Abstract
Background:
Identification of membrane proteins differentially expressed on tumor cells is a key step in drug development. The carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) is a cell adhesion protein belonging to the immunoglobulin superfamily. Here, we explore the prognostic role CEACAM6 expression on patient outcome in cancer.
Methods:
A systematic search for studies evaluating the association between tumor expression of CEACAM6 and overall survival (OS) and disease-free survival (DFS) was performed. Hazard ratios (HR) were pooled in a meta-analysis using generic inverse variance and random effect modeling. Subgroup analyses were conducted based on tumor type and method of HR extraction.
Results:
Sixteen studies met the inclusion criteria. CEACAM6 expression was associated with worse OS [HR = 1.96, 95% confidence interval (CI) = 1.51–2.53], and DFS (HR = 2.49, 95% CI = 2.01–3.07) with subgroup analysis showing no significant differences between disease site subgroups.
Conclusions:
High expression of CEACAM6 is associated with worse OS and DFS in different malignancies. CEACAM6 is a target for the future development of novel therapeutics.
Keywords: CEACAM6, disease outcome, immunotherapy, meta-analysis
Background
Cancer immunotherapy has gained momentum with the development of immune checkpoint inhibitors. 1 Inhibition of PD1 or PD-L1 has shown clinical benefit in different tumor types. 2 Inhibition of these pathways which suppress the immune response and maintain immunologic tolerance plays a key role in cancer treatment. In addition, other coinhibitory checkpoints like Lymphocyte Activating 3 (LAG3), T-cell immunoreceptor with Ig and ITIM domains (TIGIT), B- and T-lymphocyte attenuator (BTLA), or T-cell immunoglobulin domain and mucin domain 3 (TIM3) have been considered as appropriate targets. Agents against these targets are currently in clinical development with early signs of clinical activity. 3 Most of these strategies either block inhibitory signals or stimulate activating receptors such as OX40, with the goal to augment the immune response through the activation of the innate or adaptive immunity. 4 Indeed, most of these targets are membrane receptors either expressed in immune cells or in tumor cells, so antibodies can easily bind to these proteins. Other mechanisms to take advantage of the immune system in order to target tumor cells have reached the clinical setting. These include vaccines or cell therapy, among others. 1 For any of the previously described treatments, identification of proteins differentially expressed on tumor cells including those on the membrane is a necessary step in drug development.
To avoid the immune response, tumor cells can change the degree of expression of specific antigens on the cell surface with the main objective to repress the activation of immune cells that would otherwise recognize and attack the transformed cells. One of those membrane proteins is the carcinoembryonic antigen-related cell adhesion molecule (CEACAM) family, which are highly glycosylated proteins from the immunoglobulin superfamily. 5 They comprise 12 proteins capable of homo- or hetero-dimerization with other CEACAM members 6 or to other membrane proteins such as integrins. 7 They play a role in nontransformed tissues, from phagocytosis or signal transduction, to regulation of cell–cell recognition and adhesion. 8 Moreover, they have been described as bacterial pathogen receptors, 9 and their implication in proliferation, invasiveness, or apoptotic resistance has been described in relation to the oncogenic processes. 10 In addition, these proteins have been proposed as immunotherapy targets in different tumors. 11
CEACAM6 is a member of the carcinoembryonic antigen molecules, widely distributed in epithelial and myeloid cells. 12 It acts as an intercellular cell adhesion molecule to maintain tissue architecture through interactions with other CEACAM proteins. 13 It has been described as a modulator of cancer progression due to its effects on differentiation and cell growth, resistance to anoikis, treatment resistance, invasiveness and metastasis. 14 Indeed, CEACAM6 is upregulated preferentially at the apical/luminal membranes of many tumors. 12 This effect was first observed in leukemia 15 and subsequently in colorectal, 16 pancreatic, 17 gastric, 18 lung, 19 and many others.20–23
Recently, CEACAM6 has been suggested as a target for different cancer immunotherapies given the fact that its membrane expression is highly specific of tumor cells. 11 In models of non-small cell lung cancer, different antibodies against CEACAM6 block cell viability, invasion, and migration through inhibition of Src/FAK phosphorylation. 24 In models of pancreatic adenocarcinoma, the use of monoclonal antibodies (mAb) against this protein sensitizes cells to cytotoxicity using secondary antibodies. 25 A different immunotherapy strategy is the development of humanized single-chain antibody variable fragments (scFv) based on CEACAM6, 26 or specific CEACAM6-single domain antibodies (sdAB) that have demonstrated to inhibit cell growth. 27 CEACAM6 has also been used to create antibody–drug conjugates (ADC), using a tubulin inhibitor as a payload, showing preclinical efficacy in pancreatic cancer. 28
Beyond the involvement of CEACAM6 in invasion and metastasis, its role as an immune regulator has led to the development of antibodies that inhibit CEACAM6 with the aim to boost the immune response. CEACAM6 has been considered as a novel immune checkpoint, as its inhibition activates the T-cell response. An increase in CEACAM6 expression in multiple myeloma inhibited cytotoxic T-cell reactivity, therefore reducing the immune effect against tumor cells, and treatment with anti-CEACAM6 monoclonal antibodies augmented the T CD8+ activity against malignant cells. 29
In this context, CEACAM6 has been proposed as a predictor of survival and recurrence in different tumors given its involvement in cancer transformation and the new role in immune regulation. 10 In this article, we report a systematic review and meta-analysis of the prognostic association of CEACAM6 overexpression with outcome in various malignancies. We hypothesize that CEACAM6 overexpression is associated with a worse prognosis.
Methods
This systematic review and meta-analysis was performed in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) 30 and was conducted following the Cochrane Handbook for Systematic Reviews of Interventions recommendations. 31 This study was registered in PROSPERO (registration number: CRD42021266217)
Search strategy
The systematic search of the studies was carried out using Cochrane Central Register of Controlled Trials, MEDLINE (Host: PubMed), Scopus, and Web of Science from inception to 11 May 2021. The following keywords were used: ‘CEACAM6’ OR ‘CD66c’ OR ‘CEAL’ OR ‘NCA’ OR ‘KOR-SA3544 antigen’ OR ‘NCA-50/90’ OR ‘carcinoembryonic antigen-related cell adhesion molecule 6’ OR ‘Carcinoembryonic Antigen Related Cell Adhesion Molecule 6’ OR ‘Normal Cross-Reacting Antigen’ OR ‘Cluster Of Differentiation 66c’ OR ‘CD66c Antigen’ OR ‘Normal Cross-Reacting Antigen’ OR ‘Carcinoembryonic Antigen-Related Cell Adhesion Molecule 6 (Non-Specific Cross Reacting Antigen)’ OR ‘Non-Specific Crossreacting Antigen’ AND ‘CANCER’. To improve the sensitivity of the search strategy, we reviewed citation lists of included articles, as well as previous systematic reviews or meta-analyses. The full electronic search strategy is detailed in the Supplemental Figure ‘electronic search strategy’.
Study selection
Eligibility studies included (1) studies of humans (adults and children); (2) patients with hematological or solid tumors; (3) reporting of a hazard ratio (HR) for overall survival (OS) and/or disease-free survival (DFS; defined as the length of time from primary treatment of an early-stage cancer to death or any signs or symptoms of recurrent cancer) or survival curves allowing estimation of the HR for OS or DFS; (4) English language publication. Case reports, conference abstracts, and letters to editors were excluded. The titles identified by the initial search were evaluated, and potentially relevant publications were retrieved in full. Two authors (MB and EMGM) independently reviewed the full articles for eligibility. Disagreements were resolved by consensus.
Data extraction and risk of bias assessment
The following data were extracted: name of first author, year of publication, tumor type, detection method, agent used, cutoff to define positive expression, sample size, percentage of positive CEACAM6, and outcome.
The outcomes of interest were OS and DFS in patients both with and without CEACAM6 expression as defined by individual studies. The hazard ratio (HR) for OS was extracted whenever available. In cases where the HR was not reported, it was estimated from survival curves using the methods described by Parmar et al. 32 We applied a hierarchal approach to the collection of HRs, preferring those reported from multivariable analyses to univariable HR, and preferring both over HRs estimated from survival plots.
The Quality in Prognosis Studies (QUIPS) tool was used to evaluate the risk of bias in six domains: study participation (sampling bias), study attrition (attrition bias), prognostic factor measurement, outcome measurement (ascertainment bias), confounding measurement and accounting and analysis and reporting. Studies were considered to have a low, moderate, or high risk of bias, if they satisfied five to six, three to four, or one to two of the six domains, respectively. 33
Data extraction was conducted by two independent reviewers (MB and EMGM), and quality assessment was conducted by two independent reviewers (ICR and CAB). Disagreements were solved by consensus or with discussion with a third reviewer (EA)
Data synthesis and statistical analysis
Data were reported descriptively where appropriate. Extracted data were pooled using RevMan 5.4 analysis software (Cochrane Collaboration, Copenhagen, Denmark). In light of substantial clinical heterogeneity (e.g. different tumor types), estimates for HRs were pooled and weighted by generic inverse variance and computed by random effect modeling irrespective of statistical heterogeneity. Statistical heterogeneity was reported using the Cochran’s Q. Inconsistence was estimated using I2 which was considered not important (<30%), moderate (30–50%), substantial (50–75%), or considerable (>75%). In addition, the corresponding p values for Cochran’s Q and I2 statistics were considered. Subgroup analyses were performed for different disease sites and to compare studies with low risk of bias with those with moderate risk of bias. Differences between the subgroups were assessed using methods described by Deeks et al. 34 Sensitivity analysis was performed to exclude studies which assessed CEACAM6 by methods other than immunohistochemistry. Publication bias was assessed using visual inspection of the funnel plot for the most commonly reported outcome (OS). All of the statistical tests were two-sided, and statistical significance was defined as p < 0.05. No corrections were applied for multiple statistical testing.
Results
Selection of studies
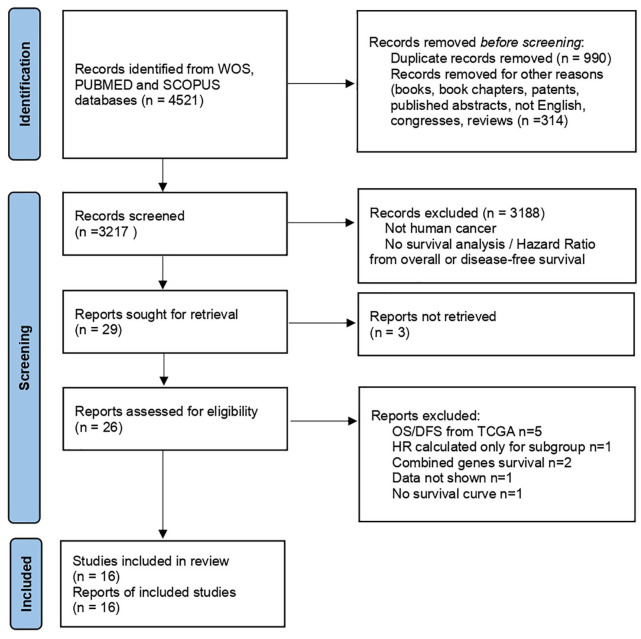
Our search identified 26 articles from which 16 were included in the review. The other 10 studies were excluded due to different reasons: exclusively use of genomic data from the TCGA database,35–39 data limited to a specific cancer subgroup, 40 data from combined analysis of different genes,41,42 or studies not reporting survival information.43,44 Eligible studies included in the analysis were retrospective studies published between 2003 and 2020 and comprised 2441 patients.17,21,45–58 Figure 1 shows the study selection schema. CEACAM6 was reported as expressed in 1,535 patients (65%). The characteristics of the included studies are shown in Table 1. For OS, three studies evaluated pancreatic cancer, four gastric cancer, two colorectal cancer, and one each for lung adenocarcinoma, breast cancer, gallbladder cancer, and osteosarcoma. For DFS, two studies evaluated colorectal cancer, and one each for lung adenocarcinoma, breast cancer, intrahepatic cholangiocarcinoma, acute lymphoblastic leukemia (ALL), and gastric cancer.
Figure 1.
PRISMA of the study selection process for CEACAM6.
Table 1.
Characteristics of the studies.
| Tumor | Detection method | Agent used | Positive cutoff | n | % CEACAM6 + | |
|---|---|---|---|---|---|---|
| Chen et al. 48 | Pancreatic | IHC | ab: monoclonal (Abcam) | positive | 99 | 91 |
| Duxbury et al. 17 | Pancreatic | IHC | ab: monoclonal By114 (Imgenex) | positive staining > 3 (0–3) | 89 | 91 |
| Gebauer et al. 49 | Pancreatic | IHC | ab: clone 9A6 (Sigma) | 2+ in > 70% cells, +3 > 30% cells (0,1+, 2+, 3+ intensity) | 137 | 72 |
| Deng et al. 50 | Gastric | IHC | ab: polyclonal (Sigma) | positive staining > 2 (0–3) | 75 | 69 |
| Roy et al. 51 | Gastric | IHC | ab: clone9A6 (abcam) | median | 106 | 50 |
| Ru et al. 52 | Gastric | IHC | ab: (abcam) | positive reviewed by independent pathologists | 436 | 51 |
| Zang et al. 46 | Gastric | IHC | ab: (abcam) | positive staining > 3 (0–3) | 160 | 59 |
| Jantscheff et al. 53 | Colorectal | IHC | ab: mab 13H10 | positive staining > 2 (0–3) | 243 | 69 |
| Kim et al. 45 | Colorectal | IHC | ab: (R&D) | median | 143 | 54 |
| Han et al. 54 | Lung adenocarcinoma | IHC | ab: 9A6 (Santa cruz) | positive staining > 2 (0–3) | 51 | 86 |
| Kobayashi et al. 55 | Lung adenocarcinoma | IHC | ab: polyclonal (Aviva Systems Biology) | >40% positive carcinoma cells | 115 | 46 |
| Ieta et al. 21 | Intrahepatic cholangiocarcinoma | RT-PCR | Specific primers | CEACAM6/GAPDH < 2 relative mRNA expression | 23 | 57 |
| Kalina et al. 56 | Acute lymphoblastic leukemia | Flow cytometry | ab: KOR-SA3544 labeled to FITC (Immunotech) | >20% | 254 | 43 |
| Maraqa et al. 47 | Breast | IHC | ab: clone9A6 (abcam) | positive staining > 1 (0–3) | 351 | 81 |
| Tian et al. 57 | Gall bladder | IHC | ab: ab78029 (Abcam) | positive cell ratio > 50% | 68 | 71 |
| Wang et al. 58 | Osteosarcoma | IHC | ab: ab154614 (Abcam) | Staining index (SI) score, >6 (0–12) | 91 | 52 |
CEACAM6, carcinoembryonic antigen-related cell adhesion molecule 6; FITC, fluorescein-5-isothiocyanate; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IHC, immunohistochemistry; RT-PCR, reverse transcription polymerase chain reaction; SI, staining index.
Risk of bias
The risk of bias, assessed using the QUIPS tool, was low for 10 articles and moderate for 6 (Table S1). The funnel plot was generally symmetrical suggesting low risk of publication bias.
Overall survival
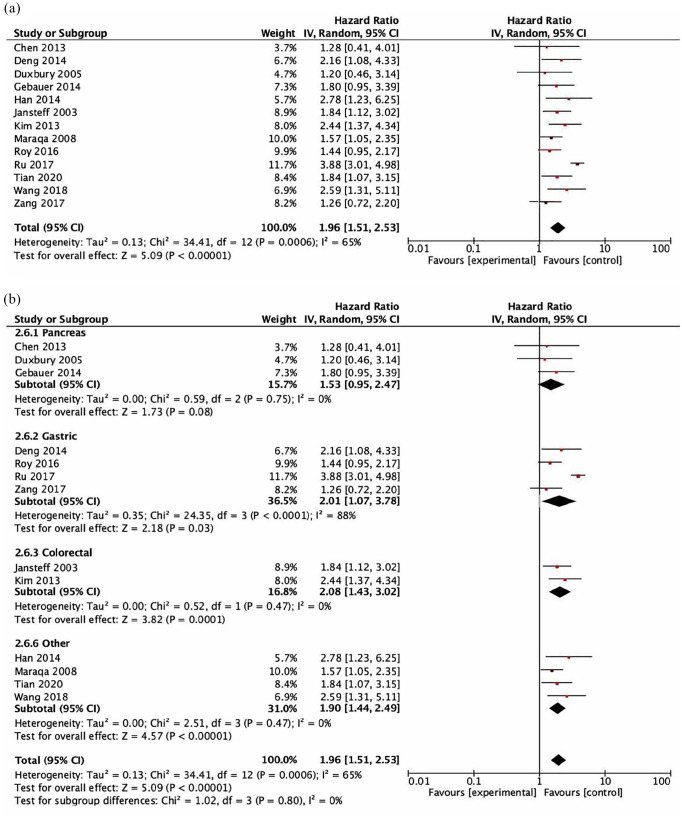
Data for the association between CEACAM6 and OS were reported or calculated in eight studies. All studies utilized immunohistochemistry to assess CEACAM6. CEACAM6 expression was associated with worse OS [HR = 1.96, 95% confidence interval (CI) = 1.51–2.53, p < 0.001, see Figure 2(a)]. Heterogeneity was significant (Cochran Q p < 0.001, I 2 = 65%). Subsequent subgroup analyses demonstrated that there were no significant differences between cancer type subgroups (subgroup difference p = 0.8, Figure 2(b)) or between the methods used for the calculation of HR between subgroups (subgroup difference p = 0.91, Supplemental Figure 1). In addition, study quality did not impact on results. There was no difference in the estimate for the association between CEACAM6 and OS for studies with a low risk of bias (HR = 2.13, 95% CI = 1.53–2.97) and those with a moderate risk of bias (HR = 1.65, 95% CI = 1.28–2.12; p for difference = 0.23).
Figure 2.
Forest plots showing hazard ratios for overall survival: CEACAM6 overall (a) and by subgroups based on disease site (b). Hazard ratios for each study are represented by squares: the size of the square represents the weight of the study in the meta-analysis; the horizontal line passing through the square represents the 95% confidence interval. All statistical tests were two-sided. The diamonds represent the estimated pooled effect. Test for overall effect based on z-test. All p values are two-sided. (a) CEACAM6 OS Overall. (b) CEACAM6 OS by disease site.
CI, confidence interval; OR, odds ratio.
Disease-free survival
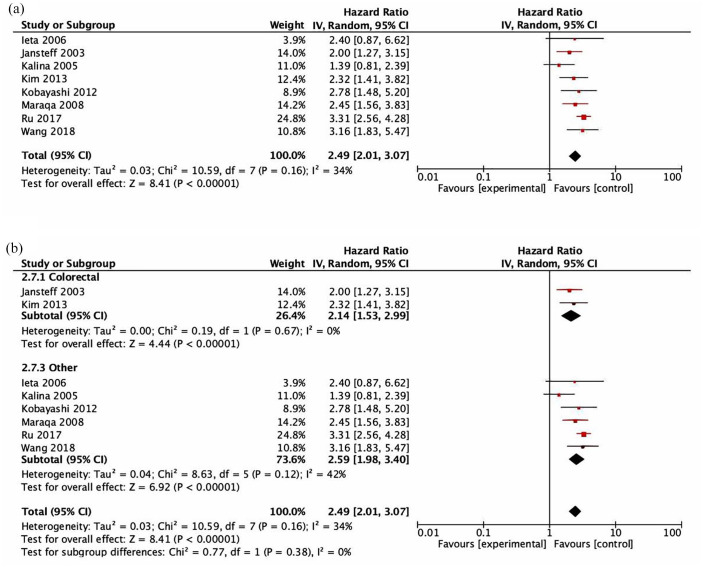
Data for the association between CEACAM6 and DFS were reported or calculated in eight studies. CEACAM6 expression was associated with detrimental DFS (HR = 2.49, 95% CI = 2.01–3.07, p < 0.001, see Figure 3(a)). The test for heterogeneity was not significant (Cochran Q p = 0.16, I2 = 34%). Sensitivity analysis excluding the two studies which did not use immunohistochemistry to assess DFS21,56 showed similar results (HR = 2.79, 95% CI = 2.35–3.30, p < 0.001). The subgroup analysis showed that there were no significant differences between cancer type subgroups (subgroup difference p = 0.38, Figure 3(b)) or between methods used for the HR calculation of the different subgroups (subgroup difference p = 0.61, Supplemental Figure 2). There was no difference in the estimate for the association between CEACAM6 and DFS for studies with a low risk of bias (HR = 2.75, 95% CI = 2.23–3.39) and those with a moderate risk of bias (HR = 2.22, 95% CI = 1.42–3.47; p for difference = 0.40).
Figure 3.
Forest plots showing hazard ratios for disease-free survival (DFS): CEACAM6 overall (a) and by subgroups based on disease site (b). Hazard ratios for each study are represented by squares: the size of the square represents the weight of the study in the meta-analysis; the horizontal line passing through the square represents the 95% confidence interval. All statistical tests were two-sided. The diamonds represent the estimated pooled effect. Test for overall effect based on z-test. All p values are two-sided. (a) CEACAM6 DFS overall. (b) CEACAM6 DFS by disease site.
CI, confidence interval; OR, odds ratio.
Discussion
In the present study, we describe the association of CEACAM6 expression with outcome in cancer by performing a meta-analysis of published data. Our results show a large association between the expression of this protein and detrimental survival across a wide range of malignancies.
CEACAM6, a carcinoembryonic antigen molecule, is a highly glycosylated protein from the immunoglobulin superfamily, principally expressed in the cellular membrane. 5 Its presence is observed in nontransformed tissues, and it has a key role in different functions from phagocytosis or signal transduction, to regulation of cell–cell recognition and adhesion. 8
Data have linked the expression of this protein with the oncogenic transformation at different levels, from differentiation and migration, to cell proliferation and survival. 14 Overall, the CEACAM family of proteins have been involved in several functions related to cancer, and some members have been proposed to act as tumor suppressors 59 and others as oncogenes. 60 CEACAM5 is the only member of the CEA family accepted as a tumor marker of recurrence in cancer patients, 13 and different clinical trials of chimeric antigen T cells (CAR-T) have used CEACAM5 as the target. 11 CEACAM6 has emerged as the most specific marker for a number of aggressive cancers, 10 as its expression is greater than CEACAM5 in many tumors. 20 CEACAM6 has been identified as a target candidate for CAR-T therapy 61 and recent data show efficacy of an anti-tumor CEACAM6 vaccine combined with PD-1 or PD-L1 inhibitors. 62 In addition, expression of CEACAM6 on cancer cells has been suggested as inhibitory of the immune response, and antibodies against CEACAM6 can boost the T-cell response, thereby potentially possessing activity consistent with checkpoint blockade. 29 In this context, a phase I clinical trial is ongoing using the anti-CEACAM6 antibody BAY1834942 in patients with advanced solid tumors. 63 Although this approach is novel, it shows great potential for clinical development especially if this family of antibodies can be combined with anti-PD-1 or PD-L1 inhibitors. 64
Our results show that expression of CEACAM6 was associated with worse OS and DFS with all studies reporting effect sizes consistent with adverse outcome (albeit not always with statistical significance in each study). This resulted in statistical heterogeneity for the OS, but not DFS analysis. For our OS analysis, pancreatic cancer studies less frequently observed statistically significant associations with worse outcome despite similar effect size as other disease sites. This is likely explained by low statistical power; two out of three of these studies analyzed survival data from very few CEACAM6 negative patients (<10% for study population). For our DFS analysis, studies in intrahepatic cholangiocarcinoma and acute lymphoblastic leukemia also did not identify statistically significant results for CEACAM6 and worse outcome. Of note, these studies did not use immunohistochemistry (IHC) for the evaluation of CEACAM6, but utilized reverse transcription polymerase chain reaction (RT-PCR) or flow cytometry, respectively. This may explain this observation. However, among other studies, no significant differences were observed between IHC and RT-PCR suggesting that method of evaluation is not a sensitive variable for outcome assessment. Similarly, we did not identify differences between tumor types, although the number of cancer subtypes included in the analysis was limited. In addition, we did not observe differences between the methods used for data extraction. Similar magnitude of effect was observed for studies in which data were extracted directly and those in which data were estimated from survival plots. Finally, when we evaluated the quality of the studies, we observed that most studies showed low risk for bias reinforcing the confidence in our results.
These analyses have several important implications. CEACAM6 is associated with worse OS and DFS, suggesting its potential use as a target for therapeutic intervention. As a membrane-bound protein, the application of different immunotherapy strategies is of great value, and development of effective antibodies against CEACAM6 for the treatment of different tumors is ongoing.24–29,61 In addition, it could be used as a prognostic biomarker and some studies have evaluated the prognostic value of CEACAM6 serum levels in patients with cancer.65–67 However, our study aimed to explore the prognostic role of CEACAM6 in human cancers, rather than in blood. With the potential role of CEACAM6 as immune checkpoint inhibitor in tumors, we focused this evaluation only on primary cancers. The prognostic effect of serum CEACAM6 may be an indirect measure of tumor volume and assessment of this independently of disease burden is not possible with study summary data. Future studies of blood CEACAM6 would be of value.
This analysis has limitations that are intrinsic to the type of data available and the analysis performed. First, as a literature-based analysis, individual patient data were not available. As such, the analysis would be subject to publication bias and relied on summary data. While inspection of the funnel plot did not suggest substantial publication bias, the risk of residual bias cannot be excluded. Second, there is no accepted and validated method for assessment of CEACAM6 expression. Therefore, there may be substantial heterogeneity, which may not be fully accounted for by our use of random-effects modeling. Finally, HR were not reported in every study and had to be estimated in a number of studies. While subgroup analysis did not suggest a difference between methods of HR extraction, this may have introduced further heterogeneity to the reported results.
In conclusion, our analyses show that overexpression of CEACAM6 is associated with a worse OS and DFS in different tumor types. Given its role in immune modulation, our data suggest that the development of strategies targeting this membrane protein could have potential for therapeutic benefit.
Supplemental Material
Supplemental material, sj-docx-1-tam-10.1177_17588359211072621 for Prognostic value of the immune target CEACAM6 in cancer: a meta-analysis by Miguel Burgos, Iván Cavero-Redondo, Celia Álvarez-Bueno, Eva María Galán-Moya, Atanasio Pandiella, Eitan Amir and Alberto Ocaña in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-2-tam-10.1177_17588359211072621 for Prognostic value of the immune target CEACAM6 in cancer: a meta-analysis by Miguel Burgos, Iván Cavero-Redondo, Celia Álvarez-Bueno, Eva María Galán-Moya, Atanasio Pandiella, Eitan Amir and Alberto Ocaña in Therapeutic Advances in Medical Oncology
Supplemental material, sj-jpg-1-tam-10.1177_17588359211072621 for Prognostic value of the immune target CEACAM6 in cancer: a meta-analysis by Miguel Burgos, Iván Cavero-Redondo, Celia Álvarez-Bueno, Eva María Galán-Moya, Atanasio Pandiella, Eitan Amir and Alberto Ocaña in Therapeutic Advances in Medical Oncology
Supplemental material, sj-jpg-2-tam-10.1177_17588359211072621 for Prognostic value of the immune target CEACAM6 in cancer: a meta-analysis by Miguel Burgos, Iván Cavero-Redondo, Celia Álvarez-Bueno, Eva María Galán-Moya, Atanasio Pandiella, Eitan Amir and Alberto Ocaña in Therapeutic Advances in Medical Oncology
Footnotes
Author contributions: Miguel Burgos: Conceptualization; Formal analysis; Investigation; Methodology; Software; Validation; Visualization; Writing – original draft; Writing – review & editing.
Iván Cavero-Redondo: Methodology; Software; Supervision.
Celia Álvarez-Bueno: Methodology; Software.Eva María Galán-Moya: Formal analysis; Validation.
Atanasio Pandiella: Supervision.
Eitan Amir: Formal analysis; Investigation; Methodology; Supervision; Validation; Writing – original draft; Writing – review & editing.
Alberto Ocaña: Conceptualization; Resources; Supervision; Visualization; Writing – original draft; Writing – review & editing.
Conflict of interest statement: The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: AO is currently an employee of Symphogen, Copenhagen, Denmark. There is no conflict of interest to declare in relation to this article. EA declares person fees from Sandoz, Novartis and Exact Sciences outside of this work.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work has been supported by Instituto de Salud Carlos III (PI19/00808), ACEPAIN, Diputación de Albacete, CIBERONC and CRIS Cancer Foundation (to AO). MB is funded by University of Castilla-La Mancha (UCLM). EMGM holds a Distinguished Researcher contract from the UCLM. This work has been supported by Junta de Comunidades de Castilla-La Mancha (SBPLY/19/180501/ 000173) (to EMGM and AO).
ORCID iDs: Miguel Burgos  https://orcid.org/0000-0001-6674-0798
https://orcid.org/0000-0001-6674-0798
Eva María Galán-Moya  https://orcid.org/0000-0001-5758-7592
https://orcid.org/0000-0001-5758-7592
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Miguel Burgos, Translational Research Unit, Translational Oncology Laboratory, Albacete University Hospital, Albacete, SpainCentro Regional de Investigaciones Biomédicas, Castilla-La Mancha University (CRIB-UCLM), Albacete, SpainDepartment of Nutrition, Food Science and Physiology, School of Pharmacy and Nutrition, University of Navarra, Pamplona, Spain.
Iván Cavero-Redondo, Health and Social Care Research Center, Universidad de Castilla-La Mancha, Cuenca, SpainRehabilitation in Health Research Center (CIRES), Universidad de las Américas, Santiago, Chile.
Celia Álvarez-Bueno, Health and Social Care Research Center, Universidad de Castilla-La Mancha, Cuenca, Spain.
Eva María Galán-Moya, Centro Regional de Investigaciones Biomédicas, Castilla-La Mancha University (CRIB-UCLM), Albacete, SpainFaculty of Nursing, Castilla-La Mancha University (UCLM), Albacete, Spain.
Atanasio Pandiella, Instituto de Biología Molecular y Celular del Cáncer (IBMCC-CIC), Salamanca, SpainInstituto de Investigación Biomédica de Salamanca (IBSAL), Salamanca, SpainCIBERONC, Salamanca, SpainConsejo Superior de Investigaciones Científicas (CSIC), Salamanca, Spain.
Eitan Amir, Division of Medical Oncology and Hematology, Princess Margaret Cancer Centre, and Department of Medicine, University of Toronto, 610 University Avue, 700U, 7-721, Toronto, ON, M5G 2M9, Canada.
Alberto Ocaña, Hospital Clínico San Carlos and CIBERONC, 28040 Madrid, SpainCentro Regional de Investigaciones Biomédicas, Castilla-La Mancha University (CRIB-UCLM), Albacete, Spain.
References
- 1. Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science 2018; 359: 1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aldea M, Andre F, Marabelle A, et al. Overcoming resistance to tumor-targeted and immune-targeted therapies. Cancer Discov 2021; 11: 874–899. [DOI] [PubMed] [Google Scholar]
- 3. Lee JB, Ha S-J, Kim HR. Clinical insights into novel immune checkpoint inhibitors. Front Pharmacol 2021; 12: 681320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fu Y, Lin Q, Zhang Z, et al. Therapeutic strategies for the costimulatory molecule OX40 in T-cell-mediated immunity. Acta Pharm Sin B 2020; 10: 414–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Paxton RJ, Mooser G, Pande H, et al. Sequence analysis of carcinoembryonic antigen: identification of glycosylation sites and homology with the immunoglobulin supergene family. Proc Natl Acad Sci USA 1987; 84: 920–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oikawa S, Sugiyama M, Kuroki M, et al. Extracellular N-domain alone can mediate specific heterophilic adhesion between members of the carcinoembryonic antigen family, CEACAM6 and CEACAM8. Biochem Biophys Res Commun 2000; 278: 564–568. [DOI] [PubMed] [Google Scholar]
- 7. Camacho-Leal P, Zhai AB, Stanners CP. A co-clustering model involving alpha5beta1 integrin for the biological effects of GPI-anchored human carcinoembryonic antigen (CEA). J Cell Physiol 2007; 211: 791–802. [DOI] [PubMed] [Google Scholar]
- 8. Kuespert K, Pils S, Hauck CR. CEACAMs: their role in physiology and pathophysiology. Curr Opin Cell Biol 2006; 18: 565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hauck CR, Agerer F, Muenzner P, et al. Cellular adhesion molecules as targets for bacterial infection. Eur J Cell Biol 2006; 85: 235–242. [DOI] [PubMed] [Google Scholar]
- 10. Beauchemin N, Arabzadeh A. Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) in cancer progression and metastasis. Cancer Metastasis Rev 2013; 32: 643–671. [DOI] [PubMed] [Google Scholar]
- 11. Han Z-W, Lyv Z-W, Cui B, et al. The old CEACAMs find their new role in tumor immunotherapy. Invest New Drugs 2020; 38: 1888–1898. [DOI] [PubMed] [Google Scholar]
- 12. Hammarström S. The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues. Semin Cancer Biol 1999; 9: 67–81. [DOI] [PubMed] [Google Scholar]
- 13. Johnson B, Mahadevan D. Emerging role and targeting of carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) in human malignancies. Clin Cancer Drugs 2015; 2: 100–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rizeq B, Zakaria Z, Ouhtit A. Towards understanding the mechanisms of actions of carcinoembryonic antigen-related cell adhesion molecule 6 in cancer progression. Cancer Sci 2018; 109: 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Berling B, Kolbinger F, Grunert F, et al. Cloning of a carcinoembryonic antigen gene family member expressed in leukocytes of chronic myeloid leukemia patients and bone marrow. Cancer Res 1990; 50: 6534–6539. [PubMed] [Google Scholar]
- 16. Schölzel S, Zimmermann W, Schwarzkopf G, et al. Carcinoembryonic antigen family members CEACAM6 and CEACAM7 are differentially expressed in normal tissues and oppositely deregulated in hyperplastic colorectal polyps and early adenomas. Am J Pathol 2000; 156: 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Duxbury MS, Matros E, Clancy T, et al. CEACAM6 is a novel biomarker in pancreatic adenocarcinoma and PanIN lesions. Ann Surg 2005; 241: 491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Y, Zang M, Li J, et al. CEACAM6 promotes tumor migration, invasion, and metastasis in gastric cancer. Acta Biochim Biophys Sin 2014; 46: 283–290. [DOI] [PubMed] [Google Scholar]
- 19. Singer BB, Scheffrahn I, Kammerer R, et al. Deregulation of the CEACAM expression pattern causes undifferentiated cell growth in human lung adenocarcinoma cells. PLoS ONE 2010; 5: e8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blumenthal RD, Leon E, Hansen HJ, et al. Expression patterns of CEACAM5 and CEACAM6 in primary and metastatic cancers. BMC Cancer 2007; 7: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ieta K, Tanaka F, Utsunomiya T, et al. CEACAM6 gene expression in intrahepatic cholangiocarcinoma. Br J Cancer 2006; 95: 532–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ameur N, Lacroix L, Roucan S, et al. Aggressive inherited and sporadic medullary thyroid carcinomas display similar oncogenic pathways. Endocr Relat Cancer 2009; 16: 1261–1272. [DOI] [PubMed] [Google Scholar]
- 23. Cameron S, de Long LM, Hazar-Rethinam M, et al. Focal overexpression of CEACAM6 contributes to enhanced tumourigenesis in head and neck cancer via suppression of apoptosis. Mol Cancer 2012; 11: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu S-J, Arundhathi A, Wang H-C, et al. Migration and invasion of NSCLC suppressed by the downregulation of Src/focal adhesion kinase using single, double and tetra domain anti- CEACAM6 antibodies. Transl Oncol 2021; 14: 101057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Duxbury MS, Ito H, Ashley SW, et al. CEACAM6 as a novel target for indirect type 1 immunotoxin-based therapy in pancreatic adenocarcinoma. Biochem Biophys Res Commun 2004; 317: 837–843. [DOI] [PubMed] [Google Scholar]
- 26. Riley CJ, Engelhardt KP, Saldanha JW, et al. Design and activity of a murine and humanized anti-CEACAM6 single-chain variable fragment in the treatment of pancreatic cancer. Cancer Res 2009; 69: 1933–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cheng T-M, Murad YM, Chang C-C, et al. Single domain antibody against carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) inhibits proliferation, migration, invasion and angiogenesis of pancreatic cancer cells. Eur J Cancer 2014; 50: 713–721. [DOI] [PubMed] [Google Scholar]
- 28. Strickland LA, Ross J, Williams S, et al. Preclinical evaluation of carcinoembryonic cell adhesion molecule (CEACAM) 6 as potential therapy target for pancreatic adenocarcinoma. J Pathol 2009; 218: 380–390. [DOI] [PubMed] [Google Scholar]
- 29. Witzens-Harig M, Hose D, Jünger S, et al. Tumor cells in multiple myeloma patients inhibit myeloma-reactive T cells through carcinoembryonic antigen-related cell adhesion molecule-6. Blood 2013; 121: 4493–4503. [DOI] [PubMed] [Google Scholar]
- 30. Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol 2009; 62: 1006–1012. [DOI] [PubMed] [Google Scholar]
- 31. Cochrane handbook for systematic reviews of interventions, https://handbook-5-1.cochrane.org/ (accessed 5 July 2021).
- 32. Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med 1998; 17: 2815–2834. [DOI] [PubMed] [Google Scholar]
- 33. Hayden JA, van der Windt DA, Cartwright JL, et al. Assessing bias in studies of prognostic factors. Ann Intern Med 2013; 158: 280–286. [DOI] [PubMed] [Google Scholar]
- 34. Deeks JJ, Higgins JPT, Altman DG. Analysing and presenting results. In: Higgins JPT, Green S, ed. Cochrane Handbook for Systematic Reviews of Interventions 4.2.5. Chichester, UK: John Wiley & Sons; 2006. [Google Scholar]
- 35. Zhu R, Ge J, Ma J, et al. Carcinoembryonic antigen related cell adhesion molecule 6 promotes the proliferation and migration of renal cancer cells through the ERK/AKT signaling pathway. Transl Androl Urol 2019; 8: 457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Arabzadeh A, McGregor K, Breton V, et al. EphA2 signaling is impacted by carcinoembryonic antigen cell adhesion molecule 1-L expression in colorectal cancer liver metastasis in a cell context-dependent manner. Oncotarget 2017; 8: 104330–104346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Du H, Li Y, Sun R, et al. CEACAM6 promotes cisplatin resistance in lung adenocarcinoma and is regulated by microRNA-146a and microRNA-26a. Thorac Cancer 2020; 11: 2473–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ho TH, Serie DJ, Parasramka M, et al. Differential gene expression profiling of matched primary renal cell carcinoma and metastases reveals upregulation of extracellular matrix genes. Ann Oncol 2017; 28: 604–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pandey R, Zhou M, Islam S, et al. Carcinoembryonic antigen cell adhesion molecule 6 (CEACAM6) in pancreatic ductal adenocarcinoma (PDA): an integrative analysis of a novel therapeutic target. Sci Rep 2019; 9: 18347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tsang JYS, Kwok YK, Chan KW, et al. Expression and clinical significance of carcinoembryonic antigen-related cell adhesion molecule 6 in breast cancers. Breast Cancer Res Treat 2013; 142: 311–322. [DOI] [PubMed] [Google Scholar]
- 41. Aydin B, Arga KY. Co-expression network analysis elucidated a core module in association with prognosis of non-functioning non-invasive human pituitary adenoma. Front Endocrinol 2019; 10: 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Beta-catenin CEACAM6 expression and prognostic value in elderly breast cancer tissues, http://www.lifesciencesite.com/lsj/life1002/220_B00971life1002_1600_1602.pdf (accessed 19 July 2021).
- 43. Jaso J, Thomas DA, Cunningham K, et al. Prognostic significance of immunophenotypic and karyotypic features of Philadelphia positive B-lymphoblastic leukemia in the era of tyrosine kinase inhibitors. Cancer 2011; 117: 4009–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kiyokawa N, Iijima K, Tomita O, et al. Significance of CD66c expression in childhood acute lymphoblastic leukemia. Leuk Res 2014; 38: 42–48. [DOI] [PubMed] [Google Scholar]
- 45. Kim KS, Kim J-T, Lee S-J, et al. Overexpression and clinical significance of carcinoembryonic antigen-related cell adhesion molecule 6 in colorectal cancer. Clin Chim Acta 2013; 415: 12–19. [DOI] [PubMed] [Google Scholar]
- 46. Zang M, Zhang Y, Zhang B, et al. CEACAM6 promotes tumor angiogenesis and vasculogenic mimicry in gastric cancer via FAK signaling. Biochim Biophys Acta 2015; 1852: 1020–1028. [DOI] [PubMed] [Google Scholar]
- 47. Maraqa L, Cummings M, Peter MB, et al. Carcinoembryonic antigen cell adhesion molecule 6 predicts breast cancer recurrence following adjuvant tamoxifen. Clin Cancer Res 2008; 14: 405–411. [DOI] [PubMed] [Google Scholar]
- 48. Chen J, Li Q, An Y, et al. CEACAM6 induces epithelial-mesenchymal transition and mediates invasion and metastasis in pancreatic cancer. Int J Oncol 2013; 43: 877–885. [DOI] [PubMed] [Google Scholar]
- 49. Gebauer F, Wicklein D, Horst J, et al. Carcinoembryonic antigen-related cell adhesion molecules (CEACAM) 1, 5 and 6 as biomarkers in pancreatic cancer. PLoS ONE 2014; 9: e113023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Deng X, Liu P, Zhao Y, et al. Expression profiling of CEACAM6 associated with the tumorigenesis and progression in gastric adenocarcinoma. Genet Mol Res 2014; 13: 7686–7697. [DOI] [PubMed] [Google Scholar]
- 51. Roy RK, Hoppe MM, Srivastava S, et al. CEACAM6 is upregulated by Helicobacter pylori CagA and is a biomarker for early gastric cancer. Oncotarget 2016; 7: 55290–55301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ru G-Q, Han Y, Wang W, et al. CEACAM6 is a prognostic biomarker and potential therapeutic target for gastric carcinoma. Oncotarget 2017; 8: 83673–83683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jantscheff P, Terracciano L, Lowy A, et al. Expression of CEACAM6 in resectable colorectal cancer: a factor of independent prognostic significance. J Clin Oncol 2003; 21: 3638–3646. [DOI] [PubMed] [Google Scholar]
- 54. Han HS, Son S-M, Yun J, et al. MicroRNA-29a suppresses the growth, migration, and invasion of lung adenocarcinoma cells by targeting carcinoembryonic antigen-related cell adhesion molecule 6. FEBS Lett 2014; 588: 3744–3750. [DOI] [PubMed] [Google Scholar]
- 55. Kobayashi M, Miki Y, Ebina M, et al. Carcinoembryonic antigen-related cell adhesion molecules as surrogate markers for EGFR inhibitor sensitivity in human lung adenocarcinoma. Br J Cancer 2012; 107: 1745–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kalina T, Vaskova M, Mejstrikova E, et al. Myeloid antigens in childhood lymphoblastic leukemia: clinical data point to regulation of CD66c distinct from other myeloid antigens. BMC Cancer 2005; 5: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tian C, Zhang B, Ge C. Effect of CEACAM6 silencing on the biological behavior of human gallbladder cancer cells. Oncol Lett 2020; 20: 2677–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang Z, Luo C, Wang H, et al. CEACAM6 is associated with osteosarcoma metastasis and facilitates epithelial-mesenchymal transition in osteosarcoma cells. Onco Targets Ther 2018; 11: 3159–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xu J, Liu B, Ma S, et al. Characterizing the tumor suppressor role of CEACAM1 in multiple myeloma. Cell Physiol Biochem 2018; 45: 1631–1640. [DOI] [PubMed] [Google Scholar]
- 60. Blumenthal RD, Hansen HJ, Goldenberg DM. Inhibition of adhesion, invasion, and metastasis by antibodies targeting CEACAM6 (NCA-90) and CEACAM5 (carcinoembryonic antigen). Cancer Res 2005; 65: 8809–8817. [DOI] [PubMed] [Google Scholar]
- 61. Schäfer D, Tomiuk S, Küster LN, et al. Identification of CD318, TSPAN8 and CD66c as target candidates for CAR T cell based immunotherapy of pancreatic adenocarcinoma. Nat Commun 2021; 12: 1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li Y, Zhu X, You J, et al. Efficacy of bivalent CEACAM6/4-1BBL genetic vaccine combined with anti-PD1 antibody in HCT116 xenograft mice. Preprint. Epub ahead of print 4 August 2021. DOI: 10.21203/rs.3.rs-618292/v1. [DOI] [Google Scholar]
- 63. Bayer. An open-label, phase 1, first-in-human, dose escalation and expansion study to evaluate the safety, tolerability, pharmacokinetics, pharmacodynamics and tumor response profile of the anti-CEACAM6 antibody BAY1834942 in patients with advanced solid tumors (Clinical Trial Registration NCT03596372), clinicaltrials.gov, https://clinicaltrials.gov/ct2/show/NCT03596372 (2021, accessed 14 July 2021).
- 64. Willuda J, Boehm H-H, Pinkert J, et al. Abstract LB-075: increased T cell-activation resulting from the combination of the anti-CEACAM6 function-blocking antibody BAY 1834942 with checkpoint inhibitors targeting either PD-1/PD-L1 or TIM-3. Cancer Res 2019; 79: LB-075. [Google Scholar]
- 65. Kurlinkus B, Ger M, Kaupinis A, et al. CEACAM6’s role as a chemoresistance and prognostic biomarker for pancreatic cancer: a comparison of CEACAM6’s diagnostic and prognostic capabilities with those of CA19-9 and CEA. Life 2021; 11: 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhao Z-S, Li L, Wang H-J, et al. Expression and prognostic significance of CEACAM6, ITGB1, and CYR61 in peripheral blood of patients with gastric cancer. J Surg Oncol 2011; 104: 525–529. [DOI] [PubMed] [Google Scholar]
- 67. Steiner N, Hajek R, Nachbaur D, et al. Levels of CEACAM6 in peripheral blood are elevated in patients with plasma cell disorders: a potential new diagnostic marker and a new therapeutic target? Dis Markers 2019; 2019: 1806034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tam-10.1177_17588359211072621 for Prognostic value of the immune target CEACAM6 in cancer: a meta-analysis by Miguel Burgos, Iván Cavero-Redondo, Celia Álvarez-Bueno, Eva María Galán-Moya, Atanasio Pandiella, Eitan Amir and Alberto Ocaña in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-2-tam-10.1177_17588359211072621 for Prognostic value of the immune target CEACAM6 in cancer: a meta-analysis by Miguel Burgos, Iván Cavero-Redondo, Celia Álvarez-Bueno, Eva María Galán-Moya, Atanasio Pandiella, Eitan Amir and Alberto Ocaña in Therapeutic Advances in Medical Oncology
Supplemental material, sj-jpg-1-tam-10.1177_17588359211072621 for Prognostic value of the immune target CEACAM6 in cancer: a meta-analysis by Miguel Burgos, Iván Cavero-Redondo, Celia Álvarez-Bueno, Eva María Galán-Moya, Atanasio Pandiella, Eitan Amir and Alberto Ocaña in Therapeutic Advances in Medical Oncology
Supplemental material, sj-jpg-2-tam-10.1177_17588359211072621 for Prognostic value of the immune target CEACAM6 in cancer: a meta-analysis by Miguel Burgos, Iván Cavero-Redondo, Celia Álvarez-Bueno, Eva María Galán-Moya, Atanasio Pandiella, Eitan Amir and Alberto Ocaña in Therapeutic Advances in Medical Oncology