Abstract
Background:
Gastric cancer is the fourth most common human malignancy and the second reason for cancer morbidity worldwide. LncRNA HOTAIR has recently emerged as a promoter of metastasis in various cancer types, including GC, through the EMT process. However, the exact mechanism of HOTAIR in promoting EMT is unknown. Aberrant expression of the miR-200 family has been linked to the occurrence and development of various types of malignant tumors. This study investigates the correlation between the HOTAIR and miR-200 family gene expression patterns in GC cell lines. We investigated the miR-200 and HOTAIR due to their common molecular features in the EMT process.
Methods:
AGS and MKN45 cell lines were transfected with si-HOTAIR, along with a negative control. The effect of HOTAIR knockdown was also analyzed on cell viability and also on the expression of miR-200 family members, including miR-200a, -200b, and -200c, in cell lines using qRT-PCR. Statistical analysis was performed to find the potential correlation between the expression level of HOTAIR and miRs.
Results:
Our results showed significant increased miR-200 family expression level in transfected AGS and MKN45 GC cells (fold changes > 2; p < 0.001). Moreover, a negative correlation was observed between HOTAIR and miR-200 expression levels in GC cell lines (p < 0.05).
Conclusion:
Our findings showed a significant association between miR-200 family and HOTAIR expression levels in GC cell lines. Taken together, the HOTAIR-miR-200 axis seems to play a vital role in human GC, suggesting a potential therapeutic target in future GC treatment.
Key Words: Gene expression, Long noncoding RNA, HOTAIR, MicroRNAs
INTRODUCTION
Gastric cancer is considered as one of the most common human malignancies and cancer morbidities worldwide[1]. Various treatment approaches such as surgery, chemotherapy and targeted therapy have continuously been developed to manage the GC patients, although the GC prognosis has still remained poor[2-5]. Tumor metastasis is an intricate process in which cancer cells move toward secondary organs far from the original tumor site. Metastatic cancer cells maintain their epithelial characteristics to some extents, showing mesenchymal characteristics such as invasion or distraction. This biological process is called EMT, which is characterized by the lack of E-cadherin expression, as well as N-cadherin and vimentin upregulation[6].
EMT is a process that is regulated by a number of signaling pathways and transcriptional/post-transcriptional factors[7], involving both miRNA and lncRNA families[8]. Several studies have shown the involvement of epigenetic modifications, including histone modifications, DNA methylation, lncRNAs, and miRNAs in the EMT process of cancer cells[9-12]. Therefore, the accumulation of genetic and epigenetic modifications can contribute to the etiology of GC[13]. It has been demonstrated that the major part of the human genome (98%) is non-coding DNA, which does not code for amino acids. MiRNAs and lncRNAs are two main ncRNA families and are able to control basic cellular processes through various mechanisms[14]. NcRNAs are classified into two major categories: structural ncRNAs and regulatory ncRNAs[15]. LncRNAs containing more than 200 nucleotides[15], have different roles in the regulation of gene expression[16]. Furthermore, miRNAs are short endogenous ncRNAs of around 18–25 nucleotides length[17,18] that can suppress target mRNAs involved in various biological pathways[19].
Among lncRNAs, the lncRNA HOTAIR is a featured one recognized in 2007[20]. This lncRNA can repress different target genes, such as miRNAs, through an epigenetic process by recruiting polycomb repressive complex 2. Its role in different cancers, particularly gastric tumors, has been proposed previously. Silencing of HOTAIR has been suggested to prevent GC cell migration, invasion, and metastasis and inverses the EMT process in GC cells[5]. The involvement of HOTAIR in EMT process through epigenetically silencing of miRNAs, such as miR-34a, has previously been reported[5]. Nevertheless, this EMT regulatory action of HOTAIR on GC cells through miRNAs has not completely been understood yet.
Among miRNAs, miR-200 family includes featured members acting as EMT regulatory factors due to their ability to cease Wnt/β-catenin pathway in GC cells[21,22]. The transcripts go through dynamic regulatory epigenetic changes associated with EMT or MET phenotypes during tumor progression[9]. It has been indicated that the miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2 proteins[22]. By this mechanism, miR-200 can inhibit the EMT process, which is involved in carcinogenesis.
Based on the epigenetic regulatory effect of HOTAIR on different miRNAs and its potential role in EMT process and due to the regulatory effect of miR-200 on EMT markers, our study was designed to find a correlation between the expression of HOTAIR and miR-200 members. The current study was aimed to find the silencing effect of HOTIAR on miR-200 members involved in EMT pathway. We found that HOTAIR knockdown increased the miR-200 family level in GC cell lines. Our findings provide new insights into the molecular mechanisms of lncRNAs-mediated regulation of miRNAs involved in the EMT pathway. To our knowledge, this is the first study that reveals a correlation between the HOTAIR and miR-200 family in GC.
MATERIALS AND METHODS
Cell lines and culture conditions
AGS and MKN45 human GC cell lines were prepared from the Pasteur Institute of Iran (Tehran, Iran). Cells were cultured in RPMI 1640 medium (Gibco, USA), supplemented with 10% FBS (Invitrogen, USA), and 1% penicillin and streptomycin (Invitrogen). The cells were then incubated at 37 °C, 5% CO2, and saturated humidity. The cells were sub-cultured when grown to suitable confluence. Cell growth was monitored using an inverted microscope. Cells in the logarithmic growth phase were cultured for further analysis.
Knockdown of HOTAIR in AGS and MKN45 cell lines
For HOTAIR gene silencing, AGS and MKN45 cells were transfected with 50 nM of si-HOTAIR; siNC was provided from Sigma (UK). The siRNAs included HOTAIR siRNA–SASI (Hs02_00380445) and negative control (MISSION® siRNA Universal Negative Control #1, SIGMA/SIC001). AGS and MNK45 cells were grown in 24-well plates to 50% confluence and subsequently transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. The experiment was repeated twice. Cells were plated until they reach 70-90% confluency at the time of transfection (AGS: 60 × 103 cells and MKN45: 110 × 103 cells) and then harvested at 48 h after transfection. Finally, the HOTAIR gene knockdown was assessed by real-time PCR.
Proliferation assay
Cell viability assay was performed using MTT kit (Atocell, England), which is used to measure cellular metabolic activity as an indicator of cell viability based on the manufacturer’s guidelines. The cell lines were plated on a 96-well plate and after 24 h of seeding, AGS and MKN45 cells were transfected with 50 nM of si-HOTAIR along with a siNC and MOCK. MTT solution was added to each well after 48 hours and incubated in humidified atmosphere with 5% CO2 at 37 °C for 4 h. Colorimetric detection was carried out at 490 nm using an ELISA-reader (Bio-Rad, USA). Four replicate wells were set up in each group, and experiments were repeated two times.
RNA extraction and cDNA synthesis from miRNAs/total RNAs
Total RNAs were extracted from the cultured cells using RNX (CinnaGen, Iran) based on the manufacturer’s guidelines. The extracted RNA was treated with DNaseI (Sigma) and then was stored at -80 °C. Using a Nanodrop spectrophotometer (Epoch, BioTek- USA), RNA purity was determined based on A260/280 nm absorbance ratio. In addition, RNA integrity was assessed using electrophoresis on a 1% agarose gel containing SafeStain (CinnaGen). Then RNA was reverse transcribed into cDNA using cDNA Synthesis Kit (Takara, Japan) following the manufacturer’s guidelines. The qPCR BONmiR kit (Cat no. BN-0011.17) was purchased from BonBiotech Company (Iran). The kit was applied to synthesize cDNA from all miRNAs, and SNORD gene was used as the internal control gene. Instead of using oligo dT and random hexamer, a relatively long sequence (50-70 nucleotides) called specific RTP (Provided from BonBiotech, Iran) was employed to trap miRNA and SNORD gene. In the end, the final amplification from RNAs attached to these pieces was carried out. The mixture was exposed to 16 °C for 10 minutes to bind the miRNAs to RTPs. It was then exposed to 42 °C for 40 minutes to completely amplify the components attached to the RTP. Tests were performed in duplicates.
Real-time PCR
Real-time PCR was performed using SYBR-Green PCR Mix (Takara) in an ABI 7100 system (Applied Biosystems, USA). The total reaction volume was 20 μl containing 1 μl of cDNA, 0.5 μl of forward primer (10 μM), 0.5 μl of reverse primer (10 μM), and 10 μl of 2× SYBR Green PCR super master mix. Conditions for PCR included denaturation at 95 °C for 5 min, 40 cycles of 5 sec at 95 °C, and annealing/extension at 60 °C for 30 s. The housekeeping gene HPRT1 was used as the internal control for HOTAIR gene, and SNORD gene was used as the internal control for miR-200 family. Primers were designed by GenScript online tool (https://www.genscript.com/tools/pcr-primers-designer) and Primer-Blast (https://www.ncbi.nlm.nih. gov/tools/primer-blast/) according to the cDNA sequences extracted from the Gene bank. The primer sequences are shown in the Table 1. To validate the real-time PCR data, melt curves were plotted, and the accuracy of the curves was confirmed for each analyzed gene and primer dimer fragments. Finally, the expression of genes was assessed by 2- Ct method (Livak method; Ct: Ct gene of interest - Ct internal control) to measure the expression level of genes in each sample analyzed. The efficiency of the primers was determined through standard curves plotted by raw data.
Table 1.
Primers used in this study
| Target gene | Primer name | Sequence |
|---|---|---|
| HOTAIR | F R |
5’-GAAAGGTCCTGCTCCGCTTC-3’ 5’-TCCTCTCGCCGCCGTCTG-3’ |
| HPRT1 | F R |
5’-GGACTTTGCTTTCCTTGGTCAG-3’ 5’-GTCAAGGGCATATCCTACAACA-3’ |
| miR-200a | F | 5’-AACGCTAACACTGTCTGGT-3’ |
| miR-200b | F | 5’-TCATCCGCTAATACTGC-3’ |
| miR-200c | F | 5’-AGACCGCTAATACTG-3’ |
| SNORD47 | F | 5’- ATC ACT GTA AAA CCG TT-3’ |
| MiR-200 Family | Universal common reverse | [BN-0011.17.5] |
Family miR-200, SNORD47 Primers Reference: Exclusively designed by Bon Yakhteh Company, Tehran, Iran; HOTAIR, HPRT1 Primers Reference: GenScript online tool; F, forward; R, reverse
Statistical analysis
Statistical analysis was performed using Graphpad Prism 8 software. Data were shown as mean ± standard deviation. Student’s t-test (unpaired) and two-way ANOVA were used for data analysis. The difference was statistically significant at p < 0.05. Gene expression analysis for the HOTAIR and miR-200 family genes was repeated two times for each sample.
RESULTS
Downregulation of HOTAIR expression levels in AGS and MKN45 GC by si- HOTAIR
The knockdown of HOTAIR led into the decreased number of cells. There was a statistically significant reduction of si-HOTAIR-transfected cells growth compared to siNC and MOCK groups (for AGS = 0.28 and for MKN45 = 0.23; p < 0.0001; Fig. 1). After the treatment of AGS and MKN45 GC cell lines with si-HOTAIR, the expression level of HOTAIR was evaluated by qRT-PCR. As shown in Figure 2A, the expression analysis of HOTAIR in AGS and MKN45 transfected by si-HOTAIR showed the decreased expression level of HOTAIR in AGS and MKN45 (fold change = 0.42 and fold change = 0.57; p = 0.0156 and p < 0.05, respectively).
Fig. 1.

Effect of knockdown on cell viability. MTT assay was performed to evaluate the cell proliferation in si-HOTAIR-transfected AGS and MKN45 cell lines. The data show a statistically significant decrease in the growth of si-HOTAIR-transfected cells compared to siNC and MOCK groups (****p < 0.0001 vs. NC)
Fig. 2.
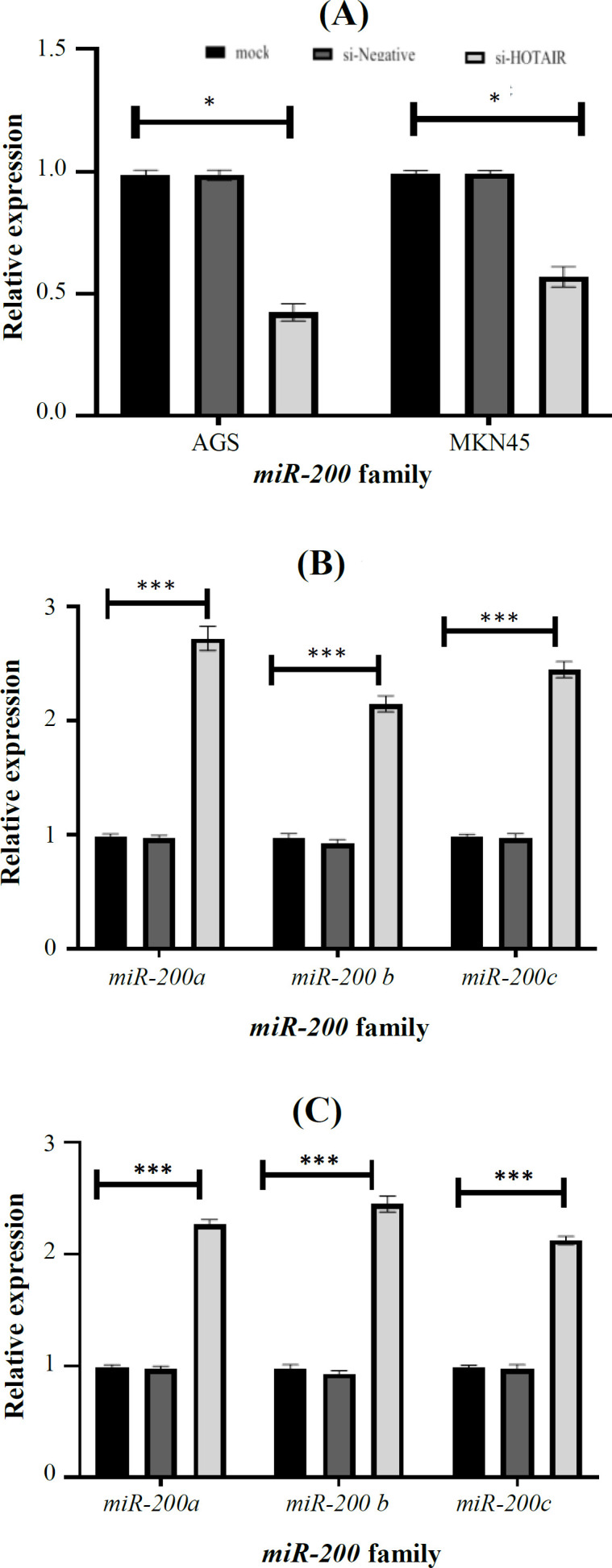
RT-qPCR indicating the downregulation of HOTAIR expression by si-HOTAIR. (A) Expression levels of HOTAIR in AGS and MKN45 cells transfected with si-HOTAIR; (B) expression levels of miR-200c/-200a/-200b in AGS cells transfected with si-HOTAIR; (C) expression levels of miR-200a/-200b/-200c in MKN45 cells transfected with si-HOTAIR. Data are expressed as mean ± SD. *p < 0.05 and ***p < 0.001
Altered gene expression of miR-200 family after HOTAIR downregulation
We hypothesized that HOTAIR can induce EMT through the downregulation of miR-200 family members targeting the E-cadherin repressors ZEB1 and ZEB2 proteins (Fig. 3). To examine the possible regulatory role of HOTAIR on miR-200 family expression in AGS and MKN45 GC cell lines, the expression levels of three members of this family, including miR-200a/-200b/-200c, were analyzed. Gene expression was evaluated in AGS and MKN45 transfected with si-HOTAIR compared to the negative controls. RT-qPCR analysis of miR-200 family expression levels was performed, as well. Our data revealed that the expression of miR-200 family was upregulated in both AGS and MKN45. In other words, the knockdown of HOTAIR led to the increased level of all three genes of the miR-200 family (Table 2). As shown in AGS cell line, the most differentiated expression miRNA was the miR-200a (fold change: 3; p < 0.001), while the least differentiated one was miR-200b gene (fold change: 2.1; p < 0.001), as represented in Figure 2B). Also, in MKN45 cell line, miR-200b gene showed the most change (fold change: 2.4; p < 0.001), while miR-200c gene reached the least change of expression (fold change: 2.1; p < 0.001; Fig. 2C).
Fig. 3.
Potential mechanism involving HOTAIR and miR-200 family members in the EMT process
Table 2.
Fold change and p value of expression analysis of miR-200 family in the loss of function study of HOTAIR in different GC cell lines
| p value | AGS | MKN45 | Gene |
|---|---|---|---|
| 0.001 | FG: 3 | FG: 2.3 | miR-200a |
| 0.001 | FG: 2.1 | FG: 2.4 | miR-200b |
| 0.001 | FG: 2.4 | FG: 2.1 | miR-200c |
FG, fold change
Correlations between the expression levels of HOTAIR and miR-200 family
Using Pearson's correlation coefficient, we found a correlation between the expression levels of HOTAIR and miR-200 family members in GC cell lines. Statistical significant inverse correlations were observed between HOTAIR and miR-200 family expression in GC cell lines. The expression levels of HOTAIR and miR-200a gene had a significant negative correlation in AGS cell line (p < 0.001; r = -0.9934; Fig. 4A). Furthermore, the expression level of miR-200a/c and HOTAIR were negatively correlated in MKN45 cell line (r = -0.9797 [Fig. 4B] and r = -0.9800 [Fig. 4C], respectively p < 0.05).
Fig. 4.
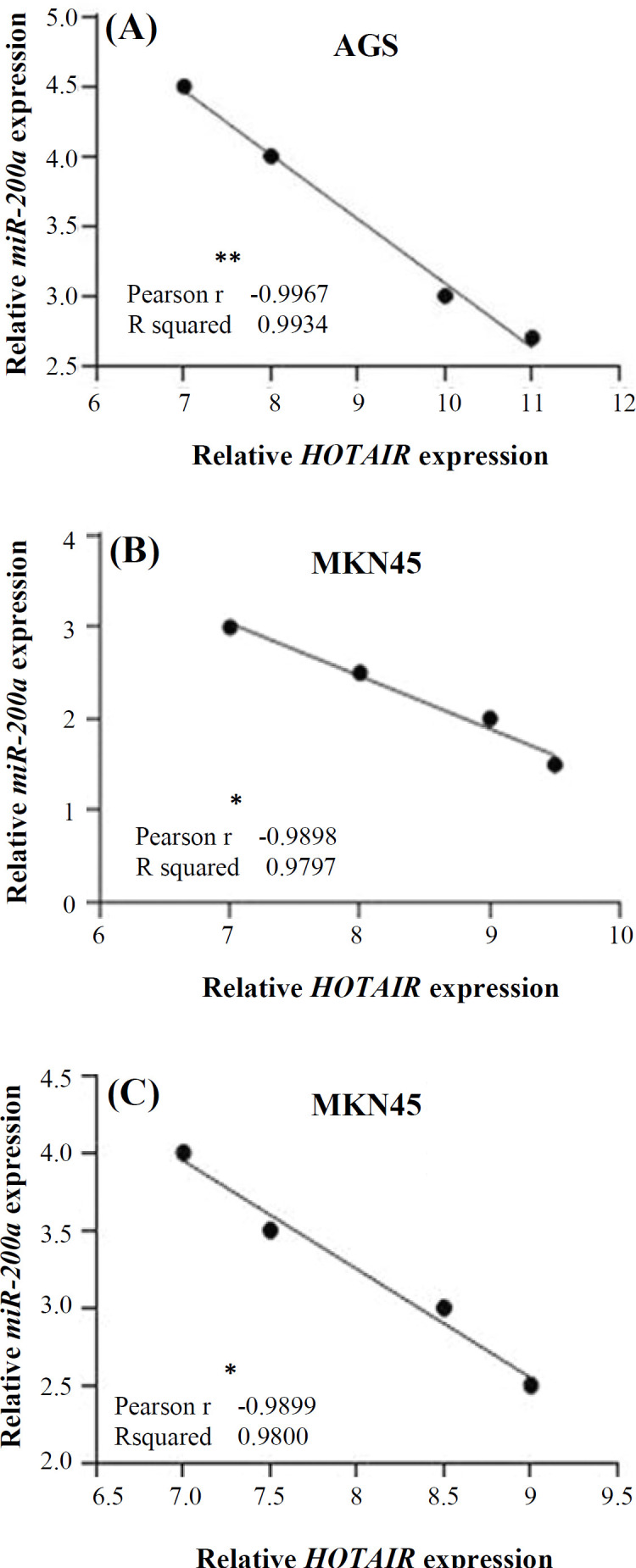
Correlation between the expression level of HOTAIR and miR-200 in AGS and MKN45 GC cell lines. Pearson’s correlation coefficient revealed the expression levels of HOTAIR and (A) miR-200a in AGS cell line (**p < 0.01). (B and C) miR-200a/-200c in MKN45 cell line were negatively correlated (*p < 0.05)
DISCUSSION
GC is known as one of the most lethal malignancies. Several studies have investigated the role of various genes in the progression of this type of cancer[6-10,23]. LncRNAs have recently been identified as novel regulators of transcriptional and epigenetic networks[24]. These RNAs achieve their biological functions through their interactions with multiple signaling molecules, including proteins[25] and miRNAs[26]. Although being expressed in special types of cells and at specific developmental stage, lncRNAs are not often associated with different kinds of cancers[27].
Since the introduction of HOTAIR by Rinn et al.[20] in 2007, numerous studies have reported that the overexpression of lncRNA HOTAIR is associated with various cancer types[28-30], suggesting the role of HOTAIR as an oncogene in a variety of human cancers. Previous studies have revealed that HOTAIR is upregulated in GC tumor tissues[23,31-33]. Our results confirmed that HOTAIR is a likely therapeutic target in the treatment of GC. However, the function of HOTAIR in GC has remained largely unknown. HOTAIR has been indicated to be able to induce EMT through repressing target genes[27,34]. In this study, we found the potential mechanism of action of HOTAIR in the downregulation of miR-200 family members as one of the featured markers of EMT.
MicRNAs have been proved to function as important regulatory RNA molecules in tumorigenesis processes[35]. MiR-200 is one of the major positive regulators in the maintenance of the epithelial phenotype via the repression of ZEB1[36]. The miR-200 family is involved in carcinogenesis through the regulation of EMT and is regulated through DNA methylation as a gene-silencing approach[37]. A few studies have indicated that miR-200 family can be downregulated in epithelial cancers, particularly GC[38-41]. The role of miR-200 has previously been reported as a tumor suppressor factor for the inhibition of the EMT process and tumor growth of GC through targeting ZEB1 and ZEB2[22].
The role of HOTAIR in EMT regulation of GC has demonstrated in various investigations[5,34,42]. HOTAIR epigenetically represses numerous factors including miR-34a; and contributes to the process of GC cell EMT[5,41] by recruiting the PRC2[5]. Previous surveys have also indicated the role of HOTAIR[27,34] and miR-200 family[9,41] in promoting EMT program in GC. As HOTAIR can silence the expression of miR-200 family, we hypothesize a correlation between HOTAIR and miR-200 in GC. Our findings suggested a novel mechanism mediating shifts between EMT and MET programs through HOTAIR and miR-200 members. Therefore, miR-200 and HOTAIR were investigated in this study due to their common molecular features in EMT process.
Our results showed that HOTAIR knockdown increases the expression level of miR-200a, miR-200b, and miR-200c in both AGS and MKN45 GC cell lines. In this study, we explored the correlation between HOTAIR knockdown and miR-200 family expression.
Our data signified that the level of miR-200 family in GC cell lines is closely associated with the HOTAIR expression level. A negative correlation was also observed between HOTAIR and miR-200 expression levels in GC cell lines. As miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2 proteins[9], our results displays that HOTAIR induces the migration and metastasis of GC cells through miR-200 family, thereby affecting EMT. We speculate that HOTAIR has an effect on the EMT through this mechanism (Fig. 3). However, further studies are needed to reveal the exact regulatory mechanism of HOTAIR on this miR family. Overexpression of HOTAIR as well as interaction analyses between HOTAIR and regulatory factors/elements can help the researchers to find more about this axis.
Our investigation highlights the need for understanding the mechanism involving biological processes and the regulatory interactions between ncRNAs and coding transcripts, which may be beneficial to GC treatment. HOTAIR-miR-200 axis seems to play an important role in human GC, reflecting a promising therapeutic target in future GC treatment. To our knowledge, this is the first study reporting the HOTAIR-miR-200 axis in GC. However, the exact mechanism should be explored by more surveys in future.
CONFLICT OF INTEREST.
None declared.
ACKNOWLEDGMENTS
We acknowledge the Iranian Council of Stem Cell Technology, Shahid Chamran university of Ahvaz, and Islamic Azad University for financial support of this study.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer. 2015;136(5):e359–e386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Newton AD, Datta J, Loaiza-Bonilla A, Karakousis GC, Roses RE. Neoadjuvant therapy for gastric cancer: current evidence and future directions. Journal of gastrointestinal oncology. 2015;6(5):534. doi: 10.3978/j.issn.2078-6891.2015.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu QJ, Ito S, Yanagihara K, Mimori k. Molecular mechanism of peritoneal dissemination in gastric cancer. Journal of cancer metastasis and treatment. 2018;4:39. [Google Scholar]
- 4.Shimizu D, Kanda M, Kodera Y. Emerging evidence of the molecular landscape specific for hematogenous metastasis from gastric cancer. World journal of gastrointestinal oncology. 2018;10(6):124–136. doi: 10.4251/wjgo.v10.i6.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Sun M, Xia R, Zhang EB, Liu X-h, Zhang ZH, Xu TPp, De W, Liu BRr, Wang ZX. Linc HOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell death and disease. 2015;6(7) doi: 10.1038/cddis.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang Y, Massagué J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118(3):277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 7.Serrano-Gomez SJ, Maziveyi M, Alahari SK. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Molecular cancer. 2016;15:18. doi: 10.1186/s12943-016-0502-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo F, Kerrigan BCP, Yang D, Hu L, Shmulevich I, Sood AK, Xue F, Zhang W. Post-transcriptional regulatory network of epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions. Journal of hematology and oncology. 2014;7:19. doi: 10.1186/1756-8722-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davalos V, Moutinho C, Villanueva A, Boque R, Silva P, Carneiro F, Esteller M. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene. 2012;31(16):2062–2074. doi: 10.1038/onc.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiesslich T, Pichler M, Neureiter D. Epigenetic control of epithelial-mesenchymal-transition in human cancer. Molecular and clinical oncology. 2013;1(1):3–11. doi: 10.3892/mco.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nature medicine. 2013;19(11):1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa FF. Epigenomics in cancer management. Cancer management and research. 2010;2:255–265. doi: 10.2147/CMR.S7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nobili S, Bruno L, Landini I, Napoli , Bechi P, Tonelli F, Rubio CA, Mini E, Nesi G. Genomic and genetic alterations influence the progression of gastric cancer. World journal of gastroenterology. 2011;17(3):290–293. doi: 10.3748/wjg.v17.i3.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue M, Zhuo Y, Shan B. MicroRNAs, long noncoding RNAs, and their functions in human disease. Methods in molecular biology. 2017;1617:1–25. doi: 10.1007/978-1-4939-7046-9_1. [DOI] [PubMed] [Google Scholar]
- 15.Dahariya S, Paddibhatla I, Kumar S, Raghuwanshi S, Pallepati A, Gutti RK. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Molecular immunology. 2019;112:82–92. doi: 10.1016/j.molimm.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 16.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ha M, Kim VN. Regulation of microRNA biogenesis. Nature reviews molecular cell biology. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 18.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136(4):669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 19.Ahadi A. Dysregulation of miRNAs as a signature for diagnosis and prognosis of gastric cancer and their involvement in the mechanism underlying gastric carcinogenesis and progression. IUBMB life. 2020;72(5):884–898. doi: 10.1002/iub.2259. [DOI] [PubMed] [Google Scholar]
- 20.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su J, Zhang A, Shi Z, Ma F, Pu P, Wang T, Zhang J, Kang Ch, Zhang Q. MicroRNA-200a suppresses the Wnt/β-catenin signaling pathway by interacting with β-catenin. International journal of oncology. 2012;40(4):1162–1170. doi: 10.3892/ijo.2011.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cong N, Du P, Zhang A, Shen F, Su J, Pu P, Wang T, Zjang J, Kang Ch, Zhang Q. Downregulated microRNA-200a promotes EMT and tumor growth through the wnt/β-catenin pathway by targeting the E-cadherin repressors ZEB1/ZEB2 in gastric adenocarcinoma. Oncology reports. 2013;29(4):1579–1587. doi: 10.3892/or.2013.2267. [DOI] [PubMed] [Google Scholar]
- 23.Hajjari M, Behmanesh M, Sadeghizadeh M, Zeinoddini M. Up-regulation of HOTAIR long non-coding RNA in human gastric adenocarcinoma tissues. Medical oncology. 2013;30(3):670. doi: 10.1007/s12032-013-0670-0. [DOI] [PubMed] [Google Scholar]
- 24.Sui C-j, Zhou Y-m, Shen W-f, Dai BH, Lu JJ, Zhang MF, Yang JM. Long noncoding RNA GIHCG promotes hepatocellular carcinoma progression through epigenetically regulating miR-200b/a/429. Journal of molecular medicine. 2016;94(11):1281–1296. doi: 10.1007/s00109-016-1442-z. [DOI] [PubMed] [Google Scholar]
- 25.Ferre F, Colantoni A, Helmer-Citterich M. Revealing protein–lncRNA interaction. Briefings in bioinformatics. 2016;17(1):106–116. doi: 10.1093/bib/bbv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing miRNA–lncRNA interactions. Methods in molecular biology. 2016;1402:271–286. doi: 10.1007/978-1-4939-3378-5_21. [DOI] [PubMed] [Google Scholar]
- 27.Lee NK, Lee JH, Park CH, Yu D, Lee Y, Cheong J-H, Noh SH, Lee SK. Long non-coding RNA HOTAIR promotes carcinogenesis and invasion of gastric adenocarcinoma. Biochemical and biophysical research communications. 2014;451(2):171–178. doi: 10.1016/j.bbrc.2014.07.067. [DOI] [PubMed] [Google Scholar]
- 28.Battistelli C, Cicchini C, Santangelo L, Tramontano A, Grassi L, Gonzalez FJ, Nonno V, Grassi G, Amicone L, Tripodi M. The Snail repressor recruits EZH2 to specific genomic sites through the enrollment of the lncRNA HOTAIR in epithelial-to-mesenchymal transition. Oncogene. 2017;36(7):942–955. doi: 10.1038/onc.2016.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang JX, Han L, Bao Z-S, Wang Y-Y, Chen LY, Yan W, Liu N, YU SZ, Pu PY, You YP, Jiang T, Kang CS. HOTAIR, a cell cycle–associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro-oncology. 2013;15(12):1595–1603. doi: 10.1093/neuonc/not131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer biology and medicine. 2015;12(1):1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan W, Liu L, Wei J, Ge Y, Zhang J, ChenH , Zhou L, Yuan Q, Zhou Ch, Yang M. A functional lncRNA HOTAIR genetic variant contributes to gastric cancer susceptibility. Molecular carcinogenesis. 2016;55(1):90–96. doi: 10.1002/mc.22261. [DOI] [PubMed] [Google Scholar]
- 32.Zhao W, Dong S, Duan B, Chen P, Shi L, Gao H, Qi H. HOTAIR is a predictive and prognostic biomarker for patients with advanced gastric adenocarcinoma receiving fluorouracil and platinum combination chemotherapy. American journal of translational research. 2015;7(7):1295–1302. [PMC free article] [PubMed] [Google Scholar]
- 33.Bhan A, Mandal SS. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochimica et biophysica acta. 2015;1856(1):151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song Y, Wang R, Li L-W, Wang Y-F, Wang Q-X, Zhang Q. Long non-coding RNA HOTAIR mediates the switching of histone H3 lysine 27 acetylation to methylation to promote epithelial-to-mesenchymal transition in gastric cancer. International journal of oncology. 2019;54(1):77–86. doi: 10.3892/ijo.2018.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corté H, Manceau G, Blons H, Laurent-Puig P. MicroRNA and colorectal cancer. Digestive and liver disease. 2012;44(3):195–200. doi: 10.1016/j.dld.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 36.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz Th. A reciprocal repression between ZEB1 and members of the miR‐200 family promotes EMT and invasion in cancer cells. EMBO reports. 2008;9(6):582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neves R, Scheel C, Weinhold S, Honisch E, Iwaniuk KM, Trompeter H-I, Niederacher D, Wernet P, Uhrberg M. Role of DNA methylation in miR-200c/141 cluster silencing in invasive breast cancer cells. BMC research notes. 2010;3:219. doi: 10.1186/1756-0500-3-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Men D, Liang Y, Chen L. Decreased expression of microRNA-200b is an independent unfavorable prognostic factor for glioma patients. Cancer epidemiology. 2014;38(2):152–156. doi: 10.1016/j.canep.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 39.Paterson EL, Kazenwadel J, Bert AG, Khew-Goodall Y, Ruszkiewicz A, Goodall GJ. Down-regulation of the miRNA-200 family at the invasive front of colorectal cancers with degraded basement membrane indicates EMT is involved in cancer progression. Neoplasia. 2013;15(2):180–IN22. doi: 10.1593/neo.121828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soubani O, Ali AS, Logna F, Ali Sh, Philip PA, Sarkar FH. Re-expression of miR-200 by novel approaches regulates the expression of PTEN and MT1-MMP in pancreatic cancer. Carcinogenesis. 2012;33(8):1563–1571. doi: 10.1093/carcin/bgs189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ning X, Shi Z, Liu X, Zhang A, Han L, Jiang Km, Kang Ch, Zhang Q. DNMT1 and EZH2 mediated methylation silences the microRNA-200b/a/429 gene and promotes tumor progression. Cancer letters. 2015;359(2):198–205. doi: 10.1016/j.canlet.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 42.Xu ZY, Yu QM, Du YA, Yang LT, Dong RZ, Huang L, Yu PF, Cheng XD. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. International journal of biological sciences. 2013;9(6):587–597. doi: 10.7150/ijbs.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]



