Abstract
Pathology tissue slides are taken as the gold standard for the diagnosis of most cancer diseases. Automatic pathology slide diagnosis is still a challenging task for researchers because of the high-resolution, significant morphological variation, and ambiguity between malignant and benign regions in whole slide images (WSIs). In this study, we introduce a general framework to automatically diagnose different types of WSIs via unit stochastic selection and attention fusion. For example, a unit can denote a patch in a histopathology slide or a cell in a cytopathology slide. To be specific, we first train a unit-level convolutional neural network (CNN) to perform two tasks: constructing feature extractors for the units and for estimating a unit’s non-benign probability. Then we use our novel stochastic selection algorithm to choose a small subset of units that are most likely to be non-benign, referred to as the Units Of Interest (UOI), as determined by the CNN. Next, we use the attention mechanism to fuse the representations of the UOI to form a fixed-length descriptor for the WSI’s diagnosis. We evaluate the proposed framework on three datasets: histological thyroid frozen sections, histological colonoscopy tissue slides, and cytological cervical pap smear slides. The framework achieves diagnosis accuracies higher than 0.8 and AUC values higher than 0.85 in all three applications. Experiments demonstrate the generality and effectiveness of the proposed framework and its potentiality for clinical applications.
Keywords: Whole slide image, computer-aided diagnosis, stochastic selection, units of interest, attention fusion
1. Introduction
Many cancer diseases depend on the microscopic examination of the tissue slide for the definitive diagnosis [1, 2, 3]. Yet it is still a very challenging task to diagnose tissue slides even for professional pathologists. The difficulties for the tissue slide diagnosis mainly lie in the following aspects: 1) The dimension of the slide tissue is enormous. It is almost impossible for pathologists to carefully inspect the details of all regions under high-resolution, thereby leading to missed diagnosis [4]. 2) The tissue morphology varies greatly. Meanwhile, some benign and malignant regions or cells appear very similar. Differentiation between benign and malignant regions can be difficult on many tissue samples [5]. 3) For some tissue diagnosis, there is no one uniform standard. Different hospitals and institutes have their own slide making and diagnosis criterion, thus leading to a large degree of inter- and intra-observer variation [6]. 4) Those tissues that are not well-processed would introduce severe artifacts [7]. 5) The labor- and time-intensive diagnosis also increases pathologists’ chance of error-making [8]. Computer-aided diagnosis (CAD) provides an alternative for the tissue slide diagnosis. Because of the computer’s tireless and objectiveness traits, the analysis results via CAD can boost the accuracy and robustness of the tissue slide diagnosis.
However, due to the limits on limitation of computation capability, CAD on tissue slides mostly focuses on the region of interests (ROIs) before 2015 [9, 10, 11]. These analyses are mainly devoted to cell-level tasks, such as cell detection, segmentation, and retrieval [12, 13, 14, 15]. There are also a few studies on regarding patch-based classification diagnosis [16, 17, 18, 19]. Nevertheless, from the viewpoint of practical assistance, computer-aided analysis of the whole slide images (WSIs) would be more most beneficial to pathologists, as their routine work is to inspect WSIs. Since 2016, studies on pathology image analysis start to focus on the WSI [20, 21, 22, 23]. In this paper, we propose a framework to analyze whole tissue images and provide the automatic diagnosis we propose an automatic framework to diagnose whole slide tissue images.
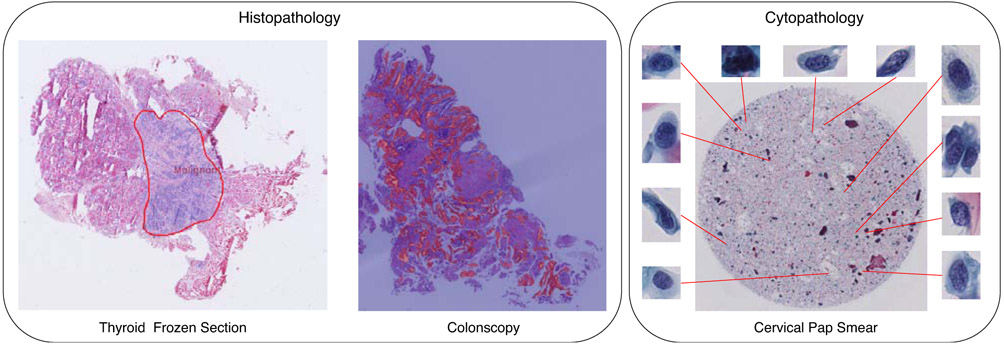
Currently, most reported WSI analysis studies are focused on one particular tissue slide, such as breast, lung, and gastric tissue [24, 22, 25, 26, 27]. However, these methods are not validated on other tissue slides leading to a lack of generalization ability. In this study, we propose a general framework for different tissue types. This framework can not only work on histological tissue slides, but also be applied to cytological slides. Fig. 1 shows WSI examples belonging to three different tissue types evaluated in the experiments.
Figure 1:
Experimental whole slide image examples of three different tissues. In this study, we apply the proposed framework to both histological and cytological applications. Histological applications include the thyroid frozen section and colonoscopy tissue slide on the left of the figure, in which patches of the slide are taken as the processing units. The cytological application deals with cervical pap smear and takes the cell inside the slide as the processing unit.
Because of the high-resolution characterization of the WSI, currently, it is almost impossible to process the WSI directly. The standard procedure to analyze WSI includes the following steps: 1) splitting a WSI into multiple units. 2) performing unit-level representation learning. 3) fusion of the representations of units to form a fixed-length WSI descriptor. 4) WSI diagnosis based upon the WSI descriptor [21, 28, 29, 30]. Since 2012, the deep neural networks gain overwhelming success in the ImageNet recognition challenge [31, 32], and convolutional neural networks (CNNs) achieve state-of-the-art performance in numerous image analysis tasks [33, 34, 35, 36]. Besides natural image applications, the biomedical image domain also takes great advantage of CNN models, across X-ray, CT, MRI, as well as digital pathology slides [37, 38, 39, 40]. To make use of CNN model’s outstanding representation learning capability, and to avoid the time-consuming hand-crafted feature engineering, we propose to use CNN model to learn the representation of the processing unit in digital pathology slides. In cytological slides, we take the cell as the basic processing unit and generate cell-based representation. Compared with analyzing histopathological slides, an extra step for cytological WSIs is to firstly detect the cells in the WSI before conducting unit representation learning. When we finish crop patches/cells in the WSIs, the following procedures are the same for analyzing both histological and cytological WSIs.
For most malignant cases, the malignant regions or the number of abnormal cells account only a small part. Directly fusing all units’ representation in the WSI is not an optimal solution [41]. The direct effect is that these few suspicious units’ representations would be overwhelmed by a large number of benign patches’ representations, thus causing great difficulty in differentiating malignant slides. There are studies introducing selection mechanisms to overcome this issue. For instance, Liao et al. propose to select the top five nodules based on the detection confidence for the automatic diagnosing lung cancer from Computed Tomography (CT) scans [42]. In this study, we propose to select a subset of the suspicious units in a stochastic manner for the WSI feature fusion to avoid the feature attenuation issue. We take advantage of the fine-tuned unit classification model, which can estimate units’ probabilities belonging to different categories. The unit selection is mainly based on each unit’s non-benign probability, namely the slide’s probability of not being benign. Those units with high non-benign probabilities are more likely to be chosen, and we denote these selected units as Units of Interest (UOI).
The attention model is one of the most popular concepts in the machine learning field in the last five years. It is widely used in speech recognition [43], natural language processing (NLP) [44, 45], and vision recognition [46, 47, 48]. The main functionality of the attention model is to capture the significance of different parts of the input sample, and thus enabling the model to focus on a few key parts of the input and ignore unrelated regions. As to the WSI classification, the attention model fits well with capturing the weights of different units for the final decision-making. For those most malignant units, we expect the attention model to acquire high weights, while for those benign ones, lower weights are expected to be assigned. Instead of directly applying the attention model on the units in the WSI, we propose to apply the attention model upon the unit’s representation extracted from the CNN model, aiming to learn the weights of these units for the WSI feature fusion to enhance the effects of those most suspicious units. In this study, we choose the self-attention mechanism, which has achieved great success in a variety of tasks [45, 46], to capture the weights of the units’ representations for the WSI representation fusion.
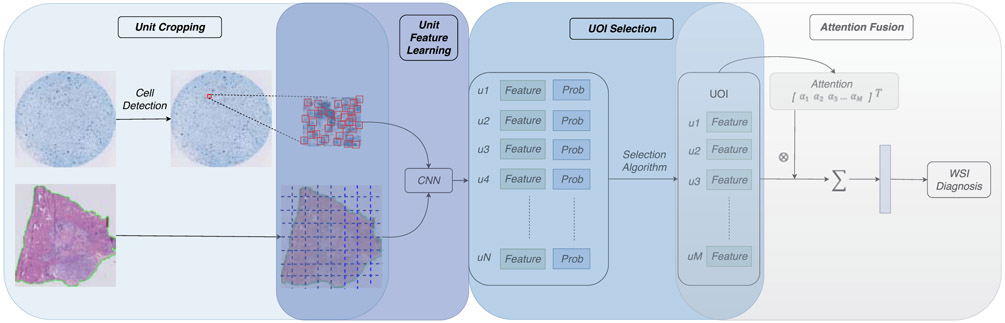
In this paper, we propose a general framework for the whole slide pathology image diagnosis using unit stochastic selection and attention fusion. Fig. 2 presents the flowchart of the proposed framework. The main contributions in this paper are:
Figure 2:
The general framework of the whole slide pathology image diagnosis using unit stochastic selection and attention fusion. The first step conducts unit cropping, including patch cropping in histological slides and cell cropping in cytological slides. Then the CNN classifier is fine-tuned and used to infer all cropped units’ features and diagnosis probabilities. After that, we propose a stochastic selection algorithm to pick up a small subset of Units Of Interest (UOI), denoted as u1,…,uM, from all the units denoted as u1,…,uN, based on the estimation of their diagnosis probabilities, where N>>M. Next, we apply the attention mechanism to learn the significant weights, denoted as α1,…,αM, of UOI to fuse a descriptor for the WSI. Finally, the diagnosis of the WSI is performed based upon the fixed-length descriptor.
We propose a general framework for the WSI analysis. This framework can be applied to both histological and cytological applications.
We introduce a novel unit stochastic selection algorithm for the WSI model training, aiming to focus on suspicious units in the slide and improve the robustness of the WSI model.
We adopt the attention model to capture the weights of the selected Units Of Interest (UOI) to form more discriminating WSI representation.
Extensive experiments on three different types of pathology slides demonstrate the generality and effectiveness of the proposed framework via various fusion methods.
2. Methods
The proposed framework for the WSI diagnosis can be roughly divided into four parts: 1) Processing units cropping, 2) Unit feature learning, 3) Units Of Interest (UOI) selection, 4) Attention-based WSI representation fusion. We will introduce these four parts in the following in detail.
2.1. Processing Units Cropping
The size of both histological and cytological slides is extremely large. Typically each WSI could have a full spatial resolution larger than 50, 000 × 50, 000 pixels at ×40 magnification. Currently, it is not feasible to directly take the gigapixel WSI as a whole and feed it into the deep neural network. The widely used manner is to split the WSI into multiple small units, individually process each unit, and then fuse units’ outcome for the final diagnosis. Adopting this manner, we need first to split the WSI into multiple processing units. In the histological WSI, patches are the basic processing units, while in the cytological WSI, cells are the basic processing units. Therefore, we need to use different manners to crop the units in cytological and histological WSIs, respectively.
2.1.1. Patch Cropping in Histological Slides
In a histopathology WSI, there usually exists a large part of background regions surrounding the tissue region. These background area does not carry any diagnostic relevant information. Additionally, they would increase the computation burden when taking them into account. To exclude the background area, we locate the tissue regions in the histopathology WSIs using the tissueloc package [49]. As can be seen in the bottom left image of Fig. 2, the tissue region is surrounded by the green contour, and only those region inside and intersected with the green contour would be used for the histopathology WSI analysis.
After locating the tissue regions, we split the whole WSI with a fixed length of stride in both vertical and horizontal directions. To reduce the computation cost in the WSI analysis, we choose the non-overlapping manner for units splitting. After the splitting process, the WSI would be divided into multiple units. We only keep those units that are either entirely inside the tissue regions or intersected with the tissue contour but with at least 75% of their pixels inside the tissue contour.
2.1.2. Cell Cropping in Cytological Slides
In cytological WSIs, the cells inside are the primary processing units. The prerequisite for any further analysis of cytological WSIs is to detect cells inside the slide. Nevertheless, there are many variations in the cell’s size and appearance, especially for those malignant ones. Compared with healthy cells, abnormal cells, which are the leading cause of cervical cancer, would usually manifest more morphology variations [50]. There are two main reasons for performing cell cropping other than cell segmentation in our framework. Firstly, because of the substantial heterogeneity of cells, accurately segmenting the boundary of the cells is still a challenging task. Additionally, the cell segmentation task is much more time-consuming and error-prone than the cell center detection task.
We propose an approximation method to crop the processing unit in the cytopathology slide by detecting the center of each cell and setting a fixed size to crop all the cells instead of detecting cell boundaries. To conduct the detection of cell centers, we adapt the segmentation network U-Net and train the network by providing the mask of cell centers instead of the binary mask of cell regions. To boost the robustness of cell center detection in different image resolutions, we resize the images and their corresponding masks using a set of scaling ratios in the training phase.
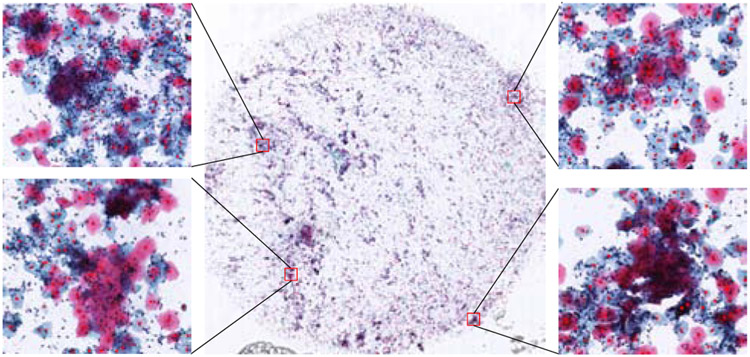
For a test WSI, we first split the whole image using the non-overlapping splitting manner into multiple patches. Then for each patch, we predict the local maxima coordinates based on the U-Net and regard those coordinates as cell centers. Then, we crop the cell units from the WSI based on these predicted cell centers with a fixed size in the vertical and horizontal direction. The cytological WSI would be divided into multiple units after this cropping process.. Fig. 3 demonstrates a sample of cell center detection result.
Figure 3:
Cell center detection results. The large image in the middle is the original slide, and the four ROIs displayed around show the results of cell center detection. From the sampled ROIs, almost all the cells are accurately detected.
2.2. Unit Feature Learning with CNN
After cropping the units, we take advantage of the CNN model to learn their representation. To train the CNN model, first we need to collect enormous labels for the units. The way to annotate WSIs is based on the specific pathology application. For the thyroid tissue slides, we make the annotation in a relatively coarse manner by drawing broad contours to cover regions of the same category. Then we crop the units from these drawn contour surrounded regions and give the units the same label as the contour’s. As for the colon tissue slides, pixel-wise annotation is provided because of its much smaller slide size. For the cervical WSIs, we give the same label to the cropped units as the cells.
Instead of training the CNN classifier from scratch, we train the WSI unit-level feature extractor based on the million-scale ImageNet pre-trained model, which can provide better parameter initialization and meanwhile speed the training process. With the fine-tuned CNN model, we apply it to all the cropped units in a WSI. For each unit, besides its feature representation, we can also obtain each unit’s probabilities belonging to different categories, which are the basis for the following unit selection.
2.3. UOI Selection
From a normal-sized WSI, we can obtain more than one thousand processing units. However, even for a malignant WSI, there exist large regions being benign. When fusing units’ features for a WSI, the features from benign units would weaken the overall WSI representation discrimination capability, especially when the benign region accounting the majority area of the WSI. With the fine-tuned CNN model, we can estimate the probability of units belonging to different categories. For all pathology diagnoses, including binary and multi-classification, we can calculate the non-benign probability of each unit by summing its probabilities belonging to all the non-benign categories. We propose to select a subset of UOI from all the cropped units of the WSI based on the unit’s non-benign probability.
In order to strength the robustness of the WSI diagnosis, we propose to introduce stochasticity in the WSI UOI selection process. In the training stage, we first set a minimum unit selection Nmin and a maximum unit selection number Nmax. For each WSI, we randomly select a value k ∈ [Nmin, Nmax] as the WSI’s current unit selection number. In addition, we sort all units based on their non-benign probabilities in descending order. Next, we split the selection procedure into two sections. We would first select N1 processing units from the front N1+ units of the sorted sequence. Then we select k − N1 units from the remaining sorted units ranked after N1+. In both selections, the chance of each unit to be selected is based on its non-benign probability. After the selection, we would obtain k UOI from the WSI and units with higher non-benign probability are more likely to be selected. With this two-layer selection stochasticity, each WSI can have multiple different unit combinations. During the testing stage, we would just select (Nmin + Nmax)/2 units with highest non-benign probabilities from the WSI for the diagnosis to remove randomness. The UOI selection algorithm is described as in Algorithm 1.
2.4. Attention-based WSI Representation Fusion
According to the diagnostic experiences of expert pathologists, they would mainly focus on those suspicious regions, and their final diagnosis on a WSI is mainly based on a few suspicious small areas in a vast WSI. Instead of equally treating all selected UOI in the fusion process, we adopt the attention mechanism to learn different weights for the UOI, with the assumption that the unit with a higher weight tends to be more informative for the WSI diagnosis. The attention we adopt in this study is self-attention.
Suppose that there are k selected units used for the WSI representation, f1, … , fk represent features of these units, where , and the number of diagnosis categories is c. We set and as the attention model’s parameters.
| Algorithm 1: WSI UOI Stochastic Selection Algorithm | |
|---|---|
|
|
Then the weight wi for each unit can be calculated by:
| (1) |
where the hyperbolic tangent function tanh(·) is used for the non-linearity transformation, and the usage of the softmax is to ensure the weights of all patches sum to 1.0. After we obtain each unit’s weight, the WSI’s representation can be calculated as:
| (2) |
After obtaining each WSI’s representation by Eq. 2, we adopt the multilayer perceptron (MLP) [51, 52] to carry out the diagnosis for each WSI. Note in the WSI classification model training process, the parameters in Eq. 1 and MLP are optimized together.
3. Experiments
To evaluate the performance of the proposed WSI diagnosis framework, we apply the framework on three different WSI applications: thyroid frozen section, colon tissue, and cervical pap smear. In this section, we will first introduce the common experimental setup. Then we separately describe the detailed experimental settings as well as the result analysis on these three applications.
3.1. Common Experimental Setup
Fig. 2 illustrates the whole pipeline of the proposed framework. The parameters in this framework vary in different WSI applications. However, some parameters are the same on both cytological and histological slides. We will cover the common parameter settings in this section.
3.1.1. Unit Cropping
For the histopathology images with a large number of background tissues, the first step is to locate the tissue regions and then to split the tissue regions to image patches. The parameters of tissue localization, as well as the level and size of cropped units from the WSI, depend on the specific histopathology application. We will introduce the parameter setting in their diagnosis section.
For cytological slides, we would first apply the U-Net to detect the cell centers. During the U-Net training phase, we set the initial learning rate as 0.02, and the batch size as 32. We also set a scaling ratio list of [0.125, 0.130, 0.15, 0.18, 0.20] to resize the original image and its corresponding mask, to enhance the network’s robustness on the cell center detection.
3.1.2. Unit Feature Extraction
We choose two most widely used neural networks, VGG16bn [53] and ResNet50 [34], to train unit-based classification model for the feature extraction. We change the final fully-connected layer based on the number of diagnosis categories in the pathology application and fine-tune the ImageNet pre-trained model.
During the fine-tuning phase of the unit classification model, we adopt stochastic gradient descent (SGD) with a momentum of 0.9 and a weight decay of 5.0e-4. The initial learning rate is set as 0.01 and decay by 0.6 after every two epochs. We train the model for 10 epochs using a batch size of 32. We evaluate the model on the validation set after each epoch and save the model with the highest accuracy for feature extraction. We adopt standard data augmentation techniques, including random rotation between 0° and 12°, horizontal flip, vertical flip, color jitter, and normalization through subtracting mean and dividing by standard deviation.
For a testing WSI, after we split it into multiple units, we apply the CNN model to these units and take the results of the CNN model’s penultimate layer as the feature representation for these units. Besides, we can obtain each unit’s probabilities belonging to all categories, by applying the Softmax to output logits of the CNN model.
3.1.3. UOI Selection
Because of the significant variations among the size of different pathology slides, and even for the slides from the same tissue, it is hard to propose a general strategy to set the parameters for the selection of UOI. Currently, we set the parameters in the UOI selection mainly based on the average number of units in the WSIs of a specific application. For the WSI with more than 3,000 units, we would choose around 300 to 500 units. For those WSIs with a unit number between 1,000 to 3,000, the selected number of UOI is set to be around 100 to 300. While for those WSIs with less than 1,000 units, we would usually select 1/3 to 1/4 of the average number of UOI for the WSI model.
3.1.4. WSI Unit Fusion
There are quite a few attention mechanisms proposed in the last few years. We mainly propose to validate the effectiveness of applying attention mechanisms for the WSI diagnosis. Based on this idea, we compare the average pooling fusion, which equally treats all selected UOI and average them to obtain the WSI descriptor, with the self-attention fusion in the experiments. In addition, we compare with the concatenation of pooling features with attention features, termed as “concat” fusion manner.
We train the WSI model using SGD with a batch size of 32 for 100 epochs. The initial learning rate is set as 1.0e-3, and the learning rate is decayed in an epoch-wise step-down manner until 0.0. We take the model with the best accuracy on the validation set to evaluate the performance on the testing set.
3.1.5. Cross Validation
Because of the hardship of collecting a large number of WSI samples, we propose to use cross-validation to run the experiments multiple times with different train/val splittings to avoid over-fitting and bias issues in the evaluation process.
3.2. Thyroid Frozen Section Diagnosis
3.2.1. Thyroid Dataset
We successively collect two batches of thyroid slides. The first collected slides contain 114 benign, 50 uncertain, and 181 malignant slides. The second collected slides include 83 benign, 7 uncertain, and 165 malignant slides. We take the first collected slides for training and the second collected slides for testing. We run the 5-fold cross-validation on the training slides and split the slides into train/val with a ratio of 4:1. The testing slides are kept the same for different cross-validations.
3.2.2. Unit Model Training
Because of the vast size of thyroid frozen sections, typically larger than 50, 000 × 50, 000, and some even larger than 100, 000 × 100, 000, it is time-consuming for fully pixel-wise annotation of the slide. We use a relatively coarse manner to annotate the thyroid frozen sections. We request the pathologists to annotate a few contours on each training slide but to make sure the tissue types inside each contour to be the same. With this annotation manner for the training slides, we only crop the unit from these annotated contours and set the type of the unit the same as the contour’s. While for the testing slides, we would use the unit cropping methods, as described in Section 2.1. Namely, we first locate the tissue regions via the “tissueloc” package with all default parameters and then split and keep only those units inside or intersected with the tissue region for the WSI diagnosis.
Considering the huge size of thyroid frozen sections as well as its pyramidal storage format, we crop unit with a size of 224 × 224 from level 3 of the slide, corresponding to the image size of 1792 × 1792 in level 0 of the slide. For an 80, 000 × 80, 000 WSI, the number of cropped units would be close to 2000. Considering the background region in the slide, the number of units cropped from a WSI is between 1,000 to 2,000. As for the unit-level CNN model training, the average number of unit numbers on five splittings for train/val for benign/uncertain/benign are as described below: the training has 20, 000 benign, 6, 000 uncertain, and 10, 000 malignant, and the validation has 5, 000 benign, 1, 500 uncertain, and 2, 500 malignant units. We perform the unit-level classification with these cropped units and use the parameters mentioned in Section 3.1.2. The unit-level classification accuracy on the validation units on both VGG16bn and ResNet50 is around 0.78.
3.2.3. WSI Model Training
Based on the previous introduction, the average number of units cropped from the WSI is between 1,000 to 2,000. We choose the UOI selection parameters as follows: Nmin = 128, Nmax = 192, N1 = 40, N1+ = 60. For the diagnosis of thyroid frozen sections, we train the pooling-based, the attention-based, and the concatenation-based WSI models as described in Section 3.1.4.
3.2.4. WSI Diagnosis Performance
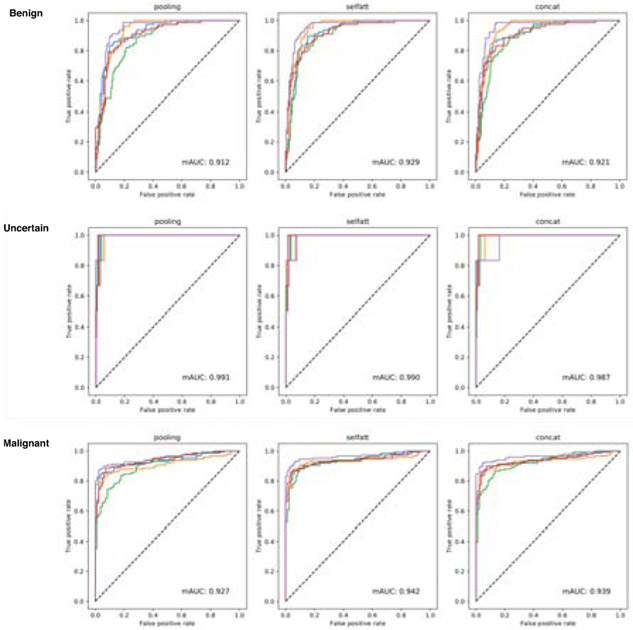
To evaluate the diagnosis performance of thyroid slides, we calculate precision and recall for all categories as well as the overall accuracy. Besides, we draw the ROC curve for three different categories in the thyroid diagnosis. We show the averaged results of 5-fold cross-validation in Table 1, and we draw five ROC curves for each cross-validation in Fig. 4. Based on the evaluation results from Table 1, we can see the self-attention fusion method can obtain better accuracy than other two unit feature fusion manners in the thyroid diagnosis on both VGG16bn and ResNet50 feature extractor. Comparing the two different feature extractors, VGG16bn obtains superior accuracy over the ResNet50 model. In addition, we can see that the recall value of the uncertain category is close to 1.0. However, its precision is very low, which means uncertain samples would be accurately predicted as uncertain. However, slides of other categories would be predicted as uncertain. While for the malignant category, its precision value is higher than the recall value, which means malignant samples tend to be predicted as other categories instead of the reverse. For the diagnosis of the thyroid nodules, predicting malignant samples to uncertain is much safer than the reverse. Because the uncertain cases can still go through further examinations while it is irreversible if the misdiagnosed malignant sample has been resected.
Table 1:
Thyroid slides diagnosis results using two CNN models (VGG16bn and ResNet50) for unit feature extraction and three fusion methods. The self-attention fusion method based on VGG16bn feature extractor obtain the best accuracy among all the configurations.
| CNN Model | WSI Label | pooling | selfatt | concat | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Precision (%) | Recall (%) | Acc (%) | Precision (%) | Recall (%) | Acc (%) | Precision (%) | Recall (%) | Acc (%) | ||
| VGG16bn | Benign | 76.7 | 77.4 | 82.7 | 80.3 | 79.5 | 85.1 | 78.6 | 78.2 | 83.9 |
| Uncertain | 31.8 | 100 | 31.0 | 96.7 | 29.9 | 96.7 | ||||
| Malignant | 94.1 | 85.0 | 95.7 | 87.6 | 95.2 | 86.3 | ||||
| ResNet50 | Benign | 77.6 | 67.5 | 79.9 | 82.2 | 75.6 | 84.4 | 82.3 | 74.9 | 84.1 |
| Uncertain | 28.9 | 100 | 29.4 | 96.7 | 27.6 | 100 | ||||
| Malignant | 88.8 | 85.6 | 93.7 | 88.5 | 94.3 | 88.0 | ||||
Figure 4:
The receiver operating characteristic (ROC) curves of thyroid slides diagnosis on pooling, self-attention, and concatenation fusion manners using the VGG16bn model as the unit feature extractor. From top to bottom are the ROC curves of thyroid’s three diagnosis categories, including Benign, Uncertain, and Malignant. The self-attention fusion attains higher AUC values compared to other two fusion manners.
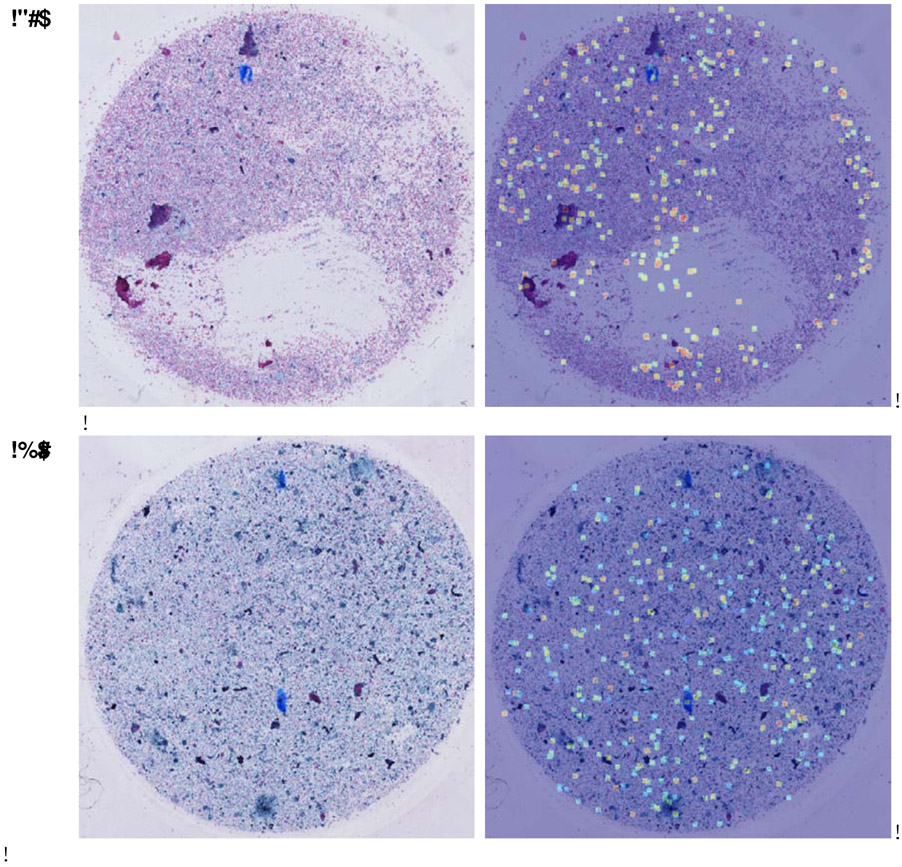
In Fig. 5, we show two demos of the selected UOI and their weights overlayed on the original slides. Based on these high-lighted areas in the right images, we can see that the UOI selection algorithm can accurately pick up the suspicious units. Additionally, those units with a more heterogeneous appearance show higher weights, which demonstrate the effectiveness of the attention mechanism on learning the weights of units.
Figure 5:
Thyroid slides unit selection and attention weights visualization demos. The images shown on the left is the down-scaled thyroid slides, and the images shown on the right is the selected UOI with attention weights overlayed on the original slide. The selected UOI are mainly located in the malignant regions, and those most serious units have higher attention weights.
As for the time cost of the thyroid slide diagnosis, it mainly contains the time used on unit feature extractor and slide classifier (including UOI selection, unit feature fusion, and classification). On average, the unit feature extraction per thyroid slide takes 49.0923s, and a single slide classification takes 0.0024s. The average time consumption of diagnosing a thyroid slide is 49.0923s.
3.3. Colonoscopy Tissue Slide Diagnosis
3.3.1. Colon Dataset
The colonoscopy tissue slides are obtained from the “Digestive-System Pathological Detection and Segmentation Challenge 2019”2. This challenge provides a total of 660 training tissue slides with an average size of 3000 × 3000, in which 410 of them are diagnosed as negative, and the rest 250 are diagnosed as positive. The testing set is not openly provided to the public. Based on this setting, we take the provided training slides as the whole dataset for WSI diagnosis framework evaluation. We carry out 5-fold cross-validation on the available slides and split the train/val dataset with a ratio of 4:1. Thus there would be 528 slides used for training and 132 slides used for validation.
3.3.2. Unit Model Training
The colon slide image has an average size of 3, 000 × 3, 000, which is very different from most commonly seen WSI with the average size of 50, 000×50, 000, and the format of the provided slides is the jpeg rather than the commonly used pyramid storage format. We choose a slightly different way to crop the units for the unit-level classification model training. Instead of using the “tissueloc” package to locate the tissue region as the first step, we omit this step in the colon slide analysis. Nevertheless, in the unit cropping process, we would add a checking procedure to determine if a cropped image is the background or the tissue. In this checking procedure, we first convert the cropped RGB image to a grayscale image. Then we calculate the average gray value of the image and compare it with a preset threshold value of 220. We take those cropped units with an average gray value of less than 220 as the tissue units and take them for the following analysis.
In addition to the slides, this dataset also provides positive tissue segmentation masks. We take advantage of the segmentation mask to separate the cropped units into positive and negative categories. We set the size of the cropped unit as 448×448 and splitting the units in a non-overlapping manner. As the segmentation mask has the same size as the slide, we crop the mask unit using the same manner. We take the unit as positive when the malignant area in its corresponding mask is larger than 5%, and the rest units are taken as negative. We would resize all the cropped images to 224 × 224 for the unit-model training.
In different cross-validation splitting, there is slight difference in the number of units in train/val and negative/positive categories. On average, the number of negative and positive patches in training is about 20, 000 and 10, 000, and the number of negative and positive patches in the validation is about 5, 000 and 2, 500, respectively. The colon unit-based classification model training is entirely in keeping with the settings mentioned in Section 3.1.2. The unit-level classification accuracy on the validation units on both ResNet50 and VGG16bn is around 0.90, and ResNet50 has slightly better accuracy compared with VGG16bn.
3.3.3. WSI Model Training
As the relatively small size of the colon tissue slide, with the non-overlapping cropping manner of patch size 448 × 448, the average number of patches in a WSI is around 50. The parameters for the UOI selection algorithm in Algorithm 1 for the colon application are set as follows: Nmin = 10, Nmax = 18, N1 = 8, N1+ = 12. After the setting of UOI selection, the training of three different feature fusion manners for the WSI model are carried out as described in Section 3.1.4.
3.3.4. WSI Diagnosis Performance
We show the colon slides diagnosis results in Table. 2 and Fig. 6. Same as the thyroid evaluation, the values in Table. 2 are averaged on five splittings, and Fig. 6 has five ROC curves of three feature fusion manners. On the colon dataset, the results of using pooling and self-attention with both VGG16bn and ResNet50 as the feature extractor are very close to 97.4%. The concatenation manner obtains the accuracy of 97.9% and 98.3% using VGG16bn and ResNet50 as the feature extractor, respectively. These results are better than the pooling and selfatt fusion manners. We also calculate the mean AUC of the five ROC curves in Fig. 6 and obtain average value as high as 0.997, which demonstrate the accurate diagnosis of the proposed framework on colon slides.
Table 2:
Colon slides diagnosis results using two CNN models (VGG16bn and ResNet50) for unit feature extraction and three fusion methods. Pooling and self-attention fusion method present very close performance on the colon dataset. Concatenation fusion manner shows superior performance compared to pooling and selfatt.
| CNN Model | WSI Label | pooling | selfatt | concat | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Precision (%) | Recall (%) | Acc (%) | Precision (%) | Recall (%) | Acc (%) | Precision (%) | Recall (%) | Acc (%) | ||
| VGG16bn | Negative | 96.1 | 98.0 | 97.3 | 95.7 | 98.0 | 97.4 | 98.5 | 98.0 | 97.9 |
| Positive | 98.8 | 97.6 | 98.8 | 97.3 | 96.9 | 97.6 | ||||
| ResNet50 | Negative | 98.8 | 97.3 | 97.3 | 98.8 | 97.3 | 97.3 | 99.3 | 98.0 | 98.3 |
| Positive | 95.7 | 98.0 | 95.7 | 98.0 | 96.9 | 98.8 | ||||
Figure 6:
The receiver operating characteristic (ROC) curves of colon tissue slides diagnosis on pooling, self-attention, and concatenation manner using the VGG16bn model as the unit feature extractor. Three different unit feature fusion methods show very similar curves for the colon application.
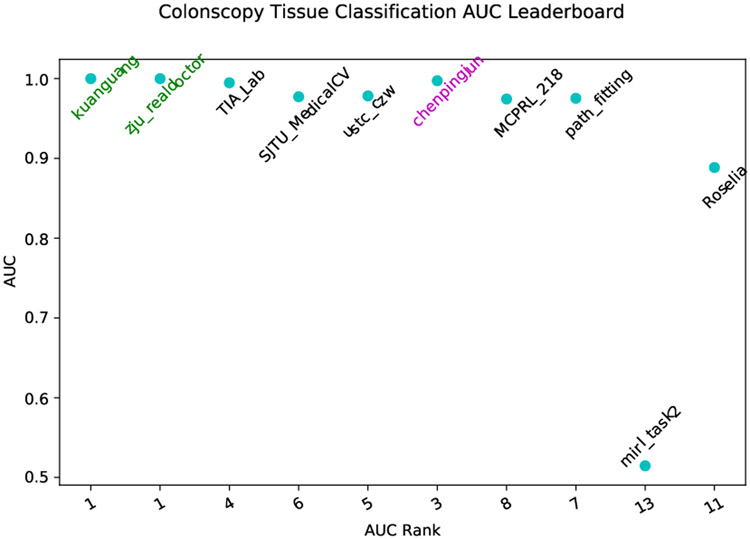
Since the colon dataset is publicly released for the MICCAI2019 Grand Pathology Challenge, we make use of the proposed framework to attend the challenge. Fig. 7 shows our result (with team name chenpingjun) compared to other contestants. The proposed framework with selfatt fusion manner ranked third place on the Colonoscopy tissue classification track obtaining an AUC value of 0.997 on a private testing dataset. The released AUC in the challenge leaderboard is consistent with our results via the cross-validation manner.
Figure 7:
Comparison of the top 10 contestants in Colonoscopy tissue classification challenge. The proposed method (chenpingjun), obtains AUC value of 0.997, and ranks th third place.
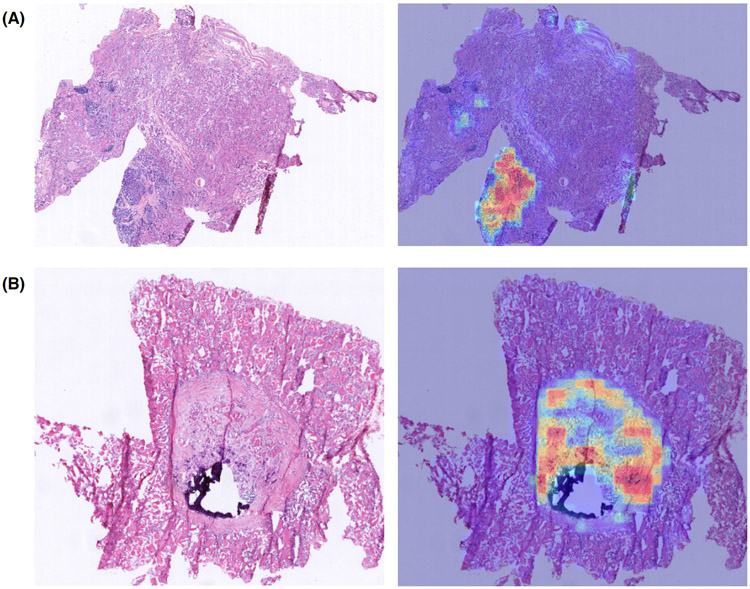
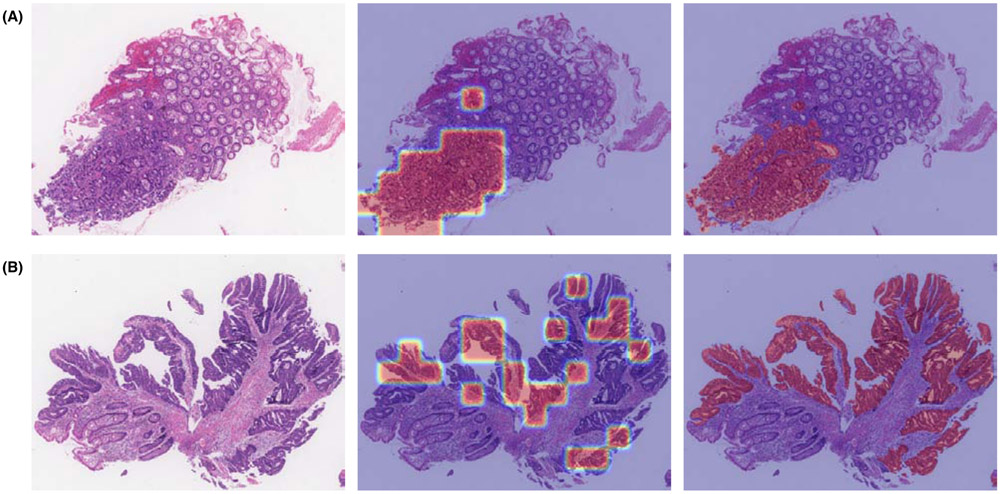
The self-attention mechanism does not show obvious superiority over pooling methods on the colon slides. Probably because there is no obvious weight difference of the units in colon slides for the diagnosis, which can be illustrated in Fig. 8. In Fig. 8, the left image is the input colon slide, and the middle one is the selected UOI with weights overlaying on the original colon slide, and the right one is the segmentation mask overlaying on the original colon slide. Comparing the middle and the right images, we can see that the selection algorithm can accurately pick up the malignant regions. As for the attention learned weights of these selected UOI, all the weights are nearly the same. However, the concatenation manner performs better than both pooling and selfatt manners in the colon tissue application. The reason could lie in the feature concatenation increases the colon WSI feature’s dimensionality and its discrimination capability, and thus to be more robust.
Figure 8:
Colon slides UOI selection and attention weights visualization demos. (A) and (B) are two demo colon slides. For each demo, from left to right, are the testing colon slide, the selected UOI with attention weights overlayed on the testing slide, and the expert annotated segmentation mask overlayed on the testing slide. Compared with the segmentation mask, the UOI selection algorithm can accurately select those positive regions. As we can see on the middle image, the weight values on these selected units are very close.
The average feature extraction per colonoscopy image is 7.6622s, and the classification per slide takes 0.0021s. The overall average time cost for diagnosing a colonoscopy image with the proposed framework takes 7.6643s.
3.4. Cervical Pap Smear Diagnosis
3.4.1. Cervical Dataset
The cervical pap smear dataset contains 264 positive slides and 108 negative slides in total. We perform 4-fold cross-validation to evaluate the performance of the proposed framework and split the slides into train/test with a ratio of 3:1. Thus, we have 198 positive slides and 81 negative slides for training and 66 positive slides and 27 negative slides for testing.
3.4.2. Cell Center Detection
For the U-Net based cell center detection, we use a total of 1, 661 images cropped from multiple cervical slides. We annotate the cell centers of these images. The average number of cells in each image is around 15. We split all these images into training and validation with a ratio of 4:1, and then we obtain 1, 329 images for training and 332 images for validation.
In the U-Net model training phase, we perform extensive data augmentations, including rotation in the range of [0°, 360°], horizontal flip, and vertical flip. As we will rescale both the image and corresponding mask by a series of ratios smaller than 0.2, we also set the smallest image size to 400 × 400 to avoid the too small resized image and mask. If the size of the rescaled image is smaller than 400 × 400, we pad the image on the right and bottom borders with a reflection of the image. Then, we randomly crop a sub-image with a size of 256 × 256 from the padded image for the U-Net model training.
After we obtain each test image’s detection probability map using the trained U-Net model, we first set 0.14 as the threshold to assign all the pixels with lower values to 0.0. Then, we locate all the local maximal peaks and take them as the detected cell centers.
3.4.3. Unit Model Training
For cervical pap smear slides, we crop the cells based on the detected cell centers. To estimate the appropriate cropping size for cell-based patches, we calculate the mean and standard deviation of all labeled cells’ size. We get an average size of 71.97 for the height of positive cells and an average size of 71.15 for the height of negative cells at level 0 of the slide. Meanwhile, the average size for the width of positive cells is 120.94, a little higher than 118.51 for negative cells. The standard deviation is around 44 for both negative and positive cells. Therefore, we set the crop size of 224 × 224 from level 1 of the slide for the unit-level classifier training and feature extraction.
In different cross-validation experiments, there exists a slight difference between each training and testing datasets. Based on the cell annotations, we split the dataset into seven classes: CCSM, Microorganism, Negative, Gland-abnormal, ASCUS, LSIL, and high-grade positive. CCSM includes the neck canal cells and squamous epithelium metaplasia cells. Microorganism stands for cervicovaginal microorganisms. Negative class is composed of all the other negative cell types. Gland-abnormal represents for the abnormalities for gland cells. ASCUS stands for atypical squamous cells of undetermined significance[54]. LSIL is short for low-grade squamous intraepithelial lesions, which means that the cervical cells show mildly abnormal changes. High-grade positive class includes all the other highly abnormal cells, including high-grade squamous intraepithelial lesion[55], squamouscell carcinoma[56] and the other positive cell types. The average number of units on train/val for different classes are as described below: the training dataset has 3,129 CCSM, 771 Microorganism, 15,640 Negative, 1,584 Gland-abnormal, 9,755 ASCUS, 9,968 LSIL and 13,537 Positive units, and the validation has 783 CCSM, 198 Microorganism, 3,912 negative, 397 Gland-abnormal, 2,439 ASCUS, 2,492 LSIL and 3,385 positive units.
With these cropped units, we perform the cell level classification training based on the settings mentioned in Section 3.1.2. On the validation units, VGG16bn obtains classification accuracy of 77.8%, and ResNet50 obtains classification accuracy of 73.4%.
3.4.4. WSI Model Training
The average number of units detected in one cervical WSI is more than 10, 000. For cervical pap smear dataset, we set Nmin = 300, Nmax = 360, N1 = 200 and N1+ = 240 in the UOI selection algorithm for the WSI model training. In the testing phase, we choose the top 330 units with high non-benign probabilities in all the processing units. All the experimental settings for WSI training and testing are described in detail in Section 3.1.4.
3.4.5. WSI Diagnosis Performance
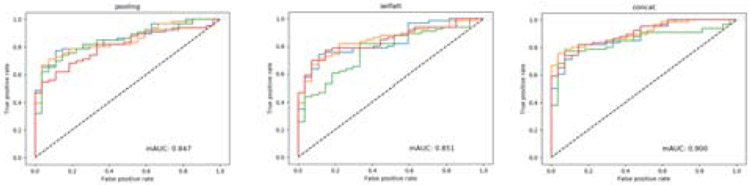
We show the diagnosis results of cervical slides in Table. 3. We use 4-fold cross-validation on the cervical dataset ,and four ROC curves are shown in Fig. 9 using three unit feature fusion manners. On the cervical dataset, we get better accuracy via ResNet50 than VGG16bn. We obtain the best accuracy of 83.0% using the concatenation fusion mechanism and ResNet50. The mean AUC calculated in Fig. 9 is 0.847 for pooling and 0.851 for self-attention, and the AUC results shows that both pooling and self-attention achieve very similar performance in cervical pap smear classification. But the mean AUC of the concatenation feature fusion method obtains 0.900, which is much higher than both the pooling and self-attention manners.
Table 3:
Cervical pap smear diagnosis results using two CNN feature extraction models (VGG16bn and ResNet50) and three fusion methods. The concat fusion attains the best performance among the three unit feature fusion methods, and unit feature extractor ResNet50 obtains better accuracy compared to VGG16bn.
| CNN Model | WSI Label | pooling | selfatt | concat | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Precision (%) | Recall (%) | Acc (%) | Precision (%) | Recall (%) | Acc (%) | Precision (%) | Recall (%) | Acc (%) | ||
| VGG16bn | Negative | 81.4 | 59.4 | 79.6 | 77.7 | 61.7 | 78.4 | 62.4 | 86.1 | 80.9 |
| Postive | 77.2 | 91.1 | 80.3 | 89.8 | 93.3 | 78.8 | ||||
| ResNet50 | Negative | 44.4 | 85.7 | 81.7 | 66.6 | 66.6 | 80.6 | 80.6 | 55.5 | 83.0 |
| Positive | 96.9 | 81.0 | 86.3 | 86.3 | 84.0 | 94.3 | ||||
Figure 9:
The receiver operating characteristic (ROC) for cervical pap smear slides using pooling, self-attention, and concatenation fusion manners with the ResNet50 as the feature extractor. Both pooling and self-attention obtain AUC value close to 0.850. The concatenation fusion outperforms both pooling and self-attention fusion mechanisms and obtains AUC value of 0.900.
The main reason that the self-attention mechanism performs similarly with the pooling in the cervical pap smear slides is that the weight difference in most processing units is not significant; thus, most units almost equally contribute to the representation fusion. From Fig.10, we can see that the color difference in all the chosen UOI differs slightly, which means that the weights overlaying on the selected UOI are similar. Same with the colon slide diagnosis, the concatenation fusion, which is the combination of pooling and self-attention, surpasses both of them. The concatenation manner increases the dimensionality of the WSI feature built upon pooled feature and attentive feature, which may help improve the WSI’s feature discrimination and increase its prediction accuracy.
Figure 10:
Cervical pap smear slides unit selection and attention weights visualization. The left image is the original slide, and the right image is the image with UOI attention weights overlayed on top of the original slide. Based on these two samples, the attention weights for selected UOI do not differ much, with few of them have a slightly darker color than the other units.
The time cost for cytopathology slides via the proposed framework contains three aspects: cell detection, cell feature extraction, and slide classification. On the cervical slides, the average time cost of cell detection per slide is 30.5244s, cell feature extraction per slide takes 10.4159s, and slide classification per slide takes 0.0027s. The total time cost for diagnosing a cervical pap smear slide is 40.9430s.
4. Discussion
Framework Generalization:
The main highlight of this framework is its generalization capability in that it can apply to both histological and cytological diagnostic applications. We evaluate this framework on two histological applications and one cytological application to demonstrate this point. Besides, the two histological applications represent two very different kinds of WSIs with huge dimension variations. Among them, the thyroid slide has an average slide dimension of 80, 000 × 80, 000, and the colon slide has an average dimension of 3, 000 × 3, 000. On all the three diagnostic tasks, the best accuracy is higher than 0.80, and the AUC value is higher than 0.85. These results validate the effectiveness and robustness of the proposed framework. The framework can be applied to WSIs with a broad range of dimensions and also different pathology domains, thereby potentially being applied to other tissue types.
Cell Center Detection:
For cytological slide diagnosis, we propose to detect the cell centers rather than to detect their exact boundary. There are two main advantages of this approximation. First, it can significantly decrease the prepossessing time and labeling work for pathologists. Compared with labeling the cell centers, annotating cell boundaries is more tedious and error-prone. Second, because of the different height and width for various cells, cell-based units may have different sizes. To feed them into the deep neural network, we need to resize all the units to the same size. However, this would affect the unit resolution, thus influence the performance of the unit-based classification model and further influence the unit extracted features. By detecting the cell centers, cropping cell-based units with a fixed size can avoid the resolution issue.
Unit Feature Learning Method:
In the current framework, we train the unit-level feature extractor using the fully-supervised scheme. Two CNN classification models, VGG16bn and ResNet50, are evaluated in the experiments. There is no noticeable performing difference between these two CNN models. The annotation manner is also different based on a specific pathology task. For instance, the thyroid application utilizes contour-based coarse annotation, the colon slide adopts pixel-wise annotation, and the cervical slide uses the cell-based labeling. For the fully-supervised training, annotation is always a significant burden for biomedical image applications. The unit-level feature extractor is one replaceable component of the whole framework, and substituting it with other feature extraction methods will not affect the usability of the framework. Also, other learning manners, such as semi-supervised, self-supervised, and unsupervised training methods, can be introduced into this framework to substitute the fully-supervised scheme.
Patch Size Setting:
In the unit-based WSI diagnostic framework, the cropped size of the unit is always an issue to consider. As the input for most CNN model is 224 × 224, we always propose to crop the unit with this size but from different levels of WSI based on its pyramidal storage. In the thyroid task, we crop the unit from level 3, which means the size of the cropped thyroid unit corresponds to 1792×1792 in level 0 of the slide. In the colon application, we crop the unit with a size of 448 × 448 and then resize to 224, which corresponds to level 1 cropping, and the cervical unit is also cropped from level 1.
Cropping from a higher level would result in fewer units, which would decrease the computing consumption. However, the resolution of the cropped unit would be lower compared with those cropped from a lower level. While a unit cropped from a low level would have a small context window that may hinder the diagnosis. The principle we use in the setting of the patch size is primarily based on whether the pathologists can make the diagnosis on the cropped unit. When the chosen level is too high, the cropped unit cannot be recognized after being resized to 224 × 224 , we need to decrease the cropping level. Similarly, if the chosen level corresponded unit has too little context information that pathologists cannot diagnose, we need to increase the cropped level to enable the cropped unit to have more context information.
Motivation on UOI Selection:
For large-size WSIs, like the thyroid and cervical applications, the number of the processing units would usually be more substantial than 1, 000. Nevertheless, the fact is that most of the units, even for malignant or positive WSIs, are benign and do not contain much diagnosis information. Adding these less informative units into the fusion process would weaken the differentiation capability of the WSI descriptor. Thus we propose to use the unit’s estimated non-benign probability to select a subset of UOI.
The advantages of the UOI selection mechanism mainly lie in the following three aspects. First, removing large amounts of benign units would increase the differentiating ability of the WSI descriptor. Second, this procedure can improve the robustness of the network. Each WSI would have multiple different UOI combinations with the stochasticity introduced in the selection algorithm, thereby strengthening the generalization ability of the WSI representation fusion. Third, we can reduce the computational costs by selecting a subset of all the units.
UOI Selection Parameter Settings:
Currently, we empirically set the parameters of the UOI selection algorithm, and we mainly follow the principle described in Section. 3.1.3. The main reference for the parameter setting is the average number of cropped units in the WSI of a particular application. For those WSIs with more than 1, 000 units, we would set the number of selected UOI to be around 100-500. While for those WSIs with less than 1, 000 units, we propose to select UOI with a relatively larger ratio to increase the robustness. We propose the theoretical principle of setting UOI selection parameters as our future study.
5. Conclusion
In this study, we propose a fully automatic framework for the whole slide pathology image diagnosis based on unit stochastic selection and attention fusion. The main contribution of this work is the generality of the proposed WSI diagnostic framework. This framework can apply to both histological and cytological applications covering a broad range of slide dimensions. Additionally, the UOI selection algorithm and the attention fusion are proposed to extract the suspicious units in the WSI and capture the significance of the UOI for the final diagnosis. We achieve diagnosis accuracies higher than 0.8 and AUC values higher than 0.85 on all three applications. In future investigations, we will explore the unit feature extraction with a small amount of or no annotations, the UOI selection parameter setting, and other unit feature fusion manners.
Acknowledgment
This study is supported by the US National Institutes of Health (R01 AR065479-02).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interests
The authors declare no conflicts of interest.
References
- [1].Rorke LB, Pathologic diagnosis as the gold standard, Cancer 79 (4) (1997) 665–667. [DOI] [PubMed] [Google Scholar]
- [2].Sardanelli F, Giuseppetti GM, Panizza P, Bazzocchi M, Fausto A, Simonetti G, Lattanzio V, Del Maschio A, Sensitivity of mri versus mammography for detecting foci of multifocal, multicentric breast cancer in fatty and dense breasts using the whole-breast pathologic examination as a gold standard, American Journal of Roentgenology 183 (4) (2004) 1149–1157. [DOI] [PubMed] [Google Scholar]
- [3].Jeelani S, Ahmed QM, Lanker AM, Hassan I, Jeelani N, Fazili T, Histopathological examination of nail clippings using pas staining (hpepas): gold standard in diagnosis of onychomycosis, Mycoses 58 (1) (2015) 27–32. [DOI] [PubMed] [Google Scholar]
- [4].Giles WH, Maclellan RA, Gawande AA, Ruan DT, Alexander EK, Moore FD, Cho NL, False negative cytology in large thyroid nodules, Annals of surgical oncology 22 (1) (2015) 152–157. [DOI] [PubMed] [Google Scholar]
- [5].Tizhoosh HR, Pantanowitz L, Artificial intelligence and digital pathology: Challenges and opportunities, Journal of pathology informatics 9 (2018) 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Elmore JG, Barnhill RL, Elder DE, Longton GM, Pepe MS, Reisch LM, Carney PA, Titus LJ, Nelson HD, Onega T, et al. , Pathologists diagnosis of invasive melanoma and melanocytic proliferations: observer accuracy and reproducibility study, bmj 357 (2017) j2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Komura D, Ishikawa S, Machine learning methods for histopathological image analysis, Computational and structural biotechnology journal 16 (2018) 34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Glaser AK, Reder NP, Chen Y, McCarty EF, Yin C, Wei L, Wang Y, True LD, Liu JT, Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens, Nature biomedical engineering 1 (7) (2017) 0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dong F, Irshad H, Oh E-Y, Lerwill MF, Brachtel EF, Jones NC, Knoblauch NW, Montaser-Kouhsari L, Johnson NB, Rao LK, et al. , Computational pathology to discriminate benign from malignant intraductal proliferations of the breast, PloS one 9 (12) (2014) e114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kayser K, Borkenfeld S, Djenouni A, Christian Manning J, Kaltner H, Kayser G, Gabius H-J, Digital pathology: how far are we from automated tissue-based diagnosis?, Analytical Cellular Pathology 2014. [Google Scholar]
- [11].Veta M, Pluim JP, Van Diest PJ, Viergever MA, Breast cancer histopathology image analysis: A review, IEEE Transactions on Biomedical Engineering 61 (5) (2014) 1400–1411. [DOI] [PubMed] [Google Scholar]
- [12].CireŞan DC, Giusti A, Gambardella LM, Schmidhuber J, Mitosis detection in breast cancer histology images with deep neural networks, in: International Conference on Medical Image Computing and Computer-assisted Intervention, 2013, pp. 411–418. [DOI] [PubMed] [Google Scholar]
- [13].Meijering E, Cell segmentation: 50 years down the road [life sciences], IEEE Signal Processing Magazine 29 (5) (2012) 140–145. [Google Scholar]
- [14].Qi X, Xing F, Foran DJ, Yang L, Robust segmentation of overlapping cells in histopathology specimens using parallel seed detection and repulsive level set, IEEE Transactions on Biomedical Engineering 59 (3) (2011) 754–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhang X, Liu W, Dundar M, Badve S, Zhang S, Towards large-scale histopathological image analysis: Hashing-based image retrieval, IEEE Transactions on Medical Imaging 34 (2) (2014) 496–506. [DOI] [PubMed] [Google Scholar]
- [16].Irshad H, Veillard A, Roux L, Racoceanu D, Methods for nuclei detection, segmentation, and classification in digital histopathology: a reviewcurrent status and future potential, IEEE reviews in biomedical engineering 7 (2013) 97–114. [DOI] [PubMed] [Google Scholar]
- [17].Spanhol FA, Oliveira LS, Petitjean C, Heutte L, A dataset for breast cancer histopathological image classification, IEEE Transactions on Biomedical Engineering 63 (7) (2015) 1455–1462. [DOI] [PubMed] [Google Scholar]
- [18].Pan X, Li L, Yang H, Liu Z, Yang J, Zhao L, Fan Y, Accurate segmentation of nuclei in pathological images via sparse reconstruction and deep convolutional networks, Neurocomputing 229 (2017) 88–99. [Google Scholar]
- [19].Shi X, Sapkota M, Xing F, Liu F, Cui L, Yang L, Pairwise based deep ranking hashing for histopathology image classification and retrieval, Pattern Recognition 81 (2018) 14–22. [Google Scholar]
- [20].Xu Y, Jia Z, Wang L-B, Ai Y, Zhang F, Lai M, Eric I, Chang C, Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features, BMC bioinformatics 18 (1) (2017) 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhu X, Yao J, Zhu F, Huang J, Wsisa: Making survival prediction from whole slide histopathological images, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017, pp. 7234–7242. [Google Scholar]
- [22].Ren J, Hacihaliloglu I, Singer EA, Foran DJ, Qi X, Adversarial domain adaptation for classification of prostate histopathology whole-slide images, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, 2018, pp. 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].BenTaieb A, Hamarneh G, Predicting cancer with a recurrent visual attention model for histopathology images, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, 2018, pp. 129–137. [Google Scholar]
- [24].Bayramoglu N, Kannala J, Heikkilä J, Deep learning for magnification independent breast cancer histopathology image classification, in: 2016 23rd International conference on pattern recognition (ICPR), 2016, pp. 2440–2445. [Google Scholar]
- [25].Lin H, Chen H, Dou Q, Wang L, Qin J, Heng P-A, Scannet: A fast and dense scanning framework for metastastic breast cancer detection from whole-slide image, in: 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), 2018, pp. 539–546. [Google Scholar]
- [26].Wang X, Chen H, Gan C, Lin H, Dou Q, Huang Q, Cai M, Heng P-A, Weakly supervised learning for whole slide lung cancer image classification, Medical Imaging with Deep Learning. [DOI] [PubMed] [Google Scholar]
- [27].Wang S, Zhu Y, Yu L, Chen H, Lin H, Wan X, Fan X, Heng P-A, Rmdl: Recalibrated multi-instance deep learning for whole slide gastric image classification, Medical image analysis 58 (2019) 101549. [DOI] [PubMed] [Google Scholar]
- [28].Hou L, Samaras D, Kurc TM, Gao Y, Davis JE, Saltz JH, Patch-based convolutional neural network for whole slide tissue image classification, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, pp. 2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Campanella G, Silva VWK, Fuchs TJ, Terabyte-scale deep multiple instance learning for classification and localization in pathology, arXiv preprint arXiv:1805.06983. [Google Scholar]
- [30].Campanella G, Hanna MG, Geneslaw L, Miraflor A, Silva VWK, Busam KJ, Brogi E, Reuter VE, Klimstra DS, Fuchs TJ, Clinicalgrade computational pathology using weakly supervised deep learning on whole slide images, Nature medicine 25 (8) (2019) 1301–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, et al. , Imagenet large scale visual recognition challenge, International journal of computer vision 115 (3) (2015) 211–252. [Google Scholar]
- [32].Krizhevsky A, Sutskever I, Hinton GE, Imagenet classification with deep convolutional neural networks, in: Advances in neural information processing systems, 2012, pp. 1097–1105. [Google Scholar]
- [33].LeCun Y, Bengio Y, Hinton G, Deep learning, nature 521 (7553) (2015) 436–444. [DOI] [PubMed] [Google Scholar]
- [34].He K, Zhang X, Ren S, Sun J, Deep residual learning for image recognition, in: Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 770–778. [Google Scholar]
- [35].Long J, Shelhamer E, Darrell T, Fully convolutional networks for semantic segmentation, in: Proceedings of the IEEE conference on computer vision and pattern recognition, 2015, pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- [36].Ren S, He K, Girshick R, Sun J, Faster r-cnn: Towards real-time object detection with region proposal networks, in: Advances in neural information processing systems, 2015, pp. 91–99. [DOI] [PubMed] [Google Scholar]
- [37].Wang Z, Li H, Zhang Q, Yuan J, Wang X, Magnetic resonance fingerprinting with compressed sensing and distance metric learning, Neurocomputing 174 (2016) 560–570. [Google Scholar]
- [38].Sui X, Zheng Y, Wei B, Bi H, Wu J, Pan X, Yin Y, Zhang S, Choroid segmentation from optical coherence tomography with graphedge weights learned from deep convolutional neural networks, Neurocomputing 237 (2017) 332–341. [Google Scholar]
- [39].Shen D, Wu G, Suk H-I, Deep learning in medical image analysis, Annual review of biomedical engineering 19 (2017) 221–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, Van Der Laak JA, Van Ginneken B, Sánchez CI, A survey on deep learning in medical image analysis, Medical image analysis 42 (2017) 60–88. [DOI] [PubMed] [Google Scholar]
- [41].Shi X, Xing F, Guo Z, Su H, Liu F, Yang L, Structured orthogonal matching pursuit for feature selection, Neurocomputing 349 (2019) 164–172. [Google Scholar]
- [42].Liao F, Liang M, Li Z, Hu X, Song S, Evaluate the malignancy of pulmonary nodules using the 3-d deep leaky noisy-or network, IEEE transactions on neural networks and learning systems 30 (11) (2019) 3484–3495. [DOI] [PubMed] [Google Scholar]
- [43].Chorowski JK, Bahdanau D, Serdyuk D, Cho K, Bengio Y, Attention-based models for speech recognition, in: Advances in neural information processing systems, 2015, pp. 577–585. [Google Scholar]
- [44].Zhou P, Shi W, Tian J, Qi Z, Li B, Hao H, Xu B, Attention-based bidi-rectional long short-term memory networks for relation classification, in: Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 2: Short Papers), 2016, pp. 207–212. [Google Scholar]
- [45].Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I, Attention is all you need, in: Advances in neural information processing systems, 2017, pp. 5998–6008. [Google Scholar]
- [46].Zhang H, Goodfellow I, Metaxas D, Odena A, Self-attention generative adversarial networks, arXiv preprint arXiv:1805.08318. [Google Scholar]
- [47].Zhang Z, Chen P, McGough M, Xing F, Wang C, Bui M, Xie Y, Sapkota M, Cui L, Dhillon J, et al. , Pathologist-level interpretable whole-slide cancer diagnosis with deep learning, Nature Machine Intelligence 1 (5) (2019) 236. [Google Scholar]
- [48].Ilse M, Tomczak JM, Welling M, Attention-based deep multiple instance learning, arXiv preprint arXiv:1802.04712. [Google Scholar]
- [49].Chen P, Yang L, tissueloc: Whole slide digital pathology image tissue localization., J. Open Source Software 4 (33) (2019) 1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Plissiti ME, Nikou C, Charchanti A, Automated detection of cell nuclei in pap smear images using morphological reconstruction and clustering, IEEE Transactions on information technology in biomedicine 15 (2) (2010) 233–241. [DOI] [PubMed] [Google Scholar]
- [51].Rosenblatt F, Principles of neurodynamics. perceptrons and the theory of brain mechanisms, Tech. rep, Cornell Aeronautical Lab Inc; Buffalo NY: (1961). [Google Scholar]
- [52].Hastie T, Tibshirani R, Friedman J, Franklin J, The elements of statistical learning: data mining, inference and prediction, The Mathematical Intelligencer 27 (2) (2005) 83–85. [Google Scholar]
- [53].Simonyan K, Zisserman A, Very deep convolutional networks for large-scale image recognition, arXiv preprint arXiv:1409.1556. [Google Scholar]
- [54].Schiffman M, Adrianza ME, Ascus-lsil triage study. design, methods and characteristics of trial participants., Acta cytologica 44 (5) (2000) 726–742. [DOI] [PubMed] [Google Scholar]
- [55].Barreth D, Schepansky A, Capstick V, Johnson G, Steed H, Faught W, Atypical squamous cellscannot exclude high-grade squamous intraepithelial lesion (asc-h): A result not to be ignored, Journal of obstetrics and gynaecology Canada 28 (12) (2006) 1095–1098. [DOI] [PubMed] [Google Scholar]
- [56].Vizcaino AP, Moreno V, Bosch FX, MUNoz N, Barros-Dios XM, Borras J, Parkin DM, International trends in incidence of cervical cancer: Ii. squamous-cell carcinoma, International journal of cancer 86 (3) (2000) 429–435. [DOI] [PubMed] [Google Scholar]