Abstract
An important gap in our understanding of the epidemiology of amebiasis is what determines the outcome of Entamoeba histolytica infections. To investigate the possible existence of invasive and noninvasive strains as one factor, the ability to differentiate individual isolates of E. histolytica is necessary. Two new loci containing internal repeats, locus 1-2 and locus 5-6, have been isolated. Each contains a single repeat block with two types of related direct repeats arranged in tandem. Southern blot analysis suggests that both loci are multicopy and may themselves be arranged in tandem arrays. Three other previously reported, internally repetitive loci containing at least two repeat blocks each with one or more related repeat units were also investigated. PCR was used to study polymorphism at each of these loci, which was detected to various degrees in each case. Variation was seen in the total number of bands obtained per isolate and their sizes. Nucleotide sequence comparison of loci 1-2 and 5-6 in five axenic isolates revealed differences in the number of repeat units, which correlated with the observed PCR product size variation, and in repeat sequence. Use of multiple loci collectively allowed differentiation of a majority of the 13 isolates studied, and we believe that these loci have the potential to be used as polymorphic molecular markers for investigating the epidemiology of E. histolytica and the potential existence of genetically distinct invasive and noninvasive strains.
The acceptance of Entamoeba histolytica and Entamoeba dispar as distinct species (2, 11) has had a major impact on our views of amebiasis, in particular its clinical management and epidemiology. It is likely that at least 90% of the infections previously ascribed to E. histolytica are actually E. dispar, while only the remaining 10% are infected with E. histolytica in its new sense. However, it also appears that many E. histolytica infections never progress to become symptomatic and are spontaneously lost. This observation raises some important questions. Are the organisms that produce invasive, symptomatic disease genetically distinct from those that give rise to asymptomatic infections? Or do all E. histolytica isolates have the potential to become invasive? Do certain invasive isolates show tropism for specific organs, with some preferentially ending up in the intestinal wall while others reach extraintestinal sites? To address the possibility of a relationship between parasite variation and infection outcome the ability to differentiate isolates of E. histolytica is necessary.
Our present knowledge of intraspecies variation in E. histolytica is limited. Isoenzyme analysis provided the first markers (25), but it now appears that isoenzyme patterns are not fixed (5) and therefore that many ‘zymodeme’ assignments are unreliable (16). A limited number of DNA markers have been shown to exhibit intraspecies diversity. Variation has been observed in the number of rRNA transcription units present on the extrachromosomal ribosomal DNA circles; only one rRNA gene copy has been seen in some strains, while the majority have two (27). Variation has also been detected in the noncoding families of short tandem repeats found both upstream and downstream of the rRNA genes (20, 21, 26). However, variability in the occurrence and instability in the length of some of these sequences limits their use for isolate identification (4). PCR amplification of the Strain-Specific Gene (6) or Tr (27), which is present upstream of one rRNA transcription unit and contains tandemly repeated internal elements, has revealed considerable variation in the number of repeats among strains of E. histolytica (8). However, the complete absence of this locus in certain strains (27) makes it a poor candidate for intraspecies typing.
At present, the most polymorphic gene of E. histolytica is that encoding the serine-rich E. histolytica protein (SREHP or K2) a surface antigen with tandem 8- and 12-amino-acid repeats (17, 28). Repeat number, sequence, and restriction enzyme site polymorphisms have been reported among different E. histolytica isolates (8, 14). However, more than one-third of the isolates tested gave the same restriction fragment pattern (8). The chitinase gene also encodes tandem repeats of a degenerate 7-amino-acid sequence (10), and a report on the use of chitinase repeat polymorphisms to distinguish isolates of E. histolytica has been published recently (14). However, there still exists a need for additional reliable polymorphic E. histolytica DNA markers.
The use of microsatellite locus analysis has gained considerable popularity as a tool for detecting intra- and interspecies variations in a number of organisms, including protozoan parasites such as Trypanosoma (22), Leishmania (24), and Plasmodium (1) spp. Using a method designed to isolate microsatellite loci, we have obtained two new polymorphic DNAs containing tandemly repeated sequences from E. histolytica. We present here the preliminary characterization of the two loci and the interstrain variations they display. In addition, three other loci showing the presence of tandemly repeated sequences have been studied for their potential as polymorphic markers for use in investigating the molecular epidemiology of E. histolytica.
MATERIALS AND METHODS
E. histolytica isolates.
Except for HM-1:IMSS clone 9, the axenic isolates were provided by John Ackers (London School of Hygiene and Tropical Medicine) (Table 1). All axenic isolates were cultured in the casein-free medium YI-S (12) with 15% heat-inactivated adult bovine serum (Sigma-Aldrich).
TABLE 1.
Origin of E. histolytica isolates
| Isolate | Origin | Culture type | Clinical information |
|---|---|---|---|
| HM-1:IMSS clone 9 | Mexico | Axenic | Isolated from rectal ulcer; patient with dysentery |
| 200:NIH | (uncertain) | Axenic | Patient with amebic dysentery |
| H-303:NIH | VietNam (?) | Axenic | Patient with amebic empyema and dysentery |
| IULA:0593:2 | Venezuela | Axenic | Patient with amebic colitis |
| IULA:1092:1 | Venezuela | Axenic | Patient with amebic colitis |
| 8691 | Bangladesh | Xenic | Patient with amebic colitis |
| 4530 | Bangladesh | Xenic | Patient with amebic colitis |
| 1320300 | Bangladesh | Xenic | Patient with amebic colitis |
| 48286 | Bangladesh | Xenic | Patient with amebic colitis |
| 2596 | South Africa | Xenic | Patient asymptomatic; serology positive by antigen gel diffusion test |
| 26.253C | South Africa | Xenic | Patient asymptomatic; serology positive by antigen gel diffusion test |
| 37.0C | South Africa | Xenic | Convalescent amebic liver abscess patient; serology positive by antigen gel diffusion test |
| 39.384C | South Africa | Xenic | Patient asymptomatic; serology positive by antigen gel diffusion test |
Xenic isolates were obtained from two sources (Table 1). Four samples were from Rashidul Haque of the International Centre for Diarrhoeal Disease Research, Bangladesh, via Aura Aguirre (London School of Hygiene and Tropical Medicine), while four others were provided by Terry Jackson of the Medical Research Council of South Africa, Durban. The South African isolates were from a patient who had recovered from amebic liver abscess (39.0C) or close family contacts of such patients who were asymptomatic at the time of isolation. All xenic strains were originally isolated in Robinson's medium (23); there is no evidence that culture conditions or media have any effect on the markers studied.
Isolation of nucleic acids.
DNA was isolated as previously described (7, 9), dissolved in 10 mM Tris-Cl (pH 8.5) and passed over a Microspin S-200 HR column (Amersham Pharmacia Biotech, Inc). RNA was removed by the addition of RNase A (Promega) to 0.05 μg ml−1.
Isolation of repeated DNA containing sequences.
A nonradioactive method designed for rapid isolation of microsatellite sequences (13, 22) was adapted. Genomic DNA of isolate HM-1:IMSS (ca. 500 ng) was digested for 2 h with a restriction enzyme, either AluI or RsaI (10 U/20-μl reaction) (Gibco-BRL), followed by incubation at 65°C for 15 min to inactivate the enzyme and passage through a S-200 column to remove the salts.
5′-Phosphorylated 24-mer (5′-pAGTCCGGATCCAAGCAAGAGCACA-3′) and nonphosphorylated 20-mer (5′-CTCTTGCTTGGATCCGGACT-3′) oligonucleotides with overlapping complementary sequences containing a BamHI site were used to generate an adapter. Then, 2.5 pmol of adapter was ligated to approximately 250 ng of digested DNA with T4 DNA ligase at 14°C.
Ligated fragments (equivalent to ca. 50 ng of DNA) were annealed to 20 pmol of a biotinylated microsatellite oligonucleotide [GATGATCCGACGCAT(CA)12, GATGATCCGACGCAT(CT)12, (CAA)12, (CTT)12, (CAT)12, (CTA)12, or (TAA)12] by denaturing at 95°C for 10 min and annealing at 60°C for 1 min; the hybrids were then bound to 100 μg of streptavidin-coated magnetic beads (Dynabeads KilobaseBinder kit; Dynal). Following incubation at room temperature for 3 h the Dynabead-DNA complexes were washed twice (10 mM Tris-Cl, pH 7.5; 1 mM EDTA; 2.0 M NaCl) and resuspended in 50 μl of TE buffer (10 mM Tris-Cl, pH 8.0; 1 mM EDTA; pH 8.0). The captured product was used as a template for PCR amplification using the 20-mer adapter oligonucleotide under standard conditions. Amplified products were analyzed on a 1.8% agarose gel (Gibco-BRL) using amplified adapter-ligated but unselected digested DNA as a control.
After electrophoresis, PCR products appearing to be enriched by the selection process were gel purified and cloned into the vector pGEM-T Easy (Promega). Recombinant plasmids were sequenced using an ABI PRISM 377 (Perkin-Elmer) and Thermo-Sequenase II dye terminator cycle sequencing kit (Amersham Pharmacia Biotech).
PCR product size polymorphisms and nucleotide sequence comparison.
Primers were designed from repeat flanking region sequences of all the loci. The genomic DNA of E. histolytica was amplified using the primers listed in Table 2 and 30 cycles of 1 min at 94°C, 1 min at the primer-dependent annealing temperature, and 2 min at 72°C, with a final extension of 5 min at 72°C. Amplified products were analyzed using 2.4% NuSieve 3:1 agarose gels (FMC) in 1× Tris-boric acid-EDTA buffer (TBE).
TABLE 2.
Oligonucleotide primers
| Primer | Primer sequence (5′ to 3′) |
|---|---|
| R1 | CTG GTT AGT ATC TTC GCC TGT |
| R2 | CTT ACA CCC CCA TTA ACA AT |
| R3 | GCT ATG GTC GGT ATC GAT ATC |
| R4 | CCT TAG GTC ACT GGT TCG AA |
| R5 | CTA AAG CCC CCT TCT TCT AT |
| R5A | CTA AAG CCC CCT TCT TCT ATA ATT |
| R6 | GTG CTA ATA ACG CCA GGG TC |
| R6A | CTC AGT CGG TAG AGC ATG GT |
| R7 | CTT TAC TTC TCT TTT ACC ACG |
| R8 | CGT GGT AAA AGA GAA GTA AAG |
| R9 | CTA CAT CTA CAG TCC TCC GCT |
| R10 | CTT ACT TCT CTT TAC CAC GAC |
| R11 | GTC GTG GTA AAG AGA AGT AAG |
| R16 | AAG CTT CCT TAG CTC AGC TG |
| R17 | TAA AAG GGG GAA GAA TAG GAA |
| R18 | GGT TTC ATG GTG TAG TTG GT |
| R19 | ACC AAC TAC ACC ATG AAA CC |
PCR products from all five axenic isolates of E. histolytica were cloned pGEM-T Easy vector (Promega) and sequenced as described above.
Southern blot analysis.
Genomic DNA of isolate HM-1:IMSS clone 9 was digested overnight with 10 U each of restriction enzymes AluI, DdeI, DraI, EcoRI, and RsaI (Gibco-BRL or MBI Fermentas), and fragments were separated by electrophoresis using 0.8% agarose gels in 1× TBE buffer and transferred to BiodyneA membranes (Gibco-BRL) using standard methods. [α-32P]dCTP-labeled double-stranded DNA probes were prepared by using the Rediprime II random prime labeling system (Amersham Pharmacia Biotech). Filters were hybridized overnight at 65°C in a solution of 1 M NaCl–1% sodium dodecyl sulfate–10% dextran sulfate and then washed to a final stringency of 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate at 65°C before autoradiography at −70°C.
The nucleotide sequence data reported here have been submitted to the GenBank database under accession numbers AF276055 to AF276065.
RESULTS
Isolation of repeated DNA containing sequences from the E. histolytica genome.
To try and obtain DNA fragments containing microsatellites, we employed a nonradioactive method based on affinity capture of single-stranded restriction fragments annealed to biotinylated microsatellite oligonucleotides, with attachment to streptavidin-coated magnetic beads (Dynal), followed by adapter-mediated PCR (13, 22). A total of twenty two PCR fragments ranging in size from 250 to 700 bp were gel purified from the total amplification products of AluI or RsaI restriction fragments annealed to one of seven biotinylated oligonucleotides. These fragments were chosen on the basis of their apparent enrichment compared to control amplification products and were cloned, sequenced, and examined for the presence of microsatellites.
No products contained sequences corresponding to the microsatellite oligonucleotides used in their capture. Furthermore, the majority of the sequences did not reveal any tandemly repeated DNAs. However, two clones (clone 1 and clone 4) derived from an approximately 480-bp fragment, obtained from AluI restriction fragments annealed to the (CTT)12 oligonucleotide, showed the presence of internal tandem repeats. The repeats seen in clones 1 and 4 were distinct. Two other clones (clone 1′ and clone 5) derived from AluI restriction fragments annealed to (TAA)12 contained the same type of repeats as clone 4. Clones 1′ and 5 contained fragments of approximately 480 and 450 bp, respectively. Further analysis was carried out on clone 1, which represents locus 5-6, and clone 4, which represents locus 1-2.
Characterization of loci 1-2 and 5-6.
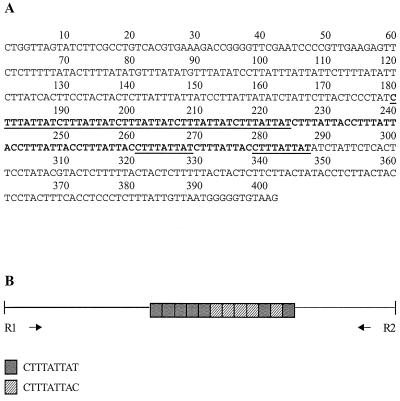
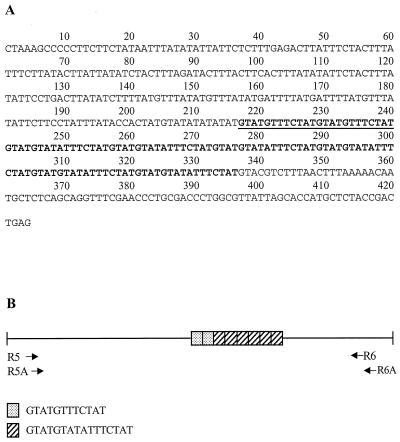
The complete sequence of the locus 1-2 clone (Fig. 1A), not including the adapter sequence, is 402 bp long and contains a single repeat block with two related direct repeats arranged in tandem (Fig. 1B). In addition to the major repeat block, tandem duplications of 8 to 12 bp are present in the flanking regions. The complete sequence of the locus 5-6 clone (Fig. 2A), not including the adapter sequence, is 424 bp long and contains a single repeat block (Fig. 2B). As in locus 1-2, other tandem duplications in the regions flanking the repeat block are also evident. BLAST search results revealed no identity of either locus 1-2 or locus 5-6 to any previously reported E. histolytica sequences.
FIG. 1.
Locus 1-2. (A) Nucleotide sequence. The main block of internal tandem repeats is in boldface. Underlined regions indicate one of the two types of repeat units. (B) Schematic representation. The two types of internal tandem repeats and their arrangement with respect to each other are shown. Tandem duplications in the flanking regions are not shown. The positions of the amplification primers are indicated.
FIG. 2.
Locus 5-6. (A) Nucleotide sequence. The main block of internal tandem repeats is in boldface. Underlined regions indicate one of the two types of repeat units. (B) Schematic representation. The two types of internal tandem repeats and their arrangement with respect to each other is shown. Tandem duplications in the flanking regions are not shown. The positions of the amplification primers are indicated. Two primer pairs were designed for locus 5-6 (Table 2). Amplification products generated by primers R5 and R6 were cloned, sequenced, and aligned for intrastrain nucleotide sequence comparisons (Fig. 4B), while the primer pair R5A-R6A was used for studying interstrain PCR product size polymorphisms (Fig. 3B).
PCR product size polymorphisms at loci 1-2 and 5-6.
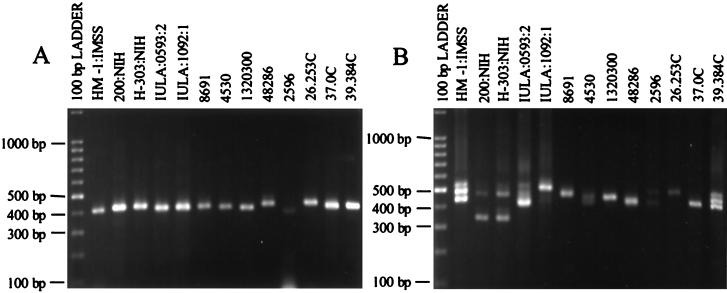
Primers were designed in the regions flanking the repetitive blocks for both locus 1-2 and locus 5-6, and the PCR amplification products were analyzed on 2.4% NuSieve agarose gels to look for fragment size polymorphism among the 13 E. histolytica isolates (Fig. 3).
FIG. 3.
Polymorphic DNA analysis of Entamoeba histolytica isolates. (A) Locus 1-2. Amplification products were generated using primers R1 and R2 at an annealing temperature of 53°C. (B) Locus 5-6. Amplification products were generated using primers R5A and R6A at an annealing temperature of 56°C. Isolate origins: HM-1:IMSS (Mexico); 200:NIH (uncertain); H-303:NIH (VietNam); IULA:1092:1 and IULA:0593:2 (Venezuela); 8691, 4530, 1320300, and 48286 (Bangladesh); 2596, 26.253, 37.0C, and 39.384C (South Africa).
Amplification of locus 1-2 gave the expected product of ca. 400 bp in isolate HM-1:IMSS clone 9 (Fig. 3A). All of the E. histolytica isolates gave a single major product. The four South African isolates gave the most variable patterns.
Amplification of locus 5-6 gave the expected product of ca. 420 bp in isolate HM-1:IMSS clone 9 (Fig. 3B), but two additional bands of ca. 480 and 520 bp were also seen. This locus is highly polymorphic. Variation is seen in the total number of bands per isolate and their sizes even within the same geographic area.
Nucleotide sequence analysis and characterization of the observed size polymorphism.
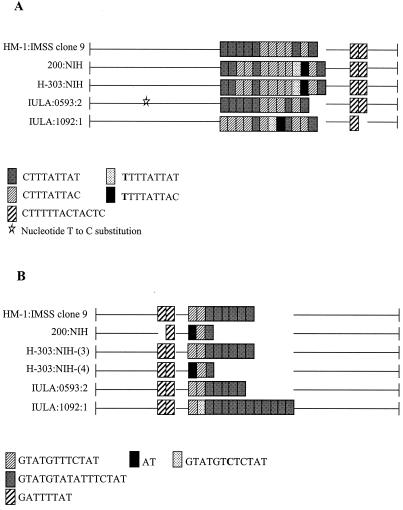
In order to study the underlying nature of the observed size polymorphisms, the amplification products of all five axenic isolates at locus 1-2 and locus 5-6 were cloned and sequenced. This analysis revealed differences in the number and sequence of the repeat units, as well as sequence variation in the regions flanking the repeat blocks (Fig. 4).
FIG. 4.
Schematic representation of locus structure in five axenic isolates of E. histolytica. Variations in number, sequence, and arrangement of repeat units are shown. Gaps have been introduced to optimize alignment. (A) Locus 1-2. (B) Locus 5-6.
There was very little variation in the total number of repeat units among the five samples at locus 1-2 (Fig. 4A). This is consistent with the slight differences in PCR product size observed in Fig. 3A. However, considerable variation existed between the isolates in the relative numbers of repeat units of type 1 versus type 2. In contrast to locus 1-2, there was considerable variation in the total number of units of one repeat type among the five strains at locus 5-6 (Fig. 4B). This high degree of variation is also reflected in the PCR product size comparison (Fig. 3B).
The locus 5-6 amplification products from isolate H-303:NIH revealed two distinct fragments of ca. 320 and 450 bp, respectively. Cloning of the PCR products from this locus also resulted in two inserts which differed in size by ca. 100 bp [designated H-303:NIH-(3) and H-303:NIH-(4); Fig. 4B].
Shared single-base alterations within the repeat blocks were seen at two positions in isolates 200:NIH and H-303:NIH at locus 1-2 (Fig. 4A). Another two single base changes were seen only within the repeat block of isolate IULA:1092:1 at locus 1-2, and this isolate was also missing a single copy of a 12-bp duplication in the 3′-flanking region. A single base change in the 5′-flanking region was present in isolate IULA:0593:2.
At locus 5-6 a single base change was seen within the repeat block (Fig. 4B) for isolate IULA:1092:1. Additionally, both isolate 200:NIH and H-303:NIH-(4) appeared to be missing the initial 10 bp of the first 12-bp repeat unit (GTATGTTTCTAT). A difference was also evident in the flanking regions of the repeat units in that isolate 200:NIH was missing a single copy of an 8-bp tandem duplication at the 5′ end. While it is possible that single nucleotide differences are PCR amplification artifacts, it is highly unlikely that shared nucleotide differences among isolates and repeats or repeat number variations could have this origin.
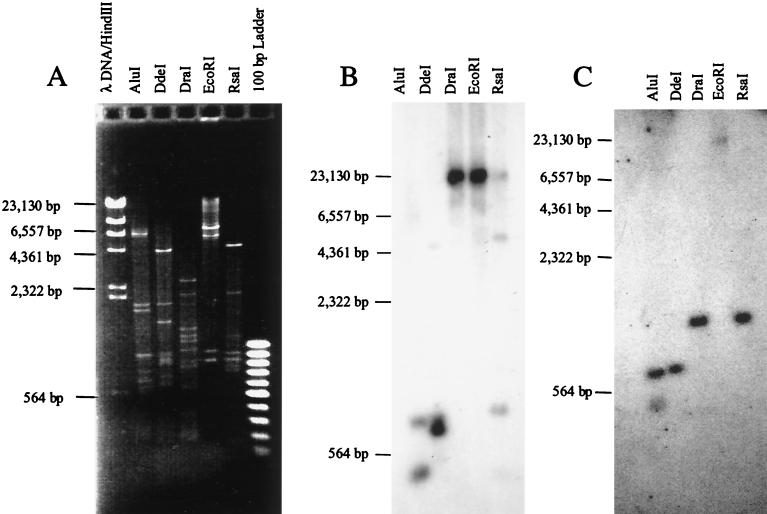
Southern blot analysis.
Southern hybridization analysis was performed using HM-1:IMSS clone 9 genomic DNA digested with five enzymes (Fig. 5) and either locus 1-2 or locus 5-6 as specific probes. The specificity of the probes was ensured by using PCR products produced from plasmid DNAs (clone 4 and clone 1, respectively).
FIG. 5.
Southern blot analysis. (A) Genomic DNA of E. histolytica isolate HM-1:IMSS digested with restriction enzymes and stained with ethidium bromide. (B) Blot of gel in panel A hybridized with a locus 1-2 specific 32P-labeled probe. (C) Blot of gel in panel A hybridized with a locus 5-6 specific 32P-labeled probe. Some of the faint bands seen in Fig. 5B may result from slight cross-hybridization to fragments of the abundant extrachromosomal circular DNA seen in the ethidium bromide-stained gel (Fig. 5A).
With the locus 1-2 specific probe, the band of ca. 400 bp in the AluI lane (Fig. 5B) was expected since the clone was obtained from an AluI restriction fragment of about the same size. It was a surprise to find that the probe gave intense hybridization signals at ⩾23 kb with the DraI-digested DNA since this enzyme cuts frequently in E. histolytica DNA and usually produces much smaller fragments, as seen in Fig. 5A. It is notable that AluI, DdeI, and RsaI give major fragments of about the same size and that DraI and EcoRI both give very large fragments.
Similarly, a band of ca. 400 bp was expected in the AluI lane with the locus 5-6 specific probe (Fig. 5C), since the clone was obtained from an AluI restriction fragment of about this size. Once again hybridization with this probe gave major fragments of about the same size with AluI and DdeI and with DraI and RsaI. EcoRI again produces a very large fragment of ⩾23 kb. Taken together, these data indicate that loci 1-2 and 5-6 exist in long tandem arrays.
Characterization of other loci containing internal repeats.
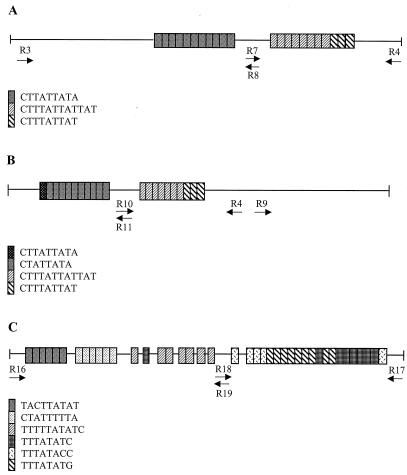
A number of other DNA elements containing internal tandem repeats have been reported in E. histolytica. No attempts have been made to study their potential for the detection of intraspecies polymorphisms. We selected three of these internally repetitive loci for study: a 978-bp element described by Michel et al. (19; GenBank accession number M77091; our designation, locus 3-4), a 931-bp DNA element isolated by J. Rosales-Encina and D. Eichinger (personal communication; GenBank accession number AF265348; our designation, locus 9-4), and a 964-bp element reported by Huang et al. (15; our designation, locus 16-17). Schematic representations of the repeat arrangements seen at these loci are given in Fig. 6A, B, and C, respectively.
FIG. 6.
Schematic representation of repeat arrangements at three other loci. (A) Locus 3-4. (B) Locus 9-4. (C) Locus 16-17. Only the major blocks of internal tandem repeats are shown. The positions of the primers for whole- and half-locus amplification are shown for all three loci.
There is a high degree of identity between the two repeat blocks of locus 3-4 and those of locus 9-4. The ten CTATTATA tandem repeats of locus 9-4 differ from the 11 CTTATTATA tandem repeats of locus 3-4 only in the absence of a single nucleotide (T) at the second position of each unit. The repeat unit CTTTATTATTAT in locus 9-4 is identical to the 12-bp repeat units of locus 3-4 with the only difference being the total number of units seen, i.e., locus 3-4 has eight units, while locus 9-4 has only seven. In fact, this high degree of identity between the two loci is apparent in the flanking regions as well. The sequences from positions 1 to 540 and positions 541 to 931 of locus 9-4 are very similar to the nucleotide stretches spanning positions 401 to 977 and positions 1 to 400 in locus 3-4, respectively (data not shown).
On comparing the sequences of loci 1-2 and 5-6 with those of loci 3-4 and 9-4 we find that the repeat unit CTTTATTAT, which occurs a total of seven times in locus 1-2, is identical to the three 9-bp units present in the second repeat blocks of both loci 3-4 and 9-4. The repeat units of locus 5-6, however, were quite unique, as are the repeat flanking regions of both loci. The nucleotide sequence of locus 16-17 is completely different from that of the other loci. There are six major types of internal repeats, with some being arranged in tandem only, while others exist as both tandem and solitary copies (Fig. 6C). Besides these, duplications of 5 to 8 bp are also seen interspersed among these repeats (not shown).
PCR product size polymorphisms at loci 3-4, 9-4 and 16-17.
Primers were designed to amplify all three loci, and the products were analyzed on 2.4% NuSieve agarose gels to look for size polymorphisms among the 13 E. histolytica isolates. In each case primers were also designed in the regions between the two main repeat blocks to look additionally for size variations in each half of the locus (Fig. 6).
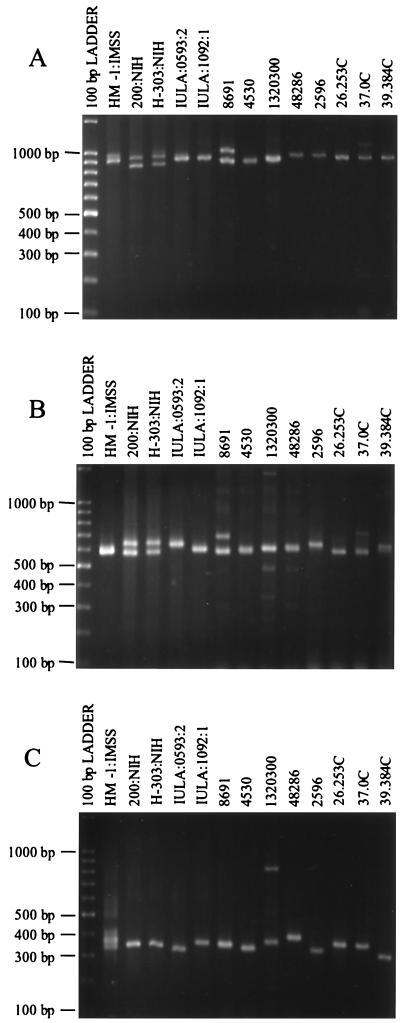
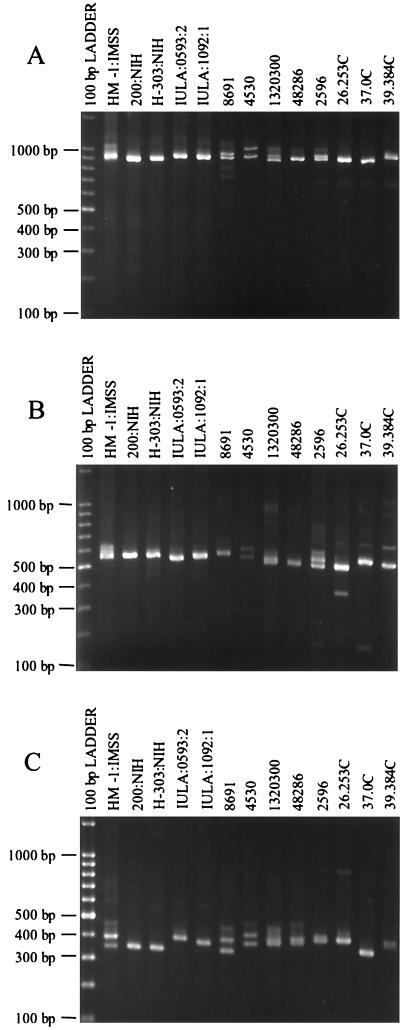
Isolate HM-1:IMSS clone 9 gave a double amplification product of ca. 900 bp at locus 3-4 (Fig. 7A). Most E. histolytica isolates gave single major products with little size variation. Isolates 200:NIH and H-303:NIH, however, show a second band of equal intensity at ca. 850 bp, while isolates 8691 and 37.0C display a second band of ca. 1 kb. Amplification of the two half-loci presented a very different pattern, with much more variation seen than with the whole locus (Fig. 7B and 7C).
FIG. 7.
Polymorphic DNA analysis of E. histolytica isolates. (A) Locus 3-4. Amplification products were generated using primers R3 and R4 at an annealing temperature of 55°C. (B) Half-locus 3-8. Amplification products were generated using primers R3 and R8 at an annealing temperature of 50°C. (C) Half-locus 7-4. Amplification products were generated using primers R7 and R4 at an annealing temperature of 50°C. Isolate origins: HM-1:IMSS (Mexico); 200:NIH (uncertain); H-303:NIH (VietNam); IULA:1092:1 and IULA:0593:2 (Venezuela); 8691, 4530, 1320300, and 48286 (Bangladesh); 2596, 26.253, 37.0C, and 39.384C (South Africa).
Amplification of locus 9-4 also gave two products between ca. 900 bp and 1 kb in isolate HM-1:IMSS clone 9 (Fig. 8A). Isolates 200:NIH and H-303:NIH again show two bands of equal intensity. As before, amplification of both half-loci (Fig. 8B and C) produced a greater variety of banding patterns than was seen at the whole locus.
FIG. 8.
Polymorphic DNA analysis of Entamoeba histolytica isolates. (A) Locus 9-4. Amplification products were generated using primers R9 and R4 at an annealing temperature of 55°C. (B) Half-locus 9-11. Amplification products were generated using primers R9 and R11 at an annealing temperature of 50°C. (C) Half-locus 10-4. Amplification products were generated using primers R10 and R4 at an annealing temperature of 50°C. Isolate origins: HM-1:IMSS (Mexico); 200:NIH (uncertain); H-303:NIH (VietNam); IULA:1092:1 and IULA:0593:2 (Venezuela); 8691, 4530, 1320300, and 48286 (Bangladesh); 2596, 26.253, 37.0C, and 39.384C (South Africa).
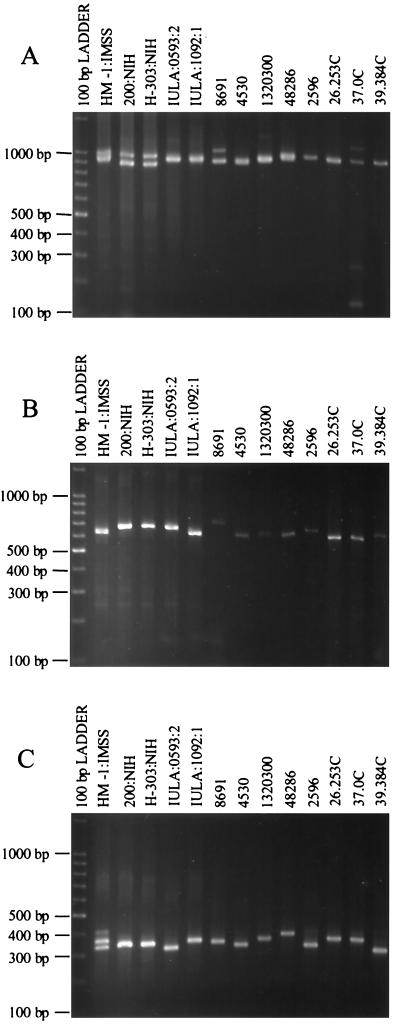
Amplification of locus 16-17 gave the expected product of ca. 900 bp in isolate HM-1:IMSS clone 9 (Fig. 9A). Many of the E. histolytica isolates gave single major products with little size variation among them, although isolate 4530 gave two clear products of equal intensity and certain others gave two bands very close in size. Amplification of the two half-loci again produced a highly polymorphic array of bands (Fig. 9B and C).
FIG. 9.
Polymorphic DNA analysis of E. histolytica isolates. (A) Locus 16-17. Amplification products were generated using primers R16 and R17 at an annealing temperature of 55°C. (B) Half-locus 16-19. Amplification products were generated using primers R16 and R19 at an annealing temperature of 54°C. (C) Half-locus 18-17. Amplification products were generated using primers R18 and R17 at an annealing temperature of 54°C. Isolate origins: HM-1:IMSS (Mexico); 200:NIH (uncertain); H-303:NIH (VietNam); IULA:1092:1 and IULA:0593:2 (Venezuela); 8691, 4530, 1320300, and 48286 (Bangladesh); 2596, 26.253, 37.0C, and 39.384C (South Africa).
DISCUSSION
We were unsuccessful in our attempt to clone microsatellite loci using a modification of a method that was successful in other organisms. We did, however, isolate two novel loci containing internal tandem repeats (1-2 and 5-6). Whether this reflects an absence or a reduced population of the classical di- and trinucleotide microsatellites in the E. histolytica genome or simply their relative abundance is at present too early to say. A (GA)27 stretch in an expressed sequence tag has been reported by Azam et al. (3), suggesting that microsatellites do exist in this organism. Three other, previously reported internally repetitive loci were also studied.
The genomic organization of the loci was investigated by Southern blotting. The large fragments generated by DraI and/or EcoRI detected with the locus 1-2 and 5-6 specific probes and the presence of major fragments of the same size with two or more restriction enzymes suggests that both loci are arranged in long tandem arrays. The fact that some enzymes generate very large fragments also was reported by Michel et al. (19) for locus 3-4, and these authors also suggested that it was tandemly arrayed. Similarly, Southern hybridization with locus 16-17 indicated that this element was tandemly repeated (15). The successful amplification of locus 9-4 using the primer orientation indicated in Fig. 6B suggests that this element is in tandem arrays also.
Significant levels of identity exist between the sequences of loci 3-4 and 9-4, in the repeat domains as well as the flanking regions. Similarity has also been noted between some of the repeat units of locus 1-2 and those of loci 3-4 and 9-4. Two other internally repetitive DNA elements were reported independently by Lohia et al. (18) and Willhoeft and Tannich (30) which also bear remarkable similarity in their repeat domains to loci 3-4 and 9-4. However, complete sequence alignment of all five DNA sequences shows enough variation to suggest that they are distinct members of the same family of repeat containing DNA elements (data not shown). That loci 3-4 and 9-4 are indeed distinct is clearly seen from comparing Fig. 7B and Fig. 8B.
Size variations within the repeated domains were studied, and all loci studied showed PCR product length polymorphism. At most loci, amplification results in two or three bands in at least some isolates. Present evidence suggests that the Entamoeba genome is tetraploid (29). It is possible that the multiple bands we observe reflect polymorphism among homologous loci on allelic chromosomes. Multiple amplification products have also been reported for the SREHP gene in a number of E. histolytica isolates (8), a finding consistent with the isolation of cDNAs containing variable numbers of internal tandem repeats (17). SREHP gives the pattern expected of a single copy gene when analyzed by Southern blotting indicating that the length differences are allelic variations. Alternatively, the presence of multiple bands in this study could be explained by the existence of these repeat loci at multiple locations in the Entamoeba genome, each with a characteristic PCR product size.
The observed size variation was further characterized by nucleotide sequence comparison of five isolates at loci 1-2 and 5-6. Isolates 200:NIH and H-303:NIH are easily differentiated by PCR from most of the other isolates but cannot reliably be separated from each other by gel electrophoresis. However, DNA sequence comparison at locus 5-6 revealed that the two isolates are distinguished by the absence of one copy of an 8-bp tandem duplication from strain 200:NIH; this difference is too small to be detected on gel electrophoresis. Thus, PCR product sizes alone may not discriminate among all distinct isolates, at least under these conditions.
From our results it appears that a number of loci showing size polymorphism are present in E. histolytica. The degree of diversity seen varies, with some loci showing more polymorphism and thus having a greater potential for detecting interstrain polymorphism among E. histolytica isolates than others. Despite being from geographically restricted regions, the Bangladeshi and South African samples could be differentiated with ease. For the most part variations in PCR product size appear to be a result of differences in the numbers of tandem repeat units. While no single locus discriminates between all the samples, the collective use of multiple loci allows differentiation of a majority of the E. histolytica isolates.
We believe that the polymorphisms we describe here have potential as tools to answer many of the outstanding questions surrounding the epidemiology of E. histolytica. We are currently studying samples from a broader geographic range to validate the general utility of these loci and are examining samples from infected family groups and amebiasis outbreaks for shared polymorphisms. In the present work, all of the isolates came from individuals who had invasive disease or were likely infected by someone who had invasive disease. Whether invasive and noninvasive strains of E. histolytica exist remains to be established, but hopefully the polymorphic loci described here will be useful in answering this important question.
ACKNOWLEDGMENTS
We thank John Ackers and Aura Aguirre (London School of Hygiene and Tropical Medicine), Terry Jackson (The Medical Research Council of South Africa, Durban), and Rashidul Haque (The International Center for Diarrhoeal Diseases Research, Bangladesh) for providing the DNA samples used in this study and John Ackers for reading the manuscript.
REFERENCES
- 1.Anderson T J C, Su X Z, Bockarie M, Lagog M, Day K P. Twelve microsatellite markers for characterisation of Plasmodium falciparum from finger-prick blood samples. Parasitology. 1999;119:113–125. doi: 10.1017/s0031182099004552. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. WHO/PAHO/UNESCO report. A consultation with experts on amoebiasis. Epidemiol Bull PAHO. 1997;18:13–14. [PubMed] [Google Scholar]
- 3.Azam A, Paul J, Sehgal D, Prasad J, Bhattacharya S, Bhattacharya A. Identification of novel genes from Entamoeba histolytica by expressed sequence tag analysis. Gene. 1996;181:113–116. doi: 10.1016/s0378-1119(96)00484-2. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharya S, Bhattacharya A, Diamond L S. Entamoeba histolytica extrachromosomal circular ribosomal DNA: analysis of clonal variation in a hypervariable region. Exp Parasitol. 1992;74:200–204. doi: 10.1016/0014-4894(92)90047-e. [DOI] [PubMed] [Google Scholar]
- 5.Blanc D S, Sargeaunt P G. Entamoeba histolytica zymodemes: exhibition of γ and δ bands only of glucose phosphate isomerase and phosphoglucomutase may be influenced by starch content in the medium. Exp Parasitol. 1991;72:87–90. doi: 10.1016/0014-4894(91)90124-f. [DOI] [PubMed] [Google Scholar]
- 6.Burch D J, Li E, Reed S, Jackson T F H G, Stanley S L., Jr Isolation of a strain-specific Entamoeba histolytica cDNA clone. J Clin Microbiol. 1991;29:696–701. doi: 10.1128/jcm.29.4.696-701.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark C G. DNA purification from polysaccharide-rich cells, p. D-3.1–D-3.2. In: Lee J J, Soldo A T, editors. Protocols in protozoology. Vol. 1. Lawrence, Kans: Allen Press; 1992. [Google Scholar]
- 8.Clark C G, Diamond L S. Entamoeba histolytica: a method for isolate identification. Exp Parasitol. 1993;77:450–455. doi: 10.1006/expr.1993.1105. [DOI] [PubMed] [Google Scholar]
- 9.Clark C G, Diamond L S. The Laredo strain and other Entamoeba histolytica-like amoebae are Entamoeba moshkovskii. Mol Biochem Parasitol. 1991;46:11–18. doi: 10.1016/0166-6851(91)90194-b. [DOI] [PubMed] [Google Scholar]
- 10.de la Vega H, Specht C A, Semino C E, Robbins P W, Eichinger D, Caplivski D, Ghosh S, Samuelson J. Cloning and expression of chitinases of Entamoebae. Mol Biochem Parasitol. 1997;85:139–147. doi: 10.1016/s0166-6851(96)02817-4. [DOI] [PubMed] [Google Scholar]
- 11.Diamond L S, Clark C G. A redescription of Entamoeba histolytica Schaudinn, 1903 (Emended Walker, 1911) separating it from Entamoeba dispar Brumpt, 1925. J Eukaryot Microbiol. 1993;40:340–344. doi: 10.1111/j.1550-7408.1993.tb04926.x. [DOI] [PubMed] [Google Scholar]
- 12.Diamond L S, Clark C G, Cunnick C C. YI-S, a casein-free medium for axenic cultivation of Entamoeba histolytica, related Entamoeba, Giardia intestinalis and Trichomonas vaginalis. J Eukaryot Microbiol. 1995;42:277–278. doi: 10.1111/j.1550-7408.1995.tb01579.x. [DOI] [PubMed] [Google Scholar]
- 13.Fischer D, Bachmann K. Microsatellite enrichment in organisms with large genomes (Allium cepa L.) BioTechniques. 1998;24:796–802. doi: 10.2144/98245st03. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh S, Frisardi M, Ramirez-Avila L, Descoteaux S, Sturm-Ramirez K, Newton-Sanchez O A, Santos-Preciado J I, Ganguly C, Lohia A, Reed S, Samuelson J. Molecular epidemiology of Entamoeba spp.: evidence of a bottleneck (demographic sweep) and transcontinental spread of diploid parasites. J Clin Microbiol. 2000;38:3815–3821. doi: 10.1128/jcm.38.10.3815-3821.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang M, Chang K P, Albach R A. A 964 bp repetitive DNA in Entamoeba histolytica is associated with linear “chromosomal” DNAs of variable sizes. Arch Med Res. 1997;28(Suppl.):S1–S4. [PubMed] [Google Scholar]
- 16.Jackson T F H G, Suparsad S. Zymodeme stability of Entamoeba histolytica and E. dispar. Arch Med Res. 1997;28(Suppl.):S304–S305. [PubMed] [Google Scholar]
- 17.Köhler S, Tannich E. A family of transcripts (K2) of Entamoeba histolytica contains polymorphic repetitive regions with highly conserved elements. Mol Biochem Parasitol. 1993;59:49–58. doi: 10.1016/0166-6851(93)90006-j. [DOI] [PubMed] [Google Scholar]
- 18.Lohia A, Haider N, Biswas B B. Characterisation of a repetitive DNA family from Entamoeba histolytica containing Saccharomyces cerevisiae ARS consensus sequences. Gene. 1990;96:197–203. doi: 10.1016/0378-1119(90)90253-n. [DOI] [PubMed] [Google Scholar]
- 19.Michel B, Alagón A, Lizardi P, Zurita M. Characterization of a repetitive DNA element from Entamoeba histolytica. Mol Biochem Parasitol. 1992;51:165–168. doi: 10.1016/0166-6851(92)90213-4. [DOI] [PubMed] [Google Scholar]
- 20.Mittal V, Bhattacharya A, Bhattacharya S. Organization of repeated sequences in the region downstream to rRNA genes in the rDNA episome of Entamoeba histolytica. Arch Med Res. 1992;23:17–18. [PubMed] [Google Scholar]
- 21.Mittal V, Sehgal D, Bhattacharya A, Bhattacharya S. A second short repeat sequence detected downstream of rRNA genes in the Entamoeba histolytica rDNA episome. Mol Biochem Parasitol. 1992;54:97–100. doi: 10.1016/0166-6851(92)90098-5. [DOI] [PubMed] [Google Scholar]
- 22.Oliveira R P, Broude N E, Macedo A M, Cantor C R, Smith C L, Pena S D J. Probing the genetic population structure of Trypanosoma cruzi with polymorphic microsatellites. Proc Natl Acad Sci USA. 1998;95:3776–3780. doi: 10.1073/pnas.95.7.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson G L. The laboratory diagnosis of human parasitic amoebae. Trans R Soc Trop Med Hyg. 1968;62:285–294. doi: 10.1016/0035-9203(68)90170-3. [DOI] [PubMed] [Google Scholar]
- 24.Russell R, Iribar M P, Lambson B, Brewster S, Blackwell J M, Dye C, Ajioka J W. Intra and inter-specific microsatellite variation in the Leishmania subgenus Viannia. Mol Biochem Parasitol. 1999;103:71–77. doi: 10.1016/s0166-6851(99)00117-6. [DOI] [PubMed] [Google Scholar]
- 25.Sargeaunt P G. The reliability of Entamoeba histolytica zymodemes in clinical diagnosis. Parasitol Today. 1987;3:40–43. doi: 10.1016/0169-4758(87)90211-0. [DOI] [PubMed] [Google Scholar]
- 26.Sehgal D, Bhattacharya A, Bhattacharya S. Analysis of a polymorphic locus present upstream of rDNA transcription units in the extrachromosomal circle of Entamoeba histolytica. Mol Biochem Parasitol. 1993;62:129–130. doi: 10.1016/0166-6851(93)90187-3. [DOI] [PubMed] [Google Scholar]
- 27.Sehgal D, Mittal V, Ramachandran S, Dhar S K, Bhattacharya A, Bhattacharya S. Nucleotide sequence organization and analysis of the nuclear ribosomal DNA circle of the protozoan parasite Entamoeba histolytica. Mol Biochem Parasitol. 1994;67:205–214. doi: 10.1016/0166-6851(94)00129-4. [DOI] [PubMed] [Google Scholar]
- 28.Stanley S L, Jr, Becker A, Kunz-Jenkins C, Foster L, Li E. Cloning and expression of a membrane antigen of Entamoeba histolytica possessing multiple tandem repeats. Proc Natl Acad Sci USA. 1990;87:4976–4980. doi: 10.1073/pnas.87.13.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willhoeft U, Tannich E. The electrophoretic karyotype of Entamoeba histolytica. Mol Biochem Parasitol. 1999;99:41–53. doi: 10.1016/s0166-6851(98)00178-9. [DOI] [PubMed] [Google Scholar]
- 30.Willhoeft U, Tannich E. Fluorescence microscopy and fluorescence in situ hybridization of Entamoeba histolytica nuclei to analyse mitosis and the localization of repetitive DNA. Mol Biochem Parasitol. 2000;105:291–296. doi: 10.1016/s0166-6851(99)00181-4. [DOI] [PubMed] [Google Scholar]