Using a novel DNA methylation measure, we find that children growing up in more disadvantaged families and neighborhoods exhibit a faster pace of biological aging.
Abstract
BACKGROUND AND OBJECTIVES:
Children who grow up in socioeconomic disadvantage face increased burden of disease and disability throughout their lives. One hypothesized mechanism for this increased burden is that early-life disadvantage accelerates biological processes of aging, increasing vulnerability to subsequent disease. To evaluate this hypothesis and the potential impact of preventive interventions, measures are needed that can quantify early acceleration of biological aging in childhood.
METHODS:
Saliva DNA methylation and socioeconomic circumstances were measured in N = 600 children and adolescents aged 8 to 18 years (48% female) participating in the Texas Twin Project. We measured pace of biological aging using the DunedinPoAm DNA methylation algorithm, developed to quantify the pace-of-aging–related decline in system integrity. We tested if children in more disadvantaged families and neighborhoods exhibited a faster pace of aging as compared with children in more affluent contexts.
RESULTS:
Children living in more disadvantaged families and neighborhoods exhibited a faster DunedinPoAm-measured pace of aging (r = 0.18; P = .001 for both). Latinx-identifying children exhibited a faster DunedinPoAm-measured pace of aging compared with both White- and Latinx White–identifying children, consistent with higher levels of disadvantage in this group. Children with more advanced pubertal development, higher BMI, and more tobacco exposure exhibited faster a faster DunedinPoAm-measured pace of aging. However, DunedinPoAm-measured pace of aging associations with socioeconomic disadvantage were robust to control for these factors.
CONCLUSIONS:
Children growing up under conditions of socioeconomic disadvantage exhibit a faster pace of biological aging. DNA methylation pace of aging might be useful as a surrogate end point in evaluation of programs and policies to address the childhood social determinants of lifelong health disparities.
What’s Known on This Subject:
Children who grow up in socioeconomically disadvantaged families face increased burden of disease throughout their lives. One hypothesis is that early-life disadvantage accelerates biological aging, but measures of biological aging in children that are sensitive to socioeconomic inequalities are lacking.
What This Study Adds:
Using a novel DNA methylation measure, we find that children growing up in more disadvantaged families and neighborhoods exhibit a faster pace of biological aging. DNA methylation measures might be useful to help project long-term health impacts from childhood interventions.
Individuals exposed to social adversity in childhood experience higher burden of aging-related disease later in life.1 Children in low socioeconomic status (SES) families experience a suite of material hardships and psychological stressors that increase risk for later-life health problems, including cardiovascular disease, Type 2 diabetes, cancer, dementia, and a shorter life span.2–4 These childhood socioeconomic gradients in adult-onset disorders partly reflect socioeconomic gradients in health problems that begin during childhood, including obesity, asthma, and stress-related mental health problems.5–7 However, even after accounting for childhood-onset health problems and adult SES, adult health continues to be graded by childhood SES.2,6,8
One hypothesis to explain social inequalities in health is that early-life disadvantage accelerates biological processes of aging.9,10 Biological aging is the gradual and progressive decline in system integrity that occurs with advancing age.11 It is theorized to originate with the accumulation of cellular-level changes, including telomere attrition and epigenetic alterations, which, in turn, drive deterioration in tissues and organ systems.12 Biological aging in this framework is distinct from programmed development. However, damage accumulation during development may accelerate aging.13 Analyses of telomere length suggest that cellular aging is already variable in childhood14,15 and may be accelerated by early-life adversity.16–18
New uncertainty about the validity of telomere length measurement as a biomarker of aging has complicated interpretation of observations that disadvantaged children tend to have shorter telomeres than their more-privileged peers.19,20 At the same time, DNA methylation–based measures known as epigenetic clocks have emerged as candidate biomarkers of aging in humans and other species.21 In some studies of adults, lower SES is associated with more advanced biological aging, as measured by epigenetic clocks, although effect sizes are small.22 In contrast, the authors of a recent meta-analysis concluded that SES was unrelated to either telomere length or epigenetic clocks in children.23 It is therefore currently unknown whether there are biomarkers of aging that reflect the socioeconomic gradient in children’s health.
Here, we use an alternative approach to the cellular-level measurement of biological aging in childhood, focusing on the pace of aging. The epigenetic clocks were developed from an analysis in which chronologically older individuals were compared with younger ones. A novel DNA methylation measure, DunedinPoAm, was developed from an analysis of rate of longitudinal change in organ-system integrity occurring in middle-adulthood in a cohort of individuals who were all the same chronological age.24 Whereas epigenetic clocks quantify the amount of aging that has already occurred up to the time of measurement, DunedinPoAm quantifies how fast an individual is aging.25 In other words, whereas epigenetic clocks tell you what time it is, pace-of-aging measures tell you how fast the clock is ticking. In children whose aging trajectories are only beginning to diverge, differences in accumulated aging (clock time) may be small. Pace-of-aging measures, which measure divergence in trajectories, might therefore be more sensitive to social determinants of health early in the life span. Indeed, in a cohort of 18-year-olds, DunedinPoAm recorded faster pace of aging in those with histories of childhood poverty and victimization, whereas several epigenetic clocks did not detect differences between these groups.24
In this article, we report the first analysis of DunedinPoAm-measured pace of aging in children. We analyzed saliva DNA methylation from 600 White- or Latinx-identifying children aged 8 to 18 from the population-based Texas Twin Project to examine whether family- and neighborhood-level cumulative socioeconomic disadvantage was associated with a faster methylation pace of aging. We also examined whether children’s racial and/or ethnic identities were associated with methylation pace of aging. For comparison, we repeated the analysis using several epigenetic clocks.26–30
Methods
Sample
Participants included 600 (285 girls) children and adolescents from 328 unique families aged 8 to 18 years (mean = 12.68, SD = 3.02) from the Texas Twin Project.31 The Texas Twin Project is an ongoing longitudinal study that includes the collection of salivary samples. Saliva samples were selected to be assayed for DNA methylation by using EPIC arrays if participants self-identified their race/ethnicity as White and/or Latinx and had contributed cortisol data (not reported here). After we excluded 8 participants during DNA methylation preprocessing, there were n = 457 participants who identified as only White, n = 77 who identified as only Latinx, and n = 61 who identified as Latinx and White. We capitalize these terms to highlight that racial and/or ethnic identities are social constructions that are not based on innate biosocial boundaries but may have biosocial effects through people’s lived experiences.32 The University of Texas Institutional Review Board granted ethical approval.
DNA Methylation and DunedinPoAm
Saliva samples were collected during a laboratory visit by using Oragene kits (DNA Genotek, Ottawa, Ontario, Canada). DNA extraction and methylation profiling were conducted by the Edinburgh Clinical Research Facility (Edinburgh, United Kingdom). The Infinium MethylationEPIC BeadChip Kit (Illumina, Inc, San Diego, CA) was used to assess methylation levels at 850 000 methylation sites. Methylation profiles were residualized for cell composition, array, and slide; all samples came from the same batch (see Supplemental Information for details). Comparison of DunedinPoAm-measured pace of aging in blood and saliva by using out-of-sample data sets is reported in Supplemental Fig 3. The analysis indicated good correspondence of DunedinPoAm measurements across blood and saliva.
See Table 1 for a description of DunedinPoAm-measured pace of aging calculation and the Supplemental Information for a description of epigenetic clocks calculation, which were converted to age-acceleration residuals for analysis by regressing participants’ computed epigenetic-clock age values on their chronological ages and predicting residual values.
TABLE 1.
Description of Study Measures
| Description | |
|---|---|
| DunedinPoAm-measured pace of aging | DunedinPoAm-measured pace of aging was calculated on the basis of the published algorithm24 by using codes available at https://github.com/danbelsky/DunedinPoAm38. Briefly, DunedinPoAm was developed from DNA methylation analysis of pace of aging in the Dunedin Study birth cohort. Pace of aging is a composite phenotype derived from analysis of longitudinal change in 18 biomarkers of organ-system integrity measured when Dunedin Study members were all 26, 32, and 38 y of age.25 Elastic-net regression machine learning analysis was used to fit pace of aging to Illumina 450k DNA methylation data generated from blood samples collected when participants were aged 38 y. The elastic-net regression produced a 46-CpG algorithm. Increments of DunedinPoAm-measured pace of aging correspond to years of physiologic change occurring per 12-mo of chronological time. The Dunedin Study mean was 1 (ie, the typical pace of aging among 38-y-olds in that birth cohort). Thus, a 0.01 increment of DunedinPoAm-measured pace of aging corresponds to a percentage point increase or decrease in an individual’s pace of aging relative to the Dunedin Study birth cohort at midlife. |
| Family-level socioeconomic disadvantage | The family-level measure was computed from parent reports of household income, parental education, occupation, history of financial problems, food insecurity (based on the US Household Food Security Survey Module54), father absence, residential instability (changes in home address), and family receipt of public assistance. These were aggregated to form a composite measure of household-level cumulative socioeconomic disadvantage (mean = −0.08, SD = 0.89), which is slightly below the larger sample’s mean described in ref 55 and is coded such that higher scores reflect greater disadvantage. |
| Neighborhood-level socioeconomic disadvantage | The neighborhood-level measure was composed from tract-level US Census data, according to the method described in ref 55. Briefly, participant addresses were linked to tract-level data from the US Census Bureau American Community Survey averaged over five years (https://www.census.gov/programs-surveys/acs). A composite score of neighborhood-level socioeconomic disadvantage was computed from tract-level proportions of residents reported as unemployed, living below the federal poverty threshold, having <12 y of education, not being employed in a management position, and single mothers. The average neighborhood-level socioeconomic disadvantage (mean = −0.09, SD = 0.87) is slightly below the larger sample’s mean described in ref 55 and is coded such that higher scores reflect greater disadvantage. |
| Tobacco exposure | We measured tobacco exposure from (1) participant self-report of tobacco use, (2) a poly-DNAm smoking score (mean = 0.00, SD = 0.3356), and (3) methylation of the AHRR gene (mean = 0.00, SD = 0.03; cg0557592157). |
| BMI | We measured BMI from in-laboratory measurements of height and weight transformed to sex- and age-normed z scores according to the method published by the US Centers for Disease Control and Prevention (mean = 0.30, SD = 1.32; https://www.cdc.gov/growthcharts/percentile_data_files.htm). |
| Puberty | We measured pubertal development using children’s self-reports on the Pubertal Development Scale.58 The scale is used to assess the extent of development across 5 sex-specific domains (for both: height, body hair growth, skin changes; for girls: onset of menses, breast development; for boys: growth in body hair, deepening of voice). A total pubertal status score was computed as the average response (1 = “not yet begun” to 4 = “has finished changing”) across all items (mean = 2.39, SD = 0.93). Pubertal development was residualized for age, sex, and an age-by-sex interaction. We also examined menarcheal status in girls (menses, n = 153 girls; no menses, n = 134 girls). |
Cumulative Socioeconomic Disadvantage
We measured children’s socioeconomic disadvantage at the family and neighborhood levels of analysis (Table 1).
Health Behavior Covariates
Smoking and obesity are socially patterned health behavior exposures that are more common in children from families and neighborhoods of lower SES.7,33 They are also associated with differential DNA methylation patterns across the genome.34–36 We therefore considered tobacco exposure and BMI in our analysis (Table 1).
Pubertal Development
Puberty tends to begin at younger ages in children growing up in conditions of early-life disadvantage, and meta-analytic evidence suggests that this is driven by experiences of threat but not deprivation or socioeconomic disadvantage.23,37,38 In our sample, associations of socioeconomic disadvantage and puberty were positive but not statistically different from 0 (family level: r = 0.08, 95% confidence interval [CI] −0.02 to 0.18, P = .10; neighborhood level: r = 0.05, 95% CI −0.05 to 0.16, P = .29). Puberty is also associated with a range of DNA methylation changes.39,40 We therefore considered children’s pubertal development in our analysis (see Table 1).
Analysis
We tested associations of childhood socioeconomic disadvantage and racial and/or ethnic identity with the pace of biological aging using regression analysis. To account for nonindependence of data on siblings, we fitted regressions using linear mixed models implemented with the lme4 R package. To ensure that our primary mixed-effects models appropriately corrected for nonindependence of the sample, we reran models with 1 randomly selected twin per pair and also reran models with clustered SEs in Mplus software. The coefficients were similar across all analytic strategies, increasing confidence in the robustness of our results (see Results section of the Supplemental Information).
We report parameter estimates with bootstrapped 95% CIs (computed with 500 simulations by using lme4’s confint.merMod function). Continuous measures were standardized for analysis to mean = 0.00 and SD = 1.00, allowing for interpretation of effect sizes in the metrics of Pearson’s r in the case of continuously distributed exposure variables and Cohen’s d in the case of nominal variables (ie, race/ethnicity). All models were adjusted for sex and children’s chronological age.
To test if associations between socioeconomic disadvantage and pace of biological aging were accounted for by social gradients in tobacco exposure and/or obesity, we conducted covariate-adjusted regressions to evaluate sensitivity of results. We conducted a parallel analysis to evaluate independence of associations from pubertal development.
For comparison, we repeated the pace-of-aging analysis using age-acceleration residuals from 5 published epigenetic clocks.
Results
DunedinPoAm-measured pace of aging from salivary DNA was approximately normally distributed in children and adolescents in the Texas Twin Project (before correction for the cell composition of saliva samples, the mean DunedinPoAm-measured pace of aging was 0.79 [SD = 0.05]). DunedinPoAm-measured pace of aging was similar in boys and girls (d = −0.03, 95% CI −0.11 to 0.06, P = .48). DunedinPoAm-measured pace of aging was similar for younger and older children (r = 0.02, 95% CI −0.08 to 0.11, P = .75). The relatively young and restricted age range of our sample may also explain why DunedinPoAm-measured pace of aging did not differ by age. In previous research, investigators have found that the pace of aging does increase with age but that this acceleration happens later in the life course.24,41
Socioeconomic Disadvantage and Latinx Identity Are Associated With Faster Methylation Pace of Aging
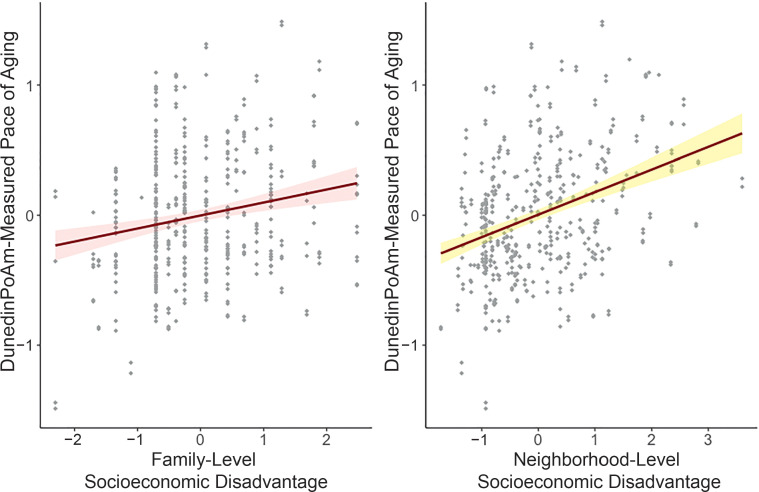
We first tested if children growing up in more socioeconomically disadvantaged circumstances exhibited faster methylation pace of aging. We conducted a separate analysis of socioeconomic disadvantage measured for children’s families and their neighborhoods. At both levels of analysis, children growing up under conditions of greater socioeconomic disadvantage exhibited a faster pace of aging, as measured by DunedinPoAm (family level: r = 0.18, 95% CI 0.08 to 0.27, P = .001; neighborhood level: r = 0.18, 95% CI 0.07 to 0.28, P = .001; Fig 1).
FIGURE 1.
Associations between family- and neighborhood-level socioeconomic disadvantage and DunedinPoAm-measured pace of aging. DunedinPoAm-measured pace of aging and socioeconomic disadvantage values are in SD units. Higher values indicate a methylation profile of faster biological aging. Regression is estimated from a linear mixed-effects model that accounts for nesting of children within families. The shaded areas represent the smoothed lower and upper 95% CIs.
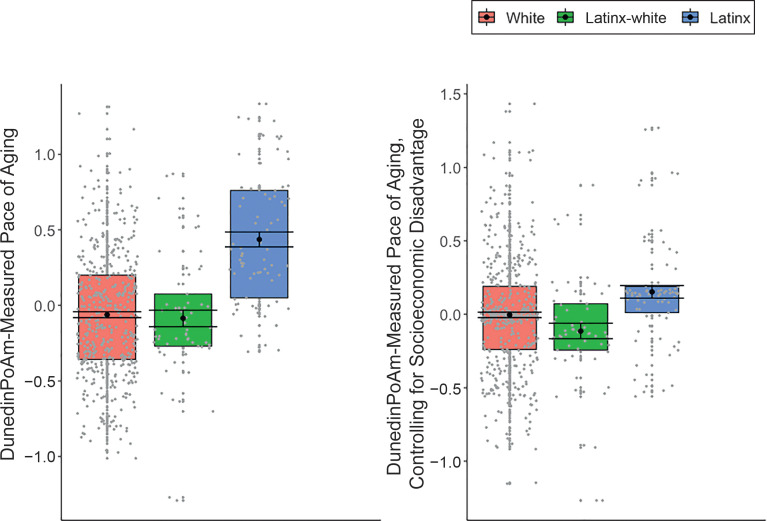
We next tested if children identifying as Latinx (12.9% of sample) and Latinx-White (10.3% of sample) exhibited a faster DunedinPoAm-measured pace of aging as compared with children identifying solely as White (76.8% of sample). Latinx-identifying children exhibited a faster DunedinPoAm-measured pace of aging compared with both White-identifying children (d = −0.21, 95% CI −0.32 to −0.09, P = .001) and Latinx White–identifying children (d = −0.16, 95% CI −0.27 to −0.04, P = .008; Fig 2).
FIGURE 2.
DunedinPoAm-measured pace of aging in children identifying as White, Latinx, and Latinx-White. DunedinPoAm-measured pace of aging values are in SD units. Higher values indicate a methylation profile of faster biological aging. Regression is estimated from linear mixed-effects model that accounts for nesting of children within families. The boxplot reveals group differences in the mean DunedinPoAm-measured pace of aging (black circle), SEs of the mean (error bars), and the first and third quartiles (lower and upper hinges). Group differences were significant at the α = .05 threshold, without adjustment for differences in socioeconomic disadvantage between groups (left panel), but were no longer significantly different from 0 when controlling for family- and neighborhood-level disadvantage (right panel).
Latinx children tended to be exposed to higher rates of family- and neighborhood-level cumulative socioeconomic disadvantage compared with White-identifying children (family level: d = −0.33, 95% CI −0.47 to −0.18, P < .001; neighborhood level: d = −0.50, 95% CI −0.55 to −0.30, P < .001) and Latinx White–identifying children (family level: d = −0.21, 95% CI −0.35 to −0.07, P = .004; neighborhood level: d = −0.29, 95% CI = −0.35 to −0.11, P < .001). We therefore tested if racial/ethnic group differences in DunedinPoAm-measured pace of aging were statistically explained by socioeconomic differences between the groups. Adjusting for group differences in family- and neighborhood-level disadvantage largely accounted for differences in methylation pace of aging between Latinx-identifying children and children identifying as White only (d = −0.07, 95% CI −0.22 to 0.08, P = .41) or Latinx-White (d = −0.09, 95% CI −0.23 to 0.05, P = .23; Fig 2); associations were no longer statistically different from 0. Statistical adjustment for racial and/or ethnic identity only modestly attenuated associations of socioeconomic disadvantage with DunedinPoAm-measured pace of aging (family level: r = 0.16, 95% CI 0.05 to 0.25, P = .003; neighborhood level: r = 0.14, 95% CI 0.03 to 0.26, P = .02).
Associations Between DunedinPoAm-Measured Pace of Aging and Socioeconomic Disadvantage Were Robust to Health Behavior and Developmental Covariates
Smoking
To test confounding of socioeconomic disadvantage associations with pace of aging by smoking, we conducted 2 sets of analyses. First, we repeated the regression analysis, excluding self-reported smokers. There were few self-reported smokers in the sample (n = 11 children [1.8% of the sample]). Results were unchanged after we excluded these participants. Second, we repeated the regression analysis, adding covariate adjustment for DNA methylation measures of tobacco exposure (genome-wide DNA methylation scores [poly-DNAm] and AHRR smoking scores). Poly-DNAm smoking profiles were positively associated with DunedinPoAm-measured pace of aging (r = 0.19, 95% CI 0.11 to 0.28, P < .001). Covariate adjustment for DNA methylation measures of tobacco exposure only modestly attenuated associations at family and neighborhood levels (family level: r = 0.16, 95% CI 0.06 to 0.25, P = .001; neighborhood level: r = 0.15, 95% CI 0.05 to 0.25, P = .003).
BMI
To test confounding of socioeconomic disadvantage associations with pace of aging by obesity and/or overweight, we repeated the regression analysis, adding covariate adjustment for BMI. Children with higher BMI had a faster DunedinPoAm-measured pace of aging (r = 0.26, 95% CI 0.18 to 0.35, P < .001). Covariate adjustment for BMI modestly attenuated associations of DunedinPoAm-measured pace of aging with socioeconomic disadvantage (family level: r = 0.14, 95% CI 0.04 to 0.23, P = .005; neighborhood level: r = 0.13, 95% CI 0.02 to 0.25, P = .03).
Pubertal Development
To test confounding of socioeconomic disadvantage associations with pace of aging by accelerated pubertal development, we repeated the regression analysis, adding covariate adjustment for pubertal development. We considered 2 measures of puberty, the Pubertal Development Scale and, in girls, menarcheal status. DunedinPoAm-measured pace of aging was weakly associated with self-reported pubertal development; however, the effect size was not statistically different from 0 (Pubertal Development Scale: r = 0.07, 95% CI −0.02 to 0.15, P = .08). DunedinPoAm indicated somewhat faster aging in girls who had experienced their first menses compared with those who had not; the effect size was small but statistically different from 0 (d = 0.19, 95% CI 0.00 to 0.38, P = .04).
Covariate adjustment for the Pubertal Development Scale modestly attenuated associations of socioeconomic disadvantage with DunedinPoAm-measured pace of aging (family level: r = 0.17, 95% CI 0.07 to 0.27, P = .001; neighborhood level: r = 0.16, 95% CI 0.05 to 0.27, P = .003). Results were similar for covariate adjustment for menarcheal status among girls (unadjusted family level: r = 0.13, 95% CI 0.00 to 0.26, P = .06; unadjusted neighborhood level: r = 0.18, 95% CI 0.06 to 0.34, P = .01; adjusted family level: r = 0.14, 95% CI −0.01 to 0.27, P = .07; adjusted neighborhood level: r = 0.17, 95% CI 0.04 to 0.33, P = .02).
In contrast to results for DunedinPoAm, epigenetic clocks were not associated with socioeconomic disadvantage and, in 2 cases, associations were in the opposite direction expected (range of r = −0.03 to 0.05; Table 2). See the Supplemental Information for full results and correlations between epigenetic clocks (Supplemental Fig 4).
TABLE 2.
Associations Between Socioeconomic Disadvantage and Epigenetic Indices of Aging
| Family-Level Socioeconomic Disadvantage | Neighborhood-Level Socioeconomic Disadvantage | |||||
|---|---|---|---|---|---|---|
| r | 95% CI | P | r | 95% CI | P | |
| DunedinPoAm24 | 0.18 | 0.08 to 0.27 | .001 | 0.18 | 0.07 to 0.28 | .001 |
| Horvath acceleration27 | 0.02 | −0.05 to 0.08 | .61 | 0.02 | −0.05 to 0.11 | .46 |
| Hannum acceleration26 | −0.03 | −0.08 to 0.04 | .43 | 0.03 | −0.04 to 0.11 | .29 |
| PedBE acceleration30 | −0.01 | −0.07 to 0.05 | .82 | 0.05 | −0.01 to 0.11 | .09 |
| PhenoAge acceleration28 | 0.01 | −0.05 to 0.07 | .69 | 0.03 | −0.05 to 0.12 | .39 |
| GrimAge acceleration29 | 0.03 | −0.02 to 0.09 | .28 | 0.05 | −0.01 to 0.13 | .11 |
Standardized regression coefficients (r) and 95% CIs are calculated by regressing DNA methylation measures on family-level socioeconomic disadvantage and neighborhood-level socioeconomic disadvantage separately. All models included covariate adjustment for the child’s age and sex.
Discussion
We analyzed saliva DNA methylation data from children and adolescents participating in the Texas Twin Project to test associations between childhood socioeconomic disadvantage and the pace of biological aging using a novel DNA methylation measure, DunedinPoAm. We found that children and adolescents growing up in more disadvantaged families and neighborhoods exhibited a faster pace of aging as measured by DunedinPoAm. Children with higher BMI and more advanced pubertal development for their age tended to have a faster DunedinPoAm-measured pace of aging, but these covariates did not account for observed socioeconomic disparities. Our results suggest that the DunedinPoAm DNA methylation measure of the pace of biological aging, originally validated in adults,24 could potentially serve as a salivary biomarker sensitive in real time to social determinants of health experienced during childhood.
Questions remain about when in the life course biological processes of aging begin.42 Observations of variation in cellular-level measurements of aging in pediatric samples suggest that quantification of biological aging in children may be possible.16,30,43 In our study, children from socioeconomically disadvantaged families and neighborhoods showed a faster pace of aging on the basis of a DNA methylation measure that tracks faster declines in organ-system integrity in midlife adults and forecasts mortality. This result could reflect at least 2 phenomena. First is that socially disadvantaged children in our study showed patterns of biological wear and tear similar to adults who have faster pace of aging. Second is that the DunedinPoAm algorithm detected molecular signatures of exposures, including early-life adversities, that were causes of faster pace of aging in the adults in the original study. Despite this ambiguity, our results reveal a molecular continuity between social inequalities in childhood and health disparities in adulthood that can inform future studies of etiology and intervention.
In our analysis of racial and/or ethnic group differences, we found that Latinx-identifying children exhibited a faster DunedinPoAm-measured pace of aging compared with children identifying as White and Latinx-White. These differences were statistically accounted for by the greater socioeconomic disadvantage experienced by the Latinx children. Thus, our findings are consistent with observations that racial and/or ethnic socioeconomic disparities are an important contributor to racial and/or ethnic disparities in health.44 Importantly, racial and/or ethnic disparities in adult health typically persist, although reduced, across all levels of SES, for example, because of race-based discrimination.32,45,46
In contrast to findings for DunedinPoAm-measured pace of aging, an analysis of several published DNA methylation clocks yielded null associations with socioeconomic disadvantage. These clocks revealed the expected associations with children’s chronological age (r = 0.66 to 0.81 in an age range of 8–18 years). However, the difference between participants’ epigenetic ages and their chronological ages (ie, epigenetic-age acceleration) did not vary with socioeconomic disadvantage, consistent with results from a recent meta-analysis revealing that threat, but not deprivation, was associated with epigenetic clocks in childhood samples.23 Measures of epigenetic-age acceleration also appear to be less sensitive to racial and/or ethnic group differences in childhood and adolescence.47 These results suggest that pace-of-aging measures, such as DunedinPoAm, may prove more sensitive to health-damaging effects of socioeconomic deprivation and of racialized disparities in SES, particularly in studies focused on the early life course.
We acknowledge limitations. First, we measured methylation in saliva DNA, which comes from a mixture of buccal cells and leukocytes, whereas the measures we analyzed were developed from blood or other tissues. We used DNA methylation algorithms to make statistical adjustment for the cellular composition of the saliva samples. Increasing confidence in our findings, they replicate results for DunedinPoAm and epigenetic clocks from blood DNA methylation measured in 18-year-olds.24 Additionally, our analysis indicated good correspondence of DunedinPoAm measurements across blood and saliva (Supplemental Information).
Second, our observational design included DNA methylation data from a single time point, precluding a test of how change in socioeconomic circumstances might affect DNA methylation measures. Ultimately, not only longitudinal repeated-measures studies but also natural experiment studies and randomized controlled trials of social programs are needed to establish causal effects of social disadvantage on DunedinPoAm-measured pace of aging and to establish DunedinPoAm as a mediator of the process through which childhood disadvantage leads to aging-related health conditions.48,49
Finally, the biology that causes variation in DunedinPoAm-measured pace of aging and the epigenetic clocks remains poorly understood. Epigenetic changes are understood to be core features of the biological process of aging.12,50 Yet, the methylation measurements we studied are only correlates of the unobserved processes of biological aging, not direct observations of it.51
Within the bounds of these limitations, our findings have implications for theory and future research. Theory and evidence from animal models suggest epigenetic changes are a mediator of early-life adversity’s effects on aging-related health decline.52 However, human studies following-up specific mechanisms identified in animals have yielded equivocal results.53 Our results contribute evidence toward proof of concept for the hypothesis that early adversity increases risk for adult disease through acceleration of the pace of biological aging.
In future research, investigators can use DunedinPoAm to test how aging processes may be accelerated in at-risk young people and if and how such accelerated aging may be modified, in particular, by program and policies that are aimed at reducing social and material disadvantage. Childhood interventions to improve equitable access to nutritious food, lower family stress, enhance neighborhood safety, and increase greenspace have the potential to improve concurrent and lifelong health.6 However, childhood interventions antedate the onset of adult disease by decades. This long gap has motivated interest in biological measures that can serve as surrogate end points for assessing the effectiveness of programs and policies aimed at improving lifelong health by promoting positive child development. Our results suggest that salivary DNA methylation measures of pace of aging may provide a surrogate or intermediate end point for understanding the health impacts of such interventions. Such applications may prove particularly useful for evaluating the effectiveness of health-promoting interventions in at-risk groups.
Acknowledgments
We thank all participants of the Texas Twin Project.
Glossary
- CI
confidence interval
- poly-DNAm
genome-wide DNA methylation
- SES
socioeconomic status
Footnotes
Dr Raffington conceptualized and designed the study, analyzed the data, drafted the manuscript, and critically revised the manuscript; Dr Belsky conceptualized and designed the study, drafted the manuscript, and critically revised the manuscript; Drs Tucker-Drob and Harden conceptualized and designed the study, collected the data, and critically revised the manuscript; Dr Malanchini collected the data and critically revised the manuscript; Mr Kothari analyzed the data; and all authors reviewed the manuscript and approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
FINANCIAL DISCLOSURE: The authors have indicated they have no financial relationships relevant to this article to disclose.
FUNDING: Supported by National Institutes of Health grants R01HD083613 and R01HD092548 and the Jacobs Foundation. Dr Raffington is supported by the German Research Foundation. Dr Belsky is supported by Russell Sage Foundation Biology and Social Science grant 1810-08987, National Institute on Aging grants R01AG066887 and R01AG061378, and the Canadian Institute for Advanced Research Child and Brain Development Network. Drs Tucker-Drob and Harden are faculty research associates of the Population Research Center at The University of Texas at Austin, which is supported by a grant (5-R24-HD042849) from the Eunice Kennedy Shriver National Institute of Child Health and Human Development. Drs Belsky, Tucker-Drob, and Harden are fellows of the Jacobs Foundation. The funders or sponsors did not participate in the work. Funded by the National Institutes of Health (NIH).
POTENTIAL CONFLICT OF INTEREST: The authors have indicated they have no potential conflicts of interest to disclose.
COMPANION PAPER: A companion to this article can be found online at www.pediatrics.org/cgi/doi/10.1542/peds.2021-049939.
References
- 1. Snyder-Mackler N, Burger JR, Gaydosh L, et al. Social determinants of health and survival in humans and other animals. Science. 2020;368(6493):eaax9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hayward MD, Gorman BK. The long arm of childhood: the influence of early-life social conditions on men’s mortality. Demography. 2004;41(1):87–107 [DOI] [PubMed] [Google Scholar]
- 3. Makenbach JP. Health Inequalities: Europe in Profile. London, United Kingdom: UK Presidency of the EU; 2006 [Google Scholar]
- 4. Miech RA, Caspi A, Moffitt TE, Wright BRE, Silva PA. Low socioeconomic status and mental disorders: a longitudinal study of selection and causation during young adulthood. Am J Sociol. 1999;104(4):1096–1131 [Google Scholar]
- 5. Kuntz B, Rattay P, Poethko-Müller C, Thamm R, Hölling H, Lampert T. Social inequalities in health of children and adolescents in Germany. Results of the cross-sectional KiGGS Wave 2 study. Journal of Health Monitoring. 2018;3(3):17–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pollack CE, Blackford AL, Du S, Deluca S, Thornton RLJ, Herring B. Association of receipt of a housing voucher with subsequent hospital utilization and spending. JAMA. 2019;322(21):2115–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Datar A, Chung PJ. Changes in socioeconomic, racial/ethnic, and sex disparities in childhood obesity at school entry in the United States. JAMA Pediatr. 2015;169(7):696–697 [DOI] [PubMed] [Google Scholar]
- 8. Campbell F, Conti G, Heckman JJ, et al. Early childhood investments substantially boost adult health. Science. 2014;343(6178):1478–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Belsky DW, Caspi A, Cohen HJ, et al. Impact of early personal-history characteristics on the pace of aging: implications for clinical trials of therapies to slow aging and extend healthspan. Aging Cell. 2017;16(4):644–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marini S, Davis KA, Soare TW, et al. Adversity exposure during sensitive periods predicts accelerated epigenetic aging in children. [published correction appears in Psychoneuroendocrinology. 2020;121:104829]. Psychoneuroendocrinology. 2020;113:104484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kirkwood TBL. Understanding the odd science of aging. Cell. 2005;120(4):437–447 [DOI] [PubMed] [Google Scholar]
- 12. López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gavrilov LA, Gavrilova NS. The reliability-engineering approach to the problem of biological aging. Ann N Y Acad Sci. 2004;1019:509–512 [DOI] [PubMed] [Google Scholar]
- 14. Middeldorp CM. Editorial: childhood stress and psychopathology: it’s not too early to look at biological aging. J Am Acad Child Adolesc Psychiatry. 2020;59(1):38–39 [DOI] [PubMed] [Google Scholar]
- 15. Humphreys KL, Esteves K, Zeanah CH, Fox NA, Nelson CA III, Drury SS. Accelerated telomere shortening: tracking the lasting impact of early institutional care at the cellular level. Psychiatry Res. 2016;246:95–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shalev I. Early life stress and telomere length: investigating the connection and possible mechanisms: a critical survey of the evidence base, research methodology and basic biology. Bioessays. 2012;34(11):943–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Belsky J, Shalev I. Contextual adversity, telomere erosion, pubertal development, and health: two models of accelerated aging, or one? Dev Psychopathol. 2016;28(4, pt 2):1367–1383 [DOI] [PubMed] [Google Scholar]
- 18. Mitchell C, McLanahan S, Schneper L, Garfinkel I, Brooks-Gunn J, Notterman D. Father loss and child telomere length. Pediatrics. 2017;140(2):e20163245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mather KA, Jorm AF, Parslow RA, Christensen H. Is telomere length a biomarker of aging? A review. J Gerontol A Biol Sci Med Sci. 2011;66(2):202–213 [DOI] [PubMed] [Google Scholar]
- 20. Sanders JL, Newman AB. Telomere length in epidemiology: a biomarker of aging, age-related disease, both, or neither? Epidemiol Rev. 2013;35(1):112–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19(6):371–384 [DOI] [PubMed] [Google Scholar]
- 22. Fiorito G, McCrory C, Robinson O, et al. ; BIOS Consortium; Lifepath Consortium . Socioeconomic position, lifestyle habits and biomarkers of epigenetic aging: a multi-cohort analysis. Aging (Albany NY). 2019;11(7):2045–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Colich NL, Rosen ML, Williams ES, McLaughlin KA. Biological aging in childhood and adolescence following experiences of threat and deprivation: a systematic review and meta-analysis. Psychol Bull. 2020;146(9):721–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Belsky DW, Caspi A, Arseneault L, et al. Quantification of the pace of biological aging in humans through a blood test, the DunedinPoAm DNA methylation algorithm. Elife. 2020;9:e54870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Belsky DW. Reply to Newman: quantification of biological aging in young adults is not the same thing as the onset of obesity. Proc Natl Acad Sci U S A. 2015;112(52):E7164–E7165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Horvath S. DNA methylation age of human tissues and cell types. [published correction appears in Genome Biol. 2015;16(1):96]. Genome Biol. 2013;14(10):R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Levine ME, Lu AT, Quach A, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY). 2018;10(4):573–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lu AT, Quach A, Wilson JG, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY). 2019;11(2):303–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McEwen LM, O’Donnell KJ, McGill MG, et al. The PedBE clock accurately estimates DNA methylation age in pediatric buccal cells. Proc Natl Acad Sci U S A. 2020;117(38):23329–23335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harden KP, Tucker-Drob EM, Tackett JL. The Texas Twin Project. Twin Res Hum Genet. 2013;16(1):385–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Goosby BJ, Cheadle JE, Mitchell C. Stress-related biosocial mechanisms of discrimination and African American health inequities. Annu Rev Sociol. 2018;44(1):319–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Centers for Disease Control and Prevention. Cigarette smoking and tobacco use among people of low socioeconomic status. Available at: http://www.cdc.gov/tobacco/disparities/low-ses/index.htm [Google Scholar]
- 34. Joehanes R, Just AC, Marioni RE, et al. Epigenetic signatures of cigarette smoking. Circ Cardiovasc Genet. 2016;9(5):436–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Joubert BR, Felix JF, Yousefi P, et al. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am J Hum Genet. 2016;98(4):680–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wahl S, Drong A, Lehne B, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541(7635):81–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Braithwaite D, Moore DH, Lustig RH, et al. Socioeconomic status in relation to early menarche among black and white girls. Cancer Causes Control. 2009;20(5):713–720 [DOI] [PubMed] [Google Scholar]
- 38. Obeidallah DA, Brennan RT, Brooks-Gunn J, Kindlon D, Earls F. Socioeconomic status, race, and girls’ pubertal maturation: results from the project on human development in Chicago neighborhoods. J Res Adolesc. 2000;10(4):443–464 [Google Scholar]
- 39. Almstrup K, Lindhardt Johansen M, Busch AS, et al. Pubertal development in healthy children is mirrored by DNA methylation patterns in peripheral blood. Sci Rep. 2016;6:28657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aylwin CF, Toro CA, Shirtcliff E, Lomniczi A. Emerging genetic and epigenetic mechanisms underlying pubertal maturation in adolescence. J Res Adolesc. 2019;29(1):54–79 [DOI] [PubMed] [Google Scholar]
- 41. Robine JM. Age Patterns in Adult Mortality. In: Rogers RG, Crimmins EM, eds.. International Handbook of Adult Mortality. Dordrecht, Netherlands: Springer Netherlands; 2011:207–226 [Google Scholar]
- 42. Gladyshev VN. The ground zero of organismal life and aging. Trends Mol Med. 2021;27(1):11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kinzina ED, Podolskiy DI, Dmitriev SE, Gladyshev VN. Patterns of aging biomarkers, mortality, and damaging mutations illuminate the beginning of aging and causes of early-life mortality. Cell Rep. 2019;29(13):4276–4284.e3 [DOI] [PubMed] [Google Scholar]
- 44. Williams DR, Priest N, Anderson NB. Understanding associations among race, socioeconomic status, and health: patterns and prospects. Health Psychol. 2016;35(4):407–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Braveman PA, Cubbin C, Egerter S, Williams DR, Pamuk E. Socioeconomic disparities in health in the United States: what the patterns tell us. Am J Public Health. 2010;100(suppl 1):S186–S196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Colen CG, Ramey DM, Cooksey EC, Williams DR. Racial disparities in health among nonpoor African Americans and Hispanics: the role of acute and chronic discrimination. Soc Sci Med. 2018;199:167–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sumner JA, Colich NL, Uddin M, Armstrong D, McLaughlin KA. Early experiences of threat, but not deprivation, are associated with accelerated biological aging in children and adolescents. Biol Psychiatry. 2019;85(3):268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. O’Donnell KJ, Chen L, MacIsaac JL, et al. DNA methylome variation in a perinatal nurse-visitation program that reduces child maltreatment: a 27-year follow-up. Transl Psychiatry. 2018;8(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Muennig P, McEwen B, Belsky DW, Noble KG, Riccio J, Manly J. Determining the optimal outcome measures for studying the social determinants of health. Int J Environ Res Public Health. 2020;17(9):3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kennedy BK, Berger SL, Brunet A, et al. Geroscience: linking aging to chronic disease. Cell. 2014;159(4):709–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Raj K, Horvath S. Current perspectives on the cellular and molecular features of epigenetic ageing. Exp Biol Med (Maywood). 2020;245(17):1532–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cunliffe VT. The epigenetic impacts of social stress: how does social adversity become biologically embedded? Epigenomics. 2016;8(12):1653–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cecil CAM, Zhang Y, Nolte T. Childhood maltreatment and DNA methylation: a systematic review. Neurosci Biobehav Rev. 2020;112:392–409 [DOI] [PubMed] [Google Scholar]
- 54. US Department of Agriculture Economic Research Service. U.S. Household Food Security Survey Module: Three-Stage Design, With Screeners. Washington, DC: US Department of Agriculture; 2012 [Google Scholar]
- 55. Engelhardt LE, Church JA, Harden KP, Tucker-Drob EM. Accounting for the shared environment in cognitive abilities and academic achievement with measured socioecological contexts. Dev Sci. 2019;22(1):e12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sugden K, Hannon EJ, Arseneault L, et al. Establishing a generalized polyepigenetic biomarker for tobacco smoking. Transl Psychiatry. 2019;9(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bojesen SE, Timpson N, Relton C, Davey Smith G, Nordestgaard BG. AHRR (cg05575921) hypomethylation marks smoking behaviour, morbidity and mortality. Thorax. 2017;72(7):646–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Petersen AC, Crockett L, Richards M, Boxer A. A self-report measure of pubertal status: reliability, validity, and initial norms. J Youth Adolesc. 1988;17(2):117–133 [DOI] [PubMed] [Google Scholar]