Abstract
Kidney disease represents a global health burden of increasing prevalence and is an independent risk factor for cardiovascular disease. Myeloid cells are a major cellular compartment of the immune system; they are found in the healthy kidney and in increased numbers in the damaged and/or diseased kidney, where they act as key players in the progression of injury, inflammation, and fibrosis. They possess enormous plasticity and heterogeneity, adopting different phenotypic and functional characteristics in response to stimuli in the local milieu. Although this inherent complexity remains to be fully understood in the kidney, advances in single-cell genomics promise to change this. Specifically, single-cell RNA sequencing (scRNA-seq) has had a transformative effect on kidney research, enabling the profiling and analysis of the transcriptomes of single cells at unprecedented resolution and throughput, and subsequent generation of cell atlases. Moving forward, combining scRNA- and single-nuclear RNA-seq with greater-resolution spatial transcriptomics will allow spatial mapping of kidney disease of varying etiology to further reveal the patterning of immune cells and nonimmune renal cells. This review summarizes the roles of myeloid cells in kidney health and disease, the experimental workflow in currently available scRNA-seq technologies, and published findings using scRNA-seq in the context of myeloid cells and the kidney.
Keywords: chronic kidney disease, basic science, dendritic cells, kidney disease, macrophage, monocytes, myeloid cells, scRNA-sequencing, sequence analysis, RNA
Introduction
The kidney is a complex organ that performs diverse functions essential for physiologic homeostasis and, consequently, patients with kidney disease often present with significant complications and comorbidities (1,2). The manifestations of kidney disease are wide ranging, encompassing acute kidney injury (AKI), autoimmune disorders, and rejection of kidney transplants, all of which are directly or indirectly immune mediated (3). Due to an increased prevalence rate, kidney disease is having a major effect on global health, both as a direct cause of morbidity and mortality, and as a major risk factor for cardiovascular disease (1).
Myeloid cells are the most abundant nucleated hematopoietic cells in the body, encompassing monocytes, macrophages, dendritic cells (DCs), and granulocytes. Myeloid cells make up a critical arm of the immune system and have diverse functions (4). In the kidney, macrophages and DCs have, most notably, been shown to influence organ homeostasis, injury, and reparative processes after diverse insults. Their function, phenotypic spectrum, and interplay with other immune and nonimmune cells is complex and incompletely understood (3,5).
Our understanding of the immune system has advanced through the application of technologies that allow single-cell resolution, primarily microscopy and flow cytometry. The number of parameters that can be measured simultaneously with these technologies is limited and reliant on prior knowledge of which antigens are present. Over recent years, progress in single-cell RNA sequencing (scRNA-seq), which aims to measure the expression levels of genes in cells in a comprehensive way, has transformed our ability to profile immune cells (2,6,7).
Roles of Myeloid Cells in Kidney Health and Disease
Myeloid cells have an adaptive nature, demonstrating transient responses, controlled differentiation, (re)location into tissues, and plasticity in response to various environmental stimuli, whether they are homeostatic or inflammatory (4). This represents a complication regarding their study within the kidney, because this major hemofiltration organ offers a challenging milieu that is not spatially uniform, and differences in immune cell types in the renal cortex and medulla have been reported (8,9).
Macrophages are the most abundant myeloid cell type in the kidney and, together with DCs, coordinate inflammatory responses to freely filtered antigenic material and safeguard the kidney from infection (8). Importantly, they are also involved in initiation and progression of renal disease and in the subsequent tissue regeneration. Due to this diversity, it is generally suggested that macrophages are divided into proinflammatory (M1 or “classically activated”) and tissue reparative (M2 or “alternatively activated”) phenotypes. However, this characterization is oversimplified with not only macrophages, but also DCs possessing inherent plasticity. Indeed, there is robust evidence for disease-associated myeloid cells sharing signature marker genes of both pro- and anti-inflammatory characteristics (4,10).
Immune cells are typically defined by different cell-surface markers, although different myeloid cell types have many markers in common; therefore, several markers will often be required to identify the cell type of interest. In addition, markers can differ between species (Table 1). Immunohistochemistry and flow cytometry are often used to identify both intra- and extracellular markers (6). In mice, kidney resident macrophages (CD11bloF4/80hi) are of dual hematopoietic origin, arising from the yolk sac and from definitive hematopoiesis, with Ly6Clo “patrolling” monocytes (CD14loCD16hi in humans) (11–13). Under inflammatory conditions, Ly6Chi monocytes (CD14hiCD16− in humans) infiltrate the kidney, differentiating into (CD11bhiF4/80lo) macrophages (11–13). Despite being limited to around 16 parameters and problems that arise from spectral overlap, flow cytometry, in particular, has been very useful and represents the most basic single-cell technology, but it is unlikely to provide the full phenotypic spectrum of myeloid cells (6,7,10).
Table 1.
Examples of markers commonly used to identify populations of myeloid cell types in mice and humans
| Markers | Mouse | Human |
|---|---|---|
| Monocyte | CD11b, Ly6C, CD192 (CCR2), CX3CR1 | CD11b, CD14, CD16 |
| Macrophage | CD11b, F4/80, CD68, CD64 | CD11b, CD68, CD163 |
| Proinflammatory | CD86, MHCII | CD86, HLA-DR |
| Anti-inflammatory | CD206 (MRC1), IL-4R | CD206 (MRC1), IL-4R |
| Tissue resident | CD192 (CCR2), CD206 (MRC1), CD68, CD74, CD81 | CD192 (CCR2), CD68, CD74, CD81 |
| Dendritic cell | CD11c, CD11b, MHCII | CD11c, CD11b, HLA-DR |
| Conventional | CX3CR1, CD103 | CD1c (BDCA1), HLA-DR |
| Plasmacytoid | Ly-49Q, CD317 (Bst2) | CD123 (IL-3Rα), CD303 (BDCA2) |
| Neutrophil | CD11b, Ly6G, CD18 | CD11b, CD16, CD32, CD66b |
| Eosinophil | CD170 (Siglec-F), CD193 (CCR3) | CD329 (Siglec-8), CD193 (CCR3) |
| Basophil | CD123 (IL-3Rα), FcεRIα | CD123 (IL-3Rα), FcεRIα |
| Mast cell | CD117 (c-Kit), FcεRIα | CD32, CD117 (c-Kit), FcεRIα |
Detection of myeloid (CD45+) cell types by flow cytometry. Combinations and level of marker expression can vary with subtype/state of activation. Ly6C, lymphocyte antigen 6; CCR2, C-C chemokine receptor type 2; CX3CR1, CX3 chemokine receptor 1; MHCII, major histocompatibility complex II; HLA-DR, human leukocyte antigen–DR isotype; MRC1, mannose receptor C-type 1; IL4R, interleukin-4 receptor; BDCA1, blood dendritic cell antigen 1; Bst2, bone marrow stromal cell antigen 2; IL-3Rα, interleukin-3 alpha receptor; BDCA2, blood dendritic cell antigen 2; Ly6G, lymphocyte antigen 6 complex locus G6D; CCR3, C-C chemokine receptor type 3; FcεRIα, Fc epsilon alpha receptor I.
Overview of Single-Cell Sequencing Technologies
High-throughput gene expression techniques, such as microarray and bulk RNA-seq, have provided a better understanding of complex organ transcriptomes; however, their RNA input requirements have limited studies to pooled populations. As the kidney contains >20 distinct cell types (including epithelial, endothelial, mesenchymal, and immune populations), the resulting gene expression profile of renal tissue reflects only an average for these heterogeneous constituents (14). Within the kidney, the proximal tubule cells dominate in number, and can thus obscure the expression profile of rarer populations. scRNA-seq technologies have addressed these limitations by enabling the objective investigation of transcriptomes of individual cells in a given sample, with generation of cell atlases of mammalian renal components. Notably, the kidney research community has been invested in open access and has made these data freely available via websites (Table 2). This permits the unbiased assessment of cellular heterogeneity, identification of new cell states and subtypes, and dynamic cellular transitions at very high resolution and accuracy (6,14,15).
Table 2.
Single-cell atlases
| Species (genotype) | Disease/Model | Method(s) | Website Link |
|---|---|---|---|
| Mouse (C57BL6/J) | Fibrosis (R-UUO) | scRNA-seq | Gene Atlas of Reversible Unilateral Uretic Obstruction Model: http://www.ruuo-kidney-gene-atlas.com |
| Mouse (C57BL6/J) | Healthy (aging, 1–30 months) | Bulk RNA-seq, scRNA-seq, | Tubula Muris Senis: https://tabula-muris-senis.ds.czbiohub.org |
| Mouse (C57BL6/J) | Acute injury (LPS-induced endotoxemia) | scRNA-seq | Mouse Kidney Single-cell Expression: https://connect.rstudio.iu.edu/content/18/ |
| Mouse (CFW) | Acute injury (IRI) | scRNA-seq | Mouse IRI scRNA: https://research.cchmc.org/PotterLab/scIRI/ |
| Mouse (C57BL6/J) | Healthy, fibrosis (FAN) | scRNA-seq, snATAC-seq | Susztaklab Kidney Biobank: https://susztaklab.com |
| Human | Healthy | ||
| Mouse (C57BL6/J) | Healthy, fibrosis (UUO), acute injury (IRI) | scRNA-seq, snRNA-seq | Kidney Interactive Transcriptomics (KIT): http://humphreyslab.com/SingleCell/ |
| Human | Allograft rejection, diabetes | ||
| Human | Healthy (including fetal) | scRNA-seq | Kidney Cell Atlas: https://www.kidneycellatlas.org |
| Human | Healthy (including fetal) | scRNA-seq | Nephrocell: http://nephrocell.miktmc.org |
| Human | Healthy (fetal) | scRNA-seq | The Human Nephrogenesis Atlas: https://sckidney.flatironinstitute.org |
Single-cell expression atlases from studies discussed in this review and of interest. R-UUO, reversible unilateral ureteral obstruction; scRNA-seq, single-cell RNA sequencing; IRI, ischemic reperfusion injury; FAN, folic acid nephropathy; snATAC-seq, single-nuclear assay for transposase-accessible chromatin using sequencing; snRNA-seq, single-nuclear RNA sequencing.
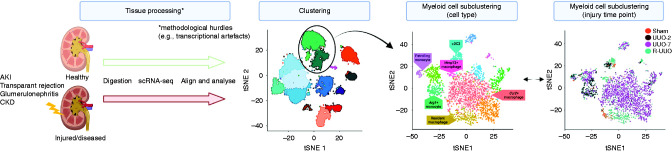
To date, a number of scRNA-seq techniques have been proposed, and the development of new protocols is a very active area of research (as reviewed by Li and Humphreys [16]). Generally, all scRNA-seq experiments share a common workflow: sample preparation, single-cell capture, reverse transcription and transcriptome amplification, library preparation, sequencing, and analysis (Figure 1).
Figure 1.
Single cell-RNA sequencing can map myeloid cell heterogeneity in the kidney. For dense tissues like the kidney, cell dissociation, typically achieved using both physical disaggregation and enzymatic digestion, is the most important step; it directly affects the molecular profiles of cells and can introduce stress-induced transcriptional artifacts (9,17). This is particularly limiting when looking at kidney disease samples where tissue is subject to inflammatory stress, although this may be overcome by excluding low-quality cells, for example, on the basis of high mitochondrial RNA content (18). Cell isolation and capture is the most unique hurdle to overcome for scRNA-seq. The use of FACS allows for the selection of cells on the basis of surface markers and is, therefore, useful when isolating a specific subset of cells for sequencing, including CD45 for immune cells. Adapted from ref. 10, with permission. R-UUO, reversible unilateral ureteral obstruction; tSNE, t-distributed stochastic neighbor embedding; UUO, unilateral ureteral obstruction.
Single-nuclear RNA-seq (snRNA-seq) is an increasingly popular alternative to scRNA-seq. Here, nuclei are isolated from cells and used for droplet-based sequencing (14). snRNA-seq has been reported to have comparable gene detection to scRNA-seq in the adult kidney and to eliminate dissociation-induced transcriptional stress responses in the inflamed fibrotic kidney. Moreover, it is compatible with frozen samples but captures immune cells with lower efficiency (19). The reason for this remains unclear. As such, it might be necessary to isolate CD45+ cells, which only account for 2%–17% of the total kidney cell population (20), to obtain the greatest possible insight into renal leukocytes using this method.
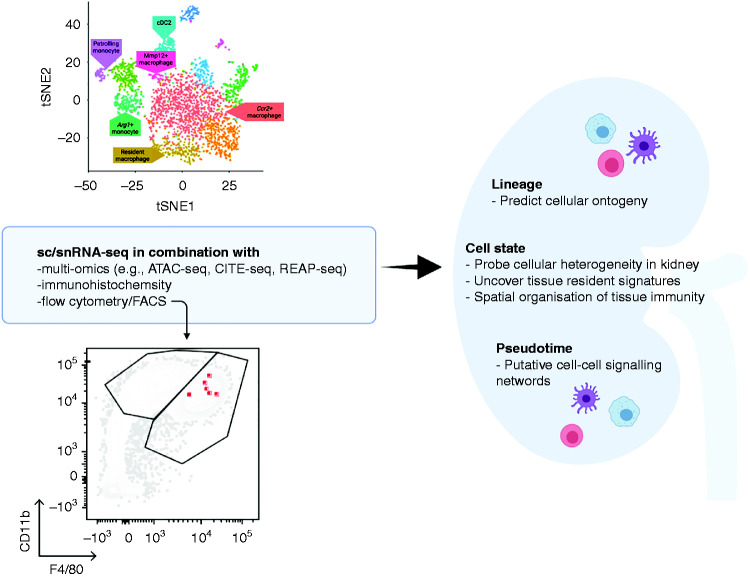
Numerous other single-cell techniques are under development to allow the direct measurement of microRNA, proteomic, or metabolic data, which scRNA-seq and snRNA-seq approaches do not provide. Most notably, simultaneous expression profiling of transcripts and cell surface proteins—such as RNA expression and protein (REAP) sequencing (21) and cellular indexing of transcriptomes and epitopes (CITE) by sequencing (22)—is now possible, permitting the correlation of protein expression with transcriptomic data. In addition, assay for transposase-accessible chromatin (ATAC) with sequencing allows profiling of chromatic accessibility, enabling investigation of the epigenetic heterogeneity (23). It has recently been used in combination with snRNA-seq to identify unique cell states within the proximal tubule and thick ascending limb in the human kidney (24), and key chromatin remodeling events and gene expression dynamics associated with kidney development in mice (Figure 2) (25).
Figure 2.
Synergistic use of single cell-RNA sequencing and other historical and cutting edge technologies can be utilized to fully characterize renal immune populations. For example, immune cells may be mapped onto flow cytometry plots ATAC-seq, assay for transposase-accessible chromatin using sequencing; CITE-seq, cellular indexing of transcriptomes and epitopes by sequencing; REAP-seq, RNA expression and protein sequencing; tSNE, t-distributed stochastic neighbor embedding.
Due to the kidney having distinct regional differences, anatomic localization and intercellular interactions of cells are important, yet these factors are largely lost when analyzing dissociated single cells. Interactions may be somewhat gleaned from receptor-ligand pair analyses for chemokines and cytokines; however, direct visualization of the spatial relationships among cells is needed to fully answer such questions. This further means that it is not possible to discriminate leukocyte populations that infiltrate or reside in the kidney from those that may be circulating through the kidney vasculature (6,15). Combining scRNA-seq and snRNA-seq with spatial transcriptomics may improve this aspect. Indeed, Ferriera et al. (26) recently used these complementary technologies to spatially map the transcriptomic signature in mouse AKI models and demonstrated how this may be applied to human samples. They identified patterns of immune and epithelial cell colocalization and, although limited by resolution, these methodologies will likely improve with future work.
scRNA-Seq Studies in Kidney Health
A comprehensive cellular anatomy of the normal kidney is crucial to completely understand the development and resolution of kidney disease; however, to date, attempts to fully map immune populations in the kidney in health are limited. In 2018, Park et al. (2) provided the first and largest-scale murine kidney cell scRNA-seq so far from healthy mouse kidney. They identified major cell subtypes that comprise the nephron and previously known renal immune cell types, including resident macrophages and neutrophils. However, it is possible that some myeloid subtypes were undetected, perhaps due to immune cell types contributing a small proportion in the uninjured kidney and/or due to sample preparation because approximately 25% of cells did not pass quality control in this study.
A recent key study used scRNA-seq with flow and mass cytometry to define the global immune landscape in healthy human adult and fetal kidneys, profiling cells across the life span (9). Mononuclear phagocytes (MNPs), natural killer (NK) cells, and T cells were most prevalent, and four subsets of MNPs, neutrophils, mast cells, and plasmacytoid DCs (pDCs) were identified within the mature kidney myeloid compartment. The MNP cluster was dominated by two monocyte-derived macrophage populations: one was transcriptionally similar to CD14+ classic monocytes, and the other to CD16+ nonclassic monocytes. It also contained a conventional DC (cDC) population and tissue-macrophage population that was skewed toward an anti-inflammatory M2 transcriptome expressing CD206. In addition, researchers demonstrated anatomic localization of immune cells by referencing their work to publicly available bulk RNA-seq data generated from known kidney regions, revealing differential distribution of immune subsets in the medulla and pelvis compared with the cortex. Analysis of ligand-receptor interactions, indicating epithelial and immune cell crosstalk, was predicted to localize antibacterial macrophages and neutrophils to regions of the kidney most susceptible to ascending infection from the urinary tract. This is in line with previous work that showed a high interstitial sodium concentration of the medulla stimulates the production of chemokines by tubular epithelial cells, which helps position macrophages to counter the immunologic threat posed by ascending bacterial infection (8). The capability to orchestrate localization of immune cells to specific regions was absent in fetal kidneys, with postnatal acquisition of transcriptional signatures indicative of roles in inflammation and immune defense (9). Another recent study by Menon et al. (27) identified resident immune cells in nondiseased human samples from tumor-nephrectomy, surveillance, and preperfusion biopsies; these included two myeloid clusters, although they were not defined into subtypes.
Dynamic changes in the immune system are a hallmark of aging (28). Recently, scRNA-seq was performed on >350,000 cells from several organs and tissues taken from C57BL/6J mice belonging to age groups ranging from 1 to 30 months (29). After computation of the overall diversity score, two clusters of renal macrophages were identified for which composition changed significantly with age. One cluster was mostly composed of cells from 1- and 3-month-old mice enriched with an anti-inflammatory signature (e.g., Cq1a, Cd74, Cd81). Conversely, the other cluster was primarily composed of cells from 18-, 21-, 24-, and 30-month-old mice that resembled a proinflammatory macrophage state (e.g., Intgal, Msrb1). At present, such comprehensive aging studies in humans are missing, but these will be important to undertake in future due to the prevalence of kidney disease increasing with age (1).
Although studies have been carried out on both human and mouse tissue, there are shortcomings in acquiring enough fresh human samples, highlighting the importance of overcoming translational issues. Studies have identified well-defined markers of myeloid cell types across murine tissues, but they lack conservation across species. A well-known example is F4/80 (encoded by adgre1), which is used to identify tissue-resident macrophages in mice but is not expressed by macrophage populations in humans. Furthermore, markers that were historically used to differentiate DCs from macrophages, such as CD11c and MHCII, are now known to be commonly expressed by tissue macrophages (28,30). Zimmerman et al. (31) used scRNA-seq to identify a cross-species kidney macrophage–specific marker. CD45+ cells, with the lymphocyte population excluded, were isolated from one mouse, rat, pig, and human kidney, revealing a cluster of cells which express C1q. Other scRNA-seq analyses of whole kidney tissue from mice (2,10) and humans (32) have also identified this C1q-expressing cluster. In the C1q-expressing cluster, novel surface markers Cd74 and Cd81 were identified as potential candidate markers of kidney-resident macrophages across species (31). These were validated by flow cytometry analyses showing CD74+CD81+ cells were more abundant in resident (CD11bloF4/80hi) compared with infiltrating (CD11bhiF4/80lo) macrophages, and they confirmed minimal exchange with bone marrow–derived cells using the parabiotic mouse model. It remains unknown if these candidates vary throughout the time course of inflammation. Indeed, C1qa expression varied over 12 myeloid subsets in our reversible unilateral ureteral obstruction (R-UUO) study (10). In future, it will be important to further investigate the effect of the mouse strain to contextualize findings. Indeed, inbred laboratory mouse strains can be highly divergent in their immune response patterns; for example, after unilateral ischemic reperfusion injury (IRI) 129/Sv mice have been shown to have fewer infiltrating leukocytes than C57BL6/J mice (33).
scRNA-Seq Studies in Kidney Injury and Disease
A phase-dependent influx of myeloid cells and changes in their phenotype occur during the time course of kidney diseases (34). To define the cellular landscape in the progression of AKI to chronic kidney disease (CKD), do Valle Duraes et al. (35) used scRNA-seq in a unilateral IRI model with or without immediate contralateral nephrectomy to study kidney regeneration or fibrosis, respectively. Seven main clusters were identified, including resident and inflammatory macrophages, neutrophils/monocytes, DC/monocytes, NK cells, T cells, and B cells. The healthy kidney was dominated by resident macrophages, whereas, after injury, inflammatory macrophages significantly expanded and resident macrophages disappeared, regardless of the model used or postinjury time point. The mixed populations of neutrophils/monocytes and DC/monocytes also expanded after injury. In another study, snRNA-seq was used with a bilateral IRI model of AKI (36). Although proximal tubules were the focus here, six leukocyte clusters were identified: three macrophage subtypes, DCs, T cells, and B cells. Ligand-receptor analysis performed on the combined leukocyte cluster suggested changes in Ccl2 (tubulointerstitium) to Ccr2 (leukocytes) signaling over time; fibroblasts and endothelial cells were the first cell types to signal leukocytes, followed by leukocyte-leukocyte signaling and increasing Ccl2-Ccr2 signaling from proximal tubules that failed to repair at 2 days and 6 weeks postinjury. The first and second macrophage cluster are likely resident and inflammatory macrophages on the basis of markers such as F13a1 and Vcan. The third cluster expressed high levels of Mmp12, a macrophage-specific metalloproteinase, and Gpnmb, a negative regulator of inflammation proposed to promote M2 polarization. This and the DC cluster were most abundant at 2 and 6 weeks postinjury, when the kidney is in the reparative phase. Our group recently used scRNA-seq in the mouse R-UUO model, in which regression of established tubulointerstitial fibrosis, a hallmark of CKD, occurs after reversal of obstruction (10). Interestingly, we also reported a macrophage cluster that was solely in kidneys that had undergone UUO reversal; it was also characterized by expression of Mmp12 and scavenger receptors, including Gpnmb and Mrc1. We have previously reported a similar Mmp12+ macrophage in liver during the resolution of liver disease (37). Although these Mmp12+ cells mapped to macrophages on the Immunological Genome Project database, they morphologically resembled monocytes and expressed high levels of Ccr2 but low levels of F4/80 by immunofluorescence. Furthermore, their transcriptome most closely aligned to monocytes infiltrating the kidney during recovery from IRI. We found that bone marrow–derived monocytes that were fed FITC-labeled collagen increased their expression of Mmp12 and Gpnmb, suggesting phagocytosis of collagen may induce expression of these markers.
In our characterization of myeloid cells in renal injury and repair, we identified a further eleven myeloid cell subsets in the R-UUO model, including some with a novel phenotype. This was uniquely done by integrating droplet- and plate-based scRNA-seq with index linkage to map the subsets onto monocyte and macrophage gates on flow cytometry (10). Three monocyte clusters were identified: patrolling and inflammatory subtypes, and a unique “profibrotic” subtype expressing Arg1 that was exclusively present at UUO day 2 (acute injury phase). These Arg1+ cells expressed markers of Ly6C+ inflammatory monocytes, including Ccr2 and F13a1, although not Ly6c2. Moreover, they were shown to express hypoxia, profibrotic, and proinflammatory genes, along with genes encoding extracellular matrix components and extracellular matrix crosslinkers. Together with a number of ligand-receptor pairs revealed between Arg1+ monocytes and mesenchymal cells, these data suggest Arg1+ cells could be derived from recruited Ly6C+ monocytes that become activated acutely toward a profibrotic phenotype in the hypoxic and inflammatory environment of the injured kidney. Indeed, there are waves of monocyte ingress during chronic progressive kidney injury (35). Interestingly, in a model of renal injury due to sepsis, a subcluster of macrophages showed increased Arg1 expression in the later postinjury time points, albeit with markers of alternative macrophage activation, such as Mrc1 (38). We further tracked the fate of circulating immune cells recruited to the kidney using paired blood exchange. It holds advantages over parabiosis because the donor cells persist in large numbers in the circulation for a relatively short time, thus, permiting the tracking of cells at multiple time points after injury or during resolution of disease. By combining paired blood exchange, flow cytometry, and pseudotime analyses, we showed early recruitment of donor monocytes to the obstructed kidney at UUO day 2; at UUO day 7, they transitioned to CCR2hi macrophages with a transcriptome almost identical to resident macrophages. Other work is indicative of CCR2+ cells being detrimental: in an IRI model with CCR2 inactivation, there was a reduction in the expansion of kidney F4/80 macrophages and severity of renal fibrosis (5). It remains to be determined if these novel populations are present at all stages in the context of CKD; for example, are the Mmp12+ macrophages present at early points of injury where endogenous kidney repair mechanisms are active and attempting repair, or only when a scarred matrix is present with no ongoing injury stimulus?
Dhillon et al. (39) performed scRNA-seq analysis on healthy and fibrotic mouse kidneys from a folic acid nephropathy model. Fourteen immune clusters were identified, compared with only five in their previous study (2), including macrophages, subclusters of DCs (CD11b+/−, pDC), granulocytes, and lymphocytes. Another study compared scRNA-seq and snRNA-seq on obstructed (fibrotic) mouse kidney at UUO day 14. Both methods had comparable gene detection; however, compared with scRNA-seq, snRNA-seq had a reduced dissociation bias and dissociation-induced transcriptional stress responses (19). One macrophage cluster was identified here, although researchers identified a novel “dedifferentiated” proximal tubule cluster that expressed numerous secreted proinflammatory cytokines, including Ccl2, macrophage proliferative cytokine IL-34, and neutrophil chemoattractants Cxcl1 and Cxcl2. This study, again, indicates that snRNA-seq is not as capable as scRNA-seq at detecting immune populations in the damaged and inflamed kidney. Indeed, the same group performed snRNA-seq on cryopreserved human diabetic kidney samples (40). Diabetic kidneys were found to have increased leukocyte numbers compared with control kidneys, as expected. Monocytes, plasma cells, T cells, and B cells were detected; however, a macrophage population was not, despite the role of macrophages in type 2 diabetes being well documented (41). Researchers further compared their diabetic samples with publicly available PBMC datasets because the leukocyte numbers in control samples were small. Increased inflammatory markers were observed in both infiltrating diabetic CD14+ monocytes and CD16+ monocytes.
Recently, Kuppe et al. (42) generated a single-cell map of the human kidney, with a focus on the tubulointerstitium and fibrosis development in CKD. However, in the CD10− (nonproximal tubular) sorted cells, three macrophage subtypes, monocytes, DCs, and mast cells were identified, as were NK, T, and B cells. Researchers proposed a working model of the pathways involved in renal fibrosis, but immune cells, which play important roles in profibrotic signaling, were absent from this model, despite being captured. Further extending our understanding of immune cells in different pathologic states, a study described the immune landscape of the human kidney in patients with lupus nephritis, a frequent complication of systemic lupus erythematosus (43). Kidney samples from patients with lupus nephritis and from healthy controls were analyzed using scRNA-seq. This revealed twenty-one immune clusters, including numerous subsets of myeloid cells in both proinflammatory responses and inflammation-resolving responses: inflammatory CD16+ macrophages, phagocytic CD16+ macrophages, tissue-resident macrophages, M2-like CD16+ macrophages, cDCs, and pDCs. Tissue-resident macrophages were the dominant cluster in healthy kidneys.
Myeloid cells are also increasingly recognized as major players in transplant rejection. Dangi et al. (18) used scRNA-seq to dissect the contribution of myeloid cell subsets to kidney transplant rejection in a mouse model. Thirteen clusters were identified as immune cell types, including two macrophage subsets, monocytes, pDCs, cDCs, and a transitioning monocyte/macrophage population. As expected, the number of cells in all immune cell clusters was the highest in rejecting kidneys, followed by tolerized kidneys, and it was markedly lower and often negligible in naive kidneys. A macrophage cluster was the second most abundant behind T cells. Interestingly, among the top cluster-defining genes, Axl was exclusively expressed only by the graft-infiltrating myeloid cell clusters macrophage cluster 1 and 2, and the macrophage/monocyte cluster, but not by any other cell types. Axl1+ macrophages were found to be significantly higher in rejected versus tolerized kidneys, and Axl promoted intragraft differentiation of inflammatory macrophages. In another scRNA-seq study, Wu et al. (32) analyzed a kidney allograft biopsy from a recipient undergoing acute rejection. They identified two distinct monocyte populations, likely a proinflammatory and classic or intermediate, further supporting a role of myeloid cells in kidney rejection in humans.
Conclusions
Over the past few years, single-cell genomics (with scRNA-seq leading the way) has provided researchers with a new experimental toolbox to dissect the roles and interactions of individual cell types in health and disease. It is allowing the re-evaluation of our understanding of myeloid cell biology in complex organs, including aspects of cellular ontogeny, differentiation, homeostasis, and range of activation states. This is especially important in the kidney, where heterogeneity and plasticity of cells within the myeloid compartment is reflected under both homeostatic and disease conditions. Integration of datasets from separate experiments using human samples will be important; humans are genetically heterogeneous and have variable environmental exposures, and procurement of enough samples can be a challenge. This also highlights the need to continue to determine the translatability of our findings. The application of scRNA-seq in both basic and translational research is expected to grow exponentially with the continued advancements and standardization of experimental and analytic methods.
Disclosures
L. Denby reports receiving research funding from GlaxoSmithKline and Regulus Therapeutics. The remaining author has nothing to disclose.
Funding
R.M.B. Bell is supported by funding from the British Heart Foundation PhD Studentship Grant FS/19/55/34890. L. Denby is supported by a Kidney Research UK Senior Fellowship Grant SF_001_20181122.
Acknowledgment
The figures were created with BioRender.com.
Author Contributions
R.M.B. Bell was responsible for visualization; R.M.B. Bell and L. Denby wrote the original draft and reviewed and edited the manuscript; and L. Denby conceptualized the study, was responsible for funding acquisition and resources, and provided supervision.
References
- 1.GBD Chronic Kidney Disease Collaboration : Global, regional, and national burden of chronic kidney disease, 1990-2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 395: 709–733, 2020. 10.1016/S0140-6736(20)30045-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park J, Shrestha R, Qiu C, Kondo A, Huang S, Werth M, Li M, Barasch J, Suszták K: Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360: 758–763, 2018. 10.1126/science.aar2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tecklenborg J, Clayton D, Siebert S, Coley SM: The role of the immune system in kidney disease. Clin Exp Immunol 192: 142–150, 2018. 10.1111/cei.13119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassler K, Schulte-Schrepping J, Warnat-Herresthal S, Aschenbrenner AC, Schultze JL: The myeloid cell compartment-cell by cell. Annu Rev Immunol 37: 269–293, 2019. 10.1146/annurev-immunol-042718-041728 [DOI] [PubMed] [Google Scholar]
- 5.Xu L, Sharkey D, Cantley LG: Tubular GM-CSF promotes late MCP-1/CCR2-mediated fibrosis and inflammation after ischemia/reperfusion injury. J Am Soc Nephrol 30: 1825–1840, 2019. 10.1681/ASN.2019010068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stewart BJ, Ferdinand JR, Clatworthy MR: Using single-cell technologies to map the human immune system - Implications for nephrology. Nat Rev Nephrol 16: 112–128, 2020. 10.1038/s41581-019-0227-3 [DOI] [PubMed] [Google Scholar]
- 7.Stubbington MJT, Rozenblatt-Rosen O, Regev A, Teichmann SA: Single-cell transcriptomics to explore the immune system in health and disease. Science 358: 58–63, 2017. 10.1126/science.aan6828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berry MR, Mathews RJ, Ferdinand JR, Jing C, Loudon KW, Wlodek E, Dennison TW, Kuper C, Neuhofer W, Clatworthy MR: Renal sodium gradient orchestrates a dynamic antibacterial defense zone. Cell 170: 860–874.e19, 2017. 10.1016/j.cell.2017.07.022 [DOI] [PubMed] [Google Scholar]
- 9.Stewart BJ, Ferdinand JR, Young MD, Mitchell TJ, Loudon KW, Riding AM, Richoz N, Frazer GL, Staniforth JUL, Vieira Braga FA, Botting RA, Popescu DM, Vento-Tormo R, Stephenson E, Cagan A, Farndon SJ, Polanski K, Efremova M, Green K, Del Castillo Velasco-Herrera M, Guzzo C, Collord G, Mamanova L, Aho T, Armitage JN, Riddick ACP, Mushtaq I, Farrell S, Rampling D, Nicholson J, Filby A, Burge J, Lisgo S, Lindsay S, Bajenoff M, Warren AY, Stewart GD, Sebire N, Coleman N, Haniffa M, Teichmann SA, Behjati S, Clatworthy MR: Spatiotemporal immune zonation of the human kidney. Science 365: 1461–1466, 2019. 10.1126/science.aat5031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conway BR, O’Sullivan ED, Cairns C, O’Sullivan J, Simpson DJ, Salzano A, Connor K, Ding P, Humphries D, Stewart K, Teenan O, Pius R, Henderson NC, Bénézech C, Ramachandran P, Ferenbach D, Hughes J, Chandra T, Denby L: Kidney single-cell atlas reveals myeloid heterogeneity in progression and regression of kidney disease. J Am Soc Nephrol 31: 2833–2854, 2020. 10.1681/ASN.2020060806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurts C, Ginhoux F, Panzer U: Kidney dendritic cells: Fundamental biology and functional roles in health and disease. Nat Rev Nephrol 16: 391–407, 2020. 10.1038/s41581-020-0272-y [DOI] [PubMed] [Google Scholar]
- 12.Mass E, Ballesteros I, Farlik M, Halbritter F, Günther P, Crozet L, Jacome-Galarza CE, Händler K, Klughammer J, Kobayashi Y, Gomez-Perdiguero E, Schultze JL, Beyer M, Bock C, Geissmann F: Specification of tissue-resident macrophages during organogenesis. Science 353: aaf4238, 2016. 10.1126/science.aaf4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SE, Pollard JW, Frampton J, Liu KJ, Geissmann F: A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336: 86–90, 2012. 10.1126/science.1219179 [DOI] [PubMed] [Google Scholar]
- 14.Park J, Liu CL, Kim J, Susztak K: Understanding the kidney one cell at a time. Kidney Int 96: 862–870, 2019. 10.1016/j.kint.2019.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rao DA, Arazi A, Wofsy D, Diamond B: Design and application of single-cell RNA sequencing to study kidney immune cells in lupus nephritis. Nat Rev Nephrol 16: 238–250, 2020. 10.1038/s41581-019-0232-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Humphreys BD: Single cell technologies: Beyond microfluidics. Kidney 360 2: 1196–1204, 2021. 10.34067/KID.0001822021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Flanagan CH, Campbell KR, Zhang AW, Kabeer F, Lim JLP, Biele J, Eirew P, Lai D, McPherson A, Kong E, Bates C, Borkowski K, Wiens M, Hewitson B, Hopkins J, Pham J, Ceglia N, Moore R, Mungall AJ, McAlpine JN, Shah SP, Aparicio S; CRUK IMAXT Grand Challenge Team : Dissociation of solid tumor tissues with cold active protease for single-cell RNA-seq minimizes conserved collagenase-associated stress responses. Genome Biol 20: 210, 2019. 10.1186/s13059-019-1830-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dangi A, Natesh NR, Husain I, Ji Z, Barisoni L, Kwun J, Shen X, Thorp EB, Luo X: Single cell transcriptomics of mouse kidney transplants reveals a myeloid cell pathway for transplant rejection. JCI Insight 5: e141321, 2020. 10.1172/jci.insight.141321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu H, Kirita Y, Donnelly EL, Humphreys BD: Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: Rare cell types and novel cell states revealed in fibrosis. J Am Soc Nephrol 30: 23–32, 2019. 10.1681/ASN.2018090912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nistala R, Meuth A, Smith C, Annayya A: Reliable and high efficiency extraction of kidney immune cells. J Vis Exp 114: 54368, 2016. 10.3791/54368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterson VM, Zhang KX, Kumar N, Wong J, Li L, Wilson DC, Moore R, McClanahan TK, Sadekova S, Klappenbach JA: Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol 35: 936–939, 2017. 10.1038/nbt.3973 [DOI] [PubMed] [Google Scholar]
- 22.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Smibert P: Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 14: 865–868, 2017. 10.1038/nmeth.4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ: Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523: 486–490, 2015. 10.1038/nature14590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muto Y, Wilson PC, Ledru N, Wu H, Dimke H, Waikar SS, Humphreys BD: Single cell transcriptional and chromatin accessibility profiling redefine cellular heterogeneity in the adult human kidney. Nat Commun 12: 2190, 2021. 10.1038/s41467-021-22368-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miao Z, Balzer MS, Ma Z, Liu H, Wu J, Shrestha R, Aranyi T, Kwan A, Kondo A, Pontoglio M, Kim J, Li M, Kaestner KH, Suszták K: Single cell regulatory landscape of the mouse kidney highlights cellular differentiation programs and disease targets. Nat Commun 12: 2277, 2021. 10.1038/s41467-021-22266-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferreira RM, Sabo AR, Winfree S, Collins KS, Janosevic D, Gulbronson C, Cheng Y, Casbon L, Barwinska D, Ferkowicz MJ, Xuei X, Zhang C, Dunn KW, Kelly KJ, Sutton TA, Hato T, Dagher PC, El-Achkar TM, Eadon MT: Integration of spatial and single-cell transcriptomics localizes epithelial cell –immune cross-talk in acute kidney injury. JCI Insight 6: e147703, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menon R, Otto EA, Hoover P, Eddy S, Mariani L, Godfrey B, Berthier CC, Eichinger F, Subramanian L, Harder J, Ju W, Nair V, Larkina M, Naik AS, Luo J, Jain S, Sealfon R, Troyanskaya O, Hacohen N, Hodgin JB, Kretzler M; Kidney Precision Medicine Project Kpmp; Nephrotic Syndrome Study Network (NEPTUNE) : Single cell transcriptomics identifies focal segmental glomerulosclerosis remission endothelial biomarker. JCI Insight 5: e133267, 2020. 10.1172/jci.insight.133267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salei N, Rambichler S, Salvermoser J, Papaioannou NE, Schuchert R, Pakalniškytė D, Li N, Marschner JA, Lichtnekert J, Stremmel C, Cernilogar FM, Salvermoser M, Walzog B, Straub T, Schotta G, Anders HJ, Schulz C, Schraml BU: The kidney contains ontogenetically distinct dendritic cell and macrophage subtypes throughout development that differ in their inflammatory properties. J Am Soc Nephrol 31: 257–278, 2020. 10.1681/ASN.2019040419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tabula Muris Consortium : A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583: 590–595, 2020. 10.1038/s41586-020-2496-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clatworthy MR: How to find a resident kidney macrophage: The single-cell sequencing solution. J Am Soc Nephrol 30: 715–716, 2019. 10.1681/ASN.2019030245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zimmerman KA, Bentley MR, Lever JM, Li Z, Crossman DK, Song CJ, Liu S, Crowley MR, George JF, Mrug M, Yoder BK: Single-cell RNA sequencing identifies candidate renal resident macrophage gene expression signatures across species. J Am Soc Nephrol 30: 767–781, 2019. 10.1681/ASN.2018090931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu H, Malone AF, Donnelly EL, Kirita Y, Uchimura K, Ramakrishnan SM, Gaut JP, Humphreys BD: Single-cell transcriptomics of a human kidney allograft biopsy specimen defines a diverse inflammatory response. J Am Soc Nephrol 29: 2069–2080, 2018. 10.1681/ASN.2018020125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu X, Li N, Shushakova N, Schmitt R, Menne J, Susnik N, Meier M, Leitges M, Haller H, Gueler F, Rong S: C57BL/6 and 129/Sv mice: Genetic difference to renal ischemia-reperfusion. J Nephrol 25: 738–743, 2012. 10.5301/jn.5000053 [DOI] [PubMed] [Google Scholar]
- 34.O’Sullivan J, Finnie SL, Teenan O, Cairns C, Boyd A, Bailey MA, Thomson A, Hughes J, Bénézech C, Conway BR, Denby L: Refining the mouse subtotal nephrectomy in male 129S2/SV mice for consistent modeling of progressive kidney disease with renal inflammation and cardiac dysfunction. Front Physiol 10: 1365, 2019. 10.3389/fphys.2019.01365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.do Valle Duraes F, Lafont A, Beibel M, Martin K, Darribat K, Cuttat R, Waldt A, Naumann U, Wieczorek G, Gaulis S, Pfister S, Mertz KD, Jianping L, Roma G, Warncke M: Immune cell landscaping reveals a protective role for regulatory T cells during kidney injury and fibrosis. JCI Insight 5: e130651, 2020. 10.1172/jci.insight.130651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirita Y, Wu H, Uchimura K, Wilson PC, Humphreys BD: Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. Proc Natl Acad Sci U S A 117: 15874–15883, 2020. 10.1073/pnas.2005477117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramachandran P, Pellicoro A, Vernon MA, Boulter L, Aucott RL, Ali A, Hartland SN, Snowdon VK, Cappon A, Gordon-Walker TT, Williams MJ, Dunbar DR, Manning JR, van Rooijen N, Fallowfield JA, Forbes SJ, Iredale JP: Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proc Natl Acad Sci U S A 109: E3186–E3195, 2012. 10.1073/pnas.1119964109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janosevic D, Myslinski J, McCarthy TW, Zollman A, Syed F, Xuei X, Gao H, Liu YL, Collins KS, Cheng YH, Winfree S, El-Achkar TM, Maier B, Melo Ferreira R, Eadon MT, Hato T, Dagher PC: The orchestrated cellular and molecular responses of the kidney to endotoxin define a precise sepsis timeline. eLife 10: e62270, 2021. 10.7554/eLife.62270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dhillon P, Park J, Hurtado Del Pozo C, Li L, Doke T, Huang S, Zhao J, Kang HM, Shrestra R, Balzer MS, Chatterjee S, Prado P, Han SY, Liu H, Sheng X, Dierickx P, Batmanov K, Romero JP, Prósper F, Li M, Pei L, Kim J, Montserrat N, Suszták K: The nuclear receptor ESRRA protects from kidney disease by coupling metabolism and differentiation. Cell Metab 33: 379–394.e8, 2021. 10.1016/j.cmet.2020.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson PC, Wu H, Kirita Y, Uchimura K, Ledru N, Rennke HG, Welling PA, Waikar SS, Humphreys BD: The single-cell transcriptomic landscape of early human diabetic nephropathy. Proc Natl Acad Sci U S A 116: 19619–19625, 2019. 10.1073/pnas.1908706116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calle P, Hotter G: Macrophage phenotype and fibrosis in diabetic nephropathy. Int J Mol Sci 21: 2806, 2020. 10.3390/ijms21082806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuppe C, Ibrahim MM, Kranz J, Zhang X, Ziegler S, Perales-Patón J, Jansen J, Reimer KC, Smith JR, Dobie R, Wilson-Kanamori JR, Halder M, Xu Y, Kabgani N, Kaesler N, Klaus M, Gernhold L, Puelles VG, Huber TB, Boor P, Menzel S, Hoogenboezem RM, Bindels EMJ, Steffens J, Floege J, Schneider RK, Saez-Rodriguez J, Henderson NC, Kramann R: Decoding myofibroblast origins in human kidney fibrosis. Nature 589: 281–286, 2021. 10.1038/s41586-020-2941-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arazi A, Rao DA, Berthier CC, Davidson A, Liu Y, Hoover PJ, Chicoine A, Eisenhaure TM, Jonsson AH, Li S, Lieb DJ, Zhang F, Slowikowski K, Browne EP, Noma A, Sutherby D, Steelman S, Smilek DE, Tosta P, Apruzzese W, Massarotti E, Dall’Era M, Park M, Kamen DL, Furie RA, Payan-Schober F, Pendergraft WF 3rd, McInnis EA, Buyon JP, Petri MA, Putterman C, Kalunian KC, Woodle ES, Lederer JA, Hildeman DA, Nusbaum C, Raychaudhuri S, Kretzler M, Anolik JH, Brenner MB, Wofsy D, Hacohen N, Diamond B; Accelerating Medicines Partnership in SLE network : The immune cell landscape in kidneys of patients with lupus nephritis [published correction appears in Nat Immunol 20: 1404, 2019]. Nat Immunol 20: 902–914, 2019. 10.1038/s41590-019-0398-x [DOI] [PMC free article] [PubMed] [Google Scholar]