Abstract
Rationale
Two distinct subphenotypes have been identified in acute respiratory distress syndrome (ARDS), but the presence of subgroups in ARDS associated with coronavirus disease (COVID-19) is unknown.
Objectives
To identify clinically relevant, novel subgroups in COVID-19–related ARDS and compare them with previously described ARDS subphenotypes.
Methods
Eligible participants were adults with COVID-19 and ARDS at Columbia University Irving Medical Center. Latent class analysis was used to identify subgroups with baseline clinical, respiratory, and laboratory data serving as partitioning variables. A previously developed machine learning model was used to classify patients as the hypoinflammatory and hyperinflammatory subphenotypes. Baseline characteristics and clinical outcomes were compared between subgroups. Heterogeneity of treatment effect for corticosteroid use in subgroups was tested.
Measurements and Main Results
From March 2, 2020, to April 30, 2020, 483 patients with COVID-19–related ARDS met study criteria. A two-class latent class analysis model best fit the population (P = 0.0075). Class 2 (23%) had higher proinflammatory markers, troponin, creatinine, and lactate, lower bicarbonate, and lower blood pressure than class 1 (77%). Ninety-day mortality was higher in class 2 versus class 1 (75% vs. 48%; P < 0.0001). Considerable overlap was observed between these subgroups and ARDS subphenotypes. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RT-PCR cycle threshold was associated with mortality in the hypoinflammatory but not the hyperinflammatory phenotype. Heterogeneity of treatment effect to corticosteroids was observed (P = 0.0295), with improved mortality in the hyperinflammatory phenotype and worse mortality in the hypoinflammatory phenotype, with the caveat that corticosteroid treatment was not randomized.
Conclusions
We identified two COVID-19–related ARDS subgroups with differential outcomes, similar to previously described ARDS subphenotypes. SARS-CoV-2 PCR cycle threshold had differential value for predicting mortality in the subphenotypes. The subphenotypes had differential treatment responses to corticosteroids.
Keywords: ARDS, COVID-19, latent class analysis, phenotyping
At a Glance Commentary
Scientific Knowledge on the Subject
Acute respiratory distress syndrome (ARDS) is a frequent sequela of severe coronavirus disease (COVID-19). Although hyper- and hypoinflammatory subphenotypes defined by unique clinical characteristics and biomarkers have been consistently identified among patients with non–COVID-19–related ARDS, and clinical outcomes among severely ill patients with COVID-19 are variable, the existence of distinct subgroups among patients with COVID-19–related ARDS remains unknown.
What This Study Adds to the Field
Using latent class analysis, we identified two subgroups among a cohort of 483 patients with COVID-19–related ARDS. Class 2 patients had higher inflammatory markers and lactate and corresponded with the previously identified hyperinflammatory subphenotype, whereas class 1 corresponded with the hypoinflammatory subphenotype. Class 2 had significantly higher 90-day mortality than class 1 (75% vs. 48%; P < 0.0001). Differential response to corticosteroid treatment was observed, with decreased mortality in steroid-treated patients in class 2 but not class 1. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) PCR cycle threshold was a predictor of mortality in class 1, but not class 2, suggesting distinct drivers of mortality among classes.
A substantial proportion of patients with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection develop the acute respiratory distress syndrome (ARDS) (1–4). Despite emerging treatment options for coronavirus disease (COVID-19), such as dexamethasone and IL-6 receptor antagonists, mortality among patients with COVID-19–related ARDS remains high (5, 6). Although some prognostic features in severe COVID-19 have been identified, the determinants of the individual variability in treatment response and outcomes remain incompletely understood (1–3).
ARDS not associated with COVID-19 is characterized by clinical and biological heterogeneity, which has been implicated as a contributing factor in negative clinical trials aimed at developing treatment strategies (7, 8). To address this heterogeneity, prior work has consistently identified two subphenotypes in non–COVID-19–related ARDS defined by differential profiles of inflammation, organ failure, and outcomes, as well as responses to randomized treatment interventions (9–11). Recently, these two subphenotypes (hyper- and hypoinflammatory) were identified in a small cohort of 39 patients with COVID-19–related ARDS (12). However, it remains unclear whether distinct biological and clinical subgroups exist in COVID-19–related ARDS and whether these subgroups are distinct from previously described non–COVID-19– related ARDS subphenotypes.
The primary objective of this study was to identify novel subgroups in COVID-19–related ARDS using minimally biased clustering methods and to evaluate the clinical and prognostic relevance of any resulting subgroups using 90-day outcome data. Using a previously described clinical classifier model, we also set out to compare any identified COVID-19–related ARDS subgroups with the previously described hypo- and hyperinflammatory ARDS subphenotypes (13).
Methods
Study Population
Among adult (age ⩾ 18) patients admitted to the ICUs at two hospitals affiliated with NewYork-Presbyterian Hospital/Columbia University Irving Medical Center (NYP/CUIMC) from March 2, 2020, through April 30, 2020, patients were included in this study if they fulfilled the following criteria: 1) had a positive PCR test for SARS-CoV-2, 2) required invasive mechanical ventilation (IMV), and 3) met criteria for ARDS, using the Berlin definition based on a review of clinical data and chest radiographs assessed by critical care physicians (4). Exclusion criteria included death before ICU admission, tracheostomy before ICU admission, or endotracheal intubation before transfer from an outside hospital. The latter group was excluded owing to lack of consistent available clinical data within 24 hours of intubation for patients transferred already on IMV and to mitigate potential selection bias of which patients were transferred during the early surge of the pandemic in New York City. Unless stated otherwise, all laboratory, ventilatory, and vital sign variables used for analysis were recorded on the initial day of IMV. The study was approved by the institutional review board at CUIMC.
Data Collection
Electronic medical records were reviewed for data on demographic, clinical, and laboratory variables for all included patients. Vital signs, ventilatory measurements, and routine laboratory studies that resulted within 24 hours of initiation of IMV were used for analysis. For blood-based biomarkers (IL-6, B-type natriuretic peptide, high-sensitivity C-reactive protein, D-dimer, ferritin, lactate dehydrogenase, lactate, high-sensitivity cardiac troponin T, procalcitonin, international normalized ratio, and activated partial thromboplastin time), data were extracted when measured as part of clinical care. For patients without IL-6 measured clinically, prospectively stored samples were tested, methods of which are described below.
Interventions delivered throughout hospitalization were recorded, including targeted treatments for COVID-19 (e.g., corticosteroids, IL-6 receptor antagonists), receipt of tracheostomy, vasopressor use, renal replacement therapy, and advanced therapies for ARDS (neuromuscular blockade, ventilation in the prone position, and extracorporeal membrane oxygenation).
The primary outcome of this study was vital status at 90 days after intubation. Secondary outcomes included vital status at 28 and 180 days after intubation, ICU length of stay, and ventilator-free days at 90 days after intubation. For the primary outcome, follow-up time was right censored on July 29, 2020, so that each patient had at least 90 days of observation.
Latent Class Analysis
Clinical data including demographics, vital signs, and laboratory values were evaluated as class-defining variables for latent class analysis (LCA). Demographics and vital signs were used in accordance with LCA for prior ARDS subphenotyping (9–11). The study design enabled inclusion of several novel laboratory and respiratory variables that were not used in prior LCA studies, such as lactate, troponin, procalcitonin, ferritin, and ventilatory ratio (14). Variables with >25% missingness (Table E1 in the online supplement), those with >15% of values outside the upper limit of assay detection, and highly correlated variables were excluded from the LCA (see the online supplement for details). Clinical outcomes and illness severity scores were not included as class-defining variables, and modeling was performed agnostic of these variables. The variables used for LCA are summarized in Table E2.
For LCA, we fit four sequential models consisting of 1–4 classes. The LCA algorithms used were similar to those in our prior studies and are described further in the online supplement (9–11). The best-fitting model was selected based on the Bayesian information criteria, entropy, and the Vuong-Lo-Mendall-Ruben test (15). Once the best model was selected, an individual patient’s class assignment was determined by the highest probability of class membership (⩾0.5). LCA was performed using Mplus8 editor version 1.7.
Clinical Classifier Model
To compare the overlap of the newly identified COVID-19–related ARDS subgroups with previously described ARDS subphenotypes, we estimated the probabilities of belonging to the hyperinflammatory subphenotype using a previously validated clinical classifier model (16). Given that COVID-19–related ARDS has a uniform etiology and the demographics of the study population were markedly different from those in which the clinical classifier models were developed, we applied a model composed of vital signs and laboratory values only. In the original study, this model had an area under the receiver operator characteristics curve of 0.94 for discriminating ARDS subphenotypes (16). The variables used in the clinical classifier model are described in Table E2. A probability of ⩾0.5 was used to assign patients to the hyperinflammatory subphenotype, and <0.5 to the hypoinflammatory subphenotype.
Variable of Importance for Mortality
Given the uncertain validity of standard critical care scoring systems such as Acute Physiology and Chronic Health Evaluation-II and Sequential Organ Failure Assessment (SOFA) scores in COVID-19, to evaluate the additional prognostic value of the identified LCA classes in comparison with cohort-specific mortality predictors in COVID-19–related ARDS, we used a supervised learning approach to identify the most important variables in predicting mortality in our cohort (12, 17). The rationale for developing these models was to identify the most important variables, among those used in the LCA modeling, in predicting mortality in this cohort. The models were not intended as a clinical prediction tool. Specifically, we used XGBoost, a gradient boosted tree algorithm, to train a model to predict mortality at Day 90 from intubation (primary outcome). Predictor variables used in this model are described in Table E2. Variable importance was calculated using the split gain for each variable across the different trees in the model. The analyses were performed using the R package XGBoost and algorithmic procedures are detailed in the online supplement.
Biospecimen Procedures
Biospecimens were obtained from the Columbia University Biobank, which is supported by the Irving Institute for Clinical and Translational Research and Columbia University’s Clinical and Translational Science Award. For patients who did not have IL-6 measured clinically, IL-6 was quantified in prospectively banked serum samples using the Human IL-6 Quantikine ELISA Kit (R&D Systems), the same assay that was used on clinical samples. SARS-CoV-2 PCR testing was performed on nasopharyngeal swab samples. PCR was performed in the clinical microbiology laboratory at CUIMC using platforms that amplify the Envelope (E) gene within the SARS-CoV-2 genome (cobas 6800 [Roche Molecular Systems] and Infinity [Cephid Inc]). Cycle threshold (CT)—the number of PCR cycles required to reach the fluorescent threshold for a positive test—was recorded if PCR tests were positive within 7 days of intubation (18–21). Most CT values (60%) were within 24 hours of intubation and 80% were within 3 days.
Statistical Analysis
Comparison between groups was performed using t tests, Wilcoxon-rank sum, or Pearson’s chi-square tests, depending on the distribution of the variable. Kaplan-Meier survival curves, censored at Day 90, were plotted to compare survival across groups. For evaluation of the prognostic value of individual variables and for exploration of treatment interaction between subphenotypes and corticosteroid exposure, logistic regression models were used to estimate odds ratios, with mortality at Day 90 as the dependent variable. Analyses were performed in RStudio (version 1.1.453) using R (version 4.0.1) or in STATA/IC version 16.1 (StataCorp LLC).
Results
Of 558 screened patients, 483 were included in this analysis (Figure E1 and Table 1). The mean age was 64 (±14) years, and 164 patients (34%) were female. Most patients (59%) were Hispanic. The median time from hospitalization to initiation of IMV was 1 day (interquartile range [IQR], 0–4), and the median duration of symptoms before admission was 7 days (IQR, 4–9). Shock and multiorgan dysfunction were common, with 407 (84%) patients requiring vasopressors and 150 (31%) requiring renal replacement therapy. Most patients received targeted therapies for COVID-19, including hydroxychloroquine (395/483, 82%), corticosteroids (322/483, 67%), IL-6 receptor antagonists (141/483, 29%), and remdesivir (28/483, 6%). Neuromuscular blockade within 48 hours of intubation was administered to 160 (33%) patients, 111 (23%) received prone positioning, 10 (2%) received extracorporeal membrane oxygenation, and 142 (29%) underwent tracheostomy. At Day 90, 260 patients (54%) had died, 206 (43%) had been discharged from the hospital alive, and 17 (4%) remained hospitalized. Twenty-eight-day and 180-day mortality were 44% (211/483) and 55% (265/483), respectively. The median length of stay in the ICU was 32 days (IQR, 15–43), and median ventilator-free days was 0 (IQR, 0–57).
Table 1.
Baseline Characteristics and Therapy Use in the COVID-19 ARDS Cohort with Stratification by COVID-19–related ARDS Classes
| Variables | Total Cohort (N = 483) | Class 1 (N = 373) | Class 2 (N = 110) | P Value |
|---|---|---|---|---|
| Age, yr | 65 ± 14 | 63 ± 15 | 67 ± 12 | 0.0025 |
| Sex | ||||
| F | 164 (34) | 125 (34) | 39 (36) | 0.7922 |
| M | 319 (66) | 248 (67) | 71 (65) | |
| Race | ||||
| Black | 86 (18) | 62 (17) | 24 (22) | 0.1248 |
| White | 56 (12) | 40 (11) | 16 (15) | |
| Hispanic | 285 (59) | 231 (62) | 54 (49) | |
| Other | 31 (6) | 24 (6) | 7 (6) | |
| Unknown (declined to report) | 25 (5) | 16 (4) | 9 (8) | |
| Body mass index | 30.2 ± 7.1 | 30.5 ± 7.2 | 29.3 ± 6.4 | 0.1575 |
| Smoker (current or former) | 98 (20) | 69 (18) | 29 (26) | 0.029 |
| Preadmission symptom duration, d | 7 (4–9) | 7 (4–9) | 7 (3–9) | 0.7555 |
| Time from admission to intubation, d | 1 (0–4) | 1 (0–3) | 1 (0–5) | 0.427 |
| Temperature, °C | 37.6 ± 1.1 | 37.9 ± 1.0 | 37.5 ± 1.2 | 0.0006 |
| Heart rate, beats per min | 111 ± 22 | 107 ± 22 | 123 ± 20 | <0.0001 |
| Respiratory rate, breaths per min | 32 (26–38) | 32 (26–37) | 34 (30–40) | 0.0011 |
| Systolic blood pressure, mm Hg | 88 ± 17 | 90 ± 16 | 82 ± 17 | <0.0001 |
| White blood cells, ×10−9/L | 13.5 ± 7.5 | 12.4 ± 6.4 | 17.2 ± 9.4 | <0.0001 |
| Absolute lymphocyte count, ×10−9/L | 0.8 (0.6–1.2) | 0.8 (0.6–1.2) | 0.9 (0.6–1.3) | 0.5682 |
| Platelets, ×10−9/L | 234 (169–304) | 238 (173–306) | 212 (151–298) | 0.1106 |
| APTT, s | 40 ± 24 | 38 ± 20 | 47 ± 33 | 0.0006 |
| International normalized ratio | 1.4 ± 0.6 | 1.3 ± 0.4 | 1.7 ± 0.9 | <0.0001 |
| PaO2 /Fi O2 | 147 ± 66 | 147 ± 65 | 149 ± 71 | 0.7321 |
| Vt/PBW, ml/kg | 6.5 ± 1.0 | 6.5 ± 1.1 | 6.6 ± 0.9 | 0.4172 |
| PEEP, cm H2O | 13 (10–15) | 12 (10–15) | 13 (10–16) | 0.6558 |
| Compliance, ml/cm H2O | 29 ± 12 | 30 ± 12 | 29 ± 13 | 0.5768 |
| Ventilatory ratio | 2.1 ± 0.9 | 2.0 ± 0.9 | 2.5 ± 1.1 | <0.0001 |
| Creatinine, mg/dL | 1.5 (1.0–2.6) | 1.3 (0.9–1.9) | 2.7 (1.8–4.6) | <0.0001 |
| Bicarbonate, mmol/L | 20 ± 4.8 | 21.5 ± 4.1 | 16.1 ± 4.7 | <0.0001 |
| Glucose, mg/dL | 190 ± 111 | 176 ± 95 | 238 ± 144 | <0.0001 |
| Sodium, mmol/L | 139 ± 7.1 | 139 ± 5.7 | 142 ± 10.1 | <0.0001 |
| Bilirubin, mg/dl | 0.6 (0.4–0.9) | 0.6 (0.4–0.8) | 0.7 (0.5–1.4) | <0.0001 |
| Albumin, g/dl | 3.07 ± 0.53 | 3.14 ± 0.49 | 2.84 ± 0.60 | <0.0001 |
| Lactate, mg/dl | 1.9 (1.3–2.7) | 1.7 (1.2–2.3) | 3.6 (2.5–6.7) | <0.0001 |
| Troponin, ng/L | 38 (16–100) | 27 (13–63) | 135 (69–246) | <0.0001 |
| IL-6, pg/ml | 202 (106–385) | 190 (103–313) | 324 (142–996) | <0.0001 |
| Procalcitonin, ng/ml | 0.9 (0.3–2.4) | 0.6 (0.3–1.4) | 2.7 (1.2–15.4) | <0.0001 |
| Ferritin, mg/dl | 1,179 (664–2,324) | 1,048 (591–2,064) | 1,864 (1,083–3,856) | <0.0001 |
| Lactate dehydrogenase, U/L | 620 (475–847) | 582 (564–656) | 855 (660–1,607) | <0.0001 |
| C-reactive protein, mg/L | 232 (143–300) | 227 (142–300) | 254 (162–300) | 0.1131 |
| D-dimer, mcg/ml | 3.8 (1.7–20) | 3.2 (1.5–13.6) | 12.7 (3.6–20) | <0.0001 |
| Cycle threshold, SARS-CoV-2 PCR | 27.0 ± 5.0 | 26.9 ± 5.1 | 27.5 ± 4.6 | 0.2845 |
| Selected therapies use* | ||||
| Any corticosteroids | 322 (67) | 247 (66) | 76 (69) | 0.539 |
| Dexamethasone | 7 (1) | 6 (2) | 1 (1) | >0.99 |
| Methylprednisolone | 223 (46) | 182 (49) | 41 (37) | 0.033 |
| Hydrocortisone | 78 (16) | 46 (12) | 32 (29) | <0.001 |
| Prednisone | 15 (3) | 13 (3) | 2 (2) | 0.538 |
| IL-6 antagonist | 141 (29) | 118 (32) | 23 (21) | 0.030 |
| Hydroxychloroquine | 395 (82) | 323 (87) | 72 (65) | <0.001 |
| Remdesivir | 28 (6) | 25 (7) | 3 (3) | 0.117 |
| Vasopressor use | 407 (84) | 298 (80) | 109 (99) | <0.0001 |
| Renal replacement therapy* | 150 (31) | 108 (29) | 42 (39) | 0.056 |
| Neuromuscular blockade† | 160 (33) | 127 (34) | 33 (30) | 0.4764 |
| Prone positioning* | 111 (23) | 97 (26) | 14 (13) | 0.0059 |
| ECMO* | 10 (2) | 5 (1.3) | 5 (4.6) | 0.0882 |
| SOFA score‡ | 10 (7–12) | 9 (7–11) | 12 (10–14) | <0.0001 |
Definition of abbreviations: APTT = activated partial thromboplastin time; ARDS = acute respiratory distress syndrome; COVID-19 = coronavirus disease; ECMO = extracorporeal membrane oxygenation; PBW = predicted body weight; PEEP = positive end-expiratory pressure; SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2; SOFA = Sequential Organ Failure Assessment.
Normally distributed data are presented as mean ± SD; nonnormally distributed data are presented as median (interquartile range); and categorical data are presented as n (%). P values represent Student’s t test for normally distributed data, Wilcoxon-rank test for nonnormally distributed data, and chi-square test for categorical values.
These variables were recorded during the course of the ICU admission and not exclusively at baseline.
Variable was recorded within 48 hours of ARDS diagnosis.
If Glasgow Coma Score missing, imputed as 15.
Latent Class Analysis
Fit statistics for the LCA models are presented in Table 2. As model complexity increased, Bayesian information criteria decreased, with the greatest decrease observed between a 1-class and 2-class model. The 2-class model was a significantly better fit than the 1-class model (Vuong-Lo-Mendall-Ruben P = 0.0075), whereas the 3-class and 4-class models were not significant improvements over a model comprising one fewer class. Together, the data were indicative of the 2-class model best fitting the population.
Table 2.
Fit Statistics for the Latent Class Analysis Models
| Classes | BIC | Entropy | N1 | N2 | N3 | N4 | P Value |
|---|---|---|---|---|---|---|---|
| 1 | 39,988 | — | 483 | — | — | — | — |
| 2 | 39,646 | 0.83 | 373 | 110 | — | — | 0.0075 |
| 3 | 39,529 | 0.88 | 355 | 109 | 19 | — | 0.2479 |
| 4* | 39,456 | 0.85 | 270 | 107 | 87 | 19 | 0.6859 |
Definition of abbreviation: BIC = Bayesian information criteria.
Each row represents a single model comprising K classes. N1–N4 are the number of observations in each class and the P value represents the Vuong-Lo-Mendell-Rubin test, which tests whether a model comprising a K class is a better fit than a K-1 class.
Seventeen perturbed starting value run(s) did not converge or were rejected in the third stage.
In the 2-class model, 373/483 (77%) of the patients were assigned to class 1 and 110/483 (23%) to class 2. Indicative of robust class allocation, the mean probabilities for class assignment were 0.96 (±0.09) and 0.92 (±0.12) for COVID-19–related ARDS class 1 and COVID-19–related ARDS class 2, respectively. Demographic differences between classes are summarized in Table 1. Patients assigned to COVID-19–related ARDS class 2 were significantly older (67 vs. 63 years, P = 0.0026), and a higher proportion were Hispanic (62% vs. 49%, P = 0.016). The proportions of patients with hypertension and diabetes mellitus were similar across the two classes (Figure E2).
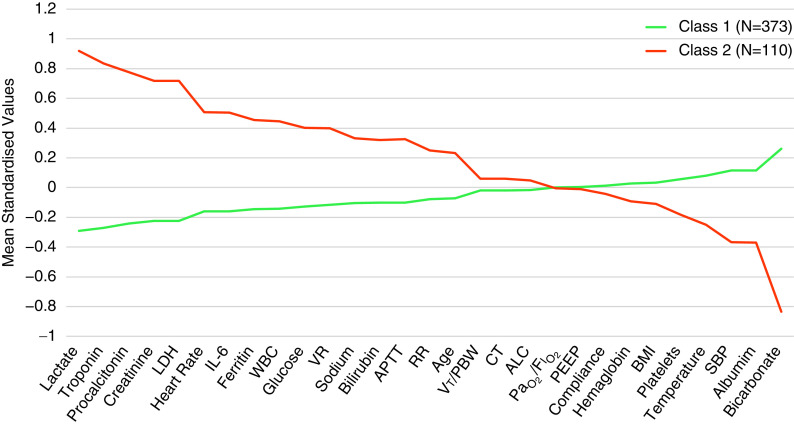
COVID-19–related ARDS class 2 was associated with higher levels of lactate, troponin, procalcitonin, and creatinine (Figure 1). Proinflammatory markers such as IL-6 and ferritin were also significantly higher in COVID-19–related ARDS class 2, whereas levels of albumin and bicarbonate were significantly lower. Among the relevant biomarkers not included in the LCA, D-dimer and C-reactive protein were higher in COVID-19–related ARDS class 2. However, the latter failed to reach statistical significance (Figure E3). Except for ventilatory ratio, which was higher in class 2, no significant differences were observed between the COVID-19–related ARDS classes in respiratory variables (Figure E4). Compared with COVID-19–related ARDS class 1, COVID-19–related ARDS class 2 patients had significantly higher mortality at 90 days and had fewer ventilator-free days (Table 3).
Figure 1.
Standardized values for continuous class-defining variables used in the latent class models. The variables are sorted from left to right in descending order for the highest values in the hyperinflammatory subphenotype. Standardized values were calculated by assigning the mean of the variables as 0 and SD as 1. ALC = absolute lymphocyte count; APTT = activated partial thromboplastin time; BMI = body mass index; CT = cycle threshold; LDH = lactate dehydrogenase; PBW = predicted body weight; PEEP = positive end-expiratory pressure; RR = respiratory rate; SBP = systolic blood pressure; VR = ventilatory ratio; WBC = white blood cell count.
Table 3.
Comparison of 90-Day Mortality and Ventilator-Free Days Censored at Day 90 in the COVID-19–related ARDS Classes and ARDS Subphenotypes
| COVID-19–related ARDS |
ARDS Subphenotypes |
|||||
|---|---|---|---|---|---|---|
| Class 1 (n = 373) | Class 2 (n = 110) | P Value | Hypo (n = 341) | Hyper (n = 142) | P Value | |
| Mortality Day 90 | 178 (48) | 82 (75) | <0.0001 | 166 (49) | 94 (66) | 0.0006 |
| Mortality Day 180 | 182 (49) | 83 (75) | <0.001 | 170 (50) | 95 (67) | 0.0010 |
| VFD | 0 (0–68) | 0 (0–1) | <0.0001 | 0 (0–67) | 0 (0–45) | 0.0003 |
Definition of abbreviations: ARDS = acute respiratory distress syndrome; COVID-19 = coronavirus disease; Hyper = hyperinflammatory subphenotype; Hypo = hypoinflammatory subphenotype; VFD = ventilator-free days.
P value for mortality represents the Pearson’s chi-square test and for VFDs represents Wilcoxon-rank test.
Data are presented as n (%) or median (interquartile range).
Comparison with ARDS Subphenotypes
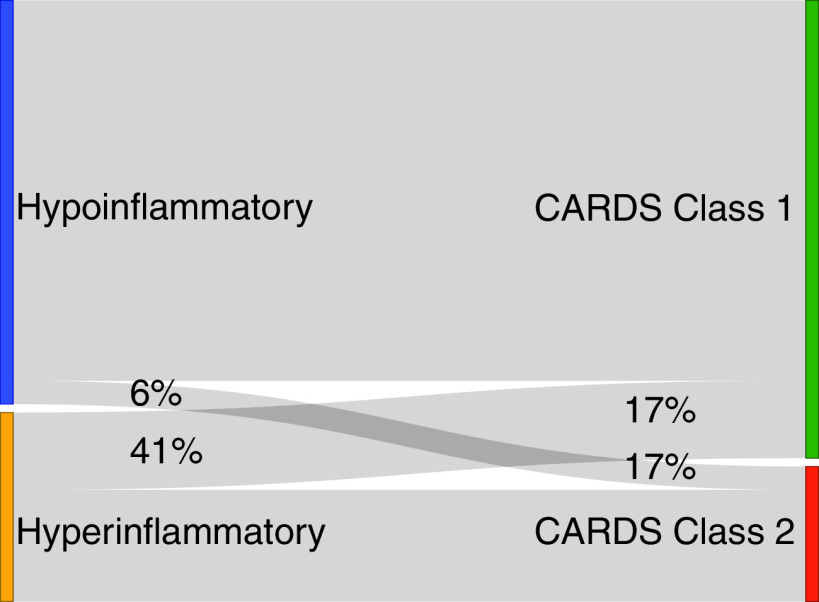
Using the clinical classifier model, 341/483 (71%) patients were classified as the hypoinflammatory ARDS subphenotype and 142/483 (29%) were classified as the hyperinflammatory ARDS subphenotype. Eighty-six percent of the COVID-19–related ARDS class 1 patients were classified to the hypoinflammatory subphenotype, and 81% of the COVID-19–related ARDS class 2 patients were classified into the hyperinflammatory subphenotype (Figure 2). Among patients who crossed over to the noncorresponding subgroup (e.g., class 1 to the hyperinflammatory subphenotype), their probability of subgroup assignment was significantly lower compared with those patients who classified into their corresponding subgroups (Figure E5), suggesting greater uncertainty in subgroup membership in these patients.
Figure 2.
Alluvial plot showing the crossover between the COVID-19–related ARDS classes and the ARDS subphenotypes (percentages represent the proportion of patients from a given subgroup that redistributed to the alternative schema’s phenotypes). ARDS = acute respiratory distress syndrome; CARDS = COVID-19–related ARDS; COVID-19 = coronavirus disease.
As expected, differences between the two ARDS subphenotypes in demographics, ventilatory variables, and biomarkers were similar in their patterns to those observed between the two COVID-19–related ARDS classes (Table 4) and to the non–COVID-19–related ARDS subphenotypes described in the literature (13, 22). Of note, unlike the COVID-19–related ARDS classes, age across the two ARDS subphenotypes was similar.
Table 4.
Differences in Baseline Characteristics and Therapy Use between the Two ARDS Subphenotypes
| Variables | Hypoinflammatory (N = 341) | Hyperinflammatory (N = 142) | P Value |
|---|---|---|---|
| Age, yr | 63 ± 15 | 64 ± 14 | 0.5505 |
| Sex | |||
| F | 120 (35) | 44 (31) | 0.4333 |
| M | 221 (65) | 98 (69) | |
| Race | |||
| Non-Hispanic Black | 53 (16) | 33 (23) | <0.0001 |
| Non-Hispanic White | 28 (8) | 28 (20) | |
| Hispanic | 223 (65) | 62 (44) | |
| Other | 22 (6) | 9 (6) | |
| Unknown (declined to report) | 15 (4) | 10 (7) | |
| Body mass index | 30.0 ± 6.8 | 30.7 ± 7.7 | 0.3139 |
| Smoker (current or former) | 60 (18) | 38 (27) | 0.0077 |
| Diabetes mellitus | 151 (44) | 61 (43) | 0.8678 |
| Hypertension | 228 (69) | 104 (73) | 0.2042 |
| Preadmission symptom duration, d | 7 (3–8) | 7 (4–9) | 0.6221 |
| Time from admission to intubation, d | 1 (0–3) | 1 (0–4) | 0.572 |
| Temperature, °C | 37.7 ± 1.0 | 37.8 ± 1.3 | 0.3539 |
| Heart rate, beats per min | 105 ± 21 | 125 ± 19 | <0.0001 |
| Respiratory rate, breaths per min | 32 (26–36) | 34 (30–41) | 0.0001 |
| Systolic blood pressure, mm Hg | 91 ± 16 | 80 ± 14 | <0.0001 |
| White blood cells, ×10−9/L | 12.9 ± 6.7 | 15.1 ± 8.9 | 0.0031 |
| Absolute lymphocyte count, ×10−9/L | 0.84 (0.6–1.2) | 0.81 (0.5–1.2) | 0.2067 |
| Platelets, ×10−9/L | 240 (178–310) | 208 (137–290) | 0.0039 |
| APTT, s | 38 ± 21 | 45 ± 30 | 0.0094 |
| International normalized ratio | 1.3 ± 0.4 | 1.6 ± 0.8 | <0.0001 |
| PaO2 /Fi O2 | 148 ± 64 | 144 ± 70 | 0.5323 |
| Vt/PBW, ml/kg | 6.5 ± 1.0 | 6.5 ± 1.1 | 0.5636 |
| PEEP, cm H2O | 12 (10–15) | 12 (10–16) | 0.6721 |
| Compliance, ml/cm H2O | 29 ± 12 | 30 ± 12 | 0.5842 |
| Ventilatory ratio | 2.0 ± 0.9 | 2.2 ± 1.0 | 0.0210 |
| Creatinine, mg/dL | 1.3 (0.9–1.8) | 2.7 (1.7–4.5) | <0.0001 |
| Bicarbonate, mmol/L | 21.8 ± 4.1 | 16.5 ± 4.3 | <0.0001 |
| Glucose, mg/dL | 185 ± 103 | 202 ± 129 | 0.1193 |
| Sodium, mmol/L | 139 ± 6 | 141 ± 9 | 0.0040 |
| Bilirubin, mg/dl | 0.6 (0.4–0.8) | 0.7 (0.5–1.2) | 0.0001 |
| Albumin, g/dl | 3.1 ± 0.5 | 2.9 ± 0.6 | <0.0001 |
| Lactate, mg/dl | 1.7 (1.2–2.3) | 2.8 (1.7–4.8) | <0.0001 |
| Troponin, ng/L | 27 (13–76) | 81 (37–181) | <0.0001 |
| IL-6, pg/ml | 185 (96–314) | 315 (145–917) | <0.0001 |
| Procalcitonin, ng/ml | 0.6 (0.3–1.4) | 2.3 (0.7–7.0) | <0.0001 |
| Ferritin, mg/dl | 1,046 (601–2,085) | 1,654 (896–2,694) | 0.0005 |
| Lactate dehydrogenase, U/L | 598 (458–773) | 717 (528–1,099) | <0.0001 |
| C-reactive protein, mg/L | 231 (142–300) | 236 (152–300) | 0.6343 |
| D-dimer, mcg/ml | 3.0 (1.5–15.9) | 9.2 (2.9–20) | <0.0001 |
| Cycle threshold | 27 ± 5.0 | 27 ± 5.0 | 0.9482 |
| Selected therapies use* | |||
| Any corticosteroids | 228 (67) | 95 (67) | 0.993 |
| Days intubation to steroids | 1 (0–7) | 1 (0–3) | 0.0134 |
| Dexamethasone | 5 (1) | 2 (1) | — |
| Methylprednisolone | 170 (50) | 53 (37) | — |
| Hydrocortisone | 42 (12) | 36 (25) | — |
| Prednisone | 11 (3) | 4 (3) | — |
| IL-6 antagonist | 101 (30) | 40 (28) | 0.750 |
| Hydroxychloroquine | 291 (85) | 104 (73) | 0.002 |
| Remdesivir | 23 (7) | 5 (4) | 0.167 |
| Vasopressor use | 269 (79) | 138 (97) | <0.0001 |
| Renal replacement therapy* | 87 (26) | 63 (45) | <0.0001 |
| Neuromuscular blockade† | 107 (32) | 53 (37) | 0.2640 |
| Prone positioning* | 88 (26) | 23 (16) | 0.0317 |
| ECMO* | 5 (1.5) | 5 (3.5) | 0.2708 |
| SOFA score‡ | 9 (7–11) | 12 (10–14) | <0.0001 |
Definition of abbreviations: APTT = activated partial thromboplastin time; ARDS = acute respiratory distress syndrome; ECMO = extracorporeal membrane oxygenation; PBW = predicted body weight; PEEP = positive end-expiratory pressure; SOFA = Sequential Organ Failure Assessment.
Normally distributed data are presented as mean ± SD; nonnormally distributed data are presented as median (interquartile range); and categorical data are presented as n (%). P values represent Student’s t test for normally distributed data, Wilcoxon-rank test for nonnormally distributed data, and chi-square or Fisher exact test for categorical values.
These variables were recorded during the course of the ICU admission and not exclusively at baseline.
Variable was recorded within 48 hours of ARDS diagnosis.
If Glasgow Coma Score missing, imputed as 15.
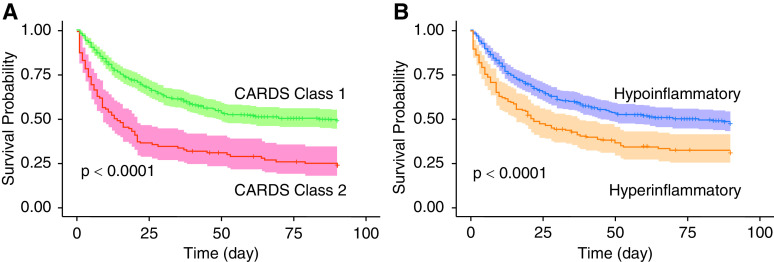
Clinical outcomes trends observed among patients in the hyperinflammatory and hypoinflammatory subphenotypes were similar to their corresponding COVID-19–related ARDS classes (Table 3). In COVID-19–related ARDS class 1 (Figure 3A) and the hypoinflammatory subphenotype (Figure 3B), there was a survival advantage observed early in the observation period that persisted to 90 days.
Figure 3.
(A and B) Kaplan-Meier survival plots censored at Day 90 stratified by COVID-19–related ARDS classes (A) and ARDS subphenotypes (B). ARDS = acute respiratory distress syndrome; CARDS = COVID-19–related ARDS; COVID-19 = coronavirus disease.
Supervised Mortality Predictor Model
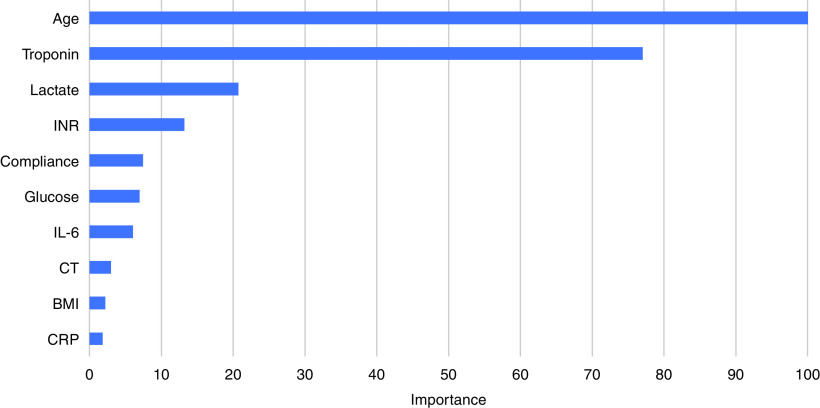
The XGBoost bootstrapped model trained to predict mortality at Day 90 had an area under the receiver operator characteristics curve of 0.84 (95% confidence interval [CI], 0.81–0.88). The top 10 most important variables in the model are presented in Figure 4 and are consistent with those described in several other studies (23–25). The several-fold higher importance values of age and troponin in comparison with other predictors in the model indicate that these two variables were the key predictors of mortality in this cohort. Among the top 10 predictor variables, age, compliance, glucose, international normalized ratio, body mass index, and CT did not feature prominently as partitioning variables in the LCA model, suggesting that the prognostic value of the LCA classes is independent of common predictors of mortality in COVID-19–related ARDS.
Figure 4.
Top 10 most important variables in the XGBoost COVID-19–related ARDS mortality predictor model. ARDS = acute respiratory distress syndrome; BMI = body mass index; COVID-19 = coronavirus disease; CRP=C-reactive protein; CT = cycle threshold; INR = international normalized ratio.
PCR CT as a Predictor of Outcome
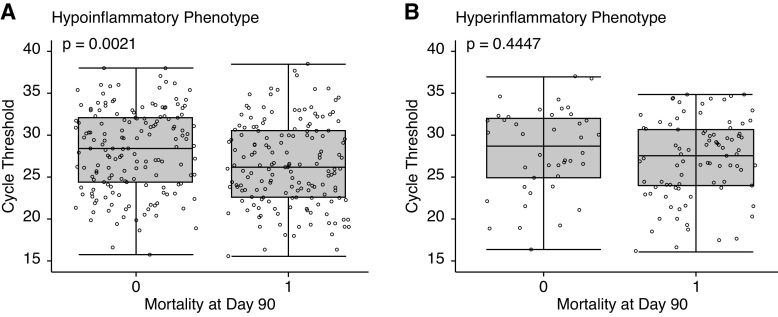
CT was an unexpected finding among the top 10 important predictor variables for mortality in COVID-19–related ARDS. When evaluated further, we found that higher CT was associated with decreased risk of mortality (odds ratio, 0.95 per unit increase in CT; 95% CI, 0.91–0.98) and remained so after adjusting for age, sex, body mass index category, race, and SOFA score (odds ratio, 0.95; 95% CI, 0.91–0.99). There was no significant difference in CT values between the hyper- and hypoinflammatory subphenotypes (P = 0.9482). When stratified by subphenotypes, CT values were similar among survivors and nonsurvivors in the hyperinflammatory subphenotype; however, CT values were significantly lower in nonsurvivors in the hypoinflammatory subphenotype (Figure 5). When the subphenotypes were dichotomized into subgroups by the population median of CT (>27.07 “CT high,” <27.07 “CT low”), no significant difference in mortality was observed in the hyperinflammatory subphenotype (CT high 66% vs. 69% CT low; P = 0.8970). In contrast, in the hypoinflammatory subphenotype, mortality was significantly lower in the CT-high subgroup than in the CT-low group (42% vs. 56%; P = 0.0149). These findings were the same when evaluated in the COVID-19–related ARDS classes (data not presented).
Figure 5.
(A and B) Boxplot of the distribution of PCR cycle threshold stratified by mortality at Day 90 in the hypoinflammatory (A) and hyperinflammatory (B) phenotypes. P value represents the Student’s t test.
Exploratory Analysis of Corticosteroid Treatment Interaction across Subgroups
In a logistic regression model, the interaction term between ARDS subphenotype and corticosteroid exposure for 90-day mortality was significant (P = 0.0295). Corticosteroid exposure was associated with a trend toward decreased mortality in the hyperinflammatory phenotype and increased mortality in the hypoinflammatory phenotype (Table 5). The model was adjusted for SOFA score to account for indication bias, and the interaction term remained statistically significant (P = 0.0410). The interaction term for corticosteroid therapy and COVID-19–related ARDS classes was also significant, for both the unadjusted (P = 0.0273) and the adjusted model (P = 0.0370).
Table 5.
Comparison of 90-Day Mortality in Subgroups Stratified by Corticosteroid in ARDS Subphenotypes and COVID-19–related ARDS Classes
| Corticosteroid Use | ARDS Subphenotype |
P Value | |
|---|---|---|---|
| Hypoinflammatory | Hyperinflammatory | ||
| Yes | 115/226 (51%) | 57/94 (61%) | 0.0295 |
| No | 51/113 (45%) | 37/48 (77%) | |
| Corticosteroid Use |
COVID-19–related ARDS Class
|
P Value | |
| Class 1 | Class 2 | ||
| Yes | 120/244 (49%) | 52/76 (68%) | 0.0273 |
| No | 58/127 (46%) | 30/34 (88%) | |
Definition of abbreviations: ARDS = acute respiratory distress syndrome; COVID-19 = coronavirus disease.
P value represents the interaction between ARDS subphenotype or COVID-19–related ARDS class and corticosteroid exposure in a logistic regression model with 90-day mortality as the dependent variable.
Discussion
In this study, we used a minimally biased approach to discover two novel subclasses in COVID-19–related ARDS. These classes have distinct biological and clinical characteristics with divergent clinical outcomes. Class 2 was associated with increased markers of inflammation, lower bicarbonate, and a higher prevalence of multiorgan failure. Using a previously validated machine learning model, we were able to classify patients with COVID-19–related ARDS into previously described hypo- and hyperinflammatory ARDS subphenotypes (9–12, 16, 26). There was consistent overlap in the characteristics and clinical outcomes between the two phenotyping schemes, in which COVID-19–related ARDS class 2 corresponded to the hyperinflammatory subphenotype.
Mortality among the entire cohort and within each subphenotype and COVID-19–related ARDS class was substantially higher than that observed in historical non–COVID-19–related ARDS studies (1–3, 6, 7). Furthermore, we observed an association of lower PCR-CT, a surrogate for viral load, with increased mortality in the cohort overall; however, this finding was largely driven by the hypoinflammatory subphenotype (19). We observed a significant interaction between the subgroups identified and corticosteroid exposure, in which treatment was associated with improved survival in the hyperinflammatory subphenotype but not the hypoinflammatory subphenotype.
In the absence of a validation cohort, the overlap of COVID-19–related ARDS classes 1 and 2 with the hypo- and hyperinflammatory subphenotypes, respectively, is a reassuring finding and lends validity to the observed differences between these subgroups. It is likely that class 2 characterizes a more severe or “specific” subset of the hyperinflammatory subphenotype given the smaller number of patients, higher prevalence of multiorgan failure, and increased mortality. In prior ARDS phenotyping studies, markers such as procalcitonin, lactate, and ferritin were not available (9–11). However, given the biological characteristics of the hyperinflammatory subphenotype, it is unsurprising that these new markers were also elevated. As these biomarkers are more widely available in clinical laboratories than research biomarkers such as IL-6, IL-8, and soluble tumor necrosis factor-1, their use may facilitate more widespread identification of ARDS subphenotypes and warrants further evaluation.
In comparison with a prior study (n = 39) describing the prevalence of the ARDS subphenotypes in COVID-19–related ARDS, the proportion of patients in the hyperinflammatory phenotype was higher in our study (12). The sample size and method for defining the phenotypes differed between the studies; nevertheless, the true prevalence of these subphenotypes in COVID-19–related ARDS and their key determinants remains unknown and needs mapping in future studies. Among these determinants, the relative influence of race and ethnicity, genetic polymorphisms, viral variants, and the use of early immunomodulators also warrant investigation.
Within this observational cohort, the main clinical value of the subphenotypes was to identify an at-risk COVID-19–related ARDS group with distinct biological characteristics. Consistent with several other studies, age was the most important variable in our supervised XGBoost mortality-predicting model (1–3, 16). Yet age was not significantly different between the two ARDS subphenotypes. Several of the top 10 variables in the supervised model were not important partitioning variables in either the COVID-19–related ARDS classes or the ARDS subphenotypes, suggesting that the prognostic information from the subphenotypes cannot be easily captured using standard prognostication approaches.
A second advantage of the presented phenotyping schema is that it stratifies the COVID-19–related ARDS population into less heterogeneous subgroups. Although the etiological insult in COVID-19 may be uniform, our data support the hypothesis that at the onset of ARDS, considerable biological heterogeneity exists within this population. It stands to reason that immunomodulatory interventions such as corticosteroids and IL-6 inhibitors may have differential efficacy across such biological subphenotypes, a heterogeneity in treatment effect. The trend toward improved mortality with corticosteroid therapy in the hyperinflammatory subphenotype and worse outcomes in the hypoinflammatory subphenotype substantiate this hypothesis.
How do we reconcile the findings of heterogeneity of treatment effect with corticosteroids in the ARDS subphenotypes with the evidence in the literature of improved outcomes in COVID-19? First, it is important to acknowledge that ours was a retrospective observational cohort and limited to nonrandomized allocation of treatment. Although we note that corticosteroid treatment was equally as common in the hyper- and hypoinflammatory subphenotypes (67% in each), the dose and type of steroids used differed across the population. The proportion of patients receiving each type of corticosteroid, however, was insufficiently large to perform subgroup analyses. Given the constraints of the study design, interpretation of the presented findings should be cautious and limited to hypothesis generation.
Within this context, in the RECOVERY (Randomised Evaluation of COVID-19 Therapy)-Dexamethasone trial, there was evidence of heterogeneity in treatment effect depending on the severity of illness, with a signal to harm in those not requiring oxygen support and the largest benefit in those who were invasively ventilated (6). In three of the largest clinical trials that tested the benefit of corticosteroids specifically in critically ill patients, no significant reduction in absolute mortality was observed: CODex (COVID-19 Dexamethasone) (P = 0.85), CAPE COVID (Community-Acquired Pneumonia: Evaluation of Corticosteroids in Coronavirus Disease) (P = 0.057), and REMAP-CAP (A Randomised, Embedded, Multi-factorial, Adaptive Platform Trial for Community-Acquired Pneumonia) (P values not available but the confidence intervals for the odds ratio of mortality crossed unity for both treatment groups) (27–29). Notably, in all these trials, there was a signal to morality benefit with corticosteroid therapy, suggesting that treating specific subphenotypes may have resulted in significantly improved mortality, and that further evaluation is warranted (30). Furthermore, in our cohort, the ratio of signal to harm in the hypoinflammatory subphenotype was small. Given the large proportion of patients in this subphenotype, it is highly likely that subsumed within it is a smaller subset of patients who may be harmed with corticosteroid use. Given the known linkage between corticosteroids and delayed viral clearance, patients with high viral load may be an interesting group to investigate in future studies.
To that end, heterogeneity of treatment effect may also help explain the modest absolute reduction in mortality in immunomodulators such as IL-6 inhibitors (5, 6, 31–33). Of note, in the recent RECOVERY trial evaluation of tocilizumab in COVID-19, mortality among mechanically ventilated patients approached 50% regardless of the treatment arm, which is similar to our cohort (5). Although overall ICU mortality in COVID-19 has declined over the course of the pandemic, among invasive mechanically ventilated patients, case fatality rates in a large meta-analysis were also similar to those described in our cohort, albeit with considerable regional variation (34). The mortality rate in our cohort was higher than that in some studies from a similar time period; however, we note that, unlike our study, these studies included all intubated patients, not just those who met ARDS criteria, and did not report 90-day mortality (35–37). Several key determinants of the variance in outcomes in COVID-19 have been described; among these, acute physiology, socioeconomic factors, and hospital strain were most pertinent to our cohort and may explain its high mortality (38). ICU strain and demand due to the surge of COVID-19 cases in New York City during the study period may have been of particular importance given their association with worse outcomes (39, 40).
This study also highlights the advantage of evaluating prognostic markers within subphenotypes. Consistent with other reports, SARS-CoV-2 PCR-CT, a surrogate for viral load, was associated with mortality in the entire cohort after adjusting for age and illness severity (18–21). Interestingly, this association with mortality was primarily observed in the hypoinflammatory subphenotype. In view of these findings, we hypothesize that viral cytotoxicity may be a primary driver of mortality in the hypoinflammatory subphenotype, whereas in the hyperinflammatory subphenotype, mortality may be driven by excessive inflammation, as evidenced by higher levels of proinflammatory markers and increased prevalence of multiorgan failure. It is also interesting to note a trend toward increased mortality with corticosteroids in the hypoinflammatory phenotype, given that their use has been implicated in delayed viral clearance in SARS-CoV-1, Middle East respiratory syndrome coronavirus, and influenza pneumonia (41–43). The subphenotype-specific association of PCR-CT with mortality reinforces the argument against a “one-size fits-all approach” to pharmacotherapies in COVID-19–related ARDS. Although PCR-CT has been related to viral load in clinical samples, the precise relationship between the quantified levels of PCR-CT and viral load is inconsistently defined across assays (44). Therefore, the findings of our study need external validation.
In prior studies, ventilatory and gas exchange variables such as PaO2 /Fi O2 and compliance have consistently been similar between the two ARDS subphenotypes. In a novel analysis in this study comparing ventilatory ratio (VR), a bedside surrogate for dead space, we observed higher values in the hyperinflammatory subphenotype than in the hypoinflammatory subphenotype (14). In contrast, the subphenotypes had similar PaO2 /Fi O2 ratios, driving pressures, and compliance, indicating that the difference in VR is likely not explained by ARDS severity alone. High dead space fractions have been reported in COVID-19–related ARDS, but the difference compared with ARDS from other etiologies and its prognostic value in COVID-19 are still areas of investigation (45, 46). The pathophysiologic and biologic mechanism underlying the difference in VR between the subphenotypes warrants further investigation. One possible hypothesis could be the higher incidence of thromboembolism in the lung microvasculature that has been reported in COVID-19–related ARDS (47). However, advanced imaging to confirm the increased prevalence of / mismatch of the lungs was not available in our cohort, and this hypothesis requires substantial further investigation.
Our study has several strengths. First, missingness among predictor variables was low. The primary outcome was 90-day mortality and therefore captured vital status over an extended time period. We were able to identify subphenotypes based on a composite of clinical and biological features using minimally biased methods that were agnostic to outcomes, and most of the biomarkers used are widely available in clinical laboratories. To our knowledge, this is one of the largest studies of COVID-19–related ARDS that has evaluated PCR-CT as a predictor of outcome.
The study also has several limitations. Our study captures data from early in the pandemic and from a single center; management strategies have since changed and overall mortality declined in this rapidly evolving pandemic. There was high demand for ICU beds during the study period and considerable hospital and ICU strain, which may limit the external validity of the study. Another limitation is the lack of a second independent cohort in which to replicate the COVID-ARDS LCA findings; however, the substantial overlap with previously validated ARDS subphenotypes reinforces the likely generalizability of these findings. PCR-CT was measured at a single time point, and thus, we were unable to test its prognostic value over time. Patients in the cohort all presented in the early part of the pandemic in which treatment approaches were less uniform than current practices, which are informed by the results of emerging clinical trials. Use of some therapies, for example, immunomodulatory drugs, may have had an influence on the circulating levels of class-defining variables and thereby had a confounding influence on the identified classes. The use of such therapies was insufficiently prevalent in our population to adequately define this phenomenon. Finally, because of resource constraints at our study site throughout the initial phase of the pandemic, prone positioning was used relatively infrequently during the study period. This practice may have influenced our outcome data, as prone positioning is known to improve mortality among patients with moderate to severe ARDS. It is notable, however, that the mortality rate observed was similar to that seen in other studies of COVID-19–related ARDS (2, 5, 6).
In summary, we identified two distinct subclasses of COVID-19–related ARDS that were remarkably similar in clinical characteristics and outcomes to previously established ARDS subphenotypes. COVID-19–related ARDS class 2 and the hyperinflammatory subphenotype were associated with adverse clinical outcomes. We observed that PCR-CT was associated with increased mortality in the hypoinflammatory subphenotype but not the hyperinflammatory phenotype. Finally, heterogeneous treatment responses were observed with corticosteroid use in the subphenotypes. Further studies should focus on replicating distinct subphenotypes within COVID-19–related ARDS as well as on identifying factors that influence differential response to treatment within severe COVID-19.
Acknowledgments
Acknowledgment
The authors thank the patients treated at Columbia University Irving Medical Center and their families. They also thank the healthcare workers who risked their own well-being to provide outstanding care during the COVID-19 pandemic.
Footnotes
Supported by CHEST Foundation Grant in COVID-19 (D.F.), NHLBI grant R35 HL140026 (C.S.C.), National Center for Advancing Translational Sciences grant UL1 TR001873 (M.R.B. and M.R.O'D.), K23 HL133489 (J.R.B.), R21 HL145506 (J.R.B.), F32AI147528 (M.J.C.), and U.S. Department of Defense grant W81XWH2110217 (M.R.B.). Biospecimens and/or data utilized for this research were obtained from the Columbia University Biobank (CUB), which is supported by the Irving Institute for Clinical and Translational Research, home to Columbia University's Clinical and Translational Science Award (CTSA) funded through Grant Number UL1TR001873.
Author Contributions: D.F., P.S., M.J.C., D.A., D.B., M.R.O'D., and C.S.C. were involved in the study conceptualization and design and data analysis and interpretation. D.F., M.J.C., D.A., A.T., M. Murn, J.F., A.R., S.Y.R.-J., M.A.A., T.S., M. Madhavan, A.G., and A.K.L. were involved in data collection and cleaning. K.D., M.V.M., and J.H. were involved in study design and data analysis and interpretation. C.A., N.H.Y., K.M.B., J.R.B., and M.R.B. were involved in data analysis and interpretation. P.S. and D.F. developed the first draft of the manuscript. P.S. and D.F. verified the underlying data. All authors reviewed and edited the final version of the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.202105-1302OC on September 20, 2021
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med . 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cummings MJ, Baldwin MR, Abrams D, Jacobson SD, Meyer BJ, Balough EM, et al. Epidemiology, clinical course, and outcomes of critically ill adults with COVID-19 in New York City: a prospective cohort study. Lancet . 2020;395:1763–1770. doi: 10.1016/S0140-6736(20)31189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grasselli G, Zangrillo A, Zanella A, Antonelli M, Cabrini L, Castelli A, et al. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy Region, Italy. JAMA . 2020;323:1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ranieri VM, Rubenfeld GD, Thompson BT, Ferguson ND, Caldwell E, Fan E, et al. ARDS Definition Task Force. Acute respiratory distress syndrome: the Berlin Definition. JAMA . 2012;307:2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 5. RECOVERY Collaborative Group. Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet . 2021;397:1637–1645. doi: 10.1016/S0140-6736(21)00676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L, et al. RECOVERY Collaborative Group Dexamethasone in hospitalized patients with Covid-19 N Engl J Med 2021384693–704.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bellani G, Laffey JG, Pham T, Fan E, Brochard L, Esteban A, et al. LUNG SAFE Investigators; ESICM Trials Group. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. JAMA . 2016;315:788–800. doi: 10.1001/jama.2016.0291. [DOI] [PubMed] [Google Scholar]
- 8. Sinha P, Calfee CS. Phenotypes in acute respiratory distress syndrome: moving towards precision medicine. Curr Opin Crit Care . 2019;25:12–20. doi: 10.1097/MCC.0000000000000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Calfee CS, Delucchi K, Parsons PE, Thompson BT, Ware LB, Matthay MA. NHLBI ARDS Network. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med . 2014;2:611–620. doi: 10.1016/S2213-2600(14)70097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Calfee CS, Delucchi KL, Sinha P, Matthay MA, Hackett J, Shankar-Hari M, et al. Irish Critical Care Trials Group. Acute respiratory distress syndrome subphenotypes and differential response to simvastatin: secondary analysis of a randomised controlled trial. Lancet Respir Med . 2018;6:691–698. doi: 10.1016/S2213-2600(18)30177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sinha P, Delucchi KL, Thompson BT, McAuley DF, Matthay MA, Calfee CS. NHLBI ARDS Network. Latent class analysis of ARDS subphenotypes: a secondary analysis of the statins for acutely injured lungs from sepsis (SAILS) study. Intensive Care Med . 2018;44:1859–1869. doi: 10.1007/s00134-018-5378-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sinha P, Calfee CS, Cherian S, Brealey D, Cutler S, King C, et al. Prevalence of phenotypes of acute respiratory distress syndrome in critically ill patients with COVID-19: a prospective observational study. Lancet Respir Med . 2020;8:1209–1218. doi: 10.1016/S2213-2600(20)30366-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sinha P, Delucchi KL, McAuley DF, O’Kane CM, Matthay MA, Calfee CS. Development and validation of parsimonious algorithms to classify acute respiratory distress syndrome phenotypes: a secondary analysis of randomised controlled trials. Lancet Respir Med . 2020;8:247–257. doi: 10.1016/S2213-2600(19)30369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sinha P, Calfee CS, Beitler JR, Soni N, Ho K, Matthay MA, et al. Physiologic analysis and clinical performance of the ventilatory ratio in acute respiratory distress syndrome. Am J Respir Crit Care Med . 2019;199:333–341. doi: 10.1164/rccm.201804-0692OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sinha P, Calfee CS, Delucchi KL. Practitioner’s guide to latent class analysis: methodological considerations and common pitfalls. Crit Care Med . 2021;49:e63–e79. doi: 10.1097/CCM.0000000000004710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sinha P, Churpek MM, Calfee CS. Machine learning classifier models can identify acute respiratory distress syndrome phenotypes using readily available clinical data. Am J Respir Crit Care Med . 2020;202:996–1004. doi: 10.1164/rccm.202002-0347OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Raschke RA, Agarwal S, Rangan P, Heise CW, Curry SC. Discriminant accuracy of the SOFA score for determining the probable mortality of patients with COVID-19 pneumonia requiring mechanical ventilation. JAMA . 2021;325:1469–1470. doi: 10.1001/jama.2021.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Magleby R, Westblade LF, Trzebucki A, Simon MS, Rajan M, Park J, et al. Impact of SARS-CoV-2 viral load on risk of intubation and mortality among hospitalized patients with coronavirus disease 2019. Clin Infect Dis . 2020:ciaa851. doi: 10.1093/cid/ciaa851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rao SN, Manissero D, Steele VR, Pareja J. A systematic review of the clinical utility of cycle threshold values in the context of COVID-19. Infect Dis Ther . 2020;9:573–586. doi: 10.1007/s40121-020-00324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pujadas E, Chaudhry F, McBride R, Richter F, Zhao S, Wajnberg A, et al. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir Med . 2020;8:e70. doi: 10.1016/S2213-2600(20)30354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. WHO Working Group on the Clinical Characterisation and Management of COVID-19 infection. A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect Dis . 2020;20:e192–e197. doi: 10.1016/S1473-3099(20)30483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sinha P, Delucchi KL, Chen Y, Zhuo H, Abbott J, Wang C, et al. Latent class analysis-derived subphenotypes are generalisable to observational cohorts of acute respiratory distress syndrome: a prospective study. Thorax . 2021:thoraxjnl-2021-217158. doi: 10.1136/thoraxjnl-2021-217158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Karthikeyan A, Garg A, Vinod PK, Priyakumar UD. Machine learning based clinical decision support system for early COVID-19 mortality prediction. Front Public Health . 2021;9:626697. doi: 10.3389/fpubh.2021.626697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ma X, Ng M, Xu S, Xu Z, Qiu H, Liu Y, et al. Development and validation of prognosis model of mortality risk in patients with COVID-19. Epidemiol Infect . 2020;148:e168. doi: 10.1017/S0950268820001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Subudhi S, Verma A, Patel AB, Hardin CC, Khandekar MJ, Lee H, et al. Comparing machine learning algorithms for predicting ICU admission and mortality in COVID-19. NPJ Digit Med . 2021;4:87. doi: 10.1038/s41746-021-00456-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Famous KR, Delucchi K, Ware LB, Kangelaris KN, Liu KD, Thompson BT, et al. ARDS Network. Acute respiratory distress syndrome subphenotypes respond differently to randomized fluid management strategy. Am J Respir Crit Care Med . 2017;195:331–338. doi: 10.1164/rccm.201603-0645OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tomazini BM, Maia IS, Cavalcanti AB, Berwanger O, Rosa RG, Veiga VC, et al. COALITION COVID-19 Brazil III Investigators. Effect of dexamethasone on days alive and ventilator-free in patients with moderate or severe acute respiratory distress syndrome and COVID-19: the CoDEX randomized clinical trial. JAMA . 2020;324:1307–1316. doi: 10.1001/jama.2020.17021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dequin P-F, Heming N, Meziani F, Plantefève G, Voiriot G, Badié J, et al. CAPE COVID Trial Group and the CRICS-TriGGERSep Network. Effect of hydrocortisone on 21-day mortality or respiratory support among critically ill patients with COVID-19: a randomized clinical trial. JAMA . 2020;324:1298–1306. doi: 10.1001/jama.2020.16761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Angus DC, Derde L, Al-Beidh F, Annane D, Arabi Y, Beane A, et al. Writing Committee for the REMAP-CAP Investigators. Effect of hydrocortisone on mortality and organ support in patients with severe COVID-19: The REMAP-CAP COVID-19 corticosteroid domain randomized clinical trial. JAMA . 2020;324:1317–1329. doi: 10.1001/jama.2020.17022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Prescott HC, Rice TW. Corticosteroids in COVID-19 ARDS: evidence and hope during the pandemic. JAMA . 2020;324:1292–1295. doi: 10.1001/jama.2020.16747. [DOI] [PubMed] [Google Scholar]
- 31. Sterne JAC, Murthy S, Diaz JV, Slutsky AS, Villar J, Angus DC, et al. WHO Rapid Evidence Appraisal for COVID-19 Therapies (REACT) Working Group. Association between administration of systemic corticosteroids and mortality among critically ill patients with COVID-19: a meta-analysis. JAMA . 2020;324:1330–1341. doi: 10.1001/jama.2020.17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Biran N, Ip A, Ahn J, Go RC, Wang S, Mathura S, et al. Tocilizumab among patients with COVID-19 in the intensive care unit: a multicentre observational study. Lancet Rheumatol . 2020;2:e603–e612. doi: 10.1016/S2665-9913(20)30277-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. The REMAP-CAP Investigators. Interleukin-6 receptor antagonists in critically ill patients with Covid-19. N Engl J Med . 2021;384:1491–1502. doi: 10.1056/NEJMoa2100433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lim ZJ, Subramaniam A, Ponnapa Reddy M, Blecher G, Kadam U, Afroz A, et al. Case fatality rates for patients with COVID-19 requiring invasive mechanical ventilation. a meta-analysis. Am J Respir Crit Care Med . 2021;203:54–66. doi: 10.1164/rccm.202006-2405OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Auld SC, Caridi-Scheible M, Blum JM, Robichaux C, Kraft C, Jacob JT, et al. and the Emory COVID-19 Quality and Clinical Research Collaborative. ICU and ventilator mortality among critically ill adults with coronavirus disease 2019. Crit Care Med . 2020;48:e799–e804. doi: 10.1097/CCM.0000000000004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ziehr DR, Alladina J, Petri CR, Maley JH, Moskowitz A, Medoff BD, et al. Respiratory pathophysiology of mechanically ventilated patients with COVID-19: a cohort study. Am J Respir Crit Care Med . 2020;201:1560–1564. doi: 10.1164/rccm.202004-1163LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schenck EJ, Hoffman K, Goyal P, Choi J, Torres L, Rajwani K, et al. Respiratory mechanics and gas exchange in COVID-19-associated respiratory failure. Ann Am Thorac Soc . 2020;17:1158–1161. doi: 10.1513/AnnalsATS.202005-427RL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Churpek MM, Gupta S, Spicer AB, Parker WF, Fahrenbach J, Brenner SK, et al. STOP-COVID Investigators. Hospital-level variation in death for critically ill patients with COVID-19. Am J Respir Crit Care Med . 2021;204:403–411. doi: 10.1164/rccm.202012-4547OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bravata DM, Perkins AJ, Myers LJ, Arling G, Zhang Y, Zillich AJ, et al. Association of intensive care unit patient load and demand with mortality rates in US Department of Veterans Affairs hospitals during the COVID-19 pandemic. JAMA Netw Open . 2021;4:e2034266. doi: 10.1001/jamanetworkopen.2020.34266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kadri SS, Sun J, Lawandi A, Strich JR, Busch LM, Keller M, et al. Association between caseload surge and COVID-19 survival in 558 U.S. hospitals, March to August 2020. Ann Intern Med . 2021;174:1240–1251. doi: 10.7326/M21-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Arabi YM, Mandourah Y, Al-Hameed F, Sindi AA, Almekhlafi GA, Hussein MA, et al. Saudi Critical Care Trial Group. Corticosteroid therapy for critically ill patients with Middle East Respiratory Syndrome. Am J Respir Crit Care Med . 2018;197:757–767. doi: 10.1164/rccm.201706-1172OC. [DOI] [PubMed] [Google Scholar]
- 42. Lee N, Allen Chan KC, Hui DS, Ng EK, Wu A, Chiu RW, et al. Effects of early corticosteroid treatment on plasma SARS-associated coronavirus RNA concentrations in adult patients. J Clin Virol . 2004;31:304–309. doi: 10.1016/j.jcv.2004.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee N, Chan PKS, Hui DSC, Rainer TH, Wong E, Choi KW, et al. Viral loads and duration of viral shedding in adult patients hospitalized with influenza. J Infect Dis . 2009;200:492–500. doi: 10.1086/600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Association for Molecular Pathology. Important issues to consider before interpreting and applying Ct values in clinical practice [updated 2021 Mar 12; accessed 2021 Jul 12]. Available from: https://www.amp.org/about/news-room/amp-blog-content/important-issues-to-consider-before-interpreting-and-applying-ct-values-in-clinical-practice/
- 45. Liu X, Liu X, Xu Y, Xu Z, Huang Y, Chen S, et al. Ventilatory ratio in hypercapnic mechanically ventilated patients with COVID-19-associated acute respiratory distress syndrome. Am J Respir Crit Care Med . 2020;201:1297–1299. doi: 10.1164/rccm.202002-0373LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morales-Quinteros L, Neto AS, Artigas A, Blanch L, Botta M, Kaufman DA, et al. PRoVENT-COVID Study Group. Dead space estimates may not be independently associated with 28-day mortality in COVID-19 ARDS. Crit Care . 2021;25:171. doi: 10.1186/s13054-021-03570-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sinha P, Bos LD. Pathophysiology of the acute respiratory distress syndrome: insights from clinical studies. Crit Care Clin . 2021;37:795–815. doi: 10.1016/j.ccc.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]