Key Points
The ADPKD computable phenotype on the basis of ICD-9/10 is an excellent screening tool to identify patients with ADPKD.
Patients who were followed in nephrology clinics had a higher sensitivity, specificity, and positive and negative predictive values.
Specificity of the ADPKD computable phenotype is comparable with other medical conditions.
Keywords: cystic kidney disease, ADPKD, computable phenotype, diagnosis, ICD code, polycystic kidney, test accuracy
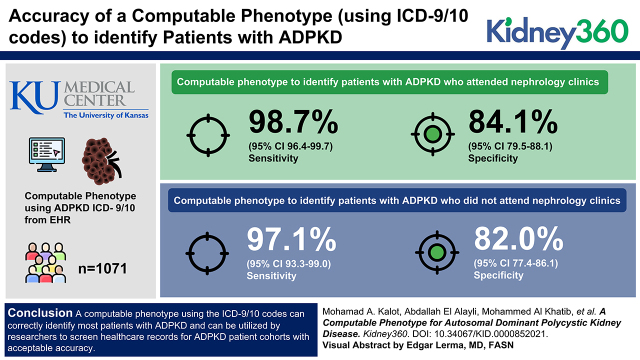
Visual Abstract
Abstract
Background
A computable phenotype is an algorithm used to identify a group of patients within an electronic medical record system. Developing a computable phenotype that can accurately identify patients with autosomal dominant polycystic kidney disease (ADPKD) will assist researchers in defining patients eligible to participate in clinical trials and other studies. Our objective was to assess the accuracy of a computable phenotype using International Classification of Diseases 9th and 10th revision (ICD-9/10) codes to identify patients with ADPKD.
Methods
We reviewed four random samples of approximately 250 patients on the basis of ICD-9/10 codes from the EHR from the Kansas University Medical Center database: patients followed in nephrology clinics who had ICD-9/10 codes for ADPKD (Neph+), patients seen in nephrology clinics without ICD codes for ADPKD (Neph−), patients who were not followed in nephrology clinics with ICD codes for ADPKD (No Neph+), and patients not seen in nephrology clinics without ICD codes for ADPKD (No Neph−). We reviewed the charts and determined ADPKD status on the basis of internationally accepted diagnostic criteria for ADPKD.
Results
The computable phenotype to identify patients with ADPKD who attended nephrology clinics has a sensitivity of 99% (95% confidence interval [95% CI], 96.4 to 99.7) and a specificity of 84% (95% CI, 79.5 to 88.1). For those who did not attend nephrology clinics, the sensitivity was 97% (95% CI, 93.3 to 99.0), and a specificity was 82% (95% CI, 77.4 to 86.1).
Conclusion
A computable phenotype using the ICD-9/10 codes can correctly identify most patients with ADPKD, and can be utilized by researchers to screen health care records for cohorts of patients with ADPKD with acceptable accuracy.
Introduction
Autosomal dominant polycystic kidney disease (ADPKD) is a genetic condition that causes bilateral renal cyst formation (1). This disease is the most common hereditary kidney disease and it affects one in 400–1000 people worldwide (2). Half of these patients will require ESKD management (3). Although genetic testing is emerging in the detection of ADPKD (4), it is limited by the number of missense mutations that need further confirmation and the diagnosis is routinely confirmed on the basis of imaging studies, such as computed tomography scans, magnetic resonance imaging, and abdominal ultrasounds, which carry the highest sensitivity in detecting ADPKD. Among these three techniques, abdominal ultrasound is the most widely used because of its low cost, availability, high sensitivity with advanced disease, and safety (4).
In the presence of health care systems that utilize electronic health records (EHR), a large amount of stored data could be easily accessed. This setting is ideal to using a computable phenotype to identify different conditions or diseases, including ADPKD. A computable phenotype is a clinical phenotype, a characteristic, or a group of several clinical features that can be automatically extracted from an EHR without health care provider interpretation or intervention (5). In this way, the computable phenotype may provide a reliable and easy way to identify patients with the condition of interest in a timely manner (6).
The World Health Organization created the International Classification of Diseases (ICD) in 1948 (7). The coding professionals transformed medical terms, procedures, and diagnoses into universal alphanumeric codes. The ICD 10th revision (ICD-10) is the most recent update and was adopted in the United States in 2013 (8). ICD codes were implemented to promote international comparability for collection, classification, processing, and presentation of health statistics (9). Because ADPKD is a relatively rare disease, using the ICD-9/10 codes to find all patients with the disease in an EHR would be very time and cost effective during recruitment for ADPKD studies (10). However, ICD codes are often inaccurate, and there is a considerable controversy regarding their value in identifying patients with specific clinical conditions. For this reason, we sought to assess test accuracy (sensitivity, specificity, positive predictive value [PPV], and negative predictive value [NPV]) and positive and negative likelihood ratio of a computable phenotype using ICD-9/10 codes in identifying patients with ADPKD.
We developed a computable phenotype using ICD-9/10 to identify patients with ADPKD. In this study, we present the test accuracy results of the computable phenotype (sensitivity, specificity, PPV, and NPV) in identifying patients with ADPKD, for patients who follow-up in nephrology clinics and those who do not. Additionally, we estimate the prevalence of ADPKD using the University of Kansas Medical Center (KUMC) database.
Methods
Study Design and Participants
We conducted a cross-sectional test accuracy study by reviewing four random samples of approximately 250 patients on the basis of ICD-9/10 codes and nephrology clinic visits, from the EHR from the KUMC database. The samples were stratified into four groups: the Neph+ and Neph− groups included patients followed in nephrology clinics who had ICD-9/10 codes for ADPKD and those who did not have ICD-9/10 codes of ADPKD, respectively; the No Neph+ and No Neph- groups included patients who were not followed in nephrology clinics, with and without ICD-9/10 codes for ADPKD, respectively.
Test Methods
We used deidentified patient information in the HERON data repository, an i2b2 data access platform (11,12), to identify patients with ADPKD using ICD-9 codes 753.12 and 753.13, and ICD-10 codes Q61.2 and Q61.3. We used the ICD-9 code 593.2 and the ICD-10 code N28.1 to label patients with renal cysts that did not have ADPKD and to enrich the sample of patients without ADPKD.
We evaluated four random samples from the deidentified dataset on the basis of the eligibility criteria for each group. At least two reviewers reviewed the medical records of each patient, in duplicate. Although we reviewed the charts after we generated the computable phenotype, the reviewers were blinded to the strata on the basis of the phenotype results. For every chart, each of the reviewers had to make a determination of whether the patient has, or does not have, ADPKD. In patients with family history of the disease, we used the unified imaging diagnosis criteria and in patients with no family history of ADPKD, the diagnosis was made if the patient has ≥10 cysts in each of the two kidneys, with kidneys measuring >13-cm-long (Table 1) (13). We used data from the last imaging available. When ADPKD status was still ambiguous, an experienced nephrologist reviewed all medical records to decide whether ADPKD is present, according to clinical criteria. If there was insufficient information to decide whether ADPKD was present, we excluded patients from the analysis. We did not consider the timing of insertion of the ICD-9/10 codes into the system.
Table 1.
Diagnostic criteria used for autosomal dominant polycystic kidney disease diagnosis
| Family History | Age, yr | Criteria |
|---|---|---|
| Positive family history (The unified imaging diagnosis criteria published in the Canadian Journal of Kidney Health and Disease.) | 15–40 | At least three unilateral or bilateral kidney cysts |
| 40–59 | At least two cysts in each kidney | |
| >60 | At least four cysts in each kidney | |
| Negative family history | Any age | Innumerable cysts in both kidneys (at least 10 cysts in each of the two kidneys) |
| Each kidney greater than 13 cm in length |
Analysis
After reviewing the charts, we collected the data and developed two separate 2×2 contingency tables after classifying patients into those with and without ADPKD and those with and without the computable phenotype. We split the results for those who attended and those who did not attend nephrology clinic. We calculated the sensitivity, specificity, NPV, and PPV with 95% confidence intervals (95% CIs). Confidence intervals for sensitivity and specificity are “exact” Clopper-Pearson CI (14). Confidence intervals for the likelihood ratios are calculated using the “Log method” as described by Altman et al. (15). We conducted a sensitivity analyses to determine whether classifying the excluded patients as having ADPKD, or as not having ADPKD, meaningfully changed the results. We calculated the prevalence of ADPKD in the available electronic medical records of the health care system. We have reported the results on the basis of the Standards for Reporting of Diagnostic Accuracy Studies (STARD) guidelines for reporting diagnostic accuracy studies (16).
Results
Our random sample included a total of 1071 patients, of which 536 were followed in the nephrology clinic and 535 were not. The average age of the patients was 63 years, 53% were males, 76% were White, and 15% were Black. The prevalence of ADPKD on the basis of positive ICD-9/10 codes in the deidentified dataset is six out of 10,000. Descriptive analyses of age, sex, and race are provided in Tables 1 and 2.
Table 2.
Descriptive analysis for patients who were followed in the nephrology clinic
| Characteristics | Autosomal Dominant Polycystic Kidney Disease | No Autosomal Dominant Polycystic Kidney Disease | Totals |
|---|---|---|---|
| Age, y, mean±SDa | 51±15 | 58±17 | 52±15 |
| Sex | |||
| F | 130 | 19 | 149 |
| M | 106 | 28 | 134 |
| Racial and ethnicity | |||
| White | 208 | 34 | 242 |
| Black | 16 | 7 | 23 |
| Native American | 0 | 1 | 1 |
| Asian | 3 | 2 | 5 |
| Other | 9 | 3 | 12 |
| Age, y, mean±SDb | 51±28 | 68±14 | 68±14 |
| Sex | |||
| F | 1 | 101 | 102 |
| M | 2 | 148 | 150 |
| Racial and ethnicity | |||
| White | 1 | 151 | 152 |
| Black | 2 | 75 | 77 |
| Native American | 0 | 1 | 1 |
| Asian | 0 | 3 | 3 |
| Other | 0 | 19 | 19 |
F, female; M, male.
Positive computable phenotype.
Negative computable phenotype.
In the Neph+ group, we found that 236 had ADPKD out of the 283 patients who had ICD-9/10 codes for ADPKD. In the Neph− group, 249 patients did not have ADPKD, out of 253 patients who did not have an ICD-9/10 codes for ADPKD. One patient had a “likely no” diagnosis (Supplemental Figure 1). The specificity did not change when considering the patient with “likely no” diagnosis as a true negative (84%).
For No Neph+ group, we found that 165 patients were correctly diagnosed with ADPKD out of the 223 patients that were diagnosed with ADPKD using the ICD-9/10 codes, two patients were diagnosed as “likely yes,” two patients as “likely no,” and 34 patients were classified as unknown due to absence of family history and renal imaging. As for group No Neph−, we found that 265 did not have the disease out of the 275 patients who did not have the ICD-9/10 codes for ADPKD, two patients were diagnosed as “likely yes,” and two patients were diagnosed as “likely no” (Supplemental Figure 2). The sensitivity did not change when considering the patients with “likely yes” diagnosis as a true positives (97%). The specificity did not change when considering the patients with “likely no” diagnosis as true negatives (82%). Tables 3, 4, and 5 summarize the 2×2 contingency tables and Table 6 summarizes the test accuracy for those were followed in the nephrology clinic and those who were not.
Table 3.
Descriptive analysis for patients who were not followed in the nephrology clinic
| Characteristics | Autosomal Dominant Polycystic Kidney Disease | No Autosomal Dominant Polycystic Kidney Disease | Totals |
|---|---|---|---|
| Age, y, mean±SDa | 59±13 | 52±24 | 57±17 |
| Sex | |||
| F | 57 | 16 | 73 |
| M | 89 | 26 | 115 |
| Racial and ethnicity | |||
| White | 142 | 39 | 181 |
| Black | 12 | 10 | 22 |
| Native American | 0 | 0 | 0 |
| Asian | 1 | 2 | 3 |
| Other | 1 | 7 | 17 |
| Age, y, mean±SDb | 70±7 | 66±15 | 66±15 |
| Sex | |||
| F | 2 | 117 | 119 |
| M | 0 | 6 | 6 |
| Racial and ethnicity | |||
| White | 4 | 207 | 211 |
| Black | 1 | 32 | 33 |
| Native American | 0 | 0 | 0 |
| Asian | 0 | 7 | 7 |
| Other | 0 | 19 | 19 |
F, female; M, male.
Positive computable phenotype.
Negative computable phenotype.
Table 4.
Contingency table displaying frequency distribution of patients who were followed in the nephrology clinic
| Computable Phenotype Status | Autosomal Dominant Polycystic Kidney Disease | No Autosomal Dominant Polycystic Kidney Disease | Totals |
|---|---|---|---|
| Positive computable phenotype | 236 | 47 | 283 |
| Negative computable phenotype | 3 | 249 | 252 |
| Totals | 239 | 296 | 535 |
Table 5.
Contingency table displaying frequency distribution of patients who were not followed in the nephrology clinic
| Computable Phenotype Status | Autosomal Dominant Polycystic Kidney Disease | No Autosomal Dominant Polycystic Kidney Disease | Totals |
|---|---|---|---|
| Positive computable phenotype | 165 | 58 | 223 |
| Negative computable phenotype | 5 | 265 | 270 |
| Totals | 170 | 323 | 493 |
Table 6.
Autosomal dominant polycystic kidney disease diagnostic test accuracy results
| Test Accuracy Results | Patients who Were Followed in The Nephrology Clinics–Effect Estimate (95% Confidence Interval) | Patients who Were Not Followed in The Nephrology Clinics–Effect Estimate (95% Confidence Interval) |
|---|---|---|
| Sensitivity | 99% (97 to 99) | 97% (93 to 99) |
| Specificity | 84% (79 to 88) | 82% (77 to 86) |
| Positive predictive value | 83% (79 to 87) | 74% (69 to 78) |
| Negative predictive value | 99% (97 to 100) | 98% (96 to 99) |
| Likelihood ratio positive | 6 (5 to 8) | 6 (4 to 7) |
| Likelihood ratio negative | 0.01 (0.00 to 0.04) | 0.04 (0.02 to 0.09) |
Discussion
Computable phenotypes are efficient to screen patients for a condition of interest, ADPKD in this study. Computable phenotypes such as any other diagnostic technique should be accurate and easy to use. The accuracy could be evaluated using sensitivity, specificity, PPV, and NPV (17). In this study, we calculated the test accuracy values for a computable phenotype comprised of ICD-9 (753.12 and 753.13) and the ICD-10 codes (Q61.2 and Q61.3) in identifying patients with ADPKD in the KUMC EHR. Overall, we found that the computable phenotype had an excellent sensitivity and NPV and an acceptable specificity and PPV. Not surprisingly, we found that patients who were followed in nephrology clinics had a higher sensitivity, specificity, PPV, and NPV compared with those who were not seen in nephrology clinics. As one would expect, these results support that nephrologists are more likely to accurately label patients as having ADPKD when they actually have the disease, compared with other specialists. However, this could be partially explained by some providers using the ICD-9/10 codes when they are referring patients to the nephrology clinic to rule out ADPKD.
Regardless of nephrology follow-up, the ADPKD computable phenotype on the basis of ICD-9/10 codes has a relatively high sensitivity of 97%–99%, compared with other computable phenotypes of other medical conditions, such as acetaminophen toxicity (94%) (18), myocardial infarction (94%), cerebrovascular disease (83%), and dementia (93%) (7). Additionally, the specificity of the ADPKD computable phenotype of 82%–84% was comparable with other medical conditions, such as acetaminophen toxicity (83%) (18), but lower than the specificity for myocardial infarction (95%), cerebrovascular disease (95%), and dementia (99%) (18,19). These results confirm the ADPKD-computable phenotype using ICD-9/10 codes is a practical tool to identify potential patients with ADPKD and to rule out ADPKD, but it is less accurate for confirming the ADPKD diagnosis.
This study is the first to comprehensively assess all aspects of test accuracy of an ADPKD-computable phenotype. Blanchette et al. reviewed records of 132 patients with an ICD-9 code for ADPKD (753.12) with a reported PPV of 95% (20). Kalatharan et al. reviewed records of 201 patients using an ICD-10 code for ADPKD (Q61.2 or Q61.3) with a reported PPV of 85% (21). These two studies did not assess the sensitivity and specificity of the computable phenotype in identifying patients with ADPKD. Our findings of a PPV of 73.4–83.4 are more comparable with the findings by Kalatharan et al., which also utilized data from a large health care system.
Our study has multiple strengths. First, this is the largest study ever done to assess the test accuracy of a computable phenotype. Additionally, we have assessed all aspects of test accuracy results (Table 4), and we have reported the results on the basis of the Standards for Reporting of Diagnostic Accuracy Studies (STARD) guidelines for reporting diagnostic accuracy studies. Finally, we have included both ICD-9 and ICD-10 codes and have compared those who were followed in the nephrology clinic and those who were not.
We note a few limitations in our study. First, our results likely underestimate the specificity estimate because we enriched our sample for patients with renal cysts. This reflects the worst-case scenario for specificity but was important to consider because that is the group that most likely gets confused with ADPKD. Another limitation was the 34 patients that had inconclusive results and were excluded from the analysis due to lack of information in the EHR to allow for categorization as having or not having ADPKD. Additionally, we did not consider the timing of insertion of the ICD-9/10 codes into the system; however, we think this reflects what providers will find in the electronic medical record and is consistent with our attempt to keep the computable phenotype simple and make it practical. Furthermore, there might be an overestimation of the sensitivity because our study did not evaluate the whole health care system and accounted for those with ADPKD who are missing a diagnosis code. However, when we evaluated a random sample of patients without ADPKD ICD-9/10 codes, and, despite enriching this group with patients who have ICD codes for renal cysts, we only identified eight additional patients with ADPKD out of 522 records evaluated. Finally, this computable phenotype was only tested in one health care system, and KUMC is considered to be a PKD referral center, so there may be bias in that there is significantly more institutional knowledge about PKD than in nonreferral centers. Future efforts should focus on testing the computable phenotype in other health care systems.
Our results show that a computable phenotype of ADPKD ICD-9/10 codes is a good tool to screen for patients and assess the feasibility of participating in ADPKD trials. This is especially important in an era with an approved treatment and many more in the pipeline that will need to be tested in trials. The computable phenotype is not accurate enough to confirm the diagnosis of ADPKD. However, it is accurate to rule out an ADPKD diagnosis. This will support a strategy focusing on patients who have ICD-9/10 for ADPKD when screening for trials and not wasting time and resources looking up patients without these codes. The final confirmation of ADPKD diagnosis relies on the patients’ radiology reports such as ultrasound, computed tomography scan, magnetic resonance imaging, and the accurate characterization of patients’ family history. One of the main challenges to automating a final ADPKD diagnosis is that information in radiology reports are summarized as open texts, which does not directly translate into ICD-9 or ICD-10 codes, or searchable elements in EHR. This could be a reason for the limited specificity of ICD-9 and ICD-10 codes in diagnosing ADPKD. Another reason may be that providers either do not know enough about the disease or how to differentiate it from simple renal cysts, or they are not familiar with the ADPKD ICD codes. Developing and evaluating algorithms that enhance the accurate detection using ICD-9/10 is an important next step to improve the specificity of this computable phenotype. The natural language processing algorithm of radiology reports and notes documenting family history could be considered and studied.
The ADPKD computable phenotype on the basis of ICD-9/10 is an excellent screening tool to identify patients with ADPKD. Assessing the accuracy of the ADPKD-computable phenotype is an important step in defining the best strategies to identify and recruit patients with ADPKD for trials at a time when many innovative interventions are being developed and will need to be tested in trials. Additional searches including specific medications and procedures could enhance the accuracy of the computable phenotype.
Disclosures
A. Yu reports having consultancy agreements with Calico, Otsuka, Navitor, and Regulus Therapeutics; reports having an ownership interest in Amgen Corp., Gilead Sciences, and Prothena; reports receiving honoraria from Elsevier and Wolters Kluwer; reports being a scientific advisor or member of the Otsuka Advisory Board; and reports having other interests/relationships with the Jared Grantham Kidney Institute, which receives royalties from Otsuka for tolvaptan, and The University of Kansas Medical Center. D. Jalal reports receiving research funding from AstraZeneca and Corvidia; reports receiving honoraria from Kansas Idea Network of Biomedical Research Excellence (K-INBRE) and Reata; and reports being a scientific advisor or member of Reata. K. McGreal reports receiving Medical Center research funding from the Sanofi Staged PKD trial sub-investigator and Reata as principle investigator on the FAlcon study. R. Mustafa reports being a scientific advisor to or member of The American College of Physicians Clinical Guideline Committee, The Canadian Society of Nephrology Clinical Practice Guidelines Committee, and The GRADE guidance group; reports having other interests/relationships with the advisory board for the renal round table, and the National Kidney Foundation Midwest. All remaining authors have nothing to disclose.
Funding
This study was supported by the Kansas PKD Research and Translation Core Center (P30 DK106912), CTSA Award UL1TR002366, and HERON.
Author Contributions
D. Jalal, M.A. Kalot, and R. Mustafa conceptualized the study; A. Alayli, M. Alkhatib, N. Husainat, M.A. Kalot, K. McGreal, and R. Mustafa were responsible for the data curation; A. Alayli, M. Alkhatib, N. Husainat, D. Jalal, M.A. Kalot, R. Mustafa, and A. Yu were responsible for the formal analysis; A. Alayli, M. Alkhatib, D. Jalal, M.A. Kalot, K. McGreal, R. Mustafa, and A. Yu were responsible for the investigation; D. Jalal, M.A. Kalot, K. McGreal, R. Mustafa, and A. Yu were responsible for the methodology; R. Mustafa was responsible for the project administration, provided supervision, and was responsible for the validation; A. Alayli, M. Alkhatib, N. Husainat, M.A. Kalot, K. McGreal, and R. Mustafa wrote original draft; and A. Alayli, D. Jalal, M.A. Kalot, K. McGreal, R. Mustafa, and A. Yu reviewed and edited the manuscript.
Acknowledgments
This study was supported by the Kansas PKD Research and Translation Core Center (P30 DK106912), CTSA Award UL1TR002366, and HERON.
Supplemental Material
This article contains the following supplemental material online at http://kidney360.asnjournals.org/lookup/suppl/doi:10.34067/KID.0000852021/-/DCSupplemental.
STARD flow diagram for a study of 536 patients from the nephrology clinic for ADPKD diagnosis status. Download Supplemental Figure 1, PDF file, 99 KB (98KB, pdf)
STARD flow diagram for a study of 535 patients who did not have nephrology visits for ADPKD diagnosis status. Download Supplemental Figure 2, PDF file, 99 KB (98KB, pdf)
References
- 1.Harris PC, Torres VE: Polycystic kidney disease. Annu Rev Med 60: 321–337, 2009. 10.1146/annurev.med.60.101707.125712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vlasschaert ME, Bejaimal SA, Hackam DG, Quinn R, Cuerden MS, Oliver MJ, Iansavichus A, Sultan N, Mills A, Garg AX: Validity of administrative database coding for kidney disease: A systematic review. Am J Kidney Dis 57: 29–43, 2011. 10.1053/j.ajkd.2010.08.031 [DOI] [PubMed] [Google Scholar]
- 3.Torres VE, Harris PC: Autosomal dominant polycystic kidney disease: The last 3 years. Kidney Int 76: 149–168, 2009. 10.1038/ki.2009.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gradzik M, Niemczyk M, Gołębiowski M, Pączek L: Diagnostic imaging of autosomal dominant polycystic kidney disease. Pol J Radiol 81: 441–453, 2016. 10.12659/PJR.894482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richesson RL, Hammond WE, Nahm M, Wixted D, Simon GE, Robinson JG, Bauck AE, Cifelli D, Smerek MM, Dickerson J, Laws RL, Madigan RA, Rusincovitch SA, Kluchar C, Califf RM: Electronic health records based phenotyping in next-generation clinical trials: A perspective from the NIH health care systems collaboratory. J Am Med Inform Assoc 20: e226–e231, 2013. 23956018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tasker RC: Why everyone should care about “computable phenotypes”. Pediatr Crit Care Med 18: 489–490, 2017. 10.1097/PCC.0000000000001115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quan H, Li B, Saunders LD, Parsons GA, Nilsson CI, Alibhai A, Ghali WA; IMECCHI Investigators : Assessing validity of ICD-9-CM and ICD-10 administrative data in recording clinical conditions in a unique dually coded database. Health Serv Res 43: 1424–1441, 2008. 10.1111/j.1475-6773.2007.00822.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Topaz M, Shafran-Topaz L, Bowles KH: ICD-9 to ICD-10: evolution, revolution, and current debates in the United States. Perspect Health Inf Manag 10: 1d, 2013 [PMC free article] [PubMed] [Google Scholar]
- 9.CDC . International Classification of Diseases, Tenth Revision (ICD-10), 2016. Available at: https://www.cdc.gov/nchs/icd/icd10.htm. Accessed April 22, 2019
- 10.Grimes DA: Epidemiologic research using administrative databases: Garbage in, garbage out. Obstet Gynecol 116: 1018–1019, 2010. 10.1097/AOG.0b013e3181f98300 [DOI] [PubMed] [Google Scholar]
- 11.Murphy SN, Weber G, Mendis M, Gainer V, Chueh HC, Churchill S, Kohane I: Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc 17: 124–130, 2010. 10.1136/jamia.2009.000893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waitman LR, Warren JJ, Manos EL, Connolly DW: Expressing observations from electronic medical record flowsheets in an i2b2 based clinical data repository to support research and quality improvement. AMIA Annu Symp Proc 2011: 1454–1463, 2011 [PMC free article] [PubMed] [Google Scholar]
- 13.Soroka S, Alam A, Bevilacqua M, Girard LP, Komenda P, Loertscher R, McFarlane P, Pandeya S, Tam P, Bichet DG: Updated Canadian expert consensus on assessing risk of disease progression and pharmacological management of autosomal dominant polycystic kidney disease. Can J Kidney Health Dis 5: 2054358118801589, 2018. 10.1177/2054358118801589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallis S: Binomial confidence intervals and contingency tests: Mathematical fundamentals and the evaluation of alternative methods. J Quant Linguist 20: 178–208, 2013. 10.1080/09296174.2013.799918 [DOI] [Google Scholar]
- 15.Altman DG, Machin D, Bryant TN, Gardner MJ, editors: Statistics with Confidence: Confidence Intervals and Statistical Guidelines, 2nd Ed., London, BMJ Books, 2000 [Google Scholar]
- 16.Cohen JF, Korevaar DA, Altman DG, Bruns DE, Gatsonis CA, Hooft L, Irwig L, Levine D, Reitsma JB, de Vet HC, Bossuyt PM: STARD 2015 guidelines for reporting diagnostic accuracy studies: Explanation and elaboration. BMJ Open 6: e012799, 2016. 10.1136/bmjopen-2016-012799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Šimundić AM: Measures of diagnostic accuracy: Basic definitions. EJIFCC 19: 203–211, 2009 [PMC free article] [PubMed] [Google Scholar]
- 18.Myers RP, Leung Y, Shaheen AAM, Li B: Validation of ICD-9-CM/ICD-10 coding algorithms for the identification of patients with acetaminophen overdose and hepatotoxicity using administrative data. BMC Health Serv Res 7: 159, 2007. 10.1186/1472-6963-7-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.So L, Evans D, Quan H: ICD-10 coding algorithms for defining comorbidities of acute myocardial infarction. BMC Health Serv Res 6: 161, 2006. 10.1186/1472-6963-6-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanchette CM, Liang C, Lubeck DP, Newsome B, Rossetti S, Gu X, Gutierrez B, Lin ND: Progression of autosomal dominant kidney disease: Measurement of the stage transitions of chronic kidney disease. Drugs Context 4: 212275, 2015. 10.7573/dic.212275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalatharan V, Pei Y, Clemens KK, McTavish RK, Dixon SN, Rochon M, Nash DM, Jain A, Sarma S, Zaleski A, Lum A, Garg AX: Positive predictive values of international classification of diseases, 10th revision coding algorithms to identify patients with autosomal dominant polycystic kidney disease. Can J Kidney Health Dis 3: 2054358116679130, 2016. 10.1177/2054358116679130 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
STARD flow diagram for a study of 536 patients from the nephrology clinic for ADPKD diagnosis status. Download Supplemental Figure 1, PDF file, 99 KB (98KB, pdf)
STARD flow diagram for a study of 535 patients who did not have nephrology visits for ADPKD diagnosis status. Download Supplemental Figure 2, PDF file, 99 KB (98KB, pdf)