Abstract
Rationale
Patients with indeterminate pulmonary nodules (IPNs) at risk of cancer undergo high rates of invasive, costly, and morbid procedures.
Objectives
To train and externally validate a risk prediction model that combined clinical, blood, and imaging biomarkers to improve the noninvasive management of IPNs.
Methods
In this prospectively collected, retrospective blinded evaluation study, probability of cancer was calculated for 456 patient nodules using the Mayo Clinic model, and patients were categorized into low-, intermediate-, and high-risk groups. A combined biomarker model (CBM) including clinical variables, serum high sensitivity CYFRA 21-1 level, and a radiomic signature was trained in cohort 1 (n = 170) and validated in cohorts 2–4 (total n = 286). All patients were pooled to recalibrate the model for clinical implementation. The clinical utility of the CBM compared with current clinical care was evaluated in 2 cohorts.
Measurements and Main Results
The CBM provided improved diagnostic accuracy over the Mayo Clinic model with an improvement in area under the curve of 0.124 (95% bootstrap confidence interval, 0.091–0.156; P < 2 × 10−16). Applying 10% and 70% risk thresholds resulted in a bias-corrected clinical reclassification index for cases and control subjects of 0.15 and 0.12, respectively. A clinical utility analysis of patient medical records estimated that a CBM-guided strategy would have reduced invasive procedures from 62.9% to 50.6% in the intermediate-risk benign population and shortened the median time to diagnosis of cancer from 60 to 21 days in intermediate-risk cancers.
Conclusions
Integration of clinical, blood, and image biomarkers improves noninvasive diagnosis of patients with IPNs, potentially reducing the rate of unnecessary invasive procedures while shortening the time to diagnosis.
Keywords: lung neoplasms, biomarkers, tumor, diagnostic imaging
At a Glance Commentary
Scientific Knowledge on the Subject
Patients with indeterminate pulmonary nodules at risk of cancer undergo high rates of invasive diagnostic procedures. Noninvasive diagnostic blood and imaging biomarkers have shown potential to aid in this context but lack the accuracy to change clinical practice.
What This Study Adds to the Field
We demonstrate that combining patient clinical information, quantitative imaging biomarkers, and blood biomarkers provides improved diagnostic accuracy.
Lung cancer is the leading cause of cancer death in the United States and worldwide. Early detection is critical to successful treatment, but the medical community is inundated with an estimated 1.5 million indeterminate pulmonary nodules (IPNs) detected annually in the United States (1). The management of IPNs suffers from an unacceptable rate of invasive diagnostic procedures on patients with benign disease and associated morbidities for many, and missed or delayed opportunities for cure for others (1–4).
The current management of patients with IPNs is driven by an assessment of the risk for cancer using a clinical risk model (5, 6), such as the widely validated Mayo Clinic model (5, 6, 7). Model or clinician risk estimates are used in concert with management guidelines issued by professional organizations such as the Lung-RADS and National Comprehensive Cancer Network guidelines (8, 9), the American College of Chest Physicians (ACCP), the British Thoracic Society (BTS) (10), and the Fleischner Society (11, 12). High-risk patients are recommended for biopsy or surgical resection, and low-risk patients are monitored with follow-up computed tomography (CT). However, the intermediate-risk group presents the greatest clinical challenge, producing the highest rate of diagnostic errors (5) and risks associated with invasive diagnostic procedures. To reduce mortality and overtreatment, a highly accurate noninvasive risk assessment strategy is critical.
To address this important clinical need, we recently investigated two biomarkers for IPN risk assessment. These biomarkers are serum CYFRA 21-1 quantified by a new, high-sensitivity free solution assay (FSA) method (13) (hs-CYFRA 21-1), and a multifeature quantitative radiomics signature that presents a quantitative approach to describing the physiology of tumor regions in three dimensions based on the CT scan (14, 15). Here we hypothesized that the combination of clinical, serum, and imaging biomarkers would enable improved noninvasive diagnostic accuracy and result in improved management of the intermediate-risk group. In this prospective collection, retrospective blinded evaluation (16) designed study, we demonstrated that a combination of hs-CYFRA 21-1 level, a quantitative radiomic signature, and the Mayo Clinic model (Mayo) risk score significantly improved the accuracy for noninvasive IPN diagnosis when compared with the current standard of care.
Methods
Patient Selection
Four case–control patient populations (“cohorts”) derived from prospectively collected specimens were used in this study (16). The training cohort was composed of patients from Vanderbilt University Medical Center (VUMC) and the Tennessee Valley Veterans Affairs Healthcare System Nashville Campus (the VUMC cohort N = 170) who consented to research between 2003 and 2017. Independent cohorts were from the University of Pittsburgh Medical Center (UPMC) (N = 99, consented 2006–2015), the Detection of Early Cancer Among Military Personnel (DECAMP) (N = 99) consortium (12 clinical centers, listed in the DECAMP Investigator Directory in the data supplement) (17) between 2013 and 2017, and the University of Colorado Denver Hospital and the Rocky Mountain Regional Veterans Affairs Medical Center (UC Denver) (N = 88) between 2010 and 2018 served as external validation cohorts. Subjects for this study were incidentally found to have an IPN at a secondary or tertiary clinical center or enrolled in a lung cancer screening program that detected an IPN between 6 and 30 mm in the largest axial diameter, were between 18 and 80 years old at the time of consent, had prospectively collected serum and CT scans (with slice thickness ⩽ 3 mm) at the time of initial nodule detection, and had follow-up, including a definitive cancer or no-cancer diagnosis. Disease outcome was biopsy proven for malignant nodules, and biopsy-proven or 2-year longitudinal imaging follow-up showing no signs of growth for benign nodules. Flow diagrams for patient inclusion are presented in Figure E1 in the online supplement. Patient age, smoking history, prior extrathoracic cancer, location of the nodule, spiculation, and nodule diameter were used to calculate a Mayo risk score (7, 18). In addition to the Mayo factors, emphysema, number of nodules in the lungs, nodule solid, part-solid, nonsolid, and family history of lung cancer were used to calculate a risk score based on the Brock University model (available only for the VUMC cohort) (19). A Mayo score of less than 0.1 was considered low risk, above 0.7 considered high risk, and 0.1–0.7 considered intermediate risk (11).
Biomarkers and Model
CT scans and serum samples had no connecting identifier to each other. Investigators performing blood measurements were completely blinded to other predictors. Investigators performing image analysis were blinded to all predictors except for the institution-provided CT slice and lobe location where the nodule of interest was located, with no information about the nature of the nodule. Neither blood nor CT scans had identifiers to outcome. For the external cohorts, all clinical data and outcome data were kept by the outside institution until after both blood and radiomics biomarker data were analyzed and the results provided to the external institution.
hs-CYFRA 21-1 levels were measured by FSA-compensated interferometric reader (CIR) in a blinded fashion in serum aliquots from the time of nodule detection as described previously (20). Nodules were segmented from CT scans for feature extraction (see Supplemental Methods E1) using the HealthMyne picture archiving and communication system (www.healthmyne.com). Segmentation of the nodule with the largest axial diameter was performed manually by a trained user and verified by trained radiologists (U.B., S.C., and K.S.). Features were evaluated for inter- and intra-user variability. Redundant features or features with high inter- and intra-user variability were removed. The radiomic risk score was derived from regression shrinkage and subset selection via the least absolute shrinkage and selection operator method. The receiver operator characteristic (ROC) curve area under the curve (AUC) with 95% confidence interval (CI) was calculated using bootstrap internal validation. For patients who underwent a diagnostic positron emission tomography (PET) scan after CT, the risk of malignancy following a diagnostic PET scan was calculated by incorporating the standardized uptake value within the nodule of interest using a four-category classification (none, faint, moderate, intense) into the Mayo score according to Herder and colleagues (21).
The combined biomarker model (CBM) integrating the Mayo risk score, hs-CYFRA 21-1 measurement, and radiomic score was calculated using a logistic regression model with flexible functional forms (restricted cubic splines allows for nonlinear effect [Supplemental Methods E2]). First, the CBM was trained on the VUMC cohort and validated on the three external cohorts. Improvement to the AUC was assessed by selecting 100 bootstrap samples and obtaining a 95% CI of the difference in AUC between Mayo alone and CBM. After assessing the validation of the approach (i.e., by seeing that the AUC for CBM was significantly improved over the Mayo model alone in each of the external cohorts), the four cohorts were pooled (n = 456), the model was fitted to this pooled cohort, and the prevalence of disease was adjusted to 0.33 (22). We internally cross-validated the discrimination and calibration performance of the procedure that we used to build the CBM model using 200 repeated threefold splitting (two-thirds of the combined data as the training set and one-third as the test set). On each of the 200 repetitions, the CBM and baseline Mayo were fitted to the training set and then calibrated to a prevalence of 0.33. For each split, 500 bootstrap samples of size 100, each with approximately 0.33 prevalence, were drawn from the one-third test set to evaluate the CBM’s likely performance on a new sample of patients from the same patient population (22, 23). To calculate reclassification, patients were grouped by malignant–benign status and then classified into low-, intermediate-, and high-risk groups using decision thresholds of 0.1 and 0.7 probability of cancer. The bias-corrected clinical net reclassification index (cNRI) was calculated for cases and control subjects separately by comparing the CBM to the Mayo score (24, 25).
Statistical Analysis
The primary endpoint is the improvement in diagnostic accuracy of IPNs when using the combined biomarker results compared with using the Mayo risk score alone. Diagnostic performance of the CBM was compared with the Mayo based on ROC AUC, sensitivity and specificity, and positive and negative diagnostic likelihood ratios (DLR+, DLR−) at the Youden Index (26) and, more relevant, DLR+ and DLR− at critical clinical decision thresholds (0.1 and 0.7). Likelihood ratio tests were used to assess whether the biomarkers alone or in combination added significantly to the Mayo-only model (25, 27, 28).
Clinical management was assessed for the VUMC and UC Denver cohorts using electronic medical record review. Only procedures relating to the nodules of interest were included. Hypothetical clinical management based on the CBM classification was determined by assuming that any patient “ruled out” (i.e., intermediate risk by Mayo with a low risk by CBM) would not undergo additional testing beyond follow-up chest CT scans, and any patient “ruled in” (i.e., intermediate risk by Mayo with a high risk by CBM) would go directly to surgery or biopsy. Any patients who were CBM high risk would be recommended for biopsy and would have had confirmation of cancer at 21 days (standard of care at VUMC). Patients who were CBM low risk would not have undergone invasive diagnostic procedures. For incorrectly reclassified intermediate-risk control subjects, one additional invasive procedure (a bronchoscopy) was assigned. A chi-square test was used to evaluate the difference between none and any invasive procedures between CBM and actual care. For incorrectly reclassified intermediate risk cases, 90 days of time to diagnosis was assigned, reflecting the standard interval for a follow-up CT scan in intermediate-risk patients. Follow-up CT scans in intermediate-risk cases were revised as follows under CBM: for those reassigned as high risk, no follow-up CT scans were assumed, and for those reassigned as low risk, one additional CT scan was assigned. Among diagnosed intermediate-risk cases, a log-rank test was used to compare time to diagnosis, and a chi-square test was used to evaluate the difference between none and any CT scans between what would be expected with CBM and without.
Results
Study Populations
The training cohort consisted of 170 patients from VUMC and the Tennessee Valley VA Healthcare Nashville Campus (Table 1). Three independent external validation cohorts consisted of 99 patients seen at UPMC, 99 patients from the DECAMP consortium lung nodule study, and 88 patients from UC Denver. The majority of IPNs (67%) from all four cohorts were in the intermediate-risk group based on the Mayo. The Mayo provided a ROC AUC of 0.76 (0.68–0.83) in the VUMC cohort, 0.59 (0.49–0.68) in DECAMP, 0.86 (0.79–0.91) in UPMC, and 0.67 (0.57–0.77) in UC Denver. The Brock University model risk was calculated for the VUMC cohort and provided an ROC AUC of 0.75 (0.67–0.83). Diagnostic PET scans were performed in 75 of the 170 VUMC patients and 40 of the 88 UC Denver patients. Within this subset, the ROC AUC of the Herder model, calculated by combining the PET avidity and the Mayo risk score, was 0.66 (0.48–0.83) in VUMC compared with 0.71 (0.54–0.88) by Mayo, representing a decrease in performance with a mean difference in AUC of −0.051 (−0.128–0.017). In the subset of the UC Denver cohort with diagnostic PET scans, the ROC AUC of the Herder model was 0.88 (0.77–0.99) versus 0.83 (0.70–0.96) by Mayo, representing an increase in performance with a mean difference in AUC of 0.046 (−0.073–0.159) (Figure E2).
Table 1.
Patient Cohort Characteristics
| VUMC | UPMC | DECAMP | Denver | |||||
|---|---|---|---|---|---|---|---|---|
| Characteristic | Benign | Cancer | Benign | Cancer | Benign | Cancer | Benign | Cancer |
| Count | 51 (30) | 119 (70) | 59 (60) | 40 (40) | 49 (50) | 50 (50) | 40 (45) | 48 (55) |
| Age, yr | 63 (55–69) | 70 (63–75) | 68 (64–74) | 68 (62–76) | 65 (61–72) | 68 (63–74) | 67 (61–72) | 66 (62) |
| Current/former smoker | 42 (82) | 115 (94) | 59 (100) | 40 (100) | 49 (100) | 50 (100) | 31 (78) | 39 (81) |
| Pack-years | 30 (4–49) | 44 (30–70) | 46 (36–70) | 40 (30–59) | 40 (30–54) | 45 (35–60) | 45 (14–54) | 45 (25–63) |
| Previous cancer | 11 (22) | 45 (37) | 2 (3) | 0 (0) | 23 (47) | 25 (50) | 5 (13) | 6 (13) |
| Located in upper lobe | 26 (51) | 77 (63) | 25 (42) | 29 (73) | 29 (59) | 26 (52) | 24 (60) | 38 (79) |
| Size, mm | 13 (8–20) | 20 (13–23) | 11 (8–14) | 21 (16–26) | 11 (8–14) | 15 (11–19) | 13 (11–19) | 19 (15–24) |
| Spiculated | 15 (29) | 50 (41) | 2 (3) | 11 (28) | 26 (53) | 22 (44 | 8 (20) | 18 (38) |
| Sex, M | 29 (57) | 70 (57) | 37 (63) | 21 (53) | 37 (76) | 39 (78) | 27 (68) | 33 (69) |
| BMI | 28 (23–33) | 27 (23–31) | 29 (25–32) | 27 (22–30) | 25 (23–28) | 25 (23–30) | 28 (26–32) | 28 (24–31) |
| Mayo model risk | ||||||||
| Risk | 29 (13–50) | 62 (36–80) | 16 (9–33) | 51 (32–75) | 44 (25–59) | 53 (34–72) | 27 (18–54) | 51 (26–77) |
| Low risk: <10% | 11 (22) | 4 (3) | 16 (27) | 1 (3) | 2 (4) | 0 (0) | 6 (15) | 4 (8) |
| Intermediate risk: 10%–70% | 33 (65) | 73 (61) | 43 (73) | 27 (68) | 35 (71) | 36 (72) | 31 (78) | 30 (63) |
| High risk: >70% | 7 (14) | 43 (36) | 0 (0) | 12 (30) | 12 (24) | 14 (38) | 3 (8) | 14 (29) |
| Cancer history | ||||||||
| Adenocarcinoma | — | 69 (58) | — | 21 (53) | — | 26 (52) | — | 34 (71) |
| Squamous cell | — | 22 (18) | — | 7 (18) | — | 11 (22) | — | 7 (15) |
| Small cell | — | 14 (12) | — | — | — | 5 (10) | — | — |
| Large cell | — | 2 (2) | — | 3 (8) | — | 2 (4) | — | — |
| Carcinoid | — | 6 (2) | — | — | — | 1 (2) | — | — |
| Unspecified NSCLC | — | 2 (2) | — | 7 (18) | — | 2 (4) | — | 2 (4) |
| Other* | — | 4 (3) | — | 2 (5) | — | 3 (6) | — | 5 (10) |
| Cancer stage | ||||||||
| I | — | 79 (67) | — | 15 (38) | — | 26 (52) | — | 33 (69) |
| II | — | 13 (11) | — | 4 (10) | — | 3 (6) | — | — |
| III | — | 11 (9) | — | 9 (23) | — | 12 (24) | — | 4 (4) |
| IV | — | 6 (5) | — | 6 (15) | — | 3 (6) | — | 6 (13) |
| Limited | — | 3 (3) | — | — | — | — | — | — |
| Extensive | — | 6 (5) | — | — | — | — | — | — |
| Missing | — | 1 (1) | — | 6 (15) | — | 6 (12) | — | 5 (10) |
Definition of abbreviations: BMI = body mass index; DECAMP = Detection of Early Lung Cancer Among Military Personnel; Denver = University of Colorado Denver; IQR = interquartile range; NSCLC = non–small cell lung cancer; UPMC = University of Pittsburgh Medical Center; VUMC = Vanderbilt University Medical Center.
Counts expressed as n (%), continuous variables expressed as median (IQR).
Other cancers include: adenosquamous (n =1), adenomatous hyperplasia (n =1), neuroendocrine (n =4), mucoepidermoid (n =1), schwannoma (n =1), adenocarcinoma and squamous cell carcinoma simultaneous primary (n =1), unknown (n =5).
Blood Biomarker
The serum concentration of hs-CYFRA 21-1 measured by FSA-CIR provided better diagnostic accuracy than the Mayo alone across all four cohorts, with an ROC AUC of 0.78 (0.71–0.85) in VUMC, 0.66 (0.56–0.74) in DECAMP, 0.86 (0.79–0.91) in UPMC, and 0.67 (0.57–0.77) in UC Denver. The combination of CYFRA and the Mayo risk through a linear logistic regression resulted in improved accuracy over both hs-CYFRA 211 and the Mayo alone, with ROC AUCs of 0.82 (0.76–0.89) in VUMC, 0.70 (0.59–0.80) in DECAMP, 0.92 (0.86–0.97) in UPMC, and 0.82 (0.73–0.90) in UC Denver (likelihood ratio test for Mayo plus CYFRA vs. Mayo alone χ2 = 25.202, DF = 2, P = 3.368 × 10−6 within the training cohort).
Radiomic Signature
The radiomics score was based on three size-related features, three shape-related features, and four textural features (Table E1). The feature selection using the least absolute shrinkage and selection operator method was repeated 20 times with random seed to ensure stable feature selection, with the top five features selected all 20 times, and all features being selected at least 5 times. The radiomics model alone outperformed the Mayo risk in all four cohorts, with ROC AUCs of 0.78 (0.70–0.86) in VUMC, 0.72 (0.62–0.82) in UPMC, 0.88 (0.82–0.95) in UPMC, and 0.67 (0.56–0.79) in UC Denver. The combination of the radiomics score and the Mayo Clinic model risk through a linear logistic regression resulted in improved predictive accuracy over both the radiomics risk and the Mayo Clinic model alone, with ROC AUCs of 0.81 (0.74–0.88) in VUMC, 0.71 (0.61–0.81) in DECAMP, 0.90 (0.83–0.96) in UPMC, and 0.70 (0.59–0.81) in UC Denver (likelihood ratio test for Mayo plus Radiomics vs. Mayo alone χ2 = 15.391, DF = 2, P = 4.55 × 10−4 within the training cohort).
The Combined Biomarker Model
The CBM includes the Mayo risk estimate as a single variable, the hs-CYFRA 21-1 value (natural log of ng/ml), and the radiomics classifier, each modeled with a restricted cubic spline in a logistic regression to yield a probability of malignancy on the basis of the three variables. The CBM was trained on the VUMC cohort, and accuracy of the locked-down model was validated in the three external cohorts (DECAMP, UPMC, and UC Denver). Addition of these biomarkers to the Mayo was assessed within the training cohort using a likelihood ratio test, with P ⩽ 1.7 × 10−7 for the CBM versus Mayo (χ2 = 36.843, DF = 4); P = 2.196 × 10−5 for CBM versus Mayo plus Radiomics score (χ2 = 21.452, DF = 2); and P = 2.966 × 10−3 for CBM versus Mayo Clinic model plus hs-CYFRA 21–1 (χ2 = 11.641, DF = 2). The CBM provided an improvement in diagnostic accuracy over the Mayo in each cohort (Figures 1B and 1C), with a mean improvement in AUC of 0.104 (95% CI: 0.098–0.111) in VUMC, 0.165 (0.155–0.175) in DECAMP, 0.087 (0.081–0.093) in UPMC, and 0.176 (0.167–0.186) in UC Denver. In the VUMC cohort only, the same comparison of the CBM versus the Brock University model demonstrated similar performance, with an improvement in AUC of 0.104 (0.958–0.112). For the subset of the cohorts with diagnostic PET scans, the CBM outperformed the Herder model in VUMC, with an ROC AUC of 0.82 (0.68–0.96) by CBM versus 0.66 (0.48–0.96) by Herder for an improvement in AUC of 0.165 (0.059–0.296), and in UC Denver with an ROC AUC of 0.92 (0.84–1.00) by CBM versus 0.88 (0.77–0.99) by Mayo for an improvement in AUC of 0.043 (−0.029– 0.123).
Figure 1.

Distribution of risk scores and receiver operating characteristics (ROC) curves for the Mayo model (Mayo) risk scores and the combined biomarker model (CBM) risk scores in the training cohort and validation cohorts. (A) Distribution of risk of malignancy for the Mayo (teal) and CBM (orange) for the training cohort (VUMC n = 170) and three validation cohorts (DECAMP n = 99, UPMC n = 99, and UC Denver n = 88). (B) ROC curves, and (C) precision/recall curves with 95% confidence interval (100 bootstrap samples) for two models. Numbers indicate area under the curve with the range of the 95% confidence interval in parentheses. DECAMP = Detection of Early Lung Cancer among Military Personnel; UC Denver = University of Colorado Denver; UPMC = Univeristy of Pittsburgh Medical Center; VUMC = Vanderbilt University Medical Center.
Calibrated Model for Clinical Implementation
After validating improved diagnostic accuracy versus the Mayo across the independent cohorts, all four cohorts were combined, and the model was recalibrated to the clinically relevant prevalence of cancer of 33% (Figures 2A–2C). Assessment of calibration revealed that the model was appropriately fitted to the population with a slope of 0.995 (mean of 500 bootstrap samples, 95% CI⩿ 0.68–1.45) and intercept of –0.045 (95% CI: −0.38–0.31). The CBM provided an improvement in diagnostic accuracy over the Mayo in this combined cohort (Figure 2B), with an average improvement in AUC of 0.124 (0.091–0.156) with a likelihood ratio test, χ2 = 134.39, DF = 4, P < 2 × 10−16. Based on Youden criteria (26), the CBM provided improved accuracy over the Mayo with respect to increased sensitivity and specificity and better DLR+/− (Table 2). More importantly, the CBM outperformed the Mayo, blood biomarker, and imaging signature alone at critical decision thresholds of 0.1 and 0.7 (based on BTS guidelines). Analysis of model performance at other common decision thresholds, including the decision thresholds suggested by the ACCP (0.05 and 0.65) are presented in Table E2.
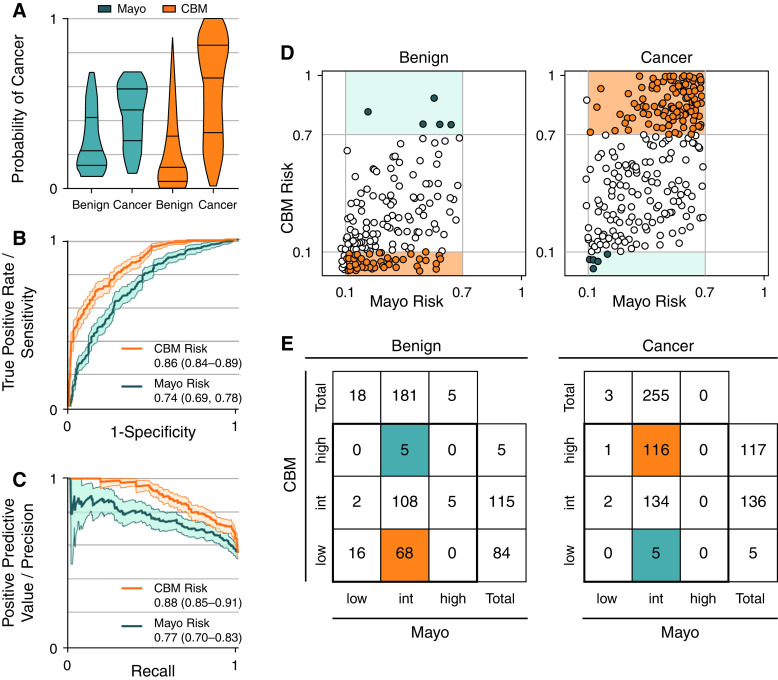
Figure 2.
Comparison of the Mayo model (Mayo) risk versus the combined biomarker model (CBM) risk in the pooled cohort (N = 456), with emphasis placed on patients ruled in and ruled out. (A) The risk distribution, (B) receiver operating characteristics curves, (C) precision-recall curves, (D) raw reclassification, and (E) reclassification frequency counts for the CBM. (D) Reclassification by the CBM. Left graph shows benign patient score distribution; right graph shows cancer patient score distribution (Mayo risk on the x-axis, CBM risk on the y-axis). Gray lines demarcate British Thoracic Society guideline thresholds of 0.1 and 0.7 risk. Intermediate-risk (int) patients who were correctly ruled in (for cancer) or ruled out (for benign) are in orange, and incorrectly ruled in or out are in teal. (E) Frequency counts show number of patients in each risk group. Note that this visual provides raw results of reclassification, and the bias-corrected clinical reclassification index (cNRI, listed in Table 2) provides an unbiased and more generalizable estimate of the reclassification rate.
Table 2.
Internal Validation Test Performance Metrics of the Mayo Risk Score and the Combined Biomarker Model
| Pooled Cohorts (N = 456) |
||
|---|---|---|
| Mayo (P.A.) | CBM | |
| ROC AUC | 0.74 (0.67–0.80) | 0.86 (0.81–0.90) |
| Difference | 0.124 (95% CI: 0.091–0.156) | |
| Precision-recall AUC | 0.77 (0.70–0.83) | 0.88 (0.85–0.91) |
| Likelihood ratio test | ||
| Residual degree of freedom | 453 | 449 |
| Residual deviance | 543.55 | 409.16 |
| Degree of freedom | — | 4 |
| Deviance | — | 134.39 |
| P value | — | <2×10−16 |
| Youden cutoff | 0.35 (0.21–0.48) | 0.36 (0.27–0.56) |
| Optimal criterion | 0.44 (0.35–0.53) | 0.59 (0.50–0.68) |
| Sensitivity | 0.75 (0.65–0.89) | 0.80 (0.69–0.90) |
| Specificity | 0.70 (0.55–0.82) | 0.82 (0.67–0.92) |
| DLR− | 0.35 (0.21–0.57) | 0.25 (0.13–0.35) |
| DLR+ | 2.49 (1.90–3.58) | 4.32 (2.67–8.52) |
| Rule-out threshold: 0.1 | ||
| Sensitivity | 0.98 (0.93–1.00) | 0.98 (0.92–1.00) |
| Specificity | 0.09 (0.00–0.22) | 0.43 (0.32–0.53) |
| DLR− | 0.13 (0.00–0.44) | 0.06 (0.00–0.18) |
| Rule-in threshold: 0.7 | ||
| Sensitivity | 0.03 (0.00–0.19) | 0.45 (0.35–0.55) |
| Specificity | 0.99 (0.94–1.00) | 0.96 (0.91–1.00) |
| DLR+ | 3.72 (0.43–inf) | 13.1 (5.57–inf) |
| Reclassification | ||
| cNRI case | — | 0.15 (0.10–0.20) |
| cNRI control | — | 0.12 (0.07–0.18) |
Definition of abbreviations: AUC = area under the ROC curve; CBM = combined biomarker model; CI = confidence interval; cNRI = bias-corrected clinical net reclassification index; DLR = diagnostic likelihood ratio; inf = infinity; Mayo = Mayo model; P.A. = prevalence adjusted; ROC = receiver operating characteristics.
The baseline Mayo clinic model calibrated to the training set, then adjusted to a prevalence of cancer of 0.33. All values reported as mean (95% CI) of the test set in the 200 repeated threefold splitting procedures.
Reclassification
Using groups defined by the BTS decision thresholds of 0.1 and 0.7, nodules were considered reclassified if their CBM score put them in a risk group different than the Mayo Clinic model. The bias-corrected clinical Net Reclassification Index (cNRI, Table 2) was 0.15 (0.10–0.20) for cases and 0.12 (0.07–0.18) for control subjects. Reclassification results are presented in Table 2 and Figure 2.
Clinical Management
The clinical follow-up, including counts of further diagnostic procedures related to the primary IPN of patients in the Mayo-based intermediate-risk group is presented in Table 3, as obtained by chart review (available only for VUMC and UC Denver subjects). Using the CBM and BTS guidelines for clinical decision-making would have resulted in a reduction in invasive procedures among intermediate-risk patients with benign nodules from 62.9% to 50.6%, or a 12.3% reduction when compared with actual care (chi-square with 1 DF = 2.52, P = 0.1127). The median time to diagnosis for intermediate cases using CBM was estimated to be 21 days compared with 60 days from clinical practice (chi-square with 1 DF = 23.40, P < 0.0001), which also would have resulted in a 29.1% reduction in patients who underwent follow-up CT scans (51.2% under standard of care and 22.6% under CBM; chi-square with 1 DF = 28.93, P < 0.0001).
Table 3.
Clinical Utility of the Combined Biomarker Model*
| VUMC + UC Denver | Mayo Intermediate, Benign Diagnosis (F/U or Biopsy Proven), n = 81 |
||
|---|---|---|---|
| Without CBM | With CBM | Improvement | |
| Procedures, n (%) | Patients | Patients | Patients |
| PET scan (one or more) | 33 (40.7%) | 28 (34.6%) | 5 (6.1%) |
| Bronchoscopy/EBUS + Nav (one or more) | 43 (53.1%) | 34 (41.9%) | 9 (11.2%) |
| Surgical resection/biopsy (one or more)† | 14 (17.3%) | 13 (16%) | 1 (1.3%) |
| Transthoracic needle aspiration (one or more) | 9 (11.1%) | 9 (11.1%) | 0 (0%) |
| Any invasive procedure‡ | 51 (62.9%) | 41 (50.6%) | 10 (12.3%) |
| Mayo Intermediate, Biopsy-proven Cancer Diagnosis, n = 165 | |||
| Follow-up CT (one or more), No. (%) | 85 (51.5%) | 37 (22.4%) | 48 (29.1%) |
| Time to diagnosis (days), median (IQR) | 60 (32–116) | 21 (21–43) | 39 |
Definition of abbreviations: CBM = combined biomarker model; CT = computed tomography; EBUS = endobronchial ultrasound; F/U = follow up; IQR = interquartile range; Mayo = Mayo model; Nav = electromagnetic navigation bronchoscopy; PET = positron emission tomography with fluorodeoxyglucose; UC Denver = University of Colorado Denver; VUMC = Vanderbilt University Medical Center.
Patient data were available in the VUMC and UC Denver cohorts. N represents the number of patients for whom clinical data were available and who were in the intermediate risk group (0.1–0.7 risk) by the prevalence-adjusted Mayo risk model.
Biopsy via bronchoscopic/surgical means, which is distinct from transthoracic/percutaneous needle aspiration.
Any invasive procedure includes bronchoscopy/EBUS plus Nav, surgical resection/biopsy, and transthoracic needle aspiration.
Discussion
This is the first study to integrate clinical, blood, and image biomarkers into a single risk model for the management of IPNs. The model uses a novel assay methodology for hs-CYFRA 21-1 biomarker quantification and novel quantitative image feature analysis. The CBM enabled improved performance relative to the current standard of care, the Mayo (Table 2), while requiring only a digital analysis of the same CT scan on which the nodule was found and a rapid, inexpensive blood test. Thus, the CBM allows risk to be determined for a patient with no extra radiation exposure or delay.
Both the blood and imaging biomarkers were first independently trained in an initial cohort (VUMC) and tested in three external validation cohorts (DECAMP, UPMC, and UC Denver). The blood biomarker (hs-CYFRA) assay is based on a label-free FSA measured by CIR (13, 20). The FSA-CIR approach enables matrix-insensitive biomarker quantification assays using a simple, rapid mix-and-read method that provides analytical sensitivities that outperform existing technologies (29). Here we chose to focus on CYFRA 21-1 as the only blood biomarker because it is repeatedly demonstrated to be the most useful single biomarker for IPN diagnosis (20, 30–33). CYFRA 21-1 is a cytokeratin fragment resulting from epithelial cell death and is a highly sensitive marker of epithelial inflammation. The high sensitivity of hs-CYFRA complements the high specificity of radiomics techniques (34). Several recent studies have demonstrated new biomarkers with added value for lung cancer diagnosis, and incorporating these and others will be the focus of future investigations (35–40).
The hypothesis of this study was that a classifier combining clinical variables with blood and imaging biomarkers could provide improved diagnostic accuracy compared with any of the constituent biomarkers. To evaluate this hypothesis, a combined risk classifier was trained on the VUMC cohort on the basis of the three biomarkers (clinical, blood, and imaging) and validated in the external cohorts (Figure 1), with significant improvement over the baseline model across all cohorts, and a shift in the population out of the intermediate-risk group (Figure E3). It is worth pointing out that the DECAMP cohort was more closely matched across all known clinical risk factors (Table 1), and the baseline clinical prediction model provided almost no discrimination between cases and control subjects. However, even in this matched population, the CBM provided improved predictive accuracy over the clinical model, with the relative improvement over the clinical baseline model of similar magnitude as the other cohorts. This result is contrasted with the UPMC cohort, where the baseline clinical prediction model provided good discrimination on its own (AUC of 0.86), but the CBM significantly improved discrimination in this cohort as well (AUC of 0.94). These results suggest that the CBM confers improved diagnostic accuracy in a range of clinical scenarios.
We then developed the model for clinical implementation. Once the CBM approach was validated, the four cohorts were combined to refit and recalibrate the model to maximize the generalizability of the model for future clinical implementation studies. All cohorts (N = 456) were combined, and the CBM signature was retrained and calibrated to a clinically relevant prevalence of cancer of 0.33 (Table 2). The prevalence of malignancy in any IPN population varies with the context. Screening contexts see from 2% in lung cancer screening trials to a high of 83% in surgical clinics (3, 41, 42). In lung nodule clinics, prevalence in past studies has ranged from the below 20% (16% in the pulmonary nodule plasma proteomic classifier study [40] and 17% in a multidisciplinary study [43]), to 25% in the community setting (44), to 48% in a tertiary care center pulmonary nodule clinic (42). We selected 0.33 as the prevalence to calibrate our model to, as it provides a middle ground between these past study results and corresponds to the expected prevalence of the population in which we intend to use this model in the future.
The potential clinical implementation of this model was evaluated by selecting risk thresholds for patient management of 0.10 (rule out) and 0.70 (rule in), as suggested by the BTS. The BTS guidelines align more closely with our clinical preference than those of the ACCP guidelines (rule in at 0.65 and rule out at 0.05). However, the same analysis presented here is included in the supplementary materials using the ACCP guidelines’ suggested thresholds.
Of the 435 nodules that were intermediate risk by the Mayo, 182 (42% of all intermediate-risk nodules) were correctly reclassified into high or low risk, with only 5 false-negative and 5 false-positive results, an incorrect reclassification rate of 2% (Figure 2). Interestingly, one of the false-negative results involved an indolent tumor that was finally resected 5 years after initial nodule detection. Therefore, a management decision based on the CBM to undergo follow-up imaging would not have been detrimental to this patient. A table of incorrectly classified patients is included in Table E3. The bias-corrected cNRI showed a strong ability to correctly reclassify intermediate-risk IPNs in both rule in (56 of 166, cNRI 0.15, 95% CI: 0.10–0.20) and rule out (53 of 142, cNRI 0.12, 95% CI: 0.07–0.18) scenarios. Based on a preliminary clinical utility analysis, management of patients with Mayo-intermediate-risk IPNs as guided by the CBM and following BTS risk thresholds suggests a possible reduction in the number of unnecessary bronchoscopies from 62.9% to 50.6% in patients with benign nodules. For patients with a malignancy, management by CBM and BTS, the estimated decrease in median time to diagnosis was from 60 days to 21 days, a duration with potential for a downward shift in cancer stage (45) and improved survival among early-stage cancers (46). These utility estimates should be viewed as speculative with further elaboration and validation in a prospective clinical trial.
The current usual care for evaluating intermediate-risk pulmonary nodules is to consider fluorodeoxyglucose-PET; however, recent studies have demonstrated that PET is not ideal in this scenario. PET is sensitive (89%) but has poor specificity (61%) for evaluating IPNs in areas of endemic fungal infections (47). PET sensitivity drops significantly as nodule size decreases, and PET can be falsely negative in partly or nonsolid nodules (48, 49) and some carcinoid tumors. In addition, PET scans are expensive, and the FDG-PET modality is not easily accessible to the global community. We posit that PET is best indicated for lung cancer staging (50), and a diagnostic biomarker test with similar added value as PET would decrease cost. In this study population, diagnostic PET was performed for 75 patients in the VUMC cohort and 40 patients in the UC Denver cohort. When incorporating the diagnostic PET avidity into risk assessment using the Herder model, there was a slight improvement over the Mayo in the UC Denver cohort, but a decrease in accuracy in the VUMC cohort. While this is an unrepresentative sample population, this agrees with past observations about the usefulness of PET within these populations (38, 51). In both populations, the CBM outperformed the Herder model.
The research presented here has several potential limitations. First, CYFRA 21-1 is a strong marker of epithelial injury in general, and therefore the presence of epithelial comorbidities (such as COPD) could confound results (52). Second, several recent studies have demonstrated improved sensitivity and specificity for IPNs by analyzing a panel of protein (53), circulating tumor cells (54), or cell-free DNA (55) biomarkers. The CBM may benefit from inclusion of additional blood biomarkers in the future. The third is because of the presence of the soil fungus Histoplasma capsulatum, endemic to the region of patients who were enrolled in the VUMC cohort, the prevalence of fungal granulomas is disproportionately larger than in the general high-risk population (47). This observation may affect the generalizability of the results, but it nonetheless demonstrates that the CBM can discriminate between some of the most challenging benign nodules. The fourth is the heterogeneity of CT scans, especially variations in the standard of care, including the scan slice thickness (0.67–3 mm) (56), as well as presence or absence of contrast agents used. However, we performed analysis of feature selection on only noncontrast CT scans and then using all CT scans, observing that the selected features and overall performance of the model warranted inclusion of both sets of CT scan data (AUC of 0.609 for a model trained on noncontrast CT scans, 0.632 for a model trained on contrast and noncontrast). Finally, several clinical models exist for use within the same context, including the widely validated Brock University model. However, in the VUMC cohort, the predictive accuracy of the Brock University model was not different from the Mayo.
Ideally, IPN management decisions are made by a multidisciplinary team of experts, in consultation with the patient through a shared decision-making process, examining the likelihood of IPN malignancy in combination with other factors, such as the potential benefit of a cure to the patient and potential risk for surgical complications. Quantitative risk models provide a solid foundation to these decision-making processes; thus, improvement of these risk models should help to improve patient care. For implementation studies of our model, the coefficients of this calibrated model are included in a downloadable calculator that is provided with the supplemental information and available at the Massion Laboratory website.
Acknowledgments
Acknowledgment
The authors thank Tracey Marsh, Ph.D., Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, for careful review of and suggestions made to strengthen the work. They also thank the DECAMP Consortium, and a list of DECAMP investigators is included in the supplemental information. They also thank Katie Dickerson of HealthMyne technical support for incredible service throughout the project.
Footnotes
This work is dedicated to Pierre Massion, our inspiration, mentor, teacher, counselor, colleague, friend, and guiding light. May we honor his legacy in our continued work to improve outcomes in lung cancer through our study of lung cancer development, biomarkers, and early detection. Every step forward will be taken down a path you have paved. May we always be inspired by patients, driven by science, and empowered by each other.
Supported by NIH (U01CA186145, U01CA200464, U01CA143062, U01CA152662-06, U01CA214165, R01CA213123, 2P30CA047904, and P50CA058187), NSF (CHE1610964), and the Lung Cancer Research Foundation.
Author Contributions: M.N.K., D.A.L., A.E.B., H.C., D.J.B., and P.P.M. designed the research. M.N.K., A.K.K., R.L.W., and D.J.B. performed blood biomarker measurements. M.N.K., D.A.L., A.B.B., S.M., S.H., U.B., C.S., M.B.S., D.J.R, and R.J.G. performed radiomics analysis. S.L.A., T.A., M.N., E.H., W.J.F., J.S., M.R., J.B., D.O.W., F.M., E.L.G., R.C.W., and P.P.M. performed patient consenting and chart review. M.N.K., S.-C.C., A.E.B., H.C., and P.P.M. performed study design, biostatistical analysis, and model interpretation. M.N.K and P.P.M. wrote the manuscript. B.D., A.E.B., H.C., and D.J.B. edited the manuscript. All authors approved the final version to be published.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.202012-4438OC on August 31, 2021
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Gould MK, Tang T, Liu I-LA, Lee J, Zheng C, Danforth KN, et al. Recent trends in the identification of incidental pulmonary nodules. Am J Respir Crit Care Med . 2015;192:1208–1214. doi: 10.1164/rccm.201505-0990OC. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin . 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3. Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, et al. National Lung Screening Trial Research Team. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med . 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lokhandwala T, Bittoni MA, Dann RA, D’Souza AO, Johnson M, Nagy RJ, et al. Costs of diagnostic assessment for lung cancer: a Medicare claims analysis. Clin Lung Cancer . 2017;18:e27–e34. doi: 10.1016/j.cllc.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 5. Ost DE, Gould MK. Decision making in patients with pulmonary nodules. Am J Respir Crit Care Med . 2012;185:363–372. doi: 10.1164/rccm.201104-0679CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gould MK, Ananth L, Barnett PG. Veterans Affairs SNAP Cooperative Study Group. A clinical model to estimate the pretest probability of lung cancer in patients with solitary pulmonary nodules. Chest . 2007;131:383–388. doi: 10.1378/chest.06-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Swensen SJ, Silverstein MD, Ilstrup DM, Schleck CD, Edell ES. The probability of malignancy in solitary pulmonary nodules: application to small radiologically indeterminate nodules. Arch Intern Med . 1997;157:849–855. [PubMed] [Google Scholar]
- 8. McKee B, McKee A, French R, Hesketh P, Wald C, Flacke S. “LUNG-RADS” a proposed standardized reporting and data system for CT lung cancer screening. J Thorac Oncol . 2012;7:S277–S278. [Google Scholar]
- 9. Wood DE, Kazerooni EA, Baum SL, Eapen GA, Ettinger DS, Hou L, et al. Lung cancer screening, version 3.2018, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw . 2018;16:412–441. doi: 10.6004/jnccn.2018.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baldwin DR, Callister ME. Guideline Development Group. The British Thoracic Society guidelines on the investigation and management of pulmonary nodules. Thorax . 2015;70:794–798. doi: 10.1136/thoraxjnl-2015-207221. [DOI] [PubMed] [Google Scholar]
- 11. Mazzone P, Powell CA, Arenberg D, Bach P, Detterbeck F, Gould MK, et al. Components necessary for high-quality lung cancer screening: American College of Chest Physicians and American Thoracic Society policy statement. Chest . 2015;147:295–303. doi: 10.1378/chest.14-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. MacMahon H, Naidich DP, Goo JM, Lee KS, Leung ANC, Mayo JR, et al. Guidelines for management of incidental pulmonary nodules detected on CT images: from the Fleischner Society 2017. Radiology . 2017;284:228–243. doi: 10.1148/radiol.2017161659. [DOI] [PubMed] [Google Scholar]
- 13. Bornhop DJ, Kammer MN, Kussrow A, Flowers RA, II, Meiler J. Origin and prediction of free-solution interaction studies performed label-free. Proc Natl Acad Sci U S A . 2016;113:E1595–E1604. doi: 10.1073/pnas.1515706113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clay R, Kipp BR, Jenkins S, Karwoski RA, Maldonado F, Rajagopalan S, et al. Computer-aided nodule assessment and risk yield (CANARY) may facilitate non-invasive prediction of EGFR mutation status in lung adenocarcinomas. Sci Rep . 2017;7:17620. doi: 10.1038/s41598-017-17659-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aerts H, Velazquez E, Leijenaar R, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun . 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mandrekar SJ, Sargent DJ. Clinical trial designs for predictive biomarker validation: theoretical considerations and practical challenges. J Clin Oncol . 2009;27:4027–4034. doi: 10.1200/JCO.2009.22.3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Billatos E, Duan F, Moses E, Marques H, Mahon I, Dymond L, et al. DECAMP investigators. Detection of early lung cancer among military personnel (DECAMP) consortium: study protocols. BMC Pulm Med . 2019;19:59. doi: 10.1186/s12890-019-0825-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deppen SA, Grogan EL. Using clinical risk models for lung nodule classification. Semin Thorac Cardiovasc Surg . 2015;27:30–35. doi: 10.1053/j.semtcvs.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McWilliams A, Tammemagi MC, Mayo JR, Roberts H, Liu G, Soghrati K, et al. Probability of cancer in pulmonary nodules detected on first screening CT. N Engl J Med . 2013;369:910–919. doi: 10.1056/NEJMoa1214726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kammer MN, Kussrow AK, Webster RL, Chen H, Hoeksema M, Christenson R, et al. Compensated interferometry measures of CYFRA 21-1 improve diagnosis of lung cancer. ACS Comb Sci . 2019;21:465–472. doi: 10.1021/acscombsci.9b00022. [DOI] [PubMed] [Google Scholar]
- 21. Herder GJ, van Tinteren H, Golding RP, Kostense PJ, Comans EF, Smit EF, et al. Clinical prediction model to characterize pulmonary nodules: validation and added value of 18F-fluorodeoxyglucose positron emission tomography. Chest . 2005;128:2490–2496. doi: 10.1378/chest.128.4.2490. [DOI] [PubMed] [Google Scholar]
- 22. Huang Y, Pepe MS. Assessing risk prediction models in case-control studies using semiparametric and nonparametric methods. Stat Med . 2010;29:1391–1410. doi: 10.1002/sim.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Van Calster B, McLernon DJ, van Smeden M, Wynants L, Steyerberg EW. Topic Group ‘Evaluating diagnostic tests and prediction models’ of the STRATOS initiative. Calibration: the Achilles heel of predictive analytics. BMC Med . 2019;17:230. doi: 10.1186/s12916-019-1466-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paynter NP, Cook NR. A bias-corrected net reclassification improvement for clinical subgroups. Med Decis Making . 2013;33:154–162. doi: 10.1177/0272989X12461856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pepe MS, Kerr KF, Longton G, Wang Z. Testing for improvement in prediction model performance. Stat Med . 2013;32:1467–1482. doi: 10.1002/sim.5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Youden WJ. Index for rating diagnostic tests. Cancer . 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 27. Demler OV, Pencina MJ, D’Agostino RB., Sr Misuse of DeLong test to compare AUCs for nested models. Stat Med . 2012;31:2577–2587. doi: 10.1002/sim.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vickers AJ, Cronin AM, Begg CB. One statistical test is sufficient for assessing new predictive markers. BMC Med Res Methodol . 2011;11:13. doi: 10.1186/1471-2288-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kammer M, Kussrow A, Carter MD, Isenberg SL, Johnson RC, Batchelor RH, et al. Rapid quantification of two chemical nerve agent metabolites in serum. Biosens Bioelectron . 2019;131:119–127. doi: 10.1016/j.bios.2019.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pujol JL, Grenier J, Daurès JP, Daver A, Pujol H, Michel FB. Serum fragment of cytokeratin subunit 19 measured by CYFRA 21-1 immunoradiometric assay as a marker of lung cancer. Cancer Res . 1993;53:61–66. [PubMed] [Google Scholar]
- 31. Seemann MD, Beinert T, Fürst H, Fink U. An evaluation of the tumour markers, carcinoembryonic antigen (CEA), cytokeratin marker (CYFRA 21-1) and neuron-specific enolase (NSE) in the differentiation of malignant from benign solitary pulmonary lesions. Lung Cancer . 1999;26:149–155. doi: 10.1016/s0169-5002(99)00084-7. [DOI] [PubMed] [Google Scholar]
- 32.Guida F, Sun N, Bantis LE, Muller DC, Li P, Taguchi A, et al. Integrative Analysis of Lung Cancer Etiology and Risk (INTEGRAL) Consortium for Early Detection of Lung Cancer assessment of lung cancer risk on the basis of a biomarker panel of circulating proteins JAMA Oncol 20184e182078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang D, Zhang X, Powell CA, Ni J, Wang B, Zhang J, et al. Probability of cancer in high-risk patients predicted by the protein-based lung cancer biomarker panel in China: LCBP study. Cancer . 2018;124:262–270. doi: 10.1002/cncr.31020. [DOI] [PubMed] [Google Scholar]
- 34. Balagurunathan Y, Schabath MB, Wang H, Liu Y, Gillies RJ. Quantitative imaging features improve discrimination of malignancy in pulmonary nodules. Sci Rep . 2019;9:8528. doi: 10.1038/s41598-019-44562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trivedi NN, Arjomandi M, Brown JK, Rubenstein T, Rostykus AD, Esposito S, et al. Risk assessment for indeterminate pulmonary nodules using a novel, plasma-protein based biomarker assay. Biomed Res Clin Pract . doi: 10.15761/brcp.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Edelsberg J, Weycker D, Atwood M, Hamilton-Fairley G, Jett JR. Cost-effectiveness of an autoantibody test (EarlyCDT-Lung) as an aid to early diagnosis of lung cancer in patients with incidentally detected pulmonary nodules. PLoS One . 2018;13:e0197826. doi: 10.1371/journal.pone.0197826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lastwika KJ, Kargl J, Zhang Y, Zhu X, Lo E, Shelley D, et al. Tumor-derived autoantibodies identify malignant pulmonary nodules. Am J Respir Crit Care Med . 2019;199:1257–1266. doi: 10.1164/rccm.201804-0628OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deppen SA, Massion PP, Blume J, Walker RC, Antic S, Chen H, et al. Accuracy of a novel histoplasmosis enzyme immunoassay to evaluate suspicious lung nodules. Cancer Epidemiol Biomarkers Prev . 2019;28:321–326. doi: 10.1158/1055-9965.EPI-18-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Patz EF, Jr, Campa MJ, Gottlin EB, Trotter PR, Herndon JE, II, Kafader D, et al. Biomarkers to help guide management of patients with pulmonary nodules. Am J Respir Crit Care Med . 2013;188:461–465. doi: 10.1164/rccm.201210-1760OC. [DOI] [PubMed] [Google Scholar]
- 40. Silvestri GA, Tanner NT, Kearney P, Vachani A, Massion PP, Porter A, et al. PANOPTIC Trial Team. Assessment of plasma proteomics biomarker’s ability to distinguish benign from malignant lung nodules: results of the PANOPTIC (pulmonary nodule plasma proteomic classifier) trial. Chest . 2018;154:491–500. doi: 10.1016/j.chest.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Grogan EL, Deppen SA, Ballman KV, Andrade GM, Verdial FC, Aldrich MC, et al. Accuracy of fluorodeoxyglucose-positron emission tomography within the clinical practice of the American College of Surgeons Oncology Group Z4031 trial to diagnose clinical stage I non-small cell lung cancer. Ann Thorac Surg . 2014;97:1142–1148. doi: 10.1016/j.athoracsur.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maiga AW, Deppen SA, Massion PP, Callaway-Lane C, Pinkerman R, Dittus RS, et al. Communication about the probability of cancer in indeterminate pulmonary nodules. JAMA Surg . 2018;153:353–357. doi: 10.1001/jamasurg.2017.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Madariaga ML, Lennes IT, Best T, Shepard JO, Fintelmann FJ, Mathisen DJ, Gaissert HA. MGH Pulmonary Clinic Collaborative. Multidisciplinary selection of pulmonary nodules for surgical resection: Diagnostic results and long-term outcomes. J Thorac Cardiovasc Surg . 2020;159:1558–1566.e3. doi: 10.1016/j.jtcvs.2019.09.030. [DOI] [PubMed] [Google Scholar]
- 44. Tanner NT, Aggarwal J, Gould MK, Kearney P, Diette G, Vachani A, et al. Management of pulmonary nodules by community pulmonologists: a multicenter observational study. Chest . 2015;148:1405–1414. doi: 10.1378/chest.15-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gould MK, Sanders GD, Barnett PG, Rydzak CE, Maclean CC, McClellan MB, et al. Cost-effectiveness of alternative management strategies for patients with solitary pulmonary nodules. Ann Intern Med . 2003;138:724–735. doi: 10.7326/0003-4819-138-9-200305060-00009. [DOI] [PubMed] [Google Scholar]
- 46. Maiga AW, Deppen SA, Pinkerman R, Callaway-Lane C, Massion PP, Dittus RS, et al. Timeliness of care and lung cancer tumor-stage progression: how long can we wait? Ann Thorac Surg . 2017;104:1791–1797. doi: 10.1016/j.athoracsur.2017.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Deppen SA, Blume JD, Kensinger CD, Morgan AM, Aldrich MC, Massion PP, et al. Accuracy of FDG-PET to diagnose lung cancer in areas with infectious lung disease: a meta-analysis. JAMA . 2014;312:1227–1236. doi: 10.1001/jama.2014.11488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nomori H, Watanabe K, Ohtsuka T, Naruke T, Suemasu K, Uno K. Evaluation of F-18 fluorodeoxyglucose (FDG) PET scanning for pulmonary nodules less than 3 cm in diameter, with special reference to the CT images. Lung Cancer . 2004;45:19–27. doi: 10.1016/j.lungcan.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 49. Veronesi G, Travaini LL, Maisonneuve P, Rampinelli C, Bertolotti R, Spaggiari L, et al. Positron emission tomography in the diagnostic work-up of screening-detected lung nodules. Eur Respir J . 2015;45:501–510. doi: 10.1183/09031936.00066514. [DOI] [PubMed] [Google Scholar]
- 50. Kandathil A, Kay FU, Butt YM, Wachsmann JW, Subramaniam RM. Role of FDG PET/CT in the eighth edition of TNM staging of non-small cell lung cancer. Radiographics . 2018;38:2134–2149. doi: 10.1148/rg.2018180060. [DOI] [PubMed] [Google Scholar]
- 51. Maiga AW, Deppen SA, Mercaldo SF, Blume JD, Montgomery C, Vaszar LT, et al. Assessment of fluorodeoxyglucose F18-labeled positron emission tomography for diagnosis of high-risk lung nodules. JAMA Surg . 2018;153:329–334. doi: 10.1001/jamasurg.2017.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu RH, Liao CZ, Luo Y, Xu WL, Li K, Chen JX, et al. Optimal cut-off values for CYFRA 21-1 expression in NSCLC patients depend on the presence of benign pulmonary diseases. Clin Chim Acta . 2015;440:188–192. doi: 10.1016/j.cca.2014.09.033. [DOI] [PubMed] [Google Scholar]
- 53. Molina R, Marrades RM, Augé JM, Escudero JM, Viñolas N, Reguart N, et al. Assessment of a combined panel of six serum tumor markers for lung cancer. Am J Respir Crit Care Med . 2016;193:427–437. doi: 10.1164/rccm.201404-0603OC. [DOI] [PubMed] [Google Scholar]
- 54. Krebs MG, Sloane R, Priest L, Lancashire L, Hou JM, Greystoke A, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol . 2011;29:1556–1563. doi: 10.1200/JCO.2010.28.7045. [DOI] [PubMed] [Google Scholar]
- 55. Pathak AK, Bhutani M, Kumar S, Mohan A, Guleria R. Circulating cell-free DNA in plasma/serum of lung cancer patients as a potential screening and prognostic tool. Clin Chem . 2006;52:1833–1842. doi: 10.1373/clinchem.2005.062893. [DOI] [PubMed] [Google Scholar]
- 56. Shafiq-Ul-Hassan M, Latifi K, Zhang G, Ullah G, Gillies R, Moros E. Voxel size and gray level normalization of CT radiomic features in lung cancer. Sci Rep . 2018;8:10545. doi: 10.1038/s41598-018-28895-9. [DOI] [PMC free article] [PubMed] [Google Scholar]