Summary
Borna disease virus 1 (BoDV-1) causes rare but often fatal encephalitis in humans. Late diagnosis prohibits an experimental therapeutic approach. Here, we report a recent case of fatal BoDV-1 infection diagnosed on day 12 after hospitalization by detection of BoDV-1 RNA in the cerebrospinal fluid. In a retrospective analysis, we detect BoDV-1 RNA 1 day after hospital admission when the cell count in the cerebrospinal fluid is still normal. We develop a new ELISA using recombinant BoDV-1 nucleoprotein, phosphoprotein, and accessory protein X to detect seroconversion on day 12. Antibody responses are also shown in seven previously confirmed cases. The individual BoDV-1 antibody profiles show variability, but the usage of three different BoDV-1 antigens results in a more sensitive diagnostic tool. Our findings demonstrate that early detection of BoDV-1 RNA in cerebrospinal fluid and the presence of antibodies against at least two different viral antigens contribute to BoDV-1 diagnosis. Physicians in endemic regions should consider BoDV-1 infection in cases of unclear encephalopathy and initiate appropriate diagnostics at an early stage.
Keywords: Borna disease virus 1, encephalitis, encephalopathy, zoonosis, recombinant protein, diagnostics, humoral immune response, antibodies, ELISA
Graphical abstract
Highlights
-
•
Borna disease virus 1 causes fatal encephalitis upon zoonotic spillover infections
-
•
An ELISA system using recombinant BoDV-1 N, X, and P proteins has been established
-
•
Antibodies against at least two different BoDV-1 antigens corroborate seroconversion
-
•
Early detection of viral RNA and antibodies could contribute to BoDV-1 diagnosis
Neumann et al. provide data on antibody responses to Borna disease virus 1 in eight cases of fatal encephalitis based on a newly developed ELISA using recombinant BoDV-1 nucleoprotein, phosphoprotein, and accessory protein X. The presence of antibodies against at least two different viral antigens contributes to BoDV-1 diagnosis.
Introduction
Borna disease virus 1 (BoDV-1), the causative agent of Borna disease in horses, sheep, and other domestic mammals, induces severe to fatal encephalitis in humans in endemic regions of southern and eastern Germany.1, 2, 3, 4, 5, 6 The bicolored white-toothed shrew (Crocidura leucodon) is the only known natural reservoir host so far.7,8 The pathogenesis of BoDV-1 infections is mainly attributed to an immunopathology mediated by virus-specific CD8+ and CD4+ T cells.9 Residence in mainly rural environments has been discussed as a risk factor for BoDV-1 infections.5 To date, there is no established therapy. In cell cultures, however, ribavirin and favipiravir (T-705) showed antiviral activity against bornaviruses.10,11 As diagnostic tools, BoDV-1-specific qRT-PCR, indirect immunofluorescence assay (iIFA), and a line blot assay have been established.5 Sensitivity of qRT-PCR is limited for cerebrospinal fluid (CSF) but reliable for biopsies or post mortem tissue samples from the brain.4,5 As iIFA is laborious and its assessment is, to some extent, subjective, while the sensitivity of the line blot assay is limited, there is a need for developing additional serological tests, such as ELISA.5 The earliest possible time for a reliable diagnosis in the course of disease and the existence of a possible window for curative treatment are poorly understood. Here, we report a case of BoDV-1 infection that was diagnosed by qRT-PCR from CSF and via seroconversion using a newly established ELISA system with recombinant BoDV-1 proteins.
Results
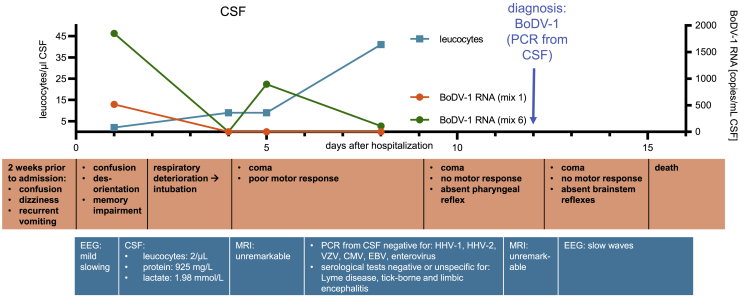
The affected individual, who was over 70 years of age, was admitted to the department of neurology of a tertiary care hospital in a BoDV-1-endemic region of southern Germany with a 2-week history of confusion, dizziness, memory impairment, and recurrent vomiting. Their medical history was insignificant, especially without immunosuppression or organ transplantation. An electroencephalogram (EEG) displayed mild diffuse slowing, and CSF parameters were non-specifically altered, indicating a blood-brain barrier disruption with a normal cell count (2 leucocytes/μL, lactate 1.98 mmol/L, protein 925 mg/L) on the day after admission (Figure 1). The next day, the individual was found unconscious and was intubated based on the assumption of severe aspiration pneumonia. The individual remained in a deep coma after stopping light sedation on day 5. CSF analysis on day 8 showed a CSF leucocyte count of 41 μL (Figure 1) and negative PCR results for herpes simplex virus, varicella-zoster virus, cytomegalovirus, Epstein-Barr virus, and enterovirus. Serological indicators of Lyme disease, tick-borne encephalitis, or autoimmune encephalitis were not detectable. On day 12, BoDV-1 infection was diagnosed by a positive qRT-PCR result2 from CSF that had been collected on day 8 after admission. At the time of the diagnosis, the individual was in a deep coma, showing no motor responses. Multiple brainstem reflexes were absent. Two brain magnetic resonance images (MRIs) on days 4 and 11 remained unremarkable. Following the individual’s reported will, we decided against experimental therapies and for palliative care. The individual died on day 15 after admission. Other available CSF samples of the individual were retrospectively examined by qRT-PCR. The first CSF obtained on day 1 after admission yielded positive results for BoDV-1 RNA ranging between 515 and 1,850 copies/mL, whereas a CSF obtained on day 4 tested negative (detection limit: 300 copies/mL; Figure 1). Post mortem, all samples from non-neuronal tissue tested negative, while samples from the frontal cortex as well as from the optical nerve and peripheral nerve tissue contained a high RNA load (Table 1). A histological work-up showed marked necrotizing lymphocytic neuritis of the peripheral femoral and small visceral nerves (data not shown).
Figure 1.
Time course of disease
A 71-year-old individual without significant past medical history living in an endemic area in southern Germany developed unspecific symptoms of BoDV-1 infection 2 weeks prior to hospitalization. The x axis shows days after hospital admission. Leucocyte count (cells/μL) in cerebrospinal fluid (CSF) is represented on the left y axis, and RNA copies (copies/mL) in CSF obtained through qRT-PCR of two target regions are represented on the right y axis. The diagnosis was made by qRT-PCR from CSF on day 12. Workup of other infectious or autoimmune causes of encephalitis remained negative. The individual died on day 15 after admission.
Table 1.
BoDV-1 qRT-PCR from post mortem tissue samples of the affected individual
| Sample | BoDV-1 RNA (copies/mL organ homogenate) |
|
|---|---|---|
| Mix 1 | Mix 6 | |
| Frontal cortex | 1.50E10 | 2.65E8 |
| Optical nerve | 3.05E9 | 7.15E7 |
| Brachial plexus | 6.15E6 | 9.80E5 |
| Femoral nerve | 3.30E4 | 1.90E4 |
| Sural nerve | 1.35E9 | 4.90E7 |
| Sartorius muscle | <3E2 | <3E2 |
| Heart muscle | <3E2 | <3E2 |
| Blood (post mortem) | <3E2 | <3E2 |
| Lung | <3E2 | <3E2 |
| Cervical lymph node | <3E2 | <3E2 |
| Spleen | <3E2 | <3E2 |
| Liver | <3E2 | <3E2 |
| Kidney | <3E2 | <3E2 |
| Urine (post mortem) | <3E2 | <3E2 |
| Stool (post mortem) | <3E2 | <3E2 |
Two targets were analyzed. Tissue samples were homogenized in approximately 1 mL 0.9% sodium chloride.
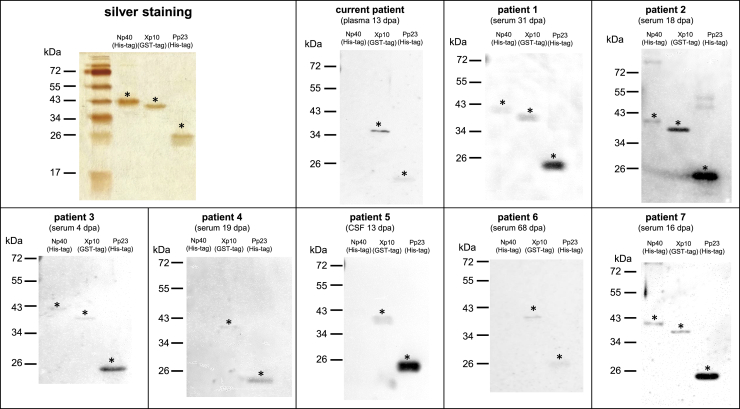
Serological tests were performed with all available serum and CSF samples of the current individual. Using iIFA, seroconversion was detected on day 13 (titer 320), whereas BoDV-1-specific antibodies were not detected in serum or CSF samples obtained earlier. To further characterize the immune response against different BoDV-1 antigens, recombinant BoDV-1 nucleoprotein (N), accessory protein X (X), and phosphoprotein (P) proteins were expressed in E. coli. Purity of these proteins was confirmed by silver staining (Figure 2). Further, the plasma sample of the current affected individual, which tested positive by iIFA, as well as selected iIFA-positive samples of 7 additional previously confirmed BoDV-1-infected individuals2,4 were evaluated by western blot using the recombinant BoDV-1 proteins (Figure 2). Samples of all affected individuals showed at least two reactive bands.
Figure 2.
Silver staining and western blots of purified recombinant BoDV-1 proteins
For silver staining, proteins were separated by SDS-PAGE using a 15% polyacrylamide gel. Lanes were loaded with 200 ng His-tagged Np40, 200 ng GST-tagged Xp10, and 100 ng His-tagged Pp23 protein. For western blots, lanes were loaded with 100 ng Np40, 200 ng Xp10, and 100 ng Pp23. Proteins were separated by SDS-PAGE using a 12% polyacrylamide gel and then blotted onto a 0.2 μm nitrocellulose membrane. Selected samples of 8 confirmed BoDV-1-infected individuals were diluted 1:100. Time of sample collection is expressed as days post admission (dpa) to hospital. Asterisks mark specific bands with the expected molecular weight expressed in kDa. All experiments were carried out once.
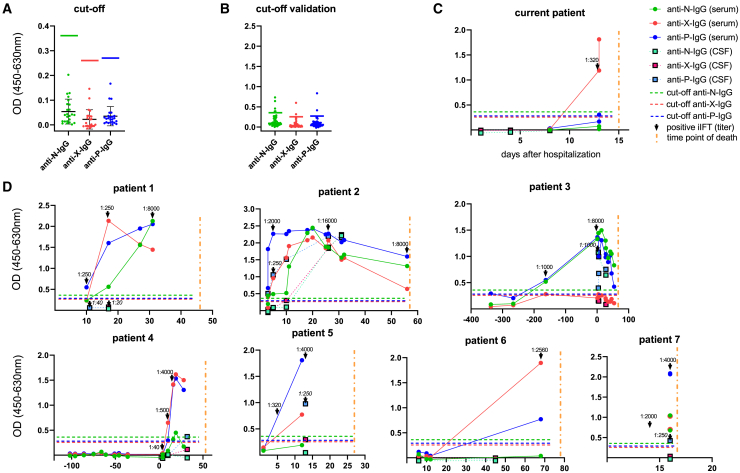
In a next step, these proteins were used to establish an ELISA. Thresholds were determined using 24 serum samples that had tested negative for BoDV-1 using iIFA (Figure 3A). For each antigen, an individual cutoff was defined as the mean plus six standard deviations. In accordance with the iIFA result, seroconversion for the current affected individual was detected by the ELISA on day 13, mainly based on reactivity against BoDV-1 X protein, whereas anti-BoDV-1-P-immunoglobulin G (IgG) was just above the cutoff in the last plasma sample obtained, and anti-BoDV-1-N-IgG was not detectable (Figure 3C).
Figure 3.
BoDV-1 IgG-ELISA
Three ELISA systems for the detection of IgG antibodies against BoDV-1 nucleoprotein (N), accessory protein X, and phosphoprotein (P) were developed. All samples were diluted 1:100 prior to testing. Dots represent serum/plasma samples (green: anti-BoDV-1-N-IgG; red: anti-BoDV-1-X-IgG; blue: anti-BoDV-1-P-IgG), and squares represent CSF samples (light green: anti-BoDV-1-N-IgG; magenta: anti-BoDV-1-X-IgG; light blue: anti-BoDV-1-P-IgG). The x axis shows days after hospital admission, and the y axis shows optical density (OD). Cutoffs for each ELISA system are indicated by horizontal lines (green: anti-BoDV-1-N ELISA; red: anti-BoDV-1-X ELISA; blue: anti-BoDV-1-P ELISA). A vertical yellow line indicates the time of death. Black arrows point to corresponding indirect immunofluorescence assay (iIFA) results. iIFA titers in CSF are indicated in italics.
(A) Determination of cutoffs as the sum of arithmetic mean (black horizontal line) plus 6 standard deviations (error line) of 24 samples, which had been characterized as negative via iIFA. Green, red, and blue horizontal lines represent cutoffs.
(B) Reactivity against BoDV-1 N, X, and P antigens in samples of affected individuals with suspected tick-borne encephalitis. Horizontal lines represent cutoffs.
(C) Time course of BoDV-1 antibodies in the current case.
(D) ELISA results for another seven confirmed and recently published BoDV-1-infected individuals. All experiments were carried out once. Interassay variabilities were calculated for N, X, and P antigens in five runs of one sample of individual 7, resulting in 30%, 10%, and 5%, respectively.
To further evaluate the sensitivity of the ELISA, longitudinal serum/plasma and CSF samples from the seven previously confirmed BoDV-1-infected individuals were tested. Both anti-BoDV-1-X-IgG and anti-BoDV-1-P-IgG were detected in the serum/plasma of all individuals during the course of the disease, while anti-BoDV-1-N-IgG did not exceed the cutoff value in two affected individuals (Figure 3D). All samples with iIFA titers above 100 were detected by IgG ELISA, showing values above the cutoff against at least one BoDV-1 antigen (Figures 3D and S1A). In the seven selected samples tested by western blot, the ELISA showed a high agreement in terms of positive antigen reactions; only a borderline-positive anti-N-IgG ELISA result for individual 4 did not show a corresponding positive band in western blot (Figures 2 and 3D). The ELISA remained negative for two CSF samples with iIFA titers of 40 and 20 (individual 1) and a serum with an iIFA titer of 40 (individual 4) collected during early stages of disease, which is in congruence with the 100-fold sample dilution used for the ELISA. In addition, iIFT titers significantly correlated with ELISA values in BoDV-1-infected individuals (Figure S2).
To further assess the specificity of the ELISA, we took advantage of an independent cohort of 56 individuals with suspected tick-borne encephalitis whose serum/plasma samples were tested for BoDV-1 IgG antibodies for diagnostic purposes. Corresponding CSF samples of all affected individuals tested negative for BoDV-1 by qRT-PCR. Regarding serum/plasma, six, two, and two samples were reactive with values above N, X, and P cutoffs as tested by ELISA (Figures 3B and S1B) but were negative or not evaluable due to unspecific background in iIFA, resulting in specificities of the single antigen ELISA tests of 89%, 96%, and 96%, respectively (Figure S2B). Specificity increased to 98% when the diagnosis was based on the presence of antibodies against at least two viral antigens. The serum that tested positive for two ELISA antigens (N and P) also showed an unspecific background in iIFA, while other positive ELISA reactions did not correlate with unspecific background iIFA patterns (Figure S1B).
To address whether the recombinant BoDV-1 proteins used in the newly established BoDV-1 ELISA cross-react with variegated squirrel bornavirus 1 (VSBV-1), the only other virus within the family Bornaviridae known to infect humans,12 plasma samples of four rabbits experimentally infected with BoDV-1 or VSBV-1 were tested in a modified version of the ELISA using an anti-rabbit-IgG secondary antibody. All infected animals showed reactivity against at least one antigen (Figure S3). Optical density (OD) values of the ELISA corresponded to the titers of iIFA. As one VSBV-1-infected rabbit tested highly positive for all three BoDV-1 proteins, a cross-reactivity of all antigens used in the BoDV-1 ELISA can be assumed.
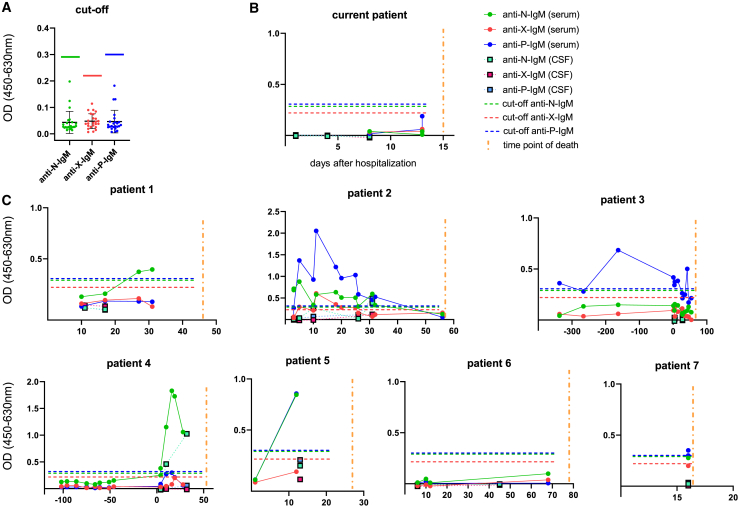
In addition, all confirmed BoDV-1-affected individuals were screened for BoDV-1-specific IgM antibodies, which were detected in available samples of six affected individuals (Figure 4). Only in one individual (individual 4) did they precede IgG detection by ELISA by six days, while the iIFA IgG titer at the time of positive IgM was 40.
Figure 4.
BoDV-1 IgM-ELISA
(A) Cutoffs for each ELISA system are indicated by horizontal lines (green: anti-BoDV-1-N ELISA; red: anti-BoDV-1-X ELISA; blue: anti-BoDV-1-P ELISA). A vertical yellow line indicates the time of death. Cutoffs were determined by 24 characterized negative samples as the sum of arithmetic mean (black horizontal line) plus 6 standard deviations (error line) Green, red and blue horizontal lines represent cutoffs.
(B and C) ELISA results for the current individual and another 7 recently published confirmed BoDV-1-infected individuals. All experiments were carried out once. Interassay variabilities were calculated for N, X, and P antigens in three runs of one sample of individual 2, resulting in 42%, 15%, and 15%, respectively.
Discussion
Our case shows that BoVD-1 RNA can be detected in CSF at early stages of the disease when CSF leucocyte counts may still be normal. However, since the viral load in CSF is usually close to the lower limit of detection of the applied qRT-PCRs, a negative result does not rule out BoDV-1 infection, as demonstrated in our case by the intermittent detection of BoDV-1 RNA in a series of consecutive CSF samples. The low negative predictive value of PCR from CSF is in accordance with the previously described cases.4,5 Post mortem, high viral RNA loads were detected in neuronal tissues of the affected individual, unequivocally confirming BoDV-1 infection. Of note, BoDV-1 was detected in the optic nerve as well as in some peripheral nerves. As the available serological tests for BoDV-1 have limitations—the interpretation of iIFA is subjective, and a line blot assay available for research purposes has limited sensitivity and detects antibodies against N and P proteins only—there is a need for the development of additional serological tests for high-throughput analyses.5
Using recombinant BoDV-1 N, X, and P proteins expressed from codon-optimized sequences in E. coli, we established three ELISA systems. In the reported case, anti-BoDV-1-X-IgG showed seroconversion and thus reflected the findings of the iIFA, whereas anti-BoDV-1-N-IgG was not detectable, and anti-BoDV-P-IgG was just above the cutoff. Testing available serum and CSF samples from seven additional affected individuals confirmed anti-BoDV-1-X-IgG as a reliable serological marker but also showed that BoDV-1 antibody profiles were remarkably variable. In this respect, all affected individuals developed IgG responses against P and X. In the current case, anti-X-IgG preceded the detection of anti-P-IgG, whereas in others, anti-P-IgG was detected first. Anti-N-IgG was detectable in only five of eight individuals and usually reached lower levels. In three out of eight affected individuals, IgG antibodies against N, P, or X proteins were already detectable in the first available serum sample after hospital admission. In one immunosuppressed individual, seroconversion occurred before hospitalization (individual 3), while in four affected individuals, BoDV-1-specific IgG was first detected 10 to 68 days after hospitalization. In general, ELISA values reflected iIFA titers in serum and CSF (Figures S1 and S2A). Higher iIFA titers corresponded to higher OD values in ELISA and to the presence of antibodies directed against more than one viral antigen during the course of disease. Overall, the results show that the usage of three different BoDV-1 antigens contributes to a more sensitive and reliable serological diagnosis of BoDV-1 infection. Inclusion of further BoDV-1 antigens such as the glycoprotein expressed in a eukaryotic system may further improve sensitivity and specificity of the ELISA.
In at least one immunocompromised individual (individual 4), detection of BoDV-1-reactive IgM preceded IgG seroconversion, as measured by ELISA, by some days, but in most affected individuals, IgM became detectable later and at lower levels compared with IgG. Thus, detection of IgM may contribute to early diagnosis in individual cases. The need for early and reliable diagnostic markers remains, in particular because most BoDV-1-infected individuals were already symptomatic at hospital admission and quickly deteriorated thereafter. In addition, one survivor of a BoDV-1 encephalitis described so far, who suffered from BoDV-1 encephalitis as part of a liver transplant, was treated early on with ribavirin.2 The crucial findings in our case are that at an early stage, neither clinical signs nor routine CSF parameters were indicative of BoDV-1 brain infection. Thus, physicians in endemic regions should initiate early BoDV-1 diagnostics in cases of unclear encephalopathy with brain-blood barrier disruption even if the CSF leucocyte count is unaltered.
Limitations of study
The present study was performed on a rather small number of samples, which is attributed to the low incidence of BoDV-1 infection. Our main conclusion—that reactivity against two viral antigens contributes to BoDV-1 diagnosis—needs to be validated in a larger panel of samples. In addition, the positive predictive value of the ELISA with its current specificity is limited due to the very low prevalence of the disease. A further limitation is the cross-reactivity of our BoDV-1 ELISA with VSBV-1 serology, which was only demonstrated for rabbit plasma samples. The modified rabbit BoDV-1 ELISA has not been formally validated with cutoffs using a panel of negative rabbit sera.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Cy3 AffiniPure Goat Anti-Human IgG (H+L) | Jackson ImmunoResearch | Cat# 109-165-003; RRID: AB_2337718 |
| Cy3 AffiniPure Goat Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch | Cat# 111-165-003; RRID: AB_2338000 |
| Polyclonal Rabbit Anti-Human IgG/HRP antibody | Agilent | Cat# P021402-2; RRID: AB_2893418 |
| Polyclonal Rabbit Anti-Human IgM/HRP antibody | Agilent | Cat# P021502-2; RRID: AB_2893505 |
| Goat Anti-Rabbit IgG (H+L)-HRP Conjugate antibody | Bio-Rad | Cat# 172-1019; RRID: AB_11125143 |
| Bacterial and virus strains | ||
| BoDV-1, 2 strains isolated from human CNS tissue | Institute of Clinical Microbiology and Hygiene, University Hospital Regensburg, Germany | N/A |
| Biological samples | ||
| Plasma samples of 2 BoDV-1 infected rabbits. The animal experiments were evaluated and approved by the ethics committee of the State Office of Agriculture, Food safety, and Fishery in Mecklenburg – Western Pomerania (LALLF M-V: LVL MV/TSD/ 7221.3-2-010/18). | Institute of Diagnostic Virology, Friedrich-Loeffler-Institut, Federal Research Institute for Animal Health, Greifswald – Insel Riems, Germany | N/A |
| Plasma samples of VSBV-1 infected rabbits. The animal experiments were evaluated and approved by the ethics committee of the State Office of Agriculture, Food safety, and Fishery in Mecklenburg – Western Pomerania (LALLF M-V: LVL MV/TSD/ 7221.3-2-010/18). | Institute of Diagnostic Virology, Friedrich-Loeffler-Institut, Federal Research Institute for Animal Health, Greifswald – Insel Riems, Germany | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| BoDV-1 nucleocapsid Np40 protein, His-tagged | Mikrogen | N/A |
| BoDV-1 phosphoprotein Pp23, His-tagged | Mikrogen | N/A |
| BoDV-1 X protein Xp10, GST-tagged | Mikrogen | N/A |
| PBS | GIBCO | Cat# 14190-094 |
| Tween 20 | Caelo | Cat# 3472 |
| Fat free milk powder | Heirler | N/A |
| RF Absorbent | Virion/Serion | Cat# Z200 |
| Mikrogen TMB | Mikrogen | Cat# 12003 |
| Mikrogen Stopp (24.9% H3PO4) | Mikrogen | Cat# 12004 |
| TRIS buffer with Tween 20, pH 8.0 | Sigma-Aldrich | Cat# T9039-10PAK |
| TaqPath™1-Step RT-qPCR Master Mix, CG | Applied Biosystems | Cat# A15299 |
| NxtScript Reaction Mix | Roche/Penzberg | Cat# 07368372103 |
| NxtScript RT 85U/μL | Roche/Penzberg | Cat# 07371527103 |
| ROX-Reference Dye | Life technologies | Cat# 12223-012 |
| RNase free DEPC water | Roth | Cat# 143.3 |
| PageRuler Prestained Protein Ladder | Thermo Fisher Scientific | Cat# 26617 |
| Roti-Block | Roth | Cat# A151.2 |
| Critical commercial assays | ||
| EZ1 Virus Mini Kit v2.0 | QIAGEN | Cat# 179799 |
| Deposited data | ||
| Dataset uploaded to Mendeley | This study | https://doi.org/10.17632/wpvkdhmgz4.1 |
| Experimental models: Cell lines | ||
| Vero cells | Institute of Clinical Microbiology and Hygiene, University Hospital Regensburg, Germany | N/A |
| Oligonucleotides | ||
| BoDV-1 Mix 1 (target: x/p gene): | Metabion; (Schlottau et al.2) | N/A |
| • BoDV1-1288-F (TAGTYAGGAGGC TCAATGGCA) | ||
| • BoDV-1-1449-R (GTCCYTCAGGA GCTGGTC) | ||
| • BoDV-1-1346-FAM (FAM-AAGAAG ATCCCCAGACACTACGACG-BHQ1) | ||
| BoDV-1 Mix 6 (target: m/g gene): | Metabion; (Schlottau et al.2) | N/A |
| • BoDV-1-2262-F (CAATYAATGCAG CYTTCAATGTCTT) | ||
| • BoDV-1-2336-R (GAATGTCYGGG CCGAGAG) | ||
| • BoDV-1-2316-FAM (FAM-CCARCA CCAATGTTCCGAAGCCG-BHQ1) | ||
| MS2: | Metabion; | N/A |
| • MS2 forward primer | Metabion; | |
| • MS2 reverse primer | Applied Biosystems; | |
| • MS2 So VIC/Tamra | (Dreier et al., 2005) | |
| Software and algorithms | ||
| GraphPad Prism V9.2.0 | GraphPad | https://www.graphpad.com |
| Other | ||
| Nunc Maxisorp Plates | Thermo Fisher Scientific | Cat# 446469 |
| μ-Plate 96 Well Black | Ibidi | Cat# 89626 |
| Nitrocellulose western blotting membrane Amersham Protran Premium 0.2 μm | Thermo Fisher Scientific | Cat# 15279794 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Markus Bauswein (markus.bauswein@ukr.de).
Materials and availability
BoDV-1 N, X, and P proteins generated in this study will be made available on request, but we may require a completed materials transfer agreement if there is potential for commercial application.
Experimental model and subject detials
Gender and age of the patients are not revealed for ethical reasons. Patients 1 to 5 of this publication correspond to patients 1 to 5 published by Niller et al.4 Patient 6 is the kidney transplant recipient 1 published by Schlottau et al.2 Patient 7 corresponds to patient 8 published by Niller et al.4 The use of retrospective patient samples has been approved by the ethical commission of the Faculty for Medicine, University of Regensburg (reference number 18-1248-101).
Method details
RNA extraction and PCR
RNA extraction and BoDV-1 specific RT-qPCR were performed following a recently published protocol.2 Briefly, total RNA from native samples was extracted using EZ1 Virus Mini Kit v2.0 on an EZ1 Advanced XL system (QIAGEN), according to the manufacturer's instructions. BoDV-1 RNA was detected using two real-time RT-qPCR assays: mix 1 and 6 were used for amplification of RNA from BoDV-1 x/phosphoprotein gene and matrix/glycoprotein gene, respectively. RT-qPCR was performed on a StepOnePlus System (Applied Biosystems). In vitro-transcribed RNA molecules were used as standards for quantification. For extraction and inhibition control, samples were spiked with MS2 phage solution and MS2 phage RT-qPCR was performed.13 All precautions of accredited laboratories, particularly separate rooms for nucleic acid extraction, amplification, and detection, were taken to avoid any cross-contamination. All samples were tested in two replicates. Positive single values of only one replicate were considered negative. RNA copies were calculated as mean of two positive replicates.
Indirect immunofluorescence assay (iIFA)
Indirect immunofluorescence assay (iIFA) was performed as previously described.2 Briefly, Vero cells persistently infected with BoDV-1 were mixed 1:2 with uninfected Vero cells and cultured overnight in 96-well microtiter plates (ibidi) to achieve confluent cell layers. Wells with uninfected Vero cells served as negative controls. After removal of the supernatant, plates were dried for 2h and fixed at 80°C for 2h. Thereafter, heat-inactivated patient samples were added in a 2-fold dilution series (starting with 1:20 for sera and 1:2 for CSF samples, dilution in TRIS buffer). After incubation for 1 h, plates were washed three times with PBS and incubated with a 1:200 dilution of Cy-3-conjugated polyclonal rabbit anti-human-IgG antibody for 1 h. For rabbit samples, a 1:500 diluted Cy3-conjugated goat anti-rabbit-IgG antibody was used. After a final washing cycle, the assays were analyzed using fluorescence microscopy. Samples with characteristic fluorescing spots in the nuclei of BoDV-1-infected Vero cells were considered positive. When similar fluorescence signals were detected in non-infected and BoDV-1-infected Vero cells, samples were considered unspecific.
Protein expression, silver staining and western blots
Sequences for BoDV-1 nucleoprotein (N), X protein and phosphoprotein (P) were codon-optimized for expression in E. coli and cloned into expression vectors pET30a, pGEX6P, and pET82, respectively. His-tagged N and P protein as well as GST-tagged X protein were purified by chromatography.
Purity of proteins was demonstrated by silver staining after SDS-PAGE according to a published protocol.14 Lanes were loaded with 200 ng of His-tagged Np40, 200 ng of GST-tagged Xp10 and 100 ng of His-tagged Pp23 protein. 5% and 15% polyacrylamide were used for stacking and separating gel, respectively.
For western blots, an SDS-PAGE was performed using a 12% polyacrylamide separating gel. Lanes were loaded with 100 ng Np40, 200 ng Xp10 and 100 ng Pp23. After separation, proteins were blotted onto a 0.2 μg nitrocellulose membrane using a wet tank transfer system. Membranes were blocked with Roti Block (containing Tween 20) and then stained with a 1:100 dilution of selected samples (plasma, serum, CSF). A horseradish-peroxidase-conjugated polyclonal rabbit anti-human-IgG diluted 1:3,000 was used as secondary antibody. Proteins were detected by chemiluminescence.
Enzyme-linked immunosorbent assay (ELISA)
Recombinant BoDV-1 proteins diluted in PBS were used to coat Nunc MaxiSorp plates (Thermo Fisher Scientific). N and X proteins were coated with 800 ng/well (in 100 μL PBS), P protein with 150 ng/well (in 100 μL PBS) at 4°C overnight and then blocked with 200 μL 5% fat-free milk powder in PBS with 0.1% Tween 20 at room temperature (RT) for 1 h. After washing three times with 200 μL PBS containing 0.1% Tween 20 (PBS-T), 100 μL of diluted patient samples were added. For IgG-ELISA, all samples (serum, plasma, CSF) were diluted 1:100 in 1% fat-free milk powder in PBS with 0.1% Tween 20. For IgM-ELISA, IgG was absorbed by pre-incubation of samples with RF Absorbent (Virion/Serion) for 15 min at RT according to the manufacturer's protocol. Plates were incubated for 1 h at RT and then washed nine times with 200 μL PBS-T. Subsequently, 50 μL horseradish-peroxidase-conjugated polyclonal rabbit anti-human-IgG diluted 1:5,000 in PBS-T were added as secondary antibody. To test for cross-reactivity with VSBV-1, samples of two BoDV-1- and two VSBV-1-infected rabbits were stained with a 1:3,000 diluted goat anti-rabbit-IgG antibody. For IgM-ELISA, 50 μL horseradish-peroxidase-conjugated polyclonal rabbit anti-human-IgM diluted 1:3,000 in PBS-T were used. After 1 h incubation at RT, plates were washed nine times with 200 μL PBS-T. 50 μL substrate solution (TMB) were added to each well, incubated for 4 min at RT, and stopped by adding 50 μL of stop solution (phosphoric acid). After 5 min of incubation, optical density was determined at 450 nm (OD450) and 630 nm (OD630) in three technical replicates using a plate reader (Microplate Reader Model 680, Bio-Rad). For evaluation, OD630 background values were subtracted from OD450 values. OD of blank was subtracted from all OD values. To control for inter-assay variability, all sample values were normalized to an external standard sample. Cut-offs for each ELISA system were determined using serum samples from 24 patients that had been tested negative by iIFA (IgG) and/or whose brain tissue had been tested negative by RT-qPCR. Cut-offs were calculated by the sum of arithmetic mean plus 6 standard deviations.
Generation of BoDV-1-/VSBV-1-reactive rabbit sera
Neonatal rabbits were inoculated within 24h after birth with either VSBV-1 or BoDV-1. All animals received 5x103 tissue infectious dose 50 (TCID50) intracerebrally. The animals were monitored daily for clinical signs and euthanized at different time points regarding a defined humane endpoint. Tissues as well as sera were collected for serological and virological analysis. The animal experiments were evaluated and approved by the ethics committee of the State Office of Agriculture, Food safety, and Fishery in Mecklenburg – Western Pomerania (LALLF M-V: LVL MV/TSD/ 7221.3-2-010/18). All procedures were carried out in approved biosafety level 3 (BSL3) facilities.
Quantification and statistical analysis
Graphs and descriptive statistical analysis were created using GraphPad Prism V9.2.0.
Acknowledgments
We thank the German Federal Ministry of Education and Research for funding of the projects “Bornavirus – Focal Point Bavaria” (grant no. 01KI2002) and ZooBoCo (grant nos. 01KI1722A and 01KI2005A).
Author contributions
Conceptualization, B.N., K.A., A.G., B.S., and M. Bauswein.; methodology, G.K., L.E., D.R., P.S., and B.A.; investigation, B.N., K.A., M.M.B., H.H.N., and M. Bauswein; writing – original draft, B.N., K.A., and M. Bauswein; writing – review & editing, D.R., K.S., M. Beer, B.M.J.L., M.P., and B.S.; funding acquisition, M.P., H.H.N., and B.S.; resources, D.R., K.S., M. Beer, K.E., M.J.R., E.S., R.A.L., and A.G.; supervision, R.A.L., M. Beer, and E.S.
Declaration of interests
P.S. is an employee of Mikrogen GmbH. E.S. is CEO and a shareholder of Mikrogen GmbH. The other authors declare no competing interests.
Published: January 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100499.
Supplemental information
Data and code availability
ELISA datasets generated in this study have been uploaded to Mendeley Data: https://doi.org/10.17632/wpvkdhmgz4.1.
References
- 1.Korn K., Coras R., Bobinger T., Herzog S.M., Lücking H., Stöhr R., Huttner H.B., Hartmann A., Ensser A. Fatal Encephalitis Associated with Borna Disease Virus 1. N. Engl. J. Med. 2018;379:1375–1377. doi: 10.1056/NEJMc1800724. [DOI] [PubMed] [Google Scholar]
- 2.Schlottau K., Forth L., Angstwurm K., Höper D., Zecher D., Liesche F., Hoffmann B., Kegel V., Seehofer D., Platen S., et al. Fatal Encephalitic Borna Disease Virus 1 in Solid-Organ Transplant Recipients. N. Engl. J. Med. 2018;379:1377–1379. doi: 10.1056/NEJMc1803115. [DOI] [PubMed] [Google Scholar]
- 3.Coras R., Korn K., Kuerten S., Huttner H.B., Ensser A. Severe bornavirus-encephalitis presenting as Guillain-Barré-syndrome. Acta Neuropathol. 2019;137:1017–1019. doi: 10.1007/s00401-019-02005-z. [DOI] [PubMed] [Google Scholar]
- 4.Niller H.H., Angstwurm K., Rubbenstroth D., Schlottau K., Ebinger A., Giese S., Wunderlich S., Banas B., Forth L.F., Hoffmann D., et al. Zoonotic spillover infections with Borna disease virus 1 leading to fatal human encephalitis, 1999-2019: an epidemiological investigation. Lancet Infect. Dis. 2020;20:467–477. doi: 10.1016/S1473-3099(19)30546-8. [DOI] [PubMed] [Google Scholar]
- 5.Eisermann P., Rubbenstroth D., Cadar D., Thomé-Bolduan C., Eggert P., Schlaphof A., Leypoldt F., Stangel M., Fortwängler T., Hoffmann F., et al. Active Case Finding of Current Bornavirus Infections in Human Encephalitis Cases of Unknown Etiology, Germany, 2018-2020. Emerg. Infect. Dis. 2021;27:1371–1379. doi: 10.3201/eid2705.204490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liesche F., Ruf V., Zoubaa S., Kaletka G., Rosati M., Rubbenstroth D., Herden C., Goehring L., Wunderlich S., Wachter M.F., et al. The neuropathology of fatal encephalomyelitis in human Borna virus infection. Acta Neuropathol. 2019;138:653–665. doi: 10.1007/s00401-019-02047-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dürrwald R., Kolodziejek J., Weissenböck H., Nowotny N. The bicolored white-toothed shrew Crocidura leucodon (HERMANN 1780) is an indigenous host of mammalian Borna disease virus. PLoS ONE. 2014;9:e93659. doi: 10.1371/journal.pone.0093659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nobach D., Bourg M., Herzog S., Lange-Herbst H., Encarnação J.A., Eickmann M., Herden C. Shedding of Infectious Borna Disease Virus-1 in Living Bicolored White-Toothed Shrews. PLoS ONE. 2015;10:e0137018. doi: 10.1371/journal.pone.0137018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nobach D., Müller J., Tappe D., Herden C. Update on immunopathology of bornavirus infections in humans and animals. Adv. Virus Res. 2020;107:159–222. doi: 10.1016/bs.aivir.2020.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Tokunaga T., Yamamoto Y., Sakai M., Tomonaga K., Honda T. Antiviral activity of favipiravir (T-705) against mammalian and avian bornaviruses. Antiviral Res. 2017;143:237–245. doi: 10.1016/j.antiviral.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 11.Jordan I., Briese T., Averett D.R., Lipkin W.I. Inhibition of Borna disease virus replication by ribavirin. J. Virol. 1999;73:7903–7906. doi: 10.1128/jvi.73.9.7903-7906.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann B., Tappe D., Höper D., Herden C., Boldt A., Mawrin C., Niederstraßer O., Müller T., Jenckel M., van der Grinten E., et al. A Variegated Squirrel Bornavirus Associated with Fatal Human Encephalitis. N. Engl. J. Med. 2015;373:154–162. doi: 10.1056/NEJMoa1415627. [DOI] [PubMed] [Google Scholar]
- 13.Dreier J., Störmer M., Kleesiek K. Use of bacteriophage MS2 as an internal control in viral reverse transcription-PCR assays. J. Clin. Microbiol. 2005;43:4551–4557. doi: 10.1128/JCM.43.9.4551-4557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrissey J.H. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal. Biochem. 1981;117:307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ELISA datasets generated in this study have been uploaded to Mendeley Data: https://doi.org/10.17632/wpvkdhmgz4.1.