Abstract
Background
There is a strong need for biomarkers to better characterize individuals with COPD and to take into account the heterogeneity of COPD. The blood protein sRAGE has been put forward as promising biomarker for COPD in general and emphysema in particular. Here, we measured plasma sRAGE levels using quantitative LC–MS and assessed whether the plasma sRAGE levels associate with (changes in) lung function, radiological emphysema parameters, and radiological subtypes of emphysema.
Methods
Three hundred and twenty-four COPD patients (mean FEV1: 63%predicted) and 185 healthy controls from the COPDGene study were selected. Plasma sRAGE was measured by immunoprecipitation in 96-well plate methodology to enrich sRAGE, followed by targeted quantitative liquid chromatography-mass spectrometry. Spirometry and HRCT scans (inspiration and expiration) with a 5-year follow-up were used; both subjected to high quality control standards.
Results
Lower sRAGE values significantly associated with the presence of COPD, the severity of airflow obstruction, the severity of emphysema on HRCT, the heterogeneous distribution of emphysema, centrilobular emphysema, and 5-year progression of emphysema. However, sRAGE values did not associate with airway wall thickness or paraseptal emphysema.
Conclusions
Rather than being a general COPD biomarker, sRAGE is especially a promising biomarker for centrilobular emphysema. Follow-up studies should elucidate whether sRAGE can be used as a biomarker for other COPD phenotypes as well.
Keywords: COPD biomarkers, Emphysema, CT phenotyping, sRAGE
Background
Chronic obstructive pulmonary disease (COPD) is characterized by persistent airflow limitation that is usually progressive and associates with an enhanced chronic airway inflammatory response in response to noxious particles or gases. The forced expiratory volume in one second (FEV1)/forced vital capacity (FVC) is used for the diagnosis and the FEV1 percent predicted is used for staging of this disease, however lung function does not provide information regarding disease activity or the underlying pathologic processes, cannot distinguish the various phenotypes of COPD such as emphysema, and is not specific for COPD. Novel biomarkers for COPD have therefore been explored in large-scale longitudinal studies like ECLIPSE, SUMMIT, SPIROMICS and COPDgene [1–5].
All these studies identified the soluble receptor for advanced glycation end-products (sRAGE) to be amongst the most promising blood biomarkers. sRAGE is the extracellular domain of the pro-inflammatory pattern recognition receptor RAGE. The freely circulating soluble form of RAGE can either be produced by alternative splicing of the AGER gene or by proteolytic cleavage of the receptor by proteases like MMP9 and ADAM10 [6]. While the membrane bound receptor has pro-inflammatory functions, sRAGE has anti-inflammatory properties by acting as a decoy receptor as well as preventing the homodimerization of RAGE, needed for down-stream signaling [7]. Recently, it became evident that RAGE signaling plays a key role in the development of COPD, contributing both to airway inflammation as well as emphysema [8–11]. Furthermore, a genetic polymorphism, rs2070600, within the AGER locus was found to be associated with circulating sRAGE levels and the risk of COPD development [12, 13]. Very robustly, all studies investigating sRAGE levels in COPD patients have found lower sRAGE levels in COPD patients [14–17] and found sRAGE to be associated with neutrophilic airway inflammation [18], acute exacerbation [17], decline in FEV1 [16], and presence of emphysema [19]. Particularly the association with emphysema is strong, showing significant correlations with carbon monoxide (CO) diffusion capacity [13, 20], and a number of quantitative HRCT measurements that reflect emphysema [20–22], as well as progression of emphysema [23].
Recently, we developed a targeted quantitative liquid chromatography-mass spectrometry (LC–MS)-based assay to detect and quantify sRAGE in blood [24]. This measurement is fully validated according to the U.S. Food and Drug Administration and European Medicines Agency guidelines and demonstrated decreased values in COPD patients, correlating with lung function and autofluorescence of advanced glycation end-products in the skin [15]. Furthermore, using this newly developed sRAGE assay, we found that smoking strongly decreases the serum levels of sRAGE within 2 h [25]. To further determine the value of sRAGE as biomarker for COPD, we now assessed the plasma sRAGE levels in the large, clinically well-characterized multi-center COPDGene cohort. The COPDGene study was chosen because we aimed to determine the role of sRAGE as a biomarker in subtypes of emphysema and the progression of emphysema over time, which could be assessed in COPDGene because this study had a 5-year follow-up, and because COPD patients were radiologically phenotyped on HRCT using a standardized classification [26].
Methods
Study participants
The study was approved by the Institutional Review Board of the National Jewish Health and all participants gave informed written consent. Former and current smokers with COPD and former and current smokers without spirometric impairment (called GOLD 0) were selected from the COPDGene study based on matching of age and body mass index (BMI) (Table 1). The GOLD 0 subject group has previously been defined as current and former smokers with a normal postbronchodilator ratio of FEV1 to forced vital capacity exceeding 0.7 and a FEV1 percentage of at least 80% predicted [27]. Subjects were between 45 and 80 years of age with a minimum of 10 pack-years smoking history (except non-smoking controls) [26]. In the COPDGene study, COPD was defined by a post-bronchodilator forced expiratory volume in the first second (FEV1) to forced vital capacity (FVC) ratio of < 0.70. Plasma was obtained from a venipuncture and P100 tube during the first (enrolling) visit on the same day as spirometry and CT scan.
Table 1.
Subject characteristics
| COPD patients | Healthy controls (GOLD 0) | p-values | |
|---|---|---|---|
| Number of subjects | 324 | 185 | NA |
| Number male subjects, % | 170 (52%) | 75 (41%) | 0.0100 |
| Age, years | 64.9 (8.0) | 65.0 (5.8) | 0.94 |
| BMI, kg/m2 | 28.4 (5.5) | 29.3 (4.9) | 0.07 |
| Number current smoker, % | 84 (26%) | 45 (24%) | 0.75 |
| Pack years | 51.4 (25.0) | 39.3 (22.0) | < 0.0001 |
| GOLD stages (I/II/III/IV) | 79/144/78/23 | NA | NA |
| FEV1, % predicted | 62.4 (22.1) | 98.0 (11.2) | < 0.0001 |
| Decrease in FEV1 in 5 years, mL | 209 (281) | 218 (215) | 0.70 |
Data presented as mean (SD) or number (% proportion), statistical significant results are depicted in bold
NA not applicable
Measurements
sRAGE was measured at baseline during the first visit of the COPDgene study. sRAGE was measured using a simplified immunoprecipitation in 96-well ELISA (IPE) methodology to enrich sRAGE coupled to targeted liquid chromatography-mass spectrometry (LC–MS) [24]. This assay measures sRAGE at clinically relevant levels between 0.1 and 10 ng/mL, which necessitates a considerable degree of sample cleanup for which anti-sRAGE antibodies (R&D Systems, Cat. No. MAB11451, clone 176902) were used. LC–MS analyses were performed with a Waters Ionkey/MS system using an ACQUITY M-Class UPLC and a XEVO mass spectrometer (Milford, MA, U.S.A.), and sRAGE was detected based on two protein-specific tryptic peptides (i.e., IGEPLVLK, VLSPQGGGPWDSVAR) originating from sRAGE’s N-terminal region which is essential for binding to most of its ligands. sRAGE values demonstrated a normal distribution in our population.
HRCT-scans were acquired using multi-detector CT scanners with at least 16 detector channels [26]. Volumetric CT acquisitions were obtained both on full inspiration (200 mAs), and at the end of normal expiration (50 mAs). Image reconstruction utilized sub-millimeter slice thickness, with smooth and edge-enhancing algorithms [26].
CT phenotyping was performed on segmented lung images, using Thirona Lung Quantification software (Thirona, http://www.thirona.eu). Total inspiratory and expiratory lung volumes, mean lung attenuation, and percentage of low attenuation areas (%LAA) below -950HU on inspiration and below -856HU on expiration were determined for the whole lung and each of the lobes independently [26]. Presence of emphysema was considered if %LAA below -950HU was larger than 6% [28], presence of heterogeneous emphysema was considered if the difference in %LAA below − 950HU between the left upper lobe (LUL) and left lower lobe (LLL) or between the right upper lobe (RUL) plus middle lobe (RML) and right lower lobe (RLL) was larger than 10%. Additionally, the 15th percentile of the lung density histogram within patients was determined (Perc15) [29]. Airway wall thickness was expressed as Pi10, defined as the square root of the wall area at the inner perimeter of a 10 mm diameter airway. This measurement provides a useful summary score of the airway wall thickness for an individual patient, and has shown to be a measure for smoking-related airway injury that can provide important information regarding longitudinal changes in airway wall thickness [30, 31].
Parallel imaging analyses of percent emphysema and percent gas trapping were performed using 3D Slicer (http://www.slicer.org/). Parametric Response Mapping (PRM) provided 3 categories: emphysema (all voxels below -950HU in the inspiratory CT and below -856 HU in the expiratory CT), air trapping (all voxels above -950HU in the inspiratory CT and below -856 HU in the expiratory CT), and normal (all voxels above both thresholds in both scans) [32]. Paraseptal emphysema was visually scored as absent, mild or substantial, and centrilobular emphysema was scored as absent, trace, mild, moderate, confluent or advanced destructive as previously described [33].
All spirometry data were collected using an EasyOne spirometer (ndd Medical Technologies, Zurich, Switzerland) and were reviewed by the pulmonary function test quality assurance core analyst of the COPDGene Study [34]. Spirometric data were typically collected at the same day as the acquired CT studies (mean time between spirometry and CT, 0.31 h).
Statistics
Data evaluation was performed using SPSS statistical software package version 23.0. p-values < 0.05 were considered statistically significant. Differences in clinical variables (age, pack years, BMI, lung function) and quantitative HRCT measures (volumes, mean densities, %LAA, Pi10, Perc15, and PRM parameters) between COPD patients and GOLD 0 were analyzed by T-Test or Mann–Whitney U, and differences between GOLD stages by Kruskal–Wallis tests. Categorical variables (sex, current smokers, centrilobular emphysema, paraseptal emphysema) were analyzed by a Chi-square test. Correlations between Srage and clinical or quantitative HRCT measures were examined by Pearson’s or Spearman’s Rank tests. The independent contribution of Srage to the variation of (5-year changes in) FEV1 and (5-year changes in) quantitative HRCT measures were analyzed by multiple linear regression adjusted for age, gender, current smoking, pack years, and BMI.
Results
Subject characteristics
Three hundred and twenty four COPD patients and 185 healthy controls (GOLD 0) were selected from the COPDGene cohort. Patient characteristics are described in Table 1. Within the COPD group, 26% were current smokers and within the healthy control group, 24% of the individuals were current smokers. Males were significantly more present in the COPD group as compared to controls: 52% vs 41% (p = 0.0100). The mean (SD) FEV1 of the COPD group was 62.4 (22.1) %predicted, indicating mild to moderate severe disease. Mean annual decline in FEV1 was similar between COPD patients and GOLD 0 controls: 42 vs 44 mL/year.
sRAGE correlation with COPD characteristics
Mean (SD) sRAGE levels were significantly lower in COPD compared to GOLD 0 controls: 1.97 (0.90) vs 2.23 (1.03) ng/mL, and more severe COPD stages demonstrated lower values compared to milder COPD stages (Fig. 1a). There was a weak but significant correlation between sRAGE and FEV1%predicted in the total and COPD population: R = 0.23 (p < 0.0001) and 0.25 (p < 0.0001), respectively. Significant but weaker correlations were also found for FEV1 (0.15, p = 0.0007) and FEV1/FVC (0.19, p < 0.0001) in the total population and for FEV1 (0.17, p = 0.0025) and FEV1/FVC (0.21, p = 0.0001) in the COPD population. In a multiple-linear regression analysis, sRAGE independently contributed to FEV1 (Table 2). sRAGE did not associate with 5-year change in FEV1, neither in ordinary nor in multiple-linear regression analyses. Additionally, we showed that the levels of sRAGE are significantly lower in subjects possessing the minor allele of the AGER polymorphism rs2070600 (Fig. 1b). This decrease was most severe in homozygotes for the minor allele (AA), but was already present in heterozygotes (GA).
Fig. 1.
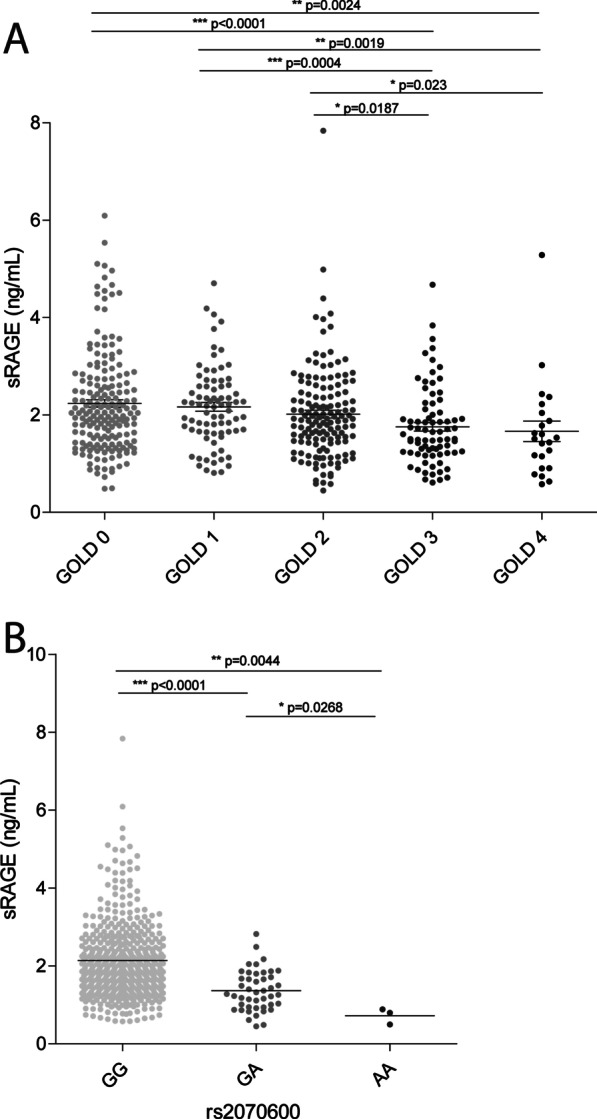
sRAGE in healthy controls (GOLD 0) and COPD patients (GOLD 1–4). A The levels of sRAGE were assessed in plasma using the simplified immunoprecipitation in 96-well ELISA (IPE) methodology coupled to targeted liquid chromatography-mass spectrometry (LC–MS). Plasma sRAGE levels were assessed in 185 healthy individuals without airway obstruction (GOLD 0), 79 GOLD stage I COPD patients, 144 GOLD stage II COPD patients, 78 GOLD stage III COPD patients and 23 GOLD stage IV COPD patients. b The plasma levels of sRAGE in subjects homozygous for the major allele of rs2070600 (GG), heterozygous (GA), or homozygous for the minor allele of rs2070600 (AA). Data is shown as individuals data points and mean ± SEM. Statistical differences were tested using a Mann–Whitney U test, *p < 0.05, **p < 0.01, ***p < 0.001. The exact p-values are indicated where appropriate
Table 2.
sRAGE explaining the variation in FEV1 and quantitative and visual HRCT measurements
| Baseline | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FEV1 | %LAA Below -950HU | Centrilobular emphysema | Pi10 | Air trapping (PRM) | ||||||
| R square: 0.199 | R square: 0.235 | R square: 0.280 | R square: 0.090 | R square: 0.186 | ||||||
| Beta | p-value | Beta | p-value | Beta | p-value | Beta | p-value | Beta | p-value | |
| sRAGE | 0.159 | 0.0001 | − 0.229 | < 0.0001 | 0.188 | < 0.0001 | − 0.046 | 0.31 | − 0.143 | 0.0009 |
| Sex | − 0.379 | < 0.0001 | − 0.143 | 0.0005 | − 0.059 | 0.14 | − 0.055 | 0.22 | − 0.159 | 0.0002 |
| Age | − 0.139 | 0.0017 | − 0.041 | 0.34 | 0.014 | 0.74 | − 0.118 | 0.0135 | 0.120 | 0.0093 |
| BMI | 0.020 | 0.62 | − 0.323 | < 0.0001 | − 0.323 | < 0.0001 | 0.120 | 0.0062 | − 0.218 | < 0.0001 |
| Current smoking | 0.067 | 0.11 | − 0.238 | < 0.0001 | − 0.090 | 0.0280 | 0.102 | 0.0269 | − 0.161 | 0.0003 |
| Pack years | − 0.220 | < 0.0001 | 0.183 | < 0.0001 | 0.387 | < 0.0001 | 0.187 | < 0.0001 | 0.195 | < 0.0001 |
| Change between baseline and 5-year follow-up | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FEV1 | %LAA Below -950HU |
Centrilobular emphysema | Pi10 | Air trapping (PRM) | ||||||
| R square: 0.051 | R square: 0.103 | R square: | R square: 0.013 | R square: 0.051 | ||||||
| Beta | p-value | Beta | p-value | Beta | p-value | Beta | p-value | Beta | p-value | |
| sRAGE | 0.020 | 0.66 | − 0.172 | 0.0003 | ND | ND | − 0.033 | 0.50 | − 0.002 | 0.97 |
| Sex | − 0.204 | < 0.0001 | − 0.084 | 0.08 | ND | ND | − 0.072 | 0.15 | 0.007 | 0.89 |
| Age | − 0.055 | 0.26 | − 0.060 | 0.23 | ND | ND | 0.033 | 0.53 | − 0.108 | 0.0433 |
| BMI | − 0.085 | 0.05 | − 0.146 | 0.0019 | ND | ND | 0.037 | 0.46 | − 0.068 | 0.17 |
| Curent smoking | 0.030 | 0.52 | 0.150 | 0.0024 | ND | ND | 0.014 | 0.79 | 0.142 | 0.0061 |
| Pack years | − 0.079 | 0.08 | 0.098 | 0.0400 | ND | ND | − 0.084 | 0.09 | 0.063 | 0.21 |
Beta standardized coefficient. Adjusted for age at enrollment, sex, BMI, current smoking, pack years. ND not determined (no 5-year follow-up scores done). sRAGE levels, BMI, smoking status and pack years were obtained at baseline, lung function parameters were obtained at baseline and at 5-year follow-up, statistical significant results are depicted in bold
HRCT analyses
Quantitative HRCT analyses demonstrated lower lung attenuation and higher airway wall thickness in COPD compared to GOLD 0 controls (Table 3). The proportion of individuals with radiological emphysema, based on %LAA below -950HU ≥ 6%, was higher in COPD patients than in GOLD 0 controls: 51% vs 11%, respectively (p < 0.0001). Within the group of individuals with %LAA below − 950HU ≥ 6% heterogeneous emphysema was present in 30% and homogeneous emphysema in 70% of the individuals (Table 4). Visually scored centrilobular emphysema (score > 0) was present in 86% of the COPD patients and 47% in the GOLD 0 controls (p < 0.0001). Paraseptal emphysema (> score 0) was present in 46% of the COPD patients and 29% of the GOLD 0 controls (p < 0.0001). The 5-year change in emphysema assessed by %LAA, Perc15 and PRM parameters was significantly higher in COPD patients than GOLD 0 controls (Table 3). In contrast, the 5-year change in airway wall thickness and air trapping was not significantly different between these two groups.
Table 3.
HRCT characteristics and correlation with sRAGE
| Radiological difference between healthy and COPD | sRAGE correlation within total group | ||||
|---|---|---|---|---|---|
| COPD patients | Healthy controls (GOLD 0) | P-value | Rho | P-value | |
| LAA below -950 HU, % | 10.37 (10.59) | 2.41 (3.00) | < 0.0001 | − 0.241 | < 0.0001 |
| 5-year change | 1.83 (4.85) | − 0.36 (2.38) | < 0.0001 | − 0.187 | 0.0001 |
| LAA % below -856 HU, % | 30.85 (18.77) | 10.08 (7.94) | < 0.0001 | − 0.186 | < 0.0001 |
| 5-year change | 3.29 (9.83) | 0.75 (6.20) | 0.0036 | − 0.115 | 0.0183 |
| Perc15, HU | 934.6 (22.8) | 914.8 (17.9) | < 0.0001 | 0.201 | < 0.0001 |
| 5-year change | − 3.65 (10.66) | 0.71 (13.06) | 0.0001 | 0.082 | 0.09 |
| PRM Emphysema, % | 8.9 (10.5) | 0.9 (1.5) | < 0.0001 | − 0.226 | < 0.0001 |
| 5-year change | 2.24 (4.85) | 0.03 (1.1) | < 0.0001 | − 0.217 | < 0.0001 |
| PRM Air trapping, % | 22.9 (12.2) | 9.2 (6.5) | < 0.0001 | − 0.147 | 0.0014 |
| 5-year change | 1.43 (7.64) | 0.87 (5.00) | 0.41 | − 0.015 | 0.75 |
| PRM Normal, % | 65.7 (20.5) | 88.3 (8.8) | < 0.0001 | 0.210 | < 0.0001 |
| 5-year change | − 3.45 (9.91) | − 0.54 (6.65) | 0.0109 | 0.109 | 0.0254 |
| Pi10, mm | 2.43 (0.52) | 1.85 (0.34) | < 0.0001 | − 0.099 | 0.0270 |
| 5-year change | 0.03 (0.33) | 0.03 (0.14) | 0.96 | − 0.033 | 0.49 |
| Centrilobular emphysema | 2 (0–5) | 0 (0–3) | < 0.0001 | − 0.197 | < 0.0001 |
| 5-year change | NA | NA | NA | NA | NA |
| Paraseptal emphysema | 0 (0–2) | 0 (0–2) | < 0.0001 | − 0.081 | 0.07 |
| 5-year change | NA | NA | NA | NA | NA |
Group data given in mean (standard deviation), or median (range)
LAA low attenuation area, Perc15 15th percentile of the lung density histogram (HU), Pi10 airway wall thickness at an internal perimeter of 10 mm, PRM parametric response mapping, NA not available, statistical significant results are depicted in bold
Table 4.
sRAGE in different subtypes of emphysema
| No Emphysema (LAA below − 950HU < 6%) | Emphysema (LAA below -950HU 6%) |
|||
|---|---|---|---|---|
| Homogeneous | Heterogeneous | |||
| No dominance − 10% < HD < 10% | Upper HD > 10% | Lower HD < − 10% | ||
| Number | 315 | 125 | 45 | 9 |
| Presence COPD | 49%1 | 86%2a | 98% | 100% |
| FEV1%pred | 84.7% (20.0)1 | 63.1% (26.5)2a | 56.9% (24.3)3 | 36.6% (10.3) |
| %LAA below -950HU | 1.97 (1.65)1 | 13.86 (7.65)2c | 23.38 (9.65) | 29.19 (11.16) |
| Centrilobular | 1 (0–4)1 | 3 (0–5) 2c | 4 (3–5)3 | 4 (2–5) |
| Paraseptal | 0 (0–2)1 | 0 (0–2)2a | 1 (0–2) | 0 (0–2) |
| sRAGE | 2.22 (1.02)1 | 1.89 (0.75)2b | 1.53 (0.73) | 1.57 (0.45) |
Data given in number, percentage, mean (standard deviation), or median (range). HD: heterogeneity difference (Upper–Lower Lobes %LAA below -950HU)
1p < 0.0001 vs emphysema
2ap < 0.05 vs heterogeneous emphysema
2bp < 0.01 vs heterogeneous emphysema
2cp < 0.0001 vs heterogeneous emphysema
3p < 0.05 vs lower lobe heterogeneous emphysema
sRAGE associations with HRCT
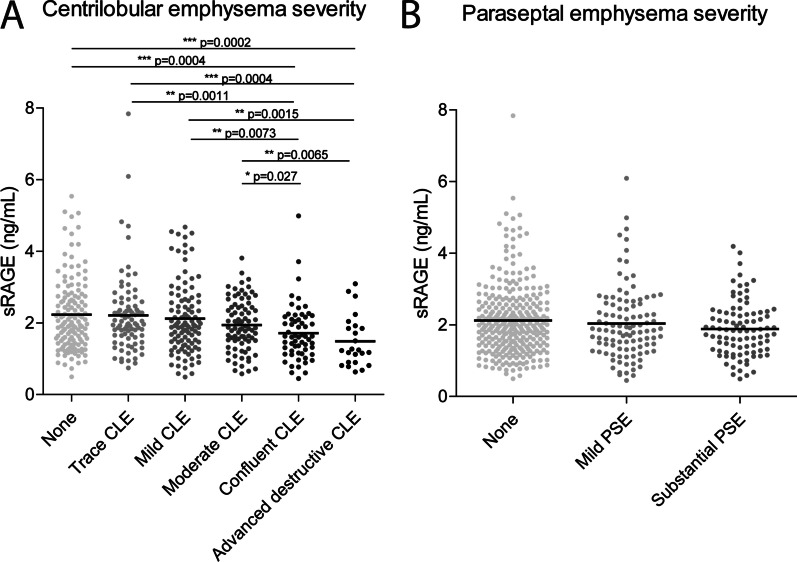
sRAGE correlated significantly with all quantitative HRCT measurements (Table 2). In multiple-linear regression analysis, lower sRAGE levels contributed independently to higher emphysema scores based on %LAA below 950HU (Table 4), and higher air trapping scores based on PRM scores (Table 4). However, there was no significant association with airway wall thickness assessed by Pi10. sRAGE levels were significantly lower in heterogeneous compared to homogeneous emphysema, but there was no difference between upper lobe versus lower lobe predominant emphysema (Table 3). sRAGE levels were significantly lower in more severe centrilobular emphysema (Fig. 2), but there was no association with paraseptal emphysema. In 5-year follow-up, lower sRAGE levels correlated significantly with changes in most emphysema parameters, but not with airway wall thickness, nor air trapping (Table 2). In a multiple-linear regression analysis, lower sRAGE values contributed independently to the 5-year progression of emphysema, but not to the change in airway thickness, nor air trapping (Table 4).
Fig. 2.
sRAGE associates with centrilobular emphysema. The levels of sRAGE were assessed in plasma using the simplified immunoprecipitation in 96-well ELISA (IPE) methodology coupled to targeted liquid chromatography-mass spectrometry (LC–MS). A The association of plasma sRAGE with centrilobular emphysema (CLE) was assessed by measuring sRAGE in plasma of 139 subjects without CLE, 85 subjects with trace CLE, 107 subjects with mild CLE, 82 subjects with moderate CLE, 60 subjects with confluent CLE and 24 subjects with advanced destructive CLE. B The association of plasma sRAGE with paraseptal emphysema (PSE) was assessed by measuring sRAGE in plasma of 299 subjects without PSE, 105 subjects with mild PSE and 93 subjects with substantial PSE. Data is shown as individuals data points and mean ± SEM. Statistical differences were tested using a Mann–Whitney U test, *p < 0.05, **p < 0.01, ***p < 0.001. The exact p-values are indicated where appropriate
Discussion
We found that lower plasma sRAGE levels were significantly associated with the presence of COPD, the AGER polymorphism rs2070600, the severity of airflow obstruction, the severity of emphysema on HRCT, heterogeneous distribution of emphysema, the centrilobular subtype of emphysema, and 5-year progression of emphysema severity. In contrast, sRAGE levels did not associate with airway wall thickness or the paraseptal subtype of emphysema. As the highest associations were observed with the severity of centrilobular emphysema, sRAGE seems to be an attractive biomarker for this radiological subtype of emphysema.
A novel finding of this study is the strong association of sRAGE with centrilobular emphysema, the most common form of smoking-related emphysema which involves lung tissue around the terminal bronchioles, at the center of the secondary lobules [35]. sRAGE demonstrated a significant association with heterogeneous emphysema, but not specifically with upper or lower lung predominance. It is unclear whether sRAGE independently contributes to the presence of heterogeneous emphysema, as heterogeneous emphysema (according to our definition) can only be present in more severely emphysematous lungs. An important additional finding of our study is the absence of a relationship with paraseptal emphysema, contributing to the idea that sRAGE is a more specific COPD sub-type biomarker. This finding is not unexpected, taking into account the completely different underlying pathology of centrilobular and paraseptal emphysema [28], whereas centrilobular emphysema is characterized by destruction of the central parts of the lung lobule, paraseptal emphysema is characterized by destruction of the lung lobules near the lobular septa and is observed mainly near the pleural surface in the upper regions of the lungs.
Because we were primarily interested in the biomarker function of plasma sRAGE in COPD and not in smoking-related effects, we did not compare COPD patients to never smokers but to a previously defined GOLD 0 group. This group encompasses former and current smokers that do not display any spirometric impairment. Our study confirms earlier reports suggesting that sRAGE shows a higher association with emphysema compared to chronic bronchitis [13, 19]. In this regard, sRAGE did not associate with Pi10, a marker reflecting airway wall thickness and associated with symptom-based chronic bronchitis [36]. Our study demonstrated also a significant association between sRAGE and FEV1, however this association was weaker compared to the association between sRAGE and emphysema. From literature it was known that emphysema independently contributes to airflow obstruction reflected by FEV1 and FEV1/FVC, a contribution that is relatively more important than that of airway wall thickness [37]. We hypothesized that sRAGE contributes indirectly to FEV1, via emphysema, and not airway wall thickness or chronic bronchitis. Also, air trapping on expiration CT scans, due to the collapse of small airways, could be mediated via such an indirect effect of emphysema. Such a hypothesis could be investigated in depth by contrasting (almost) pure chronic bronchitis and (almost) pure emphysema patients, thereby excluding the overlap between the chronic bronchitis and emphysema phenotype [38].
A recent review on the potential role of sRAGE as biomarker for COPD described several limitations of sRAGE [19], including the lack of a clinically validated assay, leading to a rather wide disparity in absolute values found between studies. The current study used a fully validated sRAGE assay following FDA guidelines [24]. By using this assay, we recently demonstrated that smoking immediately decreases sRAGE values in serum [25, 39]. Particularly in current smokers this may decrease the biomarker function of sRAGE and explain in part the variation observed in the 25% current smokers of our study. A second pitfall described in the review is the potentially confounding effect on sRAGE by a number of comorbidities [19]. We agree that this should be taken into account in larger studies than the current one. Another limitation is the scarce available information on the therapeutic modulation of RAGE signaling in human disease [19]. However, it was shown in mice that overexpression of RAGE induces inflammation and causes airspace enlargement [40], whereas RAGE knockout mice hardly develop cigarette smoke- or elastase-induced emphysema [41, 42], further supporting that RAGE signaling plays an important mechanistic role in emphysema. A further limitation of the potential biomarker role of sRAGE, not described in the abovementioned review, is its inverse relationship with disease activity of COPD, meaning that the plasma levels of sRAGE are decreasing with an increase in disease severity. This inverse relationship is probably due to the fact that free circulating sRAGE acts as a protective decoy-receptor, by binding pro-inflammatory RAGE-ligands and thus avoiding the activation of membrane bound RAGE as well as preventing the homodimerization of RAGE needed for downstream signaling of RAGE. In that perspective, we believe that the biomarker function of sRAGE can be improved by studying sRAGE in the context of its ligands, particularly the ligands that are involved in emphysematous processes.
A strength of our study is that emphysema was visually subtyped into paraseptal and centrilobular emphysema. Although visual scoring is subjective, it was performed by researchers who were blinded to any clinical or functional information, demonstrating high inter-observer agreement, comparable to other observations in literature [43, 44]. Nevertheless, there is a clear unmet need to optimize the radiological characterization of COPD, and particularly the automated quantification of subtypes of emphysema [45]. Furthermore, the plasma sRAGE levels between none, trace, mild and moderate centrilobular emphysema were quite similar, likely because these categories of emphysema are often quite localized and therefore would result in relatively little changes in circulating sRAGE levels.
Conclusion
In conclusion, this is the first study demonstrating a strong association between sRAGE and the visually scored severity of centrilobular emphysema. In contrast, there was no association with paraseptal emphysema nor with airway wall thickness. This specificity of sRAGE for centrilobular emphysema shows that endotyping of a disease needs careful and meticulous (sub)phenotyping of the disease. Further studies are needed to replicate our finding, also including more severe emphysema patients.
Acknowledgements
Not applicable.
Abbreviations
- ADAM10
A disintegrin and metalloproteinase domain-containing protein 10
- AGER
Advanced glycosylation end-product specific receptor
- BMI
Body mass index
- CLE
Centrilobular emphysema
- COPD
Chronic obstructive pulmonary disease
- CT
Computed tomography
- FDA
Food and Drug Administration
- FEV1
Forced expiratory volume in 1 s
- FVC
Forced vital capacity
- GOLD
Global initiative for chronic obstructive lung disease
- HD
Heterogeneity difference
- HRCT
High-resolution computed tomography
- HU
Hounsfield Units
- IPE
Immunoprecipitation in 96-well enzyme-linked immunosorbent assay
- LC–MS
Liquid chromatography–mass spectrometry
- LLL
Left lower lobe
- LUL
Left upper lobe
- MMP9
Matrix metallopeptidase 9
- NA
Not applicable
- ND
Not determined
- Perc15
15Th percentile of the lung density histogram
- Pi10
Square root of the wall area at the inner perimeter of a 10 mm diameter airway
- PRM
Parametric response mapping
- PSE
Paraseptal emphysema
- RAGE
Receptor for advanced glycation end-products
- RLL
Right lower lobe
- RML
Right middle lobe
- RUL
Right upper lobe
- SD
Standard deviation
- SEM
Standard error of the mean
- SNP
Single nucleotide polymorphism
- sRAGE
Soluble receptor for advanced glycation end-products
- %LAA
Percentage of low attenuation areas
- %LAA950
Percentage ratio of low-attenuation areas below a threshold of − 950 Hounsfield units
Authors' contributions
Clinical data collection: DAL, SH, RBo Analytical data collection: FK, MK, SDP Data analysis: FK, PH, EvR, JPC, NHTtH, SDP Manuscript drafting: FK, NHTtH, SDP Manuscript editing: FK, RBi, NHTtH Manuscript approval: FK, PH, RBo, EvR, JPC, DAL, MK, SH, Rbi, NHTtH, SDP. All authors read and approved the final manuscript.
Funding
SDP received funding from The Lung Foundation Netherlands (6.2.15.044JO). FK, RBi, NHTtH and SDP acknowledge the Dutch Technology Foundation STW (NWO-STW Perspectief program P12-04) and the Dutch Biomarker Development Center (BDC) for support of this work.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the PASSEL repository under the dataset identifier PASS01670, http://www.peptideatlas.org/PASS/PASS01670.
Declarations
Ethics approval and consent to participate
All clinical procedures were performed according to the standards set by the latest. Declaration of Helsinki. The study was approved by the Institutional Review Board of the National Jewish Health and all participants gave informed written consent.
Consent for publication
Not applicable.
Competing interests
The authors have nothing to disclose.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cazzola M, Puxeddu E, Ora J, Rogliani P. Evolving concepts in chronic obstructive pulmonary disease blood-based biomarkers. Mol Diagn Ther. 2019;23:603–614. doi: 10.1007/s40291-019-00413-1. [DOI] [PubMed] [Google Scholar]
- 2.Celli BR, Anderson JA, Brook R, Calverley P, Cowans NJ, Crim C, et al. Serum biomarkers and outcomes in patients with moderate COPD: a substudy of the randomised SUMMIT trial. BMJ Open Respir Res. 2019;6:e000431. doi: 10.1136/bmjresp-2019-000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faner R, Tal-Singer R, Riley JH, Celli B, Vestbo J, MacNee W, et al. Lessons from ECLIPSE: a review of COPD biomarkers. Thorax (England) 2014;69:666–672. doi: 10.1136/thoraxjnl-2013-204778. [DOI] [PubMed] [Google Scholar]
- 4.Pratte KA, Curtis JL, Kechris K, Couper D, Cho MH, Silverman EK, et al. Soluble receptor for advanced glycation end products (sRAGE) as a biomarker of COPD. Respir Res. 2021;22:127. doi: 10.1186/s12931-021-01686-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zemans RL, Jacobson S, Keene J, Kechris K, Miller BE, Tal-Singer R, et al. Multiple biomarkers predict disease severity, progression and mortality in COPD. Respir Res. 2017;18:117. doi: 10.1186/s12931-017-0597-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L, Bukulin M, Kojro E, Roth A, Metz VV, Fahrenholz F, et al. Receptor for advanced glycation end products is subjected to protein ectodomain shedding by metalloproteinases. J Biol Chem. 2008;283:35507–35516. doi: 10.1074/jbc.M806948200. [DOI] [PubMed] [Google Scholar]
- 7.Zong H, Madden A, Ward M, Mooney MH, Elliott CT, Stitt AW. Homodimerization is essential for the receptor for advanced glycation end products (RAGE)-mediated signal transduction. J Biol Chem. 2010;285:23137–23146. doi: 10.1074/jbc.M110.133827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee H, Lee J, Hong S-H, Rahman I, Yang S-R. Inhibition of RAGE attenuates cigarette smoke-induced lung epithelial cell damage via RAGE-mediated Nrf2/DAMP signaling. Front Pharmacol. 2018;9:684. doi: 10.3389/fphar.2018.00684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee H, Park J-R, Kim WJ, Sundar IK, Rahman I, Park S-M, et al. Blockade of RAGE ameliorates elastase-induced emphysema development and progression via RAGE-DAMP signaling. FASEB J. 2017;31:2076–2089. doi: 10.1096/fj.201601155R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allam VSRR, Faiz A, Lam M, Rathnayake SNH, Ditz B, Pouwels SD, et al. RAGE and TLR4 differentially regulate airway hyperresponsiveness: Implications for COPD. Allergy. 2020;76(4):1123–1135. doi: 10.1111/all.14563. [DOI] [PubMed] [Google Scholar]
- 11.Chen M, Wang T, Shen Y, Xu D, Li X, An J, et al. Knockout of RAGE ameliorates mainstream cigarette smoke-induced airway inflammation in mice. Int Immunopharmacol. 2017;50:230–235. doi: 10.1016/j.intimp.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 12.Faiz A, Rathnayake SNH, Ten Hacken NHT, Guryev V, van den Berge M, Pouwels SD. Single-nucleotide polymorphism rs2070600 regulates AGER splicing and the sputum levels of the COPD biomarker soluble receptor for advanced glycation end-products. ERJ Open Res. 2021;7:2. doi: 10.1183/23120541.00947-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng DT, Kim DK, Cockayne DA, Belousov A, Bitter H, Cho MH, et al. Systemic soluble receptor for advanced glycation endproducts is a biomarker of emphysema and associated with AGER genetic variants in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2013;188:948–957. doi: 10.1164/rccm.201302-0247OC. [DOI] [PubMed] [Google Scholar]
- 14.Gopal P, Reynaert NL, Scheijen JLJM, Engelen L, Schalkwijk CG, Franssen FME, et al. Plasma advanced glycation end-products and skin autofluorescence are increased in COPD. Eur Respir J. 2014;43:430–438. doi: 10.1183/09031936.00135312. [DOI] [PubMed] [Google Scholar]
- 15.Hoonhorst SJM, Lo Tam Loi AT, Pouwels SD, Faiz A, Telenga ED, et al. Advanced glycation endproducts and their receptor in different body compartments in COPD. Respir Res. 2016;17:46. doi: 10.1186/s12931-016-0363-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwamoto H, Gao J, Pulkkinen V, Toljamo T, Nieminen P, Mazur W. Soluble receptor for advanced glycation end-products and progression of airway disease. BMC Pulm Med. 2014;14:68. doi: 10.1186/1471-2466-14-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith DJ, Yerkovich ST, Towers MA, Carroll ML, Thomas R, Upham JW. Reduced soluble receptor for advanced glycation end-products in COPD. Eur Respir J. 2011;37:516–522. doi: 10.1183/09031936.00029310. [DOI] [PubMed] [Google Scholar]
- 18.Sukkar MB, Wood LG, Tooze M, Simpson JL, McDonald VM, Gibson PG, et al. Soluble RAGE is deficient in neutrophilic asthma and COPD. Eur Respir J. 2012;39:721–729. doi: 10.1183/09031936.00022011. [DOI] [PubMed] [Google Scholar]
- 19.Yonchuk JG, Silverman EK, Bowler RP, Agustí A, Lomas DA, Miller BE, et al. Circulating soluble receptor for advanced glycation end products (sRAGE) as a biomarker of emphysema and the RAGE axis in the lung. Am J Respir Crit Care Med. 2015;192:785–792. doi: 10.1164/rccm.201501-0137PP. [DOI] [PubMed] [Google Scholar]
- 20.Cockayne DA, Cheng DT, Waschki B, Sridhar S, Ravindran P, Hilton H, et al. Systemic biomarkers of neutrophilic inflammation, tissue injury and repair in COPD patients with differing levels of disease severity. PLoS ONE. 2012;7:e38629. doi: 10.1371/journal.pone.0038629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miniati M, Monti S, Basta G, Cocci F, Fornai E, Bottai M. Soluble receptor for advanced glycation end products in COPD: relationship with emphysema and chronic cor pulmonale: a case-control study. Respir Res. 2011;12:37. doi: 10.1186/1465-9921-12-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carolan BJ, Hughes G, Morrow J, Hersh CP, O’Neal WK, Rennard S, et al. The association of plasma biomarkers with computed tomography-assessed emphysema phenotypes. Respir Res. 2014;15:127. doi: 10.1186/s12931-014-0127-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coxson HO, Dirksen A, Edwards LD, Yates JC, Agusti A, Bakke P, et al. The presence and progression of emphysema in COPD as determined by CT scanning and biomarker expression: a prospective analysis from the ECLIPSE study. Lancet Respir Med. 2013;1:129–136. doi: 10.1016/S2213-2600(13)70006-7. [DOI] [PubMed] [Google Scholar]
- 24.Klont F, Pouwels SD, Hermans J, van de Merbel NC, Horvatovich P, Ten Hacken NHT, et al. A fully validated liquid chromatography-mass spectrometry method for the quantification of the soluble receptor of advanced glycation end-products (sRAGE) in serum using immunopurification in a 96-well plate format. Talanta. 2018;182:414–421. doi: 10.1016/j.talanta.2018.02.015. [DOI] [PubMed] [Google Scholar]
- 25.Pouwels SD, Klont F, Kwiatkowski M, Wiersma VR, Faiz A, van den Berge M, et al. Cigarette smoking acutely decreases serum levels of the chronic obstructive pulmonary disease biomarker sRAGE. Am J Respir Crit Care Med. 2018;198:1456–1458. doi: 10.1164/rccm.201807-1249LE. [DOI] [PubMed] [Google Scholar]
- 26.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, et al. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Regan EA, Lynch DA, Curran-Everett D, Curtis JL, Austin JHM, Grenier PA, et al. Clinical and radiologic disease in smokers with normal spirometry. JAMA Intern Med. 2015;175:1539–1549. doi: 10.1001/jamainternmed.2015.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynch DA, Austin JHM, Hogg JC, Grenier PA, Kauczor H-U, Bankier AA, et al. CT-definable subtypes of chronic obstructive pulmonary disease: a statement of the Fleischner Society. Radiology. 2015;277:192–205. doi: 10.1148/radiol.2015141579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stolk J, Putter H, Bakker EM, Shaker SB, Parr DG, Piitulainen E, et al. Progression parameters for emphysema: a clinical investigation. Respir Med (England) 2007;101:1924–1930. doi: 10.1016/j.rmed.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 30.Paulin LM, Smith BM, Koch A, Han M, Hoffman EA, Martinez C, et al. Occupational exposures and computed tomographic imaging characteristics in the SPIROMICS Cohort. Ann Am Thorac Soc. 2018;15:1411–1419. doi: 10.1513/AnnalsATS.201802-150OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller ER, Putman RK, Diaz AA, Xu H, San José Estépar R, Araki T, et al. Increased airway wall thickness in interstitial lung abnormalities and idiopathic pulmonary fibrosis. Ann Am Thorac Soc. 2019;16:447–454. doi: 10.1513/AnnalsATS.201806-424OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pompe E, van Rikxoort EM, Schmidt M, Rühaak J, Estrella LG, Vliegenthart R, et al. Parametric response mapping adds value to current computed tomography biomarkers in diagnosing chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2015;191:1084–1086. doi: 10.1164/rccm.201411-2105LE. [DOI] [PubMed] [Google Scholar]
- 33.Bhatt SP, Washko GR, Hoffman EA, Newell JDJ, Bodduluri S, Diaz AA, et al. Imaging advances in chronic obstructive pulmonary disease. Insights from the Genetic Epidemiology of Chronic Obstructive Pulmonary Disease (COPDGene) Study. Am J Respir Crit Care Med. 2019;199:286–301. doi: 10.1164/rccm.201807-1351SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schroeder JD, McKenzie AS, Zach JA, Wilson CG, Curran-Everett D, Stinson DS, et al. Relationships between airflow obstruction and quantitative CT measurements of emphysema, air trapping, and airways in subjects with and without chronic obstructive pulmonary disease. AJR Am J Roentgenol. 2013;201:W460–W470. doi: 10.2214/AJR.12.10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi M, Yamada G, Koba H, Takahashi H. Classification of centrilobular emphysema based on CT-pathologic correlations. Open Respir Med J. 2012;6:155–159. doi: 10.2174/1874306401206010155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim V, Davey A, Comellas AP, Han MK, Washko G, Martinez CH, et al. Clinical and computed tomographic predictors of chronic bronchitis in COPD: a cross sectional analysis of the COPDGene study. Respir Res. 2014;15:52. doi: 10.1186/1465-9921-15-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van TN, Ryujin Y, Ogawa E, Trang LTH, Kanda R, Goto K, et al. Relative contributions of emphysema and airway remodelling to airflow limitation in COPD: consistent results from two cohorts. Respirology (Carlton, VIC) 2015;20:594–601. doi: 10.1111/resp.12505. [DOI] [PubMed] [Google Scholar]
- 38.Mohamed Hoesein FAA, Schmidt M, Mets OM, Gietema HA, Lammers J-WJ, Zanen P, et al. Discriminating dominant computed tomography phenotypes in smokers without or with mild COPD. Respir Med. 2014;108:136–143. doi: 10.1016/j.rmed.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 39.Pouwels SD, Klont F, Kwiatkowski M, Wiersma VR, Faiz A, van den Berge M, et al. Reply to biswas: acute and chronic effects of cigarette smoking on sRAGE. Am J Respir Crit Care Med. 2019;199(6):806–807. doi: 10.1164/rccm.201812-2257LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stogsdill MP, Stogsdill J, Bodine BG, Fredrickson AC, Sefcik TL, Wood TT, et al. Conditional overexpression of receptors for advanced glycation end-products in the adult murine lung causes airspace enlargement and induces inflammation. Am J Respir Cell Mol Biol. 2013;49:128–134. doi: 10.1165/rcmb.2013-0013OC. [DOI] [PubMed] [Google Scholar]
- 41.Waseda K, Miyahara N, Taniguchi A, Kurimoto E, Ikeda G, Koga H, et al. Emphysema requires the receptor for advanced glycation end-products triggering on structural cells. Am J Respir Cell Mol Biol. 2015;52:482–491. doi: 10.1165/rcmb.2014-0027OC. [DOI] [PubMed] [Google Scholar]
- 42.Wolf L, Herr C, Niederstraßer J, Beisswenger C, Bals R. Receptor for advanced glycation endproducts (RAGE) maintains pulmonary structure and regulates the response to cigarette smoke. PLoS ONE. 2017;12:e0180092. doi: 10.1371/journal.pone.0180092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gietema HA, Müller NL, Fauerbach PVN, Sharma S, Edwards LD, Camp PG, et al. Quantifying the extent of emphysema: factors associated with radiologists’ estimations and quantitative indices of emphysema severity using the ECLIPSE cohort. Acad Radiol. 2011;18:661–671. doi: 10.1016/j.acra.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 44.Kim SS, Yagihashi K, Stinson DS, Zach JA, McKenzie AS, Curran-Everett D, et al. Visual assessment of CT findings in smokers with nonobstructed spirometric abnormalities in the COPDGene(®) study. Chronic Obstructive Pulmon Dis (Miami, FLA) 2014;1:88–96. doi: 10.15326/jcopdf.1.1.2013.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cavigli E, Camiciottoli G, Diciotti S, Orlandi I, Spinelli C, Meoni E, et al. Whole-lung densitometry versus visual assessment of emphysema. Eur Radiol. 2009;19:1686–1692. doi: 10.1007/s00330-009-1320-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the PASSEL repository under the dataset identifier PASS01670, http://www.peptideatlas.org/PASS/PASS01670.