Abstract
Introduction: Activating mutations in the BRAF gene have been reported in 0.8%-8% cases of NSCLC. Traditionally, diagnostics have mainly focused on detection of V600E and modalities like mutation specific IHC, allele specific real-time PCR have been utilized. This may underestimate true prevalence of the non-V600E variants. Broader panel NGS testing offers a one stop solution and may identify newer potentially targetable variants. This is a retrospective single center experience of patients with BRAF mutated NSCLC characterizing the molecular spectrum and clinicopathologic characteristics. Methods: 260 patients underwent panel based NGS testing at our center, between 2017-2020. 13 BRAF mutant cases, were detected and were clinically reviewed. Results: Thirteen cases of BRAF alterations were seen in out of 260 (5%) patients. Median age of the cohort was 62 years (range: 39-86 years) with a female predilection). Canonical BRAF V600E mutation was seen in 6 (46.2%) patients and 7 (53.8%) harbored a non-V600E alteration. Spectrum of non V600E alterations included G466E, G469A, N581I, V600_K601delins, D594G, L597Q, G649V and were commonly female (P>0.01) with a higher trend for liver metastases (P=0.09). Median PFS was 4.8 months on chemotherapy (P=0.8). All patients (13/13, 100%) were never smokers with an adenocarcinoma histology. Conclusion: This is a single center experience from an Indian NSCLC cohort and shows higher prevalence of non-V600E than V600E mutation reported in literature. This may be attributed to increased use of NGS testing revealing otherwise missed alterations on sequential single gene testing.
Keywords: BRAF, NSCLC, India, real-world, non-V600E
Introduction
Non-small-cell lung cancer (NSCLC) subtype of lung cancer accounts for 85% of all lung cancers with adenocarcinoma considered as the most common histology in NSCLC. The evolution of molecular biology has enhanced the understanding of disease biology and led to a paradigm shift in both therapeutic and prognostic landscape of this disease. The genetic risk factors can be myriad including both Li fraumeni syndrome as well as other cancer predisposition syndromes, however, they are of rare occurrence, and most cases are sporadic. Apart from other common alterations in EGFR (epidermal growth factor receptor), ALK (anaplastic lymphoma kinase) and ROS1 genes, BRAF mutations have also been reported to be actionable targets. Activating mutations in the BRAF [1] gene have been identified in myriad malignancies [2,3], commonly seen in colon, thyroid, melanoma, and some other clonal processes like Langerhans cell histiocytosis. They have also been included as a Tier1 [4] mutation in non small cell lung carcinoma (NSCLC) [5]. BRAF alterations have been reported in 0.8-8% cases of lung carcinoma [6], with ethnic differences in frequency, analogous to EGFR (epidermal growth factor receptor) mutant NSCLC.
Current NCCN (National Comprehensive Cancer Network) guidelines [5] recommend testing all lung cancers for BRAF mutations at diagnosis along with EGFR, ALK (anaplastic lymphoma kinase) and ROS1 alterations for adenocarcinoma and non-smokers with squamous histology. However, availability of adequate tissue subsequent to morphologic and immunohistochemical (IHC) diagnosis is a concern. Tissues in cases of lung core biopsies are limited not only by quantity but also tumor cellularity. Hence, sequential single gene testing for all cases may not be amenable. Broader panel based testing using next generation sequencing technology has paved the way to account for all these and keep pace with the ever evolving treatment paradigm. Diagnostic modalities which can be used for detection include mutation specific BRAF V600E IHC [7], real-time PCR (polymerase chain reaction), allele specific/ARMS (amplification refractory melting system) PCR and direct sequencing using Sanger or NGS (next generation sequencing) methods. However, since non-V600E alterations are not uncommon in the lung, IHC, PCR and Sanger based modalities are limited by the spectrum of mutations detected.
BRAF alterations in lung although rare, are associated with distinct clinicopathologic profiles as evidenced in some real world data reported in literature [8-11]. However, all these reports are limited by sample size owing to rarity of occurrence of the same. A comprehensive understanding of the disease biology, response outcomes and any potential clinical associations is warranted. This study is a single center retrospective analysis of clinicopathologic features, natural history and outcomes of this entity highlighting the need for NGS based testing in NSCLCs. To the best of our knowledge this is the 1st report from the Indian peninsula, which highlights some distinct features from that reported in literature
Methods
Patient recruitment
All NSCLC cases registered at Rajiv Gandhi Cancer Institute and Research Center, who underwent NGS based testing for BRAF alterations between January 2017-December 2020, were included in the study. The inclusion criteria were all cases on NSCLC which underwent broad panel based next generation sequencing (panel including BRAF alterations), between January 2017-December 2020 were included in this study. All patients were >18 years of age, and both genders, and all types of histologies were included in this study. Patients of NSCLC who did not undergo broad molecular profiling including testing for mutations in BRAF were excluded from the study. The demographics, clinicopathologic features, treatment details and outcomes were retrieved and recorded from the electronic medical record archives of the hospital. This study has been carried out in accordance with the Declaration of Helsinki and has been approved by the Institutional Ethics Committee (Res/SCM/2021/46).
Molecular studies
NGS was performed on formalin fixed paraffin embedded (FFPE) tumor tissue. The percentage of tumor cells relative to other cells like stromal cells, inflammatory cells and normal epithelial cells was estimated on hematoxylin and eosin stained tumor section, and an area with the maximum tumor infiltration, not less than 20% was marked for macrodissection.
Nucleic acid extraction and preparation
DNA and RNA were extracted from the formalin fixed paraffin embedded (FFPE) block after careful microdissection using SV Total Nucleic Acid Isolation System (Promega Corporation, Wisconsin, and USA). The extracted DNA was quantified using Qubit fluorometer (Thermofisher Scientific, CA, USA). The extracted RNA was quantified using Qubit fluorometer (Thermofisher Scientific, CA, USA). The RNA was reverse transcribed to complementary DNA (cDNA) using Invitrogen SuperScript IV VILO kit (Thermofisher Scientific, CA, USA).
Library preparation and sequencing
Libraries were prepared using the, Oncomine Focus Assay library preparation kit comprising 52 genes implicated in solid organ malignancies. Template preparation and enrichment were done on the Ion OneTouch Select Template Kit on Ion OneTouch 2 (all from Thermo Fisher Scientific, CA, and USA). The prepared library was checked for quality and size using TapeStation (Agilent technologies) using high sensitivity RNA kits. The prepared libraries were sequenced on the Ion PGM Sequencer or the Ion S5 Sequencer.
Data analysis
Post sequencing, the quality metrics were checked on the Torrent Suite browser (v 5.0). The variants were called by the Ion Reporter Software (v5.12) and the Oncomine Knowledge Reported using the Torrent variant caller plug-in. The overall variant call, if positive, was visualized on the integrative genomics viewer to ascertain the quality and validity of the variant called.
Response assessment
The response assessment was done in accordance with the Response Evaluation Criteria in Solid Tumors (RECIST) [12].
Statistical analysis
This is a descriptive study aimed at depicting the molecular epidemiology and clinical features of BRAF mutated NSCLC in an Indian cohort. However, indicators including incidence, prevalence could not be calculated as this is an enriched population of those patients who underwent NGS based testing, and is not representative of the population at risk. Continuous variables were expressed as median and categorical variables as frequencies (%). Comparisons of baseline characteristics between V600E and non-V600E subgroups were done using analysis of variance (ANOVA) for continuous variables and Chi-square or Fisher exact tests for categorical variables. Kaplan Meier (KM) estimating method was used to estimate the overall survival (OS) and progression free survival (PFS) and Log rank test used to compare the survival curves between groups. However owing to few case numbers, graphs could not be generated as they were not representative. All statistical analyses were done using SAS 9.4 (SAS Institute Inc., Cary, NC, USA) and conclusions were made at 5% significant levels with a P value of 0.05 being considered as statistically significant.
Results
Patient characteristics
A total of 260 NSCLC patients who underwent BRAF testing were recruited. Among these 13 (5%) were found to be harbor BRAF alterations as the main oncogenic driver. The median age of these 13 patients was 62 years (range: 39-86 years). There were 7 (53.8%) females and 6 (46.2%) males. The baseline characteristics of these patients are depicted in Table 1.
Table 1.
Comparison of clinical features between V600E and non-V600E subgroups
| Features | V600E | Non-V600E | P value |
|---|---|---|---|
| Age | Median 61 (39-76) | Median 63 (44-86) | 0.1 |
| Gender | |||
| Female | 6 | 1 | 0.01 |
| Male | 0 | 6 | |
| Smoking status | |||
| Never | 6 | 5 | 0.2 |
| Former | 0 | 2 | |
| Liver Metastases | |||
| Present | 2 | 4 | 0.09 |
| Absent | 4 | 3 | |
| Bony Metastases | |||
| Present | 3 | 2 | 0.5 |
| Absent | 3 | 5 | |
| Adrenal metastases | |||
| Present | 1 | 0 | 0.4 |
| Absent | 5 | 7 | |
| Brain Metastases | |||
| Present | 0 | 2 | 0.1 |
| Absent | 6 | 5 | |
| PDL1 | 0.3 | ||
| <1% | 1 | 4 | |
| 1-49% | 3 | 1 | |
| >50% | 0 | 0 | |
| Not done | 2 | 2 | |
| BRAF directed treatment | ---- | ||
| Yes | 1 | 1 | |
| No | 5 | 7 | |
| Median PFS on chemotherapy | 4.3 months | 4.8 months | 0.8 |
Baseline clinicopathologic characteristics associated with BRAF mutant NSCLC
The spectrum of BRAF mutation included 6 (46.2%) cases with the canonical V600E mutation and 7 (43.8%) with non V600E alterations. V600E alterations were detected in females (n=6, 100%), (P<0.01) whereas the non-V600E subgroup showed a male preponderance (6/7, 85.7%). The spectrum of non V600E alterations included G466E, G469A, N581I, V600_K601delinsE, D594G, L597Q, G649V. 11/13 (92.3%) was never smokers, and 2 were former smokers. All (13/13, 100%) cases depicted adenocarcinoma histology with varying levels of differentiation, including poorly differentiated in 6 (46.2%) cases, acinar pattern in 4 (30.7%) cases, lepidic pattern in 3 (23.1%) cases. With respect to the ECOG PS, 4 (30.7%) cases had an ECOG PS of 2, and 9 (69.3%) cases had ECOG PS 1. All cases were advanced stage (Stage IV) NSCLC, of which 11 (84.6%) patients had extra thoracic metastases, and 2 (15.4%) cases had metastases to the brain at diagnosis. 11/13 (92.3%) patients had presence of liver metastases at diagnosis (V600E vs. non V600E: 43% vs. 57%, P=0.09). Table 1 depicts the comparison of these important features between V600E and non-V600E subgroups.
With respect to co-mutations, mutations were detected in 6 patients: PIK3CA (n=1), TP53 (n=2), IDH2 (n=1), NOTCH1 (n=1), and CDK4 (n=1) genes (Table 2).
Table 2.
Table depicting the various mutations encountered in six cases of the study cohort
| Co-Mutation detected: Gene | Protein change |
|---|---|
| PIK3CA | p.Glu545Lys |
| IDH2 | p.Arg172Thr |
| NOTCH1 | p.Val1578del |
| CDK4 | p.Arg209fs |
| TP53 | p.Arg248Gln |
| TP53 | p.Cys277Phe |
Treatment details
Of the 13 patients 11 patients took treatment at our center and 2 were lost to follow up. Two patients received anti-BRAF treatment in the first line, one patient was offered vemurafenib and he showed progressive disease after 4.4 months. The other patient harbored a nonV600E mutation (V600_601delins, detected on NGS) and received combination dabrafenib-trametinib with an ongoing partial response at 6 months. The median PFS was 4.8 months for those who received chemotherapy. When comparing V600E vs. non V600E the median PFS for V600E mutant patients was 4.3 months vs. 4.8 months for non-V600E cases (P=0.8). The median OS for the entire cohort however was not reached. The rest of the patients received chemotherapy as a first line regime. Among the patients who were offered chemotherapy as first line therapy, one patient progressed in 3.3 months and was then offered anti BRAF therapy in the form of dabrafenib-trametinib. This patient showed partial response to anti-BRAF therapy and had an ongoing response for 4 months, however subsequent to this owing to inability to procure the medicine further owing to COVID19 pandemic, she was switched to chemotherapy based regime and is on continuous follow up presently. The complete details of treatment and follow up are depicted in Table 3 and Figure 1.
Table 3.
Table depicting details of treatment details and outcomes and status at last follow up of the study cohort
| Case | BRAF type | 1st line treatment | Response | 1st PFS(months) | Subsequent therapy | Overall Survival/Status |
|---|---|---|---|---|---|---|
| 1 | V600E | Vemurafenib | PD | 4.4 | pemetrexed-carboplatin | 20.3/alive |
| 2 | V600E | Gemcitabine carboplatin | PR | 4.3 | dabrafenib-trametinib | 25.13/alive |
| 3 | V600E | Gemcitabine carboplatin | PR | 7.7 | pemetrexed-carboplatin | 32.47/lfu |
| 4 | G649V | No treatment taken | ---- | ----- | -------- | 1.2/lfu |
| 5 | V600E | pemetrexed + carboplatin | PR | 3.3 | dabrafenib-trametinib | 50.6/alive |
| 6 | V600E | pemetrexed + carboplatin | PR | 5.8 | ------- | 21.33/dead |
| 7 | L597Q | pemetrexed + carboplatin | PD | 6.3 | nivolumab | 14.1/dead |
| 8 | V600E | No treatment taken | -- | -- | -- | 0.07/dead |
| 9 | D594G | pemetrexed + carboplatin | PD | 4.8 | pembrolizumab | 3.03/lfu |
| 10 | V600K601delinsE | dabrafenib + trametinib | PR | dabrafenib + trametinib | 78.83/alive | |
| 11 | N581I | Gemcitabine carboplatin | PR | 4.3 | Nivolumab | 15.2/alive |
| 12 | G469A | Pemetrexed carboplatin | PD | 2.2 | Did not follow up | 6.33/lfu |
| 13 | G466E | Surgery + Adjuvant chemo | PR | 5.4 | pemetrexed-carboplatin | 41/alive |
PD: Progressive disease, PR: partial response, lfu: lost to follow up.
Figure 1.
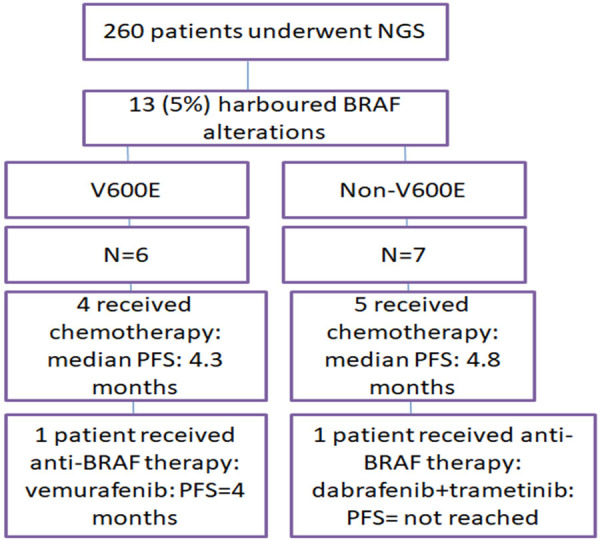
Complete details of treatment and follow up.
Discussion
This is a single center real world retrospective experience of BRAF mutant NSCLC, and from our experience it is evident that use of NGS based testing helped detect more variants in the BRAF gene, other than the canonical V600E.
Somatic mutations in BRAF gene were first reported in 2002 by Davies et al. [3], and they demonstrated an overall incidence of 8% across all malignancies and 3% in NSCLC. Paik et al. [13] reported an incidence of 3% of BRAF alterations in 18 out of 697 patients of NSCLC. In Caucasians it has been reported at a frequency of 2-5% [9], which is higher than rest of the world. However, our incidence is higher (5%), which may be attributed to multiple reasons, the overall cohort tested was smaller than these contemporary studies, the NGS testing was done on cases which tested negative for EGFR ALK and ROS1 by single gene testing. However, on comparing with the TCGA cohort of lung adenocarcinoma reported by Collisson et al. [14], they reported BRAF alterations in 9% cases (16 cases). Hence it is evident that the prevalence of BRAF mutated NSCLC has been reported variably and may not follow geographic or ethnic differences. Therefore the prevalence of V600E was 46.2% and non V600E was 43.8% in our cohort. This is distinct from reported real world data as well as from controlled trials. V600E prevalence was reported at a frequency as high as 85.7% in a study by Mu et al. in 65 Chinese patients of BRAF mutated NSCLC [9]. This may be attributed to the fact that all our patients underwent NGS based testing and not mutation specific or allele specific testing for V600E. However in Caucasians the prevalence of V600E is ~50% [8], and hence this disparity may again exist in different racial and ethnic groups. However in a few other studies by Pisapia et al. [15,16] and Noeparast et al. [17], the prevalence of non-V600E was higher when compared to V600E mutation, which concords with our findings. Another striking difference in our study was that 92.3% cases in our study were never smokers; whereas contemporary studies by Paik et al. reported 100% [13] former or current smoking status in all BRAF altered cases. However, Mu et al., reported that patients with V600E did not have any relation to smoking when compared to non V600E as well as no sex predilection (vs. non V600E being more common in males and smokers). Our study is concordant with Mu et al. [9] study that non-V600E patients were more commonly male, and two of our patients with positive smoking history also harbored non-V600E alterations. Additional evidence is from Ding et al. [10] who reported BRAF mutations in never smokers in Chinese NSCLC patients. This discrepancy maybe attributed to the fact that NGS based testing in our center is offered preferentially to non-smokers and hence may have resulted in a selection bias. As for pathologic features, majority of BRAF mutated were adenocarcinoma.
The co-mutation rate in this study was ~50% with 6 patients harboring co-mutations in the genes mentioned above. Mu et al. [9] reported a co-mutation rate of 18.5%. The discrepancy between these studies may be due to low sample size in each study and difference in distribution of BRAF mutation subtypes across ethnic groups. Co-mutations have been reported in genes like TP53, PIK3CA, and CDK4, similar to our study, all being associated with poor prognosis. In the non-V600E commutations have been reported in KRAS gene also, however we did not encounter any such case, which may be attributed to a smaller sample size. The PFS of patients with V600E and non-V600E was almost similar on chemotherapy which is similar to that reported by other real world studies by Paik et al. [13], Marchetti et al. [14] and Mu et al. [9]. When compared to wildtype, the PFS was shorter which is also concordant with already reported literature. The evidence on OS also follows a similar trend between BRAF V600E and non V600E alterations; however the same has not been reached in our cohort [18]. In a case reported by Nakanishi et al. [19], the patient showed remarkable response to pemetrexed based chemotherapy and was followed up for 97.5 months. However, in our study the median OS was not reached.
Among the two patients who received anti-BRAF treatment, the patient who was offered dabrafenib-trametinib harbored a complex indels at V600_V601 codon. The fact that he has an ongoing response after 6 months of initiation maybe attributed to the fact that the site of mutation corresponds to the site of action of the drug and may have similar functional effects downstream as V600E. However, the same requires confirmation by in-vitro functional studies, which is beyond the scope of this current study.
With respect to efficacy of immune checkpoint inhibitors in BRAF mutant NSCLC, the median PFS reported in literature for V600E and non V600E are 3.7 and 4.1 months [10], respectively. Mazieres et al. [20] also demonstrated that the PFS was higher in smokers as compared to non-smokers (4.1 vs. 1.9 months). In our report, 2 patients received nivolumab and one received pembrolizumab with a PFS of 2.2, 4.4 and 1.9 months respectively, which concords with reported literature in unselected NSCLC cases [10,20,21]. All these three patients had non-V600E alteration, and were treated with chemotherapy in first line.
These data are however preliminary and require longer follow up for definitive confirmation. The retrospective nature of this study and the biased inclusion criteria of those who underwent mutation testing may have resulted in discordant results from that already reported in literature. Our data is however is the 1st report on BRAF altered NSCLC from India and is unique in the fact that there was higher prevalence on non-V600E mutations, along with more BRAF alterations in never smokers. This highlights the fact that upfront panel based testing as also reported by Pennel et al. [22] is a boon and may soon become a recommendation in order to guide appropriate therapeutic strategies.
In our view, BRAF mutated NSCLC is a molecularly distinct entity, with distinct clinical features and prognosis. Advent of newer therapeutic modalities and the new NCCN guidelines mandates the testing for the V600E mutation in NSCLC, although the non V600E group also constitutes a definite proportion with specific characteristics. Future endeavors to these targets these may benefit this sub populations. Owing to the rarity of the occurrence, in our opinion more real-world studies incorporating data from multiple centres, as well as larger controlled trials are needed for the better understanding of the disease in question. Our data is from a single tertiary Indian cancer centre, which although limited by its sample size, clearly depicts the clinicopathological features and prognostic differences between different types of BRAF mutated NSCLC.
Disclosure of conflict of interest
None.
References
- 1.Mazieres J, Cropet C, Montané L, Barlesi F, Souquet PJ, Quantin X, Dubos-Arvis C, Otto J, Favier L, Avrillon V, Cadranel J, Moro-Sibilot D, Monnet I, Westeel V, Le Treut J, Brain E, Trédaniel J, Jaffro M, Collot S, Ferretti GR, Tiffon C, Mahier-Ait Oukhatar C, Blay JY. Vemurafenib in non-small-cell lung cancer patients with BRAF V600 and BRAF nonV600 mutations. Ann Oncol. 2020;31:289–294. doi: 10.1016/j.annonc.2019.10.022. [DOI] [PubMed] [Google Scholar]
- 2.O’Leary CG, Andelkovic V, Ladwa R, Pavlakis N, Zhou C, Hirsch F, Richard D, O’Byrne K. Targeting BRAF mutations in non-small cell lung cancer. Transl Lung Cancer Res. 2019;8:1119–1124. doi: 10.21037/tlcr.2019.10.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 4.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MH, Chang MT, Chandarlapaty S, Traina TA, Paik PK, Ho AL, Hantash FM, Grupe A, Baxi SS, Callahan MK, Snyder A, Chi P, Danila D, Gounder M, Harding JJ, Hellmann MD, Iyer G, Janjigian Y, Kaley T, Levine DA, Lowery M, Omuro A, Postow MA, Rathkopf D, Shoushtari AN, Shukla N, Voss M, Paraiso E, Zehir A, Berger MF, Taylor BS, Saltz LB, Riely GJ, Ladanyi M, Hyman DM, Baselga J, Sabbatini P, Solit DB, Schultz N. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;2017:PO.17.00011. doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Comprehensive Cancer Network. Non-small cell lung cancer. 2020 [Google Scholar]
- 6.Luk PP, Yu B, Ng CC, Mercorella B, Selinger C, Lum T, Kao S, O’Toole SA, Cooper WA. BRAF mutations in non-small cell lung cancer. Transl Lung Cancer Res. 2015;4:142–148. doi: 10.3978/j.issn.2218-6751.2014.08.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hofman V, Benzaquen J, Heeke S, Lassalle S, Poudenx M, Long E, Lantéri E, Bordone O, Lespinet V, Tanga V, Bonnetaud C, Bille Y, Ilié M, Marquette C, Barlesi F, Boutros J, Hofman P. Real-world assessment of the BRAF status in non-squamous cell lung carcinoma using VE1 immunohistochemistry: a single laboratory experience. Lung Cancer. 2020;145:58–62. doi: 10.1016/j.lungcan.2020.04.024. [DOI] [PubMed] [Google Scholar]
- 8.Barlesi F, Mazieres J, Merlio JP, Debieuvre D, Mosser J, Lena H, Ouafik L, Besse B, Rouquette I, Westeel V, Escande F, Monnet I, Lemoine A, Veillon R, Blons H, Audigier-Valette C, Bringuier PP, Lamy R, Beau-Faller M, Pujol JL, Sabourin JC, Penault-Llorca F, Denis MG, Lantuejoul S, Morin F, Tran Q, Missy P, Langlais A, Milleron B, Cadranel J, Soria JC, Zalcman G Biomarkers France contributors. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT) Lancet. 2016;387:1415–1426. doi: 10.1016/S0140-6736(16)00004-0. [DOI] [PubMed] [Google Scholar]
- 9.Litvak AM, Paik PK, Woo KM, Sima CS, Hellmann MD, Arcila ME, Ladanyi M, Rudin CM, Kris MG, Riely GJ. Clinical characteristics and course of 63 patients with BRAF mutant lung cancers. J Thorac Oncol. 2014;9:1669–1674. doi: 10.1097/JTO.0000000000000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mu Y, Yang K, Hao X, Wang Y, Wang L, Liu Y, Lin L, Li J, Xing P. Clinical characteristics and treatment outcomes of 65 patients with BRAF-mutated non-small cell lung cancer. Front Oncol. 2020;10:603. doi: 10.3389/fonc.2020.00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding X, Zhang Z, Jiang T, Li X, Zhao C, Su B, Zhou C. Clinicopathologic characteristics and outcomes of Chinese patients with non- small- cell lung cancer and BRAF mutation. Cancer Med. 2017;6:555–562. doi: 10.1002/cam4.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwartz LH, Litière S, de Vries E, Ford R, Gwyther S, Mandrekar S, Shankar L, Bogaerts J, Chen A, Dancey J, Hayes W, Hodi FS, Hoekstra OS, Huang EP, Lin N, Liu Y, Therasse P, Wolchok JD, Seymour L. RECIST 1.1-update and clarification: from the RECIST committee. Eur J Cancer. 2017;62:132–137. doi: 10.1016/j.ejca.2016.03.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paik PK, Arcila ME, Fara M, Sima CS, Miller VA, Kris MG, Ladanyi M, Riely GJ. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J. Clin. Oncol. 2011;29:2046–51. doi: 10.1200/JCO.2010.33.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malapelle U, Rossi G, Pisapia P, Barberis M, Buttitta F, Castiglione F, Cecere FL, Grimaldi AM, Iaccarino A, Marchetti A, Massi D, Medicina D, Mele F, Minari R, Orlando E, Pagni F, Palmieri G, Righi L, Russo A, Tommasi S, Vermi W, Troncone G. BRAF as a positive predictive biomarker: focus on lung cancer and melanoma patients. Crit Rev Oncol Hematol. 2020;156:103118. doi: 10.1016/j.critrevonc.2020.103118. [DOI] [PubMed] [Google Scholar]
- 16.Pisapia P, Pepe F, Malapelle U, Troncone G. BRAF mutations in lung cancer. Acta Cytol. 2019;63:247–250. doi: 10.1159/000496478. [DOI] [PubMed] [Google Scholar]
- 17.Noeparast A, Teugels E, Giron P, Verschelden G, De Brakeleer S, Decoster L, De Grève J. Non-V600 BRAF mutations recurrently found in lung cancer predict sensitivity to the combination of trametinib and dabrafenib. Oncotarget. 2017;8:60094–60108. doi: 10.18632/oncotarget.11635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchetti A, Felicioni L, Malatesta S, Grazia Sciarrotta M, Guetti L, Chella A, Viola P, Pullara C, Mucilli F, Buttitta F. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J. Clin. Oncol. 2021;29:3574–9. doi: 10.1200/JCO.2011.35.9638. [DOI] [PubMed] [Google Scholar]
- 19.Nakanishi Y, Nakagawa Y, Tsujino I, Shimizu T, Takahashi N, Hashimoto S, Masuda S. Favorable outcome with pemetrexed treatment for advanced BRAF-V600E-positive lung adenocarcinoma in a patient followed up over 8 years. J Thorac Oncol. 2018;13:e199–e202. doi: 10.1016/j.jtho.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 20.Mazieres J, Drilon A, Lusque A, Mhanna L, Cortot AB, Mezquita L, Thai AA, Mascaux C, Couraud S, Veillon R, Van den Heuvel M, Neal J, Peled N, Früh M, Ng TL, Gounant V, Popat S, Diebold J, Sabari J, Zhu VW, Rothschild SI, Bironzo P, Martinez-Marti A, Curioni-Fontecedro A, Rosell R, Lattuca-Truc M, Wiesweg M, Besse B, Solomon B, Barlesi F, Schouten RD, Wakelee H, Camidge DR, Zalcman G, Novello S, Ou SI, Milia J, Gautschi O. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann Oncol. 2019;30:1321–1328. doi: 10.1093/annonc/mdz167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herbst RS, Baas P, Kim DW, Felip E, Pérez-Gracia JL, Han JY, Molina J, Kim JH, Arvis CD, Ahn MJ, Majem M, Fidler MJ, de Castro G Jr, Garrido M, Lubiniecki GM, Shentu Y, Im E, Dolled-Filhart M, Garon EB. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet. 2016;387:1540–1550. doi: 10.1016/S0140-6736(15)01281-7. [DOI] [PubMed] [Google Scholar]
- 22.Pennell NA, Mutebi A, Zhou ZY, Ricculli ML, Tang W, Wang H, Guerin A, Arnhart T, Dalal A, Sasane M, Wu KY, Culver KW, Otterson GA. Economic impact of next-generation sequencing versus single-gene testing to detect genomic alterations in metastatic non-small-cell lung cancer using a decision analytic model. JCO Precis Oncol. 2019;3:1–9. doi: 10.1200/PO.18.00356. [DOI] [PubMed] [Google Scholar]


