Abstract
The proportion of acyclovir (ACV)-resistant herpes simplex virus (HSV) isolates in clinical specimens and laboratory isolates was determined. HSV isolates in clinical specimens and laboratory isolates were cultured in the absence or presence of 5 μg of ACV per ml. The frequency of ACV-resistant HSV was calculated as (average virus titer in the wells with ACV)/(average virus titer in the wells without ACV). The mutation frequency of HSV type 1 isolates in clinical samples (directly from patient lesions) was 7.5 × 10−4 ± 2.5 × 10−4 (mean ± standard error), and that of HSV type 2 isolates was 15.0 × 10−4 ± 4.6 × 10−4. The mutation frequencies of isolates derived in the laboratory from these clinical samples were very similar. Similarly, the 50% inhibitory concentrations for isolates in clinical samples and laboratory isolates were identical. The frequencies of ACV-resistant HSV types 1 and 2 were in a narrow range of 7.5 × 10−4 to 15.0 × 10−4 among isolates in clinical specimens and did not change for laboratory-derived pools of viral isolates.
Acyclovir (ACV) is widely used for the treatment of primary and recurrent herpes simplex virus (HSV) and varicella-zoster virus infections because of its very favorable therapeutic ratio. Since 1982, 2.0 × 106 kg of ACV and other nucleoside analogues has been distributed, with more than 50% of that amount distributed in the United States. ACV is a nucleoside analogue of guanine that is preferentially phosphorylated to ACV monophosphate by viral thymidine kinase and that is then further phosphorylated to ACV triphosphate by cellular enzymes. ACV triphosphate inhibits viral DNA polymerase and is incorporated into viral DNA, ultimately preventing elongation of viral DNA (7). HSV develops resistance predominantly (95%) as a result of mutations in genes that code for thymidine kinase, but resistance can also result from mutations in DNA polymerase (1, 3, 4, 9, 16).
ACV-resistant variants have been isolated from clinical specimens obtained before ACV was introduced (13). These variants are also readily detected in pools of laboratory strains of ACV-sensitive HSV. Mutation frequencies of 2.7 × 10−6 to 1.0 × 10−3 for HSV type 1 and 5.0 × 10−5 to 8.0 × 10−3 for HSV type 2 were detected, and these studies indicate that some proportion of HSV growing in cell culture is always resistant to ACV, even when the inoculum is considered to be susceptible to ACV by conventional plaque reduction assays (PRAs) (2, 10, 11, 15). Although this conclusion was initially obtained from studies with a small number of multiply passaged laboratory virus pools, there is now comparable information from studies with clones of a small number of clinical specimens (15).
Given the continuing strong selection pressure provided by extensive use of ACV and related compounds and concern that the level of antiviral resistance of HSV will increase, we have sought to develop additional baseline information on the prevalence of resistant mutants in clinical samples and isolates prepared from those samples. Moreover, conventional PRAs of the susceptibility of HSV to antivirals were performed with virus isolates obtained after multiple cycles of replication in tissue culture inoculated with the clinical specimen. Therefore, we also sought to determine if the prevalence of ACV-resistant mutants in the population of virions isolated in the laboratory is similar to the prevalence of mutants in the source clinical specimen obtained from the patient.
MATERIALS AND METHODS
Viral specimens.
Clinical samples from the Diagnostic Virology Laboratory of the University of Colorado Health Sciences Center were randomly chosen for study. Samples had been stored in a −70°C freezer in the prior year. ACV-resistant and ACV-sensitive strains were used as controls. HSV1S-115 is a diagnostic ACV-sensitive laboratory standard (2.5 × 107 PFU/ml) for which the 50% inhibitory concentration (IC50), as determined multiple times (in human fibroblasts), is 0.984 ± 0.4 μg/ml. HSV1R-5 (7.3 × 106 PFU/ml) is an ACV-resistant standard isolate for which the IC50 is 16.17 ± 8.1 μg/ml.
Virus isolation.
HSV isolates were prepared from clinical specimens submitted to the laboratory by inoculation into tube cultures of human embryonic lung fibroblasts (passaged between 15 and 20 times). Tube cultures were observed for the presence of the HSV cytopathic effect by optical microscopy daily for 5 days. Almost all isolates were identified within 3 days. HSV isolates were confirmed and typed with a fluorescein isothiocyanate-conjugated monoclonal antibody (PathoDx Herpes typing kit; Diagnostic Products Co., Los Angeles, Calif.).
Detection of ACV-resistant virus.
Vero cells were obtained from the American Type Culture Collection (81 CCL), and six-well plates were seeded with Vero cells (5 × 105 cells per well in 2 ml). Cells were cultured in Dulbecco's minimal essential medium (DMEM) containing 10% fetal bovine serum (FBS) at 37°C in a humidified incubator with 5% CO2 for 24 to 48 h until they were confluent. Thawed clinical specimens or virus isolates in 10-fold serial dilutions in DMEM containing 2% FBS were inoculated into quadruplicate wells (0.5 ml/well). After 1 h the inoculum was removed, 2.5 ml of a 0.4% agarose overlay containing DMEM plus FBS and no ACV or 5 μg of ACV per ml was added to duplicate wells, and the plates were incubated at 37°C. The plates were observed daily by optical microscopy until the characteristic HSV cytopathic effect was evident, the contents of the plates were fixed in 10% formalin for 1 h, and agarose plugs were removed. The cells were stained with 1.0 ml of 0.8% crystal violet in 20% ethanol for 30 s, rinsed with tap water, and dried. Plaques were counted in wells containing the lowest dilution that yielded plaques that could be easily counted with an optical microscope. The ratio of ACV-resistant HSV isolates to ACV-susceptible HSV isolates was calculated as (average virus titer in the wells with ACV)/(average virus titer in the wells without ACV). Standard ACV-resistant and ACV-sensitive strains were included in each assay.
Antiviral susceptibility testing.
Antiviral susceptibility testing was performed by PRAs with Vero cells in 12-well culture plates with a 0.4% agarose overlay (18). After the addition of HSV and adsorption (as described above), agarose containing ACV (9, 3, 1, 0.33, or 0 μg/ml) was added to triplicate wells. The cultures were incubated for 2 days, fixed with formalin, and stained with crystal violet as indicated above. The IC50 was defined as the ACV concentration that reduced the plaque number by 50% from that in the untreated control wells. The IC50 was calculated graphically from the plaque data. Standard ACV-resistant and ACV-sensitive strains were included in each assay.
Relationship between proportion of mutants in a sample and IC50.
The titers of plaque-purified resistant (IC50, 5.85 μg/ml) and sensitive (IC50, 0.61 μg/ml) HSV isolates were determined. A reconstruction experiment was performed by studying these viruses alone and mixtures of the two viruses with proportions of sensitive to resistant virus that varied from 10,000:1 to 1:500. The total virus titer of each mixture was the same. Each mixture was then subjected to simultaneous assays to determine the proportion of ACV-resistant and ACV-susceptible viruses, and the IC50 was calculated as described above.
Statistical analysis.
Student's t test was used to compare the mutation rates for the isolates in clinical specimens and those for laboratory isolates. Correlation between IC50 and mutation frequencies was calculated by regression analysis with the Microsoft Excel program.
RESULTS
Defining an assay to determine prevalence of ACV-resistant mutants.
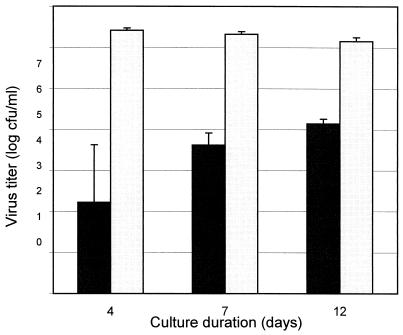
ACV-resistant mutants were detected by a modification of the procedure of Hall et al. (10). All HSV isolates or pools of HSV isolates from clinical specimens contained a variety of mutants with intermediate levels of resistance to ACV. These mutants grew slowly in the presence of the inhibitor but could be detected by a plaque assay if sufficient time had elapsed (Fig. 1). Thus, the duration of an assay used to study ACV-resistant mutants in a pool must be sufficiently long to permit detection of most mutants. Our decision to define the mutant phenotype by a 10-day assay was a result of a compromise between the results shown in Fig. 1 and practical considerations. The choice of a single concentration of ACV to define the mutant phenotype was arbitrary, since the HSV pools studied contained a continuum of mutants that varied in their susceptibilities to ACV. A total of 5 μg of ACV per ml was chosen as the concentration that is on the linear plateau phase of the ACV dose-response curve for HSV type 1 (data not shown) and permitted timely detection of most ACV-resistant mutants. This choice may not distinguish mutants with intermediate levels of resistance from those with high levels of resistance. In general, this concentration of ACV is about 5 to 10 times greater than the IC50 that defines isolates in Vero cells as susceptible (5, 14) and is close to the cutoff concentration for ACV resistance of 2.0 to 3.0 μg/ml that has generally been accepted with other cell lines in combination with clinical observations (1, 17). To confirm that the viruses growing in the presence of 5 μg of ACV per ml were resistant, three plaques of HSV type 1 were purified and their sensitivities were assessed. The IC50s for these plaques were 4.68, 7.72, and 4.36 μg/ml, respectively. Finally, the duration required for maintenance of HSV-infected cultures which lack ACV (the denominator in the calculation of the mutation rate) cannot be as long as the duration required for the detection of growth of mutants, since the plaques that develop in the absence of ACV will quickly coalesce. Consequently, we defined the total amount of input HSV on the basis of a 4-day culture, while the numbers of mutants were determined on the basis of a 10-day culture.
FIG. 1.
Plaque formation in the presence or absence of ACV as a function of time. ■■, ACV at 5 μg/ml; ▭, no ACV.
Intra- and interexperimental variances of mutation rate determination.
To examine the intraexperimental variance for mutation frequency, a sample was studied in triplicate at one time. The mean mutation frequency was 1.7 × 10−3, with a standard error of 0.42 × 10−3. This sample evaluated at three different times had a mean mutation frequency of 1.9 × 10−3, with a standard error of 0.78 × 10−3. There was no significant difference in the average mutation frequencies between intra and interexperimental data (P = 0.84).
Mutation frequencies for clinical isolates.
The average mutation frequency for HSV type 1 was 7.5 × 10−4 ± 2.5 × 10−4 (mean ± standard error) for virus in clinical specimens and 12 × 10−4 ± 4.2 × 10−4 for the corresponding laboratory isolates (Table 1). The difference in the mutation frequency between isolates in clinical specimens and laboratory isolates was not significant (P = 0.09). The average mutation frequencies for HSV type 2 in clinical specimens and laboratory isolates of 15.0 × 10−4 ± 4.6 × 10−4 and 9.3 × 10−4 ± 3.0 × 10−4, respectively, were not significantly different (P = 0.06). The difference in mutation rates between HSV type 1 and 2 isolates in clinical specimens or laboratory isolates was not significant (P = 0.08 and 0.27, respectively).
TABLE 1.
Mutation frequencies for HSV
| HSV type and isolate | Isolates in clinical sample
|
Laboratory isolates
|
||||||
|---|---|---|---|---|---|---|---|---|
| No. of PFU/ml with:
|
Mutation frequency | IC50 (μg/ml) | No. of PFU/ml with:
|
Mutation frequency | IC50 (μg/ml) | |||
| ACV | No ACV | ACV | No ACV | |||||
| Type 1 | ||||||||
| 292 | 6.0 × 102 | 3.5 × 106 | 1.7 × 10−4 | 1.39 | 1.0 × 104 | 3.3 × 107 | 3.0 × 10−4 | 0.75 |
| 305 | 1.1 × 105 | 1.9 × 108 | 5.7 × 10−4 | 1.07 | 2.7 × 107 | 3.6 × 1010 | 7.4 × 10−4 | 1.25 |
| 373 | 5.5 × 104 | 5.5 × 107 | 1.0 × 10−3 | 1.40 | 2.7 × 106 | 2.1 × 109 | 1.3 × 10−3 | 1.43 |
| 565 | 2.5 × 103 | 3.9 × 107 | 6.4 × 10−5 | 0.58 | 1.4 × 104 | 1.4 × 109 | 1.0 × 10−5 | 0.52 |
| 860 | 2.5 × 103 | 2.0 × 107 | 1.2 × 10−4 | 0.15 | 7.0 × 103 | 2.0 × 108 | 3.5 × 10−5 | 0.17 |
| 1006 | 3.1 × 105 | 2.5 × 108 | 1.3 × 10−3 | 0.83 | 6.1 × 105 | 1.9 × 1010 | 3.3 × 10−5 | 0.75 |
| 1159 | 5.5 × 102 | 1.8 × 105 | 3.1 × 10−3 | 1.44 | 2.8 × 106 | 6.3 × 108 | 4.5 × 10−3 | 1.03 |
| 1189 | 8.8 × 10 | 5.0 × 104 | 1.8 × 10−3 | 1.03 | 3.3 × 105 | 1.8 × 109 | 1.8 × 10−4 | 1.10 |
| 1335 | 3.8 × 10 | 1.4 × 105 | 2.7 × 10−4 | 0.79 | 7.5 × 10 | 2.5 × 104 | 3.0 × 10−3 | 0.81 |
| 1356 | 1.9 × 103 | 3.4 × 107 | 5.6 × 10−5 | 0.82 | 2.3 × 107 | 2.3 × 1011 | 1.0 × 10−4 | 1.04 |
| 1361 | 1.0 × 102 | 2.6 × 106 | 3.8 × 10−5 | 0.70 | 3.6 × 102 | 1.2 × 105 | 3.0 × 10−3 | 0.71 |
| 1362 | 8.3 × 102 | 4.1 × 106 | 2.0 × 10−4 | 0.63 | 2.3 × 104 | 1.9 × 109 | 1.2 × 10−5 | 0.73 |
| 1412 | 6.9 × 105 | 3.9 × 108 | 1.8 × 10−3 | 1.09 | 1.6 × 108 | 4.5 × 1010 | 3.5 × 10−3 | 0.80 |
| 1422 | 1.1 × 104 | 1.1 × 108 | 1.0 × 10−4 | 0.53 | 1.8 × 106 | 2.4 × 109 | 7.3 × 10−4 | 0.73 |
| Mean | 7.5 × 10−4 | 0.89 | 12.0 × 10−4a | 0.85b | ||||
| Standard error | 2.5 × 10−4 | 0.10 | 4.2 × 10−4 | 0.08 | ||||
| Type 2 | ||||||||
| 727 | 4.0 × 10 | 8.0 × 104 | 5.0 × 10−4 | 0.84 | 1.2 × 106 | 2.8 × 109 | 4.2 × 10−4 | 0.77 |
| 800 | 4.0 × 10 | 8.0 × 104 | 5.0 × 10−4 | 0.69 | 1.4 × 103 | 1.0 × 106 | 1.4 × 10−3 | 0.93 |
| 989 | 1.2 × 104 | 2.6 × 106 | 4.6 × 10−3 | 2.38 | 1.8 × 104 | 1.9 × 107 | 9.5 × 10−4 | 1.77 |
| 1041 | 3.8 × 10 | 2.1 × 104 | 1.8 × 10−3 | 0.88 | 5.3 × 102 | 5.0 × 105 | 1.1 × 10−3 | 1.16 |
| 1071 | 6.0 × 105 | 6.6 × 108 | 9.2 × 10−4 | 0.76 | 5.1 × 103 | 1.6 × 106 | 3.2 × 10−3 | 0.83 |
| 1081 | 6.1 × 105 | 2.9 × 108 | 2.1 × 10−3 | 1.19 | 3.6 × 106 | 1.4 × 1010 | 2.6 × 10−4 | 0.65 |
| 1282 | 8.5 × 10 | 1.0 × 105 | 8.5 × 10−4 | 0.51 | 9.3 × 103 | 1.4 × 107 | 6.6 × 10−4 | 0.82 |
| 1292 | 4.0 × 10 | 5.0 × 104 | 8.0 × 10−4 | 0.60 | 1.1 × 104 | 8.0 × 107 | 1.4 × 10−4 | 0.86 |
| 1301 | 1.5 × 105 | 1.6 × 108 | 9.6 × 10−4 | 1.54 | 1.4 × 105 | 1.6 × 108 | 8.9 × 10−4 | 1.91 |
| 1376 | 4.4 × 104 | 5.1 × 107 | 8.7 × 10−4 | 0.98 | 2.7 × 102 | 5.0 × 106 | 5.4 × 10−5 | 0.50 |
| 1405 | 3.5 × 103 | 4.6 × 107 | 7.5 × 10−5 | 0.82 | 3.4 × 103 | 9.0 × 107 | 3.8 × 10−5 | 0.56 |
| 1513 | 2.9 × 103 | 3.0 × 106 | 9.6 × 10−4 | 0.61 | 7.1 × 104 | 2.7 × 109 | 3.0 × 10−5 | 0.68 |
| 1514 | 7.5 × 103 | 7.5 × 106 | 1.0 × 10−3 | 2.24 | 7.5 × 103 | 1.0 × 107 | 7.5 × 10−4 | 1.74 |
| 1560 | 1.0 × 10 | 1.3 × 104 | 8.0 × 10−5 | 0.58 | 1.0 × 104 | 1.6 × 108 | 6.2 × 10−5 | 0.76 |
| 1624 | 1.6 × 104 | 2.5 × 106 | 6.5 × 10−3 | 1.52 | 1.0 × 102 | 2.5 × 104 | 4.0 × 10−3 | 1.34 |
| Mean | 15.0 × 10−4c | 1.08d | 9.3 × 10−4ef | 1.02g, h | ||||
| Standard error | 4.6 × 10−4 | 0.15 | 3.0 × 10−4 | 0.12 | ||||
Mutation frequency laboratory versus clinical HSV type 1, P = 0.09.
IC50 for laboratory versus clinical HSV type 1, P = 0.27.
Mutation frequency for HSV type 1 versus HSV type 2 in a clinical sample, P = 0.08.
IC50 for HSV type 1 versus HSV type 2 in a clinical sample, P = 0.16.
Mutation frequency for laboratory versus clinical HSV type 2, P = 0.06.
Mutation frequency for HSV type 1 versus HSV type 2 laboratory isolate, P = 0.27.
IC50 for laboratory versus clinical HSV type 2, P = 0.27.
IC50 for HSV type 1 versus HSV type 2 laboratory isolate, P = 0.13.
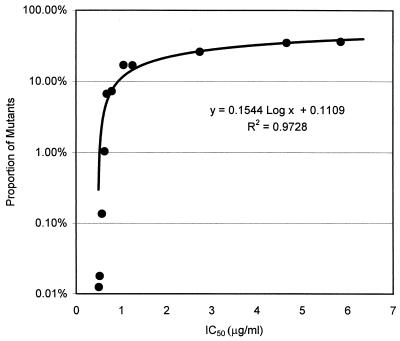
The average IC50 for HSV type 1 were 0.89 μg/ml for isolates in clinical specimens and 0.85 μg/ml for laboratory isolates. There was no significant difference between these IC50s (P = 0.27). The average IC50s for HSV type 2 isolates in clinical specimens and laboratory isolates were 1.08 and 1.02 μg/ml, respectively, and there was no significant difference (P = 0.27). The difference in average IC50s between HSV type 1 and type 2 isolates in clinical specimens or laboratory isolates was not significant (P = 0.16 and 0.13, respectively). The mutation frequency was not closely correlated with the average IC50 (R2 values, 0.1059 for HSV type 1 and 0.2314 for HSV type 2). This may be because mutation was always a low-frequency event among the isolates studied, and the IC50 determined by PRA does not change significantly until mutants make up 10% of the inoculum being tested (Fig. 2).
FIG. 2.
Relationship between proportion of mutants in a sample and IC50. The y axis is log scale.
DISCUSSION
The susceptibility of HSV to nucleoside analogues is commonly determined by PRAs (12, 17, 18). The determination by a PRA that a clinical isolate is resistant to the test drug requires that a large proportion of the HSV isolates in the pool be resistant to a defined concentration of that drug (Fig. 2). Thus, the PRA will not detect small changes in the proportion of isolates with a mutant phenotype in treated HSV lesions either during the course of therapy or after multiple courses of therapy. Such changes might be important when contacts are exposed to resistant HSV present in the peripheral lesions of an index patient. Similarly, if resistant virus enters the sensory ganglia of an index patient and if resistant virus can reactivate, then selection of resistant mutants in primary lesions might lead to an increase in the proportions of resistant HSV isolates in populations of patients who are likely to receive antiviral therapy.
In order to measure small changes in the proportion of mutant HSV isolates in a virus pool, we modified a method for the enumeration of small numbers of resistant virus in the presence of large numbers of sensitive virus (10). The application of this assay resulted in two conclusions. First, the proportion of HSV isolates in lesions that might be selected for by the application of anti-HSV therapy is approximately 7.5 × 10−4 to 15.0 × 10−4 for both HSV type 1 and HSV type 2. Published results of studies that used similar methods applied to small numbers of laboratory-passaged HSV isolates indicate that the proportions of ACV-resistant HSV mutants are 1.5 × 10−4 to 10.0 × 10−4 for HSV type 1 (8, 10) and 0.5 ×10−4 to 50.0 × 10−4 for HSV type 2 (13). These isolates had not been exposed to antiherpes virus drugs. Sarisky et al. (15) found that the mean mutation frequency for four HSV type 1 clinical isolates was similar, 3.0 × 10−4. However, they also found that four HSV type 2 clinical isolates had a mutation frequency of 8.0 × 10−3 (15). Their finding of a significantly higher mutation frequency for HSV type 2 may reflect several differences in experimental design. Perhaps most important is their use of a plaque-purified inoculum, whereas we used an inoculum that consisted of clinical isolates that closely represent the typical mix of phenotypes found in humans with HSV disease. We chose this starting point for our experiments in order to determine the resistance profiles that most closely mimic the resistance profiles for isolates in clinical situations. Sarisky et al. (15) also used MRC-5 cells and a different concentration of ACV to define mutants, and the number of clinical isolates that they examined was limited.
Second, the results presented in Table 1 demonstrate that the proportion of mutants present in a clinical specimen is not altered during the one or several cycles of replication that occur during the recovery of HSV in the laboratory in the absence of selection pressure. This being the case, the IC50s for isolates in clinical specimens and their derived isolates were found to be very similar, and either of the values would accurately reflect the presence of mutants in a clinical setting.
ACV is now widely accepted as a safe and effective treatment for the management of HSV infections in normal and immunocompromised patients. A common concern with regard to the widespread use of any antiviral agent is the emergence of resistance. Although the appearance of ACV-resistant HSV was first described in 1982, the prevalence of ACV-resistant isolates has remained stable at less than 1% among immunocompetent hosts in the subsequent 18 years (1, 5). However, these epidemiological surveys were generally performed with isolates from recurrent HSV outbreaks or from primary episodes in patients not treated with antiviral agents. Thus, they may represent isolates that had been “archived” in patients from a preantiviral era. A more sensitive estimate of the trend in antiviral resistance might be obtained by determining the frequency of mutants in patients having their first recurrence after therapy for a primary infection (Y. K. Shin et al., unpublished data). This might also better define the acquisition of resistance in treated immunocompromised patients, particularly those with AIDS or bone marrow transplants, who have a 5 to 10% incidence of resistant HSV after treatment (6, 8, 9, 14). The information on resistance phenotypes presented above provides baseline information for such an endeavor.
ACKNOWLEDGMENTS
Y. K. Shin was supported in part by the Kil Chung Hee Fellowship Fund.
We thank Patricia A. Young and other members of Virology Laboratory of the University of Colorado Health Science Center for technical help and specimens.
REFERENCES
- 1.Christophers J, Clayton J, Craske J, Ward R, Collins P, Trowbridge M, Darby G. Survey of resistance of herpes simplex virus to acyclovir in northwest England. Antimicrob Agents Chemother. 1998;42:868–872. doi: 10.1128/aac.42.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coen D M, Schaffer P A, Furman P A, Keller P M, St Clair M H. Biochemical and genetic analysis of acyclovir-resistant mutants of herpes simplex virus type 1. Am J Med. 1982;73(Suppl. 1A):351–360. doi: 10.1016/0002-9343(82)90122-x. [DOI] [PubMed] [Google Scholar]
- 3.Coen D M, Schaffer P A. Two distinct loci confer resistance to acycloguanosine in herpes simplex virus type 1. Proc Natl Acad Sci USA. 1980;77:2265–2269. doi: 10.1073/pnas.77.4.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coen D M. General aspects of virus drug resistance with special reference to herpes simplex virus. J Antimicrob Chemother. 1986;18(Suppl. B):1–10. doi: 10.1093/jac/18.supplement_b.1. [DOI] [PubMed] [Google Scholar]
- 5.Collins P, Ellis M N. Sensitivity monitoring of clinical isolates of herpes simplex virus to acyclovir. J Med Virol. 1993;41(Suppl. 1):58–66. doi: 10.1002/jmv.1890410512. [DOI] [PubMed] [Google Scholar]
- 6.Crumpacker C S, Schnipper L E, Marlowe S I, Kowalsky P N, Hershey B J, Levin M J. Resistance to antiviral drugs of herpes simplex virus isolated from a patient treated with acyclovir. N Engl J Med. 1982;306:343–346. doi: 10.1056/NEJM198202113060606. [DOI] [PubMed] [Google Scholar]
- 7.Elion G B, Furman P A, Fyfe J A, de Miranda P, Beauchamp L, Schaeffer J H. Selectivity of action of an antiherpetic agent, 9-(2-hydroxyethoxymethyl) guanine. Proc Natl Acad Sci USA. 1977;74:5716–5720. doi: 10.1073/pnas.74.12.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Englund J A, Zimmerman M E, Swierkosz E M, Goodman J L, Scholl D R, Balfour H H., Jr Herpes simplex virus resistant to acyclovir. A study in a tertiary care center. Ann Intern Med. 1990;112:416–422. doi: 10.7326/0003-4819-76-3-112-6-416. [DOI] [PubMed] [Google Scholar]
- 9.Gaudreau A, Hill E, Balfour H H, Jr, Erice A, Boivin G. Phenotypic and genotypic characterization of acyclovir-resistant herpes simplex viruses from immunocompromised patients. J Infect Dis. 1998;178:297–303. doi: 10.1086/515626. [DOI] [PubMed] [Google Scholar]
- 10.Hall J D, Coen D M, Fisher B L, Weisslitz M, Randall S, Almy R E, Gelep P T, Schaffer PA. Generation of genetic diversity in herpes simplex virus: an antimutator phenotype maps to the DNA polymerase locus. Virology. 1984;132:26–37. doi: 10.1016/0042-6822(84)90088-6. [DOI] [PubMed] [Google Scholar]
- 11.Hwang C C, Chen H H. An altered spectrum of herpes simplex virus mutations mediated by an antimutator DNA polymerase. Gene. 1995;152:191–193. doi: 10.1016/0378-1119(94)00712-2. [DOI] [PubMed] [Google Scholar]
- 12.Nugier F, Colin J N, Aymard M, Langlois M. Occurrence and characterization of acyclovir-resistant herpes simplex virus isolates: report on a two-year sensitivity screening survey. J Med Virol. 1992;36:1–12. doi: 10.1002/jmv.1890360102. [DOI] [PubMed] [Google Scholar]
- 13.Parris D S, Harrington J E. Herpes simplex virus variants resistant to high concentrations of acyclovir exist in clinical isolates. Antimicrob Agents Chemother. 1982;22:71–77. doi: 10.1128/aac.22.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Safrin S, Elbeik T, Phan L, Robinson D, Rush J, Elbaggari A, Mills J. Correlation between response to acyclovir and foscarnet therapy and in vitro susceptibility result for isolates of herpes simplex virus from human immunodeficiency virus-infected patients. Antimicrob Agents Chemother. 1994;38:246–250. doi: 10.1128/aac.38.6.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarisky R T, Nguyen T T, Duffy K E, Wittrock R J, Leary J J. Difference in incidence of spontaneous mutations between herpes simplex virus types 1 and 2. Antimicrob Agents Chemother. 2000;44:1524–1529. doi: 10.1128/aac.44.6.1524-1529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schnipper L E, Crumpacker C S. Resistance of herpes simplex virus to acycloguanosine: role of viral thymidine kinase and DNA polymerase loci. Proc Natl Acad Sci USA. 1980;77:2270–2273. doi: 10.1073/pnas.77.4.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tebas P, Scholl D, Jollick J, McHarg K, Arens M, Olivo P D. A rapid assay to screen for drug-resistant herpes simplex virus. J Infect Dis. 1998;177:217–220. doi: 10.1086/517357. [DOI] [PubMed] [Google Scholar]
- 18.Weinberg A, Bate B J, Masters H, Schneider S A, Clark J C, Wren C G, Allaman J A, Levin M J. Activity of penciclovir and acyclovir in vitro against herpes simplex virus types 1 and 2. Antimicrob Agents Chemother. 1992;36:2037–2038. doi: 10.1128/aac.36.9.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]