Abstract
Current serotyping methods classify Pasteurella multocida into five capsular serogroups (serogroups A, B, D, E, and F) and 16 somatic serotypes (serotypes 1 to 16). In the present study, we have developed a multiplex PCR assay as a rapid alternative to the conventional capsular serotyping system. The serogroup-specific primers used in this assay were designed following identification, sequence determination, and analysis of the capsular biosynthetic loci of each capsular serogroup. The entire capsular biosynthetic loci of P. multocida A:1 (X-73) and B:2 (M1404) have been cloned and sequenced previously (J. Y. Chung, Y. M. Zhang, and B. Adler, FEMS Microbiol. Lett. 166:289–296, 1998; J. D. Boyce, J. Y. Chung, and B. Adler, Vet. Microbiol. 72:121–134, 2000). Nucleotide sequence analysis of the biosynthetic region (region 2) from each of the remaining three serogroups, serogroups D, E, and F, identified serogroup-specific regions and gave an indication of the capsular polysaccharide composition. The multiplex capsular PCR assay was highly specific, and its results, with the exception of those for some serogroup F strains, correlated well with conventional serotyping results. Sequence analysis of the strains that gave conflicting results confirmed the validity of the multiplex PCR and indicated that these strains were in fact capsular serogroup A. The multiplex PCR will clarify the distinction between closely related serogroups A and F and constitutes a rapid assay for the definitive classification of P. multocida capsular types.
Pasteurella multocida is a heterogeneous species that produces septicemic or respiratory diseases in domesticated and wild animals (12). Considerable variation has been observed among strains with respect to host predilection, pathogenicity, carbohydrate fermentation, colonial morphology, and antigenic specificity (6). Serological differences were also observed in the type-specific capsular antigen of P. multocida identified by Carter (4). This resulted in the development of the indirect hemagglutination test, which now recognizes five distinct capsular serogroups (serogroups A, B, D, E, and F) (12).
Until recently, little was known of the composition of capsular material from P. multocida serogroups other than that of serogroup A, which is known to be sensitive to the action of hyaluronidase (3, 5). Nuclear magnetic resonance studies confirmed that the major polysaccharide component of the capsule was hyaluronic acid (13). The capsular material of serogroups D and F has been identified primarily through the action of mucopolysaccharidases (11). On the basis of the decapsulation profiles of P. multocida by these enzymes, it was proposed that serogroups D and F produced capsular material that contained heparin and chondroitin sulfate, respectively (11). The production of a chondroitin or chondroitin-like polysaccharide capsule by P. multocida serogroup F was recently confirmed by carbohydrate analysis (9). The monosaccharide analysis of a serogroup B P. multocida strain determined that the purified capsular polysaccharide was composed of arabinose, mannose, and galactose in a ratio of 0.5:2.0:0.8 (N. Muniandy, J. Edgar, J. B. Woolcock, and T. K. S. Mukkur, Int. Workshop Pasteurellosis Prod. Anim., 1992). The chemical composition of the serogroup E capsule remains unknown.
The biosynthetic loci of the complete capsules of P. multocida serotypes A:1 (7) and B:2 (1) have recently been identified, with gene identification within the serogroup-specific region 2 of both loci supporting previous data regarding capsule composition. Two genes within this region of the B:2 locus, bcbA and bcbB, were similar to Escherichia coli wecB and wecC, which catalyze the conversion of UDP-N-acetylglucosamine to N-acetyl-d-mannosaminuronic acid (1). The presence of these homologs in the serogroup B cap locus suggests that the mannose identified as the major type B capsular component by Muniandy et al. (Int. Workshop Pasteurellosis Prod. Anim.) exists as N-acetyl-d-mannosaminuronic acid in situ (1). Region 2 has also been partially cloned and sequenced from serogroup F, with the identification of a glycosyltransferase involved in elongation of chondroitin polymers (9). A detailed review of the composition, function, and genetics of the P. multocida A:1 and B:2 capsules has recently been published (2).
The lack of genetic knowledge regarding the capsular material of serogroups D, E, and F and the increasing need for a simple, DNA-based capsular typing method provided the impetus for the investigation described here. Oligonucleotide primers designed during sequencing of the biosynthetic loci of the capsules of serogroups A and B (1, 7) were used to determine the nucleotide sequences of the region 2 genes from the remaining three capsular serogroups (serogroups D, E, and F). Serogroup-specific sequences were then identified for use as primers in a multiplex PCR assay. This assay represents a rapid and reproducible alternative to the serological and nonserological methods currently used for the classification of P. multocida capsular types.
MATERIALS AND METHODS
Bacteria.
The P. multocida strains used in this study are described in Table 1. The P. multocida isolates from the Veterinary Pathology Laboratory were serotyped by the Veterinary Research Institute, Peradeniya, Sri Lanka. All other P. multocida serotype designations were determined by the National Animal Disease Center (NADC), Ames, Iowa (R. B. Rimler and K. A. Brogden, personal communication), or from previous publications (6, 10, 15).
TABLE 1.
History and origin of P. multocida strains used to determine specificities of the primers in the serogroup-specific PCR assays
| Strain (serogroup) | History, origin | PCR result with the following primer sets
|
||||
|---|---|---|---|---|---|---|
| CAPA | CAPB | CAPD | CAPE | CAPF | ||
| X-73 (A:1)a | Heddleston type 1 reference, chicken | + | − | − | − | − |
| P1201 (A)a | Carter type A reference, fowl | + | − | − | − | − |
| P5228 (A:1)b | United States, duck | + | − | − | − | − |
| P5229 (A:1)b | United states, duck | + | − | − | − | − |
| P5338 (A:1)b | United states, turkey | + | − | − | − | − |
| P5339 (A:1)b | United states, turkey | + | − | − | − | − |
| VP161 (A:1)c | Vietnam, chicken | + | − | − | − | − |
| VP21 (A:3)c | Australia, chicken | + | − | − | − | − |
| VP17 (A:4)c | Australia, chicken | + | − | − | − | − |
| M1404 (B:2)a | Heddleston type 2 reference, buffalo | − | + | − | − | − |
| P932 (B)a | Carter type B reference, buffalo | − | + | − | − | − |
| VP247 (B:2)c | Vietnam, pig | − | + | − | − | − |
| 0140 (B:3,4)d | Australia cattle | − | + | − | − | − |
| P1511 (B:1)a | United States, antelope | − | + | − | − | − |
| P5226 (B:4)a | Canada, bison | − | + | − | − | − |
| P5325 (B:3,4)a | United states, elk | − | + | − | − | − |
| P934 (D)ae | Carter type D reference, pig | − | − | + | − | − |
| P903 (D:11)a | Heddleston type 11 reference, pig | − | − | + | − | − |
| VP96 (D:1)c | Australia, pig | − | − | + | − | − |
| VP366 (D:3,4)c | Vietnam, pig | − | − | + | − | − |
| VP381 (D:1)c | Vietnam, pig | − | − | + | − | − |
| 0148 (E)d | South Africa, cattle | − | − | − | + | − |
| 0149 (E)d | Africa, cattle | − | − | − | + | − |
| 0150 (E)d | Africa, cattle | − | − | − | + | − |
| P1234(E)ae | Carter type E reference, cattle | − | − | − | + | − |
| P1235 (E)a | Africa, Carter type E | − | − | − | + | − |
| P4218 (F:3)ae | United states, turkey | − | − | − | − | + |
| P4988 (F:1)b | United States, deer | + | − | − | − | − |
| P5239 (F:1)b | United States, swine | + | − | − | − | − |
| P5184 (F:3,4)b | United States, sheep | + | − | − | − | − |
| P5084 (F:14, 15,12)b | United States, turkey | − | − | − | − | + |
Obtained from K. R. Rhoades and R. B. Rimler, NADC.
Obtained from K. A. Brogden, NADC.
Isolates held in the Veterinary Pathology Culture Collection, University of Queensland.
Originally obtained from Regional Veterinary Laboratory, Benalla, Victoria, Australia.
Strains were used to sequence the region 2 genes of the cap loci from serogroups D, E, and F.
PCR primers.
All primers used in the study either were synthesized by Life Technologies (Gaithersburg, Md.) or Genset Pacific (Lismore, New South Wales, Australia) or were previously synthesized for sequence determination of the P. multocida A:1 and B:2 cap loci (1, 7). The primer sequences used in the multiplex P. multocida capsular typing PCR assay are listed in Table 2.
TABLE 2.
Sequences of the oligonucleotides used in the P. multocida multiplex capsular PCR typing assay
| Serogroup | Gene | Name | Sequence (5′→3′) | Amplimer size (bp) | Position |
|---|---|---|---|---|---|
| All | KMT1 | KMT1T7 | ATCCGCTATTTACCCAGTGG | 460 | |
| KMT1SP6 | GCTGTAAACGAACTCGCCAC | ||||
| A | hyaD-hyaC | CAPA-FWD | TGCCAAAATCGCAGTCAG | 1,044 | 8846–8863a |
| CAPA-REV | TTGCCATCATTGTCAGTG | 9890–9873 | |||
| B | bcbD | CAPB-FWD | CATTTATCCAAGCTCCACC | 760 | 13621–13603b |
| CAPB-REV | GCCCGAGAGTTTCAATCC | 12863–12880 | |||
| D | dcbF | CAPD-FWD | TTACAAAAGAAAGACTAGGAGCCC | 657 | 3142–3165c |
| CAPD-REV | CATCTACCCACTCAACCATATCAG | 3789–3766 | |||
| E | ecbJ | CAPE-FWD | TCCGCAGAAAATTATTGACTC | 511 | 4387–4408d |
| CAPE-REV | GCTTGCTGCTTGATTTTGTC | 4899–4881 | |||
| F | fcbD | CAPF-FWD | AATCGGAGAACGCAGAAATCAG | 851 | 2881–2896e |
| CAPF-REV | TTCCGCCGTCAATTACTCTG | 3733–3714 |
These sequences match the coding strand of the serogroup A cap gene with GenBank accession number AF067175.
These sequences match the coding strand of the serogroup B cap gene with GenBank accession number AF169324.
These sequences match the coding strand of the serogroup D cap gene with GenBank accession number AF302465.
These sequences match the coding strand of the serogroup E cap gene with GenBank accession number AF302466.
These sequences match the coding strand of the serogroup F cap gene with GenBank accession number AF302467.
PCR conditions.
Initial PCR amplification was carried out with single primer sets from within the region 2 genes of the serogroup A and B capsule biosynthetic loci. For ease and rapidity, whole cells obtained from single colonies grown on 8% sheep blood agar were used as the template in the amplification reactions. A pipette tip was lightly touched onto a bacterial colony, and the colony was resuspended in the PCR amplification mixture (25 μl) containing each primer at a concentration of 3.2 μM, each deoxynucleoside triphosphate at a concentration of 200 μM, 1× PCR buffer, 2 mM MgCl2, and 0.5 U of Taq DNA polymerase (Gibco BRL Life Technologies). All amplifications were performed with the GeneAmp PCR system 2400 (Perkin-Elmer, Branchburg, N.J.). For products with an expected size of ≤1 kb, the following standard cycling procedure was used: an initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 30 s, and a final extension at 72°C for 5 min. The same cycling procedure was used for products of an expected size between 1 and 3 kb, except that the extension time was increased by 30 s for each additional 1 kb.
Long-template (≥3 kb) amplification used for sequence analysis of the region 2 genes from serogroups D, E, and F was performed with the eLONGase Enzyme Mix (Gibco BRL Life Technologies).
Restriction endonuclease analysis of regions demonstrating similarity between serogroups.
PCR-restriction fragment length polymorphism analysis was performed on regions 1 and 3 of the cap loci from reference type strains of all five capsular serogroups. These regions were amplified with the eLONGase Enzyme Mix (Gibco BRL Life Technologies) and oligonucleotide primers from serogroup A and B cap loci. The PCR products were then purified with the QIAquick PCR purification kit (QIAGEN Pty. Ltd., Clifton Hill, Victoria, Australia) and digested with a restriction enzyme, BamHI, EcoRI, EcoRV, HhaI, HindIII, PstI, or Sau96-I. Digestion products were visualized following agarose gel electrophoresis.
Determination of nucleotide sequences of region 2 genes of serogroups D, E, and F.
Strains P934 (serogroup D), P1234 (serogroup E), and P4218 (serogroup F) were used to determine the nucleotide sequences of the region 2 genes from the cap loci of serogroups D, E, and F. Following long-template amplification, the PCR products were purified with QIAquick PCR purification kit (QIAGEN) and cloned into the pGEM-T Easy Vector (Promega, Madison, Wis.) for sequence analysis. Nucleotide sequences were determined with the BigDye Ready Reaction DyeDeoxy Terminator Cycle Sequencing Kit (Perkin-Elmer, Foster City, Calif.), and the reactions were analyzed with a 373A DNA sequencing system at the Australian Genome Research Facility, Brisbane, Queensland, Australia.
Development of a multiplex PCR assay for capsular serogroup identification.
Following sequence determination and analysis of P. multocida capsular serogroup-specific regions, a multiplex PCR assay was developed that contained P. multocida-specific primers (14) and primer sets specific for serogroups A, B, D, E, and F. The serogroup-specific primer sets were identified according to the following criteria: (i) primer sets were to be located within genes established as unique for each of the five serogroups (hyaD, bcbD, dcbF, ecbJ, and fcbD), and (ii) amplimer length was sufficient to allow clear size discrimination. The multiplex PCR mixture contained each primer within the six primer sets at a concentration of 3.2 μM, 1 U of Taq DNA polymerase, 2 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 200 μM, and 1× PCR buffer. A standard cycling program was used, as described above. The amplified products were separated by electrophoresis in 2% agarose gels and visualized by ethidium bromide staining.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the region 2 cap loci sequences are as follows: serogroup D (strain P934), AF302465; serogroup E (strain P1234), AF302466; and serogroup F (strain P4218), AF302467.
RESULTS
Genetic organization of P. multocida capsular biosynthetic loci.
The genetic organization of cap loci from P. multocida serogroups A:1 and B:2 has been established previously. We have found a high level of similarity between serogroups A, D, and F, with positive reactions obtained with most primers throughout the P. multocida A:1 cap locus, while no amplimers were produced from serogroups B and E (data not shown). Primers specific for hyaC-hyaE (region 2) showed some variation in amplification of serogroups D and F, while PCR-restriction fragment length polymorphism analysis of the region 1 genes from serogroups A, D, and F demonstrated identical restriction profiles with BamHI, EcoRI, EcoRV, HhaI, HindIII, PstI, and Sau96-I (data not shown).
The majority of primers from the P. multocida B:2 cap locus also amplified DNA from serogroup E but not DNA from serogroup A, D, or F (data not shown). The amplification profiles also showed that while homologs of bcbI, bcbB, and bcbA were present in the serogroup E cap locus, a lower degree of similarity was observed in the intervening genes, with no amplimers demonstrated when primers specific for bcbCDEFGH were used.
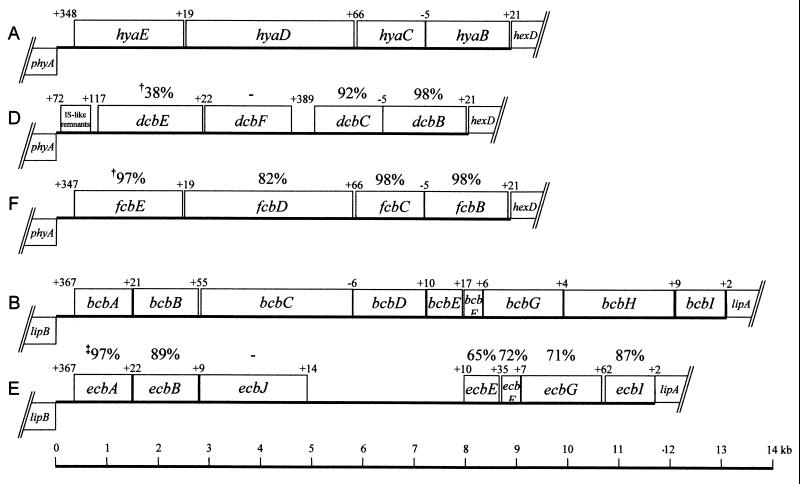
Amplification of DNA across the cap regions indicated that P. multocida serogroups A, D, and F possess similar organizations, comprising region 1 (capsule export; hexDCBA homologs), region 2 (capsule biosynthesis), and region 3 (phospholipid substitution; lipA and lipB homologs) (Fig. 1). As shown previously, P. multocida serogroup B:2 demonstrates some reassortment of region 1 and 3 genes, with lipA found to be cotranscribed with cexDCBA. Initial amplification studies and sequence determination showed a similar organization within the serogroup E cap locus (Fig. 1). The >99% identity of sequences flanking the cap loci of serogroups A and B was also demonstrated in the other three serogroups, suggesting a common chromosomal location for the cap loci of all serogroups.
FIG. 1.
Genetic organization of region 2 of the capsule biosynthetic loci of representative isolates of the five P. multocida capsular serogroups (serogroups A, B, D, E, and F). The cap loci are shown from the following strains: P. multocida A:1 strain X-73 (GenBank accession number AF067175), B:2 strain M1404 (GenBank accession number AF169324), serogroup D strain P934 (GenBank accession number AF302465), serogroup E strain P1234 (GenBank accession number AF302466), and F:3 strain P4218 (GenBank accession number AF302467). Numbers above the boxes indicates the distance (in base pairs) between the last base of the preceeding gene and the first base of the next gene. Genes depicted by boxes above the line are transcribed in the left-to-right direction, while those beneath the line are transcribed in the right-to-left direction. †, percent identity at amino acid level to A:1 capsule biosynthetic locus; ‡, percent identity at amino acid level to B:2 capsule biosynthetic locus.
Analysis of region 2 genes of serogroups D, E, and F.
In order to define further the genetic organization of the P. multocida cap loci, the region 2 genes from capsular serogroups D, E, and F were cloned and sequenced. Predicted genes from these regions were designated dcb, ecb, and fcb for serogroup D, E, and F capsule biosynthesis, respectively. Whenever possible, the letter designation was maintained between serogroup homologs. Open reading frame (ORF) sizes and homolog similarities between serogroups A, D, and F are shown in Table 3, with the similarities between the region 2 genes of serogroups B and E listed in Table 4.
TABLE 3.
Identity (similarity) between products of the region 2 capsule biosynthetic regions of P. multocida capsular serogroups A, D, and F
| Strain (serogroup) | Protein name, protein length (amino acids); % identity (similarity) to X-73 (A:1) homolog | |||
|---|---|---|---|---|
| X-73 (A:1) | HyaB, 477 | HyaC, 390 | HyaD, 972 | HyaE, 622 |
| P934 (D) | DcbB, 477; 98 (98)a | DcbC, 390; 92 (96) | DcbFb | DcbE, 603; 38 (56) |
| P4218 (F:3) | FcbB, 477; 98 (99)a | FcbC, 390; 98 (98) | FcbD, 965; 82 (90) | FcbE, 622; 97 (97) |
Percent identity (similarity) to serogroup A X-73 homolog.
This protein has <20% identity to X-73 homolog.
TABLE 4.
Identity (similarity) between products of the region 2 capsule biosynthetic regions of P. multocida serogroups B and E
| Strain (serogroup) | Protein name, protein length (amino acids); % identity (similarity) to M1404 (B:2) homolog | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| M1404 (B:2) | BcbI, 326 | BcbH, 721 | BcbG, 521 | BcbF, 124 | BcbE, 236 | BcbD, 475 | BcbC, 1,033 | BcbB, 422 | BcbA, 374 |
| P1234 (E) | EcbI, 326; 87a (91) | N/A | EcbG, 527; 71 (80) | EcbF, 126; 72 (89) | EcbE, 228; 65 (86) | EcbD, 417; 23 (44) | EcbJ/Kb | EcbB, 424; 89 (92) | EcbA, 374; 97 (98) |
Percent identity (similarity) to serogroup B M1404 homolog.
This proteiin has <20% identity to M1404 homolog.
Region 2 of serogroup D was shown to contain four genes, dcbEFCB, in the same orientation as hyaEDCB of A:1 (Fig. 1). DcbB and DcbC are homologs of HyaB and HyaC, respectively, with 98 and 92% similarities, respectively (Table 3). The 501-amino-acid product of dcbF displayed no similarity to HyaD of P. multocida A:1 but showed some similarity to bacterial glycosyltransferases (Table 5). In particular, DcbF has 54% similarity to the putative glycosyltransferase KfiC from the Escherichia coli K5 capsule biosynthetic locus. The deduced product of dcbE demonstrated 56% similarity to HyaE, although DcbE (603 amino acids) is slightly shorter than HyaE (622 amino acids) (Table 3).
TABLE 5.
Predicted functions for region 2 genes of serogroups D, E, and F
| Gene product | Size (aa)a | Homolog | % Identity (similarity)b | Predicted function |
|---|---|---|---|---|
| dcbB | 477 | hyaB | 98 (98) | Glycosyltransferase |
| dcbC | 390 | hyaC | 92 (96) | UDP-glucose dehydrogenase |
| dcbF | 501 (−19) | kfiC | 34 (54) | Glycosyltransferase |
| dcbE | 603 (−19) | hyaE | 36 (56) | Unknown function, KfiB homolog |
| fcbB | 477 | hyaB | 98 (99) | Glycosyltransferase |
| fcbC | 390 | hyaC | 98 (98) | UDP-glucose dehydrogenase |
| fcbD | 965 (−7) | pmCS | 82 (90) | Glycosyltransferase |
| fcbE | 622 | hyaE | 97 (97) | Unknown function, KfiB homolog |
| ecbA | 374 | bcbA (wecA) | 97 (98) | UDP-N-acetylglucosamine 2-epimerase |
| ecbB | 424 (+2) | bcbB (wecB) | 89 (92) | UDP-N-acetyl-d-mannoasminuronic acid dehydrogenase |
| ecbJ | 705 | Glycosyl transferase (3′), unknown (5′) | ||
| ecbK | 400 | 3′ bcbCc | 38 (55) | Unknown |
| ecbD | 417 (−67) | bcbD | 23 (44) | Unknown |
| ecbE | 228 (−8) | bcbE | 65 (86) | Unknown |
| ecbF | 126 (+2) | bcbF | 72 (89) | Unknown |
| ecbG | 527 (+6) | bcbG | 71 (80) | Unknown |
| ecbI | 326 | bcbI | 87 (91) | Unknown |
Predicted length of deduced protein (in amino acids [aa]), with differences in size from that of the homolog given in parentheses.
Percentage identity (similarity) of protein products.
The product is 392 amino acids.
The capsule biosynthesis genes of serogroup F showed significant similarity to those of P. multocida A:1, with the products of fcbB, fcbC, and fcbE exhibiting identical lengths and >97% similarities to their A:1 homologs (Table 3). The deduced product of fcbD was determined to have 90% similarity to HyaD (A:1) and pmHAS (8). In addition, FcbD had 97% similarity to the chondroitin synthase (pmCS) recently identified from the serogroup F P. multocida strain P4679 (9).
The region 2 genes of the serogroup E cap locus demonstrated a high level of similarity to those of serogroup B. The deduced products of ecbI, ecbB, and ecbA were similar in length and sequence (>92% identity) to their group B homologs (Table 4). Interestingly, there was no homolog of bcbH within the serogroup E cap locus, in which ecbI is flanked by lipA and ecbG. The sequence downstream of ecbB also differed from its serogroup B homolog. Two ORFs, ecbJ and ecbK, were identified that demonstrated little or no similarity to bcbC (Table 5). The C-terminal amino acids of EcbJ showed low levels of similarity to bacterial glycosyltransferases but no identity to any proteins encoded by the serogroup B cap locus, while the C terminus of EcbK was 55% similarity to BcbC. The remaining four ORFs within region 2 of the serogroup E cap locus (ecbDEFG) were shown to possess between 44 and 89% similarities to their serogroup B homologs, bcbDEFG (Table 4).
Development of a multiplex capsular PCR typing assay for P. multocida.
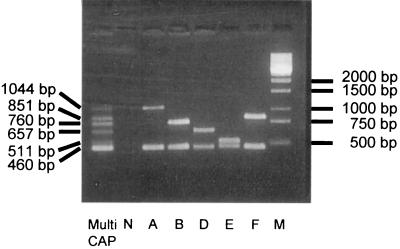
It was evident from the comparative analyses that sequences within hyaD, bcbD, dcbF, ecbJ, and fcbD were highly specific for their respective serogroups. As serogroups D and F possessed homologs of hyaB, hyaC, and, to some extent, hyaE, glycosyltransferase genes hyaD, dcbF, and fcbD represented potential serogroup-specific target sequences. The majority of the region 2 genes of serogroup E were similar to those of serogroup B. However, the sequence of ecbJ showed little similarity to sequences in GenBank and therefore represented an ideal candidate for the identification of serogroup-specific sequences. Primer sequences were then designed to generate PCR products of unique size. P. multocida-specific primers were also included as an internal control to confirm the species identification. The resultant multiplex PCR was shown to be highly specific among the reference capsular type strains, with only the P. multocida-specific product and the respective capsular serogroup-specific product being amplified (Fig. 2). The multiplex PCR was also highly specific among the majority of field isolates. However, there was some disparity between PCR type and capsular serotype within the serogroup F strains. Of the five strains previously identified as serogroup F, only strains P4218 and P5084 produced the expected serogroup F-specific PCR product. The remaining three strains (strains P4988, P5184, and P5239) were typed as serogroup A strains by the multiplex capsular PCR assay (Table 1). However, sequence analysis of the glycosyltransferase genes used for the serogroup A- and F-specific primers indicated that the strains were serogroup A, in agreement with the multiplex PCR result.
FIG. 2.
Multiplex capsular PCR typing system for P. multocida. Whole-cell PCR was performed with type strains of each of the five capsular serogroups. Lane Multi CAP, combined products from multiplex PCR of all five serogroups; lane N, a negative control; lanes A to F, multiplex PCR products from strains X-73 (A:1), M1404 (B:2), P934 (D), P1234 (E), and P4218 (F:3), respectively; lane M, DNA molecular size marker.
DISCUSSION
The identification and sequence analysis of the biosynthetic locus of the capsule of an organism can lead to a greater understanding of its capsular polysaccharide composition and can provide a genetic basis for the serological differences observed between strains. Sequence determination of the biosynthetic locus of the P. multocida serogroup A capsule by Chung et al. (7) identified components responsible for the synthesis of hyaluronic acid, consistent with hyaluronic acid being the principal component of the type A capsule. However, genetic analysis of the serogroup B biosynthetic locus revealed only three gene products with similarity to proteins known to be involved in polysaccharide biosynthesis, while six gene products had no similarity to known proteins (1). The structure of the type B capsule remains unknown. Until recently, very little was known about the compositions of the serogroup D and F capsular polysaccharides, with even less known about the serogroup E capsular polysaccharide. Therefore, determination of the sequences of the region 2 genes of the remaining serogroups was undertaken in order to gain information about the capsules of serogroups D, E, and F. This region was highly conserved between serogroups A, D, and F. Three of the four genes were homologous, with the unique gene for each group encoding a glycosyltransferase. The serogroup A (HyaD) and F (FcbD) genes were shown to encode hyaluronan and chondroitin synthases, respectively (8, 9). In serogroup D, this gene product (DcbF) was similar to the glycosyltransferase KfiC from E. coli K5 that is involved in the formation of a polysaccharide similar to heparin. The genetic similarity between the capsular biosynthetic regions of P. multocida serogroup D and E. coli K5 is consistent with the possibility that the serogroup D capsule contains heparin polymers, a proposal previously based on the decapsulation of type D strains by heparinase III (11).
Determination of the nucleotide sequence of the serogroup E biosynthesis region provided little information about the capsular polysaccharide composition. Region 2 of serogroup E contains nine genes, two of which showed similarity to genes involved in polysaccharide biosynthesis. These two genes have homologs in the P. multocida B:2 cap locus, indicating that N-acetyl-d-mannosaminuronic acid is a component of both the serogroup B and the serogroup E capsules. Of the remaining seven genes, five have homologs in the B:2 cap locus but still have no known function, one encodes a putative glycosyltransferase, and the other is unique to serogroup E. Definitive assignment of function must await further analysis and will be aided by the determination of the structures of the type B and E capsules.
Comparative analysis of the five capsular biosynthetic regions confirmed a genetic basis for the serological differences observed between strains. By using these genetic differences, we have developed a rational, DNA-based typing system for P. multocida. The multiplex capsular PCR assay provides a rapid and highly specific alternative to conventional capsular serotyping. There are currently only two laboratories worldwide that make and maintain the antisera required for capsular typing. The assay described in this report can be performed with suspected P. multocida colonies from primary isolation plates, thus reducing the time required for culture preparation. It is also highly specific for strains genetically capable of producing a serogroup-specific capsule. Notably, the PCR-based system was not affected by the geographical distribution of isolates. For example, isolates classified as serogroup A by conventional serotyping from Australia, Vietnam, and the United States all produced the appropriate amplimer with the serogroup A cap-specific primers. This assay will also help clarify the distinction between strains from closely related serogroups A and F. Indeed, the assay clearly identified three strains as serogroup A that had previously been serotyped erroneously as serogroup F, a finding confirmed by sequence analysis of the cap locus. Serogroups A and F are now known to have hyaluronic acid- and chondroitin-like polysaccharide capsules, respectively. As these are both nonimmunogenic polymers, it is unclear which antigens are responsible for the capsule-specific reactions observed in the indirect hemagglutination test. Despite the lack of knowledge regarding the specific immunogens, capsular serotyping has provided a useful system for P. multocida classification. However, with determination of the sequence of the cap locus in each serogroup, we believe that genetic cap identification by PCR will become a system for the rational and definitive typing of the P. multocida capsule.
ACKNOWLEDGMENTS
We acknowledge Susan Moss for assistance during the sequence determination. We are also grateful to Gwen Nordholm and Kim Brogden (NADC) for providing us with recent A:1 and type F P. multocida isolates.
This work was supported by the Australian Centre for International Agricultural Research, Canberra, Australia.
Footnotes
This paper is dedicated to the memory of our colleague, the late Rick Rimler, whose contribution to P. multocida research is gratefully acknowledged.
REFERENCES
- 1.Boyce J D, Chung J Y, Adler B. Genetic organisation of the capsule biosynthetic locus of Pasteurella multocida B:2. Vet Microbiol. 2000;72:121–134. doi: 10.1016/s0378-1135(99)00193-5. [DOI] [PubMed] [Google Scholar]
- 2.Boyce J D, Chung J Y, Adler B. Pasteurella multocida capsule: composition, function and genetics. J Biotechnol. 2000;83:153–160. doi: 10.1016/s0168-1656(00)00309-6. [DOI] [PubMed] [Google Scholar]
- 3.Carter G R. Some characteristics of type A strains of Pasteurella multocida. Br Vet J. 1958;114:356–357. [Google Scholar]
- 4.Carter G R. The type specific capsular antigen of Pasteurella multocida. Can J Med Sci. 1952;30:48–53. doi: 10.1139/cjms52-008. [DOI] [PubMed] [Google Scholar]
- 5.Carter G R, Annau E. Isolation of capsular polysaccharides for colonial variants of Pasteurella multocida. Am J Vet Res. 1953;14:475–478. [PubMed] [Google Scholar]
- 6.Carter G R, Chengappa M M. Recommendations for a standard system of designating serotypes of Pasteurella multocida. Am Assoc Vet Lab Diagn. 1981;24:37–42. [Google Scholar]
- 7.Chung J Y, Zhang Y M, Adler B. The capsule biosynthetic locus of Pasteurella multocida A-1. FEMS Microbiol Lett. 1998;166:289–296. doi: 10.1111/j.1574-6968.1998.tb13903.x. [DOI] [PubMed] [Google Scholar]
- 8.DeAngelis P L, Jing W, Drake R R, Achyuthan A M. Identification and molecular cloning of a unique hyaluronan synthase from Pasteurella multocida. J Biol Chem. 1998;273:8454–8458. doi: 10.1074/jbc.273.14.8454. [DOI] [PubMed] [Google Scholar]
- 9.DeAngelis P L, Padgett-McCue A J. Identification and molecular cloning of a chondroitin synthase from Pasteurella multocida type F. J Biol Chem. 2000;275:24124–24129. doi: 10.1074/jbc.M003385200. [DOI] [PubMed] [Google Scholar]
- 10.Johnson R B, Dawkins H J S, Spencer T L. Electrophoretic profiles of Pasteurella multocida isolates from animals with haemorrhagic septicaemia. Am J Vet Res. 1991;52:1644–1648. [PubMed] [Google Scholar]
- 11.Rimler R B. Presumptive identification of Pasteurella multocida serogroups A, D and F by capsule depolymerization with mucopolysaccharidases. Vet Rec. 1994;134:191–192. doi: 10.1136/vr.134.8.191. [DOI] [PubMed] [Google Scholar]
- 12.Rimler R B, Rhoades K R. Pasteurella multocida. In: Adlam C, Rutter J M, editors. Pasteurella and pasteurellosis. London, United Kingdom: Academic Press Limited; 1989. pp. 37–73. [Google Scholar]
- 13.Rosner H, Grimmecke H D, Knirel Y A, Shashkov A S. Hyaluronic acid and a (1→4)-beta-d-xylan, extracellular polysaccharides of Pasteurella multocida (Carter type A) strain 880. Carbohydr Res. 1992;223:329–333. doi: 10.1016/0008-6215(92)80032-v. [DOI] [PubMed] [Google Scholar]
- 14.Townsend K M, Frost A J, Lee C W, Papadimitriou J M, Dawkins H J. Development of PCR assays for species-and type-specific identification of Pasteurella multocida isolates. J Clin Microbiol. 1998;36:1096–1100. doi: 10.1128/jcm.36.4.1096-1100.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson M A, Rimler R B, Hoffman L J. Comparison of DNA fingerprints and somatic serotypes of serogroup B and E Pasteurella multocida isolates. J Clin Microbiol. 1992;30:1518–1524. doi: 10.1128/jcm.30.6.1518-1524.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]