Abstract
Brain morphology has been shown to be highly heritable, yet only a small portion of the heritability is explained by the genetic variants discovered so far. Here we extended the Multivariate Omnibus Statistical Test (MOSTest) and applied it to genome-wide association studies (GWAS) of vertex-wise structural magnetic resonance imaging (MRI) cortical measures from N=35,657 participants in the UK Biobank. We identified 695 loci for cortical surface area and 539 for cortical thickness, in total 780 unique genetic loci associated with cortical morphology robustly replicated in 8,060 children of mixed ethnicity from the Adolescent Brain Cognitive Development (ABCD) Study®. This reflects more than 8-fold increase in genetic discovery at no cost to generalizability compared to the commonly used univariate GWAS methods applied to region of interest (ROI) data. Functional follow up including gene-based analyses implicated 10% of all protein-coding genes and pointed towards pathways involved in neurogenesis and cell differentiation. Power analysis indicated that applying the MOSTest to vertex-wise structural MRI data triples the effective sample size compared to conventional univariate GWAS approaches. The large boost in power obtained with the vertex-wise MOSTest together with pronounced replication rates and highlighted biologically meaningful pathways underscores the advantage of multivariate approaches in the context of highly distributed polygenic architecture of the human brain.
Keywords: multivariate vertex-wise analysis, cortical surface area, cortical thickness, genome-wide association study, distributed polygenic architecture
Introduction
Variability in brain morphology is highly heritable, with twin studies estimating heritability for global measures at 89% for total surface area and 81% for mean cortical thickness1 and regional measures (adjusting for whole brain measures) at up to 46% for cortical area and 57% for thickness2. GWAS is a powerful tool for identifying genetic variants that shape the human cortex, but the full breadth of reported heritability estimates has yet to be uncovered. The most recent large-scale GWAS of brain MRI data (N=51,665) from the ENIGMA consortium identified 187 and 50 loci associated with global and regional cortical surface area and thickness, respectively3. The relatively low yield despite high heritabilities of brain morphology is likely due to high polygenicity and small effect size (discoverability) per locus4.
Both imaging genetics4 and gene expression studies5 suggest that genetic effects are distributed across cortical regions, such that variants influencing one cortical region are also likely to affect other cortical regions. Multivariate statistical methods are naturally tailored to model distributed and pleiotropic genetic effects. We recently developed a Multivariate Omnibus Statistical Test (MOSTest)6 that aggregates effects across spatially distributed phenotypes, such as cortical thickness, boosting our ability to detect variant-phenotype associations. We showed that applying MOSTest to cortical morphology region of interest (ROI) measures in the UK Biobank substantially increased loci discovery6 compared to the commonly applied mass univariate approach used by the ENIGMA consortium3, here referred to as the min-P approach. For each genetic variant tested for association with multiple phenotypes, min-P considers only the most significant p-value and corrects it for the effective number of phenotypes analyzed, thus failing to exploit distributed polygenic architecture across brain regions. In contrast, MOSTest leverages the distributed nature of genetic influences across brain regions and allows detection of genetic variants with weak effects in multiple brain regions. We have shown that the discoverability of GWAS variants underlying regional cortical area and thickness depends on the specific parcellation of cortical regions used, and that parcellations based on genetic correlations from twin studies perform better than genetically un-informed schemes4. Here we further extend MOSTest method introducing regularization of covariance matrix which provides substantial increase of discovery yield as compared to unregularized version we introduced in the previous study6. Updated method is applied to vertex-wise measures of cortical morphology showing that the combined genetic yield (number of loci discovered) for cortical area and thickness can be boosted when using MOSTest (yielding a 4.2-fold and 4.1-fold increase relative to min-P for cortical area and thickness, respectively), and boosted further when moving from a region-based approach to a more fine-grained vertex-wise approach (additional 1.9-fold and 3.0-fold increase for cortical area and thickness, respectively) without sacrificing replicability of the findings in the independent sample. Uncovering the detailed genetic architecture of cortical area and thickness will provide insight into the underlying neurobiology of the human brain, and give a better understanding of brain-related human traits, such as cognition7, as well as neurological8 and psychiatric diseases9.
Materials and Methods
Samples
For the primary analysis genotypes, T1 MRI scans, demographic and clinical data were obtained from the UK Biobank under accession number 27412. We selected White British individuals (as derived from both self-declared ethnicity and principal component analysis10) who had undergone the neuroimaging protocol and had passed genetic quality control procedures described below. The resulting sample contained 35,657 individuals with a mean age of 64.4 years (standard deviation 7.5 years), 51.7% female.
For the replication analysis we used data from the Adolescent Brain Cognitive Development (ABCD) study, with complete genetic data and baseline T1 MRI scans from data release 3.0 (NDA DOI:10.151.54/1519007) that passed the ABCD quality control procedures (N=8,060). These children had a mean age of 9.9 years (standard deviation 0.6 years), 46.9% female.
Data processing
Both for the primary and replication analyses T1-weighted structural MRI scans were processed with the FreeSurfer v5.3 standard “recon-all” processing pipeline11 to generate 1284 non-smoothed vertex-wise measures (ico3 downsampling with the medial wall removed) summarizing cortical surface area and thickness. For the primary analysis we also generated 68 ROI cortical surface area and thickness measures (based on the Desikan-Killiany parcellation). All measures were pre-residualized for age, sex, scanner site, a proxy of surface reconstruction quality (FreeSurfer’s Euler number12), the first twenty genetic principal components, and a global measure specific to each set of variables: total cortical surface area and mean cortical thickness for the regional area and thickness measurements correspondingly. Subsequently, a rank-based inverse normal transformation was applied to the residualized measures.
For discovery we used UK Biobank v3 imputed data that have undergone extensive quality control procedures as described by UK Biobank genetics team10 . In addition, we filtered out individuals with genotype missing rate >10%, variants with genotype missing rate >5%, and variants failing Hardy-Weinberg equilibrium at p=1E-9. We further removed variants with minor allele frequency below 0.5%, and imputation info score below 0.5, leaving 9 million variants.
For the replication we used ABCD genetic data that were part of data release 3.0. The data were genotyped on 646,247 genetic variants using the Affymetrix smokescreen array13. Successful genotype calls were determined based on the recommendation of Affymetrix Axiom Analysis Suite v5.0, with at least 98% call rates. Further pre-imputation quality controls included inbreeding check, sex concordance check, and cohort level missingness check. Imputation was performed using the Michigan Imputation Server14 with hrc.r1.1.2016 reference panel, Eagle v2.315 phasing and multiethnic imputation process. Best guess conversion at a threshold of 0.9 was used to convert dosage files to plink files using PLINK16. Post-imputation QC criteria were an imputation quality score greater than 0.9 and a Hardy-Weinberg threshold of 1E-6. This QC filtering was performed using PLINK16 and resulted in 13 million variants and 8060 individuals. Genetic ancestry was estimated using fastStructure17 with four ancestry factors, an individual was considered of European ancestry if its estimated posterior probability of being European was larger than 0.8. This resulted in 5060 samples classified as Europeans. There is no overlap between discovery and replication samples.
Variants were tested for association with cortical surface area and cortical thickness at each vertex and each ROI separately using the standard univariate GWAS procedure. Resulting univariate p-values and effect sizes were further combined in the MOSTest and min-P analyses to identify area- and thickness-associated loci. Replication rates for MOSTest and min-P were assessed based on independent MOSTest and min-P runs in the ABCD dataset.
MOSTest analysis
Consider N variants and M (pre-residualized) phenotypes. Let zij be a z-score from the univariate association test between ith variant and jth (residualized) phenotype and be the row vector of z-scores of the ith variant across M phenotypes. Let Z = {zij} be the N × M matrix of z-scores with variants in rows and phenotypes in columns. For each variant consider a random permutation of its genotypes and let be the matrix of z-scores from the univariate association testing between variants with permuted genotypes and phenotypes. A given number of random permutations of genotypes are done for each variant and the resulting permuted genotype vectors are tested for association with all phenotypes, therefore preserving correlation structure between phenotypes. In our discovery analysis we perform 24 random permutations of genotypes (providing ~200M z-scores to estimate distribution under null) to ensure that genome-wide significance level (5E-8) is covered with non-parametric distribution.
Let be the M × M covariance matrix of , and is its singular valued decomposition (U and V – orthogonal matrixes, S– diagonal matrix with singular values of on the diagonal). Since is symmetric, U = V, and singular valued decomposition of can be written as . Consider the regularized version of the covariance matrix , where Sr is obtained from S by keeping r largest singular values and replacing the remaining with rth largest. The MOSTest statistics for ith variant (scalar) is then estimated as , where regularization parameter r is selected separately for cortical area and thickness to maximize the yield of genome-wide significant loci. In this study we observed the largest yield for cortical surface area with r=10; the optimal choice for cortical thickness was r=20 (Figure S4). The distribution of the test statistics under null () is approximated from the observed distribution of the test statistics with permuted genotypes, using the empirical distribution in the 99.99 percentile and Gamma distribution in the upper tail, where shape and scale parameters of Gamma distribution are fitted to the observed data. The p-value of the MOSTest test statistic for the ith variant is then obtained as .
Of note, compared to the original version of the MOSTest6, regularization of the covariance matrix introduced in this study entails technical updates required for correct calibration of test statistics distribution under null. For example, analytical approximation of MOSTest test statistics distribution under null with Gamma distribution applied previously6 become invalid when covariance matrix is regularized. To handle it correctly here we use piecewise distribution with empirical distribution in the 99.99 percentile and Gamma distribution in the upper tail as described above.
While in this study the regularization parameter (r) is selected to maximize discovery yield, alternative criteria might be used. For example, r can be selected to maximize replication rate using nested cross-validation procedure. However, we expect this to have only marginal effect on the results since both discovery yield (Figure S4) and replication rate (Figure S5) remain similar in the broad range of regularization parameters.
min-P analysis
Similar to the MOSTest analysis, consider N variants and M preresidualized phenotypes. Let zij be a z-score from the univariate association test between ith variant and jth (residualized) phenotype and be the row vector of z-scores of the ith variant across M phenotypes. The min-P statistics for the ith variant is then estimated as , where Φ is a cumulative distribution function of the standard normal distribution. The distribution of the min-P test statistics under null () is approximated from the observed distribution of the test statistics with permuted genotypes, using the empirical distribution in the 99.99th percentile and Beta distribution in the upper tail, where shape parameters of Beta distribution (α and β) are fitted to the observed data. The p-value of the min-P test statistic for the ith variant is then obtained as . It is worth noting that the permutation-based method used here for multiple testing correction of the min-P results essentially represents an exact version of commonly applied approach using matrix spectral decomposition18; 19. The latter is not applicable to the MOSTest. Therefore, to provide more direct comparison of MOSTest with min-P, we use permutation-based approach for both methods.
Locus definition
Genetic loci were defined based on association summary statistics produced with MOSTest and min-P following the protocol implemented in FUMA20 with default parameters. The protocol can be summarized as the following:
Independent significant genetic variants are identified as variants with p-value<5E-8 and linkage disequilibrium (LD) r2<0.6 with each other.
A subset of these independent significant variants with LD r2<0.1 are then selected as lead variants.
For each independent significant variant all candidate variants are identified as variants with LD r2≥0.6 with the independent significant variant.
For a given lead variant the borders of the genomic locus are defined as min/max positional coordinates over all corresponding candidate variants.
Loci are then merged if they are separated by less than 250kb.
Alternatively, to facilitate comparison with the current largest brain morphology GWAS3, we also counted genetic loci applying locus definition similar to that used by ENIGMA. Briefly, the association summary statistics produced with either MOSTest or min-P were clumped with PLINK16 using p-value threshold of 5E-8 (--clump-p1) and linkage disequilibrium cutoffs of 1 Mb (--clump-kb) and r2 < 0.2 (--clump-r2). Obtained clumps of variants were considered as independent genome-wide significant genetic loci.
MiXeR analysis
MOSTest and min-P p-values were analyzed with the MiXeR tool21 to estimate the proportion of additive genetic variance explained by genome-wide significant SNPs as a function of sample size. Right censoring (MiXeR option: --z1max 5.45) was applied to mitigate extreme effects which may lead to biased estimates.
Gene-level analysis
We carried out MAGMA-based gene analyses using default settings, which entail the application of a SNP-wide mean model to GWAS summary statistics, with the use of the 1000 Genomes Phase 3 EUR reference panel. Gene-set analyses were done in a similar manner, restricting the sets under investigation to those that are part of the Gene Ontology biological processes subset (N=7343), as listed in the Molecular Signatures Database (MsigdB) v7.0. In addition, lead SNPs identified in the vertex-wise MOSTest analysis were parsed with gene-set enrichment analysis as implemented in FUMA GENE2FUNC20. In contrast to MAGMA analysis where genes are prioritized based on proximity, in this analysis genes were selected combining positional and eQTL (based on GTEx v8 data) mapping.
Replication criterion
Lead variants identified in discovery vertex-wise MOSTest and min-P analyses were tested for replication in the ABCD sample. Only variants which present in both discovery and replication data were used, resulting in 633 variants tested for cortical surface area and 487 variants for cortical thickness. The variant was considered as replicated if its nominal one-tailed p-value for association in replication cohort was < 0.05.
Results
Genetic loci discovery
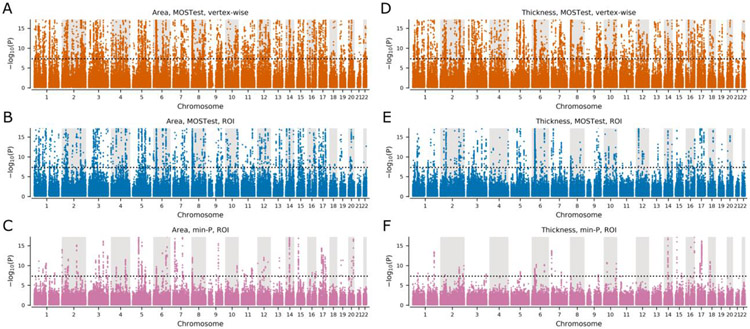
Using the vertex-wise MOSTest6, we performed a multivariate GWAS of cortical morphology, such that the significance of each locus was estimated after aggregating its effects across all vertices (1284 data points each for thickness and area). This was conducted separately for cortical surface area and thickness in 35,657 individuals from UK Biobank. Measurements from left and right hemispheres were included separately (not averaged). We identified 695 and 539 loci, respectively, equating to 780 unique loci associated with cortical morphology. Prior to performing the vertex-wise MOSTest analysis, individual cortical area and thickness measures were residualized for age, sex, scanner site, Euler number (proxy of surface reconstruction quality), the first twenty genetic principal components, and a participant-specific global measure (either total area or average thickness). Measurements from left and right hemispheres were not merged. For comparison, we repeated this procedure aggregating over 68 ROIs from the Desikan-Killiany parcellation. This resulted in the discovery of 370 loci for cortical surface area and 181 loci for cortical thickness, such that the vertex-wise MOSTest analysis provided a 1.9-fold and 3.0-fold increase in yield over the region-based MOSTest analysis, respectively. Applying the min-P approach to Desikan-Killiany ROIs resulted in further reduction in the number of loci discovered (88 for cortical surface area; 44 for cortical thickness). This represents a 4.2-fold and 4.1-fold decrease compared to the MOSTest ROI-based analysis, and a 7.9-fold and 12.3-fold decrease compared to vertex-wise MOSTest analysis, respectively. Manhattan plots are presented in Figure 1, with corresponding Q-Q plots in Figure S1.
Figure 1. Manhattan plots for cortical surface area and cortical thickness.
(A) Area, MOSTest, vertex-wise: N=695 loci. (B) Area, MOSTest, ROI: N=370 loci. (C) Area, min-P, ROI: N=88 loci. (D) Thickness, MOSTest, vertex-wise: N=539 loci. (E) Thickness, MOSTest, ROI: N=181 loci. (F) Thickness, min-P, ROI: N=44 loci. Black dotted horizontal lines show genome-wide significance threshold (P=5E-8). Loci; independent genome-wide significant (P<5E-8). Y-axes are truncated at −log10(P)=17.2 to highlight the region around genome-wide significance threshold. ROI = region of interest.
Additionally, we applied min-P approach to vertex-wise data resulting 93 loci for cortical surface area and 63 loci for cortical thickness. Numbers of loci discovered with different approaches are shown in Table S1. Specific loci discovered in each analysis are listed in Tables S2 - S9.To compare the vertex-wise MOSTest results with the most recent ENIGMA GWAS3, we also applied the ENIGMA-based definition of genetic locus. This resulted in 1598 and 1054 unique loci for cortical area and thickness respectively, and a total of 1735 unique loci for cortical morphology identified in the vertex-wise MOSTest analysis (Tables S10 and S11).
Replication analysis
Generalizability of vertex-wise MOSTest and min-P findings was assessed in replication analysis using data on 8,060 participants from the Adolescent Brain Cognitive Development (ABCD) Study as described in the Methods. This analysis revealed comparable replication rates for loci discovered with vertex-wise MOSTest and min-P both for cortical surface area (43% and 55% replicated for vertex-wise MOSTest and min-P, respectively) and cortical thickness (vertex-wise MOSTest 30%, min-P 37%). Therefore, absolute numbers of replicated loci both for cortical surface area and thickness were substantially higher for vertex-wise MOSTest than for min-P (Tables S3, S4, S6 and S7).
Substantial differences in age and ancestry between discovery and replication cohorts might reduce replication rates because genetic association analyses likely capture some ancestry- and age-specific factors. For example, it’s unlikely to capture genetic variants affecting degree of age-related cortical atrophy in the ABCD cohort. These factors might have different effect for MOSTest and min-P and thus distort comparison of replication rates. To alleviate differences between discovery and replication analyses and make comparison of replication performance more unbiased, we performed replication analysis restricting ABCD cohort to individuals of European ethnicity (N=5,060). The obtained replication rates for cortical area were 50% and 54% for MOSTest and min-P, respectively, for cortical thickness replication rates were 51% and 34% for MOSTest and min-P, respectively. Additionally, we estimated replication rates for a given number of top lead variants (N=25, 50, 80) identified in discovery phase. These results for both mixed- and European-based replication analyses are presented in Table S13.
Power analysis
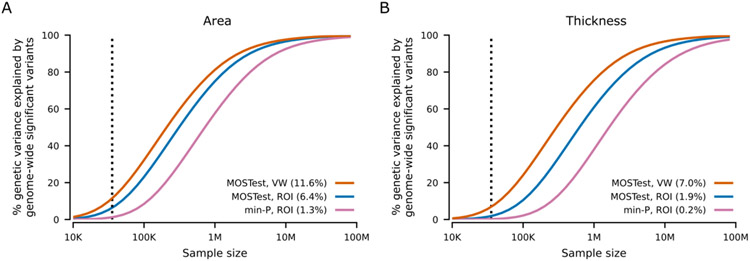
To estimate the proportion of additive genetic variance explained by genome-wide significant SNPs identified by either vertex-wise MOSTest or min-P as a function of sample size, we used the MiXeR tool21 (Figure 2). The horizontal shift of the curve indicates that the effective sample size of vertex-wise MOSTest is around 3.0-fold that of min-P. We estimate that with the current UK Biobank sample (N=35,657), 11.6% and 7.0% of the additive genetic variance in cortical surface area and thickness, respectively, can be explained by genome-wide significant loci from the vertex-wise MOSTest analysis. (Figure 2). In contrast, the min-P approach identifies 1.3% and 0.2% of the explained additive genetic variance for area and thickness, respectively (Figure 2). The power-analysis indicates that 32.2% and 24.0% of the additive genetic variance in cortical surface area and thickness, respectively, will be discovered in the full UK Biobank sample of N=100,000 using the MOSTest vertex-wise approach (Figure 2). Further, the proportion of explained variance with the min-P approach in the full UK Biobank sample is estimated to be lower than the yield of vertex-wise MOSTest in the present sample size (Figure 2).
Figure 2. Estimated percent of additive genetic variance explained by genome-wide significant SNPs as a function of sample size.
Percentages of genetic variance explained by identified SNPs (p<5E-8) from multivariate GWAS (MOSTest VW) of area (A) and thickness (B) with current sample size (N=35,657, vertical dotted line) are shown in parentheses, with MOSTest ROI and min-P ROI for comparison. VW = vertex-wise. ROI = region of interest.
Gene-level analysis
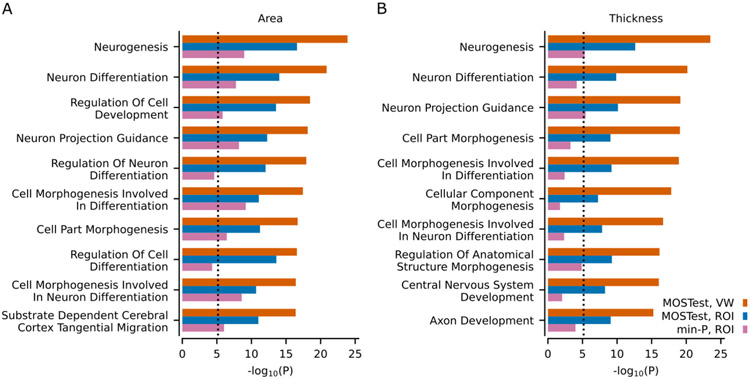
Through gene-level analyses of the vertex-wise MOSTest GWAS using MAGMA22, we found that 1647 and 1412 genes, out of a total of 19036 protein-coding genes, were significantly associated with area and thickness, respectively (Table S12). We also performed competitive gene-set analyses restricted to the Gene Ontology biological processes category (containing 7343 pathways). This resulted in 204 and 184 significant (p<0.05/7343) gene sets associated with area and thickness, respectively. The most significantly associated pathways were related to neuronal development and cell differentiation, with the top 10 shown in Figure 3. Remarkably similar pathways were highlighted in gene-set enrichment analyses using FUMA GENE2FUNC. Both for cortical area and cortical thickness, out of top 10 gene sets identified in MAGMA analysis (Figure 3) 5 gene sets are also within top 10 gene sets identified in corresponding FUMA GENE2FUNC analysis (Table S14, Table S15).
Figure 3. Gene-set analyses with MAGMA.
Results from the gene-set analysis based on multivariate GWAS on area (A) and thickness (B). Ten most significant Gene Ontology sets (N=7,343) in the vertex-wise MOSTest analysis are listed on the y-axis, in comparison with MOSTest ROI and min-P ROI. Corresponding uncorrected −log10(p-values) are shown on the x-axis. P-values were obtained using MAGMA analysis as implemented in FUMA. Vertical dotted line shows Bonferroni correction threshold (p=0.05/7343). VW = vertex-wise. ROI = region of interest.
For comparison, we also performed the same analyses on the ROI-based MOSTest and min-P GWAS summary statistics, resulting in 198 area and 66 thickness gene sets for ROI-based MOSTest and 60 area and 4 thickness gene sets for min-P. As shown in Figures S2 and S3, the vertex-wise MOSTest approach led to much greater significance for nearly all pathways identified. Interestingly, the most significant pathways identified by vertex-wise MOSTest are tightly connected with critical neurobiological processes implicated in brain development while top findings in the min-P analysis are less specific. The distributed effects of identified variants across different brain regions are also illustrated by brain maps, highlighting the mixture of effects across the cortex (Figure 4).
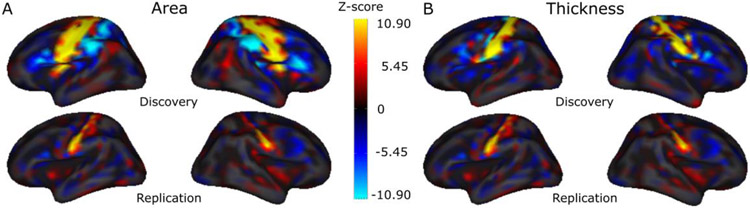
Figure 4. Lateral view of the cortex, depicting the color-coded vertex-wise z-values for the top genetic loci identified in the discovery vertex-wise MOSTest analysis.
(A): top lead variant (rs34680120, chr15:39664000, effect direction of C allele is shown, frequency of C allele = 0.94) associated with cortical surface area in discovery (top) and replication (bottom) samples. (B): top lead variant (rs8033007, chr15:39619661, effect direction of G allele is shown, frequency of G allele = 0.91) associated with cortical thickness in discovery (top) and replication (bottom) samples.
The strongest association signals identified in our discovery MOSTest analyses both for cortical surface area (lead variant: rs34680120, p<1E-300) and cortical thickness (lead variant: rs8033007, p<1E-300) are located on chromosome 15 at 15q14. Univariate vertex-wise association signals of the lead variants are presented in Figure 4. These lead variants are in strong linkage disequilibrium with each other (r2=0.33) and with rs1080066 (r2=0.50 and r2=0.30 for rs34680120 and rs8033007 respectively), which was reported as the strongest association with cortical surface area in large recent GWAS on brain morphology by ENIGMA consortium3. Regional association patterns of our strongest genetic signals both for cortical area and thickness (Figure 4) highlight cortex region around the central sulcus. The region encompasses areas responsible for all motor and sensory functions and has rapidly evolved in course of primate evolution, presumably reflecting the increasing importance of somatosensory and motor integration of hand functions23. There is also evidence that this region is strongly involved in higher cognitive functions24 and may contribute to different psychiatric disorders25; 26. Taking together this may suggest an active ongoing fine-tuning of this brain region in the human species which in turn is reflected in observed strong genetic associations altering brain morphology of the region.
Discussion
Applying the vertex-wise MOSTest method, we identified 695 loci for cortical surface area and 539 for cortical thickness, in total 780 unique genetic loci associated with cortical morphology, greatly replicated in independent sample with substantially different ethnical and age composition. This reflects an approximate 8-fold increase in discovery with no penalty on replication rate compared to the commonly applied univariate GWAS methods. Our study highlights the greatly improved yield obtained with the vertex-wise multivariate approach compared to conventional region-based univariate GWAS approach, which stems from highly distributed nature of brain morphology phenotypes, representing continuous maps per individual. The present results support the hypothesis that the genetic determinants of variability in brain morphology are extensively shared across multiple regions6. Our findings further underscore the complex molecular mechanisms shaping the human brain, which we show are largely related to neurodevelopmental processes.
Our gene-level analyses indicated that, with the current sample size, 10% of all protein-coding genes were significantly associated with brain morphology (either cortical area or thickness). Gene-set analyses for both area and thickness confirmed involvement of pathways recently reported by ENIGMA3, but with greater statistical significance. We additionally found strong evidence for the involvement of several genetic pathways regulating neuronal development and differentiation that were not identified by the min-P approach, implicating key biological processes regulating human surface area expansion and increases in thickness. This also corroborates the strong statistical signals and suggests that we are capturing true biological mechanisms that were missed by previous methodologies. These novel findings of neurobiological underpinnings associated with brain morphology provide a framework for follow-up experimental studies to identify the complex polygenic mechanisms involved in human brain development27. Further, the findings implicating neuronal development and cell differentiation can facilitate experimental studies to gain better insight into the pathobiological mechanisms of brain-related diseases including psychiatric disorders28, where we need to understand the role of polygenic mechanisms29.
Twin studies have suggested the largely independent nature of cortical surface area and thickness1. The genetic correlation between them estimated using linkage disequilibrium score regression (LDSR) is rg=−0.32 (p=6.5E-12)3. Here we identify the specific loci involved and show that these cortical phenotypes share a large proportion of genomic loci. Out of a total of 695 loci for cortical area and 539 loci for cortical thickness, 454 loci (58.2% of the total number of unique loci) were overlapping. These findings illustrate how measures of genetic correlation fail to fully capture the extent to which the genetic influences of two phenotypes are interrelated. LDSR and twin analyses depend on the consistency of effect directions across phenotypes. In contrast, the analysis performed here consider non-null loci as overlapping if they are both significant and in linkage disequilibrium, regardless of effect directions. Overlapping genetic architecture across brain regions despite the absence of strong genetic correlations are therefore plausible due to common molecular toolkits involved in neurodevelopment across brain regions30. This is in line with Allen Brain Atlas maps of the adult human brain31, showing regions with high similarity in gene expression between cortical structures consistent with the notion that the basic architecture across the entire cortex is similar or “canonical”32. This may also explain the shared genetic architecture observed for many brain-related traits and disorders33-35. Accounting for the distributed signal across the cortex (Figure 4) in a multivariate framework allowed us to boost power for discovery compared to traditional univariate approaches, such as min-P.
Compared to the current largest brain morphology GWAS (N=51,665)3, analyzing parcellation-free, vertex-wise data with MOSTest increased the yield of significant loci 4-fold for cortical surface area and 11-fold for cortical thickness, despite the lower sample size in our study (N=35K). Of note, while being generally consistent, our protocol differs in a few aspects from the previous GWAS3, where global measures were included in the principal analysis, data for cortical regions were averaged across right and left hemispheres, and the definition of genetic loci was less conservative. Using the Desikan-Killiany parcellation approximately 2.0 times more variants were identified for cortical surface area than for cortical thickness both with the min-P and the MOSTest (Table S1). In contrast, there were 1.3 times more loci for area compared to thickness when using the MOSTest for parcellation-free vertex-wise data. (Table S1). The observed difference in loci yield may be due to differing degrees of mismatch between parcellation schemes and actual architecture of the phenotypes. This seems to be particularly relevant for thickness, where variant effects obtained from an ROI parcellation scheme may be underestimated compared to the vertex-wise approach. This result may explain why parcellation schemes better reflecting the genetic architecture of the cortex improve detectability in imaging genetics studies4. It is also worth noting that smoothing of the vertex-wise data results in substantial decrease of loci yield (Figure S4). This might indicate that there is valid information in the fine spatial structure, which is getting removed when smoothing is applied. It’s therefore tempting to apply the method to the data with higher resolution (up to voxel-wise). The latter might become feasible with more advanced implementation of the method, which we currently work on.
The boost in statistical power using the multivariate vertex-wise approach is equivalent to a more than three-fold increase in effective sample size for both area and thickness (Figure 2). Our analysis suggests that the substantial gain in power provided by vertex-wise MOSTest is projected to explain approximately 32.2% and 24.0% of the additive genetic variance for cortical surface area and thickness, respectively, upon completion of UK Biobank’s target neuroimaging sample (N=100,000)36 (Figure 2). It is possible that multivariate approaches will also boost discovery of genetic associations with other human phenotypes that exhibit shared signal between traits.
To conclude, by deploying new vertex-wise MOSTest approach we have identified 780 unique loci associated with human brain morphology, highlighting its distributed polygenic architecture, and providing the foundation for functional follow-up experiments. The code implementing vertex-vise MOSTest approach is made freely available on GitHub. While our primary analysis is focused on UK Biobank, pronounced replication rates in demographically distinct ABCD cohort suggest high potential for generalizability of presented statistical framework. Flexibility of this approach allows its seamless incorporation into large-scale meta-analyses like ENIGMA37, offering unique opportunities for major advances in our understanding of the genetic determinants of brain morphology.
Supplementary Material
Highlights.
Genetic variants affecting one cortical region often affect other cortical regions.
Standard mass-univariate methods ignore the distributed nature of genetic effects.
Multivariate MOSTest method exploits distributed effects boosting genetic discovery.
Considering fine-grained vertex-wise measures improves genetic discovery further.
Obtained increase in discovery does not come at a cost of poorer generalizability.
Acknowledgements
We were funded by the Research Council of Norway (276082, 213837, 223273, 204966/F20, 229129, 249795/F20, 225989, 248778, 249795), the South-Eastern Norway Regional Health Authority (2013-123, 2014-097, 2015-073, 2016-064, 2017-004), Stiftelsen Kristian Gerhard Jebsen (SKGJ-Med-008), The European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (ERC Starting Grant, Grant Agreement No. 802998) and National Institutes of Health (R01MH100351, R01GM104400, NIDA/NCI: U24DA041123). This work was partly performed on the TSD (Tjeneste for Sensitive Data) facilities, owned by the University of Oslo, operated and developed by the TSD service group at the University of Oslo, IT-Department (USIT). (tsd-drift@usit.uio.no). Computations were also performed on resources provided by UNINETT Sigma2—the National Infrastructure for High Performance Computing and Data Storage in Norway. This work used the Extreme Science and Engineering Discovery Environment (XSEDE) including COMET and OASYS resources at the UCSD through allocation TG-IBN200001. This research has been conducted using data from UK Biobank, a major biomedical database.
ABCD Acknowledgement
Data used in the preparation of this article were obtained from the Adolescent Brain Cognitive DevelopmentSM Study (ABCD Study®) (https://abcdstudy.org), held in the NIMH Data Archive (NDA). This is a multisite, longitudinal study designed to recruit more than 10,000 children age 9-10 and follow them over 10 years into early adulthood. The ABCD Study is supported by the National Institutes of Health and additional federal partners under award numbers: U01DA041022, U01DA041028, U01DA041048, U01DA041089, U01DA041106, U01DA041117, U01DA041120, U01DA041134, U01DA041148, U01DA041156, U01DA041174, U24DA041123, and U24DA041147
A full list of supporters is available at https://abcdstudy.org/federal-partners/. A listing of participating sites and a complete listing of the study investigators can be found at https://abcdstudy.org/principal-investigators.html. ABCD Study consortium investigators designed and implemented the study and/or provided data but did not necessarily participate in analysis or writing of this report. This manuscript reflects the views of the authors and may not reflect the opinions or views of the NIH or ABCD Study consortium investigators.
The ABCD data repository grows and changes over time. The ABCD data used in this came from [NIMH Data Archive Digital Object Identifier (10.151.54/1519007)].
Footnotes
Declaration of Interests
Dr. Andreassen has received speaker’s honorarium from Lundbeck, and is a consultant to HealthLytix. Dr. Dale is a Founder of and holds equity in CorTechs Labs, Inc, and serves on its Scientific Advisory Board. He is a member of the Scientific Advisory Board of Human Longevity, Inc. and receives funding through research agreements with General Electric Healthcare and Medtronic, Inc. The terms of these arrangements have been reviewed and approved by UCSD in accordance with its conflict of interest policies. The other authors declare no competing interests.
Data and Code Availability
The data incorporated in the primary analysis were gathered from the public UK Biobank resource and will be made publicly available together with the code used to generate the data through the UK Biobank Returns Catalogue (https://biobank.ndph.ox.ac.uk/showcase/docs.cgi?id=1). ABCD study data release 3.0 is available for approved researchers in NIMH Data Archive (NDA DOI:10.151.54/1519007).
MOSTest code is freely available at https://github.com/precimed/mostest (GPLv3 license).
References
- 1.Panizzon MS, Fennema-Notestine C, Eyler LT, Jernigan TL, Prom-Wormley E, Neale M, Jacobson K, Lyons MJ, Grant MD, Franz CE, et al. (2009). Distinct genetic influences on cortical surface area and cortical thickness. Cereb Cortex 19, 2728–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eyler LT, Chen CH, Panizzon MS, Fennema-Notestine C, Neale MC, Jak A, Jernigan TL, Fischl B, Franz CE, Lyons MJ, et al. (2012). A comparison of heritability maps of cortical surface area and thickness and the influence of adjustment for whole brain measures: a magnetic resonance imaging twin study. Twin Res Hum Genet 15, 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grasby KL, Jahanshad N, Painter JN, Colodro-Conde L, Bralten J, Hibar DP, Lind PA, Pizzagalli F, Ching CRK, McMahon MAB, et al. (2020). The genetic architecture of the human cerebral cortex. Science 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Meer D, Frei O, Kaufmann T, Chen CH, Thompson WK, O'Connell KS, Monereo Sanchez J, Linden DEJ, Westlye LT, Dale AM, et al. (2020). Quantifying the Polygenic Architecture of the Human Cerebral Cortex: Extensive Genetic Overlap between Cortical Thickness and Surface Area. Cereb Cortex 30, 5597–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hawrylycz M, Miller JA, Menon V, Feng D, Dolbeare T, Guillozet-Bongaarts AL, Jegga AG, Aronow BJ, Lee CK, Bernard A, et al. (2015). Canonical genetic signatures of the adult human brain. Nat Neurosci 18, 1832–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Meer D, Frei O, Kaufmann T, Shadrin AA, Devor A, Smeland OB, Thompson WK, Fan CC, Holland D, Westlye LT, et al. (2020). Understanding the genetic determinants of the brain with MOSTest. Nat Commun 11, 3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vuoksimaa E, Panizzon MS, Chen CH, Fiecas M, Eyler LT, Fennema-Notestine C, Hagler DJ Jr., Franz CE, Jak AJ, Lyons MJ, et al. (2016). Is bigger always better? The importance of cortical configuration with respect to cognitive ability. Neuroimage 129, 356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Querbes O, Aubry F, Pariente J, Lotterie JA, Demonet JF, Duret V, Puel M, Berry I, Fort JC, Celsis P, et al. (2009). Early diagnosis of Alzheimer's disease using cortical thickness: impact of cognitive reserve. Brain 132, 2036–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rimol LM, Hartberg CB, Nesvag R, Fennema-Notestine C, Hagler DJ Jr., Pung CJ, Jennings RG, Haukvik UK, Lange E, Nakstad PH, et al. (2010). Cortical thickness and subcortical volumes in schizophrenia and bipolar disorder. Biol Psychiatry 68, 41–50. [DOI] [PubMed] [Google Scholar]
- 10.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O'Connell J, et al. (2018). The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, et al. (2006). An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31, 968–980. [DOI] [PubMed] [Google Scholar]
- 12.Rosen AFG, Roalf DR, Ruparel K, Blake J, Seelaus K, Villa LP, Ciric R, Cook PA, Davatzikos C, Elliott MA, et al. (2018). Quantitative assessment of structural image quality. Neuroimage 169, 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baurley JW, Edlund CK, Pardamean CI, Conti DV, and Bergen AW (2016). Smokescreen: a targeted genotyping array for addiction research. BMC Genomics 17, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, et al. (2016). Next-generation genotype imputation service and methods. Nat Genet 48, 1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loh PR, Danecek P, Palamara PF, Fuchsberger C, Y AR, H KF, Schoenherr S, Forer L, McCarthy S, Abecasis GR, et al. (2016). Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet 48, 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, and Lee JJ (2015). Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raj A, Stephens M, and Pritchard JK (2014). fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 197, 573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nyholt DR (2004). A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet 74, 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, and Ji L (2005). Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb) 95, 221–227. [DOI] [PubMed] [Google Scholar]
- 20.Watanabe K, Taskesen E, van Bochoven A, and Posthuma D (2017). Functional mapping and annotation of genetic associations with FUMA. Nat Commun 8, 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frei O, Holland D, Smeland OB, Shadrin AA, Fan CC, Maeland S, O'Connell KS, Wang Y, Djurovic S, Thompson WK, et al. (2019). Bivariate causal mixture model quantifies polygenic overlap between complex traits beyond genetic correlation. Nat Commun 10, 2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Leeuw CA, Mooij JM, Heskes T, and Posthuma D (2015). MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 11, e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hopkins WD, Meguerditchian A, Coulon O, Bogart S, Mangin JF, Sherwood CC, Grabowski MW, Bennett AJ, Pierre PJ, Fears S, et al. (2014). Evolution of the central sulcus morphology in primates. Brain Behav Evol 84, 19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendoza G, and Merchant H (2014). Motor system evolution and the emergence of high cognitive functions. Prog Neurobiol 122, 73–93. [DOI] [PubMed] [Google Scholar]
- 25.Fujiwara H, Hirao K, Namiki C, Yamada M, Shimizu M, Fukuyama H, Hayashi T, and Murai T (2007). Anterior cingulate pathology and social cognition in schizophrenia: a study of gray matter, white matter and sulcal morphometry. Neuroimage 36, 1236–1245. [DOI] [PubMed] [Google Scholar]
- 26.Li S, Wang S, Li X, Li Q, and Li X (2015). Abnormal surface morphology of the central sulcus in children with attention-deficit/hyperactivity disorder. Front Neuroanat 9, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silbereis JC, Pochareddy S, Zhu Y, Li M, and Sestan N (2016). The Cellular and Molecular Landscapes of the Developing Human Central Nervous System. Neuron 89, 248–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sullivan PF, and Geschwind DH (2019). Defining the Genetic, Genomic, Cellular, and Diagnostic Architectures of Psychiatric Disorders. Cell 177, 162–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gandal MJ, and Geschwind DH (2021). Polygenicity in Psychiatry-Like It or Not, We Have to Understand It. Biol Psychiatry 89, 2–4. [DOI] [PubMed] [Google Scholar]
- 30.Stiles J, and Jernigan TL (2010). The basics of brain development. Neuropsychol Rev 20, 327–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, et al. (2012). An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489, 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The Allen Institute. (n.d.). The Genetic Geography of the Brain. In. ( [Google Scholar]
- 33.Watanabe K, Stringer S, Frei O, Umicevic Mirkov M, de Leeuw C, Polderman TJC, van der Sluis S, Andreassen OA, Neale BM, and Posthuma D (2019). A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet 51, 1339–1348. [DOI] [PubMed] [Google Scholar]
- 34.Smeland OB, Frei O, Fan CC, Shadrin A, Dale AM, and Andreassen OA (2019). The emerging pattern of shared polygenic architecture of psychiatric disorders, conceptual and methodological challenges. Psychiatr Genet 29, 152–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cross-Disorder Group of the Psychiatric Genomics Consortium. Electronic address, p.m.h.e., and Cross-Disorder Group of the Psychiatric Genomics, C. (2019). Genomic Relationships, Novel Loci, and Pleiotropic Mechanisms across Eight Psychiatric Disorders. Cell 179, 1469–1482 e1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller KL, Alfaro-Almagro F, Bangerter NK, Thomas DL, Yacoub E, Xu J, Bartsch AJ, Jbabdi S, Sotiropoulos SN, Andersson JL, et al. (2016). Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat Neurosci 19, 1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson PM, Jahanshad N, Ching CRK, Salminen LE, Thomopoulos SI, Bright J, Baune BT, Bertolin S, Bralten J, Bruin WB, et al. (2020). ENIGMA and global neuroscience: A decade of large-scale studies of the brain in health and disease across more than 40 countries. Transl Psychiatry 10, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.