Abstract
The mammalian nervous system is a complex network of interconnected cells. We review emerging techniques that use the axonal transport of adeno-associated virus (AAV) vectors to dissect neural circuits. These intersectional approaches specifically target AAV-mediated gene expression to discrete neuron populations based on their axonal connectivity, including: 1) neurons with 1 defined output, 2) neurons with 1 defined input, 3) neurons with 1 defined input and 1 defined output, and 4) neurons with 2 defined inputs or outputs. The number of labeled neurons can be directly controlled to trace axonal projections and examine cellular morphology. These approaches can precisely target the expression of fluorescent reporters, optogenetic ion channels, chemogenetic receptors, disease-associated proteins, and other factors to defined neural circuits in mammals ranging from mice to macaques, and thereby provide a powerful new means to understand the structure and function of the nervous system.
Keywords: AAV, axonal transport, retrograde, anterograde, intersectional, neural circuits
Introduction
The mammalian nervous system contains anatomically distinct neuron populations connected by axon networks of immense complexity. Numerous methods have been used to trace these neural circuits, including axonally transported organic compounds conjugated to fluorescent molecules or detected by immunolabeling, axonally transported inorganic fluorescent dyes or microspheres, and recombinant gene transfer vectors generated from axonally transported viruses (Saleeba et al., 2019). Herpes-simplex virus 1 (HSV1) strain H129 is used for polysynaptic anterograde labeling of neurons (Archin and Atherton, 2002; Lo and Anderson, 2011), while rabies virus and pseudorabies virus are used for polysynaptic retrograde labeling (Strack et al., 1989; Card et al., 1998; Wickersham et al., 2007; Kim et al., 2016). HSV1 and rabies virus vectors have also been modified for monosynaptic anterograde and retrograde tracing, respectively, by deletion of viral genes that are essential for infection and replication: when these essential genes are supplied in trans to a defined neuron population, the viral tracer spreads only a single synapse beyond this neuron population (Wickersham et al., 2007; Kim et al., 2016; Zeng et al., 2017).
Adeno-associated virus (AAV) is a non-enveloped parvovirus carrying a single-stranded DNA genome of ~4.7 kb (Atchison et al., 1965). Recombinant AAV vectors are widely used for both clinical gene therapy and experimental gene delivery because they drive strong, safe, and long-lasting gene expression in non-dividing cells (Kaplitt et al., 1994; Deverman et al., 2018; Hudry and Vandenberghe, 2019). AAV vectors have several advantages relative to other viral vectors. Wild-type AAV is not pathogenic and AAV vectors are typically handled at biosafety level (BSL)-1, which does not require specialized equipment such as a biosafety cabinet. Unlike helper-dependent HSV1 and rabies virus, AAV does not replicate prior to axonal transport and a second helper virus is not required to initiate monosynaptic tracing. Many recombinant AAV genome plasmids carrying diverse promoters and transgenes are available from the Addgene repository (Table 1), and many natural and engineered AAV capsids with unique tropism have been described (Gao et al., 2004; Tervo et al., 2016; Chan et al., 2017; Davidsson et al., 2019). Manufacturers such as Addgene and the UNC Vector Core sell ready-to-use aliquots of common AAV vectors. If needed, many academic and commercial facilities can produce affordable custom AAV preparations. These advantages make AAV vectors relatively straightforward to obtain and use for neural circuit tracing. Not only can AAV vectors express fluorescent proteins in neurons to trace their axonal projections (Oh et al., 2014), but recent advances now support the use of AAV vectors for monosynaptic retrograde and anterograde labeling, in addition to specific labeling of neurons that send outputs to or receive inputs from multiple defined brain regions. This expanded toolset relies on two innovations: the directed evolution of new AAV capsids for improved axonal transport and the development of new methods for intersectional gene expression.
Table 1. Select Plasmids Available from the Addgene Repository.
Ready-to-use AAV preparations of many of these plasmids are sold directly by Addgene.
| Catalog Number | Name | Source | Notes |
|---|---|---|---|
| Recombinase AAV Plasmids | |||
| 105555 | pENN.AAV.hSyn.Cre.hGH | James M. Wilson (University of Pennsylvania) | Expresses Cre using the hSyn1 promoter. Does not contain a WPRE. |
| 51507 | AAV pmSyn1-EBFP-Cre | Hongkui Zeng (Allen Institute for Brain Science) | Expresses Cre fused to eBFP using the mouse synapsin 1 (mSyn1) promoter. Does not contain a WPRE. |
| 51669 | AAV phSyn1(S)-FlpO-bGHpA | Hongkui Zeng (Allen Institute for Brain Science) | Expresses FlpO using the hSyn1 promoter. FlpO is codon-optimized for mice. Does not contain a WPRE. |
| 149296 | pAAV-nef-CIAO2-Flp | Edward Callaway (Salk Institute) | For Cre-dependent expression of FlpO. Uses a CIAO design to reduce leak expression. |
| Cre-Dependent Fluorescent Reporter AAV Plasmids | |||
| 28306 | pAAV-FLEX-tdTomato | Edward Boyden (Massachusetts Institute of Technology) | For Cre-dependent expression of tdTomato. |
| 51502 | AAV pCAG-FLEX-EGFP-WPRE | Hongkui Zeng (Allen Institute for Brain Science) | For Cre-dependent expression of eGFP. |
| 114471 | pAAV-Ef1a-fDIO mCherry | Karl Deisseroth (Stanford University) | For Flp-dependent expression of mCherry. |
| 45185 | AAV-EF1a-BbTagBY | Joshua Sanes (Harvard University), Dawen Cai (University of Michigan) | For Cre-dependent Brainbow 3.1 labeling by coinjection with AAV #45186. |
| 45186 | AAV-EF1a-BbChT | Joshua Sanes (Harvard University), Dawen Cai (University of Michigan) | For Cre-dependent Brainbow 3.1 labeling by coinjection with AAV #45185. |
| 55650 | pAAV-hSyn Con/Fon EYFP | Karl Deisseroth (Stanford University) | For expression of eYFP requiring both Cre and Flp activity. |
| 112677 | pAAV EF1a Nuc-flox(mCherry)-EGFP | Brandon Harvey (National Institutes of Health) | For expression of nuclear-localized mCherry that is switched to nuclear-localized eGFP by Cre activity. |
| Additional Cre-Dependent AAV Plasmids | |||
| 20298 | pAAV-EF1a-double floxed-hChR2(H134R)-EYFP-WPRE-HGHpA | Karl Deisseroth (Stanford University) | For optogenetic excitation by Cre-dependent expression of channelrhodopsin-2 fused to eYFP. |
| 22222 | AAV-FLEX-Arch-GFP | Edward Boyden (Massachusetts Institute of Technology) | For optogenetic inhibition by Cre-dependent expression of archaerhodopsin-3 fused to eGFP. |
| 44361 | pAAV-hSyn-DIO-hM3D(Gq)-mCherry | Bryan Roth (University of North Carolina) | For chemogenetic excitation by Cre-dependent expression of Gq-coupled human muscarinic receptor fused to mCherry. |
| 44362 | pAAV-hSyn-DIO-hM4D(Gi)-mCherry | Bryan Roth (University of North Carolina) | For chemogenetic inhibition by Cre-dependent expression of Gi-coupled human muscarinic receptor fused to mCherry. |
| 104492 | pGP-AAV-syn-FLEX-jGCaMP7f-WPRE | Douglas Kim and GENIE project (Howard Hughes Medical Institute) | For Cre-dependent expression of the fluorescent calcium sensor jGCaMP7f. |
| 124364 | pAAV-FLEX-DTR-GFP | Eiman Azim (Salk Institute), Thomas Jessell (Howard Hughes Medical Institute) | For targeted cell ablation by Cre-dependent expression of the diphtheria toxin receptor fused to eGFP. |
| Other Plasmids | |||
| 81070 | rAAV2-retro helper | Alla Karpova (Howard Hughes Medical Institute), David Schaffer (UC Berkeley) | Helper plasmid for production of AAV2-retro. |
| 100798 | pAAV-syn-FLEX-splitTVA-EGFP-tTA | Ian Wickersham (Massachusetts Institute of Technology) | For Cre-dependent expression of rabies helper TVA protein and eGFP when coinjected with AAV #100799. |
| 100799 | pAAV-TREtight-mTagBFP2-B19G | Ian Wickersham (Massachusetts Institute of Technology) | For Cre-dependent expression of rabies helper G protein and mTagBFP2 when coinjected with AAV #100798. |
The abbreviation “FLEX” stands for “flip excision,” which is an alternative term for a Cre-dependent DIO design.
Axonal Transport of AAV Vectors
Kaspar and colleagues first reported in 2002 that AAV vectors can retrogradely transduce neurons (Kaspar et al., 2002). This occurs when AAV particles are endocytosed at an axon terminal and are retrogradely transported to the cell body within the Rab7-positive late-endosome/lysosome by the motor dynein in complex with dynactin (Castle et al., 2014b). Conversely, AAV vectors endocytosed at the cell body are most commonly trafficked directly to the nucleus and Golgi, but some particles undergo rapid anterograde axonal transport mediated by the motor kinesin-2 (Castle et al., 2014b). Although anterogradely transported AAV particles can be released at synapses and endocytosed by second-order neurons, trans-synaptic transduction by AAV vectors is weak and rarely detectable by immunolabeling. However, AAV mRNA may be detected in second-order neurons by in situ hybridization (Cearley and Wolfe, 2007; Castle et al., 2014a). The mechanisms of retrograde and anterograde axonal transport appear to be conserved among AAV serotypes (Castle et al., 2014a), although differences in endocytosis by projection neurons or at axon terminals could lead to different rates of axonal transport among serotypes.
Anterograde trans-synaptic transduction by common AAV serotypes is rarely detectable and retrograde transduction is typically weak, but directed evolution of AAV capsid libraries has identified novel AAV capsids that demonstrate enhanced axonal transport in the mammalian brain. AAV2-retro is a variant of the AAV2 capsid containing a 10-amino acid insertion in the heparin binding loop and two additional point mutations (Tervo et al., 2016). AAV2-retro demonstrates up to 100-fold greater retrograde transduction of mouse neurons than other common AAV serotypes (Tervo et al., 2016). A recent report that AAV2-retro is effective in rhesus macaques suggests that its retrograde transport is enhanced across mammalian species, further expanding its utility for neuroscience research (Weiss et al., 2020). A second new capsid with similarly enhanced retrograde transport, AAV-MNM004, was also recently identified by directed evolution (Davidsson et al., 2019).
These novel capsids can broadly map inputs to a given brain region, but in most cases they cannot target gene expression to a single defined population of neurons: AAV2-retro and AAV-MNM004 will transduce cells at the injection site as well as most neuron populations that innervate the injected region. Further, no novel capsids with enhanced anterograde trans-synaptic transduction have been identified. When more precise targeting is needed, intersectional methods can restrict gene expression to neuron populations that are defined by their axonal connectivity.
Intersectional Gene Expression
Intersectional approaches use at least two AAV vectors: the first encodes a site-specific DNA recombinase, such as Cre (“AAV-Cre”), and the second carries a genome which requires this recombinase to activate gene expression. This is called “intersectional” because only neurons transduced by both AAV vectors will express the gene of interest. A single Cre recombinase enzyme can permanently turn on gene expression from the recombinase-dependent AAV genome, so only a very small amount of the recombinase is needed. AAV-Cre can thus be delivered not only by retrograde transport, but also by trans-synaptic anterograde transport (Zingg et al., 2017).
Cre-dependent AAV genomes are commonly produced by flanking an inverted gene sequence with two pairs of Cre-recognition sequences (“lox sites”): Cre recombinase turns on gene expression by flipping the gene into the correct orientation (Sauer and Henderson, 1988; Tsien et al., 1996). This design is termed a Double Inverted Open reading frame or “DIO” (Zhang et al., 2010). Flp recombinase is also commonly used; the equivalent Flp-dependent design is termed “FlpDIO” or “fDIO” (Fenno et al., 2014).
Retrograde Expression: 1 Defined Output
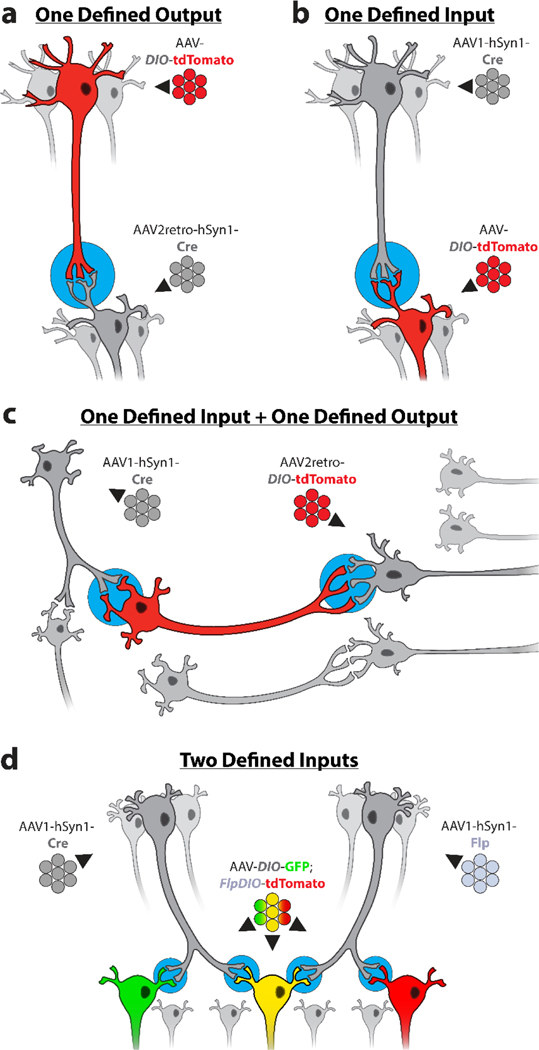
The first intersectional approach uses retrograde transport to specifically label neurons in the target brain region that send outputs to a defined brain region (Figure 1A). Cre-dependent AAV is injected in the target region and AAV-Cre is injected in the output region: retrograde transport of AAV-Cre will specifically label neurons in the target brain region that innervate the defined output region (Conner et al., 2019). Although most AAV serotypes undergo retrograde transport, we recommend using AAV2-retro-Cre for optimal efficiency. This method may be applied to bidirectional circuits if a low concentration of AAV2-retro-Cre is used, because high titers above 1013 vector genomes per mL are required for efficient anterograde trans-synaptic Cre expression (Zingg et al., 2017). Although serotypes other than AAV1 and 9 were not reported to drive trans-synaptic Cre expression even at high titers (Zingg et al., 2017, 2020), we recommend using the lowest effective dose of AAV2-retro to minimize the potential for off-target labeling in bidirectional circuits. This approach has been used to study the role of lateral hypothalamus outputs to the periaqueductal gray in predation and evasion (Li et al., 2018), to study the role of locus coeruleus outputs to the spinal dorsal horn in acute and chronic itch (Koga et al., 2020), and to study the role of peri-habenular nucleus outputs to the nucleus accumbens in depressive-like behavior caused by light exposure at night (An et al., 2020).
Figure 1. Intersectional Targeting of Defined Neural Circuits.
(A) Targeting gene expression to neurons with one defined output by retrograde transport of AAV2-retro-hSyn1-Cre. (B) Targeting gene expression to neurons with one defined input by anterograde trans-synaptic transport of AAV1-hSyn1-Cre. (C) Targeting gene expression to neurons with one defined input and one defined output by anterograde trans-synaptic transport of AAV1-hSyn1-Cre and retrograde transport of Cre-dependent AAV2-retro. (D) Targeting gene expression to neurons with two defined inputs by anterograde trans-synaptic transport of both AAV1-hSyn1-Cre and AAV1-hSyn1-Flp.
Anterograde Trans-synaptic Expression: 1 Defined Input
The second approach uses anterograde transport to specifically label neurons in the target brain region that receive inputs from a defined brain region (Figure 1B). Cre-dependent AAV is again injected in the target brain region, and AAV1-hSyn1-Cre is injected in the input region: anterograde trans-synaptic transport of AAV1-hSyn1-Cre will specifically label neurons in the target brain region that are innervated by the defined input region (Zingg et al., 2017, 2020). AAV serotype 1 and the human synapsin 1 (hSyn1) promoter are recommended for efficient anterograde trans-synaptic Cre expression (Zingg et al., 2017, 2020). An important caveat is that because AAV1 is transported retrogradely, neurons in the target brain region with outputs to the region injected with AAV1-hSyn1-Cre will also be labeled by retrograde transport of AAV1-hSyn1-Cre. This method should thus be used only in unidirectional circuits; directionality can be verified using conventional retrograde tracers such as cholera toxin subunit B or Fluoro-Gold (Saleeba et al., 2019). This approach has been used to study neurons in the bed nucleus of the stria terminalis that receive input from the parabrachial nucleus and their role in regulating stress response (Jaramillo et al., 2020), to study the cerebellar projections of pontine neurons that receive input from defined cortical regions and their role in sensorimotor processing (Henschke and Pakan, 2020), and to study the projections of neurons in the peri-habenular nucleus that receive input from retinal ganglion cells and their role in conducting light signals at night (An et al., 2020).
Multi-Synaptic Expression: 1 Defined Input and 1 Defined Output
The third approach combines both retrograde and anterograde transport to specifically label neurons that send outputs to a defined brain region and also receive inputs from another defined brain region (Figure 1C). Cre-dependent AAV2-retro is injected in the defined output region, while AAV1-hSyn1-Cre is injected in the defined input region. Neither injected brain region is labeled. Instead, neurons that innervate the defined output region are retrogradely transduced by Cre-dependent AAV2-retro, but only those neurons that are also innervated by the defined input region will receive AAV1-hSyn1-Cre and activate gene expression. It is essential to use AAV2-retro for the Cre-dependent vector because other serotypes may not drive strong retrograde gene expression. Although this approach is not strictly limited to unidirectional circuits, note that neurons with collateral outputs to both the defined input region and the defined output region may be labeled by retrograde rather than anterograde transport of AAV1-hSyn1-Cre.
This method does not restrict gene expression to a single target brain region, but instead broadly labels neurons that both innervate the defined output region and are themselves innervated by the defined input region. In addition, neurons in the output region will be labeled if they are directly innervated by the input region. To restrict gene expression to a single target brain region, a third AAV vector expressing Flp recombinase can act as a gate between AAV1-hSyn1-Cre and AAV2-retro (Fenno et al., 2014; Zingg et al., 2017). In this case, Flp-dependent AAV2-retro is injected in the defined output region, AAV1-hSyn1-Cre is injected in the defined input region, and a third Cre-dependent AAV-DIO-Flp vector is injected in the target brain region. As before, neurons that innervate the defined output region will be retrogradely transduced by Flp-dependent AAV2-retro, while neurons that are innervated by the defined input region will receive AAV1-hSyn1-Cre by trans-synaptic anterograde transport. However, because AAV-DIO-Flp is now required for gene expression by AAV2-retro, only neurons in the target brain region injected with AAV-DIO-Flp will be labeled.
Note that because very little Flp is needed to turn on gene expression, a small amount of leaky Flp expression in the absence of Cre can result in off-target labeling (Fischer et al., 2019). We recommend using the improved design for Cre-dependent gene expression recently described by Fischer and colleagues, termed Cross-over Insensitive ATG-Out or “CIAO,” in which the start codon is placed outside of non-homologous lox sites to reduce leak expression (Table 1) (Fischer et al., 2019). Use of an AAV-CIAO-Flp design will limit off-target labeling.
Multi-Synaptic Expression: 2 Defined Inputs or 2 Defined Outputs
The final approach uses both Cre and Flp recombinases to specifically label neurons in the target brain region that receive inputs from two defined brain regions (Figure 1D). The target brain region is injected with a bicistronic AAV vector carrying one Cre-dependent expression cassette and one Flp-dependent expression cassette, such as AAV-DIO-GFP;FlpDIO-tdTomato (Oh et al., 2020). AAV1-hSyn1-Cre is then injected in one defined input region and AAV1-hSyn1-Flp is injected in another defined input region. In the above example, neurons will express both GFP and tdTomato only if they receive both inputs, and thus receive both AAV-Cre and AAV-Flp by anterograde trans-synaptic transport. Note that expression of only a single gene does not provide evidence that a neuron receives only a single input, because this approach is unlikely to label every innervated neuron. Alternatively, the target brain region can be injected with an AAV vector carrying a single expression cassette that is both Cre- and Flp-dependent (Table 1) (Fenno et al., 2014); this will only label neurons that receive both inputs. As above, because AAV1-hSyn1-Cre is also transported retrogradely these approaches should be used only in unidirectional circuits. This method has been used to label neurons in the posterior medial nucleus of the thalamus that receive inputs from both motor and somatosensory cortices, and to label neurons in striatum that receive inputs from both motor cortex and the ventral posteromedial nucleus of the thalamus (Oh et al., 2020).
A similar approach can be used to retrogradely label neurons in the target brain region that send outputs to two defined brain regions. The bicistronic vector is again injected in the target brain region, while AAV2-retro-Cre is injected in one defined output region and AAV2-retro-Flp is injected in another defined output region. Neurons in the target region will express both GFP and tdTomato only if they send outputs to both regions. This approach may be applied to bidirectional circuits so long as low concentrations of AAV2-retro are used for retrograde delivery of both Cre and Flp, because efficient anterograde trans-synaptic Cre expression requires high concentrations of AAV (Zingg et al., 2017).
Controlling the Number of Labelled Neurons
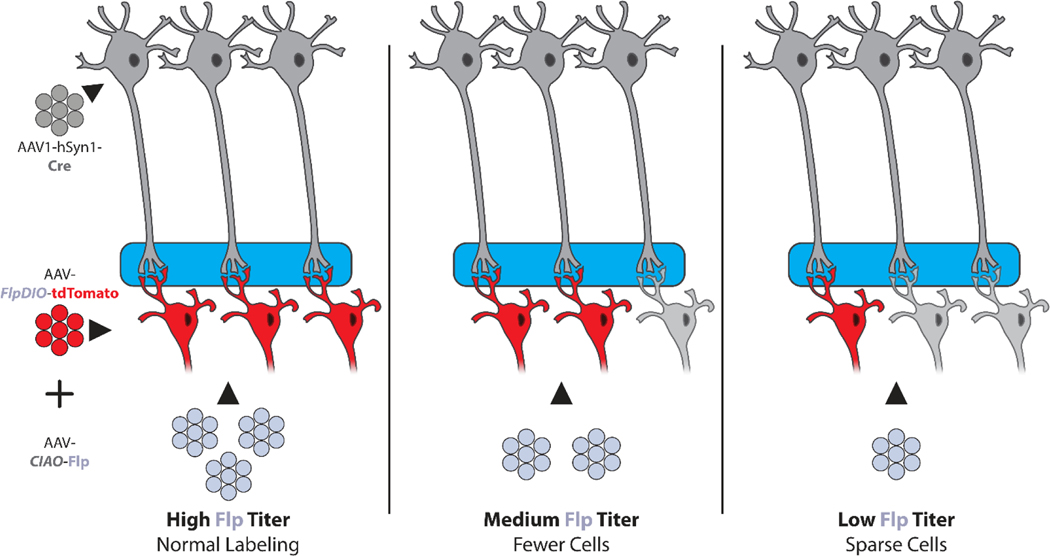
Sparse labeling may be needed to examine individual cell morphology or trace individual axons. Although reducing the dose of the Cre-dependent vector reduces the number of labeled cells, this also reduces the strength of gene expression. A superior method that preserves strong gene expression while providing direct control over the number of labeled neurons is injection of a third Cre-dependent AAV-CIAO-Flp vector, which acts as a gate between AAV-Cre and a Flp-dependent AAV vector (Lin et al., 2018; Zingg et al., 2020). For example, when labeling neurons with one defined input, AAV-DIO-tdTomato is injected in the target brain region and AAV1-hSyn1-Cre is injected in the defined input region. To control the number of labelled neurons, the same AAV1-hSyn1-Cre vector is injected in the input region, but two different vectors are injected in the target region: Cre-dependent AAV-CIAO-Flp and Flp-dependent AAV-FlpDIO-tdTomato (Table 1) (Figure 2). Trans-synaptic AAV1-hSyn1-Cre now turns on Flp expression, and Flp then turns on tdTomato expression. Because Flp is required for tdTomato expression, the number of labeled neurons is directly controlled by the dose of AAV-CIAO-Flp, which can be adjusted to achieve the desired sparseness of labeling without altering the strength of tdTomato expression (Zingg et al., 2020).
Figure 2. Controlling the Number of Labeled Neurons.
The number of labeled neurons can be directly controlled by local injection of a Cre-dependent AAV-CIAO-Flp vector, which acts as a gate between anterograde trans-synaptic delivery of AAV1-hSyn1-Cre and a Flp-dependent vector such as AAV-FlpDIO-tdTomato. Because only cells that receive AAV-CIAO-Flp can activate gene expression, reducing the dose of this vector reduces the number of labeled cells without altering the strength of gene expression.
Additional Targeting Methods
Transgenic Cre-driver mouse lines can further restrict intersectional gene expression to specific neuron subtypes. For example, in vesicular glutamate transporter 2 (VGlut2)-Cre transgenic mice or in vesicular GABA transporter (VGat)-Cre transgenic mice, AAV-FlpDIO-tdTomato is injected in the target brain region and Cre-dependent AAV1-CIAO-Flp is injected in the defined input region: innervated neurons in the target region will receive AAV1-CIAO-Flp, but only glutamatergic neurons will activate Flp and tdTomato expression in VGlut2-Cre mice, while only GABAergic neurons will activate Flp and tdTomato expression in VGat-Cre mice (Zingg et al., 2017; Li et al., 2018). This approach can also be applied to the intersectional labeling of neurons with one defined output by AAV2-retro, and it is compatible with the many characterized Cre-driver mouse lines (Harris et al., 2014), permitting intersectional labeling of neurons based on their subtype as well as their connectivity. In addition, transgenic mice that universally express a Cre-dependent reporter gene can be used instead of a Cre-dependent AAV vector (Madisen et al., 2010). This does not restrict gene expression to a single target region, but instead broadly labels the brain region injected with AAV-Cre and most or all connected brain regions (Zingg et al., 2017, 2020).
AAV-based intersectional methods can be combined with helper-dependent rabies virus vectors for monosynaptic retrograde tracing of inputs to defined neuron populations. This approach uses Cre-dependent AAV vectors to express rabies helper proteins in a population of neurons defined by Cre expression (Table 1): when helper-dependent rabies virus is injected in the target brain region, it thus replicates and initiates monosynaptic retrograde tracing only from the Cre-expressing neuron population, broadly labeling inputs to these neurons. To label inputs to a specific neuron subtype, Cre-dependent rabies helper AAVs are injected in the target brain region of transgenic Cre-driver mice, such as the VGlut2-Cre line; when helper-dependent rabies virus is locally injected in the target brain region, monosynaptic retrograde tracing will broadly map inputs to local glutamatergic neurons (Beier et al., 2015, 2019). To label inputs to neurons that project to a defined output region, Cre-dependent rabies helper AAVs are again injected in the target brain region, while AAV2-retro-Cre is injected in the defined output region; local injection of helper-dependent rabies virus in the target brain region will now label inputs to local neurons that innervate the defined output region (Beier et al., 2015, 2019; An et al., 2020). These two methods can be combined by injecting Flp-dependent rabies helper AAVs in the target brain region of Cre-driver mice, such as the VGlut2-Cre line, and injecting Cre-dependent AAV2-retro-CIAO-Flp in the defined output brain region; local injection of helper-dependent rabies virus in the target brain region will now label inputs only to those local glutamatergic neurons that also innervate the defined output region (Beier et al., 2015, 2019). Note that the injected concentration of rabies helper AAVs can directly influence the efficiency and specificity of monosynaptic retrograde tracing by helper-dependent rabies virus (Lavin et al., 2020). Optimal concentrations will vary depending on the AAV preparation and injection site and should be determined by titration following published guidelines (Lavin et al., 2020).
Other Considerations
The in vivo safety of AAV vectors is well established, but excessive Cre activity leads to off-target editing of the host genome, DNA damage, and apoptosis (Loonstra et al., 2001; Baba et al., 2005; Amin et al., 2019). This is problematic because anterograde trans-synaptic Cre expression by AAV1 is inefficient and requires a high dose of AAV1-Cre (Zingg et al., 2017). To control the level of Cre expression at the injection site, we recommend using an AAV genome that does not contain a woodchuck hepatitis virus post-transcriptional regulatory element (WPRE) (Table 1) (Zingg et al., 2020). The WPRE stabilizes mRNA and increases gene expression (Loeb et al., 1999), which results in greater trans-synaptic Cre expression in the target brain region, but may also lead to toxic overexpression of Cre at the injection site (Zingg et al., 2020). We also recommend using the neuron-specific hSyn1 promoter, which is validated for efficient trans-synaptic AAV1-Cre expression without apparent toxicity at relatively high vector concentrations above 1013 vector genomes per mL (Zingg et al., 2017, 2020), and we recommend avoiding strong pancellular promoters such as the CAG promoter, which may drive toxic overexpression of Cre (Baba et al., 2005).
Destabilized Cre (DD-Cre) can also prevent Cre toxicity by controlling Cre activity (Sando et al., 2013). DD-Cre is tagged with bacterial dihydrofolate reductase, which destabilizes Cre and triggers proteasomal degradation prior to DNA editing. Administration of the blood-brain barrier-penetrant antibiotic Trimethoprim (TMP) stabilizes DD-Cre, preventing its degradation and resulting in temporally restricted Cre activity (Sando et al., 2013). This reduces the risk of toxicity by minimizing the duration of Cre activity.
A limitation of intersectional targeting is that the input or output neurons must be spatially segregated from the target neuron population; these approaches are not practical for the study of local circuitry where AAV-Cre injection cannot reach input or output neurons without also diffusing directly to the target neurons. As noted above, AAV1-Cre should only be used for trans-synaptic anterograde labeling in unidirectional circuits because AAV1 may also be transported retrogradely. In addition, because some AAV capsids can efficiently transduce neurons from the cerebrospinal fluid (CSF) (Hordeaux et al., 2015; Castle et al., 2018), care should be taken to avoid leakage of AAV-Cre into the CSF during infusion.
Trans-synaptic anterograde spread of AAV1-Cre may be impacted by variables that are not yet fully understood due to the novelty of this approach. Synaptic vesicle release is required for trans-synaptic spread of AAV1-Cre (Zingg et al., 2020), but it is not known whether the input neuron firing rate influences the efficiency of trans-synaptic spread. AAV1 can drive anterograde trans-synaptic Cre expression over long distances, such as from input neurons in motor cortex to target neurons in lumbar spinal cord (Zingg et al., 2020), but it is not known whether the efficiency is reduced at longer distances. It also appears that AAV1-Cre does not spread trans-synaptically from most neuromodulatory projection neurons: minimal trans-synaptic labeling was reported from cholinergic input neurons, while no trans-synaptic labeling was reported from serotonergic or noradrenergic input neurons (Zingg et al., 2020). It is not known whether reduced efficacy in neuromodulatory circuits is due to reduced tropism of AAV1 for these projection neurons, lower rates of anterograde axonal transport, or differences in synaptic release. When applying this approach to a new circuit, an AAV1-Cre vector that co-expresses a fluorescent reporter can be used to label all potential input neurons, and a “color-flipping” AAV that changes the color of fluorescent reporter expression when exposed to Cre can be used to differentially label neurons in the target brain region that received AAV-Cre and those that did not (Table 1).
Discussion
These intersectional approaches have broad applications for neuroscience research. Beyond the expression of standard fluorescent proteins, intersectional methods can also be used for Brainbow labeling, in which each neuron is labeled by a stochastic combination of four different fluorescent reporters, resulting in up to 100 different colors that can be discriminated by confocal microscopy (Livet et al., 2007; Cai et al., 2013). Because each neuron is labeled a unique color, many different axons in the same circuit can be traced together without the need for sparse labeling. Brainbow uses Cre recombination to activate stochastic expression of fluorescent proteins, and thus the standard Brainbow 3.1 AAV vectors can serve as Cre-dependent AAV for intersectional labeling (Table 1). The two standard Brainbow 3.1 AAV vectors each encode two fluorescent proteins; when co-injected and activated by Cre, neurons are uniquely labeled by stochastic expression of the four total fluorescent proteins (Cai et al., 2013).
The impact of intersectional gene expression extends well beyond circuit tracing. Intersectional expression of optogenetic ion channels and chemogenetic Designer Receptors Exclusively Activated by Designer Drugs (DREADDs) permits functional interrogation of circuit activity (Table 1) (Zingg et al., 2017; Li et al., 2018; An et al., 2020; Jaramillo et al., 2020; Koga et al., 2020). Neural circuits can also be specifically ablated by intersectional expression of diphtheria toxin receptor (Table 1) (Azim et al., 2014). Activating, inhibiting, or ablating discrete neural circuits can dissect their roles in animal behavior and disease pathogenesis. In addition, intersectional expression of disease-associated proteins can model the impact and spread of pathology in specific neuron populations; this is particularly relevant for the study of Alzheimer’s disease and Parkinson’s disease, in which tau or α-synuclein pathology spreads by axonal transport within neural circuits (Wu et al., 2016; Henderson et al., 2019). AAV-based intersectional gene expression is safe and effective in diverse species and will support innovative research in large animal models including non-human primates. These powerful approaches are advancing the next generation of neuroscience research, and their development provides another important step towards understanding the complex anatomy and function of the mammalian nervous system.
Supplementary Material
Significance.
Viral vectors are widely used for experimental gene expression. Intersectional approaches use axonal transport to converge multiple viral vectors on a defined neuron population. This can label and trace neural circuits, dissect the contributions of specific circuits to animal behavior, model the spread of disease pathology, and more. These new approaches are powerful, but their novelty and complexity limit widespread adoption. We remove this barrier with an illustrated review of the history of intersectional targeting, the range of different approaches, and their emerging applications for neuroscience research.
Acknowledgments
This work was supported by the National Institutes of Health award AG066080.
Support: National Institutes of Health award AG066080 (to M.J.C.)
Footnotes
Data availability statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Conflict of Interest Statement
The authors declare that no conflict of interest exists.
References
- Amin SR, Gruszczynski C, Guiard BP, Callebert J, Launay J-M, Louis F, Betancur C, Vialou V, and Gautron S. (2019). Viral vector-mediated Cre recombinase expression in substantia nigra induces lesions of the nigrostriatal pathway associated with perturbations of dopamine-related behaviors and hallmarks of programmed cell death. J. Neurochem. 150 (3), 330–340. doi: 10.1111/jnc.14684. [DOI] [PubMed] [Google Scholar]
- An K, Zhao H, Miao Y, Xu Q, Li Y-F, Ma Y-Q, Shi Y-M, Shen J-W, Meng J-J, Yao Y-G, Zhang Z, Chen J-T, Bao J, Zhang M, and Xue T. (2020). A circadian rhythm-gated subcortical pathway for nighttime-light-induced depressive-like behaviors in mice. Nat. Neurosci. 23 (7), 869–880. doi: 10.1038/s41593-020-0640-8. [DOI] [PubMed] [Google Scholar]
- Archin NM, and Atherton SS (2002). Rapid spread of a neurovirulent strain of HSV-1 through the CNS of BALB/c mice following anterior chamber inoculation. J. Neurovirol. 8 (2), 122–135. doi: 10.1080/13550280290049570. [DOI] [PubMed] [Google Scholar]
- Atchison RW, Casto BC, and Hammon WM (1965). Adenovirus-associated Defective Virus Particles. Science 149 (3685), 754–756. doi: 10.1126/science.149.3685.754. [DOI] [PubMed] [Google Scholar]
- Azim E, Jiang J, Alstermark B, and Jessell TM (2014). Skilled reaching relies on a V2a propriospinal internal copy circuit. Nature 508 (7496), 357–363. doi: 10.1038/nature13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba Y, Nakano M, Yamada Y, Saito I, and Kanegae Y. (2005). Practical range of effective dose for Cre recombinase-expressing recombinant adenovirus without cell toxicity in mammalian cells. Microbiol. Immunol. 49 (6), 559–570. doi: 10.1111/j.1348-0421.2005.tb03753.x. [DOI] [PubMed] [Google Scholar]
- Beier KT, Gao XJ, Xie S, DeLoach KE, Malenka RC, and Luo L. (2019). Topological Organization of Ventral Tegmental Area Connectivity Revealed by Viral-Genetic Dissection of Input-Output Relations. Cell Rep. 26 (1), 159–167.e6. doi: 10.1016/j.celrep.2018.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier KT, Steinberg EE, DeLoach KE, Xie S, Miyamichi K, Schwarz L, Gao XJ, Kremer EJ, Malenka RC, and Luo L. (2015). Circuit Architecture of VTA Dopamine Neurons Revealed by Systematic Input-Output Mapping. Cell 162 (3), 622–634. doi: 10.1016/j.cell.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Cohen KB, Luo T, Lichtman JW, and Sanes JR (2013). Improved tools for the Brainbow toolbox. Nat. Methods 10 (6), 540–547. doi: 10.1038/nmeth.2450. [DOI] [PubMed] [Google Scholar]
- Card JP, Levitt P, and Enquist LW (1998). Different Patterns of Neuronal Infection after Intracerebral Injection of Two Strains of Pseudorabies Virus. J. Virol. 72 (5), 4434–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle MJ, Cheng Y, Asokan A, and Tuszynski MH (2018). Physical positioning markedly enhances brain transduction after intrathecal AAV9 infusion. Sci. Adv. 4 (11), eaau9859. doi: 10.1126/sciadv.aau9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle MJ, Gershenson ZT, Giles AR, Holzbaur ELF, and Wolfe JH (2014a). Adeno-associated virus serotypes 1, 8, and 9 share conserved mechanisms for anterograde and retrograde axonal transport. Hum. Gene Ther. 25 (8), 705–720. doi: 10.1089/hum.2013.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle MJ, Perlson E, Holzbaur EL, and Wolfe JH (2014b). Long-distance axonal transport of AAV9 is driven by dynein and kinesin-2 and is trafficked in a highly motile Rab7-positive compartment. Mol. Ther. 22 (3), 554–566. doi: 10.1038/mt.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cearley CN, and Wolfe JH (2007). A single injection of an adeno-associated virus vector into nuclei with divergent connections results in widespread vector distribution in the brain and global correction of a neurogenetic disease. J. Neurosci. 27 (37), 9928–9940. doi: 10.1523/JNEUROSCI.2185-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KY, Jang MJ, Yoo BB, Greenbaum A, Ravi N, Wu W-L, Sánchez-Guardado L, Lois C, Mazmanian SK, Deverman BE, and Gradinaru V. (2017). Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 20 (8), 1172–1179. doi: 10.1038/nn.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner JM, Bain GL, and Dulin JN (2019). Intraspinal and Intracortical Delivery of AAV Vectors for Intersectional Circuit Tracing in Non-transgenic Species. Methods Mol. Biol. 1950, 165–176. doi: 10.1007/978-1-4939-9139-6_9. [DOI] [PubMed] [Google Scholar]
- Davidsson M, Wang G, Aldrin-Kirk P, Cardoso T, Nolbrant S, Hartnor M, Mudannayake J, Parmar M, and Björklund T. (2019). A systematic capsid evolution approach performed in vivo for the design of AAV vectors with tailored properties and tropism. PNAS 116 (52), 27053–27062. doi: 10.1073/pnas.1910061116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deverman BE, Ravina BM, Bankiewicz KS, Paul SM, and Sah DWY (2018). Gene therapy for neurological disorders: progress and prospects. Nat. Rev. Drug Discov. 17 (9), 641–659. doi: 10.1038/nrd.2018.110. [DOI] [PubMed] [Google Scholar]
- Fenno LE, Mattis J, Ramakrishnan C, Hyun M, Lee SY, He M, Tucciarone J, Selimbeyoglu A, Berndt A, Grosenick L, Zalocusky KA, Bernstein H, Swanson H, Perry C, Diester I, Boyce FM, Bass CE, Neve R, Huang ZJ, and Deisseroth K. (2014). Targeting cells with single vectors using multiple-feature Boolean logic. Nat. Methods 11 (7), 763–772. doi: 10.1038/nmeth.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer KB, Collins HK, and Callaway EM (2019). Sources of off-target expression from recombinase-dependent AAV vectors and mitigation with cross-over insensitive ATG-out vectors. PNAS 116 (52), 27001–27010. doi: 10.1073/pnas.1915974116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Vandenberghe LH, Alvira MR, Lu Y, Calcedo R, Zhou X, and Wilson JM (2004). Clades of Adeno-associated viruses are widely disseminated in human tissues. J. Virol. 78 (12), 6381–6388. doi: 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JA, Hirokawa KE, Sorensen SA, Gu H, Mills M, Ng LL, Bohn P, Mortrud M, Ouellette B, Kidney J, Smith KA, Dang C, Sunkin S, Bernard A, Oh SW, Madisen L, and Zeng H. (2014). Anatomical characterization of Cre driver mice for neural circuit mapping and manipulation. Front. Neural Circuits 8, 76. doi: 10.3389/fncir.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson MX, Cornblath EJ, Darwich A, Zhang B, Brown H, Gathagan RJ, Sandler RM, Bassett DS, Trojanowski JQ, and Lee VMY (2019). Spread of α-synuclein pathology through the brain connectome is modulated by selective vulnerability and predicted by network analysis. Nat. Neurosci. 22 (8), 1248–1257. doi: 10.1038/s41593-019-0457-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henschke JU, and Pakan JM (2020). Disynaptic cerebrocerebellar pathways originating from multiple functionally distinct cortical areas. eLife 9, e59148. doi: 10.7554/eLife.59148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hordeaux J, Dubreil L, Deniaud J, Iacobelli F, Moreau S, Ledevin M, Le Guiner C, Blouin V, Le Duff J, Mendes-Madeira A, Rolling F, Cherel Y, Moullier P, and Colle M-A (2015). Efficient central nervous system AAVrh10-mediated intrathecal gene transfer in adult and neonate rats. Gene Ther. 22 (4), 316–324. doi: 10.1038/gt.2014.121. [DOI] [PubMed] [Google Scholar]
- Hudry E, and Vandenberghe LH (2019). Therapeutic AAV Gene Transfer to the Nervous System: A Clinical Reality. Neuron 101 (5), 839–862. doi: 10.1016/j.neuron.2019.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo AA, Williford KM, Marshall C, Winder DG, and Centanni SW (2020). BNST transient activity associates with approach behavior in a stressful environment and is modulated by the parabrachial nucleus. Neurobiol. Stress 13, 100247. doi: 10.1016/j.ynstr.2020.100247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplitt MG, Leone P, Samulski RJ, Xiao X, Pfaff DW, O’Malley KL, and During MJ (1994). Long-term gene expression and phenotypic correction using adeno-associated virus vectors in the mammalian brain. Nat. Genet. 8 (2), 148–154. doi: 10.1038/ng1094-148. [DOI] [PubMed] [Google Scholar]
- Kaspar BK, Erickson D, Schaffer D, Hinh L, Gage FH, and Peterson DA (2002). Targeted Retrograde Gene Delivery for Neuronal Protection. Mol. Ther. 5 (1), 50–56. doi: 10.1006/mthe.2001.0520. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Jacobs MW, Ito-Cole T, and Callaway EM (2016). Improved Monosynaptic Neural Circuit Tracing Using Engineered Rabies Virus Glycoproteins. Cell Rep. 15 (4), 692–699. doi: 10.1016/j.celrep.2016.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga K, Shiraishi Y, Yamagata R, Tozaki-Saitoh H, Shiratori-Hayashi M, and Tsuda M. (2020). Intrinsic braking role of descending locus coeruleus noradrenergic neurons in acute and chronic itch in mice. Mol. Brain 13. doi: 10.1186/s13041-020-00688-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin TK, Jin L, Lea NE, and Wickersham IR (2020). Monosynaptic Tracing Success Depends Critically on Helper Virus Concentrations. Front. Synaptic Neurosci. 12. doi: 10.3389/fnsyn.2020.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zeng J, Zhang J, Yue C, Zhong W, Liu Z, Feng Q, and Luo M. (2018). Hypothalamic Circuits for Predation and Evasion. Neuron 97 (4), 911–924.e5. doi: 10.1016/j.neuron.2018.01.005. [DOI] [PubMed] [Google Scholar]
- Lin R, Wang R, Yuan J, Feng Q, Zhou Y, Zeng S, Ren M, Jiang S, Ni H, Zhou C, Gong H, and Luo M. (2018). Cell-type-specific and projection-specific brain-wide reconstruction of single neurons. Nat. Methods 15 (12), 1033–1036. doi: 10.1038/s41592-018-0184-y. [DOI] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, and Lichtman JW (2007). Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450 (7166), 56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Lo L, and Anderson DJ (2011). A Cre-dependent, anterograde transsynaptic viral tracer for mapping output pathways of genetically marked neurons. Neuron 72 (6), 938–950. doi: 10.1016/j.neuron.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb JE, Cordier WS, Harris ME, Weitzman MD, and Hope TJ (1999). Enhanced expression of transgenes from adeno-associated virus vectors with the woodchuck hepatitis virus posttranscriptional regulatory element: implications for gene therapy. Hum. Gene Ther. 10 (14), 2295–2305. doi: 10.1089/10430349950016942. [DOI] [PubMed] [Google Scholar]
- Loonstra A, Vooijs M, Beverloo HB, Allak BA, van Drunen E, Kanaar R, Berns A, and Jonkers J. (2001). Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. PNAS 98 (16), 9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, and Zeng H. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13 (1), 133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J-Y, Han J-H, Lee H, Han Y-E, Rah JC, and Park H. (2020). Labeling Dual Presynaptic Inputs using cFork Anterograde Tracing System. Exp. Neurobiol. 29 (3), 219–229. doi: 10.5607/en20006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, Mortrud MT, Ouellette B, Nguyen TN, Sorensen SA, Slaughterbeck CR, Wakeman W, Li Y, Feng D, Ho A, Nicholas E, Hirokawa KE, Bohn P, Joines KM, Peng H, Hawrylycz MJ, Phillips JW, Hohmann JG, Wohnoutka P, Gerfen CR, Koch C, Bernard A, Dang C, Jones AR, and Zeng H. (2014). A mesoscale connectome of the mouse brain. Nature 508 (7495), 207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleeba C, Dempsey B, Le S, Goodchild A, and McMullan S. (2019). A Student’s Guide to Neural Circuit Tracing. Front. Neurosci. 13. doi: 10.3389/fnins.2019.00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sando R, Baumgaertel K, Pieraut S, Torabi-Rander N, Wandless TJ, Mayford M, and Maximov A. (2013). Inducible control of gene expression with destabilized Cre. Nat. Methods 10 (11), 1085–1088. doi: 10.1038/nmeth.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B, and Henderson N. (1988). Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. PNAS 85 (14), 5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack AM, Sawyer WB, Hughes JH, Platt KB, and Loewy AD (1989). A general pattern of CNS innervation of the sympathetic outflow demonstrated by transneuronal pseudorabies viral infections. Brain Res. 491 (1), 156–162. doi: 10.1016/0006-8993(89)90098-x. [DOI] [PubMed] [Google Scholar]
- Tervo DGR, Hwang B-Y, Viswanathan S, Gaj T, Lavzin M, Ritola KD, Lindo S, Michael S, Kuleshova E, Ojala D, Huang C-C, Gerfen CR, Schiller J, Dudman JT, Hantman AW, Looger LL, Schaffer DV, and Karpova AY (2016). A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron 92 (2), 372–382. doi: 10.1016/j.neuron.2016.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien JZ, Chen DF, Gerber D, Tom C, Mercer EH, Anderson DJ, Mayford M, Kandel ER, and Tonegawa S. (1996). Subregion- and cell type-restricted gene knockout in mouse brain. Cell 87 (7), 1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- Weiss AR, Liguore WA, Domire JS, Button D, and McBride JL (2020). Intra-striatal AAV2.retro administration leads to extensive retrograde transport in the rhesus macaque brain: implications for disease modeling and therapeutic development. Sci. Rep. 10 (1), 6970. doi: 10.1038/s41598-020-63559-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Finke S, Conzelmann K-K, and Callaway EM (2007). Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat. Methods 4 (1), 47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JW, Hussaini SA, Bastille IM, Rodriguez GA, Mrejeru A, Rilett K, Sanders DW, Cook C, Fu H, Boonen RACM, Herman M, Nahmani E, Emrani S, Figueroa YH, Diamond MI, Clelland CL, Wray S, and Duff KE (2016). Neuronal activity enhances tau propagation and tau pathology in vivo. Nat. Neurosci. 19 (8), 1085–1092. doi: 10.1038/nn.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W-B, Jiang H-F, Gang Y-D, Song Y-G, Shen Z-Z, Yang H, Dong X, Tian Y-L, Ni R-J, Liu Y, Tang N, Li X, Jiang X, Gao D, Androulakis M, He X-B, Xia H-M, Ming Y-Z, Lu Y, Zhou J-N, Zhang C, Xia X-S, Shu Y, Zeng S-Q, Xu F, Zhao F, and Luo M-H (2017). Anterograde monosynaptic transneuronal tracers derived from herpes simplex virus 1 strain H129. Mol. Neurodegener. 12. doi: 10.1186/s13024-017-0179-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gradinaru V, Adamantidis AR, Durand R, Airan RD, de Lecea L, and Deisseroth K. (2010). Optogenetic interrogation of neural circuits: technology for probing mammalian brain structures. Nat. Protoc. 5 (3), 439–456. doi: 10.1038/nprot.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingg B, Chou X-L, Zhang Z-G, Mesik L, Liang F, Tao HW, and Zhang LI (2017). AAV-Mediated Anterograde Transsynaptic Tagging: Mapping Corticocollicular Input-Defined Neural Pathways for Defense Behaviors. Neuron 93 (1), 33–47. doi: 10.1016/j.neuron.2016.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingg B, Peng B, Huang J, Tao HW, and Zhang LI (2020). Synaptic Specificity and Application of Anterograde Transsynaptic AAV for Probing Neural Circuitry. J. Neurosci. 40 (16), 3250–3267. doi: 10.1523/JNEUROSCI.2158-19.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.