Abstract
Using a serotype-specific monoclonal antibody (MAb) of dengue virus type 1 (DEN-1), 15F3-1, we identified the B-cell epitope of DEN-1 from a random peptide library displayed on phage. Fourteen immunopositive phage clones that bound specifically to MAb 15F3-1 were selected. These phage-borne peptides had a consensus motif of HxYaWb (a = S/T, b = K/H/R) that mimicked the sequence HKYSWK, which corresponded to amino acid residues 111 to 116 of the nonstructural protein 1 (NS1) of DEN-1. Among the four synthetic peptides corresponding to amino acid residues 110 to 117 of the NS1 of DEN-1, -2, -3, and -4, only one peptide, EHKYSWKS (P14M) of DEN-1, was found to bind to 15F3-1 specifically. Furthermore, P14M was shown to inhibit the binding of phage particles to 15F3-1 in a competitive inhibition assay. Histidine111 (His111) was crucial to the binding of P14M to 15F3-1, since its binding activity dramatically reduced when it changed to leucine111 (Leu111). This epitope-based peptide demonstrated its clinical diagnostic potential when it reacted with a high degree of specificity with serum samples obtained from both DEN-1-infected rabbits and patients. Based on these observations, our DEN-1 epitope-based serologic test could be useful in laboratory viral diagnosis and in understanding the pathogenesis of DEN-1.
Dengue virus (DEN) causes serious febrile illness in humans, including dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) (12). It has been estimated that 100 million cases of dengue fever (DF) occur each year, with a 5% annual case fatality rate even where appropriate supportive treatment is given (10). Primary DEN infection with any of the four serotypes (DEN-1, -2, -3, or -4) often results in a painful, debilitating, but usually nonfatal DF and subsequently leads to protection against reinfection by the same serotype. Many severe and fatal DHF and DSS cases have frequently been reported in regions where more than one serotype of DEN is circulating (9, 11). During epidemics of DHF and DSS, rapid diagnosis of the serotype(s) in patients infected with DEN is important, especially for those patients who visit physicians during the late phase of infection or those patients for whom only convalescent-phase serum samples are available. At present it is still not clear whether DHF and DSS are due to a primary or secondary DEN infection or to other immunopathologic mechanisms (9, 11). Therefore, the identification of B-cell epitopes for DEN can provide important information for the development of a safe and effective dengue vaccine and contribute to the understanding of the pathogenesis of DEN and immunological responses in DEN infection.
The B-cell epitopes of DEN-2 had been documented using overlapping synthetic peptides (PEPSCAN) to analyze the antisera (1, 14, 21), and a variety of antigenic domains of DEN-2 had been studied by antigen fragments using recombinant or enzyme cleavage proteins (18, 20, 26). Phage display, a selection technique in which a peptide or protein is expressed as a fusion with a coat protein of bacteriophage, results in a display of the fusion peptide or protein on the surface of the virion. This selection technique has been widely used to map B-cell epitopes, search for disease-specific antigen mimics, and analyze conformational epitopes or mimotopes (6, 7, 8, 19, 22, 31).
Serotype-specific B-cell epitopes of DEN have not been well documented, and to date neither a protein nor a peptide can be used in the differentiation of the four serotypes of DEN infection. Furthermore, DEN-1 was the predominant serotype for the 1987-to-1988 epidemic in Taiwan and is now the serotype most widely distributed throughout the world, including Taiwan. Therefore, we used a phage-displayed peptide library to identify the B-cell epitope for DEN-1 and further used it to diagnose DEN-1-infected patients. Our detection of DEN-1 in 20 out of 21 (95%) confirmed dengue patients but in none of 21 healthy individuals (0%) further supports the suggestion that an epitope-based peptide antigen can be developed as a convenient and efficient serologic test to identify DEN-infected patients.
MATERIALS AND METHODS
Cells and viruses.
The four dengue viruses, DEN-1 (Hawaii), DEN-2 (New Guinea C), DEN-3 (H87), and DEN-4 (H241), were provided by Duane J. Gubler of the Centers for Disease Control and Prevention, Fort Collins, Colo. These viruses were passaged in Aedes albopictus C6/36 cells. The titers for the DEN were measured by plaque assay in BHK-21 cells. The C6/36 cells were grown in RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum (FBS). BHK-21 cells were grown in RPMI 1640 medium containing 5% heat-inactivated FBS.
Antibodies.
The hybridoma cell line for serotype-specific monoclonal antibodies (MAbs) against NS1 of DEN-1 (ATCC HB47; institution no. 15F3-1) was obtained from the American Type Culture Collection (13). The cell line was grown in RPMI 1640 medium plus 10% heat-inactivated FBS. MAb 15F3-1 was affinity purified using protein G-Sepharose 4B gel. An enzyme-linked immunosorbent assay (ELISA) and Western blotting further confirmed its activity and specificity.
Human serum samples.
An active physician-based dengue surveillance system has been established in local hospitals in southern Taiwan and at National Taiwan University (NTU) Hospital in northern Taiwan. Physicians not only were trained to identify the clinical manifestations of DF, DHF, and DSS (29) but were also taught the standard protocols for collecting and transporting blood samples. All the samples were centrifuged at 800 × g for 10 min at 4°C and sent to the NTU Dengue Diagnosis Laboratory by express mail the same day. Serum and plasma samples were collected from dengue patients and tested by at least two of the following four methods: (i) virus isolation in mosquito C6/36 cells, (ii) serotype-specific reverse transcriptase-PCR (RT-PCR), (iii) dengue-specific immunoglobulin M (IgM), and (iv) a hemagluttination-inhibition test for a fourfold increase in titers of antibody to DEN in convalescent-phase serum samples (5, 16). DEN-1 patients were further confirmed by either virus isolation or serotype-specific RT-PCR. Normal serum samples from healthy adults who tested seronegative for anti-DEN antibody by a commercial capture ELISA (PanBio [Windsor, Queensland, Australia] Dengue Duo) (27, 28) were used as references to establish cutoff values.
Biopanning procedures.
Three cycles of biopanning were performed. The ELISA plate was coated with 100 μg of MAb 15F3-1/ml. After blocking with PBSB (1% bovine serum albumin [BSA] in phosphate-buffered saline [PBS]), the plate was washed rapidly five times with washing buffer (PBS plus 0.5% [wt/vol] Tween-20). The phage-displayed 12-mer peptide library (New England Biolabs, Inc. Beverly, Mass.) was diluted to 4 × 1010 phage particles for the first cycle and to 2 × 1011 phage particles for the second and third cycles, added onto the antibody-coated plate, and rocked gently for 50 min at room temperature. The plate was then washed 10 times with washing buffer. The bound phage was eluted with 100 μl of 0.2 M glycine-HCl (pH 2.2) plus 1 mg of BSA/ml and was then neutralized with 15 μl of 1 M Tris-HCl (pH 9.1). The eluted phages were amplified and titrated in Escherichia coli ER2537 culture.
Identification of phage clones by ELISA.
The ELISA plates (Falcon; Becton Dickinson, Oxnard, Calif.) were coated with 100 μl of 100-μg/ml 15F3-1 in 0.1 M NaHCO3 (pH 8.6) at room temperature for 2 h and blocked with PBSB at 4°C overnight. The serially diluted phage particles were added to the antibody-coated plates and incubated at room temperature for 1 h. The plates were washed six times with washing buffer, and 1:5,000-diluted horseradish peroxidase (HRP)-conjugated anti-bacteriophage M13 antibody (Pharmacia; catalog no. 27-9411-01) in blocking buffer was added. The plates were then incubated at room temperature for 1 h with agitation, washed six times with washing buffer, and incubated with the peroxidase substrate o-phenylenediamine dihydrochloride (OPD; Sigma). The reaction was stopped with 3 N HCl, and the plates were read using a microplate reader at 490 nm.
DNA sequencing and computer analysis.
Immunopositive phage clones were further characterized by DNA sequencing. The amplified phage was precipitated by 1/6 volume of polyethylene glycol (PEG)-NaCl (20% [wt/vol] PEG 8000 and 2.5 M NaCl). The phage pellet was resuspended in 100 μl of iodide buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 4 M NaI) with 250 μl of ethanol, and incubated at room temperature for 10 min. Phage DNA was isolated from the pellet after centrifugation at 12,000 × g for 10 min, washed in 70% ethanol, dried, and resuspended in distilled water. The DNA sequences of purified phages were determined according to the dideoxynucleotide chain termination method using an automated DNA sequencer (ABI PRISM 377; Perkin-Elmer, Norwalk, Conn.) or manually using the Sequenase kit 2.0 (United States Biomedical Corp., Cleveland, Ohio). The phage-displayed peptide sequences were aligned using MacDNASIS software (Hitachi Software Engineering Co., Ltd., Yokohama, Japan).
Western blot analysis.
Cell lysates or proteins were mixed with an equal volume of the sample buffer (50 mM Tris-HCl [pH 6.8], 100 mM dithiothreitol [DTT], 2% sodium dodecyl sulfate [SDS], 0.1% bromophenol blue, 10% glycerol), separated by SDS-polyacrylamide gel electrophoresis (PAGE), and transferred to a nitrocellulose membrane (Hybond-C Super; Amersham). The nonspecific antibody-binding sites were blocked with 5% skim milk in PBS, and the membranes were incubated with 15F3-1 as the primary antibody. The blot was then treated with HRP-conjugated goat anti-mouse immunoglobulin (Cappel Products, West Chester, Pa.) and developed with 4-chloro-1-naphthol.
Detection of DEN-1 in patient serum samples.
The ELISA plates were coated with 10 μg of individual peptide antigens/ml at 50 μl/well and incubated at 4°C for 6 h. After being washed with PBST (PBS plus 0.1% [wt/vol] Tween-20), the plates were blocked with PBSB at 4°C overnight and then incubated with the 1:100 PBSB-diluted test serum samples at room temperature for 1 h. Then 1:20,000-diluted HRP-conjugated goat anti-human IgM plus IgG (Jackson ImmunoResearch Labs, West Grove, Pa.) was added to the microtiter plates, and the procedures described under “Identification of phage clones by ELISA” above were followed.
Antibody binding and competitive inhibition assay.
The ELISA plates were coated with 10 μg of individual peptide antigens/ml or with twofold serially dilutions of these antigens at 50 μl/well and were then blocked with 1% BSA. MAb 15F3-1 was added to wells at 3 μg/ml, and wells were incubated at room temperature for 1 h. For the competitive inhibition assay, 109 phage particles were incubated with 10-fold increasing amounts of peptide EHKYSWKS (P14M), or control peptide, before being transferred to the antibody-coated plate and incubated for 1 h. The plates were incubated with HRP-conjugated anti-mouse IgG, and the procedures described under “Identification of phage clones by ELISA” above were followed.
Rabbit immunization.
Outbred laboratory rabbits were infected by intravenous (i.v.) injection with DEN-1 (Hawaii strain). Hyperimmune sera were obtained after four rounds of infection with the virus.
RESULTS
Screening of phage-displayed peptide library with a DEN-1 MAb.
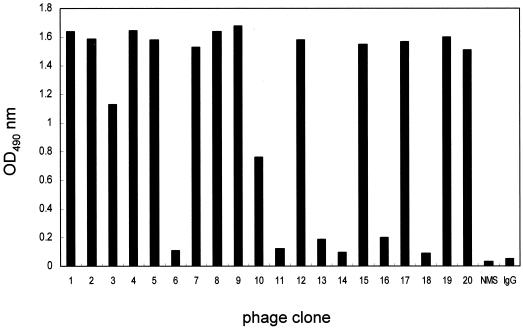
To select the immunopositive phage clones, a DEN-1 serotype-specific MAb (15F3-1) was purified from the ascites using the protein G affinity column. This MAb was shown to react specifically with NS1 of DEN-1 by both Western blotting and ELISA (data not shown). The affinity-purified antibodies were immobilized on the 96-well plates, and the bound phage clones were selected after three biopanning cycles. Fourteen of 20 selected phage clones (HB47-1, -2, -3, -4, -5, -7, -8, -9, -10, -12, -15, -17, -19, and -20), which showed significant enhancement of reactivity to antibody 15F3-1 (Fig. 1), did not bind to normal mouse serum (NMS), normal mouse IgG (Fig. 1), or 3H5 (a MAb against DEN-2 [data not shown]). To confirm that the selected phage clones bound to 15F3-1 specifically, the antibodies were incubated with 10-fold serial dilutions of both the selected phage clones and the control phage clones. Only the selected phage clones bound to 15F3-1 specifically in a dose-responsive manner (data not shown).
FIG. 1.
Selection for 15F3-1-binding phage clones. After the third round of biopanning, 20 individual phage clones were isolated; only 14 of them reacted strongly with 15F3-1.
Identification of the B-cell epitope.
The phage DNAs isolated from the first 10 immunopositive phage clones were sequenced. The inserted DNA fragment in these selected phage clones comprised 36 nucleotides. With the exception of the clone 3 insert, which contained sequence encoding 11 amino acid residues and a stop codon, the sequences of these inserts would translate into 12 amino acids (Table 1). Through alignment of phage-displayed peptide sequences using MacDNASIS software, the binding motif of antibody 15F3-1 was shown to be H-x-Y-a-W-b (a = S/T, b = K/H/R). This motif was exhibited in all 10 immunopositive phage clones (Table 1). In addition, a match between the phage-displayed peptide sequences and the published protein sequences of NS1 of flaviviruses demonstrated that the epitope for this antibody corresponded only to amino acid residues 111 to 116 of the NS1 of DEN-1 (Table 1). This is also a linear epitope, and only DEN-1 reveals such a motif. We also aligned the phage-displayed peptide sequences with other genes of DEN-1, but only NS1 exhibited this motif.
TABLE 1.
Alignment of phage-displayed peptide sequences with residues 111 to 116 of the NS1 proteins of flavivirusesa
| Virus or clone | Peptide sequence |
|---|---|
| Virus | |
| JEV | F E M G W K |
| TBEV | I K V S W K |
| DEN-2 | L R Y S W K |
| DEN-3 | L K Y S W K |
| DEN-4 | L K Y S W K |
| DEN-1 | H K Y S W K |
| Clone | |
| HB47-1 and -5 | H S Y T W K T S S V F Q |
| HB47-2 | S S D T T H V Y S W R L |
| HB47-3 | K P H V Y T W H ∗bK V P |
| HB47-4 and -8 | G M T H Y Y S W K F Q P |
| HB47-7 | F P H T Y T W K Q G D R |
| HB47-9 | N P H L Y S W K H G P K |
| HB47-12 | T Q Q H T F T W K I K A |
| HB47-15 | A E I Q H R Y T W K G W |
Synthetic peptide binding assay.
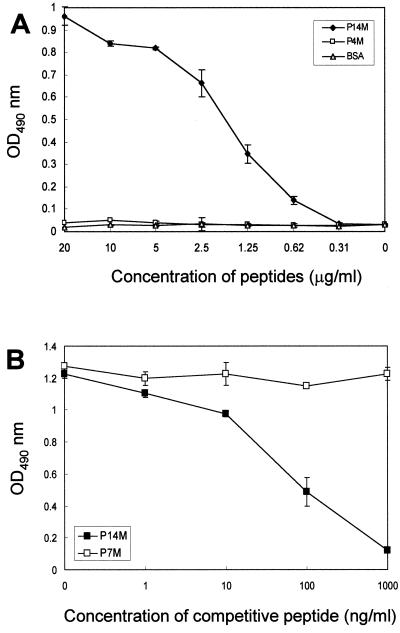
To verify that the peptide sequences corresponding to amino acid residues 110 to 117 of the NS1 of DEN-1 were indeed recognized by MAb 15F3-1, synthetic peptide binding assays were performed. The peptides were synthesized in the multiple-antigen peptide (MAP) form, because the binding efficiency of an eight-chain MAP is greater than that of a single-chain peptide (25). As shown in Fig. 2A, the synthetic peptide P14M, representing amino acid residues 110 to 117 of the NS1 of DEN-1, bound the antibody in a concentration-dependent manner. An unrelated control peptide, KGTFDPLQEPRT (P4M), and BSA revealed no such reactivity.
FIG. 2.
Characterization of the B-cell epitope of MAb 15F3-1. (A) Binding assay of synthetic peptide with MAb 15F3-1. Only the P14M peptide antigen specifically reacted with 15F3-1. (B) Competitive inhibition of phage clone binding to 15F3-1 by synthetic peptide. Binding of 15F3-1-selected phage clone (HB47-1) to 15F3-1 was inhibited only by peptide P14M.
To further confirm that the phage-displayed peptides were the epitope of 15F3-1, a peptide-competitive inhibition assay was conducted to determine whether the P14M peptide and a selected phage clone (HB47-1) competed for the same antibody-binding site. Our results showed that the binding activity of 15F3-1 with phage clone (HB47-1) was inhibited by P14M in a dose-dependent manner. One microgram of peptide P14M per milliliter was able to inhibit 90% of the binding of the phage clone to MAb 15F3-1, whereas the arbitrary control peptide SHRLHNTMPSES (P7M) had no effect at all (Fig. 2B).
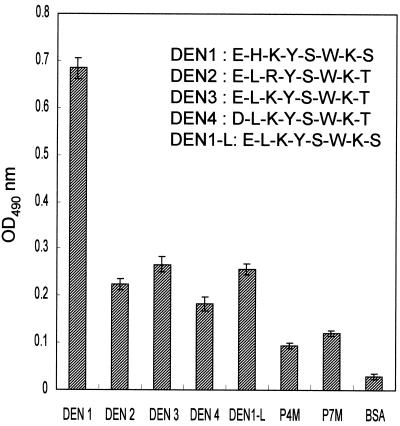
After we determined that the peptide comprising residues 111 to 116 of the NS1 of DEN-1 was the B-cell epitope of 15F3-1 by a phage-displayed random peptide library (RPL), the amino acid sequence was investigated in detail. Alignment of residues 111 to 116 of the NS1 from DEN-1, -2, -3, and -4 revealed that the amino acid sequences in this region were similar among serotypes. The major difference was that between the His111 in DEN-1 and Leu111 in DEN-2, -3, and -4 (Table 1). Because 15F3-1 was a DEN-1 serotype-specific MAb (13), the influence of His111 of DEN-1 on binding to 15F3-1 was further tested. By synthesizing the peptides containing residues 110 to 117 of the NS1 from all four DEN serotypes, their reactivities were assayed by ELISA (Fig. 3). The DEN-1 peptide (P14M) displayed a 2- to 3-times-higher reactivity with 15F3-1 than the DEN-2 peptide (ELRYSWKT [P16M]), the DEN-3 peptide (ELKYSWKT [P17M]), and the DEN-4 peptide (DLKYSWKT [P18M]). However, P16M, P17M, and P18M still managed to produce weak reactivity with 15F3-1, slightly higher than those of two unrelated control peptides, P4M (KGTFDPLQEPRT) and P7M (SHRLHNTMPSES). This experiment was repeated three times and resulted in similar patterns. In addition, the binding activity of 15F3-1 with the DEN-1-L peptide (ELKYSWKS), which had Leu instead of His as amino acid residue 111, was markedly weaker than that with the DEN-1 peptide (EHKYSWKS) (Fig. 3). These experimental observations strongly suggested that the His111 was the most important amino acid residue for binding to MAb 15F3-1.
FIG. 3.
The specificity of binding of synthetic peptides from DEN-1, -2, -3, and -4 with antibody 15F3-1 was determined by ELISA. DEN-1 peptides showed two- to sixfold-higher reactivity than DEN-2, -3, and -4 peptides, control peptides (P4M and P7M), or BSA.
Evaluation of the sensitivity and reactivity of epitope-based peptide antigen with DEN-1 patient serum samples.
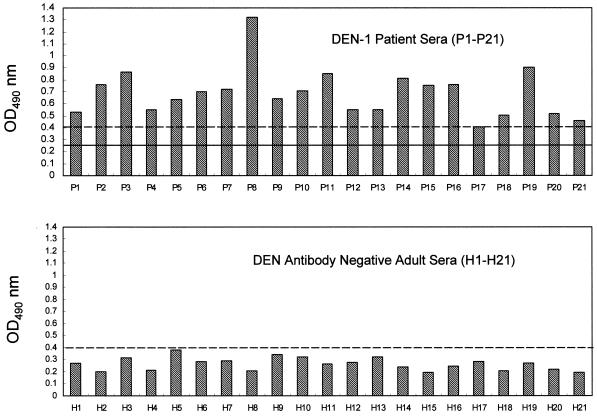
P14M was able to detect DEN-1 infection in 20 serum samples collected from 21 confirmed DEN-1 patients by ELISA (Fig. 4) and in hyperimmune serum samples of rabbits after four doses of DEN-1 Hawaii strain (data not shown). The mean optical density at 490 nm (OD490) (0.255) plus 3 times the standard deviation (0.405) was used to determine the cutoff value. The sensitivity of this epitope-based peptide serologic test was 95%. In addition, the amount of antibody varied among the different patients. In contrast, using the same epitope-based peptide serologic test, all of the serum samples obtained from 21 healthy adults were shown to be seronegative. The specificity of this test was 100% for healthy donors without DEN infection (Fig. 4).
FIG. 4.
ELISA reactivities of DEN-1 synthetic peptide P14M with serum samples from 21 DEN-1-infected patients versus 21 healthy donors. The cutoff value (dashed lines) was calculated as 0.405. Solid line, mean OD490.
DISCUSSION
In this study, we characterized the B-cell epitope of the DEN-1 mAb (15F3-1) and detected antibodies against DEN-1 from serum samples of dengue patients using the epitope-based peptide antigen. We further confirmed that the epitope for 15F3-1 was a linear epitope and that His111 played the most important role in its binding activity. In addition, experiments using this peptide antigen for viral diagnosis showed that it was capable of detecting DEN-1 in clinical human serum specimens. Therefore, it is feasible that our phage-displayed epitope and epitope-based peptide antigen can be used to develop serologic tests for laboratory diagnosis of DEN in the future.
Epitopes are divided into linear and conformational epitopes (3, 23). Linear epitopes are short stretches of the primary structure of the protein and are widely known to consist of 3 to 8 continuous amino acid residues of the primary sequence. Conformational epitopes consist of several amino acid residues that are discrete in the primary sequence but assemble to form an antigenic determinant on the tertiary structure of the native protein (3, 17, 23). Conformational epitopes are often formed from more than 10 residues separated from the primary structure (2, 4, 17). By Western blot analysis, MAb 15F3-1 detected NS1 very effectively in both native and denatured gels (data not shown) and the binding motif for 15F3-1 was located in residues 111 to 116 of the NS1 of DEN-1, indicating that the B-cell epitope of 15F3-1 is a linear epitope. Our results were similar to those of Yao et al., who also identified a linear epitope of 15F3-1 using a hexamer RPL (30).
The important role of His111 in DEN-1 versus Leu111 for DEN-2, -3, and -4 was elucidated by comparing amino acid residues 111 to 116 of NS1. Since 15F3-1 is a DEN-1-specific MAb, the crucial contribution of His111 for antibody binding was further confirmed by synthesizing four peptides corresponding to residues 110 to 117 of NS1 of DEN-1, -2, -3, and -4, with only the DEN-1 peptide, which contained His111, demonstrating greater reactivity. Such reactivity was dramatically decreased when the positively charged His111 was changed to the aliphatic side-chain Leu111 in the DEN1-L peptide. Therefore, our finding on His111 is compatible with the finding of another report that charged residues are important in the interaction of antigenic epitopes with antibodies (24).
The successful use of this peptide in detecting DEN-1 in the serum samples of confirmed dengue patients but not in those of healthy donors suggests that the epitope-based synthetic peptide P14M which we developed as an antigen could possibly be used as a serologic reagent in the diagnosis of DEN-1 patients. The ELISA using P14M as an antigen demonstrated that such a test had 95% sensitivity and 100% specificity. However, this ELISA reactivity in detecting DEN-1-infected serum samples of rabbits and human patients was not as high as that of whole virus or viral protein, which had multiple epitopes. Therefore, the identification of more epitopes of DEN-1 and the combination of more epitope-based peptide antigens to detect DEN-1 in serum samples obtained from dengue patients will definitely increase the sensitivity of this method in serologic diagnosis.
Using our epitope-based peptide antigen to detect anti-DEN antibody is relatively simple and specific and does not require paired serum samples as other, conventional tests for serological diagnosis by hemagglutination inhibition do. Therefore, it holds great promise as a routine procedure for viral diagnosis in clinical and public health laboratories. On the other hand, conventional IgM and IgG capture ELISAs, which require the preparation of DEN antigen and antibody, have been used primarily in specialized virology centers. Several reliable dengue serologic diagnosis tests are now available, but they still display difficulty in distinguishing among the four DEN serotypes and they still show a 45 to 50% cross-reaction with antibody against Japanese encephalitis virus (JEV) (15, 27, 28). We found that amino acid residues 111 to 116 of NS1 in DEN-1 and JEV were very different (Table 1). Furthermore, our additional preliminary results revealed that serum samples obtained from five JEV-hyperimmune BALB/c mice showed no ELISA reactivity with peptide P14M, which suggested that it is capable of differentiating between JEV and DEN-1 in mouse serum samples. In addition, the present preliminary work also found that this epitope-based peptide did not react with DEN-2, -3, and -4 in serum samples from patients in Taiwan infected with these serotypes; the sample size was smaller because few DEN-2 and DEN-4 were isolated here. Further investigation on the application of this epitope-based peptide to human samples, with a larger sample size, is in progress. On the other hand, our preliminary work further found that the serotype-specific epitope of DEN-2 was different from the above DEN-1 motif (unpublished data). Moreover, the method we developed can also be used to detect future DHF patients who had secondary infection with a heterologous serotype of DEN, which would minimize possible morbidity and mortality. Finally, our test will be very valuable for the further development of a serotype-specific diagnostic reagent that can be used to serologically distinguish the four serotypes in samples from dengue patients and thus help combat dengue diseases.
ACKNOWLEDGMENTS
This work was supported by grant NSC 89-2320-B-016-027 from the National Science Council of R.O.C. to H.-C.W. and by grant NHRI-CN-CL8902P from the National Health Research Institute, Department of Health, Taipei, R.O.C.
REFERENCES
- 1.Aaskov J G, Geysen H M, Mason T J. Serologically defined linear epitopes in the envelope protein of dengue 2 (Jamaica strain 1409) Arch Virol. 1989;105:209–221. doi: 10.1007/BF01311358. [DOI] [PubMed] [Google Scholar]
- 2.Appel J R, Pinilla C, Niman H, Houghten R. Elucidation of discontinuous linear determinants in peptides. J Immunol. 1990;144:976–983. [PubMed] [Google Scholar]
- 3.Barlow D J, Edwards M S, Thornton J M. Continuous and discontinuous protein antigenic determinants. Nature. 1986;322:747–748. doi: 10.1038/322747a0. [DOI] [PubMed] [Google Scholar]
- 4.Cason J. Strategies for mapping and imitating viral B-cell epitopes. J Virol Methods. 1994;49:209–220. doi: 10.1016/0166-0934(94)90045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow L, Hsu S T. MAC-ELISA for the detection of IgM antibodies to dengue type I virus (rapid diagnosis of dengue type I virus infection) Chin J Microbiol Immunol. 1989;22:278–285. [PubMed] [Google Scholar]
- 6.D'Mello F, Partidos C D, Steward M W, Howard C R. Definition of the primary structure of hepatitis B virus (HBV) pre-S hepatocyte binding domain using random peptide libraries. Virology. 1997;237:319–326. doi: 10.1006/viro.1997.8774. [DOI] [PubMed] [Google Scholar]
- 7.Folgori A, Tafi R, Meola A, Felici F, Galfre G, Cortese R, Monaci P, Alfredo N. A general strategy to identify mimotopes of pathological antigens using only random peptide libraries and human sera. EMBO J. 1994;13:2236–2243. doi: 10.1002/j.1460-2075.1994.tb06501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu Y, Shearing L N, Haynes S, Crewther P, Tilley L, Anders R F, Foley M. Isolation from phage display libraries of single chain variable fragment antibodies that recognize conformational epitopes in the malaria vaccine candidate, apical membrane antigen-1. J Biol Chem. 1997;272:25678–25684. doi: 10.1074/jbc.272.41.25678. [DOI] [PubMed] [Google Scholar]
- 9.Gubler D J. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11:480–496. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gubler D J, Clark G G. Dengue/dengue hemorrhagic fever: the emergence of a global health problem. Emerg Infect Dis. 1995;1:55–57. doi: 10.3201/eid0102.952004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halstead S B. Pathogenesis of dengue: challenges to molecular biology. Science. 1988;239:476–481. doi: 10.1126/science.3277268. [DOI] [PubMed] [Google Scholar]
- 12.Henchal E A, Putnak J R. The dengue viruses. Clin Microbiol Rev. 1990;3:376–396. doi: 10.1128/cmr.3.4.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henchal E A, Gentry M K, McCown J M, Brandt W E. Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. Am J Trop Med Hyg. 1982;31:830–836. doi: 10.4269/ajtmh.1982.31.830. [DOI] [PubMed] [Google Scholar]
- 14.Innis B L, Thirawuth V, Hemachudha C. Identification of continuous epitopes of the envelope glycoprotein of dengue type 2 virus. Am J Trop Med Hyg. 1989;40:676–687. doi: 10.4269/ajtmh.1989.40.676. [DOI] [PubMed] [Google Scholar]
- 15.Lam S K, Devine P L. Evaluation of capture ELISA and rapid immunochromatographic test for the determination of IgM and IgG antibodies produced during dengue infection. Clin Diagn Virol. 1998;10:75–81. doi: 10.1016/s0928-0197(98)00002-6. [DOI] [PubMed] [Google Scholar]
- 16.Lanciotti R S, Calisher C H, Gubler D J, Chang G J, Vorndam A V. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase polymerase chain reaction. J Clin Microbiol. 1992;30:545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laver W G, Air G M, Webster R G, Smith-Gill S J. Epitopes on protein antigens: misconceptions and realities. Cell. 1990;61:553–556. doi: 10.1016/0092-8674(90)90464-p. [DOI] [PubMed] [Google Scholar]
- 18.Megret F, Hugnot J P, Falconar A, Gentry M K, Morens D M, Murray J M, Schlesinger J J, Wright P J, Young P, Van Regenmortel M H, Deubel V. Use of recombinant fusion proteins and monoclonal antibodies to define linear and discontinuous antigenic sites on the dengue virus envelope glycoprotein. Virology. 1992;187:480–491. doi: 10.1016/0042-6822(92)90450-4. [DOI] [PubMed] [Google Scholar]
- 19.Prezzi C, Nuzzo M, Meola A, Delmastro P, Galfre G, Cortese R, Nicosia A, Monaci P. Selection of antigenic and immunogenic mimics of hepatitis C virus using sera from patients. J Immunol. 1996;156:4504–4513. [PubMed] [Google Scholar]
- 20.Roehrig J T, Bolin R A, Kelly R G. Monoclonal antibody mapping of the envelope glycoprotein of the dengue 2 virus, Jamaica. Virology. 1998;246:317–328. doi: 10.1006/viro.1998.9200. [DOI] [PubMed] [Google Scholar]
- 21.Roehrig J T, Johnson A J, Hunt A R, Bolin R A, Chu M C. Antibodies to dengue 2 virus E-glycoprotein synthetic peptides identify antigenic conformation. Virology. 1990;177:668–675. doi: 10.1016/0042-6822(90)90532-v. [DOI] [PubMed] [Google Scholar]
- 22.Scott J K, Smith G P. Searching for peptide ligands with an epitope library. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 23.Sela M. Antigenicity: some molecular aspects. Science. 1969;166:1365–1374. doi: 10.1126/science.166.3911.1365. [DOI] [PubMed] [Google Scholar]
- 24.Strauss E G, Stee D S, Schmaljohn A L, Strauss J H. Identification of antigenically important domains in the glycoproteins of Sindbis virus by analysis of antibody escape variants. J Virol. 1991;65:4654–4664. doi: 10.1128/jvi.65.9.4654-4664.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tam J P, Zavala F. Multiple antigen peptide: a novel approach to increase detection sensitivity of synthetic peptides in solid-phase immunoassays. J Immunol Methods. 1989;124:53–61. doi: 10.1016/0022-1759(89)90185-3. [DOI] [PubMed] [Google Scholar]
- 26.Trirawatanapong T, Chandran B, Putnak R, Padmanabhan R. Mapping of a region of dengue virus type-2 glycoprotein required for binding by a neutralizing monoclonal antibody. Gene. 1992;116:139–150. doi: 10.1016/0378-1119(92)90509-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaughn D W, Nisalak A, Kalayanarooj S, Solomon T, Dung N M, Cuzzubbo A, Devine P L. Evaluation of a rapid immunochromatographic test for diagnosis of dengue virus infection. J Clin Microbiol. 1998;36:234–238. doi: 10.1128/jcm.36.1.234-238.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaughn D W, Nisalak A, Solomon T, Kalayanarooj S, Nguyen M D, Kneen R, Cuzzubbo A, Devine P L. Rapid serologic diagnosis of dengue virus infection using a commercial capture ELISA that distinguishes primary and secondary infections. Am J Trop Med Hyg. 1999;60:693–698. doi: 10.4269/ajtmh.1999.60.693. [DOI] [PubMed] [Google Scholar]
- 29.World Health Organization. Dengue haemorrhagic fever: diagnosis, treatment and control. Geneva, Switzerland: World Health Organization; 1986. [Google Scholar]
- 30.Yao Z J, Kao M C, Loh K C, Chung M C. A serotype-specific epitope of dengue virus 1 identified by phage displayed random peptide library. FEMS Microbiol Lett. 1995;127:93–98. doi: 10.1111/j.1574-6968.1995.tb07455.x. [DOI] [PubMed] [Google Scholar]
- 31.Young A C, Valadon P, Casadevall A, Scharff M D, Sacchettini J C. The three-dimensional structures of a polysaccharide binding antibody to Cryptococcus neoformans and its complex with a peptide from a phage display library: implications for the identification of peptide mimotopes. J Mol Biol. 1997;274:622–634. doi: 10.1006/jmbi.1997.1407. [DOI] [PubMed] [Google Scholar]