Abstract
Borrelia burgdorferi, etiologic agent of Lyme disease, is the leading tick-borne disease in the United States with approximately 300,000 cases diagnosed annually. Disease occurs in stages beginning localized infection at the site of a tick bite and progresses to disseminated infection when antibiotic treatment is not administered in a timely manner. A multi-systemic infection develops following dissemination to numerous immunoprotective tissues, such as the heart, bladder, and joints, resulting in late Lyme disease. B. burgdorferi undergoes dynamic genetic regulation throughout mammalian infection and defining the exact role of virulence genes at distinct stages of disease is challenging. The murine model allows for the characterization of the pathogenic function of genes in B. burgdorferi, but traditional end point studies limit the ability to gather data throughout an infection study and greatly increase the required number of mice. Molecular genetic techniques to evaluate and quantitate B. burgdorferi infection are laborious and costly. To partly circumvent these issues, a codon optimized firefly luciferase, under the control of a constitutive borrelial promoter, was introduced into B. burgdorferi enabling the characterization of mutant or modified strains under in vitro growth conditions and throughout murine infection. The detection of bioluminescent B. burgdorferi is highly sensitive and allows for the repeated real-time quantitative evaluation of borrelial load during murine infection. Furthermore, bioluminescence has also been utilized to evaluate alteration in tissue localization and tissue-specific gene expression of B. burgdorferi. In this chapter, we describe the generation of bioluminescent borrelial strains along with methods for in vitro, in vivo, and ex vivo B. burgdorferi studies.
Keywords: Borrelia burgdorferi, Spirochete, Pathogen, Bioluminescence, Luciferase, In vivo imaging, Ex vivo imaging, Infection, Pathogenicity
1. Introduction
Lyme disease, caused by spirochetal pathogen Borrelia burgdorferi, is a tick-borne infection that develops into a multi-systemic infection where patients experience significant morbidity in the form of arthritis, cardiac, and neurological complications [1-3]. Antibiotic treatment of B. burgdorferi infection is highly effective during localized disease when properly diagnosed, but failure to do so allows the pathogen to disseminate to immunoprotective tissues and can develop into late Lyme disease. The 2013 CDC report of roughly 300,000 cases in the United States indicates that Lyme disease is an emerging infectious disease [4]. B. burgdorferi undergoes adaptation that requires multifaceted gene regulation to traverse a complex enzootic cycle between the Ixodes vector and mammalian host [5]. The murine model is an essential tool for the characterization of B. burgdorferi pathogenesis that cannot be accomplished utilizing an enriched artificial media, BSKII with 6% normal rabbit serum, which fails to represent physiological relevant conditions that mimic host-pathogen interactions [5-8]. Traditional end point infectivity studies have limitations when evaluating a multi-stage and long-term infection requiring a substantial increase in the number of mice per study, are laborious, and provide delayed molecular validation of phenotypic differences [9].
Bioluminescence imaging (BLI) was first published by Contag et al. demonstrating a noninvasive method to evaluate pathogens in a whole animal over time and space as the infection progresses or reaches an acute stage of disease [10]. The release of bioluminescence occurs when the Firefly luciferase (luc) enzyme oxidizes a substrate, luciferin, in the presence of ATP and oxygen [11-14]. The requirement of ATP and oxygen for the luciferase reaction indicates the viability of bacterial cells detected, unlike biofluorescent probes where protein turnover is needed to avoid emission after excitation, and thus provides additional insight into the status of infection. Additional luciferase genes from click beetle, Renilla, Guassia, and Photorhabdus luminescens, which have been used as probes in studies of various pathogens, produce light (between 480 and 580 nm) and byproducts that differ slightly from Firefly luc [15].
In vivo imaging systems (IVIS) utilize a highly sensitive couple-charged device (CCD) camera for the detection of luminescence emitted and scattered through opaque murine tissues (Perkin Elmer, [10, 16]). Numerous factors contribute to the quantifiable radiance: photon intensity per cell, presence of melanin that impedes signal, availability of necessary co-factors, tissue localization and depth of bioluminescent cells, and total number of cells in a defined region of interest (ROI) [11-14]. Experimental applications of bacterial in vivo bioluminescence can be used to monitor differences in bacterial load as the pathogen expands and disseminates within the mouse, to evaluate the effectiveness of therapeutics, and to determine the gene expression of pathogens influenced by interaction with the host. The repetitive monitoring of mice through bioluminescent emission can replace traditional molecular techniques, refine the data obtained, and greatly reduce the number of mice required for an individual experiment. Furthermore, tissue tropism for bacterial localization and gene expression can be gathered through ex vivo bioluminescence imaging.
Introduction of firefly luciferase (luc) into B. burgdorferi required codon optimization of firefly luciferase, designated Bbluc, for efficient production in a bacterium due to its AT-rich genome. Blevins et al. generated Bbluc linked to a constitutively expressed flagellar promoter (PflaB) or to an IPTG inducible promoter (PpQE30 and transformed these constructs into a B. burgdorferi B31 strain on a shuttle vector (Tables 1 and 2) [17]. Our previous work utilized bioluminescence to evaluate infection levels of wild-type and mutant B. burgdorferi strains encoding PflaB-Bbluc on a comparable borrelial shuttle vector (Tables 1 and 2) [18]. The comparison of bacterial load between borrelial strains required abundant luminescent signal for the differentiation of subtle changes in the infectivity phenotype, thus the maintenance of a multicopy of PflaB-Bbluc shuttle vector was important for success. To accomplish this task, B. burgdorferi strain ML23, lacking the infectivity-associated linear plasmid 25 (lp25), was used in conjunction with a borrelial shuttle vector encoding the lp25 gene that restores virulence, bbe22 [20-22]. Subsequently bbe22 was delivered together with PflaB-Bbluc, resulting in pBBE22luc [18]. The requirement for bbe22 during murine infection provided an in vivo selective pressure preventing the loss of pBBE22luc as the borrelial population expanded and migrated. Chan et al. incorporated a single copy of PflaB-Bbluc into bbe02, a restriction-modification system of lp25, in B. burgdorferi N40 resulting in a significantly lower but detectable bioluminescent signal in vivo when a high inoculum dose was administered (Tables 1 and 2) [23]. The advantages to a single copy of the luciferase gene are the presence of all borrelial plasmids and no risk of Bbluc loss during the murine infection; however, the low signal above background limits its application. Ideally, a luc gene incorporated into the B. burgdorferi genome with similar signal intensity to that emitted by pBBE22luc will be needed to fulfill this goal. Most recently, a bioluminescent reporter using the B. burgdorferi ospC promoter (PospC) was used to evaluate ospC expression in a temporal and tissue-specific manner using both in vivo and ex vivo imaging. These data provide further support for the complex genetic regulation that is postulated to occur throughout B. burgdorferi infection for individual genes (Tables 1 and 2) [24]. Bioluminescent imaging of B. burgdorferi infected ticks has not been published to date likely due to the following limitations: (1) low numbers of borrelial cells present; (2) requirement for a genome incorporated luciferase with quantitatively detectable signal; (3) delivery of D-luciferin to the vector; and (4) auto-fluorescing chitin that interferes with bioluminescent light emission.
Table 1.
B. burgdorferi bioluminescent plasmids
| Plasmid | Resistance | Comments/Source/Reference |
|---|---|---|
| pJSB175 | strepR | pJD7 shuttle vector carrying PflaB-Bbluc [17] |
| pJSB165 | strepR | pJD7 shuttle vector carrying PospC-Bbluc [17] |
| pJSB104 | strepR | pJD7::PpQE30-Bbluc and PflaB-lacI in tandem for inducible bioluminescence [17] |
| pJSB252 | strepR | pJD7::PpQE30-Bbluc and PflaB-lacI in divergent organization for inducible bioluminescence [17] |
| pBBE22 | luckanR | pBBE22-gate shuttle vector ncoding PflaB-Bbluc to present luciferase in multiple copies [18] |
| pJH410 | kanR | pBBE22 carrying PospC-luc reporter cassette [24] |
| pXbbe02Bbluc-aadA | strepR | Allelic exchange of a single copy of PflaB-Bbluc to displace bbe02 [23] |
Table 2.
B. burgdorferi bioluminescent strains
| Borrelia strains | Genotype/Reference |
|---|---|
| BbJSB175 | Infectious isolate BbDTR630 with pJSB175 that encodes PflaB-Bbluc [17] |
| BbJSB165 | Infectious isolate BbDTR630 with pJSB165 that encodes PospC-Bbluc [17] |
| BbJSB104 | Infectious isolate BbDTR630 with pJSB104 that encodes PpQE30-Bbluc and PflaB-lacI in tandem [17] |
| BbJSB252 | Infectious isolate BbDTR630 with pJSB252 that encodes PpQE30-Bbluc and PflaB-lacI divergently [17] |
| ML23 pBBE22luc | Clonal isolate of B. burgdorferi strain B31 lacking lp25 with pBBE22 containing bbe22 and PflaB-luc [18] |
| JS315 pBBE22luc | ML23 bbk32::StrR with pBBE22 containing bbe22 and PflaB-luc [18] |
| JF105 pBBE22luc | ML23 dbpBA::GentR with pBBE22 containing bbe22 and PflaB-luc [18] |
| ML23 pJH410 | Clonal isolate of B. burgdorferi strain B31 missing lp25 and complemented with bbe22 and PospC-luc on pBBE22 [19] |
| N40 BBE02::Bbluc-aadA | N40 D10/E9 with bbe02 disrupted by PflaB-luc and PflgB-aadA in tandem [23] |
In this chapter, we describe procedures to utilize bioluminescent B. burgdorferi under cultivation (in vitro), during experimental infection (in vivo), and from harvested infected tissues (ex vivo). The below-described methods require Biosafety Level 2 (BSL-2) or Animal Biosafety Level (ABSL-2) facilities and are in agreement with pathogen and animal-handling guidelines as described by the National Institute of Health and an institution-specific Institutional Animal Care and Use Committees (IACUC).
2. Materials
B. burgdorferi encoding Bbluc linked to promoter of interest (see Tables 1 and 2).
1× BSK media with 6% normal rabbit serum.
2 mM d-luciferin (potassium salt) in 1× PBS made fresh daily for in vitro assay.
50 mg/mL d-luciferin (potassium salt) in 1× PBS filter sterilized for in vivo experiments.
2104 EnVision Multilabel Reader (Perkin Elmer).
96-well flat-bottom white plate compatible with plate reader.
96-well flat-bottom black plate.
6–8 week old female Balb/c, C3H or C57BL/6 mice.
Electric hair clipper.
IVIS imaging system (Perkin Elmer).
Living Image software (Perkin Elmer).
XGI-8 gas anesthesia system with XIC-3 animal isolation chamber (Perkin Elmer).
Isoflurane.
Charcoal canister/pellets.
Oxygen (local gas supplier).
70% ethanol.
1 mL insulin syringes with 26G needle.
Necropsy supplies and equipment.
24 or 48-well cell culture plate.
100 mm × 15 mm petri dishes.
3. Methods
The following outlines methods for (1) in vitro luminescence assay, (2) evaluation of bioluminescence B. burgdorferi during mouse infection, and (3) ex vivo assessment of mouse tissues infected with bioluminescent B. burgdorferi. Luciferase containing plasmids and B. burgdorferi strains utilized in bioluminescence experiments are described in Tables 1 and 2.
3.1. In Vitro B. burgdorferi Bioluminescence Assay
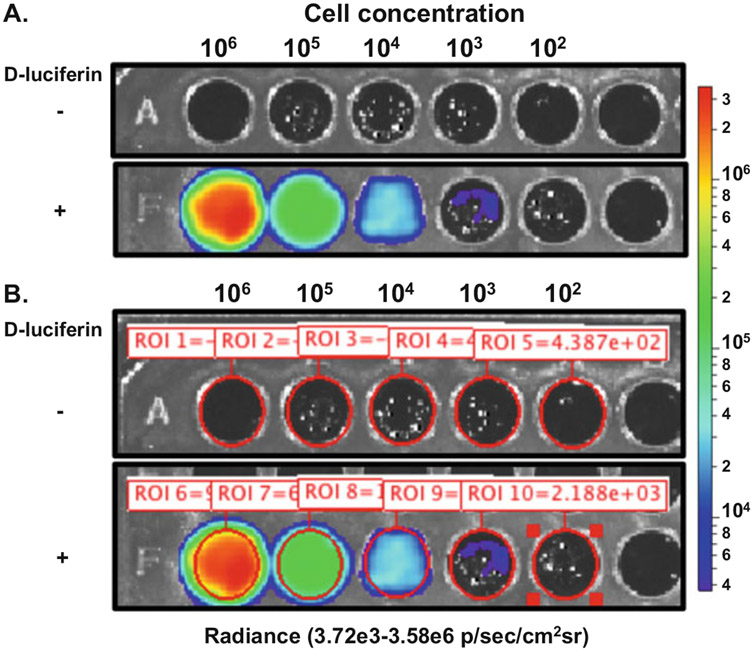
In vitro assessment of bioluminescent B. burgdorferi can be performed in one of two ways: (1) plate reader able to detect bioluminescence or (2) via IVIS (Fig. 1).
Fig. 1.
B. burgdorferi in vitro bioluminescence assay using Perkin Elmer IVIS Spectrum Imaging System. ML23 pBBE22luc was grown to mid-exponential growth phase under microaerophilic conditions. Colorimetric scale on the right indicates the intensity of bioluminescence as it relates to the different number of cells evaluated. (a) B. burgdorferi, tenfold serially diluted from 106 to 102 cells, were plated in duplicate with one set treated with 667 μM d-luciferin as indicated by the +. The row designated – was not treated with d-luciferin to serve as a background control. The image was captured after a 1 min incubation with a 1 min exposure and a binning setting of medium. (b) Region of interest (ROI) was set for each well containing B. burgdorferi as defined by the red circle and accompanying ROI number and measurement
3.1.1. Preparation of B. burgdorferi Cells for In Vitro Bioluminescence Assay
Cultivate B. burgdorferi encoding Bbluc driven by a borrelial promoter of interest under desired conditions and appropriate antibiotic selection.
Allow cultures to reach mid-exponential phase per evaluated by dark field microscopy.
Dilute all samples to 107 cells/mL in 1× BSK+6%NRS and perform serial dilutions to 10 cells/mL.
3.1.2. In Vitro B. burgdorferi Bioluminescence Plate Reader Assay
Due to the variety of plate readers available for bioluminescence assays, refer to the manufacturer’s instructions for proper use of the available model. This protocol is based on the use of the Perkin Elmer 2104 Envision Plate Reader that performs auto-injection of d-luciferin to allow for bioluminescent measurement immediately after treatment.
Transfer 100 μL of each cell concentration into a well in a white 96-well plate compatible with the plate reader. A range of 106 to 1 B. burgdorferi cell should be measured for light production. Bioluminescence should be evaluated from three independently grown cultures under the appropriate conditions and selection.
The program for dispensing 2 mM d-luciferin is as follows: 200 μL/s dispensing speed; 50 μL dispensing volume; syringe filling volume set to full; 1 plate read and one plate repeat.
A background reading is taken of each well prior to injection of the 50 μL 2 mM d-luciferin reagent, diluted to a final concentration of 667 μM, into each well.
Normalized data, adjusted for background, is exported into an Excel file for further statistical analyses. Independent cultures should be averaged and standard deviation or error calculated. Statistical significant test is determined by the experimental design of the study.
3.1.3. In Vitro B. burgdorferi Bioluminescence Assay Using the Perkin Elmer IVIS Imaging System
The following protocol is specific for the Perkin Elmer IVIS imaging system and the Living Image Software. Use caution when placing liquids into the IVIS and avoid spills as it could be harmful to the equipment.
Transfer 100 μL of each cell concentration in duplicate into a 96-well flat-bottom black plate. Samples ranging from 106 to 1 B. burgdorferi should be measured for light production. One set of samples will not be treated with 2 mM d-luciferin to serve as a background control (Fig. 1a).
Prepare the imaging system first by initializing Living Image software followed by the insertion of the black plastic sheet on the platform of the machine. Set the field of view to include all sample containing wells and as little of the surrounding space as possible. Also adjust height of subject to represent the height of the 96-well black plate, approximately 1 cm. The binning setting and exposure time will be determined by the intensity of signal from a given bioluminescent strain. Begin with binning setting of “Large” and 1 min exposure time. If image reaches saturation reduce binning or exposure time. F-stop and emission filter should remain on the default setting of 1 and open, respectively. Further detail is available in the manufacturer’s manual.
Manually pipette 50 μL of 2 mM d-luciferin to each well of the second set of samples and mix by pipetting three to four times. Allow the samples to incubate at room temperature for 1 min after pipetting the D-luciferin to allow the flash point to occur in each sample and the bioluminescent signal will quickly reduce to a plateau level emission (also known as a glow reaction).
Place the 96-well black plate within the laser grid on the IVIS platform and acquire the image. An exposure time of 1–5 min should be sufficient to acquire an image in the quantifiable range of greater than 600 counts but less than saturation (60,000 counts). Saturated images cannot be used for quantification of photons/s.
Analysis of images and quantitation of bioluminescence is outlined in Subheading 3.4.
3.2. In Vivo B. burgdorferi Bioluminescence Study
B. burgdorferi cultivation and preparation of inoculum for murine infection are standard protocols in the field [7]; therefore, their preparation will not be addressed here. Perkin Elmer IVIS is available in three models with CCD cameras that vary in sensitivity and should be taken into consideration for your specific probe design for each in vivo imaging experiment (see Note 1).
3.2.1. Preparation for the Bioluminescent B. burgdorferi In Vivo Infectivity Study
Prepare a batch of d-luciferin potassium salt at 50 mg/mL, in sterile 1× PBS, sufficient for use throughout the course of the infection period to avoid variation in concentration between time points. d-luciferin potassium salt is light sensitive and should be protected during preparation, storage, and use. Once in solution, d-luciferin should be filter sterilized using a 0.2 μm filter and placed in 1 mL aliquots prepared in light protected microcentrifuge tubes. 50 mg/mL d-luciferin potassium salt is stable for several months when stored at −20 °C (see Note 2).
Shave the ventral side of each mouse the day prior to inoculation from between the hind legs up to the bottom of the rib cage using electric hair clippers. Remove the hair from the belly without a guard revealing bare skin while being careful not to cut into the dermis. This prevents impedance of light signal due to the opaqueness due to melanin within the hair.
Organize mice into groups not larger than five mice with the intension of one mouse serving as a background control that will not be given d-luciferin (see Note 3).
3.2.2. Tracking Bioluminescent B. burgdorferi Throughout Infection
Clean the ventral side at the site of inoculation by swabbing the skin with 70% ethanol.
Intradermally inject the predetermined number of B. burgdorferi cells (see Note 4). One to 2 h after inoculation the mice should be imaged to confirm the equal delivery of B. burgdorferi cells (depending on inoculum dose) between groups.
- Prepare the XGI-8 anesthesia system. Further detail can be found in the Xenogene XGI-8 anesthesia system manual.
- Weigh F-air canisters that are filled with charcoal filters prior to each use of the XGI-8 anesthesia system. Charcoal filters must be discarded and replaced after the initial weight increases to 50 g due to use.
- Confirm that the isoflurane level is sufficient for the required imaging time. Once pressure is added to the system additional isoflurane cannot be added.
- Turn on the evacuation pump.
- Slowly turn on the valve of the oxygen tank with the regulator set at 50 psig. Turn the oxygen handle on the XGI-8 unit to the ON position.
- Turn dial of the isoflurane vaporizer to 2–2.5.
- Toggles for the IVIS flow and animal isolation chamber should be set at 0.25 lpm and 1.5 lpm, respectively, and will be turned on and off throughout the experiment as needed.
- Set up Perkin Elmer IVIS
- Open the Perkin Elmer Living Image software and select the initialization button in the acquisition control panel. Do not open the door when the light is red. A green light will appear above the door upon the completion of initialization.
- In the IVIS acquisition control panel the luminescent, photographic imaging mode, and overlay boxes are selected by default. The F/stop and emission filter should also stay on the default setting of 1 and open, respectively. Increase binning to “Large” and the measure of time from s to min. The length of exposure time may vary depending upon the intensity of the signal. Subject height of 1.5 cm is appropriate for most mouse strains and should not be modified. The default temperature of the platform is 37 °C (see Note 5).
- Prepare the IVIS platform by placing the provided black plastic mat shiny side down on the surface under the isoflurane manifold. Remove the rubber stoppers from the manifold and insert the required number for nose cones. Black shields should be placed between the nose cones to block signal from the neighboring mouse allowing quantification of an individual specimen without overlapping bioluminescence. Adjust the field of view in the software control panel to include all mice as indicated by the laser field displayed on the platform. Field of view D is needed for five mice.
- Create a folder to save the acquired images.
Intraperitoneally inject four of five mice with 5 mg of d-luciferin. The fifth mouse that is not injected with d-luciferin to serve as the background control mouse and should be marked on the tail with a marker for identification purposes.
Transfer the mice into the animal isolation chamber for treatment with isoflurane and to allow d-luciferin to circulate for 7–10 min. Securely lock the lid to the animal isolation chamber and turn on the flow of isoflurane by lifting the toggle switch. Double check the flow of oxygen in the evacuation pump gauge and isoflurane in the chamber gauge throughout the experiment.
Sedated mice can now be transferred to the IVIS platform for imaging. Turn on the flow of isoflurane to the IVIS and place each mouse in a nose cone with the ventral side facing the camera located at the top of the machine. It is recommended that the no luciferin treated mouse be in the same location on the manifold for all images to facilitate analyses and figure preparation. It is our preference to have the background mouse in the first placement on the far left (Fig. 2). When there are no mice in the isolation chamber the toggle can be turned to the off position to prevent the flow of isoflurane.
Image the mice for 1 min that will be used for quantification as described in Subheading 3.4. Images below 600 or above 60,000 counts cannot be used for ROI quantification. If the image is not in the 600–60,000 counts range at any time point, modify the exposure time or binning accordingly to reach this target (see Note 6). Save the image to a designated folder for later analyses.
Image mice for 10 min to be used for normalized images at the end of the experiment (Fig. 2). Save the image to a designated folder for later analyses.
Repeat the step 1 and 10 min imaging for the remaining sides of the mouse if so desired (Fig. 2) (see Note 7).
When imaging is complete, return mice to the cage and monitor the group as they come out of sedation. Turn off flow of isoflurane to the IVIS.
Analysis of images and quantitation of bioluminescence is outlined in Subheading 3.4.
- Shut down the IVIS and XGI-8 anesthesia system.
- Adjust the dial of isoflurane vaporizer to zero. Lock the isolation chamber and switch the toggle to the on position to allow the flow of oxygen for 5 min, purging isoflurane from the system.
- Depressurize the XGI-8 system by turning off the oxygen at the tank regulator while leaving the chamber toggle in the on position. When the regulator pressure and XGI-8 chamber gauge reaches zero the toggle can be turned off.
- Turn the handle for the oxygen pump to the off position.
- Turn off the evacuation pump.
- Weigh the F-canisters and record the weights.
- Wipe down the animal isolation chamber with 70% ethanol.
- Remove nose cones and shields from IVIS platform. Replace rubber stoppers into the manifold.
- Outside of the IVIS machine clean the nose cones and shields with 70% ethanol and store.
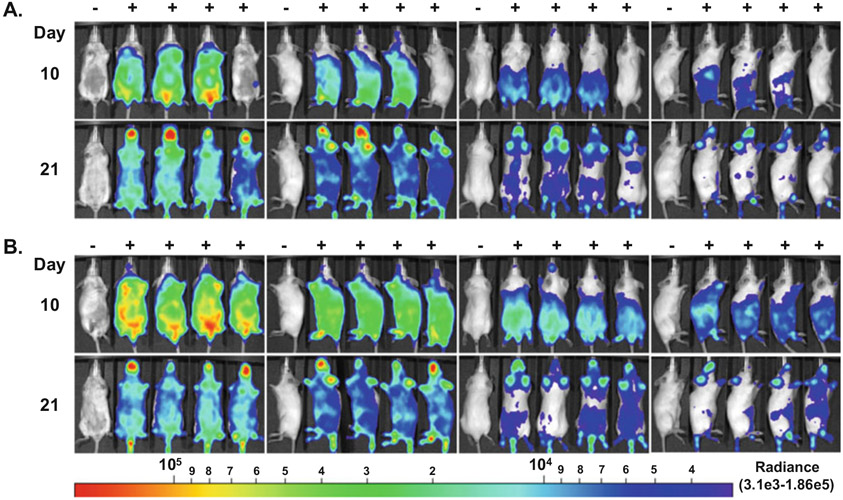
Fig. 2.
In vivo imaging of mice infected with ML23 pBBE22luc. Balb/c mice were intradermally infected with 104 (a) or 106 (b) ML23 pBBE22luc and were treated with 5 mg of d-luciferin to monitor infection indicated by the +. In vivo imaging was performed on day 10 and 21 following inoculation using the Perkin Elmer IVIS Spectrum Imaging System. Mice were imaged on all sides in the order presented from left to right for a 10 min exposure and at the large binning setting. Images were normalized to background as determined by the mouse on the far left that did not receive D-luciferin treatment and is designated with a −. All the images were set to the same radiance scale as shown below with the radiance range of 3.1e3 to 1.86e5
3.3. Ex Vivo Assessment of Bioluminescent B. burgdorferi Infected Tissues
Approximately 1 h after d-luciferin injection, for total mouse in vivo imaging, the bioluminescent signal resolves and will not overlap with further imaging of ex vivo harvested tissues described in this section. As with in vivo imaging, one mouse in each group should not undergo d-luciferin treatment to serve as a background control. It is recommended that one image mice in the same order as they are positioned in the in vivo image to allow direct comparison with the ex vivo signal. One mouse at a time is treated with d-luciferin and undergoes necropsy to capture bioluminescent signal as quickly as possible. A d-luciferin and ATP soak of tissues is used to prevent dehydration of tissues and maintain signal.
Prepare a 4 mM d-luciferin and a 2 mM ATP solution in sterile 1× PBS that will be used as a soak immediately after the tissues are harvested. Store on ice and protect from light. Prior to sacrificing an individual mouse and harvesting tissues, aliquot 500 μL of the soak solution into the wells of a cell culture plate equivalent to the number of tissues collected. For the background control tissues use 1× PBS as the soak.
In the Living Image control panel, reduce the temperature of the imaging platform from 37 °C to 23 °C to prevent dehydration of tissues during imaging. Adjust the height of subject to 1 cm and field of view to B for the approximate size of a 100 mm petri dish.
Administer a double bolus of d-luciferin via intraperitoneal injection for a final treatment of 10 mg and allow circulation for 7–10 min prior to sacrificing mouse by approved methods.
After mouse is euthanized, clean the hair with 70% ethanol and harvest tissues as quickly as possible and transfer to the soak solution for 3–5 min. Recommended tissues for ex vivo imaging are the skin flank, ear, inguinal lymph node, heart, bladder, and tibiotarsal joint.
Arrange all the harvested tissues within a petri dish with the exception of the skin flank that will be imaged separately due to the high level of emission (Fig. 3). Image the tissues for 1 and 10 min, as described for the in vivo imaging, for quantitation and normalized images, respectively (Fig. 3). Save images to a designated file.
The abundant skin flank signal will quench the visible signal of tissues with less colonization of B. burgdorferi; therefore, this particular tissue needs to be imaged separately. The skin flank should remain in the soak while the other tissues are imaged. Image the skin flank for 1 and 10 min for quantitation and normalized images, respectively. Save these images to a designated file.
Immediately after the completion of the imaging evaluation, next process the tissues for validation methodologies, such as cultivation, qPCR, and qRT-PCR (not discussed in this chapter).
Repeat for each mouse in each group being evaluated.
Analysis of images and quantitation of bioluminescence is outlined in Subheading 3.4.
Remove the black mat from the platform and wipe down with 70% ethanol.
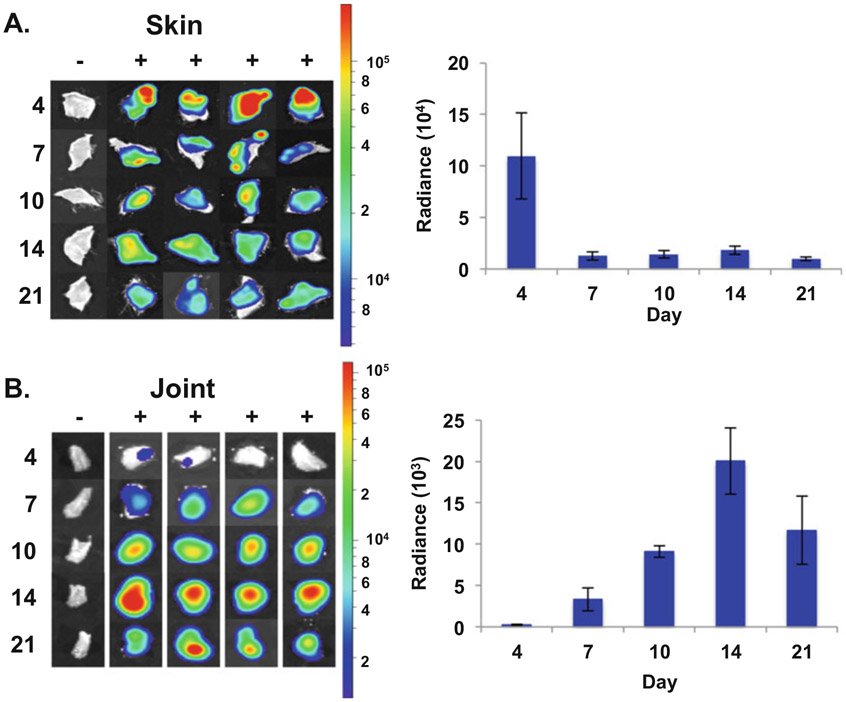
Fig. 3.
Ex vivo imaging of ML23 pBBE22luc infected murine tissues. Skin (a) and tibiotarsal joints (b) were harvested from 105 ML23 pBBE22luc infected Balb/c mice that were treated with 10 mg of d-luciferin with the exception of the background control, designated −. Bioluminescence from harvested tissues was assessed with the Perkin Elmer IVIS Spectrum Imaging System for 10 and 1 min for the normalized image and quantitation, respectively. Images were normalized for background and set on a common colorimetric scale for each tissue as shown on the right. Background radiance was subtracted from each d-luciferin treated sample per time point resulting in a normalized radiance value. Normalized radiance values were averaged and displayed in the bar graph to the right to quantify the borrelial load in the skin (a) or joint (b) over the course of a 21 day infection. Bars represent standard error
3.4. Analyses of IVIS Data
Bioluminescence will be quantified from a defined region of interest (ROI) from selected images independent of whether the data were obtained from in vitro, in vivo, or ex vivo samples (Figs. 1 and 3). For 10 min exposures to be directly compared, all images must be normalized by defining a common radiance scale where the minimum removes all background and the maximum displays the most intense signal within the series of images (Figs. 2 and 3).
3.4.1. Quantitation of IVIS Bioluminescence
Open image files to be analyzed with the browsing function. Go to file and down to browse to choose the folders containing images to be analyzed. Selected files will be displayed in the Living Image Browser window.
Highlight the desired image for analysis and select the load button. Multiple images can be analyzed together by highlighting all desired images and selecting “load as a group.”
Each image for quantitation must have a maximum count between 600 and 60,000 counts. Change the units from counts to photons for the images to be analyzed.
Using the ROI submenu in the Tool Palette to define the space to be measured by creating and placing a grid, square, circle, or free drawing that encompasses the area of interest (Fig. 1b).
Press “Measure ROI” in the ROI submenu and a new window will appear entitled “ROI measurements” consisting of total flux (photons/sec (p/s)) and average radiance (photons/sec/centimeter2/steradian(p/s/cm2/sr)). Total flux is utilized when the space measured is the same area for each well or mouse across all images analyzed and is often used for measuring the wells of 96-well plates and whole animals. Radiance accounts for differences in area and, as such, are preferred for irregular shaped ex vivo imaged tissues (see Note 8). Copy the ROI measurements into a spreadsheet and label the samples as appropriate.
In each group subtract the ROI value from the background well/mouse/tissue from those treated with d-luciferin, yielding normalized flux or radiance values. Average the normalized values, then calculate the standard error and standard deviation. Data can be graphed as scatterplots, line, or bar graphs depending on the design of each experiment (Fig. 3). Statistical analyses to determine significance will be depfimage files as described endent on the experimental design of a given study.
3.4.2. Normalization of Bioluminescent Images
Select image files as described in Subheading 3.4.1. To determine the setting for the radiance color spectrum, it is recommended that images are opened in groups that will be compared to one another (Figs. 2 and 3).
In the Tool Palette under the Adjust Image submenu, deselect the Individual box for the images in a group to be on the same colorimetric scale.
Increase the minimum Aggregate Color scale until no bioluminescent signal is visible on the no luciferin control mouse across all the images in the group. The maximum setting can be modified as needed.
To improve the appearance of pixilation change smoothing to 5 × 5 and binning to 4.
Save a .tif of the modified images to generate figures for publication.
4. Notes
Camera sensitivities vary between the Perkin Elmer IVIS imaging systems models and should be taken into account when designing an in vivo or ex vivo bioluminescent murine experiment. The Spectrum model has the most sensitive camera and is able to perform 3D reconstruction. 3D reconstruction is not available on all models and is not well suited for all pathogenic systems. Additional imaging modalities can be performed in conjunction with bioluminescence imaging.
d-luciferin in 1× PBS at 50 mg/mL can be stably stored at −20 °C for several months. The reagent typically has a vibrant yellow appearance that tends to fade and take on a gray tone when degradation has occurred. If a color change is observed the reagent should not be used for in vivo experiments, as it will yield a reduction in light emission.
There are several types of controls that have been used for in vivo bioluminescent monitoring of pathogens. We have opted for using a control mouse infected with the identical strain as those in the same image and forgoing the d-luciferin treatment, i.e., a no luciferin control. Alternatively, one mouse in each group can be infected with a similar strain lacking Bbluc or an uninfected control mouse that receives d-luciferin treatment. It is important that each image, time point, and group contains a background control mouse as the level of background bioluminescence varies throughout the course of infection and from study to study.
Intradermal versus subcutaneous injections: Both types of injections will result in a successful inoculation resulting in infection by wild-type B. burgdorferi. Subcutaneous injection emits a lower signal due to the depth of the injection relative to intradermal infection, but is quantifiable for analysis, particularly at later time points.
The binning setting determines the pixel size on the CCD camera. The larger the binning the larger the pixels, thus increased sensitivity of bioluminescent detection. Larger binning also results in a reduced spatial resolution that is acceptable due to the dispersion of the light emission that occurs as it travels through tissues.
Extended exposure times increase the sensitivity of detection, but can also saturate the image when greater than 60,000 counts are measured causing image saturation. Counts of 600 or less are considered noise; therefore, it is recommended that quantified images have counts between 600 and 60,000. The manufacturer’s instructions indicate that exposure time greater than 5 min is of little benefit for most systems. Throughout the development of in vivo imaging of bioluminescent B. burgdorferi 10 min was determined to be ideal for the longer exposure and provided substantially more sensitivity to the analysis (Skare & Hyde unpublished results).
Mice can be imaged on the ventral, dorsal, and both lateral sides. The different sides of the mouse should be imaged in the same order for the entire experiment. The intensity of the signal begins to decline over time; thus, the side imaged first cannot be accurately compared to the last image that is taken approximately 40 min after the injection of d-luciferin. If the mouse is imaged in the same pattern of ventral, lateral left, dorsal and lateral right, these images are suitable for comparison across time points.
ROI measurements provide bioluminescent measurements in the form of total flux (photons/s) and radiance (photons/s/cm2/sr). When performing in vivo imaging, a whole mouse space is defined (typically a rectangle) to use as a template for ROI measurement for every mouse and time point in a given experiment. Radiance calculates the area of the defined space for the direct comparison of areas, mice, or tissues of different sizes. Radiance is the preferred unit of measurement for ex vivo experiments as tissues differ in shape and size by tissue, mouse, and time, which can be dependent upon the amount of disease present.
Acknowledgments
The previously published images modified for this chapter were supported by Public Health Service grants R01-AI058086 (to J.T.S.) and R21-AI101740-01 (to J.A.H.) from the National Institute of Allergy and Infectious Diseases. We also acknowledge Michael Norgard and Jon Blevins for the B. burgdorferi codon optimized luc gene. We also extend our gratitude to Jeffrey Cirillo, Geoffery Kapler, and Raquel Sitcheran for generously sharing equipment and resources necessary to develop these methods for B. burgdorferi. We wish to thank Kevin Francis, Will Hauser, and Brad Taylor at Perkin Elmer for their technical support and advice.
References
- 1.Steere AC, Coburn J, Glickstein L (2004) The emergence of Lyme disease. J Clin Invest 113:1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stanek G, Wormser GP, Gray J, Strle F (2012) Lyme borreliosis. Lancet 379:461–473. doi: 10.1016/S0140-6736(11)60103-7 [DOI] [PubMed] [Google Scholar]
- 3.Shapiro ED (2014) Lyme disease. N Engl J Med 370:1724–1731. doi: 10.1056/NEJMcp1314325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mead PS (2015) Epidemiology of Lyme disease. Infect Dis Clin N Am 29:187–210. doi: 10.1016/j.idc.2015.02.010 [DOI] [PubMed] [Google Scholar]
- 5.Radolf JD, Caimano MJ, Stevenson B, Hu LT (2012) Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat Rev Microbiol 10:87–99. doi: 10.1038/nrmicro2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbour AG (1984) Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med 57:521–525 [PMC free article] [PubMed] [Google Scholar]
- 7.Zückert WR (2007) Laboratory maintenance of Borrelia burgdorferi. Curr Protoc Microbiol Chapter 12:Unit 12C.1. doi: 10.1002/9780471729259.mc12c01s4 [DOI] [PubMed] [Google Scholar]
- 8.Samuels DS (2011) Gene regulation in Borrelia burgdorferi. Annu Rev Microbiol 65:479–499. doi: 10.1146/annurev.micro.112408.134040 [DOI] [PubMed] [Google Scholar]
- 9.Miller JC (2005) Example of real-time quantitative reverse transcription-PCR (Q-RT-PCR) analysis of bacterial gene expression during mammalian infection: Borrelia burgdorferi in mouse tissues. Curr Protoc Microbiol Chapter 1D:Unit 1D.3. doi: 10.1002/9780471729259.mc01d03s00 [DOI] [PubMed] [Google Scholar]
- 10.Contag CH, Contag PR, Mullins JI et al. (1995) Photonic detection of bacterial pathogens in living hosts. Mol Microbiol 18:593–603 [DOI] [PubMed] [Google Scholar]
- 11.Hutchens M, Luker GD (2007) Applications of bioluminescence imaging to the study of infectious diseases. Cell Microbiol 9:2315–2322. doi: 10.1111/j.1462-5822.2007.00995.x [DOI] [PubMed] [Google Scholar]
- 12.Contag PR (2008) Bioluminescence imaging to evaluate infections and host response in vivo. Methods Mol Biol 415:101–118. doi: 10.1007/978-1-59745-570-1_6 [DOI] [PubMed] [Google Scholar]
- 13.Wiles S, Robertson BD, Frankel G, Kerton A (2009) Bioluminescent monitoring of in vivo colonization and clearance dynamics by light-emitting bacteria. Methods Mol Biol 574:137–153. doi: 10.1007/978-1-60327-321-3_12 [DOI] [PubMed] [Google Scholar]
- 14.Andreu N, Zelmer A, Wiles S (2011) Noninvasive biophotonic imaging for studies of infectious disease. FEMS Microbiol Rev 35:360–394. doi: 10.1111/j.1574-6976.2010.00252.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waidmann MS, Bleichrodt FS, Laslo T, Riedel CU (2011) Bacterial luciferase reporters: the Swiss army knife of molecular biology. Bioeng Bugs 2:8–16. doi: 10.4161/bbug.2.1.13566 [DOI] [PubMed] [Google Scholar]
- 16.Francis KP, Joh D, Bellinger-Kawahara C et al. (2000) Monitoring bioluminescent Staphylococcus aureus infections in living mice using a novel luxABCDE construct. Infect Immun 68:3594–3600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blevins JS, Revel AT, Smith AH et al. (2007) Adaptation of a luciferase gene reporter and lac expression system to Borrelia burgdorferi. Appl Environ Microbiol 73:1501–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyde JA, Weening EH, Chang M et al. (2011) Bioluminescent imaging of Borrelia burgdorferi in vivo demonstrates that the fibronectin-binding protein BBK32 is required for optimal infectivity. Mol Microbiol 82:99–113. doi: 10.1111/j.1365-2958.2011.07801.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skare JT, Shaw DK, Trzeciakowski JP, Hyde JA (2016) In Vivo imaging demonstrates that Borrelia burgdorferi ospC is uniquely expressed temporally and spatially throughout experimental infection. PLoS ONE 11(9):e0162501. doi: 10.1371/journal.pone.0162501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purser JE, Norris SJ (2000) Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc Natl Acad Sci U S A 97:13865–13870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labandeira-Rey M, Skare JT (2001) Decreased infectivity in Borrelia burgdorferi strain B31 is associated with loss of linear plasmid 25 or 28-1. Infect Immun 69:446–455. doi: 10.1128/IAI.69.1.446-455.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labandeira-Rey M, Seshu J, Skare JT (2003) The absence of linear plasmid 25 or 28-1 of Borrelia burgdorferi dramatically alters the kinetics of experimental infection via distinct mechanisms. Infect Immun 71:4608–4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan K, Alter L, Barthold SW, Parveen N (2015) Disruption of bbe02 by insertion of a luciferase gene increases transformation efficiency of Borrelia burgdorferi and allows live imaging in Lyme disease susceptible C3H mice. PLoS One 10:e0129532. doi: 10.1371/journal.pone.0129532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skare JT, Shaw DK, Trzeciakowski JP, Hyde JA (2016) In vivo imaging demonstrates that Borrelia burgdorferi ospC is uniquely expressed temporally and spatially throughout experimental infection. PLoS One 11:e0162501. doi: 10.1371/journal.pone.0162501 [DOI] [PMC free article] [PubMed] [Google Scholar]