Abstract
Bacteria have to cope with oxidative stress caused by distinct Reactive Oxygen Species (ROS), derived not only from normal aerobic metabolism but also from oxidants present in their environments. The major ROS include superoxide O2 −, hydrogen peroxide H2O2 and radical hydroxide HO•. To protect cells under oxidative stress, bacteria induce the expression of several genes, namely the SoxRS, OxyR and PerR regulons. Cells are able to tolerate a certain number of free radicals, but high levels of ROS result in the oxidation of several biomolecules. Strikingly, RNA is particularly susceptible to this common chemical damage. Oxidation of RNA causes the formation of strand breaks, elimination of bases or insertion of mutagenic lesions in the nucleobases. The most common modification is 8-hydroxyguanosine (8-oxo-G), an oxidized form of guanosine. The structure and function of virtually all RNA species (mRNA, rRNA, tRNA, sRNA) can be affected by RNA oxidation, leading to translational defects with harmful consequences for cell survival. However, bacteria have evolved RNA quality control pathways to eliminate oxidized RNA, involving RNA-binding proteins like the members of the MutT/Nudix family and the ribonuclease PNPase. Here we summarize the current knowledge on the bacterial stress response to RNA oxidation, namely we present the different ROS responsible for this chemical damage and describe the main strategies employed by bacteria to fight oxidative stress and control RNA damage.
Keywords: nucleotide modification, oxidative stress, 8-oxo-G, RNA oxidation, ROS, quality control of damaged RNA
Introduction
Oxidative Stress and Reactive Oxygen Species
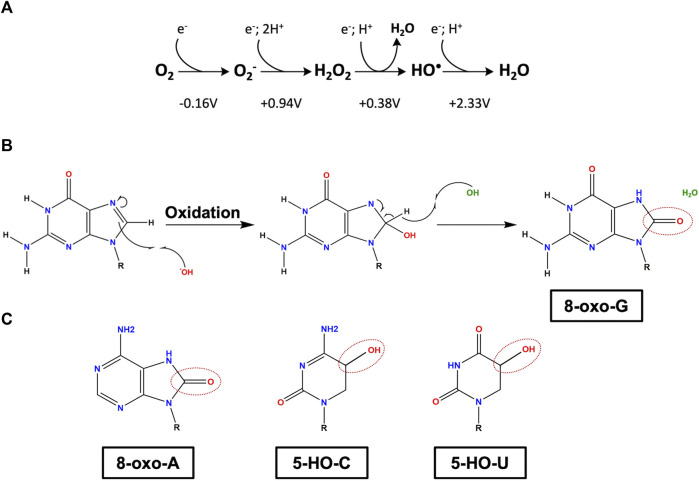
Oxygen is an abundant component of the earth’s atmosphere and a crucial player in many forms of life. However, oxygen comes with a risk that is the production of highly reactive oxygen species (ROS), which chemically modify by oxidation a variety of macromolecules (RNA, DNA, proteins, and lipids) (Markkanen 2017). Consequently, the structure and therefore the function of these macromolecules can be affected, usually resulting in cell toxicity. In bacteria and other organisms, ROS can be either generated intracellularly, as result of aerobic metabolism or exogenously from the outside environment, as consequence of local exposure to increased levels of oxidative agents (Dubbs and Mongkolsuk 2012; Fasnacht and Polacek 2021). Cells possess mechanisms to counteract oxidation and can tolerate low levels of ROS. Indeed, at low levels, ROS can act as signaling molecules controlling different cellular processes, such as quorum sensing, biofilm formation or bacterial self-destruction (Zhao and Drlica 2014; Dryden et al., 2017). However, when there is an imbalance between the amount of ROS and the ability to eliminate them, cells suffer oxidative stress (Berghoff and Klug 2012). Three naturally occurring ROS species, superoxide anion (O2−), hydrogen peroxide (H2O2), and hydroxyl radical (HO•), display different reactivities and are the ones with major relevance in aerobic environments and oxidative stress (Figure 1A).
FIGURE 1.
Main oxidative lesions found in RNA. (A) Redox state of molecular oxygen. From left to right: molecular oxygen (O2), superoxide anion (O2 −), hydrogen peroxide (H2O2), hydroxyl radical (HO•) and water (H2O). The reduction potentials are shown regarding the standard concentration of O2 to be 1M at pH 7 (adapted from Imlay 2009). (B) Reaction of a guanosine with a hydroxyl radical, yielding 8-oxo-7,8-dihydroguanosine (8-oxo-G). (C) Three modified RNA nucleobases: oxidized adenosine 8-oxo-7,8-dihydroadenosine (8-oxo-A), oxidized cytosine 5-hydroxycytosine (5-HO-C) and oxidized uridine 5-hydroxyuridine (5-HO-U). Oxidation sites are marked as dashed circles. R represents the ribose group.
Superoxide Anion O2 −
O2 − is a non-diffusible free radical formed when O2 removes one electron from a donor. It can be produced both intracellularly and exogenously (Kehrer 2000; Imlay 2009). Inside bacterial cells, the major source of O2 − is the autoxidation of enzymes like dehydrogenases, glutathione reductase and cytochromes P450 (Farr and Kogoma 1991; Gort and Imlay 1998). In eukaryotic cells, the most important source of ROS is the mitochondria via the electron transport chain system (Malla and Kwakye 2021). In phagocytes, O2 − is largely produced by NADPH oxidase and is used for destruction of pathogens (Shatwell and Segal 1996; Kong and Lin 2010). Exogenously O2 − can come from quinone-like compounds such as plumbagin and menadione or herbicidal paraquat (Farr et al., 1985; Suntres 2002; Criddle et al., 2006). Superoxide anion O2 − has a half-life in the range of microseconds and it oxidizes several targets like ascorbate and thiols (Kehrer 2000). Additionally, it can be dismutated to H2O2 and O2 through the action of superoxide dismutase (SOD) (Fridovich 1989) and act as a reducing agent of transition metals like Fe3+ (Farr and Kogoma 1991).
Hydrogen Peroxide H2O2
Hydrogen peroxide is the simplest member of the class of peroxides. The main sources of internal H2O2 are the autoxidation of flavoenzymes and the dismutation of O2 − and contrary to superoxide anion, hydrogen peroxide can easily cross the cellular membrane (Imlay 2009). There are specific environments with residual amounts of oxygen that can be hubs of H2O2 accumulation, such as the anoxic interfaces in soil, hydrothermal vents (Tapley et al., 1999; Ogino et al., 2018) and the intestine epithelium (Espey 2013). One possible theory to explain the presence of H2O2 in the intestine is its production by lactic acid bacteria (LAB) (Imlay 2009). There is evidence that LAB metabolism can use oxygen and release H2O2 to their environment (Seki et al., 2004). Released H2O2 can also cause growth arrest of other bacterial species (Pericone et al., 2000; Tong et al., 2007; Bao et al., 2017). H2O2 is more stable when compared to O2 − with a half-life in the range of minutes (Kehrer 2000), and its decomposition is mediated by peroxidases like catalase (Winterbourn 2013). The most relevant reaction in which H2O2 is involved is undoubtedly the Fenton reaction (Fenton 1894). In this reaction, H2O2 interacts with metal cofactors such as iron (Fe2+) producing the highly reactive hydroxyl radicals HO• (Shcherbik and Pestov 2019).
Hydroxyl Radicals HO•
HO• is one of the most dangerous ROS due to a half-life in the range of nanoseconds and its extreme reactivity given the high electron potential of +2.33 V (Figure 1A). It can attack most of the organic molecules in the vicinity of its formation, reacting either with proteins, lipids, or nucleic acids, especially with thiamine and guanosine (Farr and Kogoma 1991). HO• can arise from absorption of a photon by tryptophan residues and from photochemical reactions in natural waters (Vaughan and Blough 1998). However, the most relevant source of HO• in biological systems is the Fenton reaction since iron is such an important micronutrient (Liochev et al., 1999). Note that the ROS levels of the different species are interconnected. As mentioned above, O2 − reduces Fe3+ and its dismutation consequently produces H2O2; hence when the intracellular concentration of O2 − increases, the concentration of H2O2 and subsequently HO• also increases. Contrarily to O2 −, which can be eliminated by SOD, HO• cannot be eliminated by an enzymatic reaction (Reiter et al., 1995).
Reactive Oxygen Species and Cell Damage
An increase in ROS species is perilous to the cell as they cause an extensive damage to several targets, which include all biomolecules like proteins, lipids, and nucleic acids (DNA and RNA). As result of oxidation, the structure and function of these macromolecules are affected, with potentially toxic consequences that can ultimately result in cell death. Most protein oxidative modifications occur in the side chains of amino acids like oxidation of thiols and the formation of carbonyl groups affecting protein structure, conformation, and function (Dalle-Donne et al., 2006; Morzel et al., 2006; Zhang et al., 2013). Cysteine and methionine are the most susceptible amino acids to oxidation because they contain reactive sulfur atoms (Shacter 2000). In humans, most oxidized proteins are eliminated but some can accumulate and contribute to the damage associated with several diseases (Stadtman 2001). When ROS attack lipids, the products of this oxidation are lipid peroxides that mostly affect polyunsaturated fatty acids, which are critical components of cellular membranes (Niki 2009; Yin et al., 2011). Consequently, the accumulation of lipid peroxides results in changes in membrane permeability and fluidity (Dobretsov et al., 1977) and can affect ion channels, inactivate membrane transport proteins, disrupt homeostasis, and affect signaling pathways (Welsch 1987; Blair 2008; Su et al., 2019).
Nucleic acids are very susceptible to chemical damage given the reactivity of the nitrogen and oxygen atoms of the nucleobases (Simms and Zaher 2016). Among the four DNA nucleobases, guanine is the most prone to oxidation given its lower reduction potential (Steenken and Jovanovic 1997) and its oxidation can interfere with DNA metabolism like transcription and replication (Lee and Kang 2019). Contrarily to other macromolecules, damaged DNA cannot be replaced so it needs to be repaired to remain viable (Lee and Kang 2019). The main repair mechanism to correct non-bulky DNA lesions caused by oxidative stress is the base excision repair (Ignatov et al., 2017). Unlike DNA, no repair mechanisms have yet been identified for oxidized RNA, which imposes a challenge to the cell in order to keep homeostasis. This is even more important as RNA is more susceptible to oxidation than DNA (see below).
RNA Oxidation
RNA accounts for 80–90% of the total nucleic acids (Hangauer et al., 2013; Li et al., 2020), and it affects the post-transcriptional regulation of gene expression (dos Santos et al., 2018; Quendera et al., 2020). ROS can attack RNA in different ways, whether it is through formation of abasic site, strand breaks or modifications of base and sugar moieties (Liu et al., 2012; Küpfer and Leumann 2014; Tanaka and Chock 2021). When compared to DNA, RNA molecules are more prone to oxidation for several reasons: 1) Being mostly single-stranded, the nucleobases of the RNA molecules are more exposed to ROS (Hofer et al., 2005); 2) RNA is less associated with proteins compared to DNA, making it more vulnerable (Cobley et al., 2018); 3) In physiological conditions the concentration of ribonucleotides is much higher than deoxyribonucleotides; therefore, the modifications are more likely to be incorporated into RNA than in DNA (Yan and Zaher 2019) and 4) RNA lacks repair mechanisms in contrast to what happens with DNA (Küpfer and Leumann 2014).
RNA molecules can be oxidized by HO• derived from the Fenton reaction by detaching the hydrogen from the C-H bonds, resulting in several oxidation adducts, namely: 8-oxo-7,8- dihydroguanosine (8-oxo-G), 8-oxo-7,8-dihydroadenosine (8-oxo-A), 5-hydroxyuridine (5-HO-U) and 5-hydrocytosine (5-HO-C) (Simms and Zaher 2016; Yan and Zaher 2019) (Figures 1B,C). Although all four nucleobases are affected by ROS, the most common oxidation lesion found in cells is the 8-oxo-G given that guanosine is the base with the lowest reduction potential (Radak and Boldogh 2010; Shcherbik and Pestov 2019; Estevez et al., 2021). Under physiological conditions, the frequency of 8-oxo-G is approximately of 1 per 100.000 of unmodified guanosines (Shen et al., 2000). This modification has a high propensity to cause mutations in the open reading frame leading to changes in gene expression and the development of aberrant proteins (Ishii and Sekiguchi 2019; Yan and Zaher 2019). Furthermore, 8-oxo-G can dramatically reduce the rate of peptide-bond formation, strongly impairing protein synthesis (Thomas et al., 2019).
When compared to DNA, the study of RNA damage by oxidation has been neglected, and this is mainly due to the assumption that RNAs tend to have a short half-life due to a rapid turnover before causing deleterious effects (Malla and Kwakye 2021; Tanaka and Chock 2021). This neglection resulted in an underestimation of the toxic effect of RNA oxidation on cells (Li et al., 2020). Next, we will provide examples on how RNA oxidation affects different RNA species within a cell (Figure 2).
FIGURE 2.
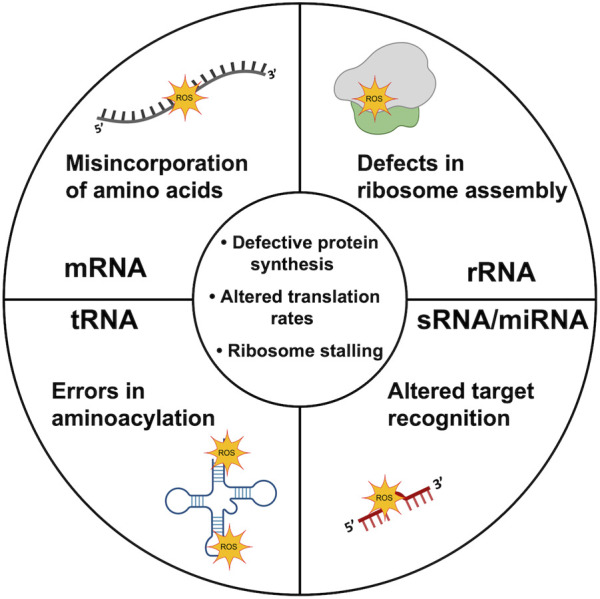
Oxidation of different RNA species. Virtually all RNA species are vulnerable to RNA oxidation. The scheme depicts major consequences of oxidation damage of rRNA, mRNA, tRNA and sRNA/miRNA molecules. In the middle of the scheme are represented defects common to oxidation of all RNA molecules. Molecule templates were retrieved from BioRender database.
Ribosomal RNA
Ribosomal RNA composes the vast majority of total RNA molecules within a cell and is responsible for the structural and functional core of the ribosome (Cech 2000; Andrade et al., 2018; Dos Santos et al., 2020). Despite rRNAs being highly structured molecules, it is well established that rRNAs are a target for oxidation, not only in bacteria but also in higher organisms, and this has been associated with diseases (Ahmad et al., 2017; Willi et al., 2018). As we could expect, oxidation-induced modifications in the rRNA will affect important functional sites of the ribosome. In Escherichia coli it was shown that both the 16S rRNA (component of the small ribosomal subunit) and the 23S rRNA (component of the large ribosomal subunit) are subject to oxidation by H2O2 in vivo, although the 23S rRNA seems more prone to nucleobase oxidation (Willi et al., 2018). Specifically, it was shown that modification of three inner residues of the 23S rRNA (A2451, C2063, U2585) located in the catalytic center of the ribosome affects multiple steps of translation, such as peptide bond formation or tRNA binding to the ribosome. Oxidation of rRNA at specific positions within the ribosome can lead to defects in ribosome assembly (Simms and Zaher 2016). In addition, it was recently shown that the absence of methylations in the 23S rRNA at specific positions could make it more susceptible to oxidative stress, suggesting that oxidative damage to RNA can be regulated by the presence of post-transcriptional nucleoside modifications like methylation (Estevez et al., 2021).
Transfer RNA
Transfer RNAs are structured molecules with great abundance and stability in the cell. These molecules decode the different codons on mRNA and allow the correct amino acid to be added to the nascent polypeptide. Certain post-transcriptional modifications normally found in tRNAs like the 5-methylaminomethyl-2-thiouridine (mnm5s2U) were also found to reduce oxidation in tRNAs (Estevez et al., 2021). Oxidation of the tRNA nucleobases is likely to induce a profound effect on the overall structure of the molecule, thus affecting protein synthesis. An in vitro study where liquid chromatography tandem mass spectrometry was applied to E. coli tRNA showed that upon oxidation induced by UV radiation, several tRNA nucleosides were modified with the 8-oxo-G adduct (Sun et al., 2018). Specifically, oxidative damage was mostly found to affect the anticodon and variable loop regions sequences of tRNA (Sun et al., 2020), which correlates with translational errors in the decoding process and aminoacylation (Yan and Zaher 2019). Zhong et al. (2015) showed that under oxidative stress caused by H2O2, the vast majority of tRNAs in E. coli were decreased in vivo. This was shown to occur due to nonspecific extended tRNA degradation, in contrast to eukaryotic cells where tRNAs are cut in halves (Thompson et al., 2008). However, another report using a different E. coli strain revealed the reduction of only tRNAGly upon oxidation also with H2O2, with the global pool of tRNA remaining unchanged (Leiva et al., 2020). Therefore, the extent of tRNAs affected by oxidation is still debatable. The reduction of tRNA levels significantly slows down the translation elongation speed (Zhu and Dai 2019). This could be beneficial for bacteria as oxidative stress increases the probability of protein misfolding, which can be toxic to the cell. Concomitantly, overexpression of tRNAs was found to protect cells against oxidative stress, presumably by decreasing ribosome jamming found upon oxidation treatment, which causes the premature dissociation of RNA from ribosomes (Zhong et al., 2015).
Messenger RNA
Messenger RNA is responsible for carrying the information encoded in DNA to synthesize proteins. The 8-oxo-G modification can affect the translation process in E. coli by interfering with the A site of mRNA codons leading to the incorporation of a different amino acid (Simms et al., 2014). Recent studies expanded the mechanism by which 8-oxo-G affects the decoding process. This adduct was found to induce a syn conformation of the mRNA molecule, causing it to pair preferentially with a tRNA carrying adenosine instead of cytidine, therefore, disrupting the translation rate (Thomas et al., 2019; Zhu and Dai 2019). From these studies it was also possible to conclude that the mRNA alterations caused by 8-oxo-G seem to affect mRNA-tRNA interactions in the ribosome decoding center. From the three main steps of the translation process, it is the elongation step that seems to be most affected by oxidative stress, with increasing levels of stalled ribosomes found on the mRNA molecule upon oxidation treatment, as observed in E. coli and cyanobacteria (Nishiyama et al., 2006; Zhong et al., 2015).
Small Non-coding RNA
Small non-coding RNAs play an important role in the regulation of gene expression (Kavita et al., 2018; Quendera et al., 2020). In the literature there is no current evidence of direct oxidation of sRNAs in prokaryotes, although it seems likely that they are also targets for ROS, as it is the case of microRNAs (miRNAs) in eukaryotes. Namely, the microRNA miR-184 is highly oxidized and enriched in 8-oxo-G nucleotides upon H2O2 treatment (Wang et al., 2015). This oxidative modification influences the function of miR-184, which caused a substantial downregulation of Bcl-xL and Bcl-w mRNAs, promoting apoptosis to a higher extent. Recently, it was also reported that the miRNA miR-1 is preferentially oxidized in cardiac hypertrophy cases (Seok et al., 2020). Therefore, it is reasonable to think that like in miRNAs, bacterial sRNAs can also be oxidized under specific conditions. Further research is necessary to clarify whether this assumption proves to be true.
However, several studies demonstrated the importance of bacterial and even archaeal sRNAs in response to oxidative stress, namely by regulating the expression of important enzymes that protect the cell from ROS attacks. For instance, several sRNAs are known to regulate catalase expression in different bacteria. In Deinococcus radiodurans, the sRNA OsiA binds directly to the catalase katA mRNA and stabilizes the transcript (Chen et al., 2019). In this bacterium it was also shown that in conditions of oxidative stress, another sRNA called OsiR is highly expressed and base-pairs with the mRNA of KatE2, resulting in increased transcript levels and increased translation of KatE2, a second catalase of this bacterium (Gao et al., 2020). In Pseudomonas stutzeri A1501, the NfiS sRNA was also found to bind to the mRNA of the catalase coding gene katB (Zhang et al., 2019). Other sRNAs can regulate the expression of superoxide dismutase, as it the case of Staphylococcus aureus RscA which represses the translation of sodA mRNA and promotes the oxidative stress response via SodM (Lalaouna et al., 2019). Nevertheless, the role of sRNA is not restricted to regulation of ROS scavenging enzymes. sRNAs have been shown to act on multiple targets (Andrade et al., 2013) and may contribute for bacterial resistance to oxidative stress, namely through regulation of the expression of alternative transcription factors (Fröhlich and Gottesman 2018).
Quality Control Pathways to Eliminate Oxidated RNA
Oxidized nucleobases incorporated into either DNA or RNA are potentially mutagenic and constitute a major challenge for cell survival. In bacteria, oxidized DNA bases (which block replication or originate mutations) can be removed by base excision repair enzymes (namely MutM, endonucleases IV and VIII) (for more information see Imlay 2013). When the oxidative damage is too profuse, SOS response is triggered, allowing RecA protein to repair DNA oxidation-induced lesions. RecA itself can also be oxidized, but this can be reversed by Msr proteins to maintain a level of reduced functional RecA necessary to carry out both efficient recombination and SOS system repair (Henry et al., 2021). In a striking contrast, no similar mechanisms of repair have yet been reported for oxidized RNA. Such pathways may exist but simply have not been recognized yet. In favor of this view, a mechanism of repair for alkylated RNA by oxidative demethylases has already been reported, proving that damages in RNA bases are repairable (Aas et al., 2003). However, two RNA-binding proteins, MutT and PNPase, have been identified to participate in surveillance mechanisms that recognize and degrade oxidized RNA.
MutT/Nudix Family Members
Incorporation of oxidized ribonucleotides during RNA synthesis can lead to mutations (Li et al., 2014). E. coli RNA polymerase is ten-times less efficient in using 8-oxo-GTP than GTP (Taddei et al., 1997) and the human RNA polymerase II presents the same discriminatory activity (Hayakawa et al., 1999). MutT protein (a member of the Nudix family) was found to hydrolyze both 8-oxo-dGTP and 8-oxo-GTP, preventing their integration into DNA and RNA, respectively (Maki and Sekiguchi 1992; Taddei et al., 1997). In Mycobacterium tuberculosis, MutT1 and ADP-ribose pyrophosphatase (ADPRase) were shown to degrade 8-oxo-dGTP and 8-oxo-GTP/GDP (Patil et al., 2013). Such enzymatic activity is well conserved and human cells have four proteins responsible for degradation of oxidized nucleotides that are MutT homologues, namely MTH1, MTH2, MTH3 and NUDT5 (reviewed in Rudd et al., 2016).
Polynucleotide Phosphorylase
Bacterial PNPase is widely conserved and a major 3′-5′ exoribonuclease involved in RNA degradation (dos Santos et al., 2018). In E. coli, PNPase was found to have high affinity for 8-oxo-G containing RNA when compared to undamaged RNA (Hayakawa et al., 2001). In agreement with this result, PNPase-deficient E. coli cells are more sensitive to hydrogen peroxide and other oxidants than the wild-type, and the levels of 8-oxo-G were found to strongly accumulate in the PNPase mutant strain, as result of deficient elimination of these defective RNA molecules. Complementation of the mutant corrects both the growth defect and reduces the levels of 8-oxo-G, suggesting a direct role of PNPase in controlling the oxidative damage (Wu et al., 2009). The importance of PNPase to fight oxidative stress has been extended to other bacterial species, such as D. radiodurans (Chen et al., 2007) and Yersinia pestis (Henry et al., 2012), where PNPase was required to protect cells under oxidative stress. Moreover, human PNPase (hPNPase) is located mainly in the mitochondria where oxidative damage is elevated, which presupposes a role for hPNPase in controlling the level of oxidized RNA (Chen et al., 2006). PNPase-knocked down HeLa cells under H2O2 treatment presented reduced growth and higher 8-oxo-G content than the control (Wu and Li 2008). Overall, PNPase is a major enzyme acting in the quality control of RNA and can discriminate among oxidized and non-oxidized forms of RNA, showing higher affinity to binding and degradation of oxidation modified RNAs in different organisms.
Another example of a protein that binds oxidized RNA is the mammalian Y box-binding protein 1 (YB-1), which specifically binds with high affinity to 8-oxo-G-containing RNA oligonucleotides (Hayakawa et al., 2002). The YB-1 domain responsible for RNA-binding is the cold shock domain and it has more than 40% homology to the major cold shock protein in E. coli, CspA (Matsumoto and Wolffe 1998). It is a multifunctional protein that can bind to both RNA and DNA and is important for transcriptional and translational regulation. When full length YB-1 was introduced and overexpressed in E. coli, bacteria became more resistant to paraquat oxidative stress (Hayakawa et al., 2002). However, when the RNA-binding domain was removed, cells were more susceptible to the oxidant. YB-1 could work as an RNA chaperone and be involved in the sequestration of oxidized RNA, preventing the translation of 8-oxo-G-containing mRNAs (Li et al., 2006). On the other hand, YB-1 could bind to oxidized ribonucleotides and recruit other factors for subsequent degradation. It is possible that both PNPase and YB-1 could have a cooperative function that protects cells against oxidized RNA, however the detailed mechanisms are still elusive (Ishii and Sekiguchi 2019).
Scavenger Enzymes Against Free Radicals in Bacteria
Bacteria (and other organisms) are only able to tolerate a certain amount of non-harmful levels of ROS. To mitigate this threat, bacteria have evolved several mechanisms to fight oxidative stress and avoid the imbalance of ROS levels that lead to cell toxicity. From these, the presence of scavenging enzymes that consume ROS (superoxide dismutases, catalases and peroxidases) are critical in self-defense mechanisms against oxidative stress in bacteria.
Superoxide Dismutase for Degradation of Superoxide
Superoxide dismutase is a family of enzymes widespread among bacteria, which act as a metalloenzyme that converts superoxide anion to hydrogen peroxide and molecular oxygen (Steinman 1988). E. coli has two cytoplasmic enzymes (Fe-SOD and Mn-SOD) and a periplasmic enzyme (Cu-Zn-SOD) (Imlay 2013). High titers of cytoplasmic SODs maintain steady-state superoxide levels at a sub-nanomolar concentration (Imlay and Fridovich 1991) in order to avoid toxicity by O2 −. Being structurally and kinetically similar, the role of Mn-SOD and Fe-SOD is to ensure that SOD activity is present under a wide range of metal bioavailability. The two isozymes are coordinately regulated in response to iron levels: when iron is abundant, Fur (Ferric uptake regulator) inhibits Mn-SOD transcription while Fe-SOD is synthesized and activated by iron; when iron levels are reduced, Fur is deactivated and there is transcription of both Mn-SOD and the sRNA RyhB, which will repress Fe-SOD mRNA (Tardat and Touati 1991; Massé and Gottesman 2002). In anaerobic conditions, only Fe-SOD synthesis persists to prepare E. coli for subsequent aeration and consequent superoxide formation. Fe-SOD enzyme is favored by evolution in anaerobic habitats due to the biological availability of iron. Regarding periplasmic SODs, they protect bacteria from superoxide that leaks from respiratory chain components to the outer part of the cytoplasmic membrane. These SODs are under the control of the RNA polymerase sigma factor RpoS (also called σ38, Sigma S or KatF), and are strongly induced when aerobic cells enter stationary phase (Imlay 2009).
Catalases and Peroxidases for Degradation of Hydrogen Peroxide
Catalases are common in most bacteria, with exceptions found in Gram-positive bacteria like streptococci, enterococci and leuconostocs (Mishra and Imlay 2012). E. coli has three main enzymes that can degrade hydrogen peroxide: alkyl hydroperoxide reductase (Ahp), catalase G (KatG) and catalase E (KatE) (Seaver and Imlay, 2001). Ahp is a two-component (AhpC-AhpF) thiol-based peroxidase that transfers electrons from NADH to H2O2, reducing it to water. KatG belongs to the catalase-peroxidase family and is only weakly expressed in exponential cells. However, the transcriptional regulator OxyR strongly induces both ahpCF and katG when cells are stressed by exogenous H2O2 (see below). KatE is strongly expressed in stationary phase cells only, since it is induced by the RpoS system (Schellhorn and Hassan, 1988). These functionally redundant enzymes are used at different conditions: peroxidases are the primary scavenger at low H2O2 concentrations, whereas catalases are used at higher levels of H2O2 or when cells are starved (Imlay, 2013). Additionally, in the E. coli periplasm, cytochrome c peroxidase (Ccp) receives electrons from the respiratory chain and transfers them directly to H2O2 (Partridge et al., 2007). Ccp instead of eliminating H2O2 allows the cell to use it as a terminal oxidant to support respiration (Khademian and Imlay, 2017). This function is beneficial only when molecular oxygen is unavailable.
Perceiving Redox Signals From Reactive Oxygen Species
To protect cells under oxidative stress, bacteria induce the expression of several genes, namely the SoxRS, OxyR and PerR systems. The activation of these multigene transcriptional pathways is critical to the bacterial response to oxidative stress and will be next presented.
SoxRS System
Bacteria have evolved transcriptional regulators capable of perceiving redox signals from ROS, and consequently regulate enzymatic antioxidant systems (Lushchak and Storey 2021). The SoxRS regulon is a response mechanism to O2 − and to redox-cycling compounds, but not to H2O2 (Gu and Imlay 2011). This system is controlled by two proteins, which activate consecutively two stages of transcription (González-Flecha and Demple 2000). SoxR is a homodimer, each with an iron-sulfur cluster [2Fe-2S] center (Hidalgo et al., 1997) and whose activity depends on its oxidation state. Only the oxidized form ([2Fe-2S]2+) is active and able to induce transcription of soxS (Gaudu and Weiss 1996). SoxS is a second transcriptional activator that induces the transcription of more than 100 genes with functions in antioxidant defense, damage repair and maintenance of central metabolisms that allow the cell to survive under oxidative stress (Nunoshiba et al., 1992; Wu and Weiss 1992). Some of the genes regulated by SoxS are sodA, nfo, zwf, fumC, acnA, fur and ntrA (González-Flecha and Demple 2000; Touati 2000; Pomposiello et al., 2003; Blanchard et al., 2007). After oxidative stress, SoxR is again reduced and inactivated through reducing systems encoded by rseC and rsxABCDGE operon. Therefore, these systems maintain SoxR in its reduced state, preventing activation of the SoxRS regulon (Koo et al., 2003). Also, when oxidative stress decreases, SoxS is rapidly degraded by proteolysis (Griffith et al., 2004). The transcription factor SoxR is widely distributed in Actinobacteria and Proteobacteria. However, while in enteric bacteria SoxR activates the expression of SoxS which in turn controls the expression of a diverse class of genes involved in the stress response, in nonenteric bacteria (like Pseudomonas aeruginosa or Streptomyces coelicolor) SoxS is absent, and SoxR directly activates a small sets of genes in response to redox-cycling agents (Chiang and Schellhorn 2012; Kobayashi et al., 2014; Mettert and Kiley 2015).
OxyR System
The OxyR system is primarily responsible for recognizing and maintaining hydrogen peroxide levels in the cell (Åslund et al., 1999). OxyR is a LysR-type transcriptional regulator, widespread in most Gram-negative bacteria but it has also been reported in some Gram-positive bacteria, such as S. aureus and S. coelicolor (Morikawa et al., 2006; Oh et al., 2007). The transcriptional activity of OxyR activity depends on its redox state, as it can either act as activator if in its oxidized form or as repressor when in reducing conditions (Christman et al., 1989; Heo et al., 2010). OxyR acts as a tetramer and specifically binds to the 5’ promoter regions of target genes at a conserved sequence motif (Toledano et al., 1994). In E. coli, the OxyR regulon spans more than 20 different genes, involved in several molecular mechanisms of adaptive response to redox stress, such as H2O2 detoxification (katE and ahpCF), heme biosynthesis (hemH), reductant supply (grxA, gorA, trxC), repression of iron import (fur, encoding Fur regulator of ferric ion uptake) and others (Zheng et al., 1999; Zheng et al., 2001a; Zheng et al., 2001b). Additionally, OxyR upregulates the expression of OxyS, a small regulatory RNA that directs peroxide stress response (González-Flecha and Demple 1999; Fröhlich and Gottesman 2018). Although OxyS does not directly affect the combat against excessive H2O2, this sRNA seems to play a role in protection from H2O2-induced mutagenesis (Altuvia et al., 1997). Additionally, depletion of OxyS has shown to result in considerably higher levels of H2O2 in E. coli (González-Flecha and Demple 1999).
Regulation by OxyR in E. coli begins by sensing the levels of H2O2 at a specific cysteine residue in the protein (C199). Under regular conditions, OxyR is present in its reduced form (Zheng et al., 1998). Increased levels of H2O2 result in the rapid oxidation of OxyR: the peroxide molecule reacts with the thiol group of C199, leading to interaction with the C208-SH to form an intramolecular disulphide bond, inducing conformational changes which alter the DNA binding properties of OxyR, allowing effective interaction with RNA polymerase (Lee et al., 2004). This molecular mechanism is then reversed through feedback regulation, since oxidized OxyR induces the expression of the grxA and gor genes, encoding glutaredoxin 1 and glutathione reductase, which act to reduce OxyR (Zheng et al., 1998). Although the OxyR system has been mostly studied in E. coli, different bacteria have evolved in the direction to adapt their particular OxyR regulon to better suit their environmental niches: they may display differences in the molecular mechanism of H2O2 regulation, the number of OxyR homologs or the type of genes present in their regulon (further reviewed in Sen and Imlay 2021).
PerR System
Despite OxyR importance, the most prevalent system for preventing peroxide oxidative stress in Gram-positive bacteria is the PerR system. PerR belongs to the ferric uptake regulator (Fur) superfamily of metalloregulatory transcription factors, firstly identified in Bacillus subtilis where it is found to be the key regulator of the H2O2 response (Bsat et al., 1998). PerR has also been identified in some Gram-negative bacteria (van Vliet et al., 1999; Li et al., 2004), found primarily in association with OxyR (Wu et al., 2006). PerR is a transcriptional factor that acts as a dimer and in response to metal ions inside the cell through metal-catalyzed oxidation (Mongkolsuk and Helmann 2002). Each subunit contains a structural site that binds irreversibly to zinc (Zn2+) and a regulatory metal binding site (Jacquamet et al., 2009). In B. subtilis, PerR binds either to manganese (Mn2+) or to ferrous iron (Fe2+), which is preferred in most conditions (Sen and Imlay 2021). The metal-bound conformation of PerR binds to DNA and this binding occurs at a specific Per box (Fuangthong and Helmann 2003) located in the promoter region or downstream from it, acting then as a repressor (Dubbs and Mongkolsuk 2012).
Regulation by PerR is based on the oxidation of ferrous iron (Fe2+) into ferric iron (Fe3+) at the regulatory site. Under non-stress conditions, binding of the regulatory metal stabilizes the conformation of the PerR dimer to better interact with DNA, which leads to the repression of the PerR regulon (Jacquamet et al., 2009). When intracellular H2O2 level increase, excess peroxide causes the oxidation of the iron ions in the regulatory sites of PerR through the Fenton reaction (Lee and Helmann 2006). PerR then undergoes conformational changes that render it unable to interact with DNA (Imlay 2015), leading to the derepression of PerR regulated genes. Most of these genes are involved in peroxide stress metabolism and protection (kat, ahpCF operon, mrgA), but some participate in metal homeostasis (hemAXCDBL operon) and surfactant production (srfA) (Chen et al., 1995; Bsat et al., 1996, 1998; Hayashi et al., 2005). Similarly to OxyR, different bacteria may have evolved to use PerR in different ways to better adapt to their environmental niche but PerR homologs tend to normally regulate similar groups of genes.
Conclusion
Aerobic organisms have developed over time the ability to maintain the levels of intracellular ROS within specific limits, to prevent ROS insults that can damage distinct classes of biomolecules. However, when self-defense mechanisms fail to protect the cell, ROS levels increase and as result of this imbalance, the cell faces oxidative stress. Due to its chemistry, RNA is highly susceptible to oxidation, which leads to the modification of nucleobases (namely, the formation of 8-oxo-G) causing RNA injury with potential mutagenic consequences. Since RNA is at the center of cellular regulatory pathways, the chemical damage of RNA triggered by ROS has a wide impact in gene expression. The importance of this impact is even stressed as RNA is highly expressed, comprising most nucleic acids within a cell, with the existence of many different classes of RNAs (mRNA, rRNA, tRNA and sRNA). Virtually, all these different RNA species are vulnerable to oxidation-induced damage, affecting translation at various levels. Not only the rate of protein synthesis is slowed but also the formation of aberrant proteins is potentiated, what may affect cell fitness and even cause cell death. The consequences of RNA damage by oxidation are widespread through all domains of life. Interestingly, in humans, the oxidation of RNA is implicated in several diseases, like cancer (Guo et al., 2020), diabetes (Cejvanovic et al., 2018), and neurodegenerative disorders (Fimognari 2015). Bacteria developed quality control mechanisms to specifically degrade oxidized RNA, in order to guarantee RNA fidelity. These surveillance mechanisms also rely on RNA-binding proteins that show higher affinity to bind oxidized RNA rather than non-damaged RNA. However, the number of proteins which were identified to participate in these mechanisms is rather small, with the best-known examples relying on PNPase and MutT. Nevertheless, it seems likely that other RNA-binding proteins participate in the surveillance of oxidized RNAs. Overall, the study of the bacterial response to oxidative stress and oxidation of RNA is an area that will certainly attract more attention in the years to come.
Acknowledgments
We would like to thank Guillem Hernandez Gras for helping in the preparation of Figure 1.
Author Contributions
JA outlined the manuscript. AS wrote the first draft with the help of AQ. AS, AQ, JS, AS and JA wrote the final manuscript. AS designed the figures. JA supervised the work. All authors edited and approved the final version of this article.
Funding
Research in our lab was supported by Project LISBOA-01-0145-FEDER-007660 (Microbiologia Molecular, Estrutural e Celular) funded by FEDER through COMPETE2020–Programa Operacional Competitividade e Internacionalização (POCI) and by FCT–Fundação para a Ciência e a Tecnologia (Portugal) through Grant PTDC/BIA-MIC/32525/2017 to JMA and Doctoral Fellowships to AFS (PD/BD/146136/2019) and APQ (PD/BD/135487/2018).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Aas P. A., Otterlei M., Falnes P. Ø., Vågbø C. B., Skorpen F., Akbari M., et al. (2003). Human and Bacterial Oxidative Demethylases Repair Alkylation Damage in Both RNA and DNA. Nature 421, 859–863. 10.1038/nature01363 [DOI] [PubMed] [Google Scholar]
- Ahmad W., Ijaz B., Shabbiri K., Ahmed F., Rehman S. (2017). Oxidative Toxicity in Diabetes and Alzheimer's Disease: Mechanisms behind ROS/RNS Generation. J. Biomed. Sci. 24, 1–10. 10.1186/s12929-017-0379-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altuvia S., Weinstein-Fischer D., Zhang A., Postow L., Storz G. (1997). A Small, Stable RNA Induced by Oxidative Stress: Role as a Pleiotropic Regulator and Antimutator. Cell 90, 43–53. 10.1016/S0092-8674(00)80312-8 [DOI] [PubMed] [Google Scholar]
- Andrade J. M., Pobre V., Arraiano C. M. (2013). Small RNA Modules Confer Different Stabilities and Interact Differently with Multiple Targets. PLoS One 8, e52866. 10.1371/journal.pone.0052866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade J. M., Santos R. F., Chelysheva I., Ignatova Z., Arraiano C. M. (2018). The RNA ‐binding Protein Hfq Is Important for Ribosome Biogenesis and Affects Translation Fidelity. EMBO J. 37. 10.15252/embj.201797631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslund F., Zheng M., Beckwith J., Storz G. (1999). Regulation of the OxyR Transcription Factor by Hydrogen Peroxide and the Cellular Thiol-Ddisulfide Status. Proc. Natl. Acad. Sci. 96, 6161–6165. 10.1073/PNAS.96.11.6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X., Yang J., De Soet J. J., Liu H., Gao X., Van Loveren C., et al. (2017). Factors Influencing the Competition between Streptococcus Oligofermentans and Streptococcus Mutans in Dual-Species Biofilms. Caries Res. 51, 507–514. 10.1159/000479044 [DOI] [PubMed] [Google Scholar]
- Berghoff B., Klug G. (2012). Small RNAs with a Role in the Oxidative Stress Response of Bacteria,” in Regulatory RNAs In Prokaryotes. Vienna: Springer Vienna, 1–14. 10.1007/978-3-7091-0218-3_1 [DOI] [Google Scholar]
- Blair I. A. (2008). DNA Adducts with Lipid Peroxidation Products. J. Biol. Chem. 283, 15545–15549. 10.1074/jbc.R700051200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard J. L., Wholey W.-Y., Conlon E. M., Pomposiello P. J. (2007). Rapid Changes in Gene Expression Dynamics in Response to Superoxide Reveal SoxRS-dependent and Independent Transcriptional Networks. PLoS One 2, e1186. 10.1371/journal.pone.0001186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bsat N., Chen L., Helmann J. D. (1996). Mutation of the Bacillus Subtilis Alkyl Hydroperoxide Reductase (ahpCF) Operon Reveals Compensatory Interactions Among Hydrogen Peroxide Stress Genes. J. Bacteriol. 178, 6579–6586. 10.1128/jb.178.22.6579-6586.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bsat N., Herbig A., Casillas‐Martinez L., Setlow P., Helmann J. D. (1998). Bacillus Subtiliscontains Multiple Fur Homologues: Identification of the Iron Uptake (Fur) and Peroxide Regulon (PerR) Repressors. Mol. Microbiol. 29, 189–198. 10.1046/j.1365-2958.1998.00921.x [DOI] [PubMed] [Google Scholar]
- Cech T. R. (2000). The Ribosome Is a Ribozyme. Science 289, 878–879. 10.1126/science.289.5481.878 [DOI] [PubMed] [Google Scholar]
- Cejvanovic V., Kjær L. K., Mørup Bergholdt H. K., Henriksen T., Weimann A., Ellervik C., et al. (2018). RNA Oxidation and Iron Levels in Patients with Diabetes. Free Radic. Biol. Med. 129, 532–536. 10.1016/j.freeradbiomed.2018.10.420 [DOI] [PubMed] [Google Scholar]
- Chen H.-W., Rainey R. N., Balatoni C. E., Dawson D. W., Troke J. J., Wasiak S., et al. (2006). Mammalian Polynucleotide Phosphorylase Is an Intermembrane Space RNase that Maintains Mitochondrial Homeostasis. Mol. Cel. Biol. 26, 8475–8487. 10.1128/mcb.01002-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Keramati L., Helmann J. D. (1995). Coordinate Regulation of Bacillus Subtilis Peroxide Stress Genes by Hydrogen Peroxide and Metal Ions. Proc. Natl. Acad. Sci. 92, 8190–8194. 10.1073/pnas.92.18.8190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Wurtmann E. J., Van Batavia J., Zybailov B., Washburn M. P., Wolin S. L. (2007). An Ortholog of the Ro Autoantigen Functions in 23S rRNA Maturation in D. Radiodurans . Genes Dev. 21, 1328–1339. 10.1101/gad.1548207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Xue D., Sun W., Han J., Li J., Gao R., et al. (2019). sRNA OsiA Stabilizes Catalase mRNA during Oxidative Stress Response of Deincoccus Radiodurans R1. Microorganisms 7, 422. 10.3390/microorganisms7100422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang S. M., Schellhorn H. E. (2012). Regulators of Oxidative Stress Response Genes in Escherichia coli and Their Functional Conservation in Bacteria. Arch. Biochem. Biophys. 525, 161–169. 10.1016/j.abb.2012.02.007 [DOI] [PubMed] [Google Scholar]
- Christman M. F., Storz G., Ames B. N. (1989). OxyR, a Positive Regulator of Hydrogen Peroxide-Inducible Genes in Escherichia coli and Salmonella typhimurium, Is Homologous to a Family of Bacterial Regulatory Proteins. Proc. Natl. Acad. Sci. 86, 3484–3488. 10.1073/PNAS.86.10.3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobley J. N., Fiorello M. L., Bailey D. M. (2018). 13 Reasons Why the Brain Is Susceptible to Oxidative Stress. Redox Biol. 15, 490–503. 10.1016/j.redox.2018.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criddle D. N., Gillies S., Baumgartner-Wilson H. K., Jaffar M., Chinje E. C., Passmore S., et al. (2006). Menadione-induced Reactive Oxygen Species Generation via Redox Cycling Promotes Apoptosis of Murine Pancreatic Acinar Cells. J. Biol. Chem. 281, 40485–40492. 10.1074/jbc.M607704200 [DOI] [PubMed] [Google Scholar]
- Dalle-Donne I., Aldini G., Carini M., Colombo R., Rossi R., Milzani A. (2006). Protein Carbonylation, Cellular Dysfunction, and Disease Progression. J. Cel. Mol. Med. 10, 389–406. 10.1111/j.1582-4934.2006.tb00407.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobretsov G. E., Borschevskaya T. A., Petrov V. A., Vladimirov Y. A. (1977). The Increase of Phospholipid Bilayer Rigidity after Lipid Peroxidation. FEBS Lett. 84, 125–128. 10.1016/0014-5793(77)81071-5 [DOI] [PubMed] [Google Scholar]
- Dos Santos R. F., Andrade J. M., Pissarra J., Deutscher M. P., Arraiano C. M. (2020). Hfq and RNase R Mediate rRNA Processing and Degradation in a Novel RNA Quality Control Process. MBio 11. 10.1128/mBio.02398-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos R. F., Quendera A. P., Boavida S., Seixas A. F., Arraiano C. M., Andrade J. M. (2018). Major 3′-5′ Exoribonucleases in the Metabolism of Coding and Non-coding RNA. Prog. Mol. Biol. Transl. Sci. 159, 101–155. 10.1016/bs.pmbts.2018.07.005 [DOI] [PubMed] [Google Scholar]
- Dryden M. S., Cooke J., Salib R. J., Holding R. E., Biggs T., Salamat A. A., et al. (2017). Reactive Oxygen: A Novel Antimicrobial Mechanism for Targeting Biofilm-Associated Infection. J. Glob. Antimicrob. Resist. 8, 186–191. 10.1016/j.jgar.2016.12.006 [DOI] [PubMed] [Google Scholar]
- Dubbs J. M., Mongkolsuk S. (2012). Peroxide-sensing Transcriptional Regulators in Bacteria. J. Bacteriol. 194, 5495–5503. 10.1128/JB.00304-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espey M. G. (2013). Role of Oxygen Gradients in Shaping Redox Relationships between the Human Intestine and its Microbiota. Free Radic. Biol. Med. 55, 130–140. 10.1016/j.freeradbiomed.2012.10.554 [DOI] [PubMed] [Google Scholar]
- Estevez M., Valesyan S., Jora M., Limbach P. A., Addepalli B. (2021). Oxidative Damage to RNA Is Altered by the Presence of Interacting Proteins or Modified Nucleosides. Front. Mol. Biosci. 8, 1–11. 10.3389/fmolb.2021.697149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr S. B., Kogoma T. (1991). Oxidative Stress Responses in Escherichia coli and Salmonella typhimurium . Microbiol. Rev. 55, 561–585. 10.1128/mmbr.55.4.561-585.199110.1128/mr.55.4.561-585.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr S. B., Natvig D. O., Kogoma T. (1985). Toxicity and Mutagenicity of Plumbagin and the Induction of a Possible New DNA Repair Pathway in Escherichia coli . J. Bacteriol. 164, 1309–1316. 10.1128/jb.164.3.1309-1316.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasnacht M., Polacek N. (2021). Oxidative Stress in Bacteria and the Central Dogma of Molecular Biology. Front. Mol. Biosci. 8, 1–13. 10.3389/fmolb.2021.671037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton H. J. H. (1894). LXXIII.-Oxidation of Tartaric Acid in Presence of Iron. J. Chem. Soc. Trans. 65, 899–910. 10.1039/CT8946500899 [DOI] [Google Scholar]
- Fimognari C. (20152015). Role of Oxidative RNA Damage in Chronic-Degenerative Diseases. Oxidative Med. Cell Longevity 2015, 1–8. 10.1155/2015/358713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridovich I. (1989). Superoxide Dismutases. J. Biol. Chem. 264, 7761–7764. 10.1016/S0021-9258(18)83102-7 [DOI] [PubMed] [Google Scholar]
- Fröhlich K. S., Gottesman S. (2018). Small Regulatory RNAs in the Enterobacterial Response to Envelope Damage and Oxidative Stress. Microbiol. Spectr. 6, 1–16. 10.1128/microbiolspec.rwr-0022-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuangthong M., Helmann J. D. (2003). Recognition of DNA by Three Ferric Uptake Regulator (Fur) Homologs in Bacillus Subtilis . J. Bacteriol. 185, 6348–6357. 10.1128/JB.185.21.6348-6357.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Chen X., Tian Y., Yan Y., Zhan Y., Zhou Z., et al. (2020). The Novel ncRNA OsiR Positively Regulates Expression of katE2 and Is Required for Oxidative Stress Tolerance in Deinococcus Radiodurans . Ijms 21, 3200. 10.3390/ijms21093200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudu P., Weiss B. (1996). SoxR, a [2Fe-2S] Transcription Factor, Is Active Only in its Oxidized Form. Proc. Natl. Acad. Sci. 93, 10094–10098. 10.1073/pnas.93.19.10094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-flecha B., Demple B. (2000). Genetic Responses to Free Radicals: Homeostasis and Gene Control. Ann. N. Y. Acad. Sci. 899, 69–87. 10.1111/j.1749-6632.2000.tb06177.x [DOI] [PubMed] [Google Scholar]
- González-Flecha B., Demple B. (1999). Role for the oxyS Gene in Regulation of Intracellular Hydrogen Peroxide in Escherichia coli . J. Bacteriol. 181, 3833–3836. 10.1128/JB.181.12.3833-3836.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gort A. S., Imlay J. A. (1998). Balance between Endogenous Superoxide Stress and Antioxidant Defenses. J. Bacteriol. 180, 1402–1410. 10.1128/jb.180.6.1402-1410.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith K. L., Shah I. M., E. Wolf R., Jr (2004). Proteolytic Degradation of Escherichia coli Transcription Activators SoxS and MarA as the Mechanism for Reversing the Induction of the Superoxide (SoxRS) and Multiple Antibiotic Resistance (Mar) Regulons. Mol. Microbiol. 51, 1801–1816. 10.1046/j.1365-2958.2003.03952.x [DOI] [PubMed] [Google Scholar]
- Gu M., Imlay J. A. (2011). The SoxRS Response of Escherichia coli Is Directly Activated by Redox-Cycling Drugs rather Than by Superoxide. Mol. Microbiol. 79, 1136–1150. 10.1111/j.1365-2958.2010.07520.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., Chen Q., Chen J., Yu J., Hu Y., Zhang S., et al. (2020). 8-Hydroxyguanosine as a Possible RNA Oxidative Modification Marker in Urine from Colorectal Cancer Patients: Evaluation by Ultra Performance Liquid Chromatography-Tandem Mass Spectrometry. J. Chromatogr. B 1136, 121931. 10.1016/j.jchromb.2019.121931 [DOI] [PubMed] [Google Scholar]
- Hangauer M. J., Vaughn I. W., McManus M. T. (2013). Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs. Plos Genet. 9, e1003569. 10.1371/journal.pgen.1003569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa H., Hofer A., Thelander L., Kitajima S., Cai Y., Oshiro S., et al. (1999). Metabolic Fate of Oxidized Guanine Ribonucleotides in Mammalian Cells. Biochemistry 38, 3610–3614. 10.1021/bi982361l [DOI] [PubMed] [Google Scholar]
- Hayakawa H., Kuwano M., Sekiguchi M. (2001). Specific Binding of 8-Oxoguanine-Containing RNA to Polynucleotide Phosphorylase Protein. Biochemistry 40, 9977–9982. 10.1021/bi010595q [DOI] [PubMed] [Google Scholar]
- Hayakawa H., Uchiumi T., Fukuda T., Ashizuka M., Kohno K., Kuwano M., et al. (2002). Binding Capacity of Human YB-1 Protein for RNA Containing 8-oxoguanine. Biochemistry 41, 12739–12744. 10.1021/bi0201872 [DOI] [PubMed] [Google Scholar]
- Hayashi K., Ohsawa T., Kobayashi K., Ogasawara N., Ogura M. (2005). The H 2 O 2 Stress-Responsive Regulator PerR Positively Regulates srfA Expression in Bacillus Subtilis. J. Bacteriol. 187, 6659–6667. 10.1128/JB.187.19.6659-6667.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry A., Shanks J., van Hoof A., Rosenzweig J. A. (2012). TheYersinia Pseudotuberculosisdegradosome Is Required for Oxidative Stress, while its PNPase Subunit Plays a Degradosome-independent Role in Cold Growth. FEMS Microbiol. Lett. 336, 139–147. 10.1111/j.1574-6968.12000.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry C., Loiseau L., Vergnes A., Vertommen D., Mérida-Floriano A., Chitteni-Pattu S., et al. (2021). Redox Controls RecA Protein Activity via Reversible Oxidation of its Methionine Residues. Elife 10. 10.7554/eLife.63747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo Y.-J., Chung I.-Y., Cho W.-J., Lee B.-Y., Kim J.-H., Choi K.-H., et al. (2010). The Major Catalase Gene ( katA ) of Pseudomonas aeruginosa PA14 Is under Both Positive and Negative Control of the Global Transactivator OxyR in Response to Hydrogen Peroxide. J. Bacteriol. 192, 381–390. 10.1128/JB.00980-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo E., Ding H., Demple B. (1997). Redox Signal Transduction: Mutations Shifting [2Fe-2S] Centers of the SoxR Sensor-Regulator to the Oxidized Form. Cell 88, 121–129. 10.1016/S0092-8674(00)81864-4 [DOI] [PubMed] [Google Scholar]
- Hofer T., Badouard C., Bajak E., Ravanat J.-L., Mattsson Å., Cotgreave I. A. (2005). Hydrogen Peroxide Causes Greater Oxidation in Cellular RNA Than in DNA. Biol. Chem. 386, 333–337. 10.1515/BC.2005.040 [DOI] [PubMed] [Google Scholar]
- Ignatov А. V., Bondarenko K. A., Makarova A. V. (2017). Non-bulky Lesions in Human DNA: The Ways of Formation, Repair, and Replication. Acta Naturae 9, 12–26. 10.32607/20758251-2017-9-3-12-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay J. A., Fridovich I. (1991). Assay of Metabolic Superoxide Production in Escherichia coli . J. Biol. Chem. 266, 6957–6965. 10.1016/s0021-9258(20)89596-9 [DOI] [PubMed] [Google Scholar]
- Imlay J. A. (2009). Oxidative Stress. EcoSal Plus 3, 231–237. 10.1128/ecosalplus.5.4.4 [DOI] [PubMed] [Google Scholar]
- Imlay J. A. (2013). The Molecular Mechanisms and Physiological Consequences of Oxidative Stress: Lessons from a Model Bacterium. Nat. Rev. Microbiol. 11, 443–454. 10.1038/nrmicro3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay J. A. (2015). Transcription Factors that Defend Bacteria against Reactive Oxygen Species. Annu. Rev. Microbiol. 69, 93–108. 10.1146/ANNUREV-MICRO-091014-104322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T., Sekiguchi M. (2019). Two Ways of Escaping from Oxidative RNA Damage: Selective Degradation and Cell Death. DNA Repair 81, 102666. 10.1016/j.dnarep.2019.102666 [DOI] [PubMed] [Google Scholar]
- Jacquamet L., Traoré D. A. K., Ferrer J.-L., Proux O., Testemale D., Hazemann J.-L., et al. (2009). Structural Characterization of the Active Form of PerR: Insights into the Metal-Induced Activation of PerR and Fur Proteins for DNA Binding. Mol. Microbiol. 73, 20–31. 10.1111/j.1365-2958.2009.06753.x [DOI] [PubMed] [Google Scholar]
- Kavita K., de Mets F., Gottesman S. (2018). New Aspects of RNA-Based Regulation by Hfq and its Partner sRNAs. Curr. Opin. Microbiol. 42, 53–61. 10.1016/j.mib.2017.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer J. P. (2000). The Haber-Weiss Reaction and Mechanisms of Toxicity. Toxicology 149, 43–50. 10.1016/S0300-483X(00)00231-6 [DOI] [PubMed] [Google Scholar]
- Khademian M., Imlay J. A. (2017). Escherichia coli Cytochrome C Peroxidase Is a Respiratory Oxidase that Enables the Use of Hydrogen Peroxide as a Terminal Electron Acceptor. Proc. Natl. Acad. Sci. USA 114, E6922–E6931. 10.1073/pnas.1701587114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Fujikawa M., Kozawa T. (2014). Oxidative Stress Sensing by the Iron-Sulfur Cluster in the Transcription Factor, SoxR. J. Inorg. Biochem. 133, 87–91. 10.1016/j.jinorgbio.2013.11.008 [DOI] [PubMed] [Google Scholar]
- Kong Q., Lin C.-l. G. (2010). Oxidative Damage to RNA: Mechanisms, Consequences, and Diseases. Cell. Mol. Life Sci. 67, 1817–1829. 10.1007/s00018-010-0277-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo M.-S., Lee J.-H., Rah S.-Y., Yeo W.-S., Lee J.-W., Lee K.-L., et al. (2003). A Reducing System of the Superoxide Sensor SoxR in Escherichia coli . EMBO J. 22, 2614–2622. 10.1093/emboj/cdg252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küpfer P. A., Leumann C. J. (2014). “Oxidative Damage on RNA Nucleobases”in Chemical Biology Of Nucleic Acids . Berlin, Heidelberg: Springer Berlin Heidelberg, 75–94. 10.1007/978-3-642-54452-1_5 [DOI] [Google Scholar]
- Lalaouna D., Baude J., Wu Z., Tomasini A., Chicher J., Marzi S., et al. (2019). RsaC sRNA Modulates the Oxidative Stress Response of Staphylococcus aureus during Manganese Starvation. Nucleic Acids Res. 47, 9871–9887. 10.1093/nar/gkz728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Lee S. M., Mukhopadhyay P., Kim S. J., Lee S. C., Ahn W.-S., et al. (2004). Redox Regulation of OxyR Requires Specific Disulfide Bond Formation Involving a Rapid Kinetic Reaction Path. Nat. Struct. Mol. Biol. 11, 1179–1185. 10.1038/nsmb856 [DOI] [PubMed] [Google Scholar]
- Lee J.-W., Helmann J. D. (20062006 44070). The PerR Transcription Factor Senses H2O2 by Metal-Catalysed Histidine Oxidation. Nature 440, 363–367. 10.1038/nature04537 [DOI] [PubMed] [Google Scholar]
- Lee T.-H., Kang T.-H. (2019). DNA Oxidation and Excision Repair Pathways. Ijms 20, 6092. 10.3390/ijms20236092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiva L. E., Pincheira A., Elgamal S., Kienast S. D., Bravo V., Leufken J., et al. (2020). Modulation of Escherichia coli Translation by the Specific Inactivation of tRNAGly under Oxidative Stress. Front. Genet. 11. 10.3389/fgene.2020.00856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Singh A. K., McIntyre L. M., Sherman L. A. (2004). Differential Gene Expression in Response to Hydrogen Peroxide and the Putative PerR Regulon of Synechocystis Sp. Strain PCC 6803. J. Bacteriol. 186, 3331–3345. 10.1128/JB.186.11.3331-3345.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Chen X., Liu Z., Ye W., Li L., Qian L., et al. (2020). Recent Advances: Molecular Mechanism of RNA Oxidation and its Role in Various Diseases. Front. Mol. Biosci. 7, 1–7. 10.3389/fmolb.2020.00184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Malla S., Shin B., Li J. M. (2014). Battle against RNA Oxidation: Molecular Mechanisms for Reducing Oxidized RNA to Protect Cells. WIREs RNA 5, 335–346. 10.1002/wrna.1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wu J., DeLeo C. (2006). RNA Damage and Surveillance under Oxidative Stress. IUBMB Life (International Union Biochem. Mol. Biol. Life) 58, 581–588. 10.1080/15216540600946456 [DOI] [PubMed] [Google Scholar]
- Liochev S. I., Benov L., Touati D., Fridovich I. (1999). Induction of the soxRS Regulon of Escherichia coli by Superoxide. J. Biol. Chem. 274, 9479–9481. 10.1074/jbc.274.14.9479 [DOI] [PubMed] [Google Scholar]
- Liu M., Gong X., Alluri R. K., Wu J., Sablo T., Li Z. (2012). Characterization of RNA Damage under Oxidative Stress in Escherichia coli . Biol. Chem. 393, 123–132. 10.1515/hsz-2011-0247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lushchak V. I., Storey K. B. (2021). Oxidative Stress Concept Updated: Definitions, Classifications, and Regulatory Pathways Implicated. EXCLI J. 20, 956–967. 10.17179/excli2021-3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki H., Sekiguchi M. (1992). MutT Protein Specifically Hydrolyses a Potent Mutagenic Substrate for DNA Synthesis. Nature 355, 273–275. 10.1038/355273a0 [DOI] [PubMed] [Google Scholar]
- Malla S., Kwakye A. (2021). RNA Oxidation: Role of Polynucleotide Phosphorylase in the Quality Control of Oxidized RNA. J. Life Sci. 3, 43–60. 10.36069/jols/20210603 [DOI] [Google Scholar]
- Markkanen E. (2017). Not breathing Is Not an Option: How to deal with Oxidative DNA Damage. DNA Repair 59, 82–105. 10.1016/j.dnarep.2017.09.007 [DOI] [PubMed] [Google Scholar]
- Massé E., Gottesman S. (2002). A Small RNA Regulates the Expression of Genes Involved in Iron Metabolism in Escherichia coli . Proc. Natl. Acad. Sci. 99, 4620–4625. 10.1073/pnas.032066599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Wolffe A. P. (1998). Gene Regulation by Y-Box Proteins: Coupling Control of Transcription and Translation. Trends Cel Biol. 8, 318–323. 10.1016/S0962-8924(98)01300-2 [DOI] [PubMed] [Google Scholar]
- Mettert E. L., Kiley P. J. (2015). Fe-S Proteins that Regulate Gene Expression. Biochim. Biophys. Acta (Bba) - Mol. Cel Res. 1853, 1284–1293. 10.1016/j.bbamcr.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S., Imlay J. (2012). Why Do Bacteria Use So many Enzymes to Scavenge Hydrogen Peroxide? Arch. Biochem. Biophys. 525, 145–160. 10.1016/j.abb.2012.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsuk S., Helmann J. D. (2002). Regulation of Inducible Peroxide Stress Responses. Mol. Microbiol. 45, 9–15. 10.1046/j.1365-2958.2002.03015.x [DOI] [PubMed] [Google Scholar]
- Morikawa K., Ohniwa R. L., Kim J., Maruyama A., Ohta T., Takeyasu K. (2006). Bacterial Nucleoid Dynamics: Oxidative Stress Response in Staphylococcus aureus . Genes Cells 11, 409–423. 10.1111/j.1365-2443.2006.00949.x [DOI] [PubMed] [Google Scholar]
- Morzel M., Gatellier P., Sayd T., Renerre M., Laville E. (2006). Chemical Oxidation Decreases Proteolytic Susceptibility of Skeletal Muscle Myofibrillar Proteins. Meat Sci. 73, 536–543. 10.1016/j.meatsci.2006.02.005 [DOI] [PubMed] [Google Scholar]
- Niki E. (2009). Lipid Peroxidation: Physiological Levels and Dual Biological Effects. Free Radic. Biol. Med. 47, 469–484. 10.1016/j.freeradbiomed.2009.05.032 [DOI] [PubMed] [Google Scholar]
- Nishiyama Y., Allakhverdiev S. I., Murata N. (2006). A New Paradigm for the Action of Reactive Oxygen Species in the Photoinhibition of Photosystem II. Biochim. Biophys. Acta (Bba) - Bioenerg. 1757, 742–749. 10.1016/j.bbabio.2006.05.013 [DOI] [PubMed] [Google Scholar]
- Nunoshiba T., Hidalgo E., Amábile Cuevas C. F., Demple B. (1992). Two-stage Control of an Oxidative Stress Regulon: the Escherichia coli SoxR Protein Triggers Redox-Inducible Expression of the soxS Regulatory Gene. J. Bacteriol. 174, 6054–6060. 10.1128/jb.174.19.6054-6060.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino T., Maegawa S., Shigeno S., Fujikura K., Toyohara H. (2018). Highly Sensitive Avoidance Plays a Key Role in Sensory Adaptation to Deep-Sea Hydrothermal Vent Environments. PLoS One 13, e0189902–10. 10.1371/journal.pone.0189902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.-Y., Shin J.-H., Roe J.-H. (2007). Dual Role of OhrR as a Repressor and an Activator in Response to Organic Hydroperoxides in Streptomyces Coelicolor . J. Bacteriol. 189, 6284–6292. 10.1128/JB.00632-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge J. D., Poole R. K., Green J. (2007). The Escherichia coli yhjA Gene, Encoding a Predicted Cytochrome C Peroxidase, Is Regulated by FNR and OxyR. Microbiology 153, 1499–1509. 10.1099/mic.0.2006/004838-0 [DOI] [PubMed] [Google Scholar]
- Patil A. G. G., Sang P. B., Govindan A., Varshney U. (2013). Mycobacterium tuberculosis MutT1 (Rv2985) and ADPRase (Rv1700) Proteins Constitute a Two-Stage Mechanism of 8-Oxo-dGTP and 8-Oxo-GTP Detoxification and Adenosine to Cytidine Mutation Avoidance. J. Biol. Chem. 288, 11252–11262. 10.1074/jbc.M112.442566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pericone C. D., Overweg K., Hermans P. W. M., Weiser J. N. (2000). Inhibitory and Bactericidal Effects of Hydrogen Peroxide Production by Streptococcus Pneumoniae on Other Inhabitants of the Upper Respiratory Tract. Infect. Immun. 68, 3990–3997. 10.1128/IAI.68.7.3990-3997.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomposiello P. J., Koutsolioutsou A., Carrasco D., Demple B. (2003). SoxRS-regulated Expression and Genetic Analysis of the yggX Gene of Escherichia coli . J. Bacteriol. 185, 6624–6632. 10.1128/JB.185.22.6624-6632.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quendera A. P., Seixas A. F., Dos Santos R. F., Santos I., Silva J. P. N., Arraiano C. M., et al. (2020). RNA-binding Proteins Driving the Regulatory Activity of Small Non-coding RNAs in Bacteria. Front. Mol. Biosci. 7, 78. 10.3389/fmolb.2020.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radak Z., Boldogh I. (2010). 8-Oxo-7,8-dihydroguanine: Links to Gene Expression, Aging, and Defense against Oxidative Stress. Free Radic. Biol. Med. 49, 587–596. 10.1016/j.freeradbiomed.2010.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter R. J., Melchiorri D., Sewerynek E., Poeggeler B., Barlow-Walden L., Chuang J., et al. (1995). A Review of the Evidence Supporting Melatonin's Role as an Antioxidant. J. Pineal Res. 18, 1–11. 10.1111/j.1600-079X.1995.tb00133.x [DOI] [PubMed] [Google Scholar]
- Rudd S. G., Valerie N. C. K., Helleday T. (2016). Pathways Controlling dNTP Pools to Maintain Genome Stability. DNA Repair 44, 193–204. 10.1016/j.dnarep.2016.05.032 [DOI] [PubMed] [Google Scholar]
- Schellhorn H. E., Hassan H. M. (1988). Transcriptional Regulation of katE in Escherichia coli K-12. J. Bacteriol. 170, 4286–4292. 10.1128/jb.170.9.4286-4292.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaver L. C., Imlay J. A. (2001). Hydrogen Peroxide Fluxes and Compartmentalization inside Growing Escherichia coli . J. Bacteriol. 183, 7182–7189. 10.1128/JB.183.24.7182-7189.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., Iida K.-i., Saito M., Nakayama H., Yoshida S.-i. (2004). Hydrogen Peroxide Production in Streptococcus Pyogenes : Involvement of Lactate Oxidase and Coupling with Aerobic Utilization of Lactate. J. Bacteriol. 186, 2046–2051. 10.1128/JB.186.7.2046-2051.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A., Imlay J. A. (2021). How Microbes Defend Themselves from Incoming Hydrogen Peroxide. Front. Immunol. 12, 1104. 10.3389/FIMMU.2021.667343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok H., Lee H., Lee S., Ahn S. H., Lee H.-S., Kim G.-W. D., et al. (2020). Position-specific Oxidation of miR-1 Encodes Cardiac Hypertrophy. Nature 584, 279–285. 10.1038/s41586-020-2586-0 [DOI] [PubMed] [Google Scholar]
- Shacter E. (2000). Quantification and Significance of Protein Oxidation in Biological Samples*. Drug Metab. Rev. 32, 307–326. 10.1081/DMR-100102336 [DOI] [PubMed] [Google Scholar]
- Shatwell K. P., Segal A. W. (1996). NADPH Oxidase. Int. J. Biochem. Cel Biol. 28, 1191–1195. 10.1016/S1357-2725(96)00084-2 [DOI] [PubMed] [Google Scholar]
- Shcherbik N., Pestov D. G. (2019). The Impact of Oxidative Stress on Ribosomes: From Injury to Regulation. Cells 8, 1379. 10.3390/cells8111379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Wu W., Hazen S. L. (2000). Activated Leukocytes Oxidatively Damage DA, RNA, and the Nucleotide Pool through Halide-dependent Formation of Hydroxyl Radical. Biochemistry 39, 5474–5482. 10.1021/bi992809y [DOI] [PubMed] [Google Scholar]
- Simms C. L., Hudson B. H., Mosior J. W., Rangwala A. S., Zaher H. S. (2014). An Active Role for the Ribosome in Determining the Fate of Oxidized mRNA. Cel Rep. 9, 1256–1264. 10.1016/j.celrep.2014.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms C. L., Zaher H. S. (2016). Quality Control of Chemically Damaged RNA. Cel. Mol. Life Sci. 73, 3639–3653. 10.1007/s00018-016-2261-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman E. R. (2001). Protein Oxidation in Aging and Age-Related Diseases. Ann. N. Y. Acad. Sci. 928, 22–38. 10.1111/j.1749-6632.2001.tb05632.x [DOI] [PubMed] [Google Scholar]
- Steenken S., Jovanovic S. V. (1997). How Easily Oxidizable Is DNA? One-Electron Reduction Potentials of Adenosine and Guanosine Radicals in Aqueous Solution. J. Am. Chem. Soc. 119, 617–618. 10.1021/ja962255b [DOI] [Google Scholar]
- Steinman H. M. (1988). “Bacterial Superoxide Dismutases” in Oxygen Radicals In Biology And Medicine (Boston, MA: Springer US, 641–646. 10.1007/978-1-4684-5568-7_101 [DOI] [PubMed] [Google Scholar]
- Su L.-J., Zhang J.-H., Gomez H., Murugan R., Hong X., Xu D., et al. (20192019). Reactive Oxygen Species-Induced Lipid Peroxidation in Apoptosis, Autophagy, and Ferroptosis. Oxidative Med. Cell Longevity 2019, 1–13. 10.1155/2019/5080843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Jora M., Solivio B., Limbach P. A., Addepalli B. (2018). The Effects of Ultraviolet Radiation on Nucleoside Modifications in RNA. ACS Chem. Biol. 13, 567–572. 10.1021/acschembio.7b00898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Limbach P. A., Addepalli B. (2020). Characterization of UVA-Induced Alterations to Transfer RNA Sequences. Biomolecules 10, 1527. 10.3390/biom10111527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntres Z. (2002). Role of Antioxidants in Paraquat Toxicity. Toxicology 180, 65–77. 10.1016/S0300-483X(02)00382-7 [DOI] [PubMed] [Google Scholar]
- Taddei F., Hayakawa H., Bouton M.-F., Cirinesi A.-M., Matic I., Sekiguchi M., et al. (1997). Counteraction by MutT Protein of Transcriptional Errors Caused by Oxidative Damage. Science 278, 128–130. 10.1126/science.278.5335.128 [DOI] [PubMed] [Google Scholar]
- Tanaka M., Chock P. B. (2021). Oxidative Modifications of RNA and its Potential Roles in Biosystem. Front. Mol. Biosci. 8, 1–13. 10.3389/fmolb.2021.685331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapley D. W., Buettner G. R., Shick J. M. (1999). Free Radicals and Chemiluminescence as Products of the Spontaneous Oxidation of Sulfide in Seawater, and Their Biological Implications. Biol. Bull. 196, 52–56. 10.2307/1543166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat B., Touati D. (1991). Two Global Regulators Repress the Anaerobic Expression of MnSOD in Escherichia coli: Fur (Ferric Uptake Regulation) and Arc (Aerobic Respiration Control). Mol. Microbiol. 5, 455–465. 10.1111/j.1365-2958.1991.tb02129.x [DOI] [PubMed] [Google Scholar]
- Thomas E. N., Simms C. L., Keedy H. E., Zaher H. S. (2019). Insights into the Base-Pairing Preferences of 8-oxoguanosine on the Ribosome. Nucleic Acids Res. 47, 9857–9870. 10.1093/nar/gkz701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D. M., Lu C., Green P. J., Parker R. (2008). tRNA Cleavage Is a Conserved Response to Oxidative Stress in Eukaryotes. Rna 14, 2095–2103. 10.1261/rna.1232808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledano M. B., Kullik I., Trinh F., Baird P. T., Schneider T. D., Storz G. (1994). Redox-dependent Shift of OxyR-DNA Contacts along an Extended DNA-Binding Site: A Mechanism for Differential Promoter Selection. Cell 78, 897–909. 10.1016/S0092-8674(94)90702-1 [DOI] [PubMed] [Google Scholar]
- Tong H., Chen W., Merritt J., Qi F., Shi W., Dong X. (2007). Streptococcus Oligofermentans Inhibits Streptococcus Mutans through Conversion of Lactic Acid into Inhibitory H2O2: a Possible Counteroffensive Strategy for Interspecies Competition. Mol. Microbiol. 63, 872–880. 10.1111/j.1365-2958.2006.05546.x [DOI] [PubMed] [Google Scholar]
- Touati D. (2000). Sensing and Protecting against Superoxide Stress inEscherichia Coli- How many Ways Are There to triggersoxRSresponse? Redox Rep. 5, 287–293. 10.1179/135100000101535825 [DOI] [PubMed] [Google Scholar]
- van Vliet A. H. M., Baillon M.-L. A., Penn C. W., Ketley J. M. (1999). Campylobacter Jejuni Contains Two Fur Homologs: Characterization of Iron-Responsive Regulation of Peroxide Stress Defense Genes by the PerR Repressor. J. Bacteriol. 181, 6371–6376. 10.1128/JB.181.20.6371-6376.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan P. P., Blough N. V. (1998). Photochemical Formation of Hydroxyl Radical by Constituents of Natural Waters. Environ. Sci. Technol. 32, 2947–2953. 10.1021/es9710417 [DOI] [Google Scholar]
- Wang J.-X., Gao J., Ding S.-L., Wang K., Jiao J.-Q., Wang Y., et al. (2015). Oxidative Modification of miR-184 Enables it to Target Bcl-xL and Bcl-W. Mol. Cel 59, 50–61. 10.1016/j.molcel.2015.05.003 [DOI] [PubMed] [Google Scholar]
- Welsch C. W. (1987). Enhancement of Mammary Tumorigenesis by Dietary Fat: Review of Potential Mechanisms. Am. J. Clin. Nutr. 45, 192–202. 10.1093/ajcn/45.1.192 [DOI] [PubMed] [Google Scholar]
- Willi J., Küpfer P., Evéquoz D., Fernandez G., Katz A., Leumann C., et al. (2018). Oxidative Stress Damages rRNA inside the Ribosome and Differentially Affects the Catalytic center. Nucleic Acids Res. 46, 1945–1957. 10.1093/nar/gkx1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterbourn C. C. (2013). The Biological Chemistry of Hydrogen Peroxide. 1st ed. Elsevier, 3–25. 10.1016/B978-0-12-405881-1.00001-X The Biological Chemistry of Hydrogen Peroxide [DOI] [PubMed] [Google Scholar]
- Wu H.-J., Seib K. L., Srikhanta Y. N., Kidd S. P., Edwards J. L., Maguire T. L., et al. (2006). PerR Controls Mn-dependent Resistance to Oxidative Stress in Neisseria Gonorrhoeae . Mol. Microbiol. 60, 401–416. 10.1111/J.1365-2958.2006.05079.X [DOI] [PubMed] [Google Scholar]
- Wu J., Jiang Z., Liu M., Gong X., Wu S., Burns C. M., et al. (2009). Polynucleotide Phosphorylase Protects Escherichia coli against Oxidative Stress. Biochemistry 48, 2012–2020. 10.1021/bi801752p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Li Z. (2008). Human Polynucleotide Phosphorylase Reduces Oxidative RNA Damage and Protects HeLa Cell against Oxidative Stress. Biochem. Biophysical Res. Commun. 372, 288–292. 10.1016/j.bbrc.2008.05.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Weiss B. (1992). Two-stage Induction of the soxRS (Superoxide Response) Regulon of Escherichia coli . J. Bacteriol. 174, 3915–3920. 10.1128/jb.174.12.3915-3920.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L. L., Zaher H. S. (2019). How Do Cells Cope with RNA Damage and its Consequences? J. Biol. Chem. 294, 15158–15171. 10.1074/jbc.REV119.006513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Xu L., Porter N. A. (2011). Free Radical Lipid Peroxidation: Mechanisms and Analysis. Chem. Rev. 111, 5944–5972. 10.1021/cr200084z [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhan Y., Yan Y., Liu Y., Hu G., Wang S., et al. (2019). The Pseudomonas Stutzeri-specific Regulatory Noncoding RNA NfiS Targets katB mRNA Encoding a Catalase Essential for Optimal Oxidative Resistance and Nitrogenase Activity. J. Bacteriol. 201. 10.1128/JB.00334-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Xiao S., Ahn D. U. (2013). Protein Oxidation: Basic Principles and Implications for Meat Quality. Crit. Rev. Food Sci. Nutr. 53, 1191–1201. 10.1080/10408398.2011.577540 [DOI] [PubMed] [Google Scholar]
- Zhao X., Drlica K. (2014). Reactive Oxygen Species and the Bacterial Response to Lethal Stress. Curr. Opin. Microbiol. 21, 1–6. 10.1016/j.mib.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Doan B., Schneider T. D., Storz G. (1999). OxyR and SoxRS Regulation of Fur . J. Bacteriol. 181, 4639–4643. 10.1128/JB.181.15.4639-4643.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Wang X., Doan B., Lewis K. A., Schneider T. D., Storz G. (2001a). Computation-directed Identification of OxyR DNA Binding Sites in Escherichia coli . J. Bacteriol. 183, 4571–4579. 10.1128/JB.183.15.4571-4579.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Wang X., Templeton L. J., Smulski D. R., Larossa R. A., Storz G. (2001b). DNA Microarray-Mediated Transcriptional Profiling of the Escherichia coli Response to Hydrogen Peroxide. J. Bacteriol. 183, 4562–4570. 10.1128/JB.183.15.4562-4570.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Åslund F., Storz G. (1998). Activation of the OxyR Transcription Factor by Reversible Disulfide Bond Formation. Science 279, 1718–1722. 10.1126/SCIENCE.279.5357.1718 [DOI] [PubMed] [Google Scholar]
- Zhong J., Xiao C., Gu W., Du G., Sun X., He Q.-Y., et al. (2015). Transfer RNAs Mediate the Rapid Adaptation of Escherichia coli to Oxidative Stress. Plos Genet. 11, e1005302–24. 10.1371/journal.pgen.1005302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M., Dai X. (2019). Maintenance of Translational Elongation Rate Underlies the Survival of Escherichia coli during Oxidative Stress. Nucleic Acids Res. 47, 7592–7604. 10.1093/nar/gkz467 [DOI] [PMC free article] [PubMed] [Google Scholar]