Abstract
Cellular ageing is one of the main drivers of organismal ageing and holds keys towards improving the longevity and quality of the extended life. Elucidating mechanisms underlying the emergence of the aged cells as well as their altered responses to the environment will help understanding the evolutionarily defined longevity preferences across species with different strategies of survival. Much is understood about the role of alterations in the DNA, including many epigenetic modifications such as methylation, in relation to the aged cell phenotype. While transcriptomes of the aged cells are beginning to be better-characterised, their translational responses remain under active investigation. Many of the translationally controlled homeostatic pathways are centred around mitigation of DNA damage, cell stress response and regulation of the proliferative potential of the cells, and thus are critical for the aged cell function. Translation profiling-type studies have boosted the opportunities in discovering the function of protein biosynthesis control and are starting to be applied to the aged cells. Here, we provide a summary of the current knowledge about translational mechanisms considered to be commonly altered in the aged cells, including the integrated stress response-, mechanistic target of Rapamycin- and elongation factor 2 kinase-mediated pathways. We enlist and discuss findings of the recent works that use broad profiling-type approaches to investigate the age-related translational pathways. We outline the limitations of the methods and the remaining unknowns in the established ageing-associated translation mechanisms, and flag translational mechanisms with high prospective importance in ageing, for future studies.
Keywords: ageing, protein biosynthesis, ribosome, RNA, translation, translational control
Introduction
Cellular ageing refers to the progressive deterioration of cellular functions over time, often leading to cell cycle arrest (senescence) or cell death [1–3]. Ageing is characterised by several well-defined hallmarks including loss of proteostasis, telomeric shortening, mitochondrial dysfunction and changes to gene expression between young and aged cells [2–4]. Cellular ageing is one of the major contributors to organismal ageing and is thought to define the longevity of species [5,6]. In this review, we first introduce known epigenetic, transcriptional, and proteomic changes of aged cells. We then link these changes with the translational dynamics of aged cells and review the three key translation control pathways implicated in cellular ageing. We further highlight important future directions and unanswered questions to be explored regarding translational control and ageing.
Epigenetic effects of cell ageing are best characterised and include DNA methylation at certain CpG sites that almost linearly correlates with cell senescence (passaging). Overall, the number of methylated sites decreases in aged human cells, however, this trend is site-dependent, with 60% of the sites hypomethylated and 40% hypermethylated in ageing [7]. The directionality of these shifts is reliable across sites, forming the foundation of epigenetic molecular clock models. In such models, CpG sites with high correlation to the ageing are pooled and can be used to predict ages of tissue samples [8,9]. Other prominent alterations include general transcriptional amplification due to the loss of histones from the chromatin, up-regulation of cryptic promoters and ‘transcriptional noise’, and alterations of histone modifications that lead to chromatin remodelling [10–13].
Epigenetic molecular clock models have been successful in the prediction of average age, but other levels of gene expression (e.g. transcription) exhibit inconsistencies in age-related signatures dependent on individual, tissue and cell type. Gene expression signatures in natural aged samples are influenced by numerous factors, including historic exposures to radiation, infectious agents, disease and (bio)chemicals [14,15]. Controlled laboratory studies using representative cell lines and model organisms are commonly used to alleviate historic variability, but carry limitations of the in vitro culture and evolutionary or longevity differences across the typical model species (e.g. Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster and Mus musculus) [5]. Perhaps the soundest approach to age-related research is an integrative one, studying across a range of organismal and cellular models, and potentially small human cohorts, to characterise the common and cell-specific signatures of ageing at various levels of gene expression [16,17].
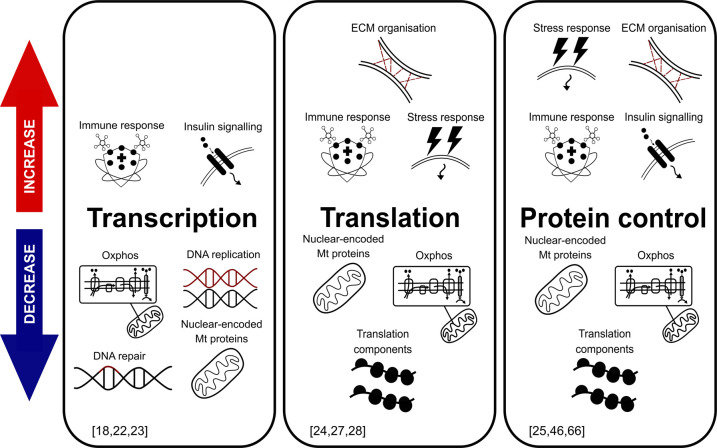
Integrative sampling approaches are, to some extent, being implemented through the study of transcriptional and proteomic changes in the aged cells. Microarray and RNA sequencing have been conducted across various species and cell types, including in vitro studies of human fibroblastic cell lines and in vivo studies of human peripheral blood cells and mouse endothelial cells [18–21]. Several transcriptomic signatures are found to be conserved across the aged cells, such as down-regulation of mitochondrial and cell division genes and up-regulation of extracellular matrix (ECM) components and apoptosis signalling genes (Figure 1) [14,18–21]. Some of the most archetypal aged transcriptome traits observed in human cell lines, multicellular models such as mouse and rat tissues and human cohorts, are the increased expression of immune response factors and reduction in the abundance of ribosome protein and ribosome biogenesis factor mRNAs (Figure 1; Table 1).
Figure 1. Overview of the cell age-related alterations in the different stages of gene expression control.
Arrows on the left indicate the relative increase or decrease in the gene expression or protein abundance associated with the ageing, as compared with the non-aged cells. Select representative review work references are shown in the bottom of the panels.
Table 1. Summary of recent works investigating transcriptional changes to gene expression in aged cells.
| Model system | Tissues studied | Method | Up-regulated | Down-regulated | Reference |
|---|---|---|---|---|---|
| C57BL6 mice (10–12 weeks vs. 14 months) | Brain, heart, kidney | Illumina short read total RNA-seq | Glutathione metabolism, Insulin signalling | Oxidative phosphorylation | [18] |
| Mouse, Human, Rats | Human: brain, kidney, muscle; Mouse: muscle, kidney, brain, heart, liver, lung, bone marrow; Rat: heart, muscle, brain, bone marrow, spinal cord | Data from 12 mice, 11 rat and 4 human microarrray studies was downloaded and used for meta-analysis | Glutathione metabolism, Immune response, Lysosome, Negative regulation of apoptosis | Oxidative phosphorylation, Mitochondrial proteins, Collagen | [19] |
| Human fibroblast cell lines (MRC-5, BJ, IMR-90, WI-38 and HFF) | - | Illumina short read RNA-seq on cells of various passages with β-galactosidase assays and immunoblotting used to confirm senescence | Lysosome, Immune response | DNA repair, RNA degradation, Oxidative phosphorylation, DNA replication, Ribosome biogenesis, Spliceosome expression | [20] |
| C57BL6 mice (8 weeks vs. 18 months) | Vascular endothelial | Illumina short read RNA-seq | PI3K/Akt signalling, ECM receptor interactions, Apoptosis | Mitotic division, Angiogenesis | [21] |
| Diversity outbred mice (6, 12 and 18 months) | Kidney tissue | Illumina short read RNA-seq | Immune and inflammatory response, DNA repair, Apoptosis regulation | Heat shock proteins | [22] |
| Human | Peripheral blood | Illumina short read RNA-seq data from 7074 human peripheral blood samples | Immune response, ECM formation, Lysosome | Mitochondrial proteins, DNA replication, DNA repair, Ribosome biogenesis | [23] |
Proteomic fluctuations in aged cells have been broadly studied, including in rat brain and liver tissues, mice lung tissue, human bone marrow and skeletal muscle and comprehensively in Caenorhabditis elegans [24–28]. In the aged tissues, increase in oxidoreductive (components of peroxisomes) [24,27] and immune response [25,26,28] proteins are commonly observed, alongside the decrease in ribosomal proteins, the latter mirroring transcriptomic alterations [25,27,28] (Table 2).
Table 2. Summary of recent works investigating protein-level changes to gene expression in aged cells.
| Model system | Tissues studied | Method | Up-regulated | Down-regulated | Reference |
|---|---|---|---|---|---|
| Nematodes | - | Liquid chromatography mass spectrometry was conducted on protein isolates from organisms at ages 1 day, 5 days and 10 days | Stress response, Unfolded protein response, mTOR signalling, Insulin signalling | Fatty acid, amino acid, carbohydrate metabolism, Peroxisome proteins, Oxidation reduction | [24] |
| Rats (6 months vs. 24 months old) | Brain and liver | Shotgun mass spectrometry was conducted on subcellular fractions including nuclear, post-nuclear fractions 1 and 2, and soluble cytosolic proteins | Extracellular matrix binding, RNA transport, Peroxisome organisation, TCA cycle | NADH dehydrogenase activity, Protein kinase activity | [25] |
| Human | Haemato-poietic stem and progenitor cells | Liquid chromatography mass spectrometry | ECM organisation, Insulin processing, Metabolic processes, Mitochondrial function | Cell cycle and DNA repair, Mitochondrial translation factors, Lymphoid development | [27] |
| Human | Skeletal muscle | Muscle biopsies from 58 participants aged between 20 to 87 years were analysed using liquid chromatography mass spectrometry | Immune response, Proteostasis, Alternative splicing | Mitochondrial functional proteins, Ribosomal proteins, Energy metabolism, Glycolysis | [28] |
| Diversity outbred mice (6, 12 and 18 months) | Kidney tissue | Mass spectrometry | Apical transporters, Immune response, Sodium reabsorption | Oxidative phosphorylation, Mitochondrial autophagy proteins, Endoplasmic reticulum membrane, Histones | [22] |
Numerous studies have shown that the correlation between mRNA and protein levels becomes progressively decoupled in ageing [22,29]. In young and aged mouse kidney samples, it was observed that several nutrient re-uptake membrane transporters showed decreased mRNA abundances compared with protein levels, whilst RNA splicing genes displayed the reverse effect [22]. Discordance between mRNA and proteins levels is also observable in aged killfish [4], macaque [30] and human brains [30,31]. This progressive decoupling is mediated by several post-transcriptional regulators including micro RNAs (miRNAs) and RNA binding proteins (RBPs), and the reduced proteostasis. In killfish and human brain, several differentially abundant proteins were found to have miRNA target sites in the transcripts encoding them. In human brains, the RBPs SFRS1, TIAL1 and AGO2 were associated with driving discordant mRNA–protein levels [4,30]. Reduction in protein degradation machinery component abundance, such as proteasomal subunits, ubiquitination proteins and Unfolded Protein Response (UPR) pathway proteins in aged human brain, is also a suggested contributor to the discordant mRNA and protein levels [4,31].
The incomplete correlation between mRNA and protein content has provoked research into translational signatures of aged cells, particularly with respect to the known translation control pathways [32–35]. Translation control mechanisms may suppress or incite the protein biosynthesis of certain mRNAs, as well as globally control protein output of the cell [32,36–47]. Translational control is rapid in nature [32,48] and common in stress response signalling, such as in hypoxic stress [49,50], heat shock [51,52], oxidative stress [52,53] and nutrient deprivation [52,54]. The control mechanisms can be enacted at any phase of translation (initiation [32,48], elongation [55,56], termination [55,57] and recycling [57]), with the phosphorylation of translation initiation or elongation factors, or their interacting proteins, being a common means of response mediation [29,32,35,38,39,55,58,59].
Rapidly accumulating ribosome profiling works begin to bring new insights into the translational control of aged cells [60–62]. Universal characteristics of translation-level responses from these works have emerged across aged (replicatively) yeast [63], mouse [64,65] and human cells [46,66], where reduced translational engagement and elongation rates were observed. Ribosome profiling studies in mouse liver, kidney [65] and skeletal muscle [64], and human skeletal muscle [66], revealed reduced translation of mitochondrial, ribosomal and translation factor transcripts in the aged tissues (Table 3). Human heart tissue also exhibited reduced translation of nuclear-encoded mitochondrial proteins, whilst translation of cytosolic ribosome components, including 14 Ribosomal Protein Small subunit (RPS) and 18 Ribosomal Protein Large subunit (RPL) transcripts, increased [46]. Studies in replicatively aged yeast [63] and rat liver [25] displayed increased translation of stress response transcripts such as MT2A, SGK1 and HSPB1. Mouse liver and kidney [65] and human heart tissues [46] displayed significant increases in extracellular matrix (ECM) component translation [64–66]. Overall, decreases in translation fidelity and efficiency have also been commonly observed in the aged cells across yeast, mice and humans [46,63,64].
Table 3. Summary of recent works investigating translation-level changes to gene expression in aged cells.
| Model system | Tissues studied | Method | Up-regulated | Down-regulated | Reference |
|---|---|---|---|---|---|
| Rats (6 months vs. 24 months old) | Brain and liver | Ribosome profiling was conducted as per Ingolia et al. [60,61] using Illumina HiSeq technologies | Immune and inflammatory response, Lipid oxidation, Stress response, Translation | Ion channel activity, Neuronal action potential, Lipid biosynthesis, Amine catabolic processes | [25] |
| Yeast | - | Replicatively aged yeast cultures were harvested at 15 and 30 hrs and underwent cycloheximide treatment before subsequent polysome and ribosome profiling | Stress response, Translation repressors | Ribosome biogenesis, Translational regulators | [63] |
| Mice | Liver, kidney and skeletal muscle | Assessed translation efficiency of specific classes of mRNAs using ribosome profiling in 3-month- and 18-month-old mice | TCA cycle, Oxidative phosphorylation, Fatty acid metabolism, Glycolysis | mTOR signalling, MAP kinase signalling, Insulin signalling, Translation components | [64] |
| Mice | Liver and kidney | Liver and kidney samples were taken across various timepoints with three biological replicates (in all except one condition) undergoing Ribo-seq | Inflammation and immune response, Lysosome, ECM organisation | Mitochondrial proteins, Redox homeostasis, Translation components | [65] |
| Human | Skeletal Muscle | Skeletal muscle biopsies were performed on three individuals aged between 40–45 and two individuals aged 80+ and the tissues were subjected to ribosome profiling with Illumina HiSeq 2500 short read sequencing | - | Mitochondrial proteins, Oxidative phosphorylation | [66] |
| Human | Heart tissue | 65 left ventricle samples from dilated cardiomyopathy (DCM) and 15 non-DCM controls were used for ribosome profiling. Footprint libraries were sequenced with Illumina HiSeq 2500 | ECM production, mTOR signalling, Translation components | Mitochondrial processes | [46] |
Several key translation control pathways are emerging in the highlights of age-associated translation control research, boosted by the global profiling methods. Understanding the full evolutionary role, biological potential, and the exact mechanisms of action of translation-mediated control pathways of the aged cells is important for outlining the routes to longevity and synthetic biology developments with specific lifespan design. In this review, we update and reflect on the Integrated Stress Response (ISR), mechanistic (mammalian) Target of Rapamycin (mTOR) and eukaryotic Elongation Factor 2 (eEF2) translation control pathways of cellular ageing. We also point out the less explored avenues of translational involvement in aged cell homeostasis and outline several priority directions for future research.
Role of the integrated stress response (ISR) in ageing
The Integrated Stress Response (ISR) is a translation control pathway effecting the initiation stage of translation. In this pathway, stress signals promote the action of stress kinases, which phosphorylate eIF2 on its alpha subunit [67]. eIF2 is the main Met-tRNAiMet carrier and a translation factor responsible for the assembly of a functional scanning complex. eIF2 is required for start codon recognition in most cases. Phosphorylation of eIF2α (to form eIF2α-P) reduces the instance of translation initiation by stabilising eIF2 interaction with its GTP exchange factor eIF2B, and thus, decreases global translation rates [32,48].
In mammals, there are four known stress kinases capable of eIF2 phosphorylation, each triggered by distinct stress stimuli. These kinases include Heme-Regulated Inhibitor (HRI), Protein Kinase R (PKR), PKR-like Endoplasmic Reticulum Kinase (PERK) and Eukaryotic translation initiation factor 2-alpha kinase 4 (EIF2AK4) (its yeast homologue is the General Control Non-depressible 2 or GCN2), which are activated under heme deprivation, viral infection, Endoplasmic Reticulum (ER) stress and amino acid deprivation, respectively [68].
It is evident that ISR activation stimulates global repression of translation, which may be favourable for increasing lifespan (Figure 2) [69,70]. An example of the protective effect of translation repression on lifespan is known from the increased longevity of Caenorhabditis elegans upon eIF2 knock-down, as well as knock-down of many other important initiation factors (notably, eIF4G) and ribosomal proteins [70].
Figure 2. Integrated Stress Response (ISR) pathways are implicated in the aged cell phenotype.
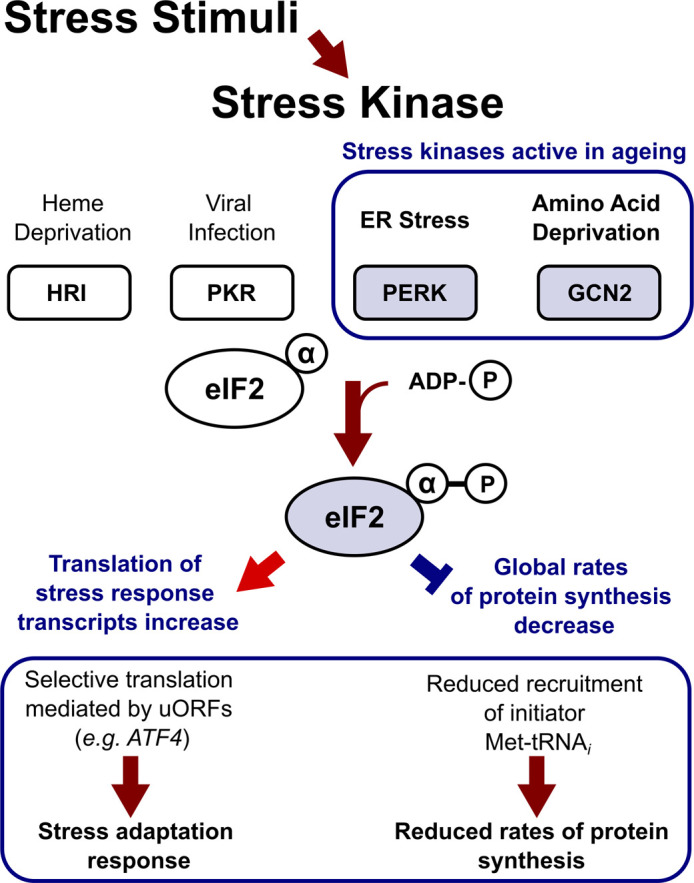
Highlighted factors (blue) are more abundant in the aged cells and exert specific activation of transcript translation (red arrow) and global reduction in protein synthesis (blue block) [48,67,71,72].
Alongside the global repression of translation observed under high eIF2α-P, ISR also induces translation of specific mRNAs involved in the stress response. Selective translation is thought to be mostly regulated via upstream Open Reading Frames (uORFs) in this case. The placement, length and amino acid sequence of the uORFs determines if main ORF engagement increases or decreases [71]. In mammals, eIF2α-P triggers increased translational expression of Activating Transcription Factor 4 mRNA (ATF4; also in yeast with its orthologue GCN4 where uORF control mechanism was discovered) [72–74], which transcriptionally induces expression of CCAAT/enhancer binding protein (C/EBP) homologous protein gene (CHOP). CHOP subsequently increases apoptotic signalling [72,73,75].
Activation of the ISR via the ER stress kinase PERK is regulated by the upstream UPR pathway. The UPR is activated in response to the accumulation of unfolded and misfolded proteins (ER stress) [76]. Localised within the ER are several ER chaperones, required for the folding and secretion of newly formed proteins, critical to assuring protein functionality. The UPR is activated in response to decreased protein folding efficiency, which activates PERK, increasing the eIF2α phosphorylation and decreasing the global protein synthesis [77]. In aged cells, the production of ER chaperone proteins is reduced, leading to the increased accumulation of unfolded or misfolded proteins. This activates PERK, stimulating ISR-mediated reduction in the protein synthesis and selective up-regulation of ER stress proteins via the dependence of their mRNAs on the uORF-controlled translation [78].
In several eukaryotic ageing models, phosphorylation of eIF2α by both PERK and GCN2/EIF2AK4 is prevalent [2,76]. In lower eukaryotes such as replicatively aged yeast (e.g. Saccharomyces cerevisiae and Schizosaccharomyces pombe), depletion of eIF2α-P negatively influenced lifespan, whilst its induction positively regulated autophagy and increased lifespan [63,79,80]. In contrast and somewhat surprisingly, in Caenorhabditis elegans, activation of the ISR was detrimental to longevity, with both pharmacological ISR inhibition and phosphorylation-defective eIF2α mutant extending the lifespan [81]. Ribosome profiling in yeast and rat brain and liver have indicated increase in the stress response transcripts translation in the aged tissues [25,63]. These results suggest ISR functions are complex and extend beyond translational down-regulation.
In the brain, low-level activation of the ISR is essential for memory formation and development. Excessive and/or chronic activation of ISR in the brain, in contrast, is associated with neurodegeneration [43,82]. Prolonged activation of the ISR is linked to Parkinson's Disease, Huntingdon's Disease, and Alzheimer's Disease (AD). This ISR involvement is not always correlative: for instance, in AD mouse models, reduction in the ISR by knock-out of eIF2α kinases PERK and EIF2AK4 partially alleviated the disease phenotype, improving synaptic plasticity and spatial memory. In addition, inhibition of the ISR via ISR inhibitor (ISRIB) has been shown to improve spatial and working memory deficits in aged mice [83–85]. ISR inhibition is a therapeutic target for neurodegenerative conditions which are often associated with organismal ageing, but the role of elevated ISR in the aged cells of higher metazoa remains unclear.
Overall it can be concluded that ISR functions may be mostly beneficial in the ageing of single-cell organisms. Substantial ISR activation and dysregulation can be detrimental to the lifespan and stimulate disease in complex multicellular animals [86]. In this regard, ISR functions in ageing may be linked to carcinogenesis, where ‘unicellular' genes are known to be exaggerated in the transcription profile [87]. It remains unclear to what extent ISR is beneficial to the various types of aged cells of multicellular organisms, an important future direction of research that needs to be investigated across different stressors and mitotic and post-mitotic cell types.
Deciphering the role of mTOR in longevity
The mechanistic (mammalian) Target of Rapamycin (mTOR) pathway is a critical translation control mechanism regulating cell growth and proliferation. The pathway is stimulated by hormones, growth factors and amino acid availability, which positively regulate the activity of mTOR complexes 1 and 2 (mTORC1 and mTORC2) [37]. mTOR complexes contain a catalytic subunit that acts as a serine/threonine protein kinase. The activation of mTOR complexes effects numerous intracellular processes, including mRNA translation, metabolism, protein degradation and cell migration [40,88,89].
mTOR1 regulates translation via two main mechanisms. In the first mechanism, mTOR1 activates ribosomal S6 kinases by phosphorylation, allowing S6K1 to activate several facilitators of translation initiation [88,90]. Among these activation targets is eIF4B, a ubiquitous component of initiation with broad mRNA scanning stimulation activity [39,88,90]. In the second mechanism, mTOR complexes phosphorylate eIF4E-Binding Proteins (4EBPs), triggering their dissociation from the sequestered eIF4E and consequent eIF4E's return to the translation-accessible pool [88,91]. The released eIF4E associates with other initiation factors, including eIF4G, and through its binding to the 5′ mRNA cap facilitates cap-dependent translation [88,92].
Adding to the overall translation stimulation effect, translational control by mTOR incites increased expression of select transcripts, in some similarity to the global and specific effects of the ISR. Terminal Oligopyrimidine (TOP) tracts have been suggested as a feature of mRNAs specifically responsive to the mTOR activation. Notable examples of TOP mRNAs include translation-related eEF1A, eEF2 and ribosomal protein S6 [40,88].
mRNAs with long-and-structured 5′UTRs have also been considered as ‘eIF4E-sensitive’ and thus more responsive to the mTOR signalling. Importantly, these transcripts include mRNAs coding for cell cycle and proliferation regulators, such as cyclins, Vascular Endothelial Growth Factor (VEGF) and MYC [88,92].
mTOR is an extensively regulated pathway, which is suppressed in various circumstances. Under stress conditions including DNA damage and hypoxia, signalling molecules including p53, AMP-activated protein kinase (AMPK) and Regulated in Development and DNA Damage responses protein 1 (REDD1) suppress mTOR complex activity through the Tuberous Sclerosis Complex (TSC) [88]. The TSC targets mTOR activating molecule, Ras homologue enriched in brain (Rheb), hydrolysing attached GTP to GDP, to disrupt its signalling properties [93].
In recent years, interest into the role of mTOR in the context of ageing has substantially grown. Studies across Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster and Mus musculus have indicated that mTOR is more active in ageing, with its activation negatively correlating with longevity [89,94–96]. Genetic knock-out of the S6K homologue gene SCH9 in replicatively aged yeast caused extensions in lifespan, firmly establishing the link between mTOR and longevity [97]. Similarly, in Caenorhabditis elegans, RNA interference (RNAi) inhibition of the Regulatory-Associated Protein of mTOR (RAPTOR) expression positively increased the animals’ lifespan [98,99]. Reduction in mTOR activity by caloric restriction or the canonical mTOR1 inhibitor, Rapamycin, has increased longevity in several model organisms, including Caenorhabditis elegans [100,101], Drosophila melanogaster [102,103] and Mus Musculus (Figure 3) [104,105].
Figure 3. mTORC1 translational control and its link to the aged cell phenotype.
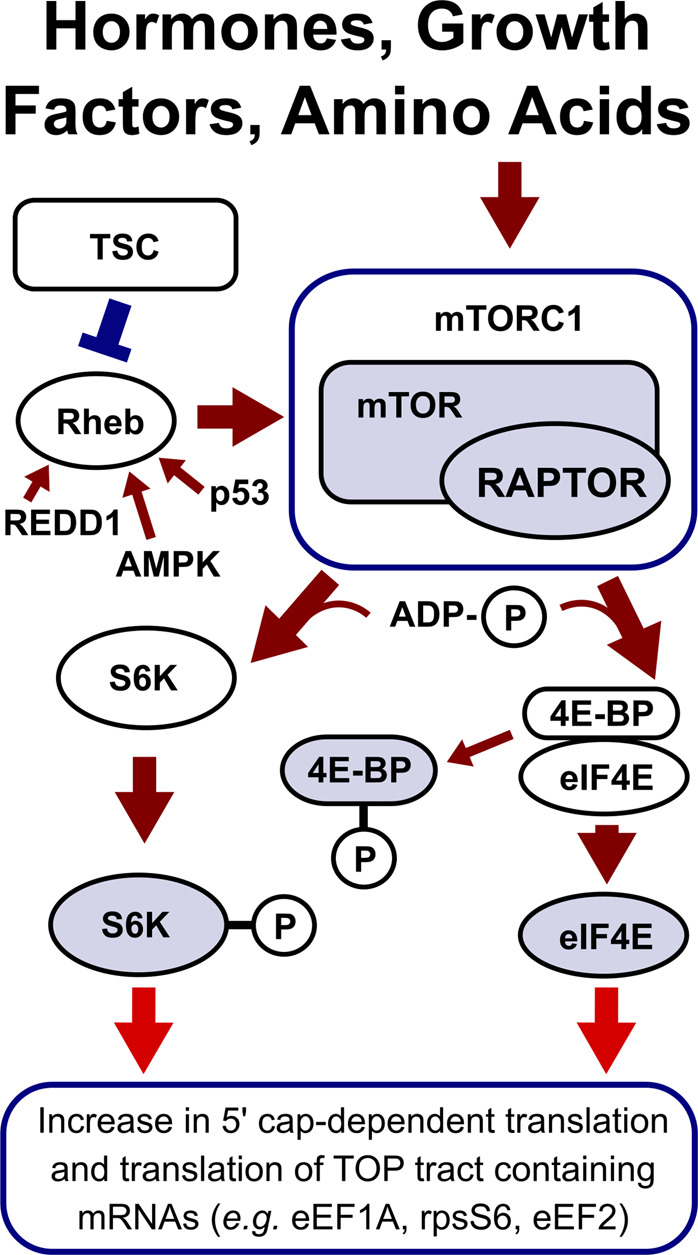
Highlighted factors (blue) are more abundant in the aged cells and enhance overall translation, with some specific stimulatory effects on mRNAs with 5′ Terminal Oligopyrimidine (TOP) tracts and long-and-structured (highly cap-and-scanning-dependent) 5′UTRs (red arrows) [88,89,92].
Ribosome profiling studies in numerous tissues and organisms have highlighted discrepancies in translational up-regulation of mTOR signalling components within aged samples. For example, ribosome profiling of human heart tissue revealed translational enrichment of mTOR signalling components [46], whilst their down-regulation was observed in mouse kidney and liver [64]. These data imply that mTOR activation at the translational level may vary across tissues, complicating the concept of blanket mTOR up-regulation accompanying ageing.
With the general knowledge that mTOR restriction positively influences lifespan in model organisms, the question has arisen whether these findings are applicable in humans. Initially, this was investigated in cell samples from patients with Hutchinson-Gilford Progeria Syndrome (HGPS), a condition characterised by premature ageing [106,107]. The study found that Rapamycin treatment effectively delayed the onset of senescence in HGPS cells and aided in dissolving progerin aggregates, which are a central attribute of the disease. The results suggest that Rapamycin treatment may be clinically beneficial for children with HGPS and may positively influence longevity, but any longer-term effects and those across tissues in an organismal setting need to be investigated.
A concern regarding the use of Rapamycin as an ageing intervention is its ability to effect mTORC2 signalling. Whilst mTORC2 is not typically susceptible to Rapamycin inhibition, chronic exposure to the compound can lead to its inhibition, as exhibited in several studies on mice [108]. mTORC2 is known to contribute to cytoskeleton organisation and insulin signalling [89]. Studies in both mice and nematodes have shown that the suppression of mTORC2 reduces lifespan [108–111], likely due to consequently-formed insulin resistance [108]. These findings indicate that caution should be taken in administering Rapamycin or analogues of the compound (rapalogs) which are capable of inhibiting both mTORC1 and mTORC2, as mTORC2 inhibition can induce detrimental effects on lifespan [89].
Treatment with Rapamycin or rapalogs for other clinical purposes such as immunosuppression has led to prolific side effects, including neutropenia, thrombocytopenia, hyperglycemia, and pneumonitis [112]. A clinical trial utilising the rapalog RAD001, which should not affect mTORC2 functionality, in elderly individuals (over 65 years), has shown an improved immune response to influenza vaccines, exhibited by increases in antibody titers compared with placebo conditions and decreases in percentages of pro-apoptotic CD4 and CD8 T-cells. Few individuals experienced adverse side effects to the low dosages (0.5 mg daily or 5 mg weekly), indicating that RAD001 may be appropriate for clinical use in older individuals [113].
To conclude, while the positive correlative link to the mTOR activation in the aged cells seems to be well-established and supported by diverse biological evidence, we are still far away from the understanding of the functional role of mTOR in ageing. Some common-sense choices are that mTOR activation can be a compensatory response of the cells required to overcome the age-related transcriptional and DNA deficiencies, or it is an inevitable consequence of the altered transcriptome of the aged cells. Cancer cells exploit the mTOR-driven activation of Rat sarcoma virus (Ras) proteins which are important for the elevated production of the oncogenes and forming the proliferative outfit of the cells, but the mTOR-induced suppression of the autophagy was also demonstrated to render the malignant cells more vulnerable by increasing the chances of ‘energy crisis' [114,115]. Thus, the existing evidence supports a view that age-related mTOR activation can be functional in some but not all cell type contexts and the evolutionary fine-tuning of the mTOR activity has been to provide the balance fit for the species-specific environment and lifespan. Consequently, it would be impossible to broadly suppress (or activate) mTOR for an overall beneficial effect on longevity, but a targeted approach accounting for the cell type and transcriptome profile would be necessary. Therefore, more detailed information on the involvement and the purpose of mTOR pathway across the aged cells of different types and in diverse environments is required in the future, a task incorporating investigations of rapid translation-based cell responses to stress factors.
Translational control by eEF2 phosphorylation in ageing
eEF2 is an elongation factor that facilitates ribosomal translocation across the codons of mRNA Open Reading Frames (ORFs). The main eEF2 regulator, eEF2 kinase (eEF2K), is activated by autophosphorylation (Thr-348 in human protein) in response to eEF2K interaction with the calcium:calmodulin complex. eEF2K can also autophosphorylate independently of calcium (Ser-500 in human protein) to become active in the calmodulin presence. Active eEF2K then phosphorylates eEF2, preventing its binding to elongating ribosomes [116,117]. By suppressing eEF2 participation in elongation, the overall rate of elongation is slowed [55,56]. eEF2K phosphorylation level is regulated by various signalling cascades. Phosphorylation of eEF2K, promoting its engagement with eEF2, is enhanced by AMPK pathway. Dephosphorylation of eEF2K is mediated by mTOR and Extracellular signal-regulated kinase (ERK) signalling [56,118].
Like the ISR, eEF2K pathway is active in stress, with eEF2K activity positively correlating with cell survival during adverse conditions. This has been observed in response to several stress stimuli, including nutrient [119], temperature [59] and genotoxic stress [120]. eEF2K activity is thought to be beneficial under stress, as it is hypothesised that the decreasing elongation rate conserves energy and improves the accuracy and fidelity of protein synthesis [56].
It has become apparent that eEF2 and its regulator eEF2K are implicated in cellular ageing (Figure 4). A ribosome profiling study in aged mouse liver showed that ribosomal occupancy in eEF2 transcripts was significantly reduced [65]. At the protein level, rat muscle samples exhibited linear decreases in eEF2 abundance across 3, 6, 12, 18 and 24 months of the animals’ age [121]. This reduced abundance of eEF2 is thought to partially contribute to the lowered translation rates in aged organisms. In addition, mTOR signalling, which is often active in ageing, to some extent regulates the activity of eEF2. mTOR signalling decreases the phosphorylation rates of eEF2K, which in turn prevents phosphorylation of eEF2, increasing its availability for translation elongation [56,89]. It thus has been proposed that suppression of eEF2K phosphorylation by mTOR signalling in ageing may negatively influence lifespan. For example, in the model organism Caenorhabditis elegans, knock-out of eEF2K orthologue efk-1 via CRISPR/Cas9 gene editing reduced translation fidelity and negatively impacted lifespan. Conversely, suppression of mTOR via Rapamycin treatment, which increased eEF2K activity, has increased translation fidelity, and extended lifespan [122]. The latter study highlights the importance of preserving translation fidelity in ageing and the likely biological function of eEF2K in the fidelity maintenance.
Figure 4. eEF2 Kinase- (eEF2K) and mTOR-mediated translational control and its link to the aged cell phenotype.
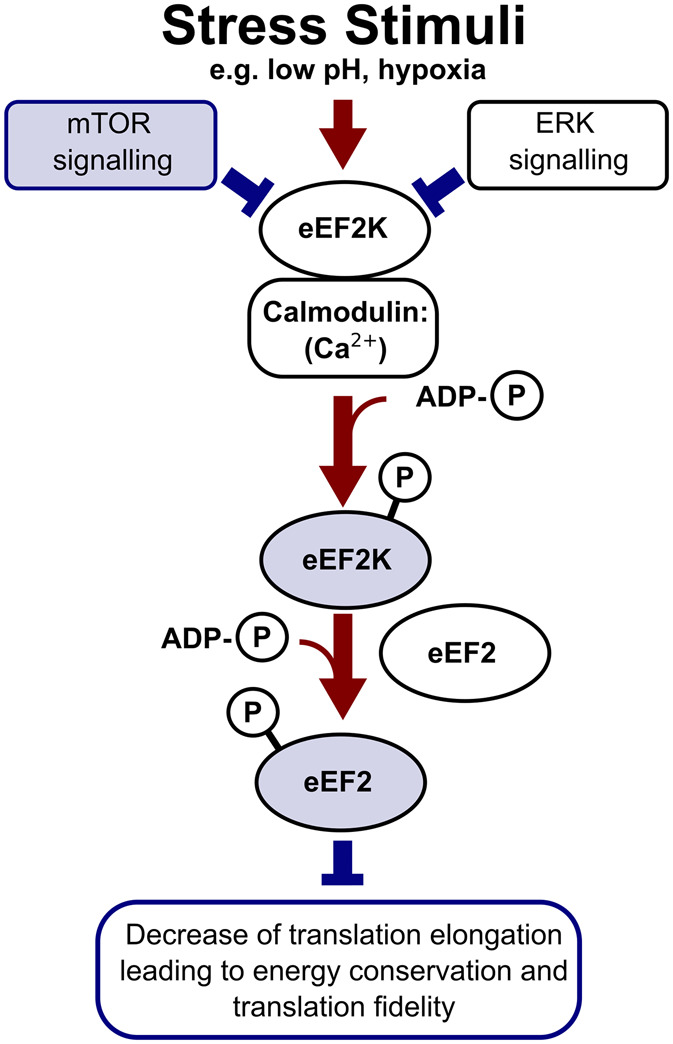
Highlighted factors and pathways (blue) are more abundant or prominent in the aged cells and generally result in the suppression of translation by reducing the translation elongation rate [56,116,117].
Yet in certain circumstances, it has been prominently demonstrated that eEF2K-induced reduction in the protein synthesis rates can be detrimental to the survivability of the cells or multicellular organisms. A pivotal study on eEF2K function in germ cells has shown that its reduced function is allowing the production of anti-apoptotic proteins and excessive survival of oocytes [123]. The increased oocyte survival in the context of species is a highly negative event, potentially leading to germline instability and embryonic defects. In contrast, in the context of neuronal function and reprogramming, it was shown that eEF2K suppression greatly reduces the Alzheimer's disease phenotype in model mice, promoting memory formation and translation-depended synaptic activity [124].
Overall, an exploration into the role of eEF2K in cellular ageing is a relatively recent research interest. Current data indicate that eEF2K activation is generally beneficial for longevity and species genetic stability, highlighting eEF2K induction as a potential therapeutic target of ageing. However, eEF2K reduces the cell capacity to rapidly respond to the stress factors, which can lead to cell death and accelerated ageing, and can be hypothesised to incur excessive DNA damage and instability and thus contribute to the carcinogenic pathways [123,124].
Future research directions and areas of interest
Evidence related to the contribution of translational dynamics to cellular ageing is becoming increasingly apparent, with several avenues prompting further exploration. Numerous examples of ribosome profiling studies have effectively characterised the aged cell translatome [63–65]. However, many of these studies took place using model organisms, with few studies conducted in humans. Existing works in human musculoskeletal tissue [66] and heart tissues [46] have highlighted aspects of the translational environment of aged cells, such as decreases in mitochondrial proteins and increases in ECM production, but there is still opportunity to broaden the range of tissues explored. There is also an increasing opportunity to utilise new techniques for investigating translation in ageing. Recent methodological advancements such as Translation Complex Profile Sequencing (TCP-seq; or its factor-selective Sel-TCP-seq variant) [125–127] may offer more detailed insight into translational control at the initiation stage for the aged cells. The TCP-seq pipeline allows for the study of 5′UTRs to characterise transcript-specific features of scanning by the small ribosomal subunits. Using TCP-seq, stalling of SSUs was observable, particularly in transcripts with long, structured 5′UTRs or uORFs, which may be of an interest to the cases of the age-related ISR and mTOR regulation [125].
In addition to the highlighted major pathways, recent studies have identified translational control through eIF5A hypusination apparent in cellular ageing. eIF5A is an elongation factor that contains a unique amino acid hypusine, synthesised by a transfer of the aminobutyl moiety from spermidine (a polyamine) to lysine 50 (human) of eIF5A [128,129]. Hypusinated eIF5A can alleviate ribosome stalling in ‘hard-to-translate’ mRNA motifs, including polyproline tracts [129,130]. Importantly, this feature of hypusinated eIF5A has been prominent in the translation of autophagic factors, such as ATG3 [130] and TFEB [131], translationally increasing their abundance. Because in ageing levels of spermidine decrease, hypusinated eIF5A becomes less available, and the autophagy is suppressed [131,132]. It is noteworthy, that a mere supplementation of spermidine in aged Drosophila brains has improved mitochondrial function and memory [128]. In aged mice, spermidine supplementation improved B-cell responses [131]. Additionally, prolonged spermidine supplementation in aged mice (6 months) improved lifespan [133]. These studies highlight the potential of spermidine as an alternative anti-ageing intervention.
Another area of interest in the field of translational control and ageing is the role of the translation rate. It has previously been shown that rates of translation decrease in cellular ageing and that the genetic knock-down of translational components is often beneficial for longevity [65,70]. Decrease of the translation rates is thought to be beneficial for several reasons. The first proposed reason is that translation is a highly energetically expensive process. By reducing translation rate in ageing, cells can redistribute energetic resources to the other processes like DNA maintenance, increasing longevity [29]. Another benefit of the reduced translation rates is the increased translation fidelity, which is considered extremely important in improving longevity [65,134]. Reducing translation rates directly rather than targeting translational control elements may be an appropriate therapeutic intervention for increasing lifespan. However, this approach is somewhat more complex, as translation must not be fully inhibited and the translation of certain genes essential for continued cellular survival must be maintained, as well as cell and tissue type-optimal translation rates must be respected.
Relating to the translation control mechanisms, another avenue of potential exploration is the therapeutic induction or reduction in the age-specific pathway mechanisms, and developments towards cell-type specific or specific stress-activated drugs. It is exciting that in all, mTOR [100,102,105], ISR [84,136] and eEF2K-based [122] age-related regulation small molecules have been successfully used to affect the pathway and increase longevity in certain circumstances (Table 4). As it becomes more apparent that the translational control has been carefully balanced by the evolution to fit the optimal longevity of the species’ individuals, cell type-selective translation effectors could be used to increase the stress resistance or decrease the proliferation programs and carcinogenicity in the critical cell types. Further research into translational control of the aged cells is needed to understand what cell types and tissues require which adjustments to extend the lifespan with minimal adverse side effects.
Table 4. Summary of recent works utilising small molecule inhibitors targeting translation control pathways to increase lifespan and ameliorate age-related functional declines.
| Intervention compound | Target pathway | Effect | Model | Outcome | Reference |
|---|---|---|---|---|---|
| ISRIB (Integrated stress response inhibitor) | ISR (specifically eIF2B) | Inhibition | Healthy normal aged mice | Reversed spatial memory deficits, Improved working memory | [84] |
| Prion-infected mice | Prevented neuronal loss, Increased survival | [135] | |||
| Rapamycin | mTOR (specifically mTORC1) | Inhibition | Human Hutchinson-Gilford Progeria Syndrome skin cells | Delayed onset of senescence, Dissolved progerin aggregates | [106,107] |
| Normal nematodes | Increased stress resistance, Lifespan extension | [100] | |||
| Normal fruit fly | Increased stress resistance, Reduced fecundity, Increased lipid levels, Lifespan extension | [102] | |||
| Genetically heterogenous mice | Lifespan extension | [105] | |||
| eEF2K | Activation | Normal nematodes | Lifespan extension | [122] | |
| RAD001 (Everolimus) | mTOR (specifically mTORC1) | Inhibition | Elderly human blood samples (65 and over) post-influenza vaccine | Increased antibody titres, Decreases in pro-apoptotic CD4 and CD8T-cells, Improved immune function | [113] |
| Spermidine | eIF5A hypusination | Activation | Normal fruit fly brain samples | Improved mitochondrial function and memory | [128] |
| Healthy normal mice | Improved B-cell responses, Reduced B-cell senescence | [131] | |||
| Healthy normal aged mice | Delayed cardiac ageing, Improved mitochondrial function, Lifespan extension | [133] |
Conclusions
Cellular ageing is a complex process which elicits alterations in gene expression at all levels. Recent research into the translation-level responses has revealed several translation control pathways implicated in lifespan and longevity. In the case of mTOR and ISR, prolonged, chronic activation of these mechanisms is detrimental to lifespan, with their inactivation by inhibitors being a potential avenue for anti-ageing therapeutics. Conversely, induction of the eEF2 inhibitor eEF2K by phosphorylation is reportedly beneficial for longevity, with its activation increasing the accuracy of protein synthesis, slowing translation rates and prolonging lifespan. Potentially, activation of this pathway may be a suitable therapeutic anti-ageing target, however more research is required in model species to comment further. In all cases, directly countering the age-specific translational alterations may not be universally beneficial, and cell type, stress and homeostatic environment, as well as the species-specific lifespan need to be carefully considered. Overall, translation control pathways are an integral part of gene expression control in aged cells and present an excellent opportunity for non-genetic correction of longevity and age-related deterioration of function or disease.
Perspectives
Cellular ageing is accompanied by the activation of several translation control pathways which regulate gene expression to affect cell survival during stress, DNA damage and proliferative rate. Translational control of the aged cells can underpin the evolutionary preferences of the species towards longevity, which is a biological function of high importance but insufficient understanding.
In the aged cells, activation of ISR and mTOR is demonstrated to impede longevity, whilst inhibition of these pathways increases lifespan. Translational control by eEF2 phosphorylation with eEF2 Kinase increases lifespan in eukaryotes such as nematodes, but its effects on more complex multicellular organisms remain uncertain.
Artificial suppression or activation of the age-specific translation control pathways has high potential for anti-ageing therapeutics. As in-depth investigation of translational responses of the cells became more universally accessible with the advent of translation profiling-type experiments, it is increasingly important to define the details of the stress and survival mechanisms of the aged cells across cell types and species with different lifespan.
Acknowledgements
The authors are grateful for the critical scientific discussions to all members of the Division of Genome Sciences and Cancer of The John Curtin School of Medical Research, The Australian National University.
Abbreviations
- AD
Alzheimer's disease
- AMPK
AMP-activated protein kinase
- ATF4
activating transcription factor 4
- CHOP
CCAAT/enhancer binding protein (C/EBP) homologous protein gene
- ECM
extracellular matrix
- eEF2K
eukaryotic elongation factor 2 kinase
- eIF2
eukaryotic initiation factor 2
- ERK
extracellular signal-regulated kinase
- GCN2
general control non-depressible 2
- HGPS
Hutchinson-Gilford Progeria Syndrome
- HRI
heme-regulated inhibitor
- ISR
integrated stress response
- ISRIB
ISR inhibitor
- miRNA
micro-RNA
- mTOR
mechanistic (mammalian) target of Rapamycin
- PERK
PKR-like endoplasmic reticulum kinase
- PKR
Protein Kinase R
- RAPTOR
Regulatory-associated protein of mTOR
- Ras
Rat sarcoma virus protein
- RBP
RNA binding protein
- REDD1
Regulated in development and DNA damage responses protein 1
- Rheb
Ras homologue enriched in brain
- RNAi
RNA interference
- (Sel)-TCP-seq
(Selective) translation complex profile sequencing
- TOP
terminal oligopyrimidine
- TSC
tuberous sclerosis complex
- uORFs
upstream open reading frames
- UPR
unfolded protein response
- VEGF
vascular endothelial growth factor
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the Australian Research Council Discovery Project Grant DP180100111 (to N.E.S.) and National Health and Medical Research Council Investigator Grant GNT1175388 (to N.E.S.).
Author Contributions
K.W. and N.E.S. contributed to the conception, drafting, revising, and final approval of the manuscript.
References
- 1.Cui, H., Kong, Y. and Zhang, H. (2012) Oxidative stress, mitochondrial dysfunction, and aging. J. Signal. Transduct. 2012, 1–13 10.1155/2012/646354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.López-Otín, C., Blasco, M.A., Partridge, L., Serrano, M. and Kroemer, G. (2013) The hallmarks of aging. Cell 153, 1194–1217 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liguori, I., Russo, G., Curcio, F., Bulli, G., Aran, L., Della-Morte, D.et al. (2018) Oxidative stress, aging, and diseases. Clin. Interv. Aging 13, 757–772 10.2147/CIA.S158513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelmer Sacramento, E., Kirkpatrick, J.M., Mazzetto, M., Baumgart, M., Bartolome, A., Di Sanzo, S.et al. (2020) Reduced proteasome activity in the aging brain results in ribosome stoichiometry loss and aggregation. Mol. Syst. Biol. 16, e9596 10.15252/msb.20209596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lidzbarsky, G., Gutman, D., Shekhidem, H.A., Sharvit, L. and Atzmon, G. (2018) Genomic instabilities, cellular senescence, and aging: in vitro, in vivo and aging-like human syndromes. Front. Med. 5, 104 10.3389/fmed.2018.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiLoreto, R. and Murphy, C.T. (2015) The cell biology of aging. Mol. Biol. Cell 26, 4524–4531 10.1091/mbc.E14-06-1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner, W. (2019) The link between epigenetic clocks for aging and senescence. Front. Genet. 10, 303 10.3389/fgene.2019.00303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, D. and Kerr, C. (2019) The epigenetics of stem cell aging comes of age. Trends Cell Biol. 29, 563–568 10.1016/j.tcb.2019.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salameh, Y., Bejaoui, Y. and El Hajj, N. (2020) DNA methylation biomarkers in aging and Age-related diseases. Front. Genet. 11, 171 10.3389/fgene.2020.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sen, P., Shah, P.P., Nativio, R. and Berger, S.L. (2016) Epigenetic mechanisms of longevity and aging. Cell 166, 822–839 10.1016/j.cell.2016.07.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pal, S. and Tyler, J.K. (2016) Epigenetics and aging. Sci. Adv. 2, e1600584 10.1126/sciadv.1600584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hänzelmann, S., Beier, F., Gusmao, E.G.. Koch, C.M., Hummel, S., Charapitsa, I.et al. (2015) Replicative senescence is associated with nuclear reorganization and with DNA methylation at specific transcription factor binding sites. Clin. Epigenet. 7, 19 10.1186/s13148-015-0057-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Booth, L.N. and Brunet, A. (2016) The aging epigenome. Mol. Cell 62, 728–744 10.1016/j.molcel.2016.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frenk, S. and Houseley, J. (2018) Gene expression hallmarks of cellular ageing. Biogerontology 19, 547–566 10.1007/s10522-018-9750-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Misra, B.B. (2020) The chemical exposome of human aging. Front. Genet. 11, 574936 10.3389/fgene.2020.574936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Podder, A., Raju, A. and Schork, N.J. (2021) Cross-species and human inter-tissue network analysis of genes implicated in longevity and aging reveal strong support for nutrient sensing. Front. Genet. 12, 719713 10.3389/fgene.2021.719713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.da Costa, J.P., Vitorino, R., Silva, G.M., Vogel, C., Duarte, A.C. and Rocha-Santos, T. (2016) A synopsis on aging-theories, mechanisms and future prospects. Ageing Res. Rev. 29, 90–112 10.1016/j.arr.2016.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brink, T.C., Demetrius, L., Lehrach, H. and Adjaye, J. (2009) Age-related transcriptional changes in gene expression in different organs of mice support the metabolic stability theory of aging. Biogerontology 10, 549–564 10.1007/s10522-008-9197-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Magalhães, J.P., Curado, J. and Church, G.M. (2009) Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics 25, 875–881 10.1093/bioinformatics/btp073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marthandan, S., Baumgart, M., Priebe, S., Groth, M., Schaer, J., Kaether, C.et al. (2016) Conserved senescence associated genes and pathways in primary human fibroblasts detected by RNA-seq. PLoS ONE 11, e0154531 10.1371/journal.pone.0154531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang, L., Liu, M., Li, Q., Shen, B., Hu, C., Fu, R.et al. (2019) Identification of differential gene expression in endothelial cells from young and aged mice using RNA-seq technique. Am. J. Transl. Res. 11, 6553–6560 PMID: [PMC free article] [PubMed] [Google Scholar]
- 22.Takemon, Y., Chick, J.M., Gyuricza, I.G., Skelly, D.A., Devuyst, O., Gygi, S.P.et al. (2020) Proteomic and transcriptomic profiling reveal different aspects of aging in the kidney. eLife 10, e62585 10.7554/eLife.62585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters, M.J., Joehanes, R., Pilling, L.C., Schurmann, C., Conneely, K.N., Powell, J.et al. (2015) The transcriptional landscape of age in human peripheral blood. Nat. Commun. 6, 8570 10.1038/ncomms9570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narayan, V., Ly, T., Pourkarimi, E., Murillo, A.B., Gartner, A., Lamond, A.I.et al. (2016) Deep proteome analysis identifies age-related processes in C. elegans. Cell Syst. 3, 144–159 10.1016/j.cels.2016.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ori, A., Toyama, B.H., Harris, M.S., Bock, T., Iskar, M., Bork, P.et al. (2015) Integrated transcriptome and proteome analyses reveal organ-specific proteome deterioration in old rats. Cell Syst. 1, 224–237 10.1016/j.cels.2015.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Angelidis, I., Simon, L.M., Fernandez, I.E.. Strunz, M., Mayr, C.H., Greiffo, F.R.et al. (2019) An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat. Commun. 10, 963 10.1038/s41467-019-08831-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hennrich, M.L., Romanov, N., Horn, P., Jaeger, S., Eckstein, V., Steeples, V.et al. (2018) Cell-specific proteome analyses of human bone marrow reveal molecular features of age-dependent functional decline. Nat. Commun. 9, 4004 10.1038/s41467-018-06353-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ubaida-Mohien, C., Lyashkov, A., Gonzalez-Freire, M., Tharakan, R., Shardell, M., Moaddel, R.et al. (2019) Discovery proteomics in aging human skeletal muscle finds change in spliceosome, immunity, proteostasis and mitochondria. eLife 8, e49874 10.7554/eLife.49874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonskikh, Y. and Polacek, N. (2017) Alterations of the translation apparatus during aging and stress response. Mech. Ageing Dev. 168, 30–36 10.1016/j.mad.2017.04.003 [DOI] [PubMed] [Google Scholar]
- 30.Wei, Y.-N., Hu, H.-Y., Xie, G.-C., Fu, N., Ning, Z.-B., Zeng, R.et al. (2015) Transcript and protein expression decoupling reveals RNA binding proteins and miRNAs as potential modulators of human aging. Genome Biol. 16, 41 10.1186/s13059-015-0608-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dick, F., Tysnes, O.-B., Alves, G.W., Nido, G.S. and Tzoulis, C. (2021) Altered transcriptome-proteome coupling indicates aberrant proteostasis in Parkinson's disease. medRxiv 10.1101/2021.03.18.21253875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hershey, J.W.B., Sonenberg, N. and Mathews, M.B. (2012) Principles of translational control: an overview. Cold Spring Harb. Perspect. Biol. 4, a011528 10.1101/cshperspect.a011528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jansen, M., De Moor, C.H., Sussenbach, J.S. and Van Den Brande, J.L. (1995) Translational control of gene expression. Pediatr. Res. 37, 681–685 10.1203/00006450-199506000-00001 [DOI] [PubMed] [Google Scholar]
- 34.Kelen, K.V.D., Beyaert, R., Inzé, D. and Veylder, L.D. (2009) Translational control of eukaryotic gene expression. Crit. Rev. Biochem. Mol. Biol. 44, 143–168 10.1080/10409230902882090 [DOI] [PubMed] [Google Scholar]
- 35.Sonenberg, N. and Hinnebusch, A.G. (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gebauer, F. and Hentze, M.W. (2004) Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 5, 827–835 10.1038/nrm1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masvidal, L., Hulea, L., Furic, L., Topisirovic, I. and Larsson, O. (2017) mTOR-sensitive translation: cleared fog reveals more trees. RNA Biol. 14, 1299–1305 10.1080/15476286.2017.1290041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janapala, Y., Preiss, T. and Shirokikh, N.E. (2019) Control of Translation at the Initiation Phase During Glucose Starvation in Yeast. IJMS 20, 4043 10.3390/ijms20164043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shirokikh, N.E. and Preiss, T. (2018) Translation initiation by cap-dependent ribosome recruitment: Recent insights and open questions. WIREs RNA 9, e1473 10.1002/wrna.1473 [DOI] [PubMed] [Google Scholar]
- 40.Roux, P.P. and Topisirovic, I. (2018) Signaling pathways involved in the regulation of mRNA translation. Mol. Cell. Biol. 38, e00070-18 10.1128/MCB.00070-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao, J., Qin, B., Nikolay, R., Spahn, C.M.T. and Zhang, G. (2019) Translatomics: the global view of translation. IJMS 20, 212 10.3390/ijms20010212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hao, P., Yu, J., Ward, R., Liu, Y., Hao, Q., An, S.et al. (2020) Eukaryotic translation initiation factors as promising targets in cancer therapy. Cell Commun. Signal. 18, 175 10.1186/s12964-020-00607-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delaidelli, A., Jan, A., Herms, J. and Sorensen, P.H. (2019) Translational control in brain pathologies: biological significance and therapeutic opportunities. Acta Neuropathol. 137, 535–555 10.1007/s00401-019-01971-8 [DOI] [PubMed] [Google Scholar]
- 44.Skariah, G. and Todd, P.K. (2021) Translational control in aging and neurodegeneration. WIREs RNA 12, e1628 10.1002/wrna.1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karamyshev, A.L. and Karamysheva, Z.N. (2018) Lost in translation: ribosome-associated mRNA and protein quality controls. Front. Genet. 9, 431 10.3389/fgene.2018.00431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Heesch, S., Witte, F., Schneider-Lunitz, V., Schulz, J.F., Adami, E., Faber, A.B.et al. (2019) The translational landscape of the human heart. Cell 178, 242–260.e29 10.1016/j.cell.2019.05.010 [DOI] [PubMed] [Google Scholar]
- 47.Simpson, L.J., Reader, J.S. and Tzima, E. (2020) Mechanical regulation of protein translation in the cardiovascular system. Front. Cell Dev. Biol. 8, 34 10.3389/fcell.2020.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spriggs, K.A., Bushell, M. and Willis, A.E. (2010) Translational regulation of gene expression during conditions of cell stress. Mol. Cell 40, 228–237 10.1016/j.molcel.2010.09.028 [DOI] [PubMed] [Google Scholar]
- 49.van den Beucken, T., Magagnin, M.G., Jutten, B., Seigneuric, R., Lambin, P., Koritzinsky, M.et al. (2011) Translational control is a major contributor to hypoxia induced gene expression. Radiother. Oncol. 99, 379–384 10.1016/j.radonc.2011.05.058 [DOI] [PubMed] [Google Scholar]
- 50.Jiang, Z., Yang, J., Dai, A., Wang, Y., Li, W. and Xie, Z. (2017) Ribosome profiling reveals translational regulation of mammalian cells in response to hypoxic stress. BMC Genomics 18, 638 10.1186/s12864-017-3996-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barraza, C.E., Solari, C.A., Marcovich, I., Kershaw, C., Galello, F., Rossi, S.et al. (2017) The role of PKA in the translational response to heat stress in saccharomyces cerevisiae. PLoS ONE 12, e0185416 10.1371/journal.pone.0185416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crawford, R.A. and Pavitt, G.D. (2019) Translational regulation in response to stress in Saccharomyces cerevisiae. Yeast 36, 5–21 10.1002/yea.3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gerashchenko, M.V., Lobanov, A.V. and Gladyshev, V.N. (2012) Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc. Natl Acad. Sci. U.S.A. 109, 17394–17399 10.1073/pnas.1120799109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gameiro, P.A. and Struhl, K. (2018) Nutrient deprivation elicits a transcriptional and translational inflammatory response coupled to decreased protein synthesis. Cell Rep. 24, 1415–1424 10.1016/j.celrep.2018.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dever, T.E. and Green, R. (2012) The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb. Perspect. Biol. 4, a013706 10.1101/cshperspect.a013706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, X., Xie, J. and Proud, C. (2017) Eukaryotic elongation factor 2 kinase (eEF2K) in cancer. Cancers 9, 162 10.3390/cancers9120162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hellen, C.U.T. (2018) Translation termination and ribosome recycling in eukaryotes. Cold Spring Harb. Perspect. Biol. 10, a032656 10.1101/cshperspect.a032656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hinnebusch, A.G. and Lorsch, J.R. (2012) The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb. Perspect. Biol. 4, a011544 10.1101/cshperspect.a011544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knight, J.R.P., Bastide, A., Roobol, A., Roobol, J., Jackson, T.J., Utami, W.et al. (2015) Eukaryotic elongation factor 2 kinase regulates the cold stress response by slowing translation elongation. Biochem. J. 465, 227–238 10.1042/BJ20141014 [DOI] [PubMed] [Google Scholar]
- 60.Ingolia, N.T., Ghaemmaghami, S., Newman, J.R.S. and Weissman, J.S. (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ingolia, N.T., Brar, G.A., Rouskin, S., McGeachy, A.M. and Weissman, J.S. (2012) The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550 10.1038/nprot.2012.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ingolia, N.T. (2014) Ribosome profiling: new views of translation, from single codons to genome scale. Nat. Rev. Genet. 15, 205–213 10.1038/nrg3645 [DOI] [PubMed] [Google Scholar]
- 63.Hu, Z., Xia, B., Postnikoff, S.D., Shen, Z.-J., Tomoiaga, A.S., Harkness, T.A.et al. (2018) Ssd1 and Gcn2 suppress global translation efficiency in replicatively aged yeast while their activation extends lifespan. eLife 7, e35551 10.7554/eLife.35551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gerashchenko, M.V., Peterfi, Z., Yim, S.H. and Gladyshev, V.N. (2021) Translation elongation rate varies among organs and decreases with age. Nucleic Acids Res. 49, e9 10.1093/nar/gkaa1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anisimova, A.S., Meerson, M.B., Gerashchenko, M.V., Kulakovskiy, I.V., Dmitriev, S.E. and Gladyshev, V.N. (2020) Multifaceted deregulation of gene expression and protein synthesis with age. Proc. Natl Acad. Sci. U.S.A. 117, 15581–15590 10.1073/pnas.2001788117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tharakan, R., Ubaida-Mohien, C., Piao, Y., Gorospe, M. and Ferrucci, L. (2021) Ribosome profiling analysis of human skeletal muscle identifies reduced translation of mitochondrial proteins with age. RNA Biol. 18, 1555–1559 10.1080/15476286.2021.1875647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adomavicius, T., Guaita, M., Zhou, Y., Jennings, M.D., Latif, Z., Roseman, A.M.et al. (2019) The structural basis of translational control by eIF2 phosphorylation. Nat. Commun. 10, 2136 10.1038/s41467-019-10167-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taniuchi, S., Miyake, M., Tsugawa, K., Oyadomari, M. and Oyadomari, S. (2016) Integrated stress response of vertebrates is regulated by four eIF2α kinases. Sci. Rep. 6, 32886 10.1038/srep32886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hipkiss, A.R. (2007) On why decreasing protein synthesis can increase lifespan. Mech. Ageing Dev. 128, 412–414 10.1016/j.mad.2007.03.002 [DOI] [PubMed] [Google Scholar]
- 70.Hansen, M., Taubert, S., Crawford, D., Libina, N., Lee, S.-J. and Kenyon, C. (2007) Lifespan extension by conditions that inhibit translation in caenorhabditis elegans. Aging Cell 6, 95–110 10.1111/j.1474-9726.2006.00267.x [DOI] [PubMed] [Google Scholar]
- 71.Wek, R.C. (2018) Role of eIF2α kinases in translational control and adaptation to cellular stress. Cold Spring Harb. Perspect. Biol. 10, a032870 10.1101/cshperspect.a032870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Teske, B.F., Wek, S.A., Bunpo, P., Cundiff, J.K., McClintick, J.N., Anthony, T.G.et al. (2011) The eIF2 kinase PERK and the integrated stress response facilitate activation of ATF6 during endoplasmic reticulum stress. MBoC 22, 4390–4405 10.1091/mbc.e11-06-0510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hussain, S.G. and Ramaiah, K.V.A. (2007) Reduced eIF2α phosphorylation and increased proapoptotic proteins in aging. Biochem. Biophys. Res. Commun. 355, 365–370 10.1016/j.bbrc.2007.01.156 [DOI] [PubMed] [Google Scholar]
- 74.Hinnebusch, A.G. (1993) Gene-specific translational control of the yeast GCN4 gene by phosphorylation of eukaryotic initiation factor 2. Mol. Microbiol. 10, 215–223 10.1111/j.1365-2958.1993.tb01947.x [DOI] [PubMed] [Google Scholar]
- 75.Derisbourg, M.J., Hartman, M.D. and Denzel, M.S. (2021) Modulating the integrated stress response to slow aging and ameliorate age-related pathology. Nat. Aging 1, 760–768 10.1038/s43587-021-00112-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chadwick, S.R. and Lajoie, P. (2019) Endoplasmic reticulum stress coping mechanisms and lifespan regulation in health and diseases. Front. Cell Dev. Biol. 7, 84 10.3389/fcell.2019.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Estébanez, B., de Paz, J.A., Cuevas, M.J. and González-Gallego, J. (2018) Endoplasmic reticulum unfolded protein response, aging and exercise: an update. Front. Physiol. 9, 1744 10.3389/fphys.2018.01744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chadwick, S.R., Fazio, E.N., Etedali-Zadeh, P., Genereaux, J., Duennwald, M.L. and Lajoie, P. (2020) A functional unfolded protein response is required for chronological aging in saccharomyces cerevisiae. Curr. Genet. 66, 263–277 10.1007/s00294-019-01019-0 [DOI] [PubMed] [Google Scholar]
- 79.Jiménez-Saucedo, T., Berlanga, J.J. and Rodríguez-Gabriel, M. (2021) Translational control of gene expression by eIF2 modulates proteostasis and extends lifespan. Aging 13, 10989–11009 10.18632/aging.203018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Steffen, K.K. and Dillin, A. (2016) A ribosomal perspective on proteostasis and aging. Cell Metab. 23, 1004–1012 10.1016/j.cmet.2016.05.013 [DOI] [PubMed] [Google Scholar]
- 81.Derisbourg, M.J., Wester, L.E., Baddi, R. and Denzel, M.S. (2021) Mutagenesis screen uncovers lifespan extension through integrated stress response inhibition without reduced mRNA translation. Nat. Commun. 12, 1678 10.1038/s41467-021-21743-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bellato, H.M. and Hajj, G.N.M. (2016) Translational control by eIF2α in neurons: Beyond the stress response: Translational Control by eIF2α in Neurons. Cytoskeleton. 73, 551–565 10.1002/cm.21294 [DOI] [PubMed] [Google Scholar]
- 83.Jan, A., Jansonius, B., Delaidelli, A., Bhanshali, F., An, Y.A., Ferreira, N.et al. (2018) Activity of translation regulator eukaryotic elongation factor-2 kinase is increased in Parkinson disease brain and its inhibition reduces alpha synuclein toxicity. Acta Neuropathol. Commun. 6, 54 10.1186/s40478-018-0554-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Krukowski, K., Nolan, A., Frias, E.S., Boone, M., Ureta, G., Grue, K.et al. (2020) Small molecule cognitive enhancer reverses age-related memory decline in mice. eLife 9, e62048 10.7554/eLife.62048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Longo, F., Mancini, M., Ibraheem, P.L., Aryal, S., Mesini, C., Patel, J.C.et al. (2021) Cell-type-specific disruption of PERK-eIF2α signaling in dopaminergic neurons alters motor and cognitive function. Mol. Psychiatry 10.1038/s41380-021-01099-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moon, S.L., Sonenberg, N. and Parker, R. (2018) Neuronal regulation of eIF2α function in health and neurological disorders. Trends Mol. Med. 24, 575–589 10.1016/j.molmed.2018.04.001 [DOI] [PubMed] [Google Scholar]
- 87.Trigos, A.S., Pearson, R.B., Papenfuss, A.T. and Goode, D.L. (2019) Somatic mutations in early metazoan genes disrupt regulatory links between unicellular and multicellular genes in cancer. eLife 8, e40947 10.7554/eLife.40947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saxton, R.A. and Sabatini, D.M. (2017) mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 10.1016/j.cell.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Papadopoli, D., Boulay, K., Kazak, L., Pollak, M., Mallette, F., Topisirovic, I.et al. (2019) mTOR as a central regulator of lifespan and aging. F1000Res. 8, 998 10.12688/f1000research.17196.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Holz, M.K., Ballif, B.A., Gygi, S.P. and Blenis, J. (2005) mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 123, 569–580 10.1016/j.cell.2005.10.024 [DOI] [PubMed] [Google Scholar]
- 91.Qin, X., Jiang, B. and Zhang, Y. (2016) 4E-BP1, a multifactor regulated multifunctional protein. Cell Cycle 15, 781–786 10.1080/15384101.2016.1151581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nandagopal, N. and Roux, P.P. (2015) Regulation of global and specific mRNA translation by the mTOR signaling pathway. Translation 3, e983402 10.4161/21690731.2014.983402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Franz, D.N. and Capal, J.K. (2017) mTOR inhibitors in the pharmacologic management of tuberous sclerosis complex and their potential role in other rare neurodevelopmental disorders. Orphanet. J. Rare Dis. 12, 51 10.1186/s13023-017-0596-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Weichhart, T. (2018) mTOR as regulator of lifespan, aging, and cellular senescence: a mini-review. Gerontology 64, 127–134 10.1159/000484629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Seto, B. (2012) Rapamycin and mTOR: a serendipitous discovery and implications for breast cancer. Clin. Transl. Med. 1, 29 10.1186/2001-1326-1-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Abdel-Magid, A.F. (2019) Rapalogs potential as practical alternatives to rapamycin. ACS Med. Chem. Lett. 10, 843–845 10.1021/acsmedchemlett.9b00215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fabrizio, P. (2001) Regulation of longevity and stress resistance by Sch9 in yeast. Science 292, 288–290 10.1126/science.1059497 [DOI] [PubMed] [Google Scholar]
- 98.Jia, K., Chen, D. and Riddle, D.L. (2004) The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development 131, 3897–3906 10.1242/dev.01255 [DOI] [PubMed] [Google Scholar]
- 99.Vellai, T., Takacs-Vellai, K., Zhang, Y., Kovacs, A.L., Orosz, L. and Müller, F. (2003) Influence of TOR kinase on lifespan in C. elegans. Nature 426, 620–620 10.1038/426620a [DOI] [PubMed] [Google Scholar]
- 100.Robida-Stubbs, S., Glover-Cutter, K., Lamming, D.W., Mizunuma, M., Narasimhan, S.D., Neumann-Haefelin, E.et al. (2012) TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 15, 713–724 10.1016/j.cmet.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kapahi, P., Kaeberlein, M. and Hansen, M. (2017) Dietary restriction and lifespan: lessons from invertebrate models. Ageing Res. Rev. 39, 3–14 10.1016/j.arr.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bjedov, I., Toivonen, J.M., Kerr, F., Slack, C., Jacobson, J., Foley, A.et al. (2010) Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 11, 35–46 10.1016/j.cmet.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schinaman, J.M., Rana, A., Ja, W.W., Clark, R.I. and Walker, D.W. (2019) Rapamycin modulates tissue aging and lifespan independently of the gut microbiota in drosophila. Sci. Rep. 9, 7824 10.1038/s41598-019-44106-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Harrison, D.E., Strong, R., Sharp, Z.D., Nelson, J.F., Astle, C.M., Flurkey, K.et al. (2009) Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 460, 392–395 10.1038/nature08221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Miller, R.A., Harrison, D.E., Astle, C.M., Baur, J.A., Boyd, A.R., de Cabo, R.et al. (2011) Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J. Gerontol. A Biol. Sci. Med. Sci 66A, 191–201 10.1093/gerona/glq178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cao, K., Graziotto, J.J., Blair, C.D., Mazzulli, J.R., Erdos, M.R., Krainc, D.et al. (2011) Rapamycin reverses cellular phenotypes and enhances mutant protein clearance in Hutchinson-Gilford progeria syndrome cells. Sci. Transl. Med. 3, 89ra58 10.1126/scitranslmed.3002346 [DOI] [PubMed] [Google Scholar]
- 107.Graziotto, J.J., Cao, K., Collins, F.S. and Krainc, D. (2012) Rapamycin activates autophagy in Hutchinson-Gilford progeria syndrome: implications for normal aging and age-dependent neurodegenerative disorders. Autophagy 8, 147–151 10.4161/auto.8.1.18331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lamming, D.W., Ye, L., Katajisto, P., Goncalves, M.D., Saitoh, M., Stevens, D.M.et al. (2012) Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 335, 1638–1643 10.1126/science.1215135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lamming, D.W., Mihaylova, M.M., Katajisto, P., Baar, E.L., Yilmaz, O.H., Hutchins, A.et al. (2014) Depletion of Rictor, an essential protein component of m TORC 2, decreases male lifespan. Aging Cell 13, 911–917 10.1111/acel.12256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chellappa, K., Brinkman, J.A., Mukherjee, S., Morrison, M., Alotaibi, M.I., Carbajal, K.A.et al. (2019) Hypothalamic mTORC2 is essential for metabolic health and longevity. Aging Cell 18, e13014 10.1111/acel.13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Soukas, A.A., Kane, E.A., Carr, C.E., Melo, J.A. and Ruvkun, G. (2009) Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in caenorhabditis elegans. Genes Dev. 23, 496–511 10.1101/gad.1775409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pallet, N. and Legendre, C. (2013) Adverse events associated with mTOR inhibitors. Expert Opin. Drug Saf. 12, 177–186 10.1517/14740338.2013.752814 [DOI] [PubMed] [Google Scholar]
- 113.Mannick, J.B., Del Giudice, G., Lattanzi, M., Valiante, N.M., Praestgaard, J., Huang, B.et al. (2014) mTOR inhibition improves immune function in the elderly. Sci. Transl. Med. 6, 268ra179 10.1126/scitranslmed.3009892 [DOI] [PubMed] [Google Scholar]
- 114.Gremke, N., Polo, P., Dort, A., Schneikert, J., Elmshäuser, S., Brehm, C.et al. (2020) mTOR-mediated cancer drug resistance suppresses autophagy and generates a druggable metabolic vulnerability. Nat. Commun. 11, 4684 10.1038/s41467-020-18504-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mossmann, D., Park, S. and Hall, M.N. (2018) mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat. Rev. Cancer 18, 744–757 10.1038/s41568-018-0074-8 [DOI] [PubMed] [Google Scholar]
- 116.Taha, E., Gildish, I., Gal-Ben-Ari, S. and Rosenblum, K. (2013) The role of eEF2 pathway in learning and synaptic plasticity. Neurobiol. Learn. Mem. 105, 100–106 10.1016/j.nlm.2013.04.015 [DOI] [PubMed] [Google Scholar]
- 117.Liu, R. and Proud, C.G. (2016) Eukaryotic elongation factor 2 kinase as a drug target in cancer, and in cardiovascular and neurodegenerative diseases. Acta Pharmacol. Sin. 37, 285–294 10.1038/aps.2015.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang, X., Regufe da Mota, S., Liu, R., Moore, C.E., Xie, J., Lanucara, F.et al. (2014) Eukaryotic elongation factor 2 kinase activity is controlled by multiple inputs from oncogenic signaling. Mol. Cell. Biol. 34, 4088–4103 10.1128/MCB.01035-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Leprivier, G., Remke, M., Rotblat, B., Dubuc, A., Mateo, A.-R.F., Kool, M.et al. (2013) The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell 153, 1064–1079 10.1016/j.cell.2013.04.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kruiswijk, F., Yuniati, L., Magliozzi, R., Low, T.Y., Lim, R., Bolder, R.et al. (2012) Coupled activation and degradation of eEF2K regulates protein synthesis in response to genotoxic stress. Sci. Signal. 5, ra40 10.1126/scisignal.2002718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mobley, C.B., Mumford, P.W., Kephart, W.C., Haun, C.T., Holland, A.M., Beck, D.T.et al. (2017) Aging in rats differentially affects markers of transcriptional and translational capacity in soleus and plantaris muscle. Front. Physiol. 8, 518 10.3389/fphys.2017.00518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xie, J., de Souza Alves, V., von der Haar, T., O'Keefe, L., Lenchine, R.V., Jensen, K.B.et al. (2019) Regulation of the elongation phase of protein synthesis enhances translation accuracy and modulates lifespan. Curr. Biol. 29, 737–749.e5 10.1016/j.cub.2019.01.029 [DOI] [PubMed] [Google Scholar]
- 123.Chu, H.-P., Liao, Y., Novak, J.S., Hu, Z., Merkin, J.J., Shymkiv, Y.et al. (2014) Germline quality control: eEF2K stands guard to eliminate defective oocytes. Dev. Cell 28, 561–572 10.1016/j.devcel.2014.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Beckelman, B.C., Yang, W., Kasica, N.P., Zimmermann, H.R., Zhou, X., Keene, C.D.et al. (2019) Genetic reduction of eEF2 kinase alleviates pathophysiology in Alzheimer's disease model mice. J. Clin. Invest. 129, 820–833 10.1172/JCI122954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Archer, S.K., Shirokikh, N.E., Beilharz, T.H. and Preiss, T. (2016) Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature 535, 570–574 10.1038/nature18647 [DOI] [PubMed] [Google Scholar]
- 126.Shirokikh, N.E., Archer, S.K., Beilharz, T.H., Powell, D. and Preiss, T. (2017) Translation complex profile sequencing to study the in vivo dynamics of mRNA–ribosome interactions during translation initiation, elongation and termination. Nat. Protoc. 12, 697–731 10.1038/nprot.2016.189 [DOI] [PubMed] [Google Scholar]
- 127.Wagner, S., Herrmannová, A., Hronová, V., Gunišová, S., Sen, N.D., Hannan, R.D.et al. (2020) Selective translation complex profiling reveals staged initiation and co-translational assembly of initiation factor complexes. Mol. Cell 79, 546–560.e7 10.1016/j.molcel.2020.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liang, Y., Piao, C., Beuschel, C.B., Toppe, D., Kollipara, L., Bogdanow, B.et al. (2021) eIF5A hypusination, boosted by dietary spermidine, protects from premature brain aging and mitochondrial dysfunction. Cell Rep. 35, 108941 10.1016/j.celrep.2021.108941 [DOI] [PubMed] [Google Scholar]
- 129.Zimmermann, A., Carmona-Gutierrez, D. and Madeo, F. (2021) Spermidine supplementation in rare translation associated disorders. CST 5, 29–32 10.15698/cst2021.03.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lubas, M., Harder, L.M., Kumsta, C., Tiessen, I., Hansen, M., Andersen, J.S.et al. (2018) eIF5A is required for autophagy by mediating ATG3 translation. EMBO Rep. 19, e46072 10.15252/embr.201846072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang, H., Alsaleh, G., Feltham, J., Sun, Y., Napolitano, G., Riffelmacher, T.et al. (2019) Polyamines control eIF5A hypusination, TFEB translation, and autophagy to reverse B cell senescence. Mol. Cell 76, 110–125.e9 10.1016/j.molcel.2019.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Madeo, F., Eisenberg, T., Pietrocola, F. and Kroemer, G. (2018) Spermidine in health and disease. Science 359, eaan2788 10.1126/science.aan2788 [DOI] [PubMed] [Google Scholar]
- 133.Eisenberg, T., Abdellatif, M., Schroeder, S., Primessnig, U., Stekovic, S., Pendl, T.et al. (2016) Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat. Med. 22, 1428–1438 10.1038/nm.4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Martinez-Miguel, V.E., Lujan, C., Espie-Caullet, T., Martinez-Martinez, D., Moore, S., Backes, C.et al. (2021) Increased fidelity of protein synthesis extends lifespan. Cell Metab. 33, 2288–2300.e12 10.1016/j.cmet.2021.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Halliday, M., Radford, H., Zents, K.A.M., Molloy, C., Moreno, J.A., Verity, N.C.et al. (2017) Repurposed drugs targeting eIF2α-P-mediated translational repression prevent neurodegeneration in mice. Brain 140, 1768–1783 10.1093/brain/awx074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rabouw, H.H., Langereis, M.A., Anand, A.A., Visser, L.J., de Groot, R.J., Walter, P.et al. (2019) Small molecule ISRIB suppresses the integrated stress response within a defined window of activation. Proc. Natl Acad. Sci. U.S.A. 116, 2097–2102 10.1073/pnas.1815767116 [DOI] [PMC free article] [PubMed] [Google Scholar]