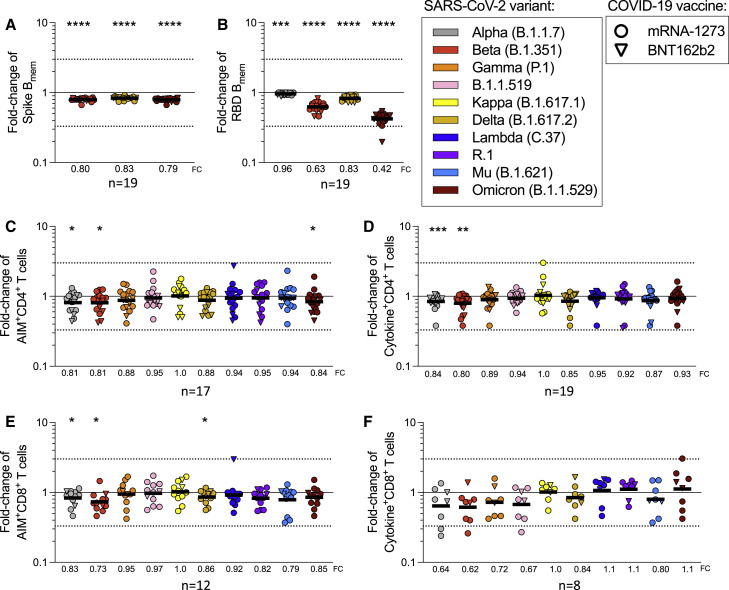
CD4+ and CD8+ T cell responses to spike MPs derived from the ancestral strain and from MPs representing Alpha, Beta, Gamma, Delta, B.1.1.519, Kappa, Lambda, and R.1 variants were measured by AIM OX40+CD137+ (CD4+ T cells) or CD69+CD137+ (CD8+ T cells) (Tarke et al., 2021b). For each subject/variant/vaccine combination, we calculated the T cell recognition fold-change relative to the ancestral sequence (variant/ancestral). Only donors with a positive spike ancestral response were included in the analysis (CD4: LOS = 0.03%, SI > 2; CD8: LOS = 0.04%, SI > 2). Figure 1 summarizes the fold-change results for all vaccine platforms combined and separately, for CD4+ (Figure 1B) and CD8+ (Figure 1D) T cells. For all variants, regardless of the vaccine platform considered, no significant decrease (fold-change <1.00 by the Wilcoxon signed rank T test compared with a hypothetical median of 1) was detected. In all cases, the geomean fold-change variation was close to 1.00 (i.e., no change). The average fold-change values, considering all 24 different vaccine/variant combinations (three vaccine platforms and eight variants), were 1.01 (range 0.84–1.3) for CD4+ and 1.1 (range 0.81–1.5) for CD8+ T cells. Thus, in fully vaccinated subjects at least 84% (CD4+) and 81% (CD8+) T cell responses detected by AIM assays were preserved across vaccine platforms.
Figure 1.
Impact of variant-associated mutations on spike-specific CD4+ and CD8+ T cell recognition
T cell responses from fully vaccinated COVID-19 vaccinees were assessed with variant spike MPs. The effect of mutations associated with each variant MP was expressed as relative (fold-change variation) to the T cell reactivity detected with the ancestral strain MP. Results from COVID-19 mRNA-1273 (n = 20, circles), BNT162b2 (n = 20, triangles), and Ad26.COV2.S (n = 12, squares) vaccinees are presented together, and separately, by vaccine platform. For fold-change (FC) calculations, only donors responding to the ancestral S MP were included.
(A) Representative gating of CD4+ T cells of a mRNA-1273 vaccine recipient responding to different SARS-CoV-2 variants’ MPs is shown.
(B) Fold-change is calculated for AIM+ CD4+ T cells relative to the ancestral strain in COVID-19 vaccinees.
(C) A representative gating example is shown for an mRNA-1273 vaccine recipient for CD8+ T cells against the SARS-CoV-2 variants in analysis.
(D) Fold-change is calculated for AIM+ CD8+ T cells relative to the ancestral strain in COVID-19 vaccinees. Coefficients of variation (CV) and the geometric mean FCs for the variants are listed in each graph and plotted as log scale. Significance of FC decreases for each variant was assessed by Wilcoxon signed rank T test compared with a hypothetical median of 1. See also Figures S1, S2, and S3 and Table S1.
At the level of individual donors, a decrease greater than an arbitrary 3-fold threshold (0.33 by fold-change, indicated by dotted lines) was only observed for two Ad26.COV2.S vaccinees and the Lambda variant, one donor for CD4+, and another for CD8+ T cells. No variant/donor combination was associated with a decrease greater than 10-fold (0.1 by fold-change). Figures S2 A and S2B show the corresponding AIM+ percentages and their relative paired comparisons based on the magnitude of the responses for each variant with the ancestral spike reactivity. Analysis of the coefficient of variation (CV) of the fold changes for each variant across vaccine platforms revealed some significant differences in the variation across vaccine platforms, particularly for Ad26.COV2.S (CD4+: mRNA-1273 versus Ad26.COV2.S p = 0.0009; BNT162b2 versus Ad26.COV2.S p = 0.0078; CD8+: mRNA-1273 versus Ad26.COV2.S p = 0.0024. Mann-Whitney with multiple comparison correction). Overall, these results indicate that both CD4+ and CD8+ T cell responses are largely conserved against variants, irrespective of the variant and vaccine platform considered.
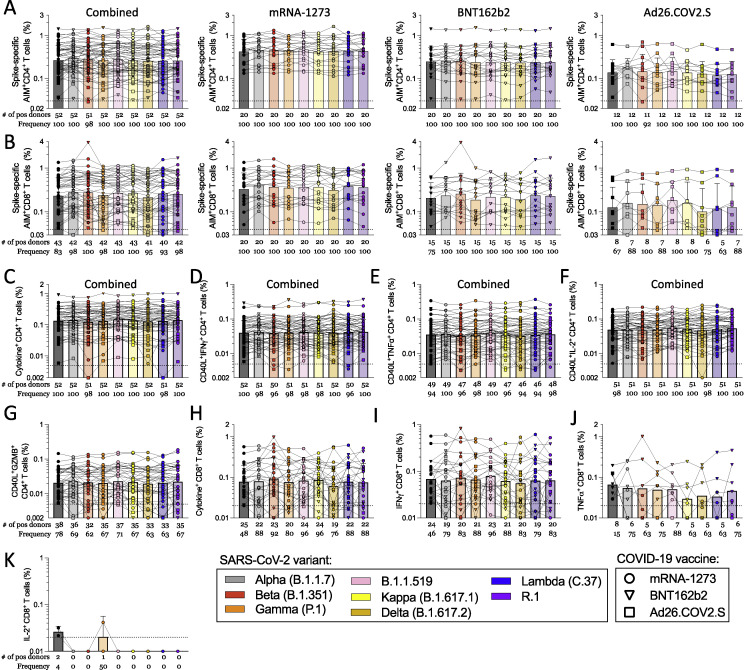
Figure S2.
Magnitude of CD4+ and CD8+ T cell responses in COVID-19 fully vaccinated individuals against ancestral and variant SARS-CoV-2 spike, related to Figures 1 and 2
AIM+ and cytokine+ T cell reactivities against MPs spanning the entire sequence of different SARS-CoV-2 variants are shown for PBMCs from fully vaccinated COVID-19 mRNA-1273 (n = 20, circles), BNT162b2 (n = 20, triangles), and Ad26.COV2.S (n = 12, squares) vaccinees analyzed by vaccine platform or combined together.
(A and B) Data for (A) AIM+ CD4+ and (B) AIM+ CD8+ T cells is shown.
(C–G) (C) The total cytokine response of all vaccinees combined was quantified by summing spike-specific CD40L expressing CD4+ T cells also expressing (D) IFNγ, (E) TNFα, (F) IL-2, or (G) granzyme B.
(H–K) For CD8+ T cells, the total cytokine response is shown (H) as calculated by the total IFNγ (I), TNFα (J), or IL-2 (K) CD8+ T cells. The frequency of response is based on the LOS (dotted line) for the ancestral response and SI > 2, while the frequency of responses across different variants is based on the number of donors responding to the ancestral spike pool. All data shown is background subtracted.
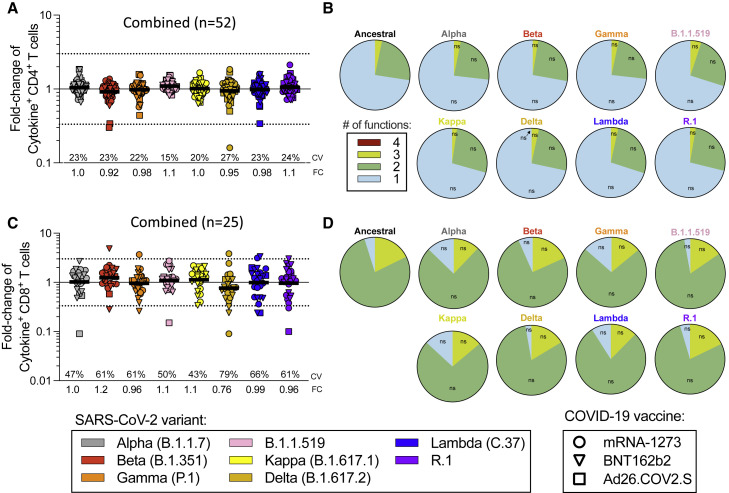
By ICS, CD4+ T cell responses to the ancestral Wuhan spike pool were observed for 52 subjects, and CD8+ T cell responses were observed for 25 subjects. Thus, combined ICS results for all vaccine platforms are presented. CD4+ T cell responses were associated with a polyfunctional response, encompassing IFNγ, TNFα, IL-2, and/or granzyme B in combination with CD40L expression (Figures 2 A, 2B, S1, and S2C–S2G). Cytokine+ CD8+ T cells were measured using IFNγ, TNFα, and IL-2 (Figures 2C, 2D, S1, and S2H–S2K). CD8+ T cell functionality was assessed based on IFNγ, TNFα, IL-2, and/or granzyme B expression. No differences in CD8+ T cell functionality were observed between ancestral and variant recognition (Figure 2D). Overall, the average fold-change values considering all variants was 1.00 (range 0.92–1.1) for CD4+ and 1.01 (range 0.76–1.2) for CD8+ T cells. The greatest decrease was to 0.76 for Delta by cytokine+ CD8+ T cells. At the level of individual CD4+ T cell responses, decreases greater than 3-fold were observed for three donors: one Ad26.COV2.S donor for Beta and Lambda, one Ad26.COV2.S donor for Beta, and one mRNA-1273 donor for Delta (Figure 2A). No decreases greater than 10-fold were observed. For CD8+ T cells, decreases greater than 10-fold were observed with one Ad26.COV2.S donor for Alpha and R.1 and one mRNA-1273 donor for Delta. Decreases in the 3- to 10-fold range were observed for four BNT162b2 donors (one for Beta, one for Gamma, one for Delta, and one for Lambda), one Ad26.COV2.S donor for Lambda, and one mRNA-1273 donor with R.1 (Figure 2C).
Figure 2.
Impact of variant-associated mutations on spike-specific CD4+ and CD8+ T cell cytokine responses
Fully vaccinated COVID-19 vaccinees were assessed with variant spike MPs and the effect of mutations associated with each variant MP is expressed as relative (FC variation) to the T cell reactivity detected with the ancestral strain MP and plotted as log scale. Results from COVID-19 mRNA-1273 (n = 20, circles), BNT162b2 (n = 20, triangles), and Ad26.COV2.S (n = 12, squares) vaccinees are presented together.
(A) Fold-change values for cytokine+CD4+ T cells are calculated based on the sum of CD4+ T cells expressing CD40L in combination with IFNγ, TNFα, IL-2, or granzyme B.
(B) The functionality of the spike-specific CD40L+ CD4+ T cell is defined by the different combinations of IFNγ, TNFα, IL-2, or granzyme B.
(C and D) (C) Fold-change values for cytokine+CD8+ T cells are calculated based on the sum of CD8+ T cells producing IFNγ, TNFα, or IL-2, and (D) the functionality of the spike-specific CD8+ T cells is calculated by looking at the different combinations of IFNγ, TNFα, IL-2, or granzyme B, excluding single positive granzyme B. All data shown are background subtracted with an SI > 2. CVs and the geometric mean FCs for the variants are listed in each graph. Significance of FC decreases for each variant was assessed by Wilcoxon signed rank T test compared with a hypothetical median of 1. See also Figures S1 and S2 and Table S1.
Considering the different assay readouts (AIM and ICS) and different donors analyzed, the fold-change was calculated in 171 instances for 8 different variants, for a total of 1,368 determinations. T cell responses with decreases greater than 3-fold were observed in 14 instances (1.02%) of variant/subject combinations tested and decreases greater than 10-fold were observed in 3 instances (0.22%) of variants/subjects tested. Thus, in almost 99% of cases, the differences in measured T cell recognition were less than 3-fold.
In the context of AIM+ T cells responses measured at time point 1 (2 weeks post-first-dose, Figure S3 ), we found a very similar pattern to what was observed at time point 2, with no substantial decreases in each of the variants analyzed at either population or individual level, except for 3 out of 280 instances (1%) with AIM+ CD8+ T cells (Figure S3B). These results confirm, in a larger dataset, that T cell responses from vaccinated subjects are largely preserved against Alpha, Beta, and Gamma (Tarke et al., 2021b). Importantly, these results extend these observations to more VOI/VOCs, including the prominent Delta variant.
Figure S3.
Fold-change values and magnitude of AIM+ T cell responses 2 weeks after the first vaccine dose, related to Figure 1
CD4+ and CD8+ T cell responses were assessed with variant spike MPs 2 weeks after the donors received the first dose of COVID-19 vaccine. The effect of mutations associated with each variant MP is expressed as relative (FC variation) to the T cell reactivity detected with the ancestral strain MP. COVID-19 mRNA-1273 (n = 19, circles), BNT162b2 (n = 20, triangles), and Ad26.COV2.S (n = 12, squares) vaccinees are presented together, and separately, by vaccine platform.
(A–D) The FC is calculated in respect of the ancestral strain in COVID-19 vaccinees for (A) AIM+ CD4+ and (B) AIM+ CD8+ T cells. The magnitude of AIM+ T cell reactivity against the spike MPs is shown for (C) CD4+ and (D) CD8+ T cells. The frequency of response is based on the LOS (dotted line) for the ancestral response and SI > 2, while the frequency of responses across different variants is based on the number of donors responding to the ancestral spike pool. CV and geometric mean of the FC for the variants are listed in each graph. Significance of FC decreases for each variant was assessed by Wilcoxon signed rank T test compared with a hypothetical median of 1.
Cross-recognition of SARS-CoV-2 variants by memory T cells
We then examined memory T cells and memory B cells 3–4 months after vaccination. At this time point, samples from eight NVX-CoV2373 vaccinated individuals were available and therefore included in the analysis. Spike-specific CD4+ T cell memory was characterized by AIM and ICS (Figures 3 A–3C, S4 A, and S4B), including memory-circulating T follicular helper cells (cTFH) (Figures 3G and S4E).
Figure 3.
Vaccinee memory T and B cell recognition of COVID-19 variants
(A and B) Fully vaccinated recipients of the COVID-19 mRNA-1273 (n = 12, circles), BNT162b2 (n = 15, triangles), Ad26.COV2.S (n = 14, squares), and NVX-CoV2373 (n = 8, diamonds) vaccines were assessed for T and B cell memory to variant spikes. FC values were calculated based on the response to the ancestral spike for subjects with a measurable response and plotted as log scale. CD4+ T cell FC values are shown for (A) AIM and (B) ICS assay.
(C–E) (C) The functional profile of spike-specific CD40L+CD4+ T cells was calculated as the percentage of cells with 1, 2, 3, or 4 functions defined by intracellular staining for IFNγ, TNFα, IL-2, or granzyme B. CD8+ T cell fold-change values are shown for the (D) AIM and (E) ICS assay.
(F) The functional profile of cytokine producing CD8+ T cells was calculated as the percentage of cells with 1, 2, 3, or 4 functions defined by intracellular staining for IFNγ, TNFα, IL-2, or granzyme B, excluding granzyme B single positive cells. p values for the functional profile of CD4+ and CD8+ T cells were calculated by Mann-Whitney.
(G–I) (G) Spike-specific cTFH+CD4+ T cells were calculated based on CXCR5+ of AIM+CD4+ T cells. SARS-CoV-2-specific memory B cells are shown to (H) spike and (I) RBD.
(J–L) Variant/ancestral FC values are shown for the (J) antibody neutralization assay as well as (K) spike and (L) RBD IgG serology. The geometric mean of the FC values is listed at the bottom of each graph. Significance of FC decreases for each variant was assessed by Wilcoxon signed rank T test compared with a hypothetical median of 1. See also Figures S1 and S4 and Table S1.
Figure S4.
Magnitude of T and B cell responses in COVID-19 vaccinated individuals 3.5 months after vaccination and antibody neutralization titer with early COVID-19 infected individuals, related to Figure 3
COVID-19 mRNA-1273 (n = 12, circles), BNT162b2 (n = 15, triangles), Ad26.COV2.S (n = 14, squares), and NVX-CoV2373 (n = 8, diamonds) vaccine recipients were assessed for T and B cell responses to variant SARS-CoV-2 spike MPs; all vaccine platforms are analyzed together.
(A and B) The magnitude of response is shown for (A) CD4+ T cells in the AIM assay and (B) the sum cytokine+CD4+ T cells, which was calculated from CD40L+ CD4 cells expressing IFNγ, TNFα, IL-2, or granzyme B.
(C and D) The magnitude of responding CD8+ T cells is shown for (C) the AIM assay and (D) the sum of cytokines, as calculated from the CD8+ T cells expressing IFNγ, TNFα, IL-2, or granzyme B, excluding single positive granzyme B.
(E) The total magnitude of spike-specific AIM+cTFH+CD4+ T cells is shown.
(F–I) The frequency of (F) spike- and (G) RBD-specific B cells among total memory B (Bmem) cells was assessed, as well as the frequency of variant-specific Bmem response within the ancestral response to (H) spike and (I) RBD.
(J) The antibody neutralization assay titer is shown for COVID-19 vaccinees.
(K and L) (K) The FC values are shown for early COVID-19 infected donors for the neutralization assay and (L) the magnitude of the neutralization titers for these donors.
(M and N) (M) Spike and (N) RBD IgG titers are shown. The frequency of response is based on the LOS (dotted line) for the ancestral response and SI > 2, while the frequency of responses across different variants is based on the number of donors responding to the ancestral spike pool. Significance of FC decreases for each variant was assessed by Wilcoxon signed rank T test compared with a hypothetical median of 1.
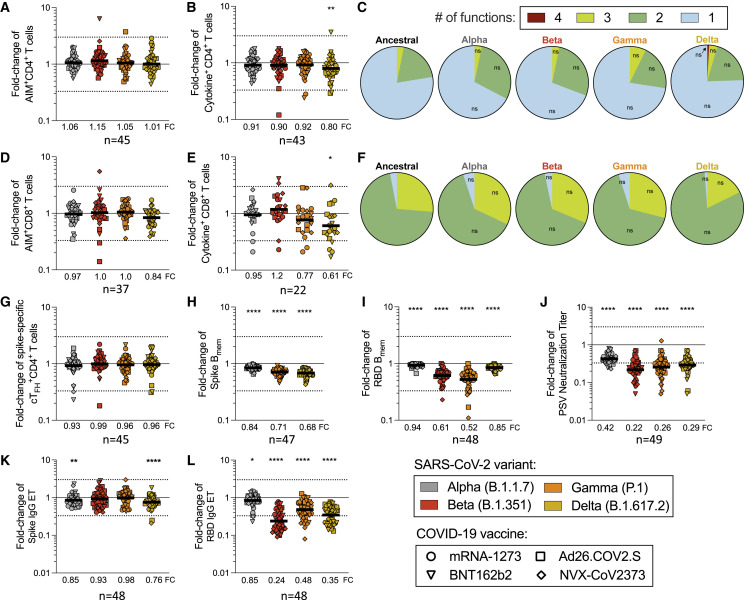
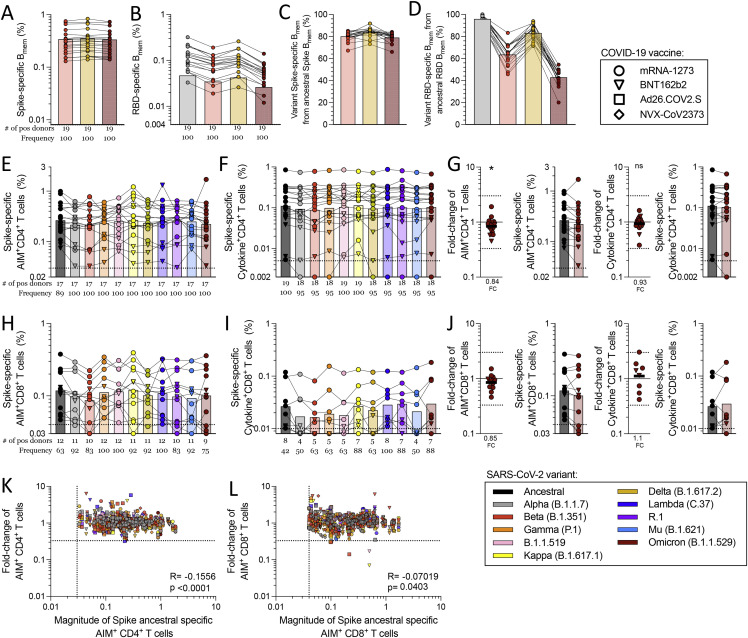
No significant decrease of memory CD4+ T cell recognition of Alpha, Beta, or Gamma variants was observed by AIM, cytokine, or cTFH metrics, with the exception of Delta cytokine+ CD4+ T cells (p = 0.0024) (Figures 3A, 3B, and 3G). Mean preservation of CD4+ T cell recognition was 0.90 (range 0.80–1.2), considering all three assays and variants. At the individual level, no substantial decreases in CD4+ T cell variant recognition were observed by AIM. Cytokine response decreases >3-fold were observed in 4 donors and 5 out of 172 instances (2.9%). cTFH memory cell recognition of variants decreased >3-fold in 4 donors and 5 out of 180 instances (2.8%) (Figure 3G).
Spike-specific CD8+ T cell memory was characterized by overall recognition retention of 0.95 by AIM and 0.88 by ICS (Figures 3D–3F, S4C, and S4D). No significant decreases of memory CD8+ T cell recognition of Alpha, Beta, or Gamma variants were observed by AIM. By ICS, CD8+ T cells retained 0.77 and 0.61 recognition of Gamma and Delta, respectively (Figures 3D–3F). At the individual level, decreases >3-fold were observed in 2 donors and 2 out of 148 instances (1.3%) by AIM, and 6 donors and 10 out of 88 instances (11.4%) by ICS, none of which were greater than 10-fold. Thus, the overall pattern of variant recognition by memory CD4+ and CD8+ T cells paralleled the peak T cell responses (Figures 1, 2, and 3). Memory CD4+ T cell recognition of these variants was largely preserved, including Delta. Some memory CD8+ T cell recognition decreases were noted by cytokine production, particularly against Delta.
We next examined the ability of spike-specific memory B cells to recognize variants Alpha, Gamma, and Delta (Figures 3H, S4F, and S4H). Significant losses in memory B cell recognition of spike for Alpha (variant/Wuhan = 0.84; p < 0.0001), Gamma (0.71; p < 0.0001), and Delta (0.68; p < 0.0001) were observed (Figure 3H). The RBD of spike is the primary target of SARS-CoV-2 neutralizing antibodies and a site of variant neutralizing antibody escape. We therefore characterized RBD-specific memory B cell recognition of variants Alpha, Beta, Gamma, and Delta (Figures 3I, S4G, and S4I). Significant decreases in RBD-specific memory B cell recognition of Alpha (0.94; p < 0.0001), Beta (0.61; p < 0.0001), Gamma (0.52; p < 0.0001), and Delta (0.85; p < 0.0001) were all noted (Figure 3I).
Neutralizing antibody titers to variants were measured and compared with a D614G reference virus for the same vaccinated individuals (Figures 3J and S4J). Neutralization decreases were significant for Alpha (p < 0.0001), Beta (p < 0.0001), Gamma (p < 0.0001), and Delta (p < 0.0001) variants (Figure 3J). The highest neutralization antibody titers were against D614G, and reductions in neutralizing titers of 2.4-fold, 4.5-fold, 3.8-fold, and 3.4-fold against Alpha, Beta, Gamma, and Delta variants were noted (Figures 3J and S4J). A similar pattern was observed for COVID-19 convalescent subjects (Figures S4K and S4L). Spike and RBD binding IgG titers in vaccinated subjects had similar trends to neutralizing antibodies but with smaller differences (Figures 3K, 3L, S4M, and S4N). In conclusion, while no significant change in T cell recognition was noted, decreases in memory B cell and neutralizing antibody recognition of all variants analyzed were apparent.
Predicted impact of SARS-CoV-2 variants on T cell epitopes
With the recent emergence of the Omicron variant, studies were immediately expanded to include Omicron. We first predicted the impact of variant mutations for CD4+ and CD8+ T cell epitopes experimentally curated in the IEDB (www.IEDB.org) (Grifoni et al., 2021; Vita et al., 2019) (Table S4). In addition to Omicron, we included a wider panel of early and late SARS-CoV-2 variants for comparison.
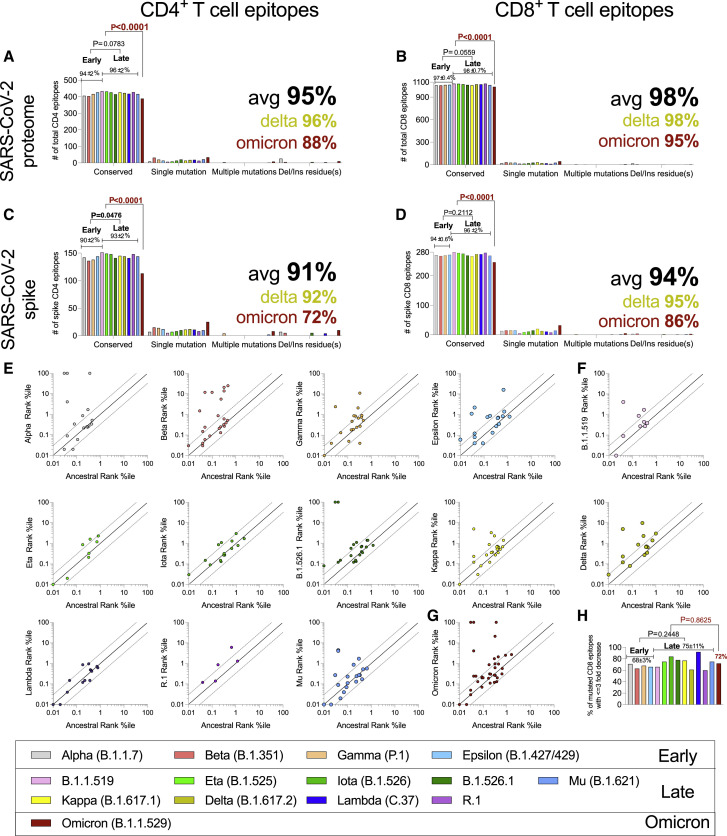
For CD4+ T cells, an average of 95% of the epitopes spanning the entire SARS-CoV-2 proteome were fully conserved (no mutations) across the variants (Figure 4 A). The Delta variant was not associated with a significant decrease (Figure 4A), while the fraction of fully conserved epitopes was reduced in Omicron (88%), compared with the other variants (p < 0.0001) (Figure 4A). A similar result was observed for CD8+ T cell epitopes, with 98% overall conservation but 95% for Omicron (p < 0.0001) (Figure 4B). Considering only spike epitopes, an average of 91% and 94% CD4+ and CD8+ T cell epitopes, respectively, were conserved in the various variants. The Omicron variant was associated with the fewest fully conserved spike epitopes for both CD4+ and CD8+ T cells (CD4+: 72%, p < 0.0001; CD8+: 86%, p < 0.0001) (Figures 4C and 4D).
Figure 4.
Sequence conservation of SARS-CoV-2 T cell epitopes in variants
(A–D) The number of epitopes fully conserved or having single or multiple mutations (including insertions/deletions) was computed across SARS-CoV-2 variants. The analysis shown represents the breakdown of conserved and mutated CD4+ (A and C) and CD8+ T cell epitopes (B and D) for all SARS-CoV-2 proteins (A and B) and spike protein only (C and D). The percentage of conserved epitopes was calculated for each variant separately. Average conservancy and standard deviations were calculated for all variants and then separately for early variants, more recent SARS-CoV-2 variants, and Omicron.
(E–H) Predicted HLA binding affinities of mutated versus ancestral sequences of CD8+ T cell epitopes, based on epitope/HLA combinations curated in the IEDB data as of July 2021. Predicted HLA binding values to the relevant HLA allelic variant were calculated using the IEDB recommended NetMHCpanEL 4.1 (Reynisson et al., 2020) algorithm. Points outside the dotted lines in each panel indicate instances where the predicted HLA binding capacity of the mutated peptide was increased (>3-fold) or decreased (<3-fold). (E) Early, (F) late, and (G) Omicron SARS-CoV-2 variants are shown. (H) Percentage of mutated CD8+ T cell epitopes associated with a 3-fold decrease in predicted binding capacity. Comparisons of epitopes conservancy across early and current variants were performed by unpaired Mann-Whitney test. Comparison with the Omicron variant was performed by one sample T test. Large font bold numbers indicate average conservation in all variants (black), Delta (ocher), and Omicron (dark red). See also Tables S3 and S4.
We further found that 82% of 9-mers encompassing the entire spike protein are conserved in Omicron, compared with 86% of the CD8 epitopes. Thus, mutations do not appear to occur more frequently in areas of spike recognized as epitopes.
To address whether Omicron mutations preferentially impacted more dominant epitopes, we divided epitopes into dominant and subdominant, based on the frequency of individual responses as previously described (Grifoni et al., 2021). This analysis indicated that the most dominant CD4+ epitopes tend to be more frequently conserved than subdominant epitopes (75% versus 64%), while a modest opposite trend was observed for CD8+ T cell epitopes (84% versus 88%) (data not shown).
The values reported in Figures 4A–4D were stringent estimates of the number of preserved epitopes because conservative substitutions and changes not impacting HLA binding can still be cross-reactively recognized. Accordingly, we examined the effect of the mutations on the predicted binding affinity of each CD8 epitope for which HLA restriction could be inferred (Figures 4E–4G). Notably, in the majority of cases, the variant-associated mutations were predicted to not impair HLA binding capacity (Figures 4E–4G). Importantly, 72% of the epitopes with Omicron variant mutations were predicted to retain similar HLA class I binding capabilities, which was not dissimilar to other SARS-CoV-2 variants (p = 0.8625; Figure 4H). In conclusion, bioinformatic analyses suggest that the majority of CD4+ and CD8+ T cell epitopes are unaffected by mutations, regardless of whether early or late variants were considered (Figure 4A–4D). These data suggest that variant evolution was not driven by T cell escape. In the case of Omicron, the number of totally conserved spike epitopes was decreased. However, the majority of Omicron epitopes (full proteome or spike) were still 100% conserved, and the majority of mutated epitopes were predicted to still be recognized by T cells.
Experimental assessment of Omicron-specific memory B and T cells
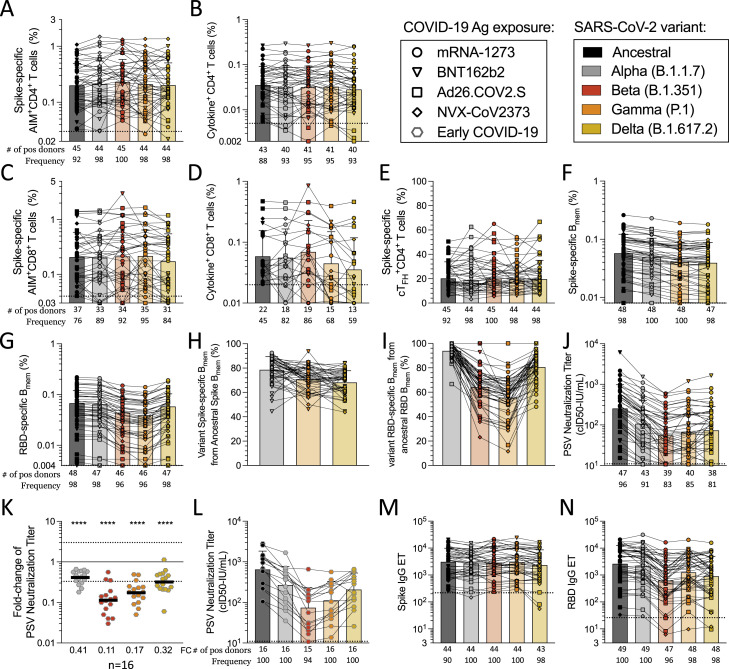
Considering that the Omicron variant contains 15 mutations in the RBD, we sought to investigate whether mRNA vaccination generated memory B cells that recognized Omicron spike and RBD. Memory B cells obtained from subjects receiving either mRNA-1273 or BNT162b2 (5–6 months post-vaccination) had significantly lower recognition of Omicron spike compared with the ancestral strain (p < 0.0001) (Figures 5 A, S5A, and S5C). Memory B cell recognition of Omicron RBD was significantly decreased, with 0.42 retained recognition (p < 0.0001) (Figures 5B, S5B, and S5D), substantially lower than Alpha, Beta, or Delta RBD binding. In sum, memory B cell recognition of Omicron RBD, known to be important for most neutralizing antibodies, was substantially reduced compared with other variants.
Figure 5.
Impact of Omicron and other variants on memory T cell and B cell recognition
The response to SARS-CoV-2 variants was assessed in individuals 5–6 months after full vaccination with mRNA-1273 (n = 12, circles) and BNT162b2 (n = 7, triangles) COVID-19 vaccines.
(A and B) The FC values are shown for (A) memory B cell responses to spike and (B) RBD Omicron compared with other variants.
(C) The FC values for CD4+ T cell responses by AIM are shown.
(D) The FC of all cytokine+CD4+ T cells is calculated from the sum of CD4+ cells expressing CD40L in combination with IFNγ, TNFα, IL-2, or granzyme B.
(E) The FC values for CD8+ T cell responses measured by AIM are shown.
(F) The FC of all cytokine+CD8+ T cells, as calculated from the sum of IFNγ, TNFα, or IL-2. The geometric mean of the FC values for each variant is listed in each graph and plotted as log scale. Significance of FC decreases for each variant was assessed by Wilcoxon signed rank T test compared with a hypothetical median of 1. See also Figure S5 and Table S1.
Figure S5.
Response to SARS-CoV-2 variants in fully vaccinated donors 5–6 months after vaccination, related to Figure 5
5–6 months after vaccination, COVID-19 mRNA-1273 (n = 12, circles) and BNT162b2 (n = 7, triangles) vaccine recipients were assessed for T cell responses to variant spikes by AIM and ICS assays.
(A and B) The frequency of (A) Omicron spike- and (B) RBD-specific B cells among total Bmem cells was assessed and compared with frequency of B cell recognizing other variants.
(C and D) In addition, frequency of Omicron-specific Bmem response within the ancestral response to (C) Spike and (D) RBD was determined.
(E) The magnitude of response is shown for AIM+CD4+ T cells.
(F) The total cytokine response for CD4+ T cells is calculated by summing the CD40L+ cells also expressing IFNγ+, TNFα+, IL-2+, or granzyme B+.
(G) The CD4+ T cell response to the Omicron variant is shown for AIM and ICS, including the FC values and magnitude, and those values are duplicated in (E) and (F) and Figures 5C and 5D.
(H and I) For CD8+ T cells, (H) the magnitude of AIM+CD8+ T cells is shown and (I) the total cytokine+CD8+ T cells calculated by summing the IFNγ+, TNFα+, IL-2+, or granzyme B+ CD8+ T cells, excluding granzyme B single positive cells.
(J) The FC values and magnitude of response is shown for the CD8+ T cell responses to the Omicron variant by AIM and ICS, and those values are duplicated in (H) and (I) and Figures 5E and 5F. The frequency of response is based on the LOS (dotted line) for the ancestral response and SI > 2, while the frequency of responses across different variants is based on the number of donors responding to the ancestral spike pool. Significance of FC decreases for each variant was assessed by Wilcoxon signed rank T test compared with a hypothetical median of 1. COVID-19 mRNA-1273 (circles), BNT162b2 (triangles), Ad26.COV2.S (squares), and NVX-CoV2373 (diamonds) vaccine recipients were assessed for T cell responses to variant spikes by AIM assay at various time points, ranging from 2 weeks after the first dose to 5–6 months after the last dose of vaccine.
(K and L) The correlation of magnitude and FC values (K) AIM+ CD4+ or (L) CD8+ T cells was analyzed for all time points combined (n = 183 donors). R and p values are the results of a Pearson correlation.
Next, we experimentally determined the impact of Omicron mutations on T cell responses in comparison with other variants in a cohort of individuals vaccinated 5–6 months before blood donation and also in parallel subsequently used for epitope mapping. The overall conservation of memory CD4+ T cell recognition of Omicron spike was 0.84 (84%) by AIM and 0.93 (93%) by ICS assay (Figure S5 G). A significant decrease was observed for Omicron by AIM, comparable in magnitude to that of Alpha or Beta variants (Figures 5C, 5E, and S5). No significant decrease was observed for CD4+ T cell recognition of Omicron by ICS; significant differences were only observed for Alpha and Beta (Figures 5D, 5F, S5E, and S5F). At the individual subject level, no AIM+ or ICS+ CD4+ T cell recognition decreases >3-fold were observed.
The preservation of memory CD8+ T cell recognition of Omicron spike was 0.85 (85%) by AIM and 1.1 (110%) by ICS (Figures 5E, 5F and S5J), with neither change being statistically significant. Significant decreases were observed for Alpha, Beta, and Delta by AIM (Figures 5E and 5F). In the context of Omicron, 3 out of the 12 positive donors analyzed for CD8+ T cell responses showed a minimal decrease at the fold-change level that nevertheless placed the response below the AIM assay limit of sensitivity; thus, the frequency of positive responders to Omicron was 75%, the lowest of all variants (Figure S5H). A loss of positive responders to Omicron was also observed by ICS (7 out of 8, 88%) (Figure S5I). For all time points analyzed in this study, we found a very weak inverse correlation between fold-change decrease and the magnitude of the spike-specific T cell responses (Figures S5K and S5L), suggesting that overall weaker responses tend to be less frequently associated with decreases in the variants. This might simply reflect weaker responses being associated with a lesser dynamic range and therefore decreases being less reliably measured. In any case, it did not support the notion that significant decreases are selectively associated with weak responses. We also examined the notion that weaker responses might be associated with individual HLA allele combinations, utilizing bioinformatic tools specifically designed to detect HLA associations (Paul et al., 2017). No specific HLA class I or class II alleles were significantly correlated to reduced variant recognition in our cohort (data not shown); however, the limited sample size was not powered to detect HLA associations, which usually require substantially larger numbers of observations. Overall, compared with other variants, no clear pattern of a loss of CD4+ or CD8+ T cell recognition of Omicron was observed by either T cell assay.
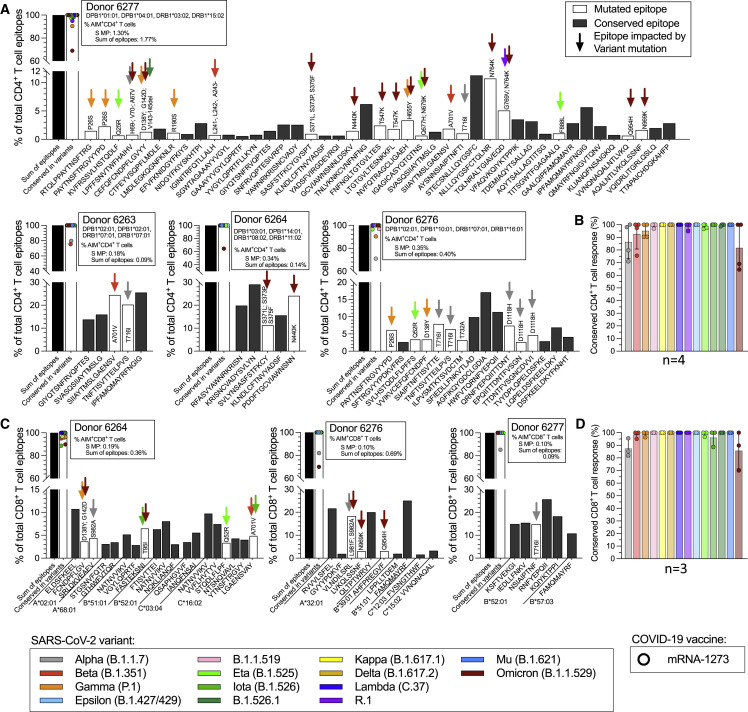
To further examine the molecular mechanism involved in the observed effects of T cell recognition of variant spike epitopes, we selected four donors for in-depth spike epitope identification studies and variant analyses (Figures 6 A–6D; Table S5). Each vaccinated donor recognized 5–42 (median 11) individual CD4+ T cell epitopes in spike (Figure 6A). Approximately 80% of the CD4+ T cell response was associated with epitopes fully conserved in Omicron, with the actual values per donor ranging from 65% to 100% (Figure 6B). Each vaccinated donor with a measurable CD8+ T cell response against the ancestral strain was found to recognize 6–19 (median 10) spike CD8+ T cell epitopes (Figure 6C). Approximately 80% of the CD8+ T cell response was associated with epitopes fully conserved in Omicron, with the values per donor ranging from 70% to 100% (Figure 6D). These results were in agreement with the bioinformatic analyses (Figure 4). In general, there was also good correspondence between the peptide pools, except with the CD8+ T cell response by donor 6,276, where the sum of the response for the individual epitopes exceeded the one observed for the megapool, suggesting that perhaps some of the identified epitopes may not be generated efficiently from the originating 15-mers contained in the megapool (Figure 6C). In summary, these epitope mapping data showed how the wide epitope repertoire associated with vaccine-induced responses counterbalances the effect of variant mutations of observed spike epitopes.
Figure 6.
Impact of SARS-CoV-2 variants on spike epitope repertoires of fully vaccinated donors 5–6 months after vaccination
The response to SARS-CoV-2 variants was assessed in individuals 5–6 months after full vaccination with mRNA-1273 (n = 4, circles).
(A–D) (A) CD4+ T cell epitope repertoires were determined for four mRNA-1273 vaccinees, and (C) CD8+ T cell epitope repertoires were determined for three mRNA-1273 vaccinees (no CD8+ T cell response was measurable for donor 6,263) by testing the inferred HLA class I restricted epitopes based on the individual HLA-A, -B, and -C typing and applying the NetMHCpan EL4.1 algorithm implemented in the IEDB with a 4th percentile cutoff. The percent of T cell response associated with conserved epitopes for each individual donor for (B) CD4+ and (D) CD8+ T cells is shown for each variant assessed. Each graph shows the total response detected with the ancestral spike MP, and the summed total response detected against each of the individual epitopes identified. The histograms show the percentage of the total response accounted from each epitope, where black bars indicate non-mutated epitopes, while mutated epitopes are represented by open bars, with color coding further indicating which variant mutations are associated with the epitope. Based on these data, the fraction of the total response to each variant that can be accounted for by non-mutated epitopes can be calculated, as also shown in the graph. See also Tables S1, S2, S3, and S5.
Discussion
Here, we analyzed adaptive immunity in vaccinated individuals to a comprehensive panel of SARS-CoV-2 variants, including Delta and Omicron, for multiple vaccines. Our data demonstrate that the vast majority of T cell epitopes are fully conserved, not only in the “early” variants previously analyzed (Collier et al., 2021; Geers et al., 2021; Keeton et al., 2021; Melo-González et al., 2021; Riou et al., 2021; Tarke et al., 2021b) but also in newer variants, suggesting that the continued evolution of variants has not been associated with increased escape from T cell responses at the population level.
At the level of the full proteome, which is relevant for natural infection, 95% of reported class II and 98% of class I epitopes were fully conserved by computational analysis based on IEDB data extracted in July 2021. In the case of Omicron, the fraction of epitopes that were fully conserved dropped to 88% for class II and 95% for class I epitopes in the whole proteome. Focused only on spike, relevant in the context of vaccination, 91% of class II and 94% of class I epitopes were still fully conserved. The fraction of totally conserved spike epitopes in Omicron dropped to 72% for class II and 86% for class I epitopes. The higher number of mutated T cell epitopes in spike was expected because many variant mutations are localized in the spike protein. Overall, the majority of T cell epitopes available in IEDB are conserved at the sequence level in all variants analyzed so far, including Omicron. It should be emphasized that an epitope mutation does not preclude cross-reactive recognition of the mutated sequence. To partially address this point, we calculated the fraction of class I epitope mutations predicted to be associated with a decrease in binding affinity to the relevant HLA. We found that HLA binding was well conserved for the majority of the mutated epitopes. The impact on HLA binding was no different for Omicron epitopes than for other variants. These observations argue against a model that mutations accumulated in Omicron might be the result of T cell immune pressure at the population level.
T cell recognition of several variants, including Delta and Omicron, was experimentally measured in donors vaccinated with mRNA-1273, BNT162b2, or Ad26.COV2.S. Variant recognition relative to the ancestral sequence was similar in the three different vaccine platforms tested, which is reassuring in terms of the potential implications for protective effects being similar regardless of the vaccine platform considered. A significant higher variability was detected with Ad26.COV2.S, possibly related to evidence that Ad26.COV2.S induces spike-specific T cells mainly targeting the S1 region of spike, while other vaccines appear to elicit a more broad spike-specific T cell response (Kim Huat et al., 2021).
The majority of memory T cells were not impacted by variants’ mutations, which is again reassuring in terms of the potential implications for T cell protective effects being similar regardless of the different vaccine cohorts considered. Significant fold-change decreases were noted in the 3.5-month memory time point for the Delta variant when utilizing cytokine production as a readout. While it is possible that this function is more impacted in mutated sequences, a consistent difference was not observed across variants.
Memory T cell responses to the various variants, including Omicron, were dissected in detail in a cohort of donors 6–7 months following vaccination. The results confirmed that the majority of both CD4+ and CD8+ T cell responses detected by the AIM assay were preserved at this late time point. CD8+ T cell response decreases were observed when utilizing the IFNγ production as a readout in certain cases. Of note, regardless of the assay, Omicron responses were largely preserved in both CD4+ and CD8+ T cells. Broadly speaking, it is plausible that any antigen-specific T cell loss smaller than 2-fold is of modest relevance, given that the T cells respond as a recall response with relatively short doubling times. From animal models, we are not aware of conditions where less than 2-fold changes in antigen-specific T cell frequencies resulted in a measurable difference in protective immunity. It is also important to note that individual decreases or lack of responses were noted in 25% or less of the individuals in the case of Omicron, suggesting that some selected HLA class profiles may be more susceptible to the impact of Omicron. In-depth epitope identification experiments revealed that both CD4+ and CD8+ T cell responses in vaccinated donors were broad, and the data further demonstrated that for each individual donor/variant combination the majority of responses identified were to epitopes that were fully conserved. These data provide a clear explanation for the limited impact of variant-associated mutations on T cell responses at the population level.
Adaptive immunity against SARS-CoV-2 consists of multiple branches (Sette and Crotty, 2021). Memory B cell recognition of variants’ spikes was reduced in all cases, but the reductions were moderate against Delta spike and Beta RBD, demonstrating substantially retained memory B cell recognition of most variants (Cho et al., 2021; Goel et al., 2021; Sokal et al., 2021a). However, memory B cell recognition of Omicron RBD was substantially reduced. Memory B cell binding to Omicron RBD is likely to be detectable at affinities insufficient for virus neutralization in vitro. This is consistent with the observations that neutralizing antibody titers against Omicron are generally low in individuals after two doses of mRNA-1273 or BNT162b2. Nevertheless, Omicron neutralizing antibody titers rapidly increase after a third immunization (Liu et al., 2021; Planas et al., 2021; Schmidt et al., 2021; Sokal et al., 2021b), most likely due to the presence of memory B cells that recognized Omicron RBD, as observed here. Memory B cells may have important contributions in protective immunity by making anamnestic neutralizing antibody responses after infection (Cameroni et al., 2021; Carreño et al., 2021; Cele et al., 2021; Doria-Rose et al., 2021; Gagne et al., 2021; Garcia-Beltran et al., 2021; Zou et al., 2021).
These data provide reasons for optimism, as most vaccine-elicited T cell responses remain capable of recognizing all known SARS-CoV-2 variants. Nevertheless, the data also underline the need for continued surveillance and the potential danger posed by continued variant evolution that could result in further reduction of T cell responses. Incorporation of additional elements eliciting broader T cell responses directed toward more conserved targets into vaccine strategies may be considered as a means to increase vaccine effectiveness against future variants.
Limitations of the study
This study has limitations. A caveat is that all experiments were performed with a robust concentration of peptides (1 μg/mL), which might underestimate the impact of mutations on T cells. It is also currently unknown what level of epitope conservation is likely to preserve functional T cell responses in vivo, and currently no rigorous correlate of protection based on T cell responses has been generated to understand the impact of T cell responses against various SARS-CoV-2 outcomes such as severe disease. Furthermore, the assays used in our study are testing peptide-based responses rather than the responses that will occur in vivo. A variant might change multiple features of epitope presentation; e.g., by mutations outside the epitope that change processing, or more globally by evolving additional immune evasion strategies. Finally, this study has not investigated subjects following natural infection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-human CD8 BUV496 (clone RPA-T8) | BD Biosciences | Cat# 612942; RRID:AB_2870223 |
| Mouse anti-human CD3 BUV805 (clone UCHT1) | BD Biosciences | Cat# 612895; RRID:AB_2870183 |
| Mouse anti-human TNF alpha eFluor450 (clone MAb11) | Life Tech | Cat# 48-7349-42; RRID:AB_2043889 |
| Mouse anti-human CD14 V500 (clone M5E2) | BD Biosciences | Cat# 561391; RRID:AB_10611856 |
| Mouse anti-human CD19 V500 (clone HIB19) | BD Biosciences | Cat# 561121; RRID:AB_10562391 |
| Mouse anti-human CD4 BV605 (clone RPA-T4) | BD Biosciences | Cat# 562658; RRID:AB_2744420 |
| Mouse anti-human IFN gamma FITC (clone 4S.B3) | Invitrogen (Thermo Fisher Scientific) | Cat# 11-7319-82; RRID:AB_465415 |
| Rat anti-human IL-2 BB700 (clone MQ1-17H12) | BD Biosciences | Cat# 566405; RRID:AB_2744488 |
| Mouse anti-human CD69 PE (clone FN50) | BD Biosciences | Cat# 555531; RRID:AB_395916 |
| Mouse anti-human CD134 (OX40) PE-Cy7 (clone Ber-ACT35) | BioLegend | Cat# 350012; RRID:AB_10901161 |
| Mouse anti-human CD137 APC (clone 4B4-1) | BioLegend | Cat# 309810; RRID:AB_830672 |
| Mouse anti-human Granzyme B AF700 (clone GB11) | BD Biosciences | Cat# 560213; RRID:AB_1645453 |
| Mouse anti-human CD154 (CD40 Ligand) APC-ef780 (clone 24-31) | eBioscience (Thermo Fisher Scientific) | Cat# 47-1548-42; RRID:AB_1603203 |
| Mouse anti-human CD4 BV605 (clone RPA-T4) | BD Biosciences | Cat# 562658; RRID:AB_2744420 |
| Rat anti-human CXCR5 (CD185) BB700 (clone RF8B2) | BD Biosciences | Cat# 566469; RRID:AB_2869769 |
| Mouse anti-human CD279 (PD-1) PE-Dazzle594 (clone EH12.2H7) | BioLegend | Cat# 329940; RRID:AB_2563659 |
| Mouse anti-human CD19 BUV563 (clone SJ25C1) | BD Biosciences | Cat# 612916; RRID:AB_2870201 |
| Mouse anti-human IgD Pacific Blue (clone IA6-2) | BioLegend | Cat# 348224; RRID:AB_2561597 |
| Mouse anti-human CD20 BV510 (clone 2H7) | BioLegend | Cat# 302340; RRID:AB_2561941 |
| Mouse anti-human IgM BV570 (clone MHM-88) | BioLegend | Cat# 314517; RRID:AB_10913816 |
| Mouse anti-human CD27 BB515 (clone M-T271) | BD Biosciences | Cat# 564642; RRID:AB_2744354 |
| Mouse anti-human IgA Vio Bright FITC (clone IS11-8E10) | Miltenyi Biotec | Cat# 130-113-480; RRID:AB_2734076 |
| Mouse anti-human CD3 PerCP (clone SK7) | BioLegend | Cat# 344814; RRID:AB_10639948 |
| Mouse anti-human CD14 PerCP (clone 63D3) | BioLegend | Cat# 367152; RRID:AB_2876693 |
| Mouse anti-human CD16 PerCP (clone 3G8) | BioLegend | Cat# 302030; RRID:AB_940380 |
| Mouse anti-human CD56 PerCP (clone 3G8) | BioLegend | Cat# 318342; RRID:AB_2561865 |
| Mouse anti-human IgG PerCP/Cyanine5.5 (clone M1310G05) | BioLegend | Cat# 410710; RRID:AB_2565788 |
| Mouse anti-human CD38 APC/Fire 810 (clone HIT2) | BioLegend | Cat# 303550; RRID:AB_2860784 |
| Bacterial and virus strains | ||
| rVSV-SARS-CoV-2 | This study | N/A |
| Biological samples | ||
| COVID-19 vaccinated donor blood samples | LJI Clinical Core | N/A |
| Convalescent COVID-19 donor blood samples | LJI Clinical Core | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Brilliant Staining Buffer Plus | BD Biosciences | Cat# 566385 |
| Live/Dead Viability Dye eFluor506 | Invitrogen (Thermo Fisher Scientific) | Cat# 65-0866-14 |
| Live/Dead Fixable Blue Stain Kit | Thermo Fisher Scientific | Cat# L34962 |
| Synthetic peptides | TC Peptide Lab | https://www.tcpeptide.com |
| Ancestral (WT) Spike Protein | AcroBiosystems | Cat# SPN-C82E9 |
| Alpha (B.1.1.7) Spike Protein | AcroBiosystems | Cat# SPN-C82E5 |
| Beta (B.1.351) Spike Protein | AcroBiosystems | Cat# SPN-C82E4 |
| Gamma (P.1) Spike Protein | AcroBiosystems | Cat# SPN-C82E6 |
| Delta (B.1.617.2) Spike Protein | AcroBiosystems | Cat# SPN-C82Ec |
| Omicron (B.1.1.529) Spike Protein | AcroBiosystems | Cat# SPN-C82Ee |
| Ancestral (WT) RBD Protein | BioLegend | Cat# 793906 |
| Alpha (B.1.1.7) RBD Protein | AcroBiosystems | Cat# SPD-C82E6 |
| Beta (B.1.351) RBD Protein | AcroBiosystems | Cat# SPD-C82E5 |
| Gamma (P.1) RBD Protein | AcroBiosystems | Cat# SPD-C82E7 |
| Delta (B.1.617.2) RBD Protein | AcroBiosystems | Cat# SPD-C82Ed |
| Omicron (B.1.1.529) RBD Protein | AcroBiosystems | Cat# SPD-C82E4 |
| Ancestral (WT) Spike Protein for ELISA | AcroBiosystems | Cat# SPN-C52H9 |
| Alpha (B.1.1.7) Spike Protein for ELISA | AcroBiosystems | Cat# SPN-C52H6 |
| Beta (B.1.351) Spike Protein for ELISA | AcroBiosystems | Cat# SPN-C52Hk |
| Gamma (P.1) Spike Protein for ELISA | AcroBiosystems | Cat# SPN-C52Hg |
| Delta (B.1.617.2) Spike Protein for ELISA | AcroBiosystems | Cat# SPN-C52He |
| Omicron (B.1.1.529) Spike Protein for ELISA | AcroBiosystems | Cat# SPN-CH2Hz |
| Ancestral (WT) RBD Protein for ELISA | AcroBiosystems | Cat# SPD-C52H1 |
| Alpha (B.1.1.7) RBD Protein for ELISA | AcroBiosystems | Cat# SPD-C52Hn |
| Beta (B.1.351) RBD Protein for ELISA | AcroBiosystems | Cat# SPD-C52Hp |
| Gamma (P.1) RBD Protein for ELISA | AcroBiosystems | Cat# SPD-C52Hr |
| Delta (B.1.617.2) RBD Protein for ELISA | AcroBiosystems | Cat# SPD-C82Hh |
| Omicron (B.1.1.529) RBD Protein for ELISA | AcroBiosystems | Cat# SPD-C522e |
| Experimental models: Cell lines | ||
| Vero | ATCC | ATCC Cat# CCL-81, RRID:CVCL_0059 |
| HEK293T | ATCC | ATCC Cat# CRL-3216, RRID:CVCL_0063 |
| Recombinant DNA | ||
| phCMV3-SARS-CoV-2 | Hastie et al., 2021 | N/A |
| Software and algorithms | ||
| GraphPad Prism 9 | GraphPad | https://www.graphpad.com/; RRID:SCR_002798 |
| FlowJo 10 | FlowJo | https://www.flowjo.com/; RRID:SCR_008520 |
| IEDB | Grifoni et al., 2021 | https://www.iedb.org; RRID:SCR_006604 |
Resource availability
Lead contact
Please refer to the lead contact (Alessandro Sette, alex@lji.org) for further information pertaining to availability of resources and reagents.
Materials availability
Upon specific request and execution of a material transfer agreement (MTA) to the lead contact or to A.G., aliquots of the peptide pools utilized in this study will be made available. Limitations will be applied on the availability of peptide reagents due to cost, quantity, demand, and availability.
Experimental model and subject details
Human sample donors
The La Jolla Institute for Immunology (LJI) Clinical Core recruited healthy adults who had received the first and, when applicable, second dose of a COVID-19 vaccination among the mRNA-1273, BNT162b2, Ad26.COV2.S or NVX-CoV2373 available vaccinations. At the time of enrollment in the study, all donors gave informed consent. The LJI Clinical Core facility has collected blood draws under IRB approved protocols (LJI; VD-214) when possible two weeks after each vaccine dose administered (timepoint 1 and timepoint 2), 3.5 months after the last dose received (timepoint 3) and/or 5-6 months after the last dose (timepoint 4). All donors had their SARS-CoV-2 antibody titers measured by ELISA, as described below. Additional information on gender, ethnicity, age and timepoint of collection of the vaccinee cohorts are summarized in Table S1. Pheresis blood donations from an additional cohort of mRNA vaccinees were provided by the contact research organization (CRO) BioIVT and collected under the same IRB approval (VD-214) at LJI.
Pseudovirus production
Recombinant SARS-CoV-2-spike pseudotyped VSV-ΔG-GFP were generated with the specific amino acid mutations listed: D614G (WT), B.1.1.7 (Alpha; 69-70 deletion, 144 deletion, N501Y, A570D, D614G, P681H), B.1.351 (Beta; L18F, D80A, D215G, 241-243 deletion, K417N, E484K, N501Y, D614G, A71V), P.1 (Gamma; L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F) and B.1.617.2 (Delta; T19R, F157-R158 deletion, L452R, T478K, D614G, P681R, D950N).
Method details
SARS-CoV-2 variants of concern selection and bioinformatic analysis
The genome sequences for the B.1.1.7, B.1.351, P.1. and B.1.427/429 variants were selected as previously described (Tarke et al., 2021b). For the additional variants selected, the sequence variations in the variant viruses were derived by comparison with Wuhan-1 (NC_045512.2). All mutated amino acids in the different variants are outlined in Table S3. To determine the impact of the selected variants on T cell epitopes, CD4 and CD8 T cell epitopes were extracted from the IEDB database (www.IEDB.org)(Vita et al., 2019) on July 8th 2021 using the following query: Organism: SARS-CoV2 (ID:2697049, SARS2), Include positive assays only, No B cells, No MHC assays, Host: Homo sapiens (human) and either MHC restriction type: Class I for CD8 epitopes or Class II for CD4 epitopes. Additional manual filtering was performed on the extracted datasets allowing only epitopes of 9-14 residues in size for class I and 13-25 residues for class II. This resulted in a total of 446 and 1092 epitopes for CD4 and CD8, respectively. The binding capacity of SARS-CoV-2 T cell epitopes, and their corresponding variant-derived peptides, was determined for their putative HLA class I restricting allele(s) in a smaller epitope subset (n=833) where information regarding allele restriction was available. Prediction analyses for class I were determined utilizing the NetMHCpan EL4.1 algorithm (Reynisson et al., 2020) implemented by the IEDB’s analysis resource(Dhanda et al., 2019; Vita et al., 2019). Predicted binding for class I analyses are expressed in terms of percentile. For each epitope-variant pair a fold-change of affinities (variant /WT) was determined, corresponding values FC >3, indicating a 3-fold or greater decrease in affinity due to the mutation, were accordingly categorized as a decrease in binding capacity, and a FC <0.3 as an increase; FCs between 0.3 and 3 were designated as neutral.
Peptide synthesis and Megapool preparation
All the peptides used in this study were synthesized as crude material (TC Peptide Lab, San Diego, CA), and then individually resuspended in dimethyl sulfoxide (DMSO) at a concentration of 10–20 mg/mL. For preparation of spike megapools sets of 15-mer peptides overlapping by 10 amino acids were synthetized to span the entire SARS-CoV-2 protein of the ancestral Wuhan sequence and a selection of the SARS-CoV-2 variants [AlphaB.1.1.7), Beta (B.1.351), Gamma (P.1), B.1.1.519, Kappa (B.1.617.1), Delta (B.1.617.2), Lambda (C37), R1, Mu (B.1.621) and Omicron (B.1.1.529)]. The Megapools (MP) for each variant were created by pooling aliquots of the corresponding individual peptides and then performing a sequential lyophilization. The resulting lyocake was subsequently resuspended in DMSO at 1 mg/mL as previously described (Grifoni et al., 2020; Tarke et al., 2021a, 2021b).
Blood isolation and HLA typing
The LJI Clinical Core performed blood collection and sample processing based on SOPs previously established and described (Dan et al., 2021; Tarke et al., 2021a).
Whole blood was collected in heparin coated blood bags, and the cellular fraction was separated from plasma by a centrifugation at 1850 rpm for 15 minutes. The plasma was consequently collected and stored at -20°C for serology assays, while the cellular fraction underwent density-gradient sedimentation to obtain the PBMCs using Ficoll-Paque (Lymphoprep, Nycomed Pharma, Oslo, Norway)(Grifoni et al., 2020). Isolated PBMCs were stored in liquid nitrogen in cryopreserved cell recovery media containing 90% heat-inactivated fetal bovine serum (FBS; Hyclone Laboratories, Logan UT) and 10% DMSO (Gibco) until cellular assays were performed. HLA typing was performed by an ASHI-accredited laboratory at Murdoch University (Western Australia) for Class I (HLA A; B; C) and Class II (DRB1, DRB3/4/5, DQA1/DQB1, DPB1), as previously described(Tarke et al., 2021a) (Table S2).
SARS-CoV-2 serology and PSV neutralization assay
SARS-CoV-2 serology was performed for all plasma samples collected as previously described(Rydyznski Moderbacher et al., 2020). Briefly, 1 ug/mL SARS-CoV-2 spike (S) Receptor Binding Domain (RBD) was used to coat 96-well half-area plates (ThermoFisher Cat#3690), which were then incubated at 4°C overnight. After blocking the plates the next day at room temperature for 2 hours with 3% milk in phosphate buffered saline (PBS) and 0.05% Tween-20, the heat-inactivated plasma was added for an additional 90-minute incubation at room temperature, followed by incubation with the conjugated secondary antibody. Plates were read on the Spectramax Plate Reader at 450 nm using the SoftMax Pro. For data analysis of SARS-CoV-2 serology, the limit of detection (LOD) was defined as 1:3 while the limit of sensitivity (LOS) was established based on uninfected subjects, using plasma from normal healthy donors that did not receive COVID-19 vaccination.
The SARS-CoV-2 pseudovirus (PSV) neutralization assay was performed for timepoint 3 samples as previously described(Mateus et al., 2021). A monolayer of VERO cells (ATCC, Cat# CCL-81) was generated by seeding 2.5x104 cells in flat clear-bottom black 96-well plates (Corning, Cat# 3904). Pre-titrated recombinant virus for each variant were incubated with serially diluted human heat- inactivated plasma at 37°C for 1-1.5 hours. Confluent VERO cell monolayers were added and incubated for 16 hours at 37°C in 5% CO2 then fixed in 4% paraformaldehyde in PBS pH 7.4 (Santa Cruz, Cat# sc-281692) with 10 μg/ml Hoechst (Thermo Scientific, Cat#62249). Cells were imaged using a Cell Insight CX5 imager to quantify the total number of cells and infected GFP expressing cells to determine the percentage of infection. Neutralization titers (inhibition dose 50-ID50) were calculated using the One-Site Fit Log IC50 model in Prism 8.0 (GraphPad), and calibrated to WHO international standard (20/268). Samples that did not reach 50% inhibition at the lowest serum dilution of 1:20 were considered as non-neutralizing and were calibrated as 10.73 IU/mL.
Flow cytometry-based T cell assays
Activation Induced Marker (AIM) and Intra Cellular Staining (ICS) assays have been separately described in detail previously (Grifoni et al., 2020; Mateus et al., 2021; Tarke et al., 2021b). In this study, we performed both assays separately at timepoint 3, while we combined them for timepoints 1, 2, and 4. To assess the best protocol for AIM+ICS assay, we carried out the three assays in parallel in the same samples (Figure S1). The best assay configuration to retain AIM marker expression and simultaneously detect cytokines required the addition of CD137 antibody to culture as described in detail below. Figure S1 shows also the comparison of this AIM+ICS protocol with the classical AIM or ICS assays; no significant differences are observed among protocols, suggesting that the combined assay can be used to simultaneously detect AIM+ cells and the cytokine profile.
In all T cell assays, PBMCs were cultured in the presence of SARS-CoV-2-specific (ancestral or variant) MPs [1 μg/ml] in 96-well U-bottom plates at a concentration of 1x106 PBMC per well. As a negative control, an equimolar amount of DMSO was used to stimulate the cells in triplicate wells and phytohemagglutinin (PHA, Roche, 1μg/ml) stimulated cells were used as positive controls. After incubation for 24 hours at 37°C in 5% CO2, cells were either stained for AIM markers only or an additional incubation of 4 hours was carried out by adding Golgi-Plug containing brefeldin A, Golgi-Stop containing monensin (BD Biosciences, San Diego, CA), and in the case of the AIM+ICS assay combined CD137 APC antibody was additionally added in culture (2:100; Biolegend Cat# 309810). In all assays, cells were stained on their surface for 30 min at 4°C in the dark. For AIM assays, cells were then acquired directly, while for both ICS and AIM+ICS assays, cells were additionally fixed with 1% of paraformaldehyde (Sigma-Aldrich, St. Louis, MO), permeabilized, and blocked for 15 minutes followed by intracellular staining for 30 min at room temperature.
All samples were acquired on a ZE5 5-laser cell analyzer (Bio-Rad laboratories) and analyzed with FlowJo software (Tree Star Inc.). The gates for AIM or cytokine positive cells were drawn relative to the negative and positive controls for each donor. A representative example of the gating strategy for AIM, ICS or AIM+ICS assays is depicted in Figure S1 . Specifically, lymphocytes were gated, followed by single cells determination. T cells were gated for being positive to CD3 and negative for a Dump channel including in the same colors CD14, CD19 and Live/Dead staining. The CD3+CD4+ and CD3+CD8+ were further gated based on OX40+CD137+ and CD69+CD137+ AIM markers, respectively. For ICS, CD3+CD4+ and CD3+CD8+ cells were further gated based on a combination of each cytokine (IFNγ, TNFα, IL-2, Granzyme B) with CD40L or FSC-A, respectively (Figure S1). To the total cytokine response and T cell functionality was calculated from Boolean gating of single cytokines or Granzyme B that was applied to CD3+CD4+ or CD3+CD8 cells. In the resulting data generated from the AIM and ICS T cell assays, the background was removed from the data by subtracting the average of the % of AIM+ or Cytokine+ cells plated in triplicate wells stimulated with DMSO. The Stimulation Index (SI) was calculated by dividing the % of AIM+ cells after SARS-CoV-2 stimulation with the average % of AIM+ cells in the negative DMSO control. An SI greater than 2 and a LOS of 0.03% or 0.04% AIM+ CD4+ or CD8+ cells, respectively, after background subtraction was considered to be a positive response based on the median twofold standard deviation of T cell reactivity in negative DMSO controls. For ICS, CD4+ T cell responses were based on the expression of CD40L in combination with IFNγ, TNFα, IL-2 or Granzyme B, the sum of the double positive represents the overall CD4+Cytokine+. CD8+ T cell responses were based on expression of IFNγ, TNFα, IL-2 or Granzyme B. In both cases, each single and multiple positive cytokines were background subtracted individually and found positive only if fulfilling the criteria of an SI greater than 2 and a LOS of 0.005% ICS+ CD4+ T cells (all timepoints) or and a LOS of 0.02% or 0.01% ICS+CD8+ cells, for timepoints 2 and 3 or 4, respectively, after background subtraction. The LOS for ICS was considered to be a positive response based on the median twofold standard deviation of T cell reactivity in negative DMSO controls for all timepoints calculated on IFNγ. The multifunctional analyses for T cells were based on the sum of the multiple responses. To note, granzyme B was considered only if in combination with either CD40L or any other cytokine, while single positive granzyme B positive T cells were not considered in this analysis.
T cell epitope identification assays
To identify SARS-CoV-2-specific T cell epitopes, two different strategies for peptide testing were applied mirroring what was previously described (Tarke et al., 2021a). In both cases, epitopes were identified by AIM assay. In the context of the CD4+T cell responses, 15-mer peptides overlapping by 10 amino acids spanning entire SARS-CoV-2 ancestral spike protein were synthesized. All peptides were synthesized as crude material (TC Peptide Lab, San Diego, CA, San Diego, CA) and individually resuspended in dimethyl sulfoxide (DMSO) at a concentration of 20 mg/mL. A portion of the 15-mer peptides were pooled into smaller mesopools of ten peptides each. All pools were resuspended at 1 mg/mL in DMSO. Each donor was tested first with the smaller mesopools to reach the single positive 15 mer. In the context of the CD8+T cell responses, predicted spike peptides based on the individual HLA-A,-B and C typing were synthetized applying a cutoff of 4%ile using the NetMHCpan EL4.1 algorithm (Reynisson et al., 2020) implemented by the IEDB’s analysis resource(Dhanda et al., 2019; Vita et al., 2019). The predicted peptides were tested in the corresponding donors to identify the CD8 spike-specific epitopes. Hence, HLA restriction was inferred based on the HLA molecules expressed in the donor tested, and predicted HLA binding capacity to the allelic variant in question.
Flow cytometry-based B cell assays
Detection of antigen-specific B cells by flow cytometry was performed using B cell probes consisting of SARS-CoV-2 viral proteins conjugated with fluorescent streptavidin, as previously described by our group (Dan et al., 2021). Spike and RBD recombinant proteins used in this study are described in the key resources table. Initially, two separate flow cytometry panels were used to identify Spike or RBD variants among recipients of the four vaccine platforms studied here. A third and a fourth panel were used to compare Omicron-specific B cell responses with the other VOCs, in PBMCs from mRNA vaccines (mRNA-1273 and BNT162b2) recipients. To enhance specificity, identification of both WT spike and WT RBD B cells was performed using two fluorochromes for each protein, prior to gating on variant B cells. For that, biotinylated WT SARS-CoV-2 spike was incubated with Streptavidin in either BV711 (BioLegend, Cat# 405241) or BV421 (BioLegend, Cat# 405225) at a 20:1 ratio (∼6:1 molar ratio) for 1 hour at 4°C. In a separate panel, biotinylated WT RBD was also conjugated with streptavidin BV711 (BioLegend, Cat# 405241) or streptavidin PE-Cy7 (BioLegend, Cat# 405206) in a 2.2:1 ratio (∼4:1 molar ratio). The streptavidin-fluorochrome conjugates used to tetramerize the SARS-CoV-2 variant proteins in the first two panels are listed as follows: Alpha (B.1.1.7) spike BUV737 (BD bioscience, Cat# 612775), Alpha RBD BV785 (Biolegend, Cat# 613013); Beta (B.1.351) RBD, BUV615 (BD bioscience, Cat# 613013),Gamma (P.1) spike, BV785 (Biolegend, Cat# 405249), Gamma RBD, BV737(BD biosciences, Cat# 612775), Delta (B.1.617.2) spike and RBD, Alexa Fluor 647 (Thermo Fischer Scientific, Cat# S21374).
For the additional two panels employed to identify Omicron-specific B cells, the colors used for the variants for both RBD and Spike were: B.1.351 BUV615, B.1.1.529 BUV737, RBD.617.2. For the RBD panel, RBD B.1.1.7 labelled with BV785 was also included in the analysis.
Streptavidin PE-Cy5.5 (Thermo Fisher Scientific, Cat# SA1018) was used as a decoy probe to minimize background by eliminating SARS-CoV-2 nonspecific streptavidin-binding B cells. Seven million PBMCs were placed in U-bottom 96 well plates and stained with a solution consisting in 5μ of biotin (Avidity, catalog no. Bir500A) to avoid cross reactivity among probes, 20 ng of decoy probe, 416 ng of spike and 20.1 ng of RBD per sample, diluted in Brilliant Buffer (BD Biosciences, Cat# 566349) and incubated for 1 hour at 4°C, protected from light. After washing with PBS, cells from both spike and RBD panels were incubated with surface antibodies diluted in Brilliant Buffer, for 30 at 4°C, protected from light. Viability staining was performed using Live/Dead Fixable Blue Stain Kit (Thermo Fisher, Cat# L34962) diluted 1:200 in PBS and incubation at 4°C for 30 minutes. Acquisition was performed on Cytek Aurora and analyses were made using Flow Jo v. 10.7.1 (BD Biosciences). The frequency of Variants-specific memory B cells was expressed as a percentage of WT spike or RBD memory B cells (Singlets, Lymphocytes, Live, CD3– CD14– CD16– CD56–CD19+ CD20+ CD38int/–, IgD– and/or CD27+ spike or RBD BV711+, spike or RBD BV421+). PBMCs from a known positive control (COVID-19 convalescent subject) and an unexposed subject were included to ensure consistent sensitivity and specificity of the assay.
Quantification and statistical analysis
Data and statistical analyses were performed in FlowJo 10 and GraphPad Prism 8.4, unless otherwise stated. Statistical details of the experiments are provided in the respective figure legends and in each methods section pertaining to the specific technique applied. Data plotted in logarithmic scales are expressed as geometric mean. In all assays, fold-change was calculated as the ratio of the variant pool/ ancestral pool for samples with a positive ancestral pool response. Significance of fold-change decreases for each variant was assessed by Wilcoxon Signed Rank T test compared to a hypothetical median of 1.
Acknowledgments
This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and Department of Health and Human Services under contract No. 75N93021C00016 to A.S. and contract No. 75N93019C00065 to A.S. and D.W. A.T. was supported by a PhD student fellowship through the Clinical and Experimental Immunology Course at the University of Genoa, Italy. We thank Gina Levi and the LJI clinical core for assistance in sample coordination and blood processing. We gratefully thank the authors from the originating laboratories responsible for obtaining the specimens, as well as the submitting laboratories where the genome data were generated and shared via GISAID and on which this research is based. We would like to thank Vamseedhar Rayaprolu and Erica Ollmann Saphire for providing the recombinant SARS-CoV-2 receptor binding domain (RBD) protein used in the ELISA assay.
Author contributions
Conceptualization, A.G., S.C., and A.S.; data curation and bioinformatic analysis, A.G. and J.S.; formal analysis, A.T., A.G., Z.Z., C.H.C., and J.M.D.; funding acquisition, S.C., A.S., and D.W.; investigation, A.T., N.M., Z.Z., C.H.C., B.G., N.I.B., E.D.Y., R.d.S.A., and A.G.; resources, S.M. and E.P.; supervision, J.S., G.F., D.W., S.C., A.S., R.d.S.A., J.M.D., C.H.C., and A.G.; writing, A.G., S.C., and A.S.
Declaration of interests
A.S. is a consultant for Gritstone Bio, Flow Pharma, Arcturus Therapeutics, ImmunoScape, CellCarta, Avalia, Moderna, Fortress, and Repertoire. S.C. has consulted for GSK, JP Morgan, Citi, Morgan Stanley, Avalia NZ, Nutcracker Therapeutics, University of California, California State Universities, United Airlines, and Roche. All the other authors declare no competing interests. L.J.I. has filed for patent protection for various aspects of T cell epitope and vaccine design work.
Inclusion and diversity
We worked to ensure sex balance in the selection of non-human subjects. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Published: January 24, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2022.01.015.
Supplemental information
Data and code availability
All the data generated in this study are available in the published article and summarized in the corresponding tables, figures, and supplemental materials.
References
- Callaway E. Beyond Omicron: what's next for COVID's viral evolution. Nature. 2021;600:204–207. doi: 10.1038/d41586-021-03619-8. [DOI] [PubMed] [Google Scholar]
- Cameroni E., Saliba C., Bowen J.E., Rosen L.E., Culap K., Pinto D., De Marco A., Zepeda S.K., di Iulio J., Zatta F., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. bioRxiv. 2021 doi: 10.1101/2021.12.12.472269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreño J.M., Alshammary H., Tcheou J., Singh G., Raskin A., Kawabata H., Sominsky L., Clark J., Adelsberg D.C., Bielak D., et al. Activity of convalescent and vaccine serum against a B.1.1.529 variant SARS-CoV-2 isolate. medRxiv. 2021 doi: 10.1101/2021.12.20.21268134. [DOI] [PubMed] [Google Scholar]
- Cele S., Jackson L., Khan K., Khoury D.S., Moyo-Gwete T., Tegally H., Scheepers C., Amoako D., Karim F., Bernstein M., et al. SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. medRxiv. 2021 doi: 10.1101/2021.12.08.21267417. [DOI] [Google Scholar]
- Chakraborty C., Sharma A.R., Bhattacharya M., Agoramoorthy G., Lee S.-S. Evolution, mode of transmission, and mutational landscape of newly emerging SARS-CoV-2 variants. mBio. 2021;12 doi: 10.1128/mBio.01140-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho A., Muecksch F., Schaefer-Babajew D., Wang Z., Finkin S., Gaebler C., Ramos V., Cipolla M., Mendoza P., Agudelo M., et al. Anti-SARS-CoV-2 receptor-binding domain antibody evolution after mRNA vaccination. Nature. 2021;600:517–522. doi: 10.1038/s41586-021-04060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier A.Y., Brown C.M., McMahan K., Yu J., Liu J., Jacob-Dolan C., Chandrashekar A., Tierney D., Ansel J.L., Rowe M., et al. 2021. Immune responses in fully vaccinated individuals following breakthrough infection with the SARS-CoV-2 Delta variant in Provincetown, Massachusetts. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371 doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda S.K., Mahajan S., Paul S., Yan Z., Kim H., Jespersen M.C., Jurtz V., Andreatta M., Greenbaum J.A., Marcatili P., et al. IEDB-AR: immune epitope database-analysis resource in 2019. Nucleic Acids Res. 2019;47:W502–W506. doi: 10.1093/nar/gkz452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose N.A., Shen X., Schmidt S.D., O’Dell S., McDanal C., Feng W., Tong J., Eaton A., Maglinao M., Tang H., et al. Booster of mRNA-1273 strengthens SARS-CoV-2 omicron neutralization. medRxiv. 2021 doi: 10.1101/2021.12.15.21267805. [DOI] [Google Scholar]
- Gagne M., Corbett K.S., Flynn B.J., Foulds K.E., Wagner D.A., Andrew S.F., Todd J.-P.M., Honeycutt C.C., McCormick L., Nurmukhambetova S.T., et al. Protection from SARS-CoV-2 Delta one year after mRNA-1273 vaccination in rhesus macaques coincides with anamnestic antibody response in the lung. Cell. 2021;185:113–130.e15. doi: 10.1016/j.cell.2021.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. medRxiv. 2021 doi: 10.1101/2021.12.14.21267755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geers D., Shamier M.C., Bogers S., den Hartog G., Gommers L., Nieuwkoop N.N., Schmitz K.S., Rijsbergen L.C., van Osch J.A.T., Dijkhuizen E., et al. SARS-CoV-2 variants of concern partially escape humoral but not T-cell responses in COVID-19 convalescent donors and vaccinees. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abj1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R.R., Painter M.M., Apostolidis S.A., Mathew D., Meng W., Rosenfeld A.M., Lundgreen K.A., Reynaldi A., Khoury D.S., Pattekar A., et al. mRNA vaccination induces durable immune memory to SARS-CoV-2 with continued evolution to variants of concern. bioRxiv. 2021 doi: 10.1101/2021.08.23.457229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Sidney J., Vita R., Peters B., Crotty S., Weiskopf D., Sette A. SARS-CoV-2 human T cell epitopes: adaptive immune response against COVID-19. Cell Host Microbe. 2021;29:1076–1092. doi: 10.1016/j.chom.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Robertson D.L., COVID-19 Genomics UK (COG-UK) Consortium SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie K.M., Li H., Bedinger D., Schendel S.L., Dennison S.M., Li K., Rayaprolu V., Yu X., Mann C., Zandonatti M., et al. Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: A global consortium study. Science. 2021;374:472–478. doi: 10.1126/science.abh2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim S.S.A., Karim Q.A. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet. 2021;398:2126–2128. doi: 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeton R., Richardson S.I., Moyo-Gwete T., Hermanus T., Tincho M.B., Benede N., Manamela N.P., Baguma R., Makhado Z., Ngomti A., et al. Prior infection with SARS-CoV-2 boosts and broadens Ad26.COV2.S immunogenicity in a variant-dependent manner. Cell Host Microbe. 2021;29:1611–1619.e5. doi: 10.1016/j.chom.2021.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Huat N.K., Er Lim J.M., Gill U.S., de Alwis R., Tan N., Nan Toh J.Z., Abbott J.E., Usai C., Ooi E.E., Low J.G.H., et al. Differential immunogenicity of homologous versus heterologous boost in Ad26.COV2.S vaccine recipients. medRxiv. 2021 doi: 10.1101/2021.10.14.21264981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Iketani S., Guo Y., Chan J.F.-W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. 2021. Striking antibody evasion manifested by the omicron variant of SARS-CoV-2. bioRxiv. [DOI] [PubMed] [Google Scholar]
- Madelon N., Heikkilä N., Sabater Royo I., Fontannaz P., Breville G., Lauper K., Goldstein R., Grifoni A., Sette A., Siegrist C.-A., et al. Omicron-specific cytotoxic T-cell responses are boosted following a third dose of mRNA COVID-19 vaccine in anti-CD20-treated multiple sclerosis patients. medRxiv. 2021 doi: 10.1101/2021.12.20.21268128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J., Dan J.M., Zhang Z., Rydyznski Moderbacher C., Lammers M., Goodwin B., Sette A., Crotty S., Weiskopf D. Low-dose mRNA-1273 COVID-19 vaccine generates durable memory enhanced by cross-reactive T cells. Science. 2021;374 doi: 10.1126/science.abj9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo-González F., Soto J.A., González L.A., Fernández J., Duarte L.F., Schultz B.M., Gálvez N.M.S., Pacheco G.A., Ríos M., Vázquez Y., et al. Recognition of variants of concern by antibodies and T cells induced by a SARS-CoV-2 inactivated vaccine. Front. Immunol. 2021;12:747830. doi: 10.3389/fimmu.2021.747830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlcochova P., Kemp S.A., Dhar M.S., Papa G., Meng B., Ferreira I.A.T.M., Datir R., Collier D.A., Albecka A., Singh S., et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599:114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S.P., Day T., Arino J., Colijn C., Dushoff J., Li M., Mechai S., Van Domselaar G., Wu J., Earn D.J.D., Ogden N.H. The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr. Biol. 2021;31:R918–R929. doi: 10.1016/j.cub.2021.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., Arlehamn C.S.L., Schulten V., Westernberg L., Sidney J., Peters B., Sette A. Experimental validation of the RATE tool for inferring HLA restrictions of T cell epitopes. BMC Immunol. 2017;18:20. doi: 10.1186/s12865-017-0204-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.-H., Porrot F., Staropoli I., Lemoine F., et al. Considerable escape of SARS-CoV-2 variant Omicron to antibody neutralization. bioRxiv. 2021 doi: 10.1101/2021.12.14.472630. [DOI] [PubMed] [Google Scholar]
- Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48:W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou C., Keeton R., Moyo-Gwete T., Hermanus T., Kgagudi P., Baguma R., Valley-Omar Z., Smith M., Tegally H., Doolabh D., et al. Escape from recognition of SARS-CoV-2 Beta variant spike epitopes but overall preservation of T cell immunity. Sci. Transl. Med. 2021 doi: 10.1126/scitranslmed.abj6824. https://www.science.org/doi/abs/10.1126/scitranslmed.abj6824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F., Muecksch F., Weisblum Y., Silva J.D., Bednarski E., Cho A., Wang Z., Gaebler C., Caskey M., Nussenzweig M.C., et al. Plasma neutralization properties of the SARS-CoV-2 Omicron variant. medRxiv. 2021 doi: 10.1101/2021.12.12.21267646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal A., Barba-Spaeth G., Fernández I., Broketa M., Azzaoui I., de La Selle A., Vandenberghe A., Fourati S., Roeser A., Meola A., et al. mRNA vaccination of naive and COVID-19-recovered individuals elicits potent memory B cells that recognize SARS-CoV-2 variants. Immunity. 2021;54:2893–2907.e5. doi: 10.1016/j.immuni.2021.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal A., Broketa M., Meola A., Barba-Spaeth G., Fernández I., Fourati S., Azzaoui I., de La Selle A., Vandenberghe A., Roeser A., et al. Immune escape of SARS-CoV-2 Omicron variant from mRNA vaccination-elicited RBD-specific memory B cells. bioRxiv. 2021 doi: 10.1101/2021.12.21.473528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatatos L., Czartoski J., Wan Y.H., Homad L.J., Rubin V., Glantz H., Neradilek M., Seydoux E., Jennewein M.F., Maccamy A.J., et al. mRNA vaccination boosts cross-variant neutralizing antibodies elicited by SARS-CoV-2 infection. Science. 2021 doi: 10.1126/science.abg9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A.T., Linster M., Tan C.W., Le Bert N., Chia W.N., Kunasegaran K., Zhuang Y., Tham C.Y.L., Chia A., Smith G.J.D., et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021;34:108728. doi: 10.1016/j.celrep.2021.108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Sidney J., Kidd C.K., Dan J.M., Ramirez S.I., Yu E.D., Mateus J., da Silva Antunes R., Moore E., Rubiro P., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021;2:100204. doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Sidney J., Methot N., Yu E.D., Zhang Y., Dan J.M., Goodwin B., Rubiro P., Sutherland A., Wang E., et al. Impact of SARS-CoV-2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep. Med. 2021;2:100355. doi: 10.1016/j.xcrm.2021.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uriu K., Kimura I., Shirakawa K., Takaori-Kondo A., Nakada T.-A., Kaneda A., Nakagawa S., Sato K., Genotype to Phenotype Japan (G2P-Japan) Consortium Neutralization of the SARS-CoV-2 Mu variant by convalescent and vaccine serum. N. Engl. J. Med. 2021;385:2397–2399. doi: 10.1056/NEJMc2114706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R., Wheeler D.K., Sette A., Peters B. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019;47:D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky R.P., Walke H.T., Fauci A.S. SARS-CoV-2 variants of concern in the United States-challenges and opportunities. JAMA. 2021;325:1037–1038. doi: 10.1001/jama.2021.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Xia H., Xie X., Kurhade C., Machado R.R.G., Weaver S.C., Ren P., Shi P.-Y. Neutralization against Omicron SARS-CoV-2 from previous non-Omicron infection. bioRxiv. 2021 doi: 10.1101/2021.12.20.473584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data generated in this study are available in the published article and summarized in the corresponding tables, figures, and supplemental materials.