Abstract
Background
Early gut microbial colonization is important for postnatal growth and immune development of the chicken. However, at present, commercial chickens are hatched and raised without adult hens, thus are cut off from the microbiota transfer between hens and chicks. In this study, we compared the gut microbiota composition between hen-reared and separately reared chicks, and its impact on the resistance to H9N2 avian influenza virus, with the motive of investigating the impact of this cutoff in microbiota transfer.
Results
We used the 16SrRNA sequencing method to assess the composition of the gut microbiota in chicks represented by three hen-reared groups and one separately reared group. We found that the diversity of gut microbes in the chicks from the three hen-reared groups was more abundant than in the separately reared group, both at the phylum and genus levels. Our findings highlight the importance of early parental care in influencing the establishment of gut microbiota in the early life of chicks. SourceTracker analysis showed that the feather and cloaca microbiota of hens are the main sources of gut microbiota of chicks. After H9N2 exposure, the viral infection lasted longer in the separately reared chicks, with the viral titers in their oropharyngeal swabs being higher compared to the hen-reared chicks at day 5 post-infection. Interestingly, our results revealed that the gut microbiota of the hen-reared chicks was more stable after H9N2 infection in comparison to that of the separately reared chicks.
Conclusions
Microbiota transfer between the hens and their chicks promotes the establishment of a balanced and diverse microbiota in the early life of the chicks and improves microbiota stability after H9N2 challenge. These findings advance our understanding of the protective role of gut microbiota in the early life of chicks and should be instrumental in improving chick rearing in the commercial poultry industry.
Video Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s40168-021-01200-z.
Keywords: Gut microbiota, Maternal microbial transmission, H9N2 avian influenza virus, Disease resistance
Background
The complex microbiome on and within a host, critically linked with health and well-being, is referred to as the “second genome” of the host [1]. This “second genome” is not innate, but is derived from microbial dispersal and affects the growth [2, 3], immunity [4], behavior and cognitive abilities of the host [5, 6].
Maternal factors present in the prenatal and postnatal environments are important for gut microbial assembly in the offspring [7]. There is mounting evidence suggesting that the establishment of the gut microbiota in early life is dependent on mother-to-child transmission. For example, before the birth of a fetus, compounds produced by the mother’s microbiota can be transferred to the fetus to promote the generation of innate immune cells [8]. As the fetus passes through the birth canal, maternal vaginal microorganisms are ingested and vertically transferred [9]. After birth, a variety of microorganisms from the mother and the environment further colonize the gut of the newborn [9]. These processes are crucial for the recruitment and establishment of the neonatal microbiota and promote the development of the immune system in the gut, and other parts of the body, for defense against pathogens [10]. Babies born by cesarean delivery and fed formula have a delayed development of their gut microbiota due to insufficient contact with their mothers, which slows the maturation of the metabolic and immune systems [11–13].
In oviparous animals (such as birds), mechanisms associated with the establishment and assembly of early-life gut microbiota may be different from those observed in mammals [14]. Specifically, embryos develop in closed eggs and experience minimal contact with microbes from ovipositors, maternal feathers, and other components of the nest environment [15–17]. After hatching, parental care behaviors that include egg incubation, saliva exchange between parents and nestlings during feeding, and prolonged periods of physical contact between the parents and offspring, facilitate the transmission of gut microbiota and subsequent colonization in chicks [18, 19].
Recent studies have emphasized the imperative role of the gut microbiota in shaping immunity against viral diseases in chickens and ducks [20, 21]. However, the influence of the maternally mediated early-life assembly of the gut microbiota on the immune system in birds remains mostly unexplored. A study reported low gut microbial diversity in hand-reared passerine chicks and inferred that parental care is crucial in shaping the gut microbiota of the chicks [19]. However, the impact of microbiota transfer and establishment in the chicks, and its role in disease resistance demands to be investigated at deeper insights. As an important farm animal, chickens have been hatched and fed by hens throughout their evolutionary history. However, recent commercial chicken breeding is based on hatcheries and the chicks are raised in the absence of adult hens. The consequences of this switch in the feeding mode on the gut microbiome and associated immunity need to be addressed.
In this study, we examined and compared the gut microbiota composition and dispersal in hen-reared and separately reared chicks under laboratory conditions. Furthermore, we compared the resistance of these two groups of chicks against the H9N2 avian influenza virus (AIV) to evaluate the effect of the maternally mediated early-life gut microbiome assembly in immune development. Our research systematically studied the influence of maternal factors on the establishment of early-life gut microbiota and immune system development in chickens, which helps deepen our understanding of the healthy development of chickens in the poultry industry.
Results
Hen-reared chicks had higher gut microbiota diversity
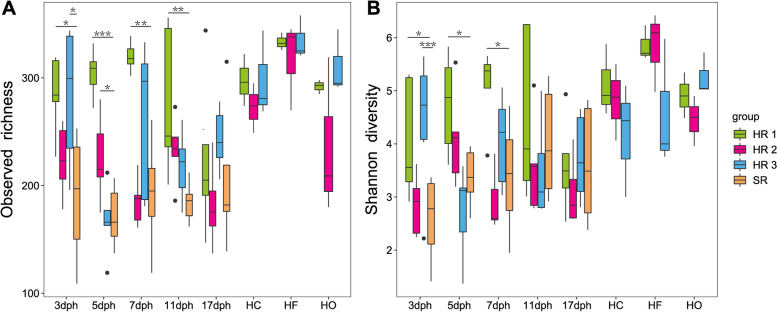
Three hen-reared (HR) groups and one separately reared (SR) group of chicks were designed for this study (Table S1). To determine whether maternal symbiotic microbes influence the gut microbiota diversity of the chicks, alpha diversity of gut microbiota in the chicks was estimated by the Shannon and Observed OTUs index (Fig. 1A, B and Table S2). Our results revealed that from days 3 to 5 post-hatching (dph), generally, gut microbiota diversity of hen-reared groups was significantly higher than that of chicks in the separately reared group (Dunnett test, P < 0.05). Gut microbiota richness of the hen-reared groups was significantly higher than the separately reared group at 3 to 11 dph (Dunnett test, P < 0.05). Age in days had an effect on the diversity and richness in the gut microbiota of the chicks of the separately reared group, with diversity and richness showing initial increase between 5 to 7 dph, followed by a subsequent decrease. For the maternal bacterial community, on average, feathers of the hens had a higher richness and diversity than the oropharyngeal and cloacal swabs (Fig. 1).
Fig. 1.
Boxplots showing the alpha diversity indices of gut microbiota from hen-reared (HR) chicks, separately reared (SR) chicks, and hens. A Observed richness and B Shannon diversity index. The horizontal bar in the boxes represents the median. The top and bottom of the boxes represent the 75th and 25th percentiles, respectively. The upper and lower whiskers extend to data not exceeding 1.5× the interquartile range from the upper edge and lower edge of the box, respectively. The asterisk represents significant difference in comparison to the separately reared group by the Dunnett test, *P < 0.05; **P <0.01;***P < 0.001. dph: days post-hatching; HC: hen cloacal swab; HF: hen feathers; HO: hen oropharyngeal swab
Hen-reared chicks had a richer initial gut microbiota composition
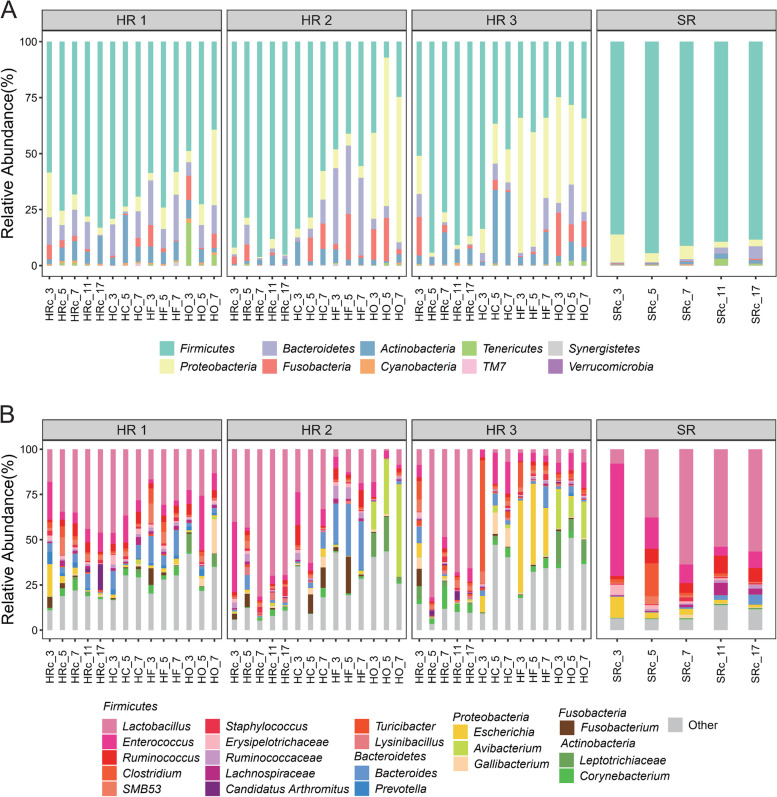
To investigate the establishment and variation of gut microbiota during chick development, we statistically analyzed the composition and abundance of bacterial taxa in the microbiota of chicks and hens. The microbial compositions of feather, oropharyngeal, and cloacal swabs of the hens are quite different. For example, at the phylum level (Fig. 2A), oropharyngeal swab samples from most of the hens had higher levels of Proteobacteria (average of 38.04%). In contrast, Firmicutes (feather: 49.36%; cloacal swab: 67.80%), Bacteroidetes (feather: 18.45%; cloacal swab: 6.41%), and Actinobacteria (feather: 6.15%; cloacal swab: 13.07%) were more prevalent in the feather and cloacal swabs. At the genus level (Fig. 2B), oropharyngeal swab samples from hens displayed abundance in Avibacterium (16.48%) and Leptotrichiaceae (9.74%), while feather samples had higher proportions of Bacteroides (13.99%) and Lactobacillus (13.27%). The cloacal swab samplers were noted to be enriched with Lactobacillus (26.71%), Enterococcus (10.88%), and Clostridium (7.82%).
Fig. 2.
Composition of the gut microbiota among the different groups. A Phylum-level and B top 20 common genus-level gut microbial communities in the hen-reared (HR) chicks, separately reared (SR) chicks, and maternal hen microbiota. HC: hen cloacal swab; HF: hen feathers; HO: hen oropharyngeal; HRc: hen-reared chick; SRc: separately reared chick
In the separately reared group, the composition of the gut microbiota of the chicks had high relative abundances of Firmicutes (86.16%, 95.5%, 91.27%) and Proteobacteria (12.45%, 4.08%, 5.91%) at 3–7 dph, with the relative abundances of other phyla being less than 1%. From 11 to 17 dph, a slight increase was noted in the abundances of Bacteroidetes (11 dph: 2.61%, 17 dph: 5.6%), Actinobacteria (11 dph: 2.26%, 17 dph: 1.62%) and Tenericutes (11 dph: 2.91%, 17 dph: 0.67%) in the separately reared group (Fig. 2A). At the genus level (Fig. 2B), Enterococcus (62.12%) was observed to be most abundant in the separately reared chick group at 3 dph, which decreased at 5 dph (17.26%), whereas Lactobacillus (37.80%), Clostridium (18.10%), and Ruminococcus (8.09%) increased. The gut microbiota was generally stable between 7 to 17 dph and was dominated by Lactobacillus (63.72–56.65%), followed by Ruminococcus (5.33–7.63%) and Enterococcus (10.31–8.98%).
Chicks in the hen-rearing group had more abundant gut microbiota, with high abundances of Firmicutes at 3 to 7 dph, including other phyla that were common in the hen gut communities — Bacteroidetes (3dph, HR 1–3: 12.22%, 1.02%, 10.23%; 5 dph, HR 1–3: 6.79%, 8.82%, 0.25%), Proteobacteria (3 dph, HR 1–3: 20.04%, 2.99%, 17.07%; 5 dph, HR 1–3: 6.47%, 3.10%, 1.73%), Actinobacteria (3 dph, HR 1–3: 2.08%, 0.49%, 3.75%; 5 dph, HR 1–3: 6.01%, 1.26%, 3.33%), and Fusobacteria (3 dph, HR 1–3: 6.52%, 3.27%, 17.23%; 5dph, HR 1–3: 3.25%, 7.37%, 0.09%) (Fig. 2A). At the genus level, chicks in the hen hen-reared groups were found to be generally enriched with Lactobacillus (HR 1–3: 18.06%, 40.15%, 8.72%), Enterococcus (HR 1–3: 20.86%, 38.90%, 6.82%), Fusobacterium (HR 1–3: 6.10%, 3.11%, 9.52%), and Bacteroides (HR 1–3: 4.69%, 0.92%, 8.81%) at 3 dph; and enriched with Lactobacillus (HR 1–3: 34.59%, 43.32%, 81.91%), SMB53 (HR 1–3: 9.72%, 4.09%, 2.35%), and Bacteroides (HR 1–3: 3.24%, 7.88%, 0.23%) at 5 dph.
Difference in the gut microbiota of chicks under different feeding modes
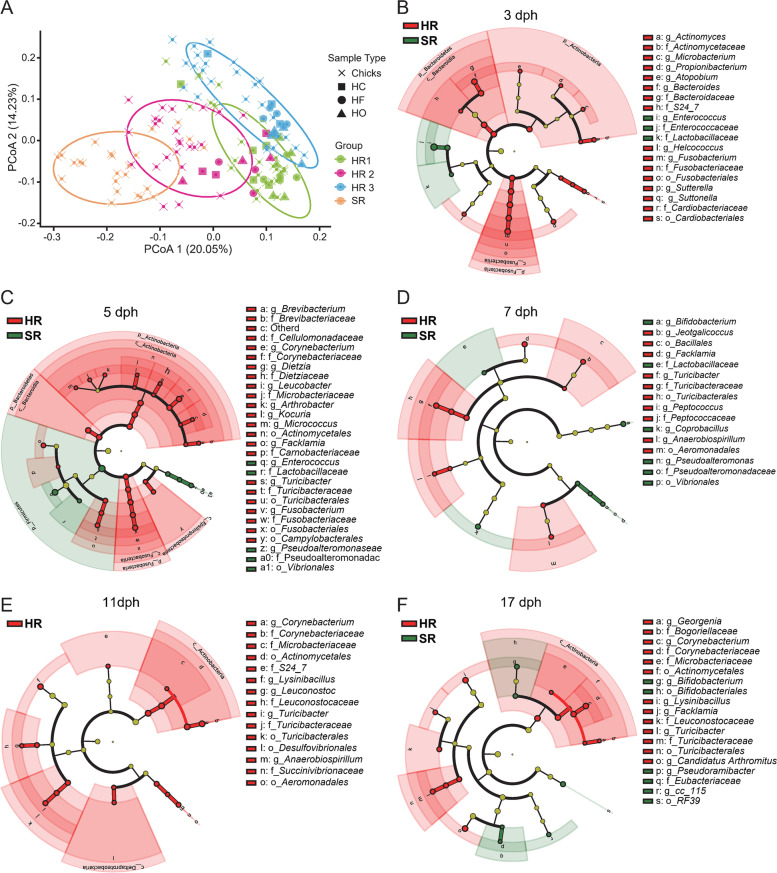
A principal coordinate analysis (PCoA), based on the Bray–Curtis distances, was used to assess the differences in bacterial community structure between the chick and the hen microbiome samples (Fig. 3A). The PCoA revealed that the samples of the hen-reared (HR) chicks were closer to their mothers. Our observations indicated that the gut microbiota of chicks in each group was influenced by the maternal bacterial communities. Moreover, the distance between the three hen-reared groups was closer (ANOSIM, R = 0.4748, P = 0.001).
Fig. 3.
Differences in the gut microbiota of chicks raised under different rearing patterns. A Principal coordinates analysis (PCoA) of the bacterial communities from the four reared group chicks and their hens. HC: hen cloacal swab; HF: hen feathers; HO: hen oropharyngeal. B–F LEfSe analysis generated differences in the abundance of the bacterial taxa of hen-reared groups (red) and separately reared group (green) chicks at different times (P < 0.05, LDA > 2)
Linear discriminant analysis effect size analysis (LEfSe) (P < 0.05, LDA > 2) identified representative gut bacterial taxa for the chicks in the hen-reared groups and the separately reared group. Consistent with the microbial composition results, the diversity of gut microbiota varied greatly in the early life of the chicks. At 3 to 5 dph (Fig. 3B, C), phyla Actinobacteria, Bacteroidetes, and Fusobacteria and their classified bacteria were the dominant genera in the hen-reared group, with only families Enterococcaceae and Lactobacillaceae being significantly enriched in the separately reared group. At 11 dph (Fig. 3E), no dominant microbiota genera were found in the separately reared group, whereas the gut microbiota of the hen-reared groups was enriched in the order Actinomycetales, genus Lysinibacillus, and families Muribaculaceae (S24-7), Leuconostocaceae, and Turicibacteraceae. At 17 dph (Fig. 3F), the family Eubacteriaceae, genus Erysipelotrichaceae CC-115, order Bifidobacteriales, and Enterococcaceae bacterium RF39 were dominant in the separately reared group, whereas the family Corynebacteriaceae, genus Lysinibacillus, and orders Actinomycetales and Turicibacteriales were prevalent in the chicks of the hen-reared groups.
Maternal gut microbes are substantial contributors to the gut microbiota of chicks in early life
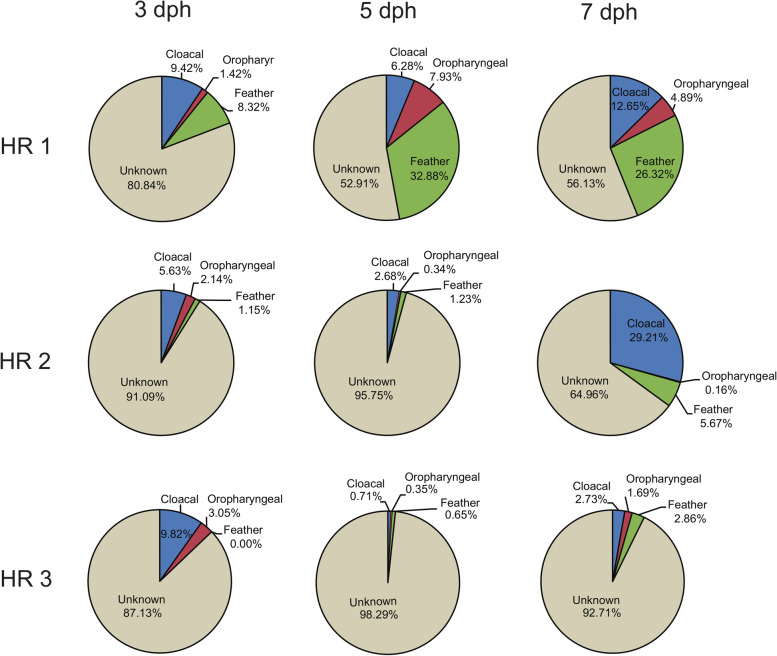
It was evident from our studies that the gut microbiota of chicks and their maternal hen’s microbiota had a considerable OTU-level similarity. Accordingly, we used Bayesian community-level source tracking to investigate the contribution of maternal sources in the gut community assembly of the chicks. The SourceTracker analysis predicted that the maternal cloacal swab and feather communities were important sources for the gut communities of the chicks (Fig. 4). At 3 dph, the chick gut microbiota communities of the three hen-reared groups were predicted to have received 9.42%, 5.63%, and 9.82% of their bacteria from respective hen cloacal swab communities, with an additional 1.15–9.32% from their hen’s feather communities. At 5 dph, the maternal microbiota of hen-reared groups 1 and 2 contributed had lower contribution to the chick’s gut microbiota, although the maternal microbiota of hen-reared group 1 was still the main source of the gut microbiota of its chicks. At 7 dph, the gut microbiota of the three hen-reared groups 1, 2, and 3 were 12.65%, 29.21%, and 2.73% from the cloaca of their hens, and 26.32%, 5.67%, and 2.86% from the hen’s feather, respectively. In contrast, the maternal oropharyngeal microbiota was not a major source of the gut microbiota for the chicks at any stage.
Fig. 4.
Community-level source-tracking models for the maternal sources of early gut community assembly. Pie charts of the mean proportions
Hen-reared chicks showed a more stable bacterial community composition after H9N2 infection
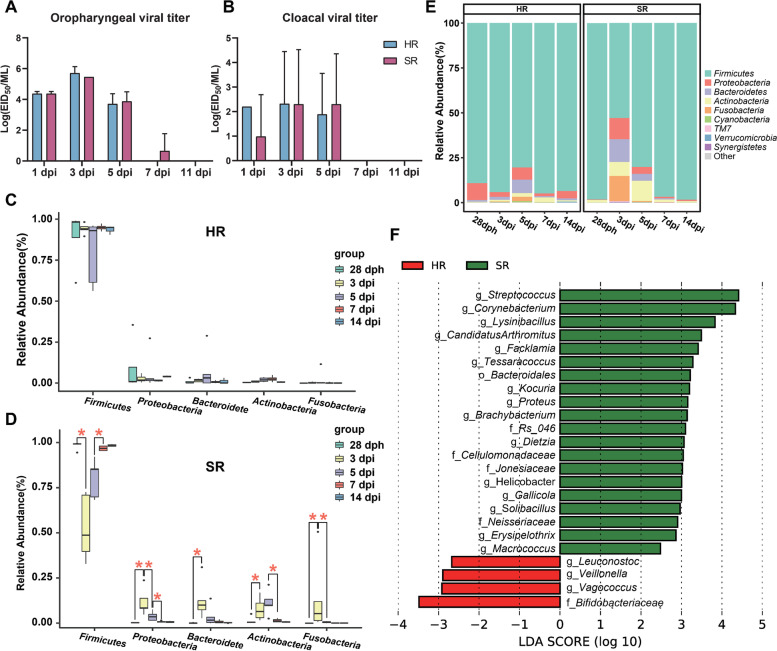
We predicted the functional profiles of the gut microbiota from the chicks using PICRUSt [22] and found that the pathways associated with membrane transport, amino acid metabolism, carbohydrate metabolism, and replication and repair were enriched in the hen-reared chicks. Most of these functional pathways can be linked to the basic life activities of the chicken. For the separately reared chicks, there was a greater enrichment of pathways associated with disease, such as infectious diseases (Figure S1). These results offered the scope to speculate that chicks with different rearing patterns might have differing resistance to infectious diseases. To assess the impact of hen rearing on disease resistance in chicks, we performed an infection experiment with the H9N2 avian influenza A virus (AIV). We selected the HR2 group of the hen-reared group of chicks (here in referred to as hen-reared group) and the separately reared group of chicks to infect with the H9N2 virus. At days 1, 3, 5, 7, and 11 post-infection (dpi), viral titers from oropharyngeal and cloacal swabs were determined (Fig. 5A, B). In both groups, the peak of viral shedding detected by oropharyngeal swabs was at 3 dpi with a decline at 5 dpi. The separately reared group of chicks still had detectable virus in one oropharyngeal swab at 7 dpi (1.95 lgEID50/mL, average of 0.65 lgEID50/mL). The viral titers assessed by cloacal swabs in the separately reared group (3.45 ± 0.71 lgEID50/mL) of chicks were slightly higher than in the hen-reared group (2.82 ± 0.53 lgEID50/mL) at 5 dpi, though this difference was not statistically significant.
Fig. 5.
Viral titers and gut microbiota of chicks infected with H9N2 virus in the hen-reared (HR) and separately reared (SR) groups of chicks. A, B Viral titers in oropharyngeal and cloacal swabs of post-H9N2-infected chickens. Chicks (28 dph) were intranasally inoculated with 10−6EID50/0.2ml of LPAIH9N2. EID50 was calculated by the Reed and Muench method. C, D Comparison of the relative abundances of the major bacterial phyla representing the gut microbiota of HR and SR groups after H9N2 infection; *P < 0.05; the horizontal bar in the boxes represents the median. The top and bottom of the boxes represent the 75th and 25th percentiles, respectively. The upper and lower whiskers extend to data not exceeding 1.5× the interquartile range from the upper edge and lower edge of the box, respectively. E Phylum-level composition of the gut microbiota in the post-H9N2-infected chickens. F LEfSe analysis of the 16SrRNA sequences of the post H9N2 infected chickens (P < 0.01, LDA > 2)
The bacterial community compositions of the cloacal swabs collected on 3, 5, 7, and 14 dpi were assessed by sequencing amplicons of the 16S rRNA gene. Before H9N2 challenge, our results showed that there were significant differences in the composition of the gut microbiomes between the hen-reared group (HR2 group) and the separately reared group (SR group) before 7dph, but there is no significant difference after 7dph (Table S3). For example, the family Fusobacteriaceae of the hen-reared group was significantly more abundant than in the separately reared group at 3 to 5 dph (Mann-Whitney U test, P < 0.05) (Table S3). There was no significant difference in the diversity (Dunnett test, P > 0.05) and composition (Mann-Whitney U test, P > 0.05) of gut microbiota between the separately reared group and hen-reared group at the day of the challenge (28 dph) (Table S2, Table S3). After H9N2 challenge, microbial classifications revealed that the composition of the chick microbiota changed more dramatically in the separately reared group after H9N2 challenge (Fig. 5C, D, and E). Within these chicks, Firmicutes were significantly reduced and Proteobacteria, Fusobacteria, Bacteroidetes, and Actinobacteria were significantly increased (Mann-Whitney U test, 28 dph vs 3 dpi, P < 0.05) at 3 dpi, whereas Fusobacteria and Proteobacteria were significantly reduced in the separately reared group at 5 dpi (Mann-Whitney U test, 3dpi vs 5 dpi, P < 0.05) (Fig. 5D, Table S4). At 7 dpi, Proteobacteria and Actinobacteria were significantly reduced and Firmicutes were significantly increased in the separately reared group (Mann-Whitney U test, 5dpi vs 7 dpi, P < 0.05) (Fig. 5D, Table S4). In contrast, the composition of the chick microbiota in the hen-reared group did not change significantly at the phylum level from 3 to 14 dpi (Fig. 5D, Table S4). Overall, the composition of chick microbiota in the hen-reared groups was observed to be more stable after H9N2 challenge (Fig. 5C, Table S4). Supervised comparison of samples using the LEfSe algorithm and logarithmic LDA (P < 0.01, LDA > 2) showed that in the hen-reared group, the cloacal swab microbiota was characterized by genera Vagococcus, Veillonella, and Leuconostoc and the family Bifidobacteriaceae, while the separately reared group were differentially enriched with the pathogenic bacterium genera Streptococcus and Corynebacterium, as well as Lysinibacillus, Facklamia, and Tessaracoccus (Fig. 5F).
Discussion
The immune system of animals rapidly develops early in life and is affected by the gut microbiome [23]. Gut microbiota in early life imprints the host immune phenotype for a long time and affects the ability to resist diseases in later phases of life [24]. Current poultry industry chick-rearing methods disrupt microbiota transmission between hens and their chicks. We explored the influence of maternal microbial sources on the early development of the chick gut microbiota in this study. Hen-reared chicks had a diverse gut microbiota, while the gut microbiome of the separately reared chicks had a much lower diversity and richness in the first few days after hatching. Our results are consistent with studies in mammals that suggest the high microbial diversity of gut communities of newborns might come from their biological mothers [25]. From the perspective of gut microbiota composition, the dominant phylum of the hen-reared chickens in the first week included Firmicutes, Bacteroidetes, Proteobacteria, Actinomycetes, and Fusobacteria, which were also abundant in the gut microbiota of the hens. The microbiota of the separately reared chicks was dominated by representatives of phylum Firmicutes at all time points examined in the present study. The composition of the gut microbiota of the hen-raised chicks and their mothers were more similar in comparison to the separately reared chicks, which suggests that hen feeding has a profound influence on the establishment of the initial gut microbiota in chicks.
A recent study on the gut microbiota of passerines demonstrated that the oral cavity microbiota of birds has a strong promoting effect on the establishment of their nestlings’ early gut microbiota as a probable consequence of the nestlings being fed by the birds through oral means [19]. Another study indicated that the similar gut microbial communities found in mothers and their nestlings may be due to vertical transmission through feeding [26]. Unlike passerines, hens do not feed their chicks through oral means; instead, the chicks peck food on the ground which is likely to be contaminated with their hen’s feces. We found that the maternal oral microbiota contributed little to the chick’s intestinal community at any stage. However, the hen gut microbiota, followed by feathers, was the most important source of a chick’s gut microbiota in early life (Fig. 4). The fact that chicks peck food contaminated with hen droppings on the ground increases the possibility of transmission of the maternal gut microbiota to the chicks. Chicks often hide under hen feathers to keep warm and thus have full contact with their hen’s feathers, which potentially results in the transfer of bacteria from the feathers to the chick gut.
Ecological theory suggests that microorganisms that pre-empt an ecological niche can affect the health of the host and the phenomenon is known as the “priority effect” [25]. In mammals, there is an overwhelming amount of evidence, associated with the development of the mammalian immune system in early life, which suggests that maternal microbes help build the gut microbiota and improve the immunity in infants [27, 28]. Birds separated from their parents, such as juvenile ostriches, fail to establish a balanced intestinal microflora that leads to increased mortality [29]. Our study revealed that the microbiota of hen-reared chicks had a greater diversity and higher proportion of potential health-associated bacteria, such as the genus Bacteroides [30], family Muribaculaceae (S24-7) [31, 32] and Fusobacteriaceae [33, 34], genus Turicibacter [29], and order Bacillales [35]. We speculate that this promises to have a beneficial impact on the development of the immune system in the chicks. PICRUSt predicted a greater risk of infectious diseases in the separately reared group of chicks (Figure S1). The H9N2 infection experiment, executed in the present study, supported this. Although the H9N2 infection disturbed the composition of the gut microbiota in both the hen-reared and separately reared groups, the hen-reared group had a relatively more stable gut microbiota post H9N2 infection (Fig. 5C, D, and E). The separately reared group showed drastic changes in the gut microbiota post H9N2 infection, with phyla Proteobacteria, Actinobacteria, Bacteroidetes, and Fusobacteria increasing significantly, while the phylum Firmicutes decreased sharply at 3 dpi. Our observations are consistent with previous findings that the infection of SPF chickens with H9N2 results in an increase in bacterial members representing the phylum Proteobacteria and a decrease in Firmicutes [36]. Members of phyla Bacteroidetes and Proteobacteria have earlier been reported to be increased in mice infected with H9N2 [37, 38]. Changes in gut microbiota can lead to disruption of mucosal immune responses in susceptible hosts [39]. Conversely, more stable gut microbiota might be an important contributory factor in conferring protection and resistance to the host against viral infections. The significant reduction in the phylum Firmicutes, which produce short-chain fatty acids, in the separately reared group may affect the bactericidal activity of alveolar macrophages and thus reduce the ability to combat secondary bacterial infections [40]. In our study, we found that the separately reared chicks had a more difficult access to rehabilitation after H9N2 infection. Viral titer positive duration was longer in the oropharyngeal swabs of chicks in the separately reared group. At 7 dpi, contrary to the chicks of the separately reared group, no viruses were detected in the oropharyngeal swabs of chicks in the hen-reared group. Viral titers in the oropharyngeal and cloacal swabs of the separately reared group were higher than in the hen-reared group at 5 dpi, though this difference was not significant.
Conclusions
In summary, our study investigated the influence of the disruption of microbiota transfer from hens to chicks under current commercial chicken breeding industry practices, where eggs are incubated in a hatchery in the absence of hens. Our analysis demonstrated that hen rearing helps chicks establish a much higher diversity of gut microbiota in early life. On the contrary, the guts of separately reared chicks were colonized by a large number of opportunistic or pathogenic bacteria (e.g., genera Enterococcus and Escherichia) in early life. Hen-reared chicks retain a relatively stable gut microbiota after H9N2 challenge. These findings advance our understanding of the role of vertical transmission in the initial establishment of neonatal intestinal microbiota of chicks and may be effective in improving the rearing of chicks in the commercial poultry industry.
Methods
Experimental animals and sample collection
In the present study, chicks were hatched from SPF White Leghorn chicken eggs. We chose the Qingyuan chicken as the hens because the White Leghorn chicken is not broody. We used three hen-reared (HR) groups and one separately reared (SR) group of chicks. For each hen-reared group, 10 fertile eggs were hatched by a brooding hen, with the newly hatched chicks raised in the presence of the hen. For the separately reared group, 10 fertile eggs were hatched in a cabinet egg incubator at 37.8 °C and 40–50% relative humidity, and the newly hatched chicks were separately reared. Chicks had free access to standard feed and water under a 12-h light and 12-h dark cycle. Cloacal swabs were collected from each chick at days 3, 5, 7, 11, and 17 post-hatching (dph) in all groups. Cloacal swabs, oropharyngeal swabs, and wing feathers were collected from each hen at 3, 5, and 7 dph. All samples were collected and kept on ice before being transported to the laboratory. The samples were immediately frozen at − 80 °C until DNA extraction.
Avian influenza virus infection experiments
H9N2 avian influenza virus (strain name: A/Chicken/Guangdong/Lz-wzp-10/2013, GenBank accession numbers OK035258 to OK035265) was propagated in 9-day-old SPF chicken embryos, titrated using the hemagglutination assay (HA), and stored at – 80 °C for later use. Chicks were infected with the H9N2 virus at 28 dph. Chicks from a randomly selected hen-reared group and the separately reared group were transferred to an isolator and were inoculated with 10−6 50% embryo infectious dose (EID50)/0.2 ml of the H9N2 virus via the ocular and nasal routes. At 3, 5, 7, and 11 days post-infection (dpi), oropharyngeal and cloacal swabs of the chicks were collected and stored in Dulbecco’s Modified Eagle’s Medium (DMEM) containing penicillin–streptomycin at a concentration of 10000u/mL to determine the viral titer. Viral titers of single samples were determined by the EID50 assay and the titer detection limit was 0.75 log10EID50/mL. The Mann-Whitney U test was used to compare the differences in viral titers between the hen-reared and separately reared groups. Cloacal swabs of the chicks were collected at 0 (28 dph), 3, 5, 7, and 14 dpi for 16S rRNA sequencing (Table S1).
DNA extraction and sequencing
Total DNA was extracted using the CTAB/SDS method. PCR amplicons for the V3 and V4 regions of the 16S rRNA gene were generated with the primers 341F (5′-CCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) [41]. TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, USA) was used to construct sequencing libraries. Constructed libraries were quantified by Qubit and Q-PCR and sequenced on the Illumina Novaseq 6000 platform. Sequence analysis of the 169 samples from the chick cloacal (n = 142), hen cloacal (n = 9), hen feather (n = 9) and hen oropharyngeal (n = 9) swab niches yielded 10,614,968 tags with an average of 62810.46 ± 5307.84 tags per sample.
16S rRNA data processing and statistical analysis
Paired-end reads were assigned to samples based on their unique barcode and were truncated by removing the barcode and primer sequences using the Cutadapt (version 1.18) pipeline [42]. Sequences containing ambiguous or low-quality bases were filtered using Trimmomatic [43]. Reads were merged using the FLASH software [44]. Clustering of operational taxonomic units (OTUs) was carried considering a sequence similarity of 97%. Chimeric sequences were removed using Vsearch [45]. We retained OTUs with abundances > 0.01% of the total abundance. Finally, we identified 418 OTUs for the downstream analysis. OTUs were annotated using GreenGenes (v.13.8, 97% identity reference set) [46] and classified at the domain, phylum, class, order, family, genus, and species levels.
Alpha diversity index (Shannon, Observed OTUs) was calculated for the normalized OTU table using the R package vegan (v.2.4.0) and phyloseq (v.1.14.0) [47]. For multiple-group comparisons, the Dunnett test was employed to calculate the significance of the alpha-diversity, with P values corrected using false discovery rate (FDR) correction. The CSS method was used to standardize the OTUs table before calculating the beta diversity. Principal coordinates analysis (PCoA) of the bacterial communities was performed using Bray–Curtis distances. Analysis of similarities (ANOSIM) was performed using the ANOSIM function implemented in the R package vegan, where R > 0 suggests that the distance within a group is less than the distance between groups and the groupings are effective.
SourceTracker was used to analyze the possible sources and proportions of the gut microbiota in the early life of the chicks [48]. Linear discriminant analysis (LDA) effect size (LEfSe) was performed using the online Huttenhower Galaxy server (http://huttenhower.sph.harvard.edu/galaxy/). Microbial functional profiles were predicted using PICRUSt [22]. The heatmaps were generated using the TBtools software [49], with the input data being standardized by the R functions scale (x, center = TRUE, scale = TRUE).
Supplementary Information
Additional file 1: Figure S1. Microbial metabolic pathways among the hen-reared (HR) and separately-reared (SR) groups. Input data was standardized with the R functions scale (x, center = TRUE, scale = TRUE).
Additional file 2: Table S1. Metadata for each sample used in this study.
Additional file 3: Table S2. Comparison of the alpha diversity (Observed OTU and Shannon index) between the pairwise groups performed using the Dunnett test.
Additional file 4: Table S3. Comparison of the relative abundances (mean ± SEM) of the major bacterial families representing the gut microbiota of the hen-reared (HR 2) group and separately-reared group at various stages from 3 dph to 28 dph.
Additional file 5: Table S4. Comparison of the relative abundances (mean ± SEM) of the major bacterial phyla representing the gut microbiota between the pairwise groups of post H9N2 infected chickens by Mann-Whitney U test.
Acknowledgements
Not applicable.
Abbreviations
- HR
Hen-reared
- SR
Separately reared
- dph
Days post-hatching
- dpi
Days post-infection
- HC
Hen cloacal
- HF
Hen feathers
- HO
Hen oropharyngeal
- PCoA
Principal coordinate analysis
- LEfSe
Linear discriminant analysis effect size
- AIV
Avian influenza virus
- HA
Hemagglutination assay
- EID50
50% embryo infectious dose
- OTUs
Operational taxonomic units
- ANOSIM
Analysis of similarities
Authors’ contributions
YS conceived, designed, and supervised the study. RB, XL, ZZ, XC, JP, RW, RY and XS did the animal experiment. XL and RB conducted bioinformatics analyses. XL and RB wrote the drafts of the manuscript. KX, AR and DMI commented on and revised the drafts of the manuscript. All authors read and approved the final draft of the manuscript.
Funding
This work was supported by the Guangdong Major Project of Basic and Applied Basic Research (2020B0301030007), Laboratory of Lingnan Modern Agriculture Project (NT2021007), the National Natural Science Foundation of China (31822056), the Guangdong Science and Technology Innovation Leading Talent Program (2019TX05N098), the 111 Project (D20008), and the Department of Education of Guangdong Province (2019KZDXM004 and 2019KCXTD001).
Availability of data and materials
The datasets generated in the current study were deposited to the NCBISRA database under the BioProject accession no. PRJNA747156.
Declarations
Ethics approval and consent to participate
Animal experiments were approved by South China Agricultural University’s Committee on the Ethics of Animal Experimentation (permit 2021f083).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaobing Li and Ran Bi contributed equally to this work.
References
- 1.Morar N, Bohannan BJ. The conceptual ecology of the human microbiome. Q Rev Biol. 2019;94(2):149–175. doi: 10.1086/703582. [DOI] [Google Scholar]
- 2.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13(4):260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwarzer M, Strigini M, Leulier F. Gut microbiota and host juvenile growth. Calcif Tissue Int. 2018;102(4):387–405. doi: 10.1007/s00223-017-0368-y. [DOI] [PubMed] [Google Scholar]
- 4.Thackray LB, Handley SA, Gorman MJ, Poddar S, Bagadia P, Briseño CG, et al. Oral antibiotic treatment of mice exacerbates the disease severity of multiple flavivirus infections. Cell Rep. 2018;22(13):3440–3453. doi: 10.1016/j.celrep.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson AL, Xia K, Azcarate-Peril MA, Rosin SP, Fine JP, Mu W, et al. Infant gut microbiome composition is associated with non-social fear behavior in a pilot study. Nat Commun. 2021;12(1):3294. doi: 10.1038/s41467-021-23281-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ezenwa VO, Gerardo NM, Inouye DW, Medina M, Xavier JB. Microbiology. Animal behavior and the microbiome. Science. 2012;338(6104):198–199. doi: 10.1126/science.1227412. [DOI] [PubMed] [Google Scholar]
- 7.Francis AP, Dominguez-Bello MG. Early-life microbiota perturbations and behavioral effects. Trends Microbiol. 2019;27(7):567–569. doi: 10.1016/j.tim.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Wang S, Egan M, Ryan CA, Boyaval P, Dempsey EM, Ross RP, et al. A good start in life is important-perinatal factors dictate early microbiota development and longer term maturation. FEMS Microbiol Rev. 2020;44(6):763–781. doi: 10.1093/femsre/fuaa030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willyard C. Could baby's first bacteria take root before birth? Nature. 2018;553(7688):264–266. doi: 10.1038/d41586-018-00664-8. [DOI] [PubMed] [Google Scholar]
- 10.Macpherson AJ, de Agüero MG, Ganal-Vonarburg SC. How nutrition and the maternal microbiota shape the neonatal immune system. Nat Rev Immunol. 2017;17(8):508–517. doi: 10.1038/nri.2017.58. [DOI] [PubMed] [Google Scholar]
- 11.Goedert JJ. Intestinal microbiota and health of adults who were born by cesarean delivery. JAMA Pediatr. 2016;170(10):1027. doi: 10.1001/jamapediatrics.2016.2310. [DOI] [PubMed] [Google Scholar]
- 12.Reyman M, van Houten MA, van Baarle D, Bosch A, Man WH, Chu M, et al. Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat Commun. 2019;10(1):4997. doi: 10.1038/s41467-019-13014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drall KM, Tun HM, Morales-Lizcano NP, Konya TB, Guttman DS, Field CJ, et al. Clostridioides difficile colonization is differentially associated with gut microbiome profiles by infant feeding modality at 3-4 months of age. Front Immunol. 2019;10:2866. doi: 10.3389/fimmu.2019.02866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39(2):372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 15.Nyholm SV. In the beginning: egg-microbe interactions and consequences for animal hosts. Philos Trans R Soc Lond B Biol Sci. 2020;375(1808):20190593. doi: 10.1098/rstb.2019.0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mallott EK, Amato KR. Host specificity of the gut microbiome. Nat Rev Microbiol. 2021. 10.1038/s41579-021-00562-3. [DOI] [PubMed]
- 17.Teyssier A, Lens L, Matthysen E, White J. Dynamics of gut microbiota diversity during the early development of an avian host: Evidence from a cross-foster experiment. Front Microbiol. 2018;9:1524. doi: 10.3389/fmicb.2018.01524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandl HB, van Dongen WF, Darolová A, Krištofík J, Majtan J, Hoi H. Composition of bacterial assemblages in different components of reed warbler nests and a possible role of egg incubation in pathogen regulation. PLoS One. 2014;9(12):e114861. doi: 10.1371/journal.pone.0114861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen CY, Chen CK, Chen YY, Fang A, Shaw GT, Hung CM, et al. Maternal gut microbes shape the early-life assembly of gut microbiota in passerine chicks via nests. Microbiome. 2020;8(1):129. doi: 10.1186/s40168-020-00896-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abaidullah M, Peng S, Kamran M. Current findings on gut microbiota mediated immune modulation against viral diseases in chicken. Viruses. 2019;11(8). 10.3390/v11080681. [DOI] [PMC free article] [PubMed]
- 21.Figueroa T, Bessière P, Coggon A, Bouwman KM, van der Woude R, Delverdier M, et al. The microbiota contributes to the control of highly pathogenic H5N9 influenza virus replication in ducks. J Virol. 2020;94(10). 10.1128/jvi.00289-20. [DOI] [PMC free article] [PubMed]
- 22.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31(9):814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warne RW, Kirschman L, Zeglin L. Manipulation of gut microbiota during critical developmental windows affects host physiological performance and disease susceptibility across ontogeny. J Anim Ecol. 2019;88(6):845–856. doi: 10.1111/1365-2656.12973. [DOI] [PubMed] [Google Scholar]
- 24.Amenyogbe N, Kollmann TR, Ben-Othman R. Early-life host-microbiome interphase: the key frontier for immune development. Front Pediatr. 2017;5:111. doi: 10.3389/fped.2017.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sprockett D, Fukami T, Relman DA. Role of priority effects in the early-life assembly of the gut microbiota. Nat Rev Gastroenterol Hepatol. 2018;15(4):197–205. doi: 10.1038/nrgastro.2017.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreisinger J, Kropáčková L, Petrželková A, Adámková M, Tomášek O, Martin JF, et al. Temporal stability and the effect of transgenerational transfer on fecal microbiota structure in a long distance migratory bird. Front Microbiol. 2017;8:50. doi: 10.3389/fmicb.2017.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al Nabhani Z, Eberl G. Imprinting of the immune system by the microbiota early in life. Mucosal Immunol. 2020;13(2):183–189. doi: 10.1038/s41385-020-0257-y. [DOI] [PubMed] [Google Scholar]
- 28.Gomez de Agüero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351(6279):1296–1302. doi: 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- 29.Videvall E, Song SJ, Bensch HM, Strandh M, Engelbrecht A, Serfontein N, et al. Early-life gut dysbiosis linked to juvenile mortality in ostriches. Microbiome. 2020;8(1):147. doi: 10.1186/s40168-020-00925-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown EM, Ke X, Hitchcock D, Jeanfavre S, Avila-Pacheco J, Nakata T, et al. Bacteroides-derived sphingolipids are critical for maintaining intestinal homeostasis and symbiosis. Cell Host Microbe. 2019;25(5):668–680. doi: 10.1016/j.chom.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Cui Y, Hu X, Liao X, Zhang Y. Chlorophyll supplementation in early life prevents diet-induced obesity and modulates gut microbiota in mice. Mol Nutr Food Res. 2019;63(21):e1801219. doi: 10.1002/mnfr.201801219. [DOI] [PubMed] [Google Scholar]
- 32.Ferrere G, Wrzosek L, Cailleux F, Turpin W, Puchois V, Spatz M, et al. Fecal microbiota manipulation prevents dysbiosis and alcohol-induced liver injury in mice. J Hepatol. 2017;66(4):806–815. doi: 10.1016/j.jhep.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 33.Lee SC, Chua LL, Yap SH, Khang TF, Leng CY, Raja Azwa RI, et al. Enrichment of gut-derived fusobacterium is associated with suboptimal immune recovery in hiv-infected individuals. Sci Rep. 2018;8(1):14277. doi: 10.1038/s41598-018-32585-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mutlu EA, Keshavarzian A, Losurdo J, Swanson G, Siewe B, Forsyth C, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in hiv infected subjects. PLoS Pathog. 2014;10(2):e1003829. doi: 10.1371/journal.ppat.1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng Y, Huang Y, Wang Y, Wang P, Song H, Wang F. Antibiotics induced intestinal tight junction barrier dysfunction is associated with microbiota dysbiosis, activated NLRP3 inflammasome and autophagy. PLoS One. 2019;14(6):e0218384. doi: 10.1371/journal.pone.0218384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yitbarek A, Weese JS, Alkie TN, Parkinson J, Sharif S. Influenza a virus subtype H9N2 infection disrupts the composition of intestinal microbiota of chickens. FEMS Microbiol Ecol. 2018;94(1). 10.1093/femsec/fix165. [DOI] [PubMed]
- 37.Zhang Q, Hu J, Feng JW, Hu XT, Wang T, Gong WX, et al. Influenza infection elicits an expansion of gut population of endogenous bifidobacterium animalis which protects mice against infection. Genome Biol. 2020;21(1):99. doi: 10.1186/s13059-020-02007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Liu X, Chen F, Zuo K, Wu C, Yan Y, et al. Avian influenza virus subtype H9N2 affects intestinal microbiota, barrier structure injury, and inflammatory intestinal disease in the chicken ileum. Viruses. 2018;10(5). 10.3390/v10050270. [DOI] [PMC free article] [PubMed]
- 39.Amoroso C, Perillo F, Strati F. The role of gut microbiota biomodulators on mucosal immunity and intestinal inflammation. Cells. 2020;9(5). 10.3390/cells9051234. [DOI] [PMC free article] [PubMed]
- 40.Sencio V, Barthelemy A, Tavares LP, Machado MG, Soulard D, Cuinat C, et al. Gut dysbiosis during influenza contributes to pulmonary pneumococcal superinfection through altered short-chain fatty acid production. Cell Rep. 2020;30(9):2934–2947. doi: 10.1016/j.celrep.2020.02.013. [DOI] [PubMed] [Google Scholar]
- 41.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal. 2011;17(1):10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 43.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Magoč T, Salzberg SL. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: A versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6(3):610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McMurdie PJ, Holmes S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, et al. Bayesian community-wide culture-independent microbial source tracking. Nat Methods. 2011;8(9):761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. Tbtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–202. 10.1016/j.molp.2020.06.009. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Microbial metabolic pathways among the hen-reared (HR) and separately-reared (SR) groups. Input data was standardized with the R functions scale (x, center = TRUE, scale = TRUE).
Additional file 2: Table S1. Metadata for each sample used in this study.
Additional file 3: Table S2. Comparison of the alpha diversity (Observed OTU and Shannon index) between the pairwise groups performed using the Dunnett test.
Additional file 4: Table S3. Comparison of the relative abundances (mean ± SEM) of the major bacterial families representing the gut microbiota of the hen-reared (HR 2) group and separately-reared group at various stages from 3 dph to 28 dph.
Additional file 5: Table S4. Comparison of the relative abundances (mean ± SEM) of the major bacterial phyla representing the gut microbiota between the pairwise groups of post H9N2 infected chickens by Mann-Whitney U test.
Data Availability Statement
The datasets generated in the current study were deposited to the NCBISRA database under the BioProject accession no. PRJNA747156.