Abstract
Recent studies have provided insights into innate and adaptive immune dynamics in coronavirus disease 2019 (COVID-19). However, the exact features of antibody responses that govern COVID-19 disease outcomes remain unclear. In this study, we analyzed humoral immune responses in 229 patients with asymptomatic, mild, moderate and severe COVID-19 over time to probe the nature of antibody responses in disease severity and mortality. We observed a correlation between anti-spike (S) immunoglobulin G (IgG) levels, length of hospitalization and clinical parameters associated with worse clinical progression. Although high anti-S IgG levels correlated with worse disease severity, such correlation was time dependent. Deceased patients did not have higher overall humoral response than discharged patients. However, they mounted a robust, yet delayed, response, measured by anti-S, anti-receptor-binding domain IgG and neutralizing antibody (NAb) levels compared to survivors. Delayed seroconversion kinetics correlated with impaired viral control in deceased patients. Finally, although sera from 85% of patients displayed some neutralization capacity during their disease course, NAb generation before 14 d of disease onset emerged as a key factor for recovery. These data indicate that COVID-19 mortality does not correlate with the cross-sectional antiviral antibody levels per se but, rather, with the delayed kinetics of NAb production.
COVID-19 is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which infects host cells via angiotensin-converting enzyme 2 (refs. 1,2). Although 80% of infections are mild or asymptomatic (World Health Organization, https://www.who.int/), patients with moderate and severe COVID-19 develop a wide range of symptoms, including respiratory, vascular and neurological complications3–5. Several studies have linked cellular and humoral immune responses to viral clearance and distinct disease trajectories4,6–9. For instance, inflammatory cytokines and chemokines, including interferons (IFNs), interleukin (IL)-1β, IL-4, IL-6 and IL-18 and CXCL9/10, are associated with worse COVID-19 outcome4,8–10. Importantly, in contrast to the marked decreases in circulating T cells observed in patients with COVID-19 (refs. 4,6,7,11), circulating B cells do not seem to decrease5,8. Additionally, several studies reported an overall increase in both anti-SARS-CoV-2 spike IgM and IgG (anti-S IgG), as well as neutralizing IgG and IgA antibodies in patients with COVID-19 (refs. 12–15). However, information about how antibody responses affect the course of COVID-19 trajectory, and how they correlate with additional host factors, viral titers and clinical outcome, is still missing.
Antiviral antibodies correlate with distinct COVID-19 outcomes
To profile the SARS-CoV-2-specific humoral immune response, 185 hospitalized patients with COVID-19, with a total of 300 samples, were enrolled in this study after admission to Yale-New Haven Hospital (YNHH) between March 18, 2020, and May 27, 2020. In parallel, we enrolled 44 non-hospitalized participants, including participants with asymptomatic and mild disease. Additionally, 16 vaccinated volunteers were included in this study. All vaccinated donors were SARS-CoV-2-negative by reverse transcription quantitative polymerase chain reaction (RT-qPCR) and enzyme-linked immunosorbent assay (ELISA)-negative for SARS-CoV-2 IgG. Finally, 105 samples of healthcare workers (HCWs) served as uninfected healthy controls (SARS-CoV-2-negative by RT-qPCR and serology). Basic demographics and clinical characteristics for each cohort are summarized in Supplementary Tables 1–3. For our initial analysis, hospitalized patients were first stratified based on disease severity into moderate and severe disease groups by levels of supplemental oxygen requirement and admission to the intensive care unit (ICU). Further investigations divided patients with COVID-19 according to clinical outcomes, stratifying patients who ultimately recovered or died from infection, as previously described8. Three hundred samples were collected during hospital stay, including sequential follow-up measurements with a range of 1–7 longitudinal time points per patient that occurred 3–60 d after the onset of symptoms. We assessed viral RNA load using nasopharyngeal swabs; levels of plasma cytokines and chemokines; leukocyte populations, profiled by flow cytometry using freshly isolated peripheral blood mononuclear cells (PBMCs); and antibody profiles, using both ELISA and neutralizations assays. See the Methods for details of the assays.
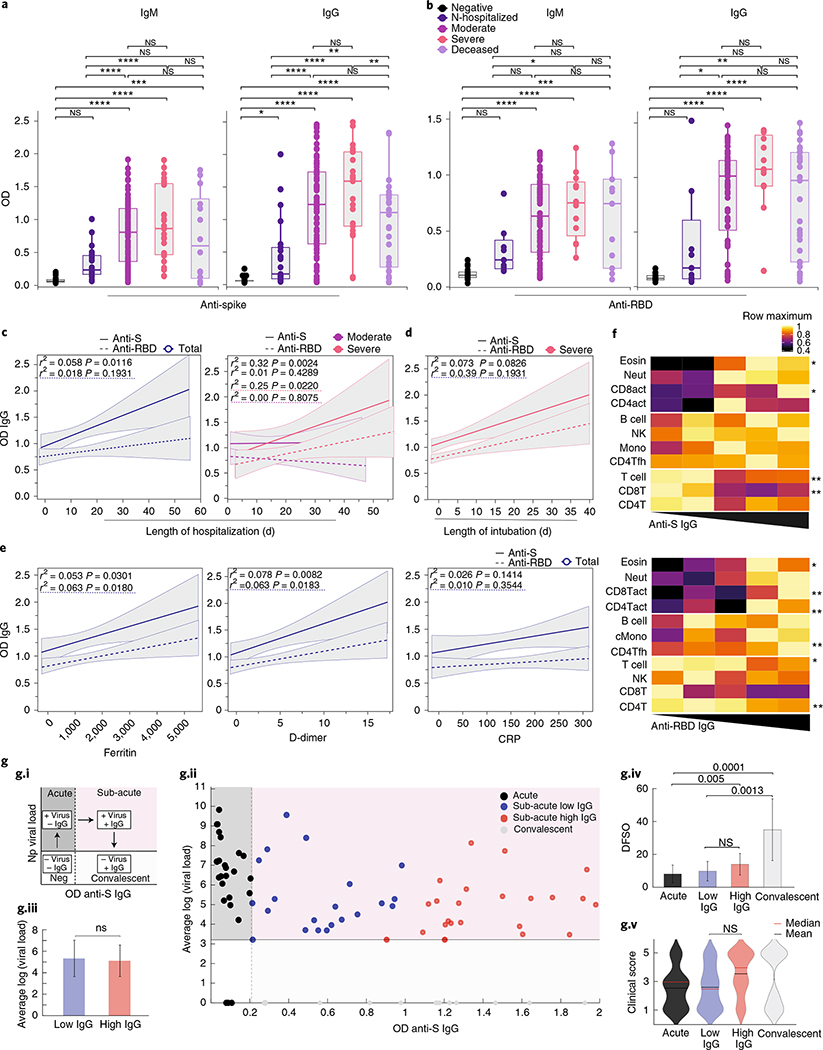
Plasma sample analysis showed that 95.7% and 97.5% of total patients hospitalized with COVID-19 had virus-specific IgG against the spike (S1) or receptor-binding domain (RBD) regions of the proteins, respectively, reaching the peak of IgG production around day 15 after symptom onset (Extended Data Fig. 1a,b). The average of anti-S or anti-RBD IgG levels from uninfected control HCW donors was used to determine the limit threshold. Although the maximum levels of anti-RBD IgG reached during the disease course were reduced in elderly patients (Extended Data Fig. 1a), no differences were observed over time (Extended Data Fig. 1f). Increased levels of anti-RBD IgG were observed in patients with obesity. No differences were observed in antibody levels between hospitalized patients of different sexes (Extended Data Fig. 1c–f). Maximum viral titers from nasopharyngeal swabs were reached approximately 11–13 d after symptom onset. Overall, maximum nasal viral RNA load did not correlate with disease severity; no differences were observed between the non-hospitalized group and the moderate and severe patient groups (Extended Data Fig. 2a). In contrast, hospitalized patients showed a significant increase in maximum anti-S and anti-RBD IgM and IgG levels compared to non-hospitalized individuals (Fig. 1a,b). Additionally, anti-S IgG levels correlated with length of hospitalization among patients with severe disease but not moderate disease (Fig. 1c). Consistent with this observation, parameters associated with worsened clinical progression, such as intubation, ferritin and D-dimer levels, were positively correlated with anti-S IgG levels (Fig. 1d,e). In contrast, anti-RBD IgG levels were not correlated with length of hospitalization (Fig. 1b–e). Thus, these data indicate that elevated anti-S IgG levels are associated with worse disease outcome in patients with severe COVID-19, confirming previous observations that antibody responses were consistently higher among hospitalized patients16–19. Notably, in hospitalized patients, deceased patients did not have higher levels of virus-specific IgG or IgM than live, discharged patients (Fig. 1a). These results indicated a fundamentally different feature of their antibody responses compared to patients with severe disease who survived the infection.
Fig. 1 |. COVID-19 severity correlates with anti-S antibodies.
a,b, Plasma reactivity to S protein and RBD in patients with COVID-19. a, Anti-S IgM and IgG. IgM (HCW, n = 21; non-hospitalized, n = 21; moderate, n = 92; severe, n = 25; deceased, n = 14). IgG (HCW, n = 87; non-hospitalized, n = 21; moderate, n = 94; severe, n = 23; deceased, n = 34). b, Anti-RBD IgM and IgG. IgM (HCW, n = 21; non-hospitalized, n = 7; moderate, n = 75; severe, n = 13; deceased, n = 11). IgG (HCW, n = 21; non-hospitalized, n = 6; moderate, n = 74; severe, n = 13; deceased, n = 31). Negative controls: HCWs. N-hospitalized, non-hospitalized. Each dot represents a single individual at their maximum antibody titer over the disease course. Significance: one-way ANOVA corrected for multiple comparisons using Tukey’s method. Boxes represent the distribution of variables with quartiles and outliers. Horizontal bars: mean values. c, Correlation and linear regression of maximum levels for each patient of virus-specific IgG and length of hospitalization over time. Left, all patients. Right, patients grouped by disease severity. Regression lines are shown as dark purple (moderate) or pink (severe). d,e, Correlation of virus-specific IgG and (d) length of intubation or (e) patients’ maximum levels of ferritin, D-dimer and CRP. Pearson’s correlation coefficients and linear regression significance are colored accordingly; shading represents 95% CI. f, Heat map correlation analysis between virus-specific IgG levels and major immune cell populations in PBMCs. Color intensity indicates the relative cell frequency. Significance: one-way ANOVA corrected for multiple comparisons using Dunnett’s method. **P < .01, *P < .05. g.i, Scheme. g.ii, Scatter plot of patients with COVID-19 with S1 IgG+ samples at their first collection time point. Dashed vertical line: threshold of S1 IgG positivity. Solid horizontal line: limit of detection. g.iii, Comparison of mean log viral loads between IgG-low and IgG-high groups using two-sample t-test (two-sided). Bars represent average ± s.d. g.iv, Days from symptom onset for each group. Bars represent average ± s.d. g.v, Violin plots of clinical scores for each cluster. Solid black lines, mean; dashed lines, median. Significance: Kruskal–Wallis corrected for multiple comparisons using Dunn’s method. CI, confidence interval; OD, optical density; NK, natural killer; Np, nasopharyngeal; NS, not significant.Source data
Owing to the correlation observed between anti-S IgG levels and disease severity, as well as previous reports describing changes in leukocyte populations in severe COVID-19, including lymphopenia and increased monocyte, neutrophil, basophil and eosinophil numbers4,6–9,11, we next assessed whether changes in virus-specific antibodies were linked to alterations in innate and adaptive circulating immune cell types. Virus-specific antibody levels negatively correlated with T cells and positively correlated with monocyte and eosinophil numbers, but no correlation was found with circulating natural killer or B cells (Fig. 1f). Furthermore, we observed a positive correlation between anti-RBD, but not anti-S, IgG levels and circulating T follicular helper CD4+ T (Tfh) cells as well as CD38+HLA-DR+TCR-activated CD4 (CD4act) T cells (Fig. 1f). To control for the potential influence of differential viral loads in our analysis of the relationship between disease severity and magnitude of S IgG response, we stratified hospitalized patients at their initial collection time points into acute (positive viral load, negative IgG), sub-acute (positive viral load, positive IgG) and convalescent (negative viral load, positive IgG) phases of COVID-19 (Fig. 1g.i). We next focused our analysis on patients within the same phase of disease (‘sub-acute’, pink region) and stratified patients into S IgG low (blue) and S1 IgG high (red) patient groups based on the 50th percentile S1 IgG value. We observed that our clustering did not produce significant differences in either viral load (Fig. 1g.iii) or days from symptom onset (DFSO) (Fig. 1g.iv) among the S ‘low’ and S1 ‘high’ groups. Lastly, we assessed whether the magnitude of the anti-S IgG response correlated with reduced disease severity among patients with matched viral loads and DFSO and found no significant differences in average clinical score between S ‘low’ and S ‘high’ groups (Fig. 1g.v).
Notably, the development of anti-S IgG responses did not correlate with general improvements in patient clinical scores, even when patients with equivalent viral loads were compared. Given these results, we conclude that anti-S IgG antibodies positively correlate with COVID-19 severity and appear to offer a limited ability to modify disease trajectory once developed during natural SARS-CoV-2 infection. Furthermore, anti-S IgG antibodies positively correlated with COVID-19 severity, along with the circulating levels of monocytes and eosinophils, but independent of circulating T cells, Tfh cells or viral load.
Delayed antibody production in lethal COVID-19
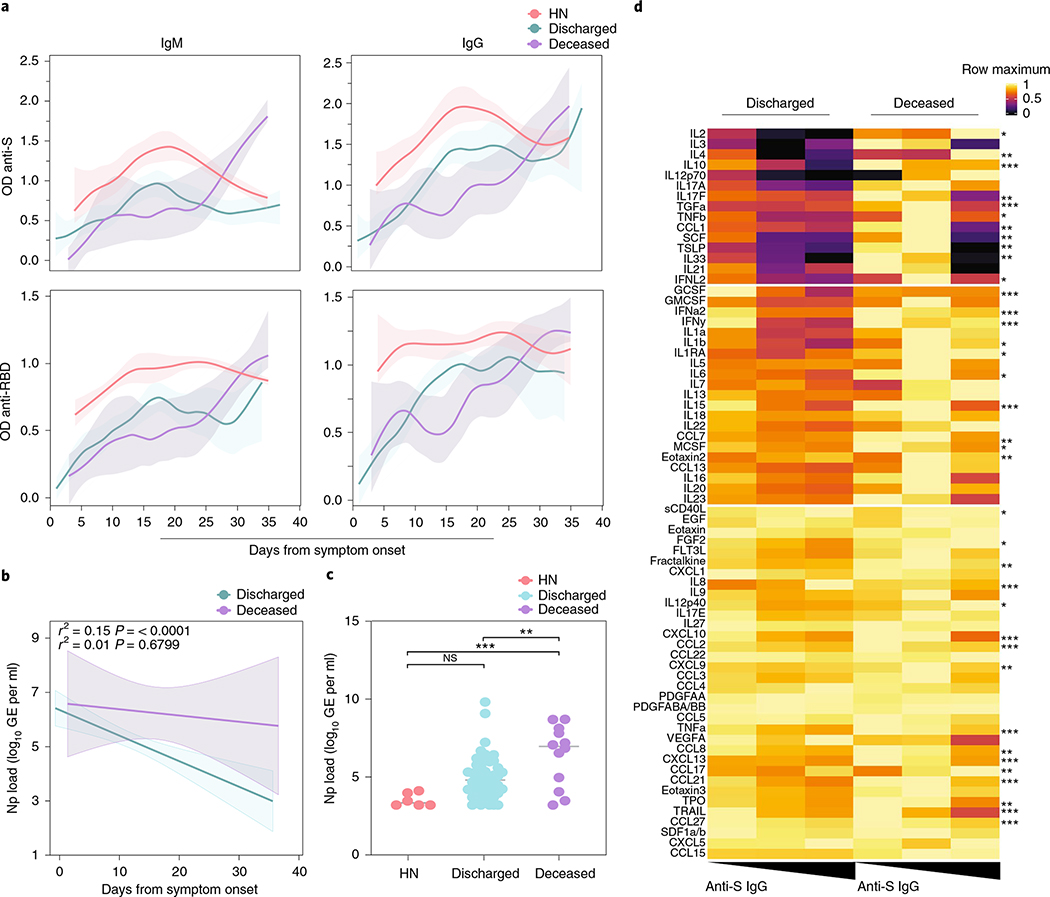
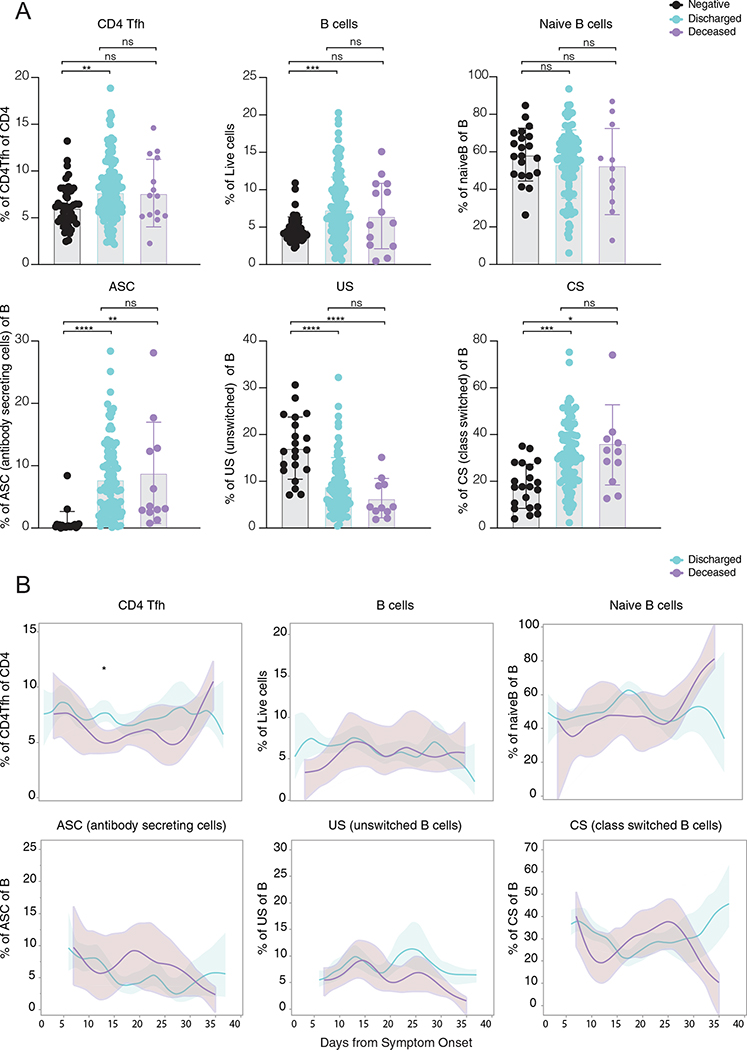
Given the lower levels of antiviral antibodies found in deceased patients, we next addressed whether the timing of antibody responses differs between severe versus lethal disease. Longitudinal analysis revealed distinct kinetics: discharged patients reached a peak of anti-S and anti-RBD IgG levels earlier than deceased patients (Fig. 2a). In contrast, deceased patients reached higher maximum levels of anti-S IgM and IgG than discharged patients in later stages of disease (Fig. 2a). Patients with high neutralizing antibody titers were included in this figure as a reference; additional analysis can be found in Fig. 3a,b. Longitudinal antibody trajectories between discharged and deceased groups were consistent with their distinct capacity to clear the virus; that is, discharged patients were more efficient in viral clearance when compared side by side with deceased patients (Fig. 2b). Additionally, lower levels of nasal viral RNA load measured at the time of maximum antibody levels were observed in discharged patients (Fig. 2c). We did not observe differences in B cell dynamics in patients with distinct clinical outcomes (Extended Data Fig. 3a,b). Despite no differences between discharged and deceased groups at aggregate levels, longitudinal analysis indicated a higher frequency of Tfh cells at DFSO 10–15 in discharged than in deceased patients with COVID-19. Thus, death from COVID-19 correlated with a delay in the development of virus-specific IgG and virus clearance.
Fig. 2 |. Serum antibody kinetics reveals distinct COVID-19 outcomes.
a, Patients’ plasma reactivity to S protein and RBD measured by ELISA. Anti-S and Anti-RBD IgM and IgG comparison in discharged or deceased patients. Longitudinal data plotted over time continuously. Regression lines are shown as light blue (discharged), purple (deceased) and red (high neutralizers). Lines indicates cross-sectional averages from each group, with shading representing 95% CI and colored accordingly. Anti-S IgM (discharged, n = 126; deceased, n = 14). Anti-S IgG (discharged, n = 127; deceased, n = 33). Anti-RBD IgM (discharged, n = 88; deceased, n = 11). Anti-S RBD (discharged, n = 87; deceased, n = 30). b,c, Viral loads measured by nasopharyngeal swabs are plotted as log10 of genome equivalents (GEs). b, Viral loads against time after symptom onset accordingly with patient outcome. Regression lines are shown as light blue (discharged) or purple (deceased), with shading representing 95% CI. Pearson’s correlation coefficients and linear regression significance are colored accordingly. c, Viral load measured in discharged, deceased and high neutralizer (HN) patients. (HN, n = 6; discharged, n = 53; deceased, n = 12). Each dot represents the viral load of a single individual at their maximum antibody titer over the disease course. One-way ANOVA corrected for multiple comparisons using Tukey’s method were used to determine significance. ***P = 0.0005, **P = 0066. d, Heat map correlation analysis between Anti-S IgG (OD450 nm) levels and plasma cytokine/chemokine measurements in discharged (n = 146) or deceased (n = 26) patients. Patients are arranged across columns based on anti-S IgG levels. Each row represents a cytokine/chemokine and is normalized by its maximum value (assigned value of 1). Color intensity indicates the relative cytokine concentration (log10) normalized against the same population across all subjects. k-means clustering was used to arrange patients and measurements. Significance was assessed by one-way ANOVA testing corrected for multiple comparisons using Tukey’s method. sCD40L, FGF2, IL-1β, IL-1RA, IL-2, IL-6, IL-12, MCSF, TNF-β, CCL1, TPO, IFN-L2 (P < .05); fractalkine, IL-4, IL-17F, CCL7, CXCL9, eotaxin2, CC17, SCF, TSLP, IL-33 (P < .01); GCSF, IFN-α, IFN-γ, IL-8, IL-10, IL-15, CXCL10, CCL2, TGF-α, TNF-α, CCL8, CXCL13, CCL21, LIF, TRAIL, CCL27 (P < 0.001). ***P < .001 **P < .01, *P < .05. CI, confidence interval; NS, not significant; OD, optical density.
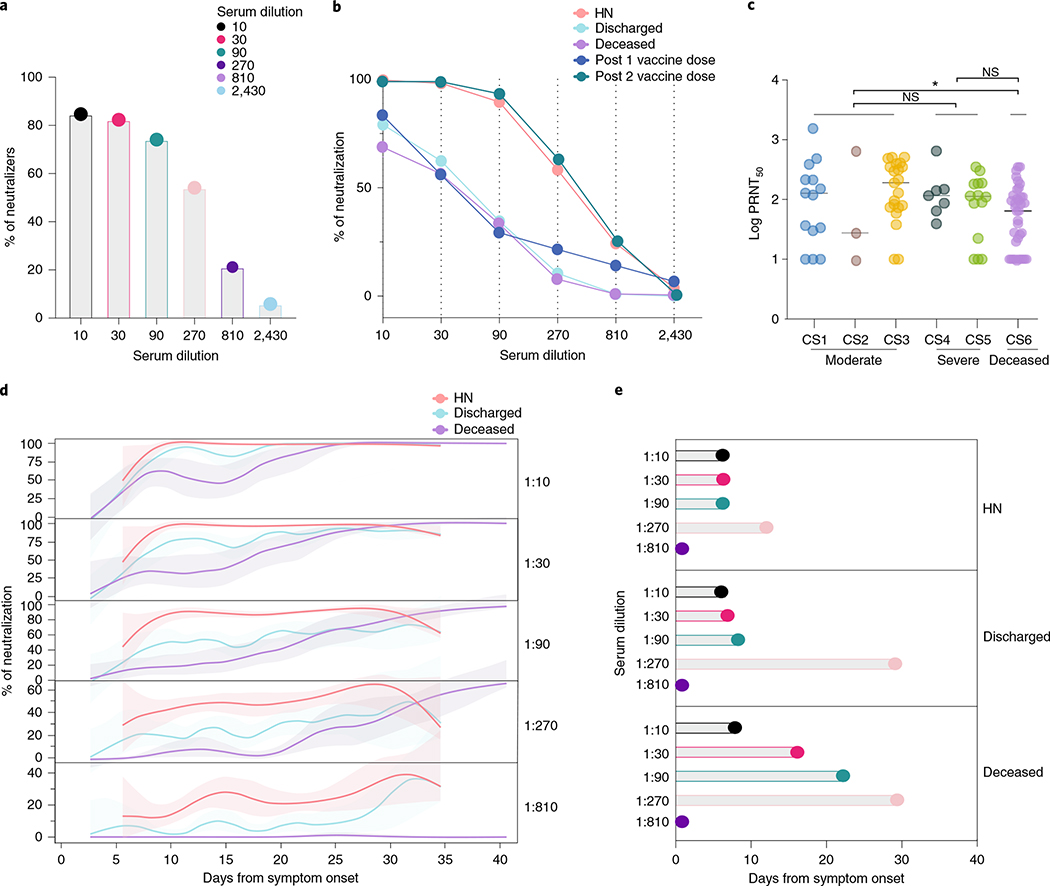
Fig. 3 |. Neutralizing antibody temporal dynamics distinguish discharged and deceased patients with COVID-19.
a–e, Longitudinal neutralization assay using wild-type SARS-CoV-2. a, Frequency of neutralizers, n = 83. b, Neutralization capacity among discharged (light blue), deceased (purple) and high neutralizer (HN) (red) patients at the experimental six-fold serially dilutions (from 1:3 to 1:2,430). HCWs, below the threshold for anti-S/RBD ELISA, were used as negative controls. Post 1 vaccine dose, 28 d after 1 vaccine dose. Post 2 vaccine dose, 7 d after 2 vaccine dose. (HCWs, n = 22; discharged, n = 41; deceased, n = 37; HN, n = 13; post 1 vaccine dose, n = 9; post 2 vaccine dose, n = 7). Pearson’s correlation analysis were used to accessed significance. HN: r2 0.785, P (two-tailed) 0.0185; discharged: r2 0.438, P (two-tailed) 0.1516; deceased: r2 0.437, P (two-tailed) 0.1524; Post 1 vaccine dose: r2 0.424, P (two-tailed) 0.1609; post 2 vaccine dose: r2 0.822, P (two-tailed) 0.0126. c, Maximum neutralization titer (PRNT50) per patient according to clinical severity scale as described in Methods. CS, clinical score. One-way ANOVA corrected for multiple comparisons using Tukey’s method was used to determine significance. *P = 0.0213. d, Longitudinal data plotted over time of neutralization capacity among discharged (light blue), deceased (purple) and HN (red) patients at the experimental six-fold serially dilutions (from 1:3 to 1:2,430). Lines indicates cross-sectional averages from each group, with shading representing 95% CI and colored accordingly. e, Average of days from symptom onset to reach 50% of neutralization at each experimental serum dilution among groups. CI, confidence interval; NS, not significant.
We next assessed a possible correlation between cytokine and chemokine levels and virus-specific antibody production. Discharged patients showed a positive correlation between anti-S IgG and several chemokines, growth factors and tissue repair mediators, including sCD40L, IL-8, CCL17 and eotaxin2 (Fig. 2d), consistent with a ‘protective signature’ that we recently observed in these patients who recovered from COVID-19 (ref. 8). Additionally, discharged patients showed a negative correlation between anti-S IgG and plasma inflammatory markers previously associated with poor disease outcomes and death, such as IFN-I, IFN-II and IFN-III and IL-1, IL-6, IL-17 and IL-10 (Fig. 2d). Deceased patients, in contrast, showed fewer correlations with anti-S IgG levels and plasma cytokines and chemokines (Fig. 2d).
Early neutralizing antibodies correlate with COVID-19 recovery
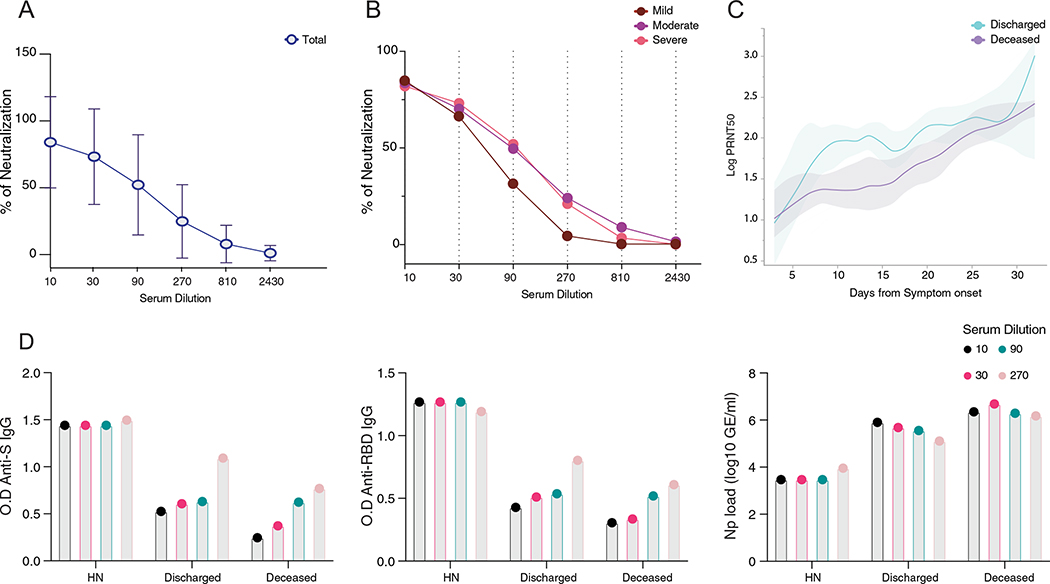
Production of anti-S/RBD IgG antibodies is generally associated with virus neutralization13 and has been linked with protection against SARS-CoV-2 infection after vaccination in animal models20,21. We next assessed the kinetics of NAbs produced against SARS-CoV-2 by performing a neutralization assay using wild-type SARS-CoV-2 in patients with a range of anti-S IgG titers (0.29–2.50 optical density at 450 nm (OD450 nm)). Patients with a single time point within the first week after symptoms onset were not included in the analysis; samples from HCWs negative for SARS-CoV-2 by RT-qPCR were used as control samples and were below the threshold for anti-S/RBD ELISA. Previous studies reported a low frequency of convalescent patients with COVID-19 with potent neutralization capacity of over 1:1,000 titers13. Indeed, although 89% of patients in our cohort showed some neutralization capacity during their disease course, 74–84% of hospitalized patients exhibited neutralizing activity only at lower dilutions (1:10–1:90 titers), and only 6–21% of patients showed neutralizing activity at higher dilutions (1:810–1:2,430 titers) (Fig. 3a,b and Extended Data Fig. 4a,b). The neutralization levels were lower among non-hospitalized participants with mild disease, even at lower dilutions of 1:90 (31%) and 1:270 (4%) titers (Extended Data Fig. 4b). Based on this stratification, we designated patients with >1:810 neutralizing titer as high neutralizers. The overall maximum neutralization levels of hospitalized patients were similar to vaccinated volunteers 28 d after receiving the first dose of the mRNA vaccines. Seven days after the second vaccine dose, neutralization titers were similar to those of infected patients with high neutralization capacity and significantly higher than the general hospitalized cohort. The plaque reduction half-maximal neutralizing titer (PRNT50) was undetectable for 17.5% of patients, whereas 19.5% of patients had a PRNT50 at 1:270 and only 1.3% of patients at 1:810. Of note, the levels of PRNT50, measured at the maximum level of neutralization over the disease course, were not significantly different among hospitalized patients stratified by disease severity (Fig. 3c). These PRNT50 patterns among hospitalized patients are consistent with recentlu reported data22. In contrast, deceased patients had reduced PRNT50 compared to patients who developed moderate, but not severe, disease (Fig. 3c).
Notably, our longitudinal analysis also revealed faster NAb kinetics, as well as a higher peak, in discharged patients than in deceased patients. Discharged patients reached 50% of neutralization at 1:90 titer around day 9 after symptom onset, whereas deceased patients peaked 1 and 2 weeks later at 1:30 and 1:90 titers, respectively (Fig. 3d,e and Extended Data Fig. 4c). We then compared maximum anti-S IgG titers, anti-RBD IgG titers and viral loads among high neutralizers, discharged patients and deceased patients, when each group reached 50% of neutralization. No significant differences were observed between discharged and deceased groups (Extended Data Fig. 4d). Despite equivalent maximum NAb titers, distinct temporal antibody dynamics were strongly linked with clinical disease outcome. The high neutralizers had maximum levels of anti-RBD IgG and NAb from the very first sampling (5 d after disease onset) and maintained high levels throughout the hospital stay (Fig. 2b). High neutralizers had lower levels of nasal viral RNA load measured at the time of maximum antibody level in comparison to deceased or discharged patients (Fig. 2c).
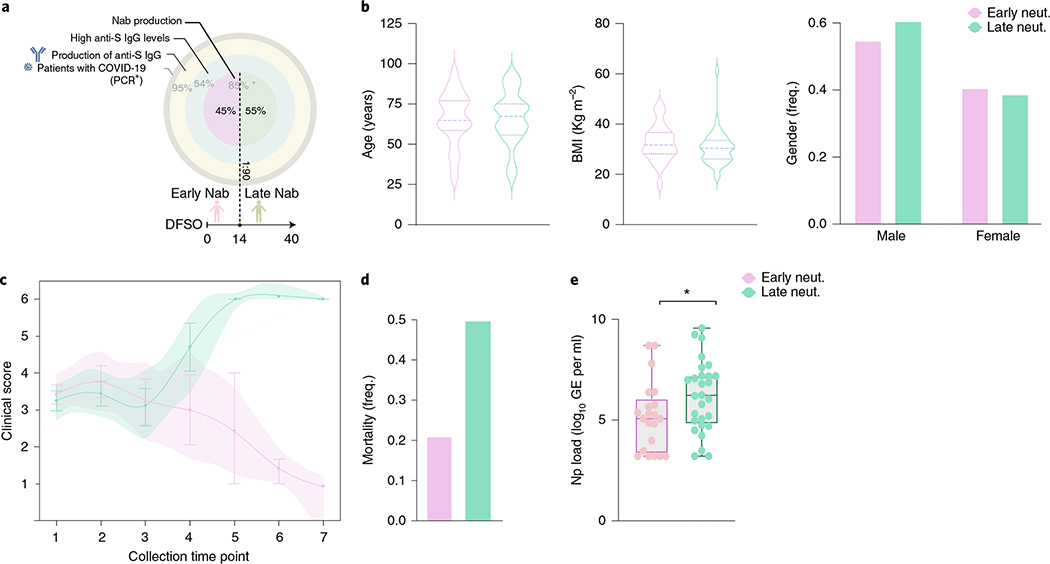
Finally, we asked whether the timing of NAb production correlates with disease trajectory. Within our cohort, 54.6% of the patients had >50% of neutralization activity at 1:90 titer but only 19.8% at 1:270. Patients were then grouped into those who developed >50% neutralization activity at 1:90 titer NAb levels before 14 d of symptom onset (early) and those who did not (late) (Fig. 4a). Early neutralization activity did not correlate with age or body mass index (BMI), and the frequency of males and females was not significantly different between early or late neutralizers (Fig. 4b). Nevertheless, early NAb production correlated with improving clinical signs and lower mortality than late neutralizers, who showed worse disease progression and higher mortality (Fig. 4c,d). Moreover, the maximum viral loads reached over the disease course were lower in early neutralizers (Fig. 4e). Together, these data indicate that clinical trajectories and outcomes do not correlate with the levels of NAb produced over the disease course but with the timing of NAb production.
Fig. 4 |. Early neutralizing antibodies correlate with better COVID-19 clinical trajectory.
Patient stratification by early NAb capacity, based on levels of anti-S IgG, PRNT50 titers and days from symptom onset. a, Cohort overview by IgG anti-S titers (three external circles) and NAb production (internal circle). Frequency of patients in each level is indicated in light gray. Frequency of early and late neutralizers stratified based on days from symptom onset at 1:90 dilution is indicated in black. *Frequency of patients with NAb capacity over the disease course in patients with high levels of anti-S IgG. b, Distribution of age, BMI and frequency of males and females between early (>50% neutralization activity in 1:90 titer before day 14 after symptom onset) or late (<50% neutralization activity in 1:90 titer before day 14 after symptom onset) neutralizers, as determined in a. c, Disease progression measured by clinical severity score for patients in each group. The lines represent the mean ± s.e.m for each group and are ordered by the collection time points for each patient, with regular collection intervals of 3–4 d. Shade represents 95% CI and is colored accordingly. d, Percentage of mortality in each group (early neutralizers, n = 27; late neutralizers, n = 45). e, Viral load measured by nasopharyngeal (Np) swabs plotted as log10 of genome equivalents (GEs) in early and late neutralizers (early neutralizers, n = 21; late neutralizers, n = 28). Each dot represents a single individual at their maximum antibody titer over the disease course. Box analysis with minimum and maximum represented for each group. Horizontal bar indicates mean values. Significance was accessed using unpaired t-test. *P (two-tailed) = 0.0467. CI, confidence interval.
Discussion
The dynamics of virus-specific antibody responses evolve rapidly throughout infection. The magnitude of humoral responses was previously correlated with disease severity in patients infected with various coronaviruses, including MERS, SARS-CoV-1 and SARS-CoV-2 (refs. 16–19) Previous work investigating SARS-CoV-2 infection also found that patients with severe COVID-19 have relatively higher levels of SARS-CoV-2-specific antibodies23–25. In addition to the magnitude of humoral responses, other investigations explored whether the diversity of SARS-CoV-2 antigenic targets might play a role in conferring protection against severe COVID-19 (refs. 26,27).
Our analyses extend these findings through longitudinal sampling of COVID-19 patient sera to suggest that differences in the kinetics of humoral response might also play a role in protection from fatal COVID-19. Specifically, our work suggests that there is a critical time window in which the neutralizing antibodies must develop to improve virological control and disease outcome.
Our temporal analyses revealed a distinct antibody kinetics between discharged and deceased patients. Deceased patients showed slower antibody dynamics, even though they reached higher levels later in the disease trajectory. A recent study performing longitudinal analysis also reported a delayed and incomplete humoral immune response in deceased patients with COVID-19 (ref. 28). Our study confirms and extends these observations by demonstrating that seroconversion kinetics between discharged and deceased patients can directly affect viral clearance with faster viral control or prolonged viral shedding, respectively. Additionally, discharged patients showed a negative correlation between anti-S IgG and plasma inflammatory markers, uncovering a potential role of antibodies in protecting these patients from immunopathology, especially in light of our earlier study describing a severe inflammatory signature in patients with COVID-19 (ref. 8). Somehow, this protective role of antiviral antibodies fails to operate in deceased patients, as we saw loss of correlation between anti-S IgG and protective tissue repair growth factors. Our data suggest that the loss of the protective role of antibodies in lethal disease is due to their late onset.
Our study demonstrated that NAb responses developed within 14 d of symptom onset correlated with recovery, whereas those induced at later time points appear to lose this protective effect. It is unclear why antibodies generated after this time point are unable to promote viral clearance and recovery in patients with COVID-19. We speculate that the virus might become inaccessible to the antibodies after a certain time point, by establishing infection within immune-privileged tissues. Alternatively, disease might be driven by late-onset, antibody-mediated immunopathology. For instance, antibodies from patients with severe COVID-19 show pro-inflammatory Fc modification signatures, including high levels of afucosylated IgG1 (ref. 29), which could potentially drive pathologic responses. Consistent with these findings and the potential role of antibodies in immunopathology, our data indicate that anti-S, but not anti-RBD, antibody levels in patients with COVID-19 correlate with disease severity, length of hospital stay, length of intubation and various clinical parameters of disease. In addition, the levels of anti-S IgG, when matched for similar viral load and days from symptom onset, correlated with COVID-19 severity. Future studies are needed to address the precise mechanism of the failure of late antibody responses and the potential immunopathological roles of anti-S IgG.
Recent postmortem tissue analysis from patients with lethal SARS-CoV-2 infection suggested a defective induction of germinal centers, including inefficient generation of Tfh cells30; these data are consistent with our observations. Nevertheless, because our analysis did not include isolation of secondary lymphoid tissues, we were unable to assess the dynamics of B cell populations in lymph nodes, and this might explain why we failed to observe differences in B cell dynamics despite our observations of general differences in the kinetics of humoral responses across patient cohorts. We observed a positive correlation between anti-S IgG and Tfh in discharged, but not deceased, patients.
The use of convalescent plasma has been proposed as an urgent therapeutic modality for patients with COVID-19. However, large clinical trials conducted to evaluate the efficacy of plasma therapy failed to observe clear benefits31–35, perhaps due to variable neutralizing capacity in the donor pool. Potent neutralizing monoclonal antibodies cloned from B cells isolated from convalescent patients were proposed to address this issue in COVID-19 therapeutics. However, a recent large trial of monoclonal antibodies also failed to observe improvements when administered to patients with late-stage severe disease or patients who were hospitalized36; similar observations were made with trials by Regeneron36–38. Additional early trials have shown promising results in individuals who were newly infected with the virus39. Our results demonstrated that anti-S or neutralizing antibodies observed within 14 d of symptom onset correlated with improved disease trajectory. These data suggest that antibody-based therapies might benefit patients most when given within this 2-week time window.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41591-021-01355-0.
Methods
Ethics statement.
This study was approved by Yale Human Research Protection Program institutional review board (FWA00002571, protocol ID 2000027690). Vaccinated volunteers were included in this study under the respective protocol, ID 2000028924. Informed consent was obtained from all enrolled patients, volunteers and HCWs, including additional cohorts from the Connecticut National Guard and vaccinated HCW volunteers.
Patients.
One hundred and eighty-five patients with COVID-19 who were admitted to YNHH between March 18, 2020, and May 27, 2020, were included in this study. Additionally, 41 non-hospitalized participants were enrolled in this study, including individuals with asymptomatic and mild disease. These participants (n = 25) were enrolled by the IMPACT group, and an additional 16 serum samples were obtained from members of the Connecticut National Guard with mild COVID-19. Additionally, 16 vaccinated volunteers were included in this study. Vaccinated donors received the mRNA vaccine (Moderna or Pfizer), and neutralization analyses were performed 28 d and 7 d after vaccine dose 1 and 2, respectively. All vaccinated donors were PCR and ELISA negative for SARS-CoV-2. HCW participants, screened serially (every 2 weeks), served as uninfected healthy controls (SARS-CoV-2-negative by RT-qPCR and serology). No statistical methods were used to predetermine sample size. Patients were scored for COVID-19 disease severity through review of electronic health records (EHRs) at each longitudinal time point. Scores were assigned by a clinical infectious disease physician according to a custom-developed disease severity scale. Moderate disease status (clinical score 1–3) was defined as: SARS-CoV-2 infection requiring hospitalization without supplementary oxygen (1); infection requiring non-invasive supplementary oxygen (<3 L min−1 to maintain SpO2 > 92%) (2); and infection requiring non-invasive supplementary oxygen (>3 L min−1 to maintain SpO2 > 92% or >2 L min−1 to maintain SpO2 > 92%) and had a high-sensitivity C-reactive protein (CRP) > 70 and received tocilizumab. Severe disease status (clinical score 4 or 5) was defined as: infection meeting all criteria for clinical score 3 and also requiring admission to the ICU and >6 L min−1 supplementary oxygen to maintain SpO2 > 92% (4) or infection requiring invasive mechanical ventilation or extracorporeal membrane oxygenation in addition to glucocorticoid or vasopressor administration (5). Clinical score 6 was assigned for deceased patients. For all patients, days from symptom onset were estimated as follows: (1) highest priority was given to explicit onset dates provided by patients; (2) next-highest priority was given to the earliest reported symptom by a patient; and (3), in the absence of direct information regarding symptom onset, we estimated a date through manual assessment of the EHR by an independent clinician. Demographic information was aggregated through a systematic and retrospective review of the EHR and was used to construct Supplementary Table 1. Symptom onset and etiology were recorded through standardized interviews with patients or patient surrogates upon enrolment in our study or, alternatively, through manual EHR review if no interview was possible owing to clinical status. The clinical data were collected using EPIC EHR May 2020 and REDCap 9.3.6 software. At the time of sample acquisition and processing, investigators were completely unaware of patient conditions. Blood acquisition was performed and recorded by a separate team. Information of patient conditions was not available until after processing and analyzing raw data by flow cytometry and ELISA. A clinical team, separate from the experimental team, performed chart reviews to determine patients’ relevant statistics. Cytokine and flow cytometry analyses were blinded. Patients’ clinical information and clinical score coding were revealed only after data collection.
Isolation of patient plasma and PBMCs.
Whole blood was collected in sodium heparin-coated vacutainers and kept on gentle agitation until processing. All blood was processed on the day of collection. Plasma samples were collected after centrifugation of whole blood at 400g for 10 min at room temperature without brake. The undiluted serum was then transferred to 15-ml polypropylene conical tubes and aliquoted and stored at −80 °C for subsequent analysis. PBMCs were isolated using Histopaque (Sigma-Aldrich, 10771–500ml) density gradient centrifugation in a Biosafety Level 2+ facility. After isolation of undiluted serum, blood was diluted 1:1 in room temperature PBS, layered over Histopaque in a SepMate tube (StemCell Technologies, 85460) and centrifuged for 10 min at 1,200g. The PBMC layer was isolated according to the manufacturer’s instructions. Cells were washed twice with PBS before counting. Pelleted cells were briefly treated with ACK lysis buffer for 2 min and then counted. Percentage viability was estimated using standard trypan blue staining and an automated cell counter (Thermo Fisher Scientific, AMQAX1000).
SARS-CoV-2-specific antibody measurements.
ELISAs were performed as previously described40. In short, Triton X-100 and RNase A were added to serum samples at final concentrations of 0.5% and 0.5 mg ml−1, respectively, and incubated at room temperature for 30 min before use to reduce risk from any potential virus in serum. Next, 96-well MaxiSorp plates (Thermo Fisher Scientific, 442404) were coated with 50 μl per well of recombinant SARS-CoV-2 S1 protein (ACROBiosystems, S1N-C52H3-100μg) at a concentration of 2 μg ml−1 in PBS and were incubated overnight at 4 °C. The coating buffer was removed, and plates were incubated for 1 h at room temperature with 200 μl of blocking solution (PBS with 0.1% Tween-20 and 3% milk powder). Serum was diluted 1:50 in dilution solution (PBS with 0.1% Tween-20 and 1% milk powder), and 100 μl of diluted serum was added for 2 h at room temperature. Plates were washed three times with PBS-T (PBS with 0.1% Tween-20) and 50 μl of horseradish peroxidase anti-human IgG antibody (GenScript, A00166, 1:5,000) or anti-human IgM peroxidase antibody (Sigma-Aldrich, A6907, 1:5,000) diluted in dilution solution added to each well. After 1 h of incubation at room temperature, plates were washed three times with PBS-T. Plates were developed with 100 μl of TMB Substrate Reagent Set (BD Biosciences, 555214), and the reaction was stopped after 15 min by the addition of 2 N sulfuric acid. Plates were then read at a wavelength of 450 nm and 570 nm.
Cytokine and chemokine measurements.
Patient serum was isolated as before, and aliquots were stored at −80 °C. Sera were shipped to Eve Technologies on dry ice, and levels of cytokines and chemokines were measured using the Human Cytokine Array/Chemokine Array 71-403 Plex Panel (HD71). All samples were measured upon the first thaw.
Viral RNA measurements.
Nasopharyngeal swab samples were collected approximately every 4 d for SARS-CoV-2 RT-qPCR analysis where clinically feasible. RNA concentrations were measured as previously described41. In brief, total nucleic acid was extracted from 300 μl of viral transport medium (nasopharyngeal swabs) using the MagMAX Viral/Pathogen Nucleic Acid Isolation Kit (Thermo Fisher Scientific) with a modified protocol and eluted into 75 μl of elution buffer. We used 5 μl of extracted nucleic acid as template in an RT-qPCR assay to detect SARS-CoV-2 RNA42, using the U.S. Centers for Disease Control and Prevention real-time RT-qPCR primer/probe sets for 2019-nCoV_N1 and 2019-nCoV_N2 and the human RNase P as an extraction control. Virus RNA copies were quantified using a ten-fold dilution standard curve of RNA transcripts that we previously generated42. The lower limit of detection for SARS-CoV-2 genomes assayed by qPCR in nasopharyngeal specimens was established as described42. In addition to a technical detection threshold, we also used a clinical referral threshold (detection limit) to either (1) refer asymptomatic HCWs for diagnostic testing at a Clinical Laboratory Improvement Amendments (CLIA)-approved laboratory or (2) cross-validate results from a CLIA-approved laboratory for SARS-CoV-2 qPCR-positive individuals upon study enrolment. Individuals above the technical detection threshold, but below the clinical referral threshold, were considered SARS-CoV-2-positive for the purposes of our research.
Flow cytometry.
Antibody clones and vendors were as follows: BB515 anti-hHLA-DR (G46-6) (1:400) (BD Biosciences), BV785 anti-hCD16 (3G8) (1:100) (BioLegend), PE-Cy7 anti-hCD14 (HCD14) (1:300) (BioLegend), BV605 anti-hCD3 (UCHT1) (1:300) (BioLegend), BV711 anti-hCD19 (SJ25C1) (1:300) (BD Biosciences), Alexa Fluor647 anti-hCD1c (L161) (1:150) (BioLegend), biotin anti-hCD141 (M80) (1:150) (BioLegend), PE-Dazzle594 anti-hCD56 (HCD56) (1:300) (BioLegend), PE anti-hCD304 (12C2) (1:300) (BioLegend), APC/Fire750 anti-hCD11b (ICRF44) (1:100) (BioLegend), PerCP/Cy5.5 anti-hCD66b (G10F5) (1:200) (BD Biosciences), BV785 anti-hCD4 (SK3) (1:200) (BioLegend), APC/Fire750 or PE-Cy7 or BV711 anti-hCD8 (SK1) (1:200) (BioLegend), BV421 anti-hCCR7 (G043H7) (1:50) (BioLegend), Alexa Fluor 700 anti-hCD45RA (HI100) (1:200) (BD Biosciences), PE anti-hPD1 (EH12.2H7) (1:200) (BioLegend), APC anti-hTIM3 (F38-2E2) (1:50) (BioLegend), BV711 anti-hCD38 (HIT2) (1:200) (BioLegend), BB700 anti-hCXCR5 (RF8B2) (1:50) (BD Biosciences), PE-Cy7 anti-hCD127 (HIL-7R-M21) (1:50) (BioLegend), PE-CF594 anti-hCD25 (BC96) (1:200) (BD Biosciences), BV711 anti-hCD127 (HIL-7R-M21) (1:50) (BD Biosciences), BV421 anti-hIL17a (N49-653) (1:100) (BD Biosciences), Alexa Fluor 700 anti-hTNFa (MAb11) (1:100) (BioLegend), PE or APC/Fire750 anti-hIFN-γ (4S.B3) (1:60) (BioLegend), FITC anti-hGranzymeB (GB11) (1:200) (BioLegend), Alexa Fluor 647 anti-hIL-4 (8D4-8) (1:100) (BioLegend), BB700 anti-hCD183/CXCR3 (1C6/CXCR3) (1:100) (BD Biosciences), PE-Cy7 anti-hIL-6 (MQ2-13A5) (1:50) (BioLegend), PE anti-hIL-2 (5344.111) (1:50) (BD Biosciences), BV785 anti-hCD19 (SJ25C1) (1:300) (BioLegend), BV421 anti-hCD138 (MI15) (1:300) (BioLegend), Alexa Fluor 700 anti-hCD20 (2H7) (1:200) (BioLegend), Alexa Fluor 647 anti-hCD27 (M-T271) (1:350) (BioLegend), PE/Dazzle594 anti-hIgD (IA6-2) (1:400) (BioLegend), PE-Cy7 anti-hCD86 (IT2.2) (1:100) (BioLegend), APC/Fire750 anti-hIgM (MHM-88) (1:250) (BioLegend), BV605 anti-hCD24 (ML5) (1:200) (BioLegend), BV421 anti-hCD10 (HI10a) (1:200) (BioLegend), BV421 anti-CDh15 (SSEA-1) (1:200) (BioLegend), Alexa Fluor 700 streptavidin (1:300) (Thermo Fisher Scientific), BV605 streptavidin (1:300) (BioLegend). In brief, freshly isolated PBMCs were plated at 1–2 × 106 cells per well in a 96-well U-bottom plate. Cells were resuspended in Live/Dead Fixable Aqua (Thermo Fisher Scientific) for 20 min at 4 °C. After a wash, cells were blocked with Human TruStain FcX (BioLegend) for 10 min at room temperature. Cocktails of desired staining antibodies were added directly to this mixture for 30 min at room temperature. For secondary stains, cells were first washed and supernatant aspirated; then, to each cell pellet, a cocktail of secondary markers was added for 30 min at 4 °C. Before analysis, cells were washed and resuspended in 100 μl of 4% paraformaldehyde for 30 min at 4 °C. After this incubation, cells were washed and prepared for analysis on an Attune NxT (Thermo Fisher Scientific). Data were analyzed using FlowJo software version 10.6 (Tree Star). The specific sets of markers used to identify each subset of cells are summarized in Extended Data Fig. 5.
Cell lines and virus.
Vero E6 kidney epithelial cells were cultured in DMEM supplemented with 1% sodium pyruvate (non-essential amino acid) and 5% FBS at 37 °C and 5% CO2. The cell line was obtained from the American Type Culture Collection and has been tested negative for contamination with mycoplasma. SARS-CoV-2 (ancestral strain, D614G) USA-WA1/2020 was obtained from BEI Resources (NR-52281) and was amplified in Vero E6 cells. Cells were infected at a multiplicity of infection of 0.01 for 3 days to generate a working stock, and, after incubation, the supernatant was clarified by centrifugation (450g × 5 min) and filtered through a 0.45-μm filter. The pelleted virus was then resuspended in PBS and then aliquoted for storage at −80 °C. Viral titers were measured by standard plaque assay using Vero E6 cells. Briefly, 300 μl of serial fold virus dilutions were used to infect Vero E6 cells in MEM supplemented with NaHCO3, 4% FBS and 0.6% Avicel RC-581. Plaques were resolved at 48 h after infection by fixing in 10% formaldehyde for 1 h followed by 0.5% crystal violet in 20% ethanol staining. Plates were rinsed in water to plaque enumeration. All experiments were performed in a Biosafety Level 3 facility with approval from the Yale Environmental Health and Safety office.
Neutralization assay.
Patient and healthy donor sera were isolated as before and then heat treated for 30 min at 56 °C. Six-fold serially diluted plasma, from 1:3 to 1:2,430, was incubated with SARS-CoV-2 (ancestral strain, D614G) for 1 h at 37 °C. The mixture was subsequently incubated with Vero E6 cells in a six-well plate for 1 h for adsorption. Then, cells were overlayed with MEM supplemented with NaHCO3, 4% FBS and 0.6% Avicel mixture. Plaques were resolved at 40 h after infection by fixing in 10% formaldehyde for 1 h, followed by staining in 0.5% crystal violet. All experiments were performed in parallel with negative control sera with an established viral concentration to generate 60–120 plaques per well.
Statistical analysis.
All analyses of patient samples were conducted using MATLAB 2020a, GraphPad Prism 8.4.3, JMP 15 and R 3.4.3. Patient heat maps were clustered using the k-means algorithm. Each row in the heat maps represents an immune cell population or relative cytokine concentration (log10) and is normalized by its maximum value (assigned value of 1). Color intensity indicates the relative cell frequency. Multiple group comparisons were analyzed by running both parametric (analysis of variance (ANOVA)) and non-parametric (Kruskal–Wallis) statistical tests. Multiple comparisons were corrected using Tukey’s method, Dunn’s method and Dunnett’s method as indicated in the figure legends. For comparisons between stable groups, two-sided, unpaired t-tests were used.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All the background information on HCWs and clinical information for patients in this study are included in Source Data Fig. 1. Additionally, all of the raw FCS files for the flow cytometry analyses are available at ImmPort (https://www.immport.org/shared/home; study ID SDY1655). Additional correspondence and requests for materials should be addressed to the corresponding author (A.I). Source data are provided with this paper.
Extended Data
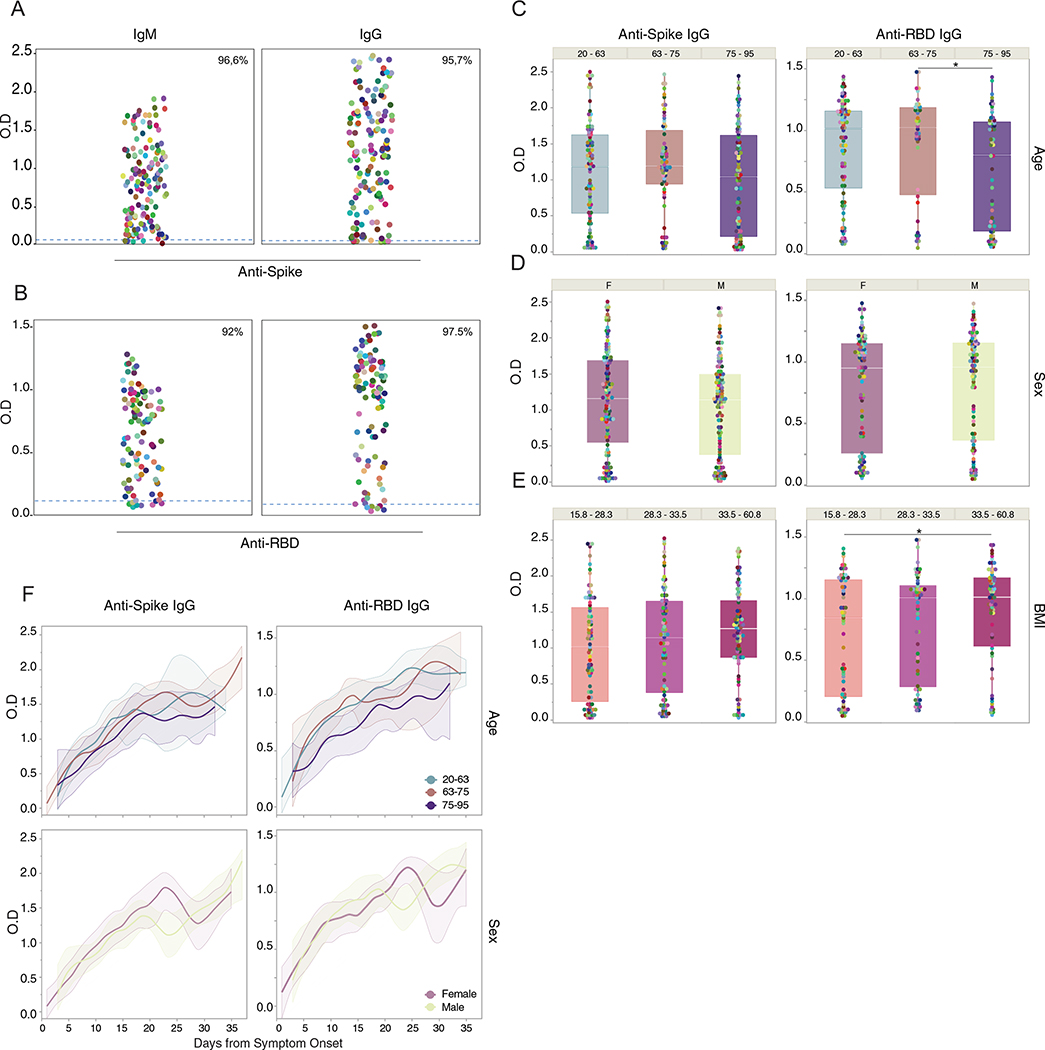
Extended Data Fig. 1 |. Correlation analysis of virus-specific antibodies and age, sex and BMI.
a-e, Plasma reactivity to S protein and RBD by ELISA. a, Anti-S IgM and IgG of total COVID-19 patients regardless of disease severity. Patients, IgM (n = 139); IgG (n = 159). b, Anti-RBD IgM and IgG of total COVID-19 patients regardless of disease severity. Patients, IgM (n = 99); IgG (n = 120). Each color dot represents a single individual at its maximum antibody titer over the disease course. Dashed line indicates HCW average values (limit threshold). HCW, Anti-S IgM (n = 21); IgG (n = 87); HCW, Anti-RBD IgM (n = 21); IgG (n = 21). IgG levels of Anti-S (left) or Anti-RBD (right) by (c) age, (d) sex and (e) BMI. Each dot represents a single individual at its maximum antibody titer over the disease course. Boxes represent variables’ distribution with quartiles and outliers. Horizontal bars indicate mean values. OD, optical density at 450 nm (OD450 nm). F, females; M, males. One-way ANOVA corrected for multiple comparisons using Tukey’s and unpaired t-test (two-tailed) were used to determine significance. Anti-RBD IgG *p = 0.0130 (age); *p = 0.0301 (BMI). f, IgG levels of Anti-S (left) or Anti-RBD by age and sex. Longitudinal analysis over time. Lines indicate cross-sectional averages from each group, with shading representing 95% CI and colored accordingly.
Extended Data Fig. 2 |. SARS-CoV-2 viral load and disease severity.
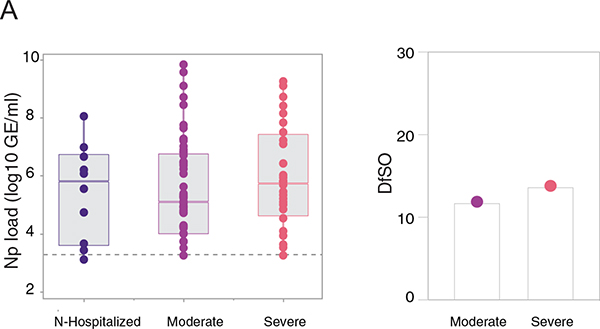
a, Left, Viral load measured by nasopharyngeal swabs plotted as log10 of genome equivalents in non-hospitalized and hospitalized, moderate and severe COVID-19 patients. (N-hospitalized, n = 10; moderate, n = 97; severe, n = 65). Each dot represents a single individual at its maximum viral titer over the disease course. Dashed line indicates threshold for positivity. Boxes represent variables’ distribution with quartiles and outliers. Horizontal bars indicates mean values. Right, Average of days from symptom onset (DfSO) comparison between groups. N-hospitalized, non-hospitalized. One-way ANOVA corrected for multiple comparisons using Tukey’s were used to determine significance.
Extended Data Fig. 3 |. Overview of cellular immune profiles in COVID-19 patients.
a,b, Immune cell subsets of interest, plotted as a percentage of a parent population as (a) aggregate and (b) continuously over time according to the days of symptom onset for discharged or deceased patients. a, Immune cell subsets comparison in discharged or deceased patients. Negative controls (HCWs) are shown in black. Each dot represents a single individual at its maximum antibody titer over the disease course. Grey bars indicate mean values. ANOVA corrected for multiple comparisons using Tukey’s were used to determine significance. b, Longitudinal data plotted over time continuously. Regression lines are shown as light blue (discharged) and purple (deceased) and indicate cross-sectional averages from each group with shading representing 95% CI and are coloured accordingly. (HCW, n = 49; Discharged, n = 118; Deceased, n = 15). CD4Tfh, follicular helper T cells. ASC, antibody secreting cells. US, unswitched. CS, class switched.
Extended Data Fig. 4 |. Virus-specific antibodies and viral load correlation with PRNT50.
a,b, Neutralization capacity among (a) total COVID-19 patients or (b) between mild (dark red), moderate (purple) and severe (pink) at the experimental sixfold serially dilutions (from 1:3 to 1:2430). Lines represent average ± standard deviations. Total patients, n = 63; Moderate, n = 45; Severe, n = 19. Pearson correlation analysis were used to accessed significance. moderate: R2 0.575, p(two-tailed) 0.0804; severe: R2 0.552, p(two-tailed) 0.0902; c, Longitudinal data plotted over time of PRNT50 between discharged (light blue) and deceased (purple). Lines indicates cross-sectional averages from each group, with shading representing 95% CI and colored accordingly. d, Levels of IgG (left) Anti-S, (middle) RBD and (right) viral load between high neutralizers, deceased and discharged patients. The indicated levels were measured at the average day from symptom onset in which each group reach 50% of neutralization at each experimental serum dilution as specified in Fig. 3f. HN, high neutralizers.
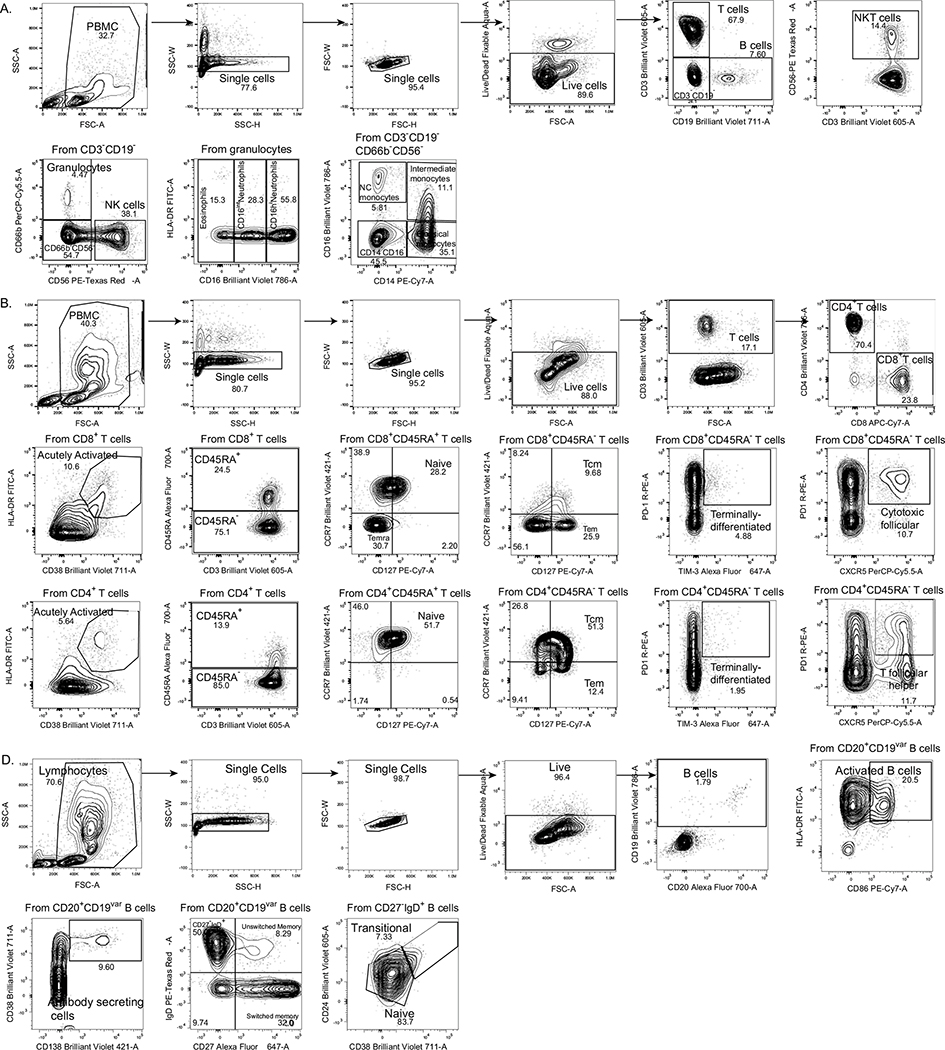
Extended Data Fig. 5 |. Gating strategies.
Gating strategies are shown for the key cell populations described in Fig. 1f and Extended Data Fig. 3. a, Leukocyte gating strategy to identify lymphocytes and granulocytes. b, T cell surface staining gating strategy to identify CD4 and CD8 T cells, TCR-activated T cells, follicular T cells, and additional subsets. c, B cell surface staining gating strategy to identify B cells subsets.
Supplementary Material
Acknowledgements
We thank M. Linehan for technical and logistical assistance and C. Wilen, D. Mucida and T. Castro for discussions. We also thank C. Wilen for kindly providing the virus. This work was supported in part by the Women’s Health Research at Yale Pilot Project Program, the Fast Grant from Emergent Ventures at the Mercatus Center, the Mathers Foundation, the Ludwig Family Foundation, the Department of Internal Medicine at the Yale School of Medicine, the Yale School of Public Health and the Beatrice Kleinberg Neuwirth Fund. IMPACT received support from the Yale COVID-19 Research Resource Fund. A.I. is an Investigator of the Howard Hughes Medical Institute. C.L. is a Pew Latin American Fellow. P.Y. is supported by the Gruber Foundation and the National Science Foundation. B.I. is supported by National Institute of Allergy and Infectious Diseases 2T32AI007517-16. C.B.F.V. is supported by NWO Rubicon 019.181EN.004.
Footnotes
Yale IMPACT Research Team
Abeer Obaid16, Alexander James Robertson5, Alice Lu-Culligan1, Alice Zhao5, Allison Nelson16, Anderson Brito5, Angela Nunez16, Anjelica Martin1, Anne E. Watkins5, Bertie Geng16, Caitlin J. Chun5, Chaney C. Kalinich5, Christina A. Harden5, Codruta Todeasa16, Cole Jensen5, Coriann E. Dorgay5, Daniel Kim1, David McDonald16, Denise Shepard10, Edward Courchaine17, Elizabeth B. White5, Eric Song1, Erin Silva16, Eriko Kudo1, Giuseppe DeIuliis16, Harold Rahming16, Hong-Jai Park16, Irene Matos16, Isabel Ott5, Jessica Nouws16, Jordan Valdez16, Joseph Fauver5, Joseph Lim18, Kadi-Ann Rose16, Kelly Anastasio19, Kristina Brower5, Laura Glick16, Lokesh Sharma16, Lorenzo Sewanan16, Lynda Knaggs16, Maksym Minasyan16, Maria Batsu16, Mary Petrone5, Maxine Kuang5, Maura Nakahata16, Melissa Linehan1, Michael H. Askenase20, Michael Simonov16, Mikhail Smolgovsky16, Natasha C. Balkcom5, Nicole Sonnert1, Nida Naushad16, Pavithra Vijayakumar16, Rick Martinello3, Rupak Datta4, Ryan Handoko16, Santos Bermejo16, Sarah Prophet21, Sean Bickerton17, Sofia Velazquez20, Tara Alpert19, Tyler Rice1, William Khoury-Hanold1, Xiaohua Peng16, Yexin Yang1, Yiyun Cao1, Yvette Strong16 and Zitong Lin5
16Yale School of Medicine, New Haven, CT, USA. 17Department of Biochemistry and of Molecular Biology, Yale University School of Medicine, New Haven, CT, USA. 18Yale Viral Hepatitis Program, Yale University School of Medicine, New Haven, CT, USA. 19Yale Center for Clinical Investigation, Yale University School of Medicine, New Haven, CT, USA. 20Department of Neurology, Yale University School of Medicine, New Haven, CT, USA. 21Department of Molecular, Cellular and Developmental Biology, Yale University School of Medicine, New Haven, CT, USA.
Competing interests
A.I. served as a consultant for Spring Discovery, Boehringer Ingelheim and Adaptive Biotechnologies. I.Y. reports being a member of the mRNA-1273 Study Group and has received funding to her institution to conduct clinical research from BioFire, MedImmune, Regeneron, PaxVax, Pfizer, GlaxoSmithKline, Merck, Novavax, Sanofi-Pasteur and Micron. All other authors declare no competing financial interests.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41591-021-01355-0.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41591-021-01355-0.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Hoffmann M et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan R et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Z et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 8, 420–422 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen G et al. Clinical and immunologic features in severe and moderate Coronavirus Disease 2019. J. Clin. Invest. 130, 2620–2629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giamarellos-Bourboulis EJ et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe 27, 992–1000 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Z et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe 27, 883–890 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucas C et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 584, 463–469 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez L et al. Systems-level immunomonitoring from acute to recovery phase of severe COVID-19. Cell Rep. Med. 1, 100078 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Valle DM et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 26, 1636–1643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathew D et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science 369, eabc8511 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnes CO et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell 182, 828–842 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robbiani DF et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584, 437–442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt F et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 217, e20201181 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z et al. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci. Transl. Med. 13, eabf1555 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y et al. A comprehensive, longitudinal analysis of humoral responses specific to four recombinant antigens of SARS-CoV-2 in severe and non-severe COVID-19 patients. PLoS Pathog. 16, e1008796 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X et al. Disease severity dictates SARS-CoV-2-specific neutralizing antibody responses in COVID-19. Signal Transduct. Target Ther. 5, 180 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu WT et al. Antibody profiles according to mild or severe SARS-CoV-2 infection, Atlanta, Georgia, USA, 2020. Emerg. Infect. Dis. 26, 2974–2978 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu KKQ et al. Comorbid illnesses are associated with altered adaptive immune responses to SARS-CoV-2. JCI Insight 10.1172/jci.insight.146242 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu J et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 369, 806–811 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Folegatti PM et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 396, 467–478 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Beltran WF et al. COVID-19-neutralizing antibodies predict disease severity and survival. Cell 184, 476–488 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long QX et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 26, 845–848 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Ripperger TJ et al. Orthogonal SARS-CoV-2 serological assays enable surveillance of low-prevalence communities and reveal durable humoral immunity. Immunity 53, 925–933 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guthmiller JJ et al. SARS-CoV-2 infection severity is linked to superior humoral immunity against the spike. mBio 10.1128/mBio.02940-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atyeo C et al. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity 53, 524–532 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Röltgen K et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci. Immunol. 5, eabe0240 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zohar T et al. Compromised humoral functional evolution tracks with SARS-CoV-2 mortality. Cell 183, 1508–1519 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abry P et al. Spatial and temporal regularization to estimate COVID-19 reproduction number R(t): promoting piecewise smoothness via convex optimization. PLoS ONE 15, e0237901 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaneko N et al. Loss of Bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell 183, 143–157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agarwal A et al. Convalescent plasma in the management of moderate covid-19 in adults in India: open label phase II multicentre randomised controlled trial (PLACID Trial). Brit. Med. J. 371, m3939 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pathak EB Convalescent plasma is ineffective for covid-19. Brit. Med. J. 371, m4072 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Holliman R, Seal DV, Archer H & Doman S Controlled trial of chemical disinfection of urinary drainage bags. Reduction in hospital-acquired catheter-associated infection. Br. J. Urol. 60, 419–422 (1987). [DOI] [PubMed] [Google Scholar]
- 34.Valk SJ et al. Convalescent plasma or hyperimmune immunoglobulin for people with COVID-19: a rapid review. Cochrane Database Syst. Rev. 5, CD013600 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Z, Wang X, Fu Y, Zhang X & Liu C Neutralizing antibodies for the treatment of COVID-19. Acta Pharm. Sin. B 11, 304–307 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eli Lilly and Company. Lilly statement regarding NIH’s ACTIV-3 clinical trial. https://www.lilly.com/news/stories/statement-activ3-clinical-trial-nih-covid19 (October 26, 2020).
- 37.May B Regeneron halts enrollment of critically ill patients in a COVID-19 antibody trial. https://www.biospace.com/article/regeneron-halts-enrollment-in-covid-19-trial-following-safety-signal-in-critically-ill-patients (BioSpace, 2020). [Google Scholar]
- 38.FDA authorizes monoclonal antibodies for treatment of COVID-19. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-monoclonal-antibodies-treatment-covid-19 (U.S. Food and Drug Administration, 2020). [Google Scholar]
- 39.Chen P et al. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. N. Engl. J. Med. 384, 229–237 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amanat F et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 26, 1033–1036 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wyllie AL et al. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. N. Engl. J. Med. 10.1056/NEJMc2016359 (2020). [DOI] [Google Scholar]
- 42.Vogels CBF et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat. Microbiol. 5, 1299–1305 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the background information on HCWs and clinical information for patients in this study are included in Source Data Fig. 1. Additionally, all of the raw FCS files for the flow cytometry analyses are available at ImmPort (https://www.immport.org/shared/home; study ID SDY1655). Additional correspondence and requests for materials should be addressed to the corresponding author (A.I). Source data are provided with this paper.