Abstract
The mammalian airways are lined by a continuous epithelial layer that is maintained by diverse populations of resident multipotent stem cells. These stem cells are responsible for replenishing the epithelium both at homeostasis and following injury, making them promising targets for stem cell and genetic-based therapies for a variety of respiratory diseases. However, the mechanisms that regulate when and how these stem cells proliferate, migrate, and differentiate remains incompletely understood. Here, we find that the high mobility group (HMG) domain transcription factor Lef-1 regulates proliferation and differentiation of murine tracheal basal cells. We demonstrate that conditional deletion of Lef-1 stalls basal cell proliferation at the G1/S transition of the cell cycle, and that Lef-1 knockout cells are unable to maintain luminal tracheal cell types in long-term air-liquid interface culture. RNA sequencing analysis revealed that Lef-1 knockout (Lef-1KO) results in downregulation of key DNA damage response and cell cycle progression genes, including the kinase Chek1. Furthermore, chemical inhibition of Chek1 is sufficient to stall basal cell self-renewal in a similar fashion as Lef-1 deletion. Notably, the cell cycle block imposed by Lef-1KO in vitro is transient and basal cells eventually compensate to proliferate normally in a Chek1-independent manner. Finally, Lef-1KO cells were unable to fully regenerate tracheal epithelium following injury in vivo. These findings reveal that Lef-1 is essential for proper basal cell function. Thus, modulating Lef-1 function in airway basal cells may have applications in regenerative medicine.
Keywords: Lef-1, Stem Cells, Proliferation, Cell Cycle, Differentiation, Self-Renewal, DNA Damage Response
Graphical Abstract
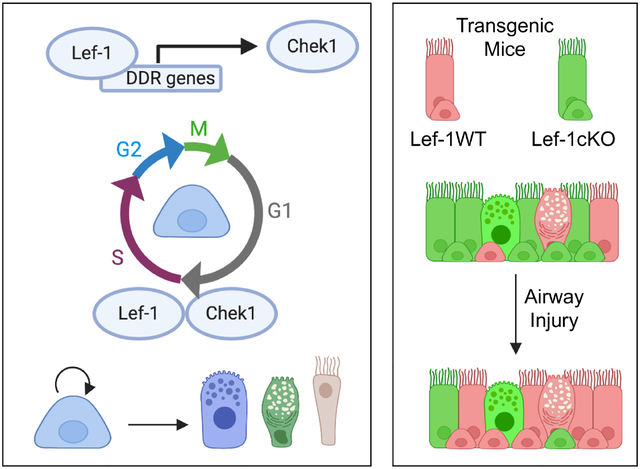
Airway basal stem cells utilize Lef-1 to regulate the expression of certain DNA damage response genes, including Chek1, and facilitate proper G1/S checkpoint cell cycle progression required for self-renewal and differentiation (left panel). Mouse tracheal epithelial cells conditionally deleted for Lef-1 (Lef-1cKO) have a selective disadvantage, compared to wild type Lef-1 (Lef-1WT) cells, to regenerate an injured epithelium.
1 |. INTRODUCTION
Resident stem cells (SCs) are responsible for the maintenance and regeneration of the tissues they reside within, and are thus promising targets for regenerative medicine. However, the mechanisms that govern SC self-renewal, survival, and differentiation remain incompletely understood. SCs often reside within a specialized microenvironment that regulates SC function1. This SC niche is composed of the surrounding mesenchyme, nerves, vasculature, and other non-SC cell types that all work in concert to control SC function by transmitting specific signals to the SCs2, 3. However, the identity of these signals, and how exactly they control SC quiescence, self-renewal, and differentiation remains incomplete.
In many developing and adult tissues, Wnt signaling pathways play critical roles in regulating SC niches and their responses to environmental insults. Canonical Wnts bind transmembrane-frizzled receptors, resulting in the stabilization of β-catenin and its subsequent localization to the nucleus. Once in the nucleus, β-catenin forms complexes with members of the T-Cell Factor/Lymphoid Enhancer Factor (TCF/Lef) family of transcription factors and proceeds to modulate transcription of genes involved in self-renewal, proliferation, and migration, including cyclin D1 and c-myc4–6. Lef-1-mediated Wnt signaling has been shown to be heavily involved in both SC maintenance and differentiation in many tissue types. In bud-forming epithelial organs, including the mammary gland, lung, teeth, and hair follicles, Lef-1 activation is necessary for SC proliferation and subsequent organ morphogenesis during development7–10. Canonical Wnt signaling is also associated with hematopoietic stem cell (HSC) pool maintenance in adult bone marrow11, 12. In other adult tissues, including the epidermis/hair follicle, Lef-1 is associated with progenitor cell lineage-restriction and terminal differentiation13. Furthermore, studies conducted in various human cancer cell lines have demonstrated that Lef-1 is involved in the survival and progression of multiple cancer types, where its increased and prolonged expression facilitates increased proliferation and invasion of tumor cells14–16. The functions of Lef-1 are therefore complex and context-dependent, necessitating closer examination of its function in pulmonary stem cells.
The lung is composed of several distinct trophic levels of epithelia, each of which has its own SC niche responsible for tissue maintenance and repair17, 18. Studies in the lung have shown that Wnt signaling promotes SC expansion and regeneration of injured tissue19, 20. However, the field’s understanding of the master regulators of Wnt signaling involved in lung SC regeneration remain incomplete. Airway basal cells (BCs) serve as the primary stem/progenitor cell population of mouse trachea and human conducting surface airway epithelium (SAE) of the cartilaginous airways21. Recent findings have also demonstrated that submucosal gland myoepithelial cells (MECs) act as a reserve stem cell population of the murine trachea and are capable of replenishing the BC compartment and subsequently differentiating into luminal cell types following severe airway injury22, 23. Interestingly, it was also reported that overexpression of Lef-1 in MECs results in spontaneous lineage commitment toward a BC phenotype with multipotent capacity for airway regeneration22. Thus, Lef-1 may play a role in airway BC self-renewal and/or lineage commitment, though this has not been formally tested.
In the present study, we demonstrate that Lef-1 facilitates self-renewal and proliferation of murine airway BCs. Conditional deletion of Lef-1 from BCs isolated from murine tracheae results in a significantly reduced ability to proliferate in culture and maintain properly differentiated airway epithelium. Lef-1 knockout (Lef-1KO) BCs fail to self-renew and arrest at the G1/S phase transition of the cell cycle. We show that Lef-1 controls genes involved in the DNA damage response/repair pathway, as well as genes involved in G1/S checkpoint regulation. Given that BCs are responsible for the maintenance of human conducting airway epithelia, these findings may be relevant to disease processes affecting the regenerative capacity of BCs.
2 |. MATERIALS AND METHODS:
2.1 |. Animal Studies
All mouse studies were approved by an Institutional Animal Care and Use Committee. Mice of the following strains were used and maintained on a C57BL/6 background: B6.129-Gt(ROSA)26Sortm1(cre/ERT2)Tyj/J (ROSA-CreERT2; The Jackson Laboratory, stock number 008463), B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J x (ROSA-TG) Cre-reporter mice (The Jackson Laboratory, stock number 007676), B6.Cg-Lef1tm1Hhx/J (Lef-1cKO; The Jackson Laboratory, stock number 030908), and ROSA26-CAG-LoxPEGFPStopLoxP(Lef-1 mice22). All transgenic mouse lines and abbreviated names used in the text can be found in Table S1 in the Supplemental Methods. Mice were induced with tamoxifen and injured with a single i.p. injection of naphthalene as described in the Supplemental Methods.
2.2 |. Primary Cell Isolation and Culture
Surface epithelial cells were isolated from resected murine tracheae using enzymatic digestion as previously described22. A detailed procedure can be found in the Supplemental Methods. Culture conditions and media used to propagate BCs were previously described22. For air-liquid interface (ALI) differentiated cultures, primary murine airway BCs were first expanded in SAGM, then seeded onto 0.33 cm2 polyester transwell membranes (Corning). Culture media was then changed to PneumaCult-ALI (Stem Cell Technologies) when polarized and apical media was removed. See Supplemental Methods for details.
2.3 |. Cell Proliferation and Competition Assays
Cell proliferation assays were performed on cultured primary mouse tracheal BCs isolated from ROSA-CreERT2:Lef-1fl/fl:ROSA-TG mice. Passage 3 (P3) BCs were treated with either 4-hydroxytamoxifen (OH-Tam) (Sigma-Aldrich) or 100% ethanol (vehicle control) for 3 days, with the media replaced every day. The cells were then plated onto 6 well dishes, and cell number was determined at 24 hrs intervals. Competition assays were performed using P3 primary tracheal BCs isolated from Lef-1cKO, Lef-1cKO:ROSA-TG, or Lef-1cKO:Lef-1cKI mice were mixed at a 9:1 ratio with ROSA-TG BCs, treated with OH-Tam as described above, and then co-cultured for 5 passages. Quantification of GFP+, tdTomato+, and GFP−tdTomato− cells was done using flow cytometry (Becton Dickinson LSR II). See Supplemental Methods for details.
2.4 |. Immunofluorescence and Fluorescent in situ Hybridization
Mouse tracheae were excised and fixed in 4% paraformaldehyde (PFA) overnight at 4°C, then washed in PBS and embedded in OCT frozen blocks. Tracheal longitudinal frozen sections were cut and used for immunofluorescent staining as described in the Supplemental Methods. Airway cell cultures (both expanding BC cultures and ALI cultures) were fixed in 4% PFA for 20 min at room temperature, then subjected to the same immunofluorescent staining protocol as for tissue sections. Detection of EdU was done using Click-iT™ Plus EdU Cell Proliferation Kit for Imaging (ThermoFisher Scientific) and the accompanying protocol. A complete list of the antibodies used can be found in Table S2 of the Supplemental Methods. For fluorescent in situ hybridization, BCs were spun onto glass microscope slides, and performed using a ViewRNA Cell Plus Assay Kit (ThermoFisher Scientific). See Supplemental Methods for details.
2.5 |. RNA Sequencing
Cultured basal cells (P4) were treated once with OH-Tam or 100% ethanol as mentioned above. Cells were collected for RNA isolation at the indicated times following treatment. Transcription profiling using RNA-Seq was performed by the University of Iowa Genomics Division using manufacturer recommended protocols. See Supplemental Methods for details.
2.6 |. Statistical Analysis
Results are reported as mean +/− SEM. Statistical analysis was conducted using Prism version 8 (GraphPad Software). The statistical tests used are stated in each figure legend. Data were considered significant at p < 0.05.
3 |. RESULTS
3.1 |. Lef-1 deletion acutely prevents BC proliferation in vitro.
Airway BCs can be trapped in a self-renewal state in culture through the inhibition of Rho-associated coiled-coil containing protein kinase (ROCK). To determine if Lef-1 is essential for BC self-renewal, we isolated primary BCs from ROSACreERT2:ROSALoxPtdTomatoStopLoxPEGFP (Lef-1WT:ROSA-TG) and ROSACreERT2:ROSALoxPtdTomatoStopLoxPEGFP:Lef-1fl/fl (Lef-1cKO:ROSA-TG) mice and cultured them for 4 passages in modified Small Airway Growth Medium containing ROCK-inhibitor (Figure 1A). The cells were then treated with either 4-hydoxytamoxifen (OH-Tam) or ethanol (vehicle control) to induce Cre and thus Lef-1 deletion and conversion of the EGFP reporter. After three days of OH-Tam treatment, the majority of the cells had begun expressing EGFP (Figure 1B). Consistent with our primary hypothesis, OH-Tam treated Lef-1cKO:ROSA-TG BCs had a significantly reduced ability to proliferate (Figure 1C). By contrast, OH-Tam-treated Lef-1WT:ROSA-TG BCs, and vehicle-treated Lef-1cKO:ROSA-TG BCs had no significant change in proliferation, suggesting that the reduction in proliferation was most likely due to the loss of Lef-1 expression (Figure 1C).
Figure 1. Deletion of Lef-1 from BCs decreases proliferative capacity.
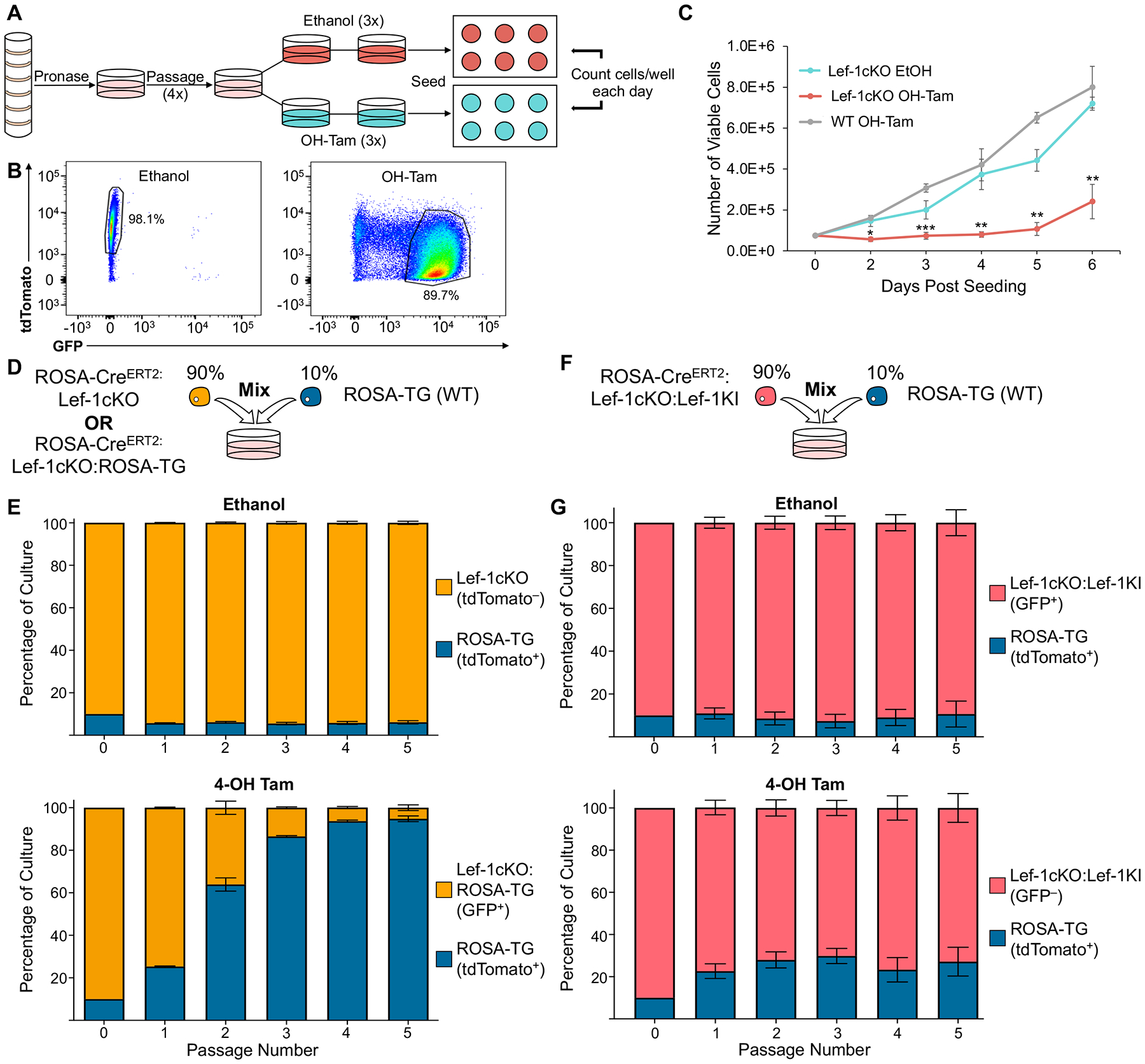
(A) Schematic of basal cell (BC) isolation and expansion from murine tracheae. To induce Lef-1 excision, cells were treated with either ethanol or 1μM 4-hydroxytamoxifen (OH-Tam) three times over the course of three days. Treated cells were then plated onto 6 well plates at a density of 75,000 cells/well. Viable cells/well were quantified from at least two separate wells each day for 6 days from each of 3 independent donor cell pools. (N=6–8 total wells quantified for each condition). (B) FACS plots of ethanol and OH-Tam treated BCs on the third day of OH-Tam treatment. BCs were treated as outlined in A. (C) Number of viable cells/day from experiments outlined in A. (D) Schematic of competition assays shown in E. (E) BCs isolated from ROSA-CreERT2:Lef-1cKO mice (for ethanol treated cultures) or ROSA-CreERT2:Lef-1cKO:ROSA-TG (for OH-Tam treated cultures) were mixed with BCs isolated from WT ROSA-TG mice at a 9:1 ratio (Lef-1cKO:WT ROSA-TG). (F) Schematic of competition assays shown in G. (G) BCs isolated from ROSA-CreERT2:Lef-1cKO:Lef-1KI mice were mixed with BCs isolated from WT ROSA-TG mice at a 9:1 ratio (Lef-1cKO:Lef-1KI : WT ROSA-TG). Mixed cultures in E and G were treated with either ethanol or OH-Tam during the first three days of culture. Cultures were then expanded until near confluence and passaged. Ratios of each BC population was quantified at each passage via flow cytometry for five passages. Three independent donor pools were evaluated in duplicate to generate the graphs in E and G (N=6 total). Graphs show means +/− SEM. Asterisks indicate statistical significance of (C) unpaired two-tailed Student’s T-test (* indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001).
To verify that Lef-1 deletion inhibits BC proliferation through a cell-autonomous process, we conducted in vitro competition assays. BCs isolated from ROSALoxPtdTomatoStopLoxPEGFP (ROSA-TG) and ROSACreERT2:Lef-1fl/fl mice (Lef-1cKO) were mixed at a 1:10 ratio (ROSA-TG:Lef-1cKO) and then treated with either ethanol or OH-Tam in mixed culture (Figure 1D). At each passage, the number of tdTomato+ (ROSA-TG) and tdTomato− (Lef-1cKO) cells were quantified using flow cytometry. As expected, the ratio of ROSA-TG cells to Lef-1cKO in OH-Tam treated cultures increased significantly with each successive passage, while this ratio remained unchanged in ethanol treated cultures (Figure 1E).
Next we assessed how Lef-1 gene deletion functionally impared Lef-1cKO BC expansion. Q-PCR and immunofluorescence were insufficiently sensitive to detect Lef-1 expression in wild type BCs and thus failed to confirm a reduction in Lef-1 gene product (at the mRNA or protein level) in Lef-1cKO BCs (data not shown). To exclude the possibility that the loss of an essential genomic locus within the deleted Lef-1 intronic sequence was the functional cause of cell cycle defects in Lef-1KO BCs, we conducted rescue experiments in which ectopic human Lef-1 (hLef-1) expression was activated from a ROSA locus at the time of mouse Lef-1 deletion. These experiments utilized a ROSACreERT2:ROSAflEGFPstop-flLef-1 knock-in transgene capable of expressing human Lef-1 in response to Cre activity. We crossed these mice with the Lef-1fl/fl mice to generate ROSACreERT2:ROSAflEGFPstop-flLef-1:Lef-1fl/fl (Lef-1cKO:Lef-1KI) mice. We then conducted competition assays with ROSA-TG cells co-cultured with Lef-1cKO:Lef-1KI cells at 1:10 ratio, respectively (Figure 1F). Following OH-Tam induction, forced expression of human Lef-1 in mouse Lef-1cKO BC led to a near complete rescue in the ability to proliferate when compared to vehicle treated cultures (Figure 1G). Collectively, these proliferation and competition assays demonstrate that Lef-1 is acutely required for self-renewal of airway BCs.
3.2 |. Lef-1 is transiently expressed in a small subset of basal cells.
Given that attempts to detect Lef-1 by immunofluorescence and Q-PCR in bulk cultures were unsuccessful, we hypothesized that Lef-1 might be transiently expressed in a small subset of cells during a critical phase of the cell cycle. Indeed, analyzing Lef-1 expression in a publicly available single-cell RNA sequencing dataset showed that Lef-1 was expressed at low-levels in a subset of BCs (Figure S1A–C)24. To this end, we performed single molecule fluorescent in situ hybridization (smFISH) on Lef-1cKO BCs that were treated with either ethanol or OH-Tam for three days. On the day following the last treatment, cells were pulsed with 5-ethynyl-2’-deoxyuridine (EdU) for two hours, then lifted from the dish and cytospun onto glass slides for smFISH localization of Lef-1. EdU pulsing was used to determine if cells in active S-phase demonstrated increased levels of Lef-1 expression. Because Lef-1 transcriptionally regulates expression of CCND1 (cyclin D1)4 and cyclin D1 is required for cell enter into S-phase, we hypothesized that BC Lef-1 expression might be highest in BCs stalled in early S-phase and thus enriched in EdU. Our results demonstrated that the ethanol-treated (WT) group contained infrequent cells that had Lef-1 mRNA signal (Figure S1D,E) above the background levels observed in OH-Tam (KO) cells (Figure S1F). Contrary to our initial hypothesis, Lef-1 signal did not appear to correlate with EdU positivity, suggesting that Lef-1 is unlikely to have a role in S-phase. Overall, these results indicate that Lef-1 is indeed expressed in BCs, albeit at low level and in a small subset of cells at any given time.
3.3 |. Lef-1 is required for maintenance of a differentiated airway epithelium in vitro.
BCs are responsible for the maintanence of a properly differentiated airway epithelium, which is comprised of multiple luminal cell types. Lef-1 is necessary for cell cycle progression in BC expansion media containing ROCK-inhibitor, two SMAD inhibitors that repress TGFBR and BMPR signaling, and a Wnt activator. To better appreciate the role Lef-1 has in homeostatic maintenance of BCs and its impact on differentiation, we performed studies on Lef-1cKO BCs in air-liquid interface (ALI) cultures. We hypothesized that deletion of Lef-1 in BCs could impair their ability to self-renew by one of two mechanisms: Lef-1 deletion could lead to a depletion of the BCs or could induce differentiation toward ciliated and/or secretory cells.
To differentiate between these two mechanisms, we mixed Cre− ROSA-TG and Lef-1cKO:ROSA-TG BCs at a 1:1 ratio and seeded them into air-liquid interface (ALI) cultures in the presence of OH-Tam for the first three days (Figure 2A). In this setting, Lef-1cKO:ROSA-TG cells progressively switched from tdTomato+ to GFP+ over the first three days, while ROSA-TG cells (WT for Lef-1 and lacking CreERT2) remained tdTomato+. By day 15 at ALI, Lef-1cKO:ROSA-TG (GFP+) cells predominated in the culture, increasing from the initial 50% to 62%, while ROSA-TG (tdTomato+) composed 38% of the cultures (Figure 2A, B). However, the number of ROSA-TG (tdTomato+) cells progressively increased over time, making up almost 80% of the differentiated cultures by 60 days at ALI (Figure 2A, B). Phenotypic characterization of these cultures for the distribution of ciliated cells (α-tubulin+), secretory goblet cells (Muc5B+), and secretory club cells (Scgb1a1+) showed that Lef-1cKO:ROSA-TG (GFP+) BCs initially had a greater capacity to differentiate into these luminal cell types, as 70–80% of all ciliated, goblet, and club cells were GFP+ after 15 days at ALI (Figure 2C–E). However, by 60 days at ALI, ~80% of all ciliated and goblet cells were derived from tdTomato+ ROSA-TG cells (Figure 2C–D). Notably, this trend was not seen with secretory club cells, as about half of all club cells were still derived from GFP+ Lef-1cKO:ROSA-TG cells at 60 days of ALI (Figure 2E). These data indicate that BCs lacking Lef-1 differentiate into various luminal cell types more rapidly than WT BCs, but fail to maintain steady luminal cell numbers over time.
Figure 2. BC Lef-1 expression facilitates the maintenance of a differentiated epithelium in vitro.
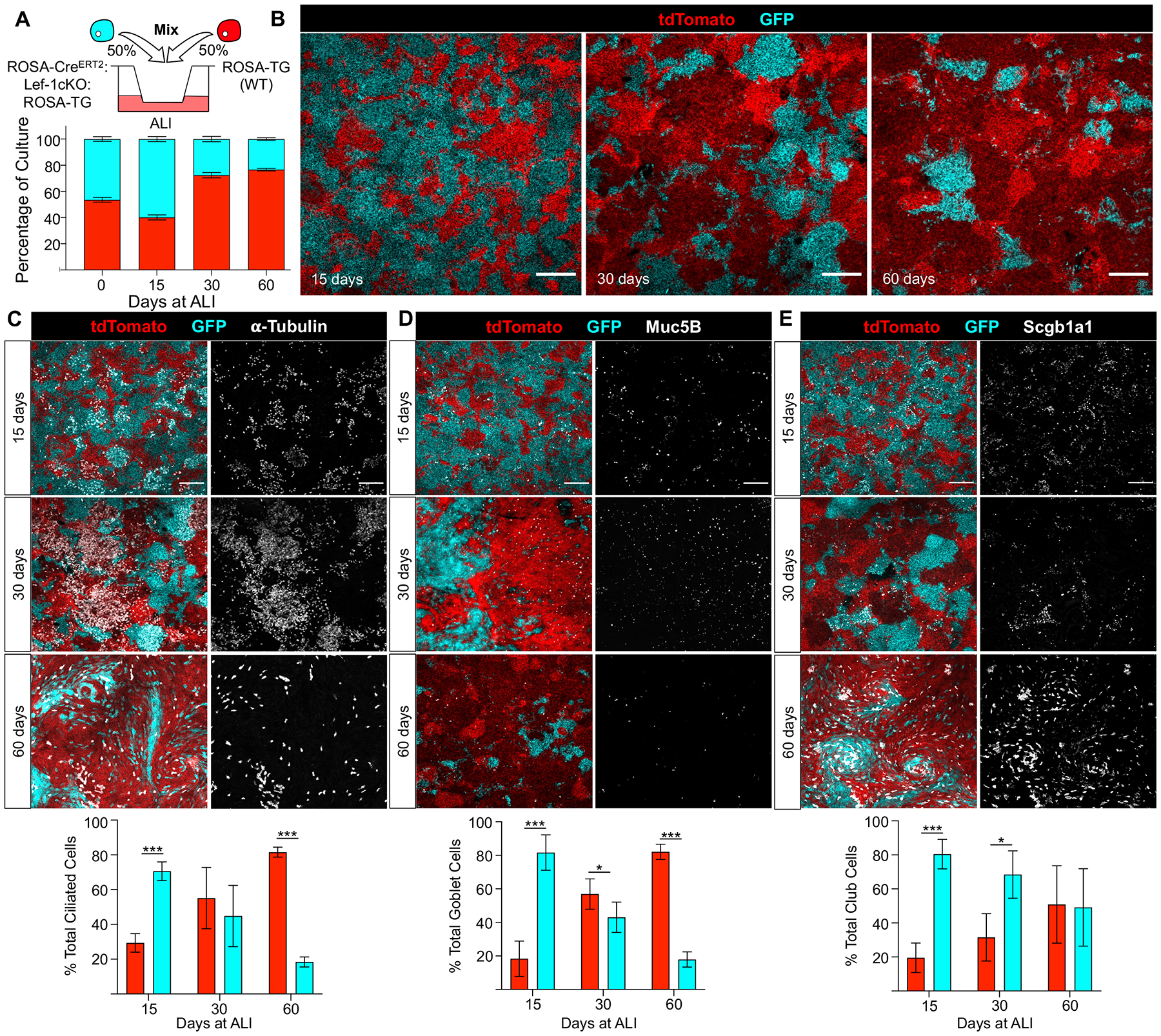
(A) Diagram of the experimental design using mixed basal cell (BC) populations seeded into air-liquid interface (ALI) cultures (top panel). Cells were mixed at a 1:1 ratio and seeded onto transwells in the presence of 1μM 4-hydroxytamoxifen (OH-Tam) for the first three days of culture in Small Airway Growth Media (SAGM). Cells were then polarized by removing the media from the apical chamber and changing to differentiation media in the basolateral chamber. Bottom panel shows the percentage of ROSA-TG (tdTomato+) and Lef-1cKO:ROSA-TG (GFP+) airway epithelial cells in the ALI culture after seeding and at 15, 30 and 60 days after polarization. The percentage of the culture that was tdTomato+ or GFP+ was quantified by determining the total area of each genotype at each timepoint (N ≥ 9 transwells from three donor pools, N ≥ 3 images/transwell were quantified) (B) Images of cultures at 15, 30, and 60 days following polarization. Scale bars, 200 μm. (C-E) Mixed ALI cultures stained for ciliated cell marker α-tubulin (C), goblet cell marker Mucin 5B (Muc5B) (D), and club cell marker Secretoglobulin 1a1 (Scgb1a1) (E), at the indicated timepoints after polarization (N ≥ 3 transwells, ≥ 2 images/transwell). Scale bars, 100 μm. Graphs show means +/− SEM. Asterisks indicate statistical significance by unpaired two-tailed Student’s T-test (* p < 0.05, ** p < 0.01, and *** p < 0.001).
We hypothesized that Lef-1 deletion in BCs induced terminal differentiation to luminal cells and thus anticipated that BCs would deplete from ALI cultures over time following OH-Tam treatment. To investigate this hypothesis, mixed ALI cultures were embedded and sectioned after 15, 30, and 60 days at ALI. Consistent with this hypothesis, phenotypic staining for BCs (Krt5+) revealed that ~60% of BCs were Lef-1 WT (tdTomato+) after 15 days at ALI (Figure S2A,D). However, the percentage of Lef-1KO BCs increased in proportion over time reaching ~75% by 60 days at ALI (Figure S2B–F). Contrary to our hypothesis, these data indicate that Lef-1 deletion does not cause BC depletion in a differentiated epithelium. Overall, these findings demonstrate that deleting Lef-1 in BCs initially drives differentiation toward luminal cells and are consistent with a requirement for Lef-1 in self-renewal and maintenance of a multipotent BC compartment.
3.3 |. BCs escape a Lef-1-dependent block in self-renewal with time in culture.
We noted in our proliferation assays (Figure 1C) that Lef-1cKO BCs began to proliferate after 6 days in culture. Similarly, our competition assays (Figure 1D,E) demonstrated that Lef-1cKO BCs reached a steady state of ~5% of the culture by passage 4. We hypothesized that Cre-mediated excision of Lef-1 was incomplete and led to survival bias of cells with one or both Lef-1 allele intact. To test this, we measured the doubling time of OH-Tam-treated Lef-1cKO BCs through multiple passages in expansion media. We then genotyped at each passage to determine the status of the Lef-1 gene deletion in the cells that proliferated. These cells continued to grow for at least four passages following OH-Tam treatment, and had a similar doubling time to passage-matched EtOH-treated cells after passage 2 (Figure 3B). Notably, genotyping revealed that the majority of recovered cells remained null for the Lef-1 gene after four passages (Figure 3C).
Figure 3. BCs escape a Lef-1-dependent block in cell cycle progression with time in culture.
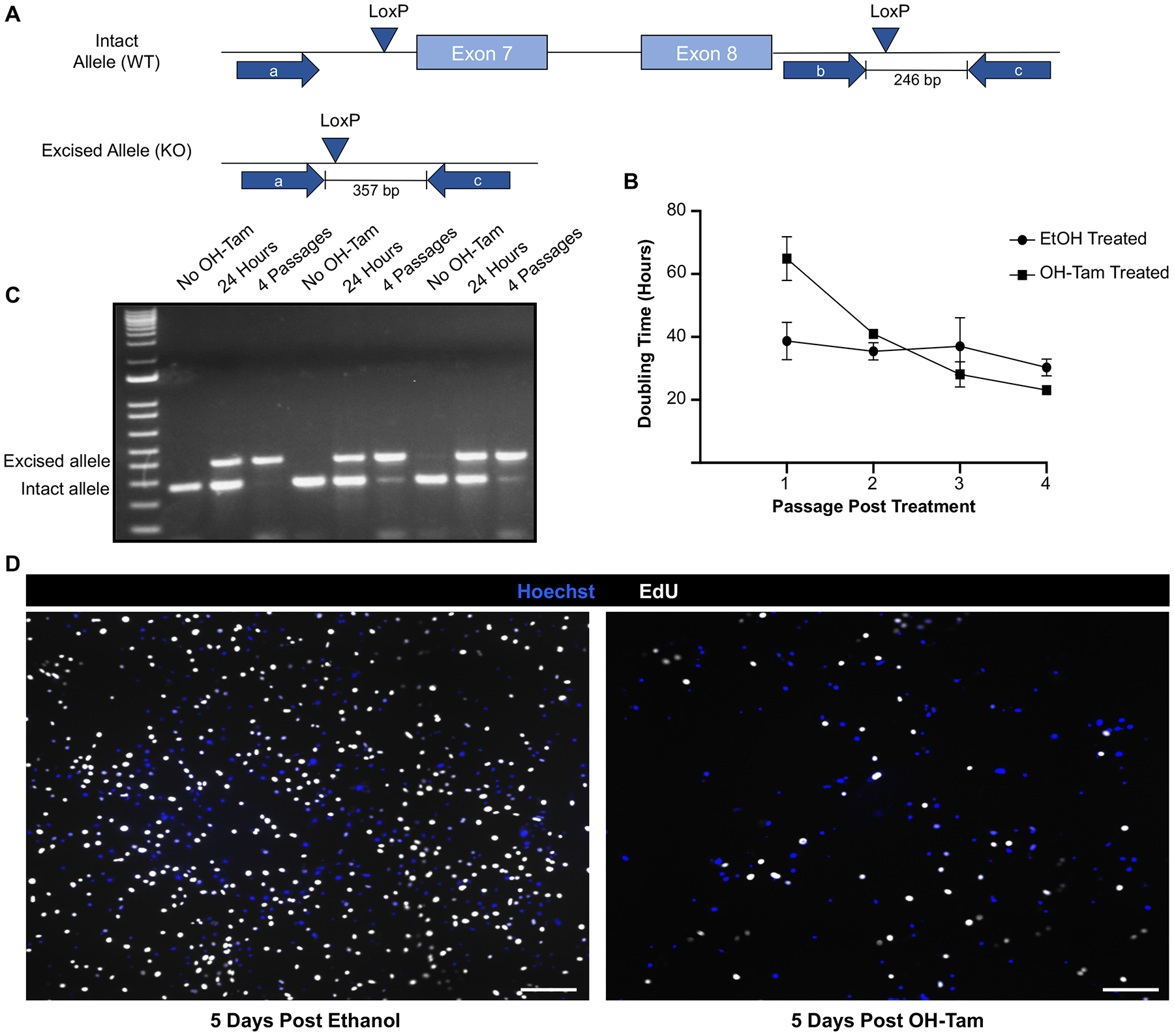
(A) Schematic of the genotyping protocol used to determine deletion status of Lef-1 in Lef-1cKO basal cells (BCs). (B) Doubling times of Lef-1cKO BCs treated with either ethanol (EtOH) or OH-Tam for three days, then passaged four times. Passaging occurred as the cultures neared confluency (85–90%). (C) DNA from Lef-1cKO BC cultures was collected either in the absence of 4-hydroxytamoxifen (OH-Tam), 24 hours following OH-Tam treatment, or 4 passages following OH-Tam treatment. Cultures were treated with OH-Tam on three consecutive days using the same experimental protocol for other studies. PCR was performed using the primer sets indicated in A. Genotyping revealed that cultures passaged 4 times following OH-Tam are predominately composed of BCs with excised Lef-1 alleles. Agarose gels show the DNA results from three separate replicates. (D) Lef-1cKO BCs were pulsed with 5-ethynyl-2’-deoxyuridine (EdU) for 12 hrs at 5 days following treatment with EtOH (top image) or OH-Tam. Images show EdU incorporation (white) with Hoechst counter stain. Scale bars, 30 μm.
We hypothesized that proliferation-competent Lef-1cKO BCs might be derived from a small subset of cells similar to a tumor that epigenetically escapes a radiation or chemical block in cell cycle progression25, 26. To approach this hypothesis, we performed a 12 hour EdU pulse in Lef-1cKO cells at an early stage of proliferative recovery. Results from these experiments demonstrated a lack of apparent clonal expansion, as EdU incorporation was evenly distributed among the cells at 5 days following OH-Tam treatment (Figure 3D). Thus, BCs are capable of overcoming a Lef-1-dependent block in proliferation with time and the mechanism of this escape occurs in the majority of Lef-1cKO BCs. Collectively, these data demonstrate that Lef-1-dependent and -independent mechanisms are involved in cell cycle progression of BCs in culture, though the mechanism of this Lef-1-independent adaption remains unknown.
3.4 |. Lef-1 facilitates BC progression through the G1/S phase of the cell cycle.
High levels of Lef-1 expression have been shown to correlate with the proliferative and invasive characteristics in variety of cancers, including human lung adenocarcinomas4, 27, 28. However, its importance in regulating self-renewal of stem cells at homeostasis is largely unexplored. Given the requirement of Lef-1 for airway BC self-renewal in culture, we evaluated whether Lef-1-deficient BCs stopped proliferating at the same point in the cell cycle. These studies in asynchronous cell culture utilized EdU incorporation to mark S phase, Cyclin D1 to mark G1 phase, and phosphorylated histone 3 (His-H3P) to mark late G2 and M phases (G2/M) in vehicle- or OH-Tam-treated Lef-1cKO BCs (Figure 4A–B). The percentage of BCs in the G1/S transition phase (Cyclin D1+EdU+) was significantly higher after Lef-1 deletion (32.4% for OH-Tam vs 8.6% for vehicle treatment groups) (Figure 4A–C). Additionally, the percentage of cells that were in the later stages of S phase (CyclinD1−EdU+) was significantly lower after Lef-1 deletion (2.2% for OH-Tam vs 22.2% for vehicle treatment groups) (Figure 4C).
Figure 4. Lef-1 deletion in BCs leads to cell cycle arrest at the G1/S boundary and altered expression of genes associated with cell cycle regulation.
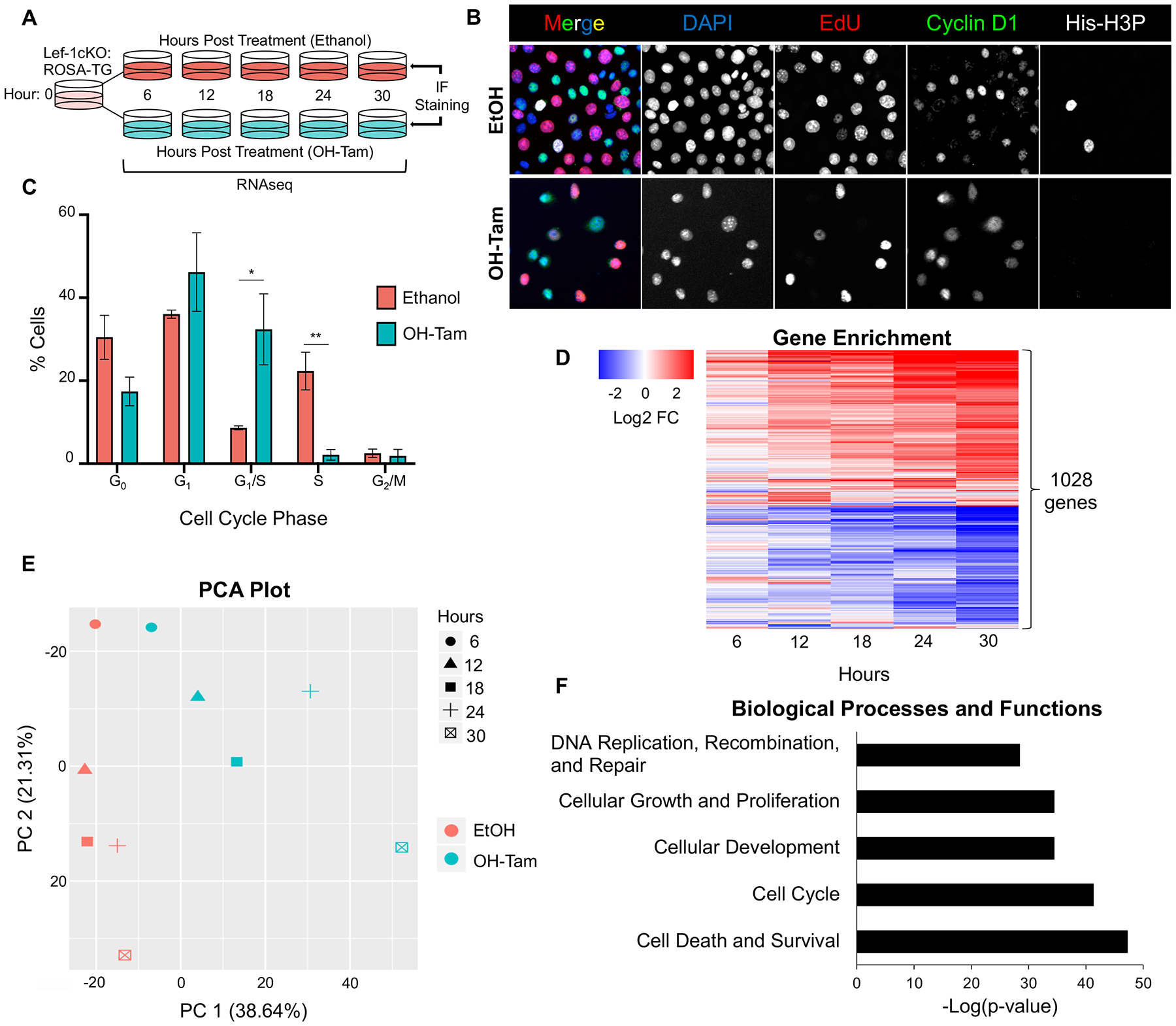
(A) Experimental schematic for cell cycle and RNA-seq analyses. Passage 4 (P4) Lef-1cKO:ROSA-TG or Lef-1cKO BCs were treated once with either 100% ethanol or 2 μM 4-hydroxytamoxifen (OH-Tam). Lef-1cKO:ROSA-TG BCs were used for RNA-seq and Lef-1cKO BCs were used for immunofluorescence (IF). Lef-1cKO BCs used for IF were pulsed with 5-Ethynyl-2´-deoxyuridine (EdU) 1 hour prior to fixing. (B) Lef-1cKO BCs were immunostained for the indicated antigens 30 hrs after ethanol or OH-Tam treatment. (C) Quantification of BCs in each indicated phase of the cell cycle as a % of total cells imaged from experiment in B. CyclinD1+EdU−His-H3P− cells were scored as being in G1, CyclinD1+EdU+His-H3P− cells were scored as being in G1/S, CyclinD1−EdU+His-H3P− cells were scored as being S phase, and CyclinD1−EdU−His-H3P+ cells were scored as being in G2/M phase. Cells negative for all markers were scored as being in G0. Graph shows means +/− SEM, N=3. Asterisks indicate statistical significance of unpaired two-tailed Student’s T-test (*p < 0.05, **p < 0.01). (D) RNA-seq experiment on N=4 independent donor cell pools for each experimental time point. Shown is a heatmap of the 1028 differentially expressed (p < 0.05, BH corrected) genes, following Lef-1 deletion, that had an absolute fold change (FC) ≥ +/−2 in expression in at least one of the indicated timepoints after treatment. (E) Principal Component Analysis (PCA) of the 21,390 genes detected. (F) Ingenuity Pathway Analysis (IPA) biological processes and functions defined by gene expression patterns showing -Log(p-values).
To better determine how Lef-1 influences cell cycle progression of airway BCs, we performed bulk RNAseq on Lef-1cKO cells at 0, 6, 12, 18, 24, and 30 hours following treatment with either OH-Tam or ethanol. Of the 21,390 protein-coding genes identified, 6,406 genes were significantly differentially expressed genes (DEGs) (Benjamini-Hochberg adjusted t-test; p<0.05) (Table S3). When normalized to their time-matched ethanol-treated controls, 1,028 genes had altered expression > 2-fold in at least one of the five timepoints analyzed (Figure 4D, Table S4). Principal-component analysis (PCA) of all 21,390 genes showed a clear divergence with time in ethanol vs OH-Tam treated cultures, with the first two PCs accounting for 59.95% of the total variance (Figure 4E). Ingenuity Pathway Analysis (IPA) was used to discover biological processes that were significantly altered in OH-Tam treated Lef-1cKO BCs (Table S5). This analysis revealed significantly altered biological processes including Cell Cycle, Cellular Growth and Proliferation, and DNA Replication and Repair (Figure 4F, Table S5A). Taken together, these data support the importance of Lef-1 in BC progression through the cell cycle.
3.5 |. Lef-1 deletion disrupts DNA damage response pathways in airway BCs.
Adult stem cells must ensure their genomic integrity for the life of the host and thus are equipped with highly effective DNA damage response (DDR) pathways. Stem cells must also sense when DNA damage is too significant to self-renew and such states have been linked to differentiation, senescence, and apoptosis29, 30. However, whether stem cell lineage-commitment and differentiation are directly controlled by DDR pathway regulation or simply a default response to excessive DNA damage remains unclear30. Given the importance of the G1/S DNA damage checkpoint in cellular replication, we sought to investigate whether disruption of Lef-1 in BCs leads to alterations in the expression of DDR genes. In support of his hypothesis, the majority of the 139 differentially expressed genes associated with DDR were downregulated following Lef-1 deletion (Table S5C). IPA also revealed lists of biological functions (Table S5A) and canonical pathways (Table S5B) that were associated with DDR and DNA replication in Lef-1cKO BCs, including Double-stranded DNA Break Repair, Excision Repair, p53 Signaling, and G1/S checkpoint regulation (Figure 5A, B). Together, these data suggest that loss of Lef-1 in BCs could perturb cellular responses to DNA damage.
Figure 5. Lef-1 regulates the DNA damage response (DDR) pathway in BCs.
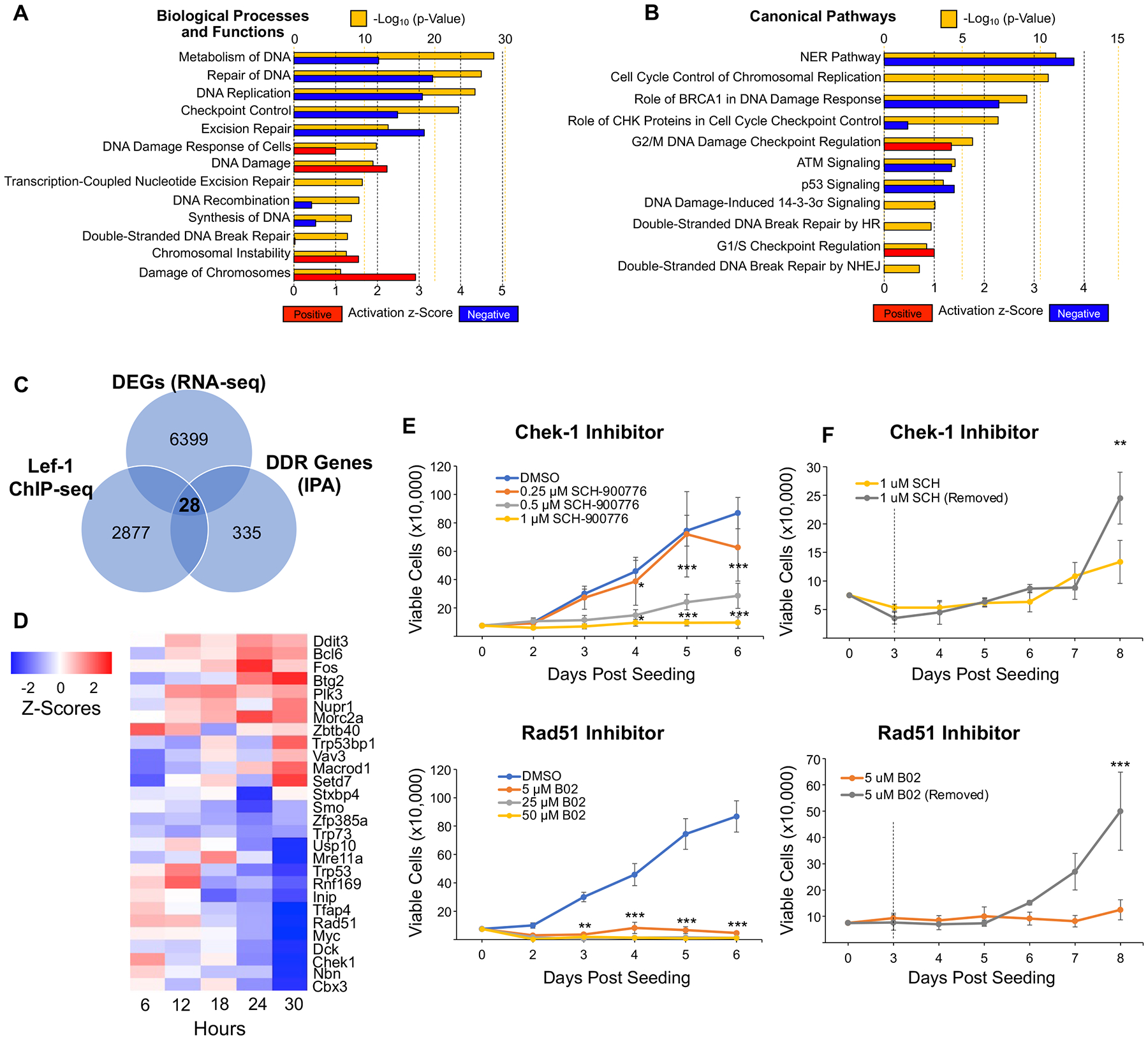
(A-B) Ingenuity Pathway Analysis (IPA) was used to identify biological processes and functions (A) and canonical pathways (B) that were significantly altered in Lef-1cKO:ROSA-TG BCs based on RNA-seq analysis. Graphs display the -Log(p-value) and activation z-score, as determined by IPA, of each process/function or pathway. Red bars indicate positive z-scores (upregulated), while blue bars indicate negative z-scores (downregulated). (C) Venn diagram of overlapping genes from three datasets: 1) Differentially expressed genes (DEGs) in Lef-1cKO BCs, 2) genes involved in DDR from IPA, and 3) genes containing Lef-1 binding sites as determined by ChIP-seq of hair follicle stem cells13. This analysis produced a list of 26 direct Lef-1 target genes involved in DDR that were also DEGs in Lef-1cKO BCs. (D) Heat map depicting Z-scores of the 26 DDR genes over the time course of OH-Tam treatment. (E-F) Proliferation assays of BCs treated with various concentrations of Chek-1 inhibitor (SCH-900776; top graphs) or Rad51 inhibitor (B02; bottom graphs). (E) BCs were continuously exposed to various concentrations of the two inhibitors. (F) BCs treated with the lowest effective concentration of inhibitor that prevented proliferation and then had the inhibitor removed from one group at three days (dotted line) to assess reversibility. Graphs show means +/− SEM, N ≥ 3 donor cell pools. Asterisks indicate statistical significance by 2-way ANOVA and Bonferroni multiple comparison test (*p < 0.05, **p < 0.01, ***p < 0.001).
To better understand how Lef-1 impacts expression of DDR gene, we utilized a previously published list of direct Lef-1 target genes obtained via chromatin immunoprecipitation sequencing (ChIP-seq) of transit amplifying cells of the hair follicle niche13. Notably, 1138 DEGs (or 18%) in our RNA-seq dataset were also found in this ChIP-seq dataset (Table S6A). Cross referencing Lef-1cKO DEGs against DDR genes and Lef-1 target genes resulted in a list of 28 potential Lef-1 target genes associated with DDR pathways (Figure 5C, Table S6B). This list included Trp53, and its family member Trp73, Myc, Mre11, Checkpoint kinase 1 (Chek1), and Rad51 (Figure 5D), all of which have been previously identified as important regulators of cell cycle checkpoint progression and DNA repair31–34. The majority (16 out of 28) of these DDR genes were downregulated in our RNA-seq dataset (Figure 5D). These data support the hypothesis that Lef-1 controls BC proliferation through the regulation of the DDR pathway.
Out of the list of 28 DDR genes, Chek1 and Rad51 were two of the most significantly downregulated genes following Lef-1 deletion. To evaluate the involvement of these genes in the regulation of BC proliferation, we treated BCs with chemical inhibitors of either Rad51 or Chek1. Inhibition of Rad51 (with B02) nearly completely blocked BC replication out to 6 days (Figure 5E). Inhibition of Chek1 (with SCH-900776) also led to suppressed proliferation at higher concentrations. To evaluate the reversibility of Rad51 and Chek1 inhibition, we treated BCs with these inhibitors, then removed the inhibitor after 3 days of treatment. Removal of B02 from BCs led to a recovery in proliferation as compared to sustained treatment with the Rad51 inhibitor (Figure 5F). However, removal of SCH-900776 from BCs produced a marginal proliferative advantage at later timepoints and there was a clear rise in proliferation by 7 day in the presence of sustained Chek1 inhibition. This observed proliferative escape from Chek1 inhibition was similar to that observed in BCs following Lef-1cKO (Figure 1C,E; Figure 3C,D), suggesting that BCs can compensate for a G1/S block imposed by inhibition of Chek1 or Lef-1.
Given that Chek1 and Rad51 expression is altered by Lef-1 deletion, we hypothesized that later-passage Lef-1cKO BCs are able to escape G1/S cell cycle blockade and proliferate normally by adapting replication-recovery pathways that are independent of Chek1 and/or Rad51. If true, proliferation-competent Lef-1cKO BCs would be resistant to Chek1 or Rad51 inhibition. Indeed, Chek1-inhibited Lef-1cKO BCs reached confluency at the same rate as DMSO-treated proliferation-competent Lef-1cKO BCs. Notably, Rad51 inhibitor-treated proliferation-competent Lef-1cKO BCs had a similar level of growth inhibition as WT BCs and failed to proliferate over six-day study period (Figure S3). These findings suggest that Lef-1 likely controls a Chek1-dependent G1/S checkpoint in BCs and that adaptation to an imposed block by inhibiting either protein enables BCs to reengage the cell cycle.
3.6 |. SMAD-Dependent TGFß Signaling Pathways are Activated in BCs Following Lef-1 Deletion Despite Sustained Dual SMAD Chemical Inhibition.
BMP and TGFß pathways have roles in regulating BC proliferation and differentiation, and these pathways can be influenced by other signaling pathways including Wnt and Notch signaling35–38. These interconnected and carefully regulated signaling cascades allow stem cell niches to respond appropriately during tissue maintenance and repair following injury39–41. Therefore, we sought to determine if ablating Lef-1-mediated Wnt signaling altered the state of other major signaling pathways in BCs. To this end, we performed IPA Upstream Analysis, which evaluates the activity of potential upstream regulators (e.g., transcription factors and kinases) that might be responsible for the observed altered expression of genes imposed by Lef-1KO. Upstream Analysis of our RNA-seq dataset revealed a prediction of activated TGFß signaling, as TGFß1, SMAD3, and SMAD4 were all predicted to be activated in Lef-1cKO BCs (Figure S4A–D, Table S7), despite the presence of dual SMAD inhibitors in the culture media. Several upstream regulators involved in cell cycle progression were predicted to be inhibited, including several members of the E2F family, Myc, and FoxM1 (Figure S4A). Notably, each of these factors was a DEG in Lef-1cKO cells, including four isoforms of E2F. Among these, SMAD3 is known to directly associate with Lef-1, and the Myc promoter is directly regulated by Lef-142, 43. These data provide further support that Lef-1 deletion imposes a block in BC cell cycle progression. The prediction of SMAD-dependent signaling activation was particularly intriguing, as SMAD signaling is known to inhibit the proliferative abilities of BCs in vivo and in vitro, and also plays a role in mucociliary differentiation38.
3.7 |. Deletion of Lef-1 in vivo reduces the regenerative capacity of BCs following airway injury.
BCs are the primary progenitor responsible for replacing SAE cells lost to injury or infection21. Therefore, we sought to determine if Lef-1 regulates BC regenerative capacity in an in vivo setting. ROSACreERT2:Rosa-TG (Lef-1 WT) and Lef-1cKO:ROSA-TG mice were induced with tamoxifen once a day for five days to trace and induce Lef-1 deletion in SAE cell types (Figure 6A). One week following the tamoxifen induction, mice were then administered a single dose of naphthalene (200 mg/kg) to induce moderate airway injury and compared a corn oil-injected uninjured control gorup22. Three weeks following the injury, tracheae were collected and processed for tissue sectioning. The uninjured control groups demonstrated no significant differences between Lef-1 WT and Lef-1cKO:ROSA-TG animals in the total number of lineage-tagged (GFP+) SAE cells (Figure 6B,C,F) or the abundance of lineage-tagged BCs (GFP+Krt5+) (Figure 6B,C,G). Notably, naphthalene-injured Lef-1cKO:ROSA-TG mice had significantly fewer lineage-tagged total SAE cells and BCs than mock injured Lef-1cKO:ROSA-TG mice (Figure 6D–G). Naphthalene-injured Lef-1 WT mice showed no significant difference in lineage-tagged SAE cell and BC abundance compared to their uninjured counterparts, indicating that the differences seen are not simply due to injury (Figure 6D–G). Overall, these findings indicate that Lef-1cKO BCs have a significantly reduced regenerative capacity following an injury. However, these results also show that Lef-1 is not required for BC survival in vivo, as the abundance of lineage-tagged BCs in mock-injured Lef-1cKO:ROSA-TG mice was similar to that of Lef-1 WT mice.
Figure 6. In vivo loss of Lef-1 reduces the regenerative capacity of BCs following airway injury.
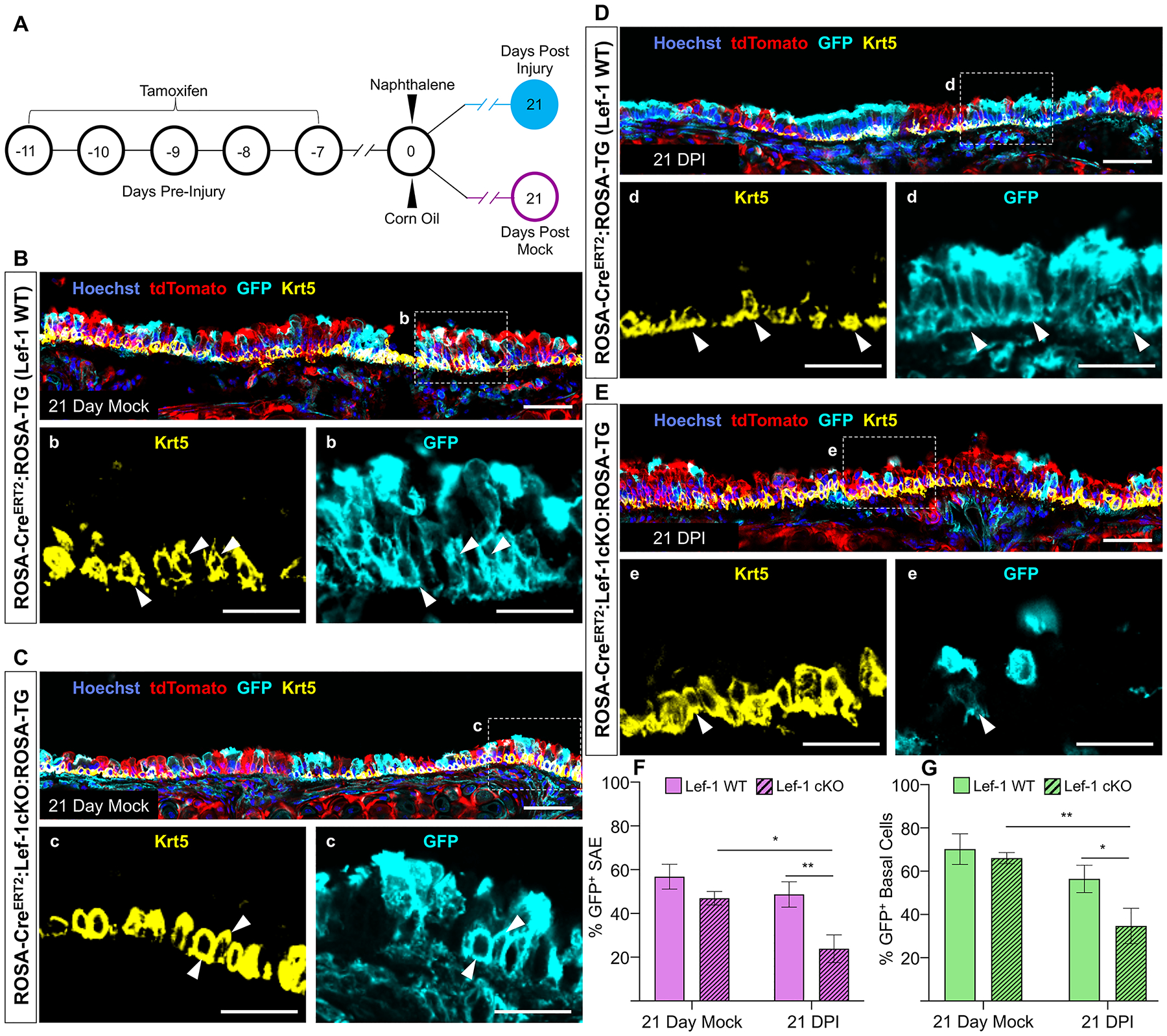
(A) Timeline of when tamoxifen inductions and naphthalene injury occurred. Injury was done using 200 mg naphthalene/kg body weight. (B-E) Confocal images of tracheal sections from mock injured (B) ROSA-CreERT2:ROSA-TG (Lef-1WT) mice and (C) Lef-1cKO:ROSA-TG mice, and naphthalene-injured (D) Lef-1WT mice and (E) Lef-1cKO:ROSA-TG mice. Sections were immunostained for tdTomato, GFP, and Krt5. Box regions in the main panels are magnified below each image. White arrowheads indicate basal cells (Krt5+) that are also lineage-traced (GFP+). Scale bars are 50 μm for main the panels and 25 μm for the magnified boxed regions. (F and G) Quantification of the percentage of lineage-traced (GFP+) cells in (F) the entire surface airway epithelium (SAE) and (G) in the Krt5+ BCs population. Graphs show means +/− SEM, N ≥ 4 independent mice. Significance was determined using 2-way ANOVA and Bonferroni multiple comparison test (*p < 0.05, **p < 0.01).
4 |. DISCUSSION
SC niches found within adult organs facilitate tissue maintenance and repair, and these processes are controlled by careful regulation of multiple signaling pathways1, 35, 37, 44. In the mammalian proximal airway, BCs have been defined as the primary resident SC, slowly cycling under homeostatic conditions but able to rapidly proliferate and differentiate into multiple luminal lineages following injury21. Though Wnt signaling has been demonstrated to be active within the proximal airway epithelia during injury repair20, its role in the individual cell types of the SAE, including BCs, has remained unclear. In the present study, we sought to elucidate the role of Lef-1, a Wnt signaling effector, within the BC compartment of the surface airway epithelium.
Ablation of Lef-1 from BCs led to transient loss in the ability to proliferation in vitro (Figure 1) with a block occurring at the G1/S phase of the cell cycle (Figure 4). Based on several criteria, this failure to progress through the cell cycle was at least in part due to cell-intrinsic functions of Lef-1. First, mixing of Lef-1cKO with WT BCs failed to rescue the proliferation defect, ruling out the possibility that Lef-1 mediates secretion of ligands that act in a paracrine manner to maintain a proliferative state. Second, activation of hLef-1 expression in Lef-1KO BCs rescued the proliferative defect. Consistent with a Lef-1-dependent G1/S transition mechanism was the downregulation of key DNA damage response pathway genes (e.g., Rad51 and Chek1), which are critical for progression through S phase during DNA replication (Figure 5). Although many of these genes were known targets of Lef-1 in other SC systems13, we cannot draw conclusions on the direct Lef-1 targets in BCs that control the cell cycle. However, chemical inhibition of Chek1 in WT BCs phenocopied Lef-1KO BCs (Figure 5) and like Lef-1cKO BCs, Chek1-inhibited BCs were eventually able to escape G1/S-blockade with time. Moreover, late-passage Lef-1cKO BCs that escape cell cycle arrest are resistant to Chek1 inhibition, suggests a functional relationship between Lef-1 and Chek1-dependent regulation in BC progression through G1/S.
We hypothesized that downregulation of critical DNA damage checkpoint genes in Lef-1cKO BCs, such as Rad51 and Chek1, might elevate the level of double-stranded DNA breaks within BCs and thus lead to terminal differentiation in culture and in vivo. However, localization of γH2AX, a histone that binds to double-stranded DNA breaks, failed to demonstrate elevations in Lef-1cKO cells at 3 days following OH-Tam treatment in culture (data not shown). Similarly, there was no change in γH2AX expression in BCs of the trachea following in vivo injury of Lef-1cKO mice (data not shown). For these reasons, we hypothesize that Lef-1 promotes cell cycle progression during late G1 phase prior to significant DNA replication that would be associated with enhanced requirements for DNA repair. Supporting this hypothesis was the finding that Lef-1 mRNA was expressed in a subset of BCs not actively synthesizing DNA (Figure S1). Furthermore, the transient nature of the proliferative block imposed by Lef-1KO or Chek1 inhibition supports the lack of significant DNA damage that would permanently impair BC self-renewal.
Given the clear block in cell cycle progression in Lef-1cKO BCs, it was somewhat surprising that there was undetectable Lef-1 protein and mRNA in bulk cultures. Only a small fraction of BCs express Lef-1 mRNA when detected by smFISH (Figure S1). Given the infrequent and transient nature of Lef-1 expression in BCs, we conclude that Lef-1 is required during a short window in late G1 phase. Our rescue experiments with hLef-1 in Lef-1cKO cells, however, lends strong support that Lef-1 is indeed required for BC self-renewal, and the loss of non-coding sequences within the deleted intron of Lef-1cKO BCs is not responsible for the observed alterations in cell cycle progression (Figure 1).
The mechanism by which BCs regain an ability to proliferate following Lef-1KO or Chek1 inhibition is currently unclear. However, several cell systems have demonstrated redundancy if TCF/Lef-1 signaling. For example, in the mouse hair follicle, both Lef-1 and TCF7 act redundantly to regulate stem cell activation and lineage progression13. Similarly, the four TCF/Lef family members (Lef-1, TCF7, TCF7L1, and TCF7L2) function in an additive and redundant manner to specifying epithelial progenitors during mouse lung development45. Thus, it is possible that other TCFs can compensate for the loss of Lef-1 and reestablish a self-renewing state. However, if any of these other TCFs was indeed compensating for the loss of Lef-1, it remains unclear why this compensation utilizes a Chek1-independent mechanism involved in the G1/S checkpoint. It should also be noted that the in vitro cell culture system used may drive the observed Lef-1-independent reprogramming that allows for cell cycle progression and this may not occur in vivo. Indeed, our in vivo injury studies in mice suggest that this compensation does not occur in vivo, with Lef-1 deletion leading to a decline in the overall capacity of BCs to regenerate luminal cells following injury. This could be due to the lack of time required for Lef-1-independent reprogramming to occur, or the fact that our in vitro culture conditions do not fully represent the in vivo BC state.
Our in vitro and in vivo studies both suggest that Lef-1 deletion is not lethal to BCs. Based on our in vivo studies, Lef-1 deletion appears to have little impact on BCs or luminal cell behavior in the absence of injury (Figure 6). However, mixed WT and Lef-1cKO in vitro ALI cultures demonstrated an increase in Lef-1cKO BCs at 60-days post-seeding (Figure S2) despite a decline in Lef-1cKO ciliated and goblet cells (Figure 2). This phenotype diverges from what was observed in vivo, where Lef-1KO BC numbers declined following injury. Unlike the in vitro ALI culture studies, Lef-1cKO BCs and their luminal descendants had a survival disadvantage following injury in comparison to WT BCs (Figure 6). These differences may reflect the need for Lef-1cKO BCs to mature toward a unipotent state capable of proliferation, one that was afforded in ALI and proliferative cultures, but overshadowed in vivo by the more rapid proliferative advantage of WT BCs following injury. Taken together, these findings suggest that Lef-1 is required for BCs to maintain a multipotent state and that multiple BC subtypes may exist within mixed cultures of tracheal BCs with differential proliferative requirements for Lef-1. Indeed, others have observed functional heterogeneity in BCs using in vivo clonal analysis and the existence of unipotent BCs that cannot differentiate into luminal cells46–48. The functions of unipotent BCs remains to be determined, but could serve a structural role in airway maintenance and repair.
5 |. CONCLUSIONS
In summary, our findings demonstrate that Lef-1 has a role in regulating the proliferation of BCs through the control of cell cycle progression (Figure 7). Using multiple cell systems, we show that Lef-1 deletion can alter the ability of multipotent BCs to self-renew and differentiate into luminal cells (Figure 7). While the direct targets of Lef-1 remain to be clearly defined and could involve transcriptional and post-transcriptional mechanisms, these studies implicate Lef-1 targets involved in the G1/S DNA damage checkpoint. Thus, modulating Lef-1 function in airway BCs may have applications in regenerative medicine.
Figure 7. Lef-1 regulates BC function through the transcriptional control of cell cycle progression and DNA damage response genes.
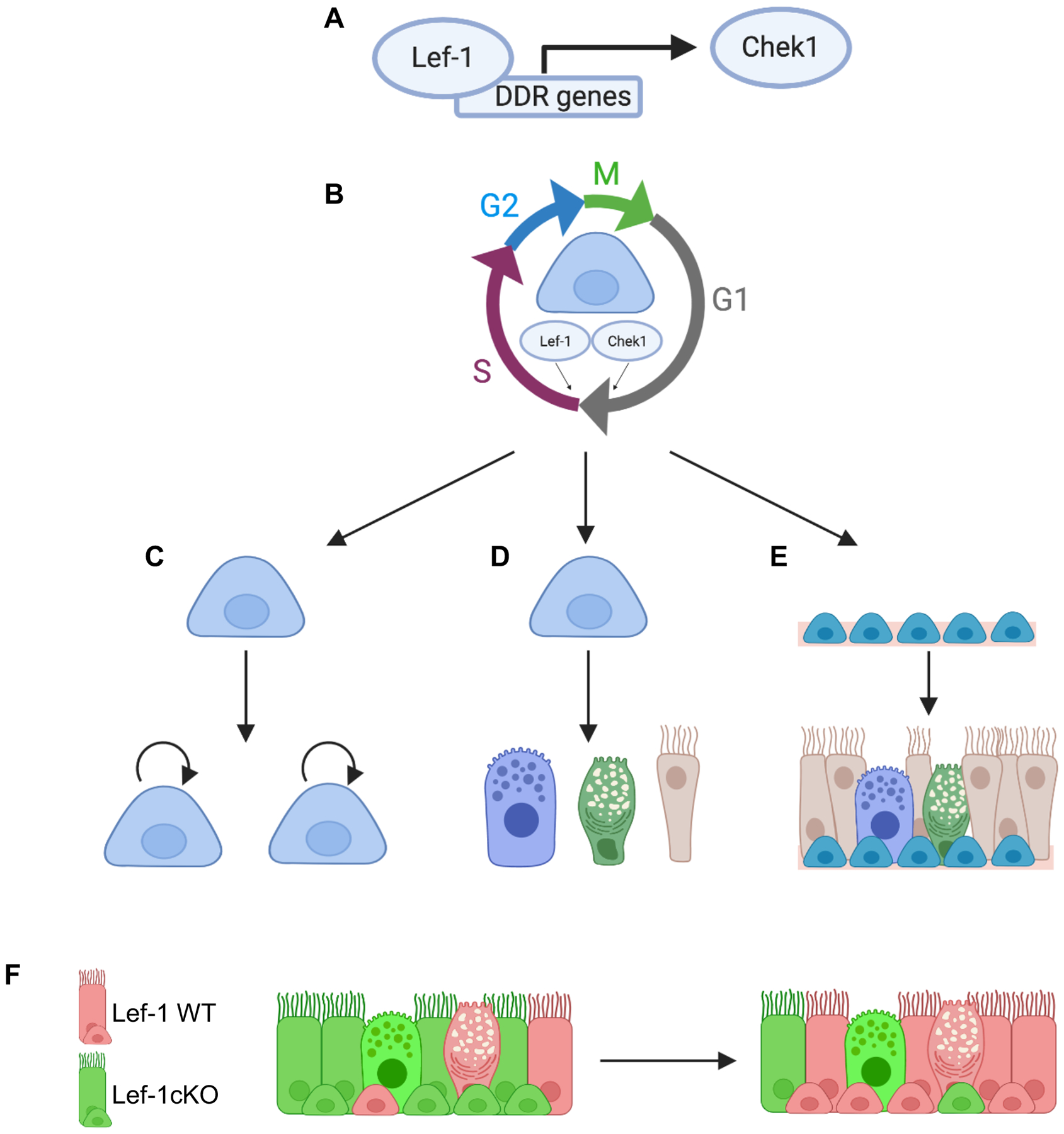
(A) Lef-1 expression in BCs regulates genes involved in DNA damage response (DDR)/DNA replication, as well as genes involved in cell cycle progression. Inhibition of Chek1 phenocopies Lef-1cKO and Lef-1cKO BCs that recovery the ability to proliferate are resistant to Chek1 inhibition. Thus, Chek1 is a candidate Lef-1-dependent factor controlling cell cycle progression in BCs. (B) Working model whereby Lef-1 and Chek1 regulate BC progression through G1/S. (C-E) Lef-1 enables BCs to (C) self-renew, (D) differentiate into multiple luminal cell lineages, and (E) regenerate airway epithelium lost to injury. (F) Lef-1cKO BCs are impaired in these capacities and outcompeted by Lef-1WT BCs in the regenerating epithelium. Figure was created using BioRender.
Supplementary Material
Supplemental Figure S1. Lef-1 is transiently expressed in a small subset of BCs. (A-C) In silico analysis of Lef-1 expression in various lung epithelial cell types. Data analysis and illustration was done using the Lung Cell Atlas interactive exploration tool (www.lungcellatlas.org). (A) tSNE plot of the various lung epithelial cell types identified in human lungs by single-cell RNA sequencing. (B) The same tSNE plot from A overlayed with a heatmap of Lef-1 expression. (C) Enrichment Z-scores of Lef-1 expression, comparing lung conducting airway epithelial cell types with parenchymal cell types. (D-G) Fluorescent in situ hybridization for Lef-1 in Lef-1cKO basal cells (BCs) revealing infrequent cells positive for Lef-1 expression. Lef-1cKO BCs were treated with either ethanol (EtOH) (D,E), or OH-Tam to induce Lef-1 deletion (F,G). Cells were pulsed with 5-ethynyl-2’-deoxyuridine (EdU) 2 hours prior to fixing in order to determine if Lef-1 was upregulated in cells going through S-phase. Dotted boxes indicate the magnified regions shown in d and e. Scale bars, 25 μm in D-G and 15 μm in d,e.
Supplemental Figure S2. Lef-1cKO BCs persist long-term in ALI culture. (A-C) Representative images of cross-sections taken from (A) 15, (B) 30, and (60) day old mixed ALI cultures detailed in Figure 2. Sections were stained for tdTomato, GFP, and Krt5 (BC marker). White arrowheads indicate GFP+ Krt5+ cells. Scale bars, 50 μm. (D-F) Quantifications of WT (tdTomato+) and Lef-1cKO (GFP+) BCs in the images represented in A-C. Graphs show means +/− SEM (N ≥ 4 transwells/time point). Asterisks indicate statistical significance by unpaired two-tailed Student’s T-test (**p < 0.01, *** p < 0.001).
Supplemental Figure S3. BCs that recover from a Lef-1-dependent block in proliferation are resistant to Chek1 inhibition. Lef-1cKO BCs that had been passaged 5 times following OH-Tam treatment were treated with the indicated chemical inhibitors (SCH 900776–Chek1 inhibitor; B02–Rad51 inhibitor). Cultures were monitored over a period of 6 days following seeding and phase contrast images were taken on the last day. Cultures were maintained in their respective inhibitors until the end of the assay. Scale bars, 30 μm.
Supplemental Figure S4. Upstream analysis reveals that TGFβ signaling is activated in Lef-1cKO basal cells, while signaling linked to cell cycle progression and proliferation is inhibited. (A and B) IPA software was used to conduct Upstream Analysis on differentially expressed genes in Lef-1cKO BCs. Analysis predicted that several key signaling regulators associated with cell cycle progression and proliferation were predicted to be inhibited (A), while signaling regulators associated with differentiation of BCs (TGFβ and SMAD) were predicted to be activated by Lef-1 deletion (B). (C-E) Z-scores were used to produce heatmaps depicting genes differentially expressed in Lef-1cKO BCs linked to TGFβ1 signaling (C), SMAD3 signaling (D), and SMAD4 signaling (E). Z-scores from 6, 18, and 30 hours post OH-Tam treatment are depicted.
SIGNIFICANCE STATEMENT.
Airway basal cells (BCs) are the primary progenitor cells of the conducting airway epithelium and thus are promising targets for durable genetic- and cell-based therapies of lung diseases such as cystic fibrosis. However, the mechanisms that govern their regenerative abilities are not fully understood. We show that Lef-1, an effector of Wnt signaling, is critical for BC cell cycle progression through G1/S and influences BC differentiation in vitro. Loss of Lef-1 significantly reduces the capacity of BC to regenerate the tracheal epithelium in vivo following injury. Manipulating Lef-1 expression in BCs may provide opportunities to modulate the regenerative capacity of BCs and their progeny.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health P30 DK054759, P01 HL152960, R01 DK047967 to JFE, and a grant from the Cystic Fibrosis Foundation to JFE. RNA-seq data presented herein were obtained at the Genomics Division of the Iowa Institute of Human Genetics which is supported, in part, by the University of Iowa Carver College of Medicine.
Footnotes
CONFLICT OF INTEREST
The authors declared no potential conflicts of interest.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author.
REFERENCES:
- 1.Fuchs E, Tumbar T, Guasch G. Socializing with the Neighbors: Stem Cells and Their Niche. Cell. 2004;116:769–778. [DOI] [PubMed] [Google Scholar]
- 2.Hsu Y-C, Pasolli HA, Fuchs E. Dynamics between Stem Cells, Niche, and Progeny in the Hair Follicle. Cell. 2011;144:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung M-I, Bujnis M, Barkauskas CE, et al. Niche-mediated BMP/SMAD signaling regulates lung alveolar stem cell proliferation and differentiation. Development. 2018;145:dev163014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shtutman M, Zhurinsky J, Simcha I, et al. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci U S A. 1999;96:5522–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao X, Mi Y, Ma Y, et al. LEF1 regulates glioblastoma cell proliferation, migration, invasion, and cancer stem-like cell self-renewal. Tumor Biology. 2014;35:11505–11511. [DOI] [PubMed] [Google Scholar]
- 6.Clevers H Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. [DOI] [PubMed] [Google Scholar]
- 7.Liu X, Driskell RR, Luo M, et al. Characterization of Lef-1 promoter segments that facilitate inductive developmental expression in skin. J Invest Dermatol. 2004;123:264–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boras-Granic K, Chang H, Grosschedl R, et al. Lef1 is required for the transition of Wnt signaling from mesenchymal to epithelial cells in the mouse embryonic mammary gland. Dev Biol. 2006;295:219–231. [DOI] [PubMed] [Google Scholar]
- 9.Sun Z, Yu W, Sanz Navarro M, et al. Sox2 and Lef-1 interact with Pitx2 to regulate incisor development and stem cell renewal. Development. 2016;143:4115–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan D, Yue Y, Zhou W, et al. Submucosal gland development in the airway is controlled by lymphoid enhancer binding factor 1 (LEF1). Development. 1999;126:4441–4453. [DOI] [PubMed] [Google Scholar]
- 11.Reya T, Duncan AW, Ailles L, et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. [DOI] [PubMed] [Google Scholar]
- 12.Duncan AW, Rattis FM, DiMascio LN, et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nature Immunology. 2005;6:314–322. [DOI] [PubMed] [Google Scholar]
- 13.Adam RC, Yang H, Ge Y, et al. Temporal Layering of Signaling Effectors Drives Chromatin Remodeling during Hair Follicle Stem Cell Lineage Progression. Cell stem cell. 2018;22:398–413.e397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bucan V, Mandel K, Bertram C, et al. LEF-1 regulates proliferation and MMP-7 transcription in breast cancer cells. Genes Cells. 2012;17:559–567. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh TH, Hsu CY, Tsai CF, et al. A novel cell-penetrating peptide suppresses breast tumorigenesis by inhibiting beta-catenin/LEF-1 signaling. Sci Rep. 2016;6:19156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zoico E, Darra E, Rizzatti V, et al. Role of adipose tissue in melanoma cancer microenvironment and progression. Int J Obes (Lond). 2018;42:344–352. [DOI] [PubMed] [Google Scholar]
- 17.Hogan BL, Barkauskas CE, Chapman HA, et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell stem cell. 2014;15:123–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lynch TJ, Engelhardt JF. Progenitor cells in proximal airway epithelial development and regeneration. Journal of cellular biochemistry. 2014;115:1637–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Goss AM, Cohen ED, et al. A Gata6-Wnt pathway required for epithelial stem cell development and airway regeneration. Nat Genet. 2008;40:862–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynch TJ, Anderson PJ, Xie W, et al. Wnt Signaling Regulates Airway Epithelial Stem Cells in Adult Murine Submucosal Glands. Stem cells (Dayton, Ohio). 2016;34:2758–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rock JR, Onaitis MW, Rawlins EL, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proceedings of the National Academy of Sciences. 2009;106:12771–12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lynch TJ, Anderson PJ, Rotti PG, et al. Submucosal Gland Myoepithelial Cells Are Reserve Stem Cells That Can Regenerate Mouse Tracheal Epithelium. Cell stem cell. 2018;22:779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tata A, Kobayashi Y, Chow RD, et al. Myoepithelial Cells of Submucosal Glands Can Function as Reserve Stem Cells to Regenerate Airways after Injury. Cell stem cell. 2018;22:668–683.e666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vieira Braga FA, Kar G, Berg M, et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nature Medicine. 2019;25:1153–1163. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Y, Tao L, Yi J, et al. The Role of Canonical Wnt Signaling in Regulating Radioresistance. Cell Physiol Biochem. 2018;48:419–432. [DOI] [PubMed] [Google Scholar]
- 26.Downward J Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. [DOI] [PubMed] [Google Scholar]
- 27.Shang D, Bi R, Han T, et al. Expression and proliferation-promoting role of lymphoid enhancer-binding factor 1 in human clear cell renal carcinoma. Cancer Invest. 2014;32:368–374. [DOI] [PubMed] [Google Scholar]
- 28.Bleckmann A, Siam L, Klemm F, et al. Nuclear LEF1/TCF4 correlate with poor prognosis but not with nuclear β-catenin in cerebral metastasis of lung adenocarcinomas. Clin Exp Metastasis. 2013;30:471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milyavsky M, Gan OI, Trottier M, et al. A Distinctive DNA Damage Response in Human Hematopoietic Stem Cells Reveals an Apoptosis-Independent Role for p53 in Self-Renewal. Cell stem cell. 2010;7:186–197. [DOI] [PubMed] [Google Scholar]
- 30.Simonatto M, Latella L, Puri PL. DNA damage and cellular differentiation: more questions than responses. J Cell Physiol. 2007;213:642–648. [DOI] [PubMed] [Google Scholar]
- 31.Yoon SW, Kim DK, Kim KP, et al. Rad51 regulates cell cycle progression by preserving G2/M transition in mouse embryonic stem cells. Stem Cells Dev. 2014;23:2700–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ullah Z, de Renty C, DePamphilis ML. Checkpoint kinase 1 prevents cell cycle exit linked to terminal cell differentiation. Mol Cell Biol. 2011;31:4129–4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang S, El-Deiry WS. p73 or p53 directly regulates human p53 transcription to maintain cell cycle checkpoints. Cancer Res. 2006;66:6982–6989. [DOI] [PubMed] [Google Scholar]
- 34.Deng R, Tang J, Ma JG, et al. PKB/Akt promotes DSB repair in cancer cells through upregulating Mre11 expression following ionizing radiation. Oncogene. 2011;30:944–955. [DOI] [PubMed] [Google Scholar]
- 35.Rock Jason R, Gao X, Xue Y, et al. Notch-Dependent Differentiation of Adult Airway Basal Stem Cells. Cell stem cell. 2011;8:639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao X, Bali AS, Randell SH, et al. GRHL2 coordinates regeneration of a polarized mucociliary epithelium from basal stem cells. The Journal of cell biology. 2015;211:669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tadokoro T, Gao X, Hong CC, et al. BMP signaling and cellular dynamics during regeneration of airway epithelium from basal progenitors. Development. 2016;143:764–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mou H, Vinarsky V, Tata PR, et al. Dual SMAD Signaling Inhibition Enables Long-Term Expansion of Diverse Epithelial Basal Cells. Cell stem cell. 2016;19:217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DiRenzo DM, Chaudhary MA, Shi X, et al. A crosstalk between TGF-β/Smad3 and Wnt/β-catenin pathways promotes vascular smooth muscle cell proliferation. Cellular signalling. 2016;28:498–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vallée A, Lecarpentier Y, Guillevin R, et al. Interactions between TGF-β1, canonical WNT/β-catenin pathway and PPAR γ in radiation-induced fibrosis. Oncotarget. 2017;8:90579–90604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voorneveld PW, Kodach LL, Jacobs RJ, et al. The BMP pathway either enhances or inhibits the Wnt pathway depending on the SMAD4 and p53 status in CRC. Br J Cancer. 2015;112:122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Labbe E, Letamendia A, Attisano L. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-beta and wnt pathways. Proc Natl Acad Sci U S A. 2000;97:8358–8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He T-C, Sparks AB, Rago C, et al. Identification of c-MYC as a Target of the APC Pathway. Science. 1998;281:1509–1512. [DOI] [PubMed] [Google Scholar]
- 44.Katoh M, Katoh M. WNT Signaling Pathway and Stem Cell Signaling Network. Clinical Cancer Research. 2007;13:4042–4045. [DOI] [PubMed] [Google Scholar]
- 45.Gerner-Mauro KN, Akiyama H, Chen J. Redundant and additive functions of the four Lef/Tcf transcription factors in lung epithelial progenitors. Proceedings of the National Academy of Sciences. 2020;117:12182–12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hong KU, Reynolds SD, Watkins S, et al. In vivo differentiation potential of tracheal basal cells: evidence for multipotent and unipotent subpopulations. American journal of physiology. Lung cellular and molecular physiology. 2004;286:L643–649. [DOI] [PubMed] [Google Scholar]
- 47.Ghosh M, Brechbuhl HM, Smith RW, et al. Context-Dependent Differentiation of Multipotential Keratin 14–Expressing Tracheal Basal Cells. American journal of respiratory cell and molecular biology. 2011;45:403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghosh M, Smith RW, Runkle CM, et al. Regulation of trachebronchial tissue-specific stem cell pool size. Stem cells (Dayton, Ohio). 2013;31:2767–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. Lef-1 is transiently expressed in a small subset of BCs. (A-C) In silico analysis of Lef-1 expression in various lung epithelial cell types. Data analysis and illustration was done using the Lung Cell Atlas interactive exploration tool (www.lungcellatlas.org). (A) tSNE plot of the various lung epithelial cell types identified in human lungs by single-cell RNA sequencing. (B) The same tSNE plot from A overlayed with a heatmap of Lef-1 expression. (C) Enrichment Z-scores of Lef-1 expression, comparing lung conducting airway epithelial cell types with parenchymal cell types. (D-G) Fluorescent in situ hybridization for Lef-1 in Lef-1cKO basal cells (BCs) revealing infrequent cells positive for Lef-1 expression. Lef-1cKO BCs were treated with either ethanol (EtOH) (D,E), or OH-Tam to induce Lef-1 deletion (F,G). Cells were pulsed with 5-ethynyl-2’-deoxyuridine (EdU) 2 hours prior to fixing in order to determine if Lef-1 was upregulated in cells going through S-phase. Dotted boxes indicate the magnified regions shown in d and e. Scale bars, 25 μm in D-G and 15 μm in d,e.
Supplemental Figure S2. Lef-1cKO BCs persist long-term in ALI culture. (A-C) Representative images of cross-sections taken from (A) 15, (B) 30, and (60) day old mixed ALI cultures detailed in Figure 2. Sections were stained for tdTomato, GFP, and Krt5 (BC marker). White arrowheads indicate GFP+ Krt5+ cells. Scale bars, 50 μm. (D-F) Quantifications of WT (tdTomato+) and Lef-1cKO (GFP+) BCs in the images represented in A-C. Graphs show means +/− SEM (N ≥ 4 transwells/time point). Asterisks indicate statistical significance by unpaired two-tailed Student’s T-test (**p < 0.01, *** p < 0.001).
Supplemental Figure S3. BCs that recover from a Lef-1-dependent block in proliferation are resistant to Chek1 inhibition. Lef-1cKO BCs that had been passaged 5 times following OH-Tam treatment were treated with the indicated chemical inhibitors (SCH 900776–Chek1 inhibitor; B02–Rad51 inhibitor). Cultures were monitored over a period of 6 days following seeding and phase contrast images were taken on the last day. Cultures were maintained in their respective inhibitors until the end of the assay. Scale bars, 30 μm.
Supplemental Figure S4. Upstream analysis reveals that TGFβ signaling is activated in Lef-1cKO basal cells, while signaling linked to cell cycle progression and proliferation is inhibited. (A and B) IPA software was used to conduct Upstream Analysis on differentially expressed genes in Lef-1cKO BCs. Analysis predicted that several key signaling regulators associated with cell cycle progression and proliferation were predicted to be inhibited (A), while signaling regulators associated with differentiation of BCs (TGFβ and SMAD) were predicted to be activated by Lef-1 deletion (B). (C-E) Z-scores were used to produce heatmaps depicting genes differentially expressed in Lef-1cKO BCs linked to TGFβ1 signaling (C), SMAD3 signaling (D), and SMAD4 signaling (E). Z-scores from 6, 18, and 30 hours post OH-Tam treatment are depicted.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.


