Abstract
X-inactive-specific transcript (Xist) is a long non-coding RNA (lncRNA) essential for X-chromosome inactivation (XCI) in female placental mammals. Thirty years after its discovery, it is still puzzling how this lncRNA triggers major structural and transcriptional changes leading to the stable silencing of an entire chromosome. Recently, a series of studies in mouse cells have uncovered domains of functional specialization within Xist mapping to conserved tandem repeat regions, known as Repeats A-to-F. These functional domains interact with various RNA binding proteins (RBPs) and fold into distinct RNA structures to execute specific tasks in a synergistic and coordinated manner during the inactivation process. This modular organization of Xist is mostly conserved in humans, but recent data point towards differences regarding functional specialization of the tandem repeats between the two species. In this review, we summarize the recent progress on understanding the role of Xist repetitive blocks and their involvement in the molecular mechanisms underlying XCI. We also discuss these findings in the light of the similarities and differences between mouse and human Xist.
Keywords: chromatin, epigenetics, lncRNA, tandem repeat, X-chromosome inactivation
Introduction
X-inactive-specific transcript (Xist) is a long non-coding RNA (lncRNA) responsible for X-Chromosome Inactivation (XCI), the dosage compensation mechanism that equalizes X gene dosage between XX females and XY males in placental mammals. The importance of Xist in XCI regulation was uncovered by loss of function studies showing that lack of dosage compensation for the X chromosome results in female-specific lethality in mice [1,2]. These pioneer studies unraveled a relevant function for this non-coding transcript with implications on survival, placing Xist at the central stage of lncRNA research.
Xist is the only gene expressed exclusively from the inactive X chromosome (Xi). This monoallelic expression is established through the concerted action of several cis- and trans-acting players during early embryonic development (reviewed in [3]). Xist characteristically covers the Xi in a process that is essential for chromosome-wide transcriptional silencing, formation of facultative heterochromatin and spatial 3D structural rearrangement of the X chromosome (reviewed in [4,5]). How a single lncRNA coordinates these major structural and transcriptional changes over an entire chromosome still remains enigmatic. XCI continues to be a stimulating research field and a ground for prolific experimentation that is constantly re-shaping our understanding of Xist function.
The Xist gene produces a spliced ∼17-to-19 kb lncRNA. A remarkable feature of Xist is the existence of several conserved tandem repeat sequences, known as Repeats A-to-F (Figure 1A). Despite their conservation, considerable variability in the number of repetitive motifs can be found in different mammalian species [6,7]. Whether this variation translates into the functional diversity of Xist function across mammals remains an open question. Interestingly, deletion analyses of mouse Xist repeats uncovered particular functions of these modules, creating a new paradigm of Xist as a multitasking lncRNA with specialized functional domains that interact with specific RNA Binding Proteins (RBPs) [8–11]. Together, these functional studies reveal that the different Xist repeats seem to act in a synergistic and coordinated manner, orchestrating the process of XCI from its inception to its end. In this review, we summarize recent progress in our understanding of the role of the different Xist repeats and their protein interactors in XCI in the mouse system and discuss how these findings compare to what we know about the human XIST RNA.
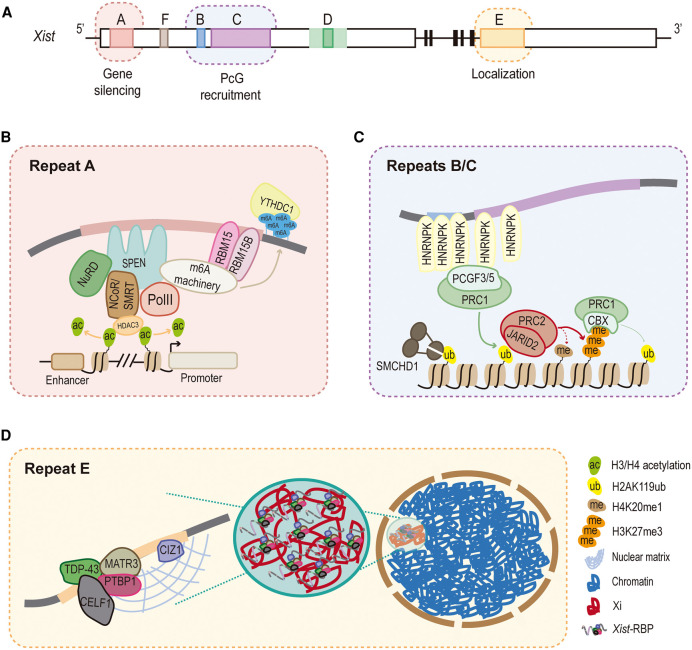
Figure 1. The role of mouse Xist repeats in XCI.
(A) Schematic representation of the mouse Xist gene with exons 1-to-7 represented by white and black rectangles, and the Repeats A-to-F highlighted in colors. For Repeat D, both the core motif (dark green) and truncated motifs (light green) are represented. (B–D) Detailed outline of the factors interacting with Repeat A and mediating gene silencing (B), with Repeats B/C and involved in PcG recruitment (C), and with Repeat E and important factors for Xist localization within the inactive X territory (D); refer to main text for details.
Repeat A — The hub for transcriptional silencing
Repeat A, mapping to the first ∼335–700 nucleotides of mouse Xist (Figure 1A), is the most conserved RNA module [6,7], composed of seven and a half repetitive motifs that form hairpin-like structures separated by single-stranded regions [12]. Work from several groups, using deletion analysis in murine embryonic stem cells (ESCs) and mouse models, demonstrated that Repeat A is essential for the transcriptional silencing of virtually all genes on the Xi [10,13–17] (Table 1).
Table 1. List of Xist deletions targeting Xist Repeats A-to-F in mouse cells and their associated phenotypes.
| Repeats | Xist RNA deletion | System | Phenotype | References |
|---|---|---|---|---|
| Repeat A | ΔSX: 5′ end of Xist including Repeat A (∼900 bp) | Mouse XY ESCs with an inducible Xist at the endogenous locus | Mild effect on Xist coating; Gene silencing failure; Recruitment of PcG complexes | [8,17,31] |
| ΔA (∼800 bp) | Mice with a Repeat A deletion at the endogenous locus | Loss of Xist expression; Gene silencing failure in embryonic and extraembryonic lineages | [14] | |
| 5′ end of Xist containing Repeat A (∼900 bp) | Mice with a CAG promoter driving Xist expression at the endogenous locus | Gene silencing failure for most, but not all, genes in embryonic and extraembryonic lineages | [15] | |
| ΔA (∼370 bp) | Mouse XY ES cells with inducible Xist cDNA TG in an autosome | Down-regulation of Xist expression; Gene silencing failure | [16] | |
| 5′ end of Xist including Repeat A (∼725 bp) | Mouse XX Cast/129 hybrid ESCs with an inducible Xist at the endogenous locus | Gene silencing failure for most genes | [10] | |
| ΔA (∼550 bp) | Mouse XX hybrid Cast/129 ESCs with a Tsix deletion | Mild effect on Xist coating; Gene silencing failure | [13] | |
| Repeats B and C | ΔXN: deletion of Repeats F, B and C (∼4000 bp) | Mouse XY ESCs with an inducible Xist cDNA TG integrated in X-linked Hprt locus | Complete loss of H3K27me3 and H2AK119ub deposition; Mild reduction in gene silencing; Smaller Xist clouds | [17,31] |
| ΔXN: deletion of Repeats F, B and C (∼4000 bp) | Mouse ESCs with inducible Xist cDNA TG on an autosome | No H3K27me3/H2AK119ub enrichment | [32] | |
| ΔXR-PID: deletion of Repeat B and a small part of Repeat C (∼600 bp) | Mouse XY Cast/129 hybrid ESCs with an inducible Xist TGs on autosomes | No H3K27me3/H2AK119ub enrichment; Reduction in gene silencing | [19] | |
| ΔF + B + C (∼3900 bp) | Mouse XY ESCs with an inducible Xist at the endogenous locus | No H3K27me3/H2AK119ub enrichment | [8] | |
| ΔB + 1/2C (∼1300 bp) | Significant decrease in H3K27me3/H2AK119ub enrichment | |||
| ΔB (∼330 bp) | Decrease in H3K27me3/H2AK119ub enrichment | |||
| ΔC (∼1700 bp) | Normal H3K27me3/H2AK119ub enrichment | |||
| ΔB + C (∼2100 bp) | No H3K27me3/H2AK119ub enrichment; Relaxation of X-linked gene silencing | |||
| ΔPID: Repeat B and partial repeat C (∼1700 bp) | Mouse XX Cast/129 hybrid ESCs with an inducible Xist at the endogenous locus | No H3K27me3/H2AK119ub enrichment; Reduction in gene silencing | [10] | |
| ΔB (∼300 bp) | Transformed tetraploid female MEFs | Diffuse Xist clouds; No H3K27me3/H2AK119ub enrichment | [9] | |
| ΔC (∼1700 bp) | Normal H3K27me3/H2AK119ub enrichment | |||
| ΔB (∼300 bp) | Mouse XX hybrid Cast/129 ESCs with a Tsix deletion | Initial H3K27me3 and H2AK119ub enrichment is not maintained; Reduction in gene silencing; Diffuse Xist clouds | [9,13] | |
| ΔB (∼300 bp) | Mouse XX hybrid Cast/129 ESCs with a MS2-tagged 129 Xist allele | Loss of Xi compaction; Impaired inactivation of late silenced genes | [41] | |
| Repeat E | ΔE (∼1300 bp) | Mouse XX hybrid Cast/129 ESCs with a Tsix deletion | Increase expression of escape genes | [48] |
| ΔE (∼1500 bp) | Mouse XY ESCs with inducible Xist cDNA TG in an autosome | No CIZ1 recruitment | [46] | |
| ΔE (∼1200 bp) | Transformed tetraploid female MEFs | Disruption of Xist coating; Reduced H3K27me3 enrichment; No CIZ1 recruitment | [47] | |
| ΔE (∼1300 bp) | Mouse XX hybrid Cast/129 ESCs with MS2-tag on Xist 129 allele | Normal initiation of XCI; Xist RNA dispersal from the Xi, defective gene silencing and decreased H3K27me3 enrichment at the maintenance stage | [11] | |
| Repeat F | X-FR mutant: deletion including Repeat F (∼700 bp) | Transformed tetraploid female MEFs with an inducible Xist transgene on an autosome | Reduced Xist expression and weak Xist clouds | [53] |
| ΔLBS region: deletion including Repeat F (∼785 bp) | Mouse XY ESCs with inducible Xist expression from the endogenous locus | Compromise recruitment of the Xi to the nuclear lamina; Gene silencing failure | [52] | |
| Deletion containing Repeat F (∼950 bp) | Transformed tetraploid female MEFs | Reduced Xist expression and weak Xist clouds | [9] | |
| ΔLBS region: deletion including Repeat F (∼850 bp) | Mouse XX Cast/129 hybrid ESCs with an inducible Xist at the endogenous locus | Weak defect in gene silencing | [10] | |
| Repeat D | ΔD (∼1100 bp) | Transformed tetraploid female MEFs | No effect on Xist coating or PcG recruitment | [9] |
Abbreviations: CAG, Cytomegalovirus-Actin-Globin; Cast/129, Mus musculus Castaneus/Mus musculus domesticus 129 strain; cDNA, complementary DNA; CIZ1, CDKN1A-Interacting Zinc finger protein 1; ESCs, Embryonic Stem Cells; H2AK119ub, ubiquitination of the histone H2A at lysine 119; H3K27me3, tri-methylation of histone H3 on lysine 27; Hprt, Hypoxanthine phosphoribosyltransferase; MS2, Bacteriophage MS2 (Emesvirus zinderi); TG, TransGene; Tsix, Xist antisense RNA; Xi, inactive X chromosome; Xist, X-inactive-specific transcript; XX: female biological sex; XY, male biological sex.
The mechanistic understanding of the transcriptional silencing imposed by Repeat A has evolved considerably in the past decade. An initial model postulated that transcriptional silencing was achieved through the interaction with Polycomb Repressive Complex 2 (PRC2), a repressive modifying chromatin complex responsible for the tri-methylation of histone H3 on lysine 27 (H3K27me3) that is enriched along the Xi [18]. This model was challenged after the identification of a different RNA module as the main responsible for PRC2 recruitment (Repeats B/C, see below) [8–10,19]. In addition, the discovery of bona-fide Repeat A interactors by unbiased proteomic approaches and mapping of RNA–protein interactions [12,20,21] allowed a more precise understanding of the molecular function of this module in XCI, which does not involve PRC2.
Among the Repeat A interactors, SPEN (SPlit ENds), a >400 kDa RBP containing a SPOC (SPEN Paralogue/Orthologue C-terminal) transcriptional repressive domain, has been identified as a major player in Xist-dependent transcriptional silencing [20,22–25]. Interaction of SPEN with Repeat A is dependent on its RNA Recognition Motifs (RRM) 2–4 [12,20,22,25] that are important to ensure Xist stability and spreading across the X chromosome undergoing inactivation [26]. Upon interaction with Xist, SPEN is recruited to both active promoters and enhancers on the X chromosome to effectively initiate gene silencing [22]. At these sites, SPEN, through its SPOC domain, acts as a scaffold to recruit repressive factors such as the NCoR/SMRT (Nuclear receptor Co-Repressor/ Silencing Mediator of Retinoic acid and Thyroid hormone receptor) complex [22]. Through its HDAC3 (Histone DeACetylase 3) subunit, the NCoR/SMRT complex is responsible for deacetylating histones H3 and H4, one of the first chromatin events during XCI [27]. However, lack of HDAC3 only has a mild impact on gene silencing [22,27], suggesting that other SPEN partners are involved in the transcriptional silencing of the Xi. Indeed, the SPOC domain of SPEN also interacts with the NuRD (Nucleosome Remodeling and Deacetylase) complex, RNA polymerase II (PolII) and associated factors, as well as the N6-methyladenosine (m6A) RNA methylation machinery [22], but the relevance of these interactions for XCI are yet to be understood (Figure 1B).
Additional SPOC-containing proteins, such as RBM15 (RNA Binding Motif protein 15) and RBM15B also interact with Repeat A [20,21,23]. These proteins play a central role in the recruitment of the m6A RNA methylation machinery. m6A is the most abundant modification in eukaryotic RNAs, influencing several RNA processing events such as stability, splicing, translation and secondary structures, in both physiological and pathological contexts (reviewed in [28]). m6A is deposited along the Xist transcript, which includes a m6A hotspot just downstream of Repeat A [21,29]. This modification is then recognized and bound by the YTHDC1 (YTH Domain Containing 1) m6A reader [21,29,30] (Figure 1B). The involvement of RBM15/15B, the m6A methylation machinery and YTHDC1 were initially suggested to have an important impact on X-linked gene silencing [21]. However, according to recent data, the role of RBM15/15B, the m6A methylation complex or the m6A hotspot region downstream of Repeat A seem to be rather modest [10,30] and clearly do not compensate for the effects of SPEN loss [22]. Further experiments will be needed to have a finer understanding of the role of m6A methylation in XCI. To summarize, Repeat A acts as a hub for the initiation of transcriptional silencing by recruiting SPEN, other SPOC-containing proteins and associated repressive factors to elicit gene silencing as Xist spreads along the X chromosome (Figure 1B).
Repeats B/C — The hub for recruitment of Polycomb group complexes
The region of mouse Xist containing Repeats B and C folds independently from the Repeat A module [29]. Repeats B and C have a very different sequence content, but both are cytosine-rich. Whereas Repeat B is composed of ∼30 CCCCAG/CCCCUG motifs mapping to nucleotides ∼2850–3050, Repeat C is composed of 14 sequence motifs of ∼115 bp mapping to nucleotides ∼3115–4730 in the mouse Xist transcript. The Repeats B/C module is important for locking the silent state of the Xi, through the recruitment of Polycomb Group (PcG) protein complexes to the Xi upon initial establishment of Xist-mediated silencing by the Repeat A/SPEN module.
The first hint for an involvement of these repeats in PcG protein recruitment came from the observation that an inducible Xist cDNA transgene lacking Repeats B and C, along with Repeat F, called ΔXN, is unable to recruit the PRC2 complex to the Xi, in contrast with a Xist mutant lacking the Repeat A (ΔA) [31]. This region was then shown to be also required for the recruitment of the Polycomb Repressive Complex 1 (PRC1), responsible for ubiquitination of the histone H2A at lysine 119 (H2AK119ub) [32]. Further deletion analysis mapped the minimal sequence for PRC1/PRC2 recruitment to a region containing Repeat B and a portion of Repeat C [19]. When individually deleted, Repeat B has a notorious impact on PcG recruitment, in contrast with the absence of a measurable effect of Repeat C [8,9]. However, PcG protein recruitment to the Xi is only found to be completely abolished when both Repeats B and C (or a substantial part of Repeat C) were missing [8,10,13] (Table 1).
How the B/C module recruits both PRC1 and PRC2 to the Xi and the corresponding sequence of events are questions that have been widely investigated. Work from different labs showed that cytosine-rich Repeats B and C act as docking sites for HNRNPK (Heterogeneous Nuclear RiboNucleoProtein K) [8,9,19,33,34]. HNRNPK interacts with the non-canonical PCGF3/PCGF5–PRC1 complex that is brought to the Xi, decorating chromatin with the H2AK119ub mark [9,19]. This chromatin modification precedes H3K27me3 enrichment on the Xi [27] and serves as the main mechanism to recruit PRC2 through the recognition of H2AK119ub by the JARID2 cofactor [31,35,36]. The H3K27me3 mark will then serve as an anchor for the recruitment of the CBX (ChromoBoX homolog) subunit of canonical PRC1 [37], thus reinforcing the PcG-mediated heterochromatinization of the Xi (Figure 1C). This is recognized as the major pathway for PcG protein recruitment to the Xi by the B/C module, but alternative mechanisms have also been proposed [9].
The dynamics of PcG recruitment mediated by Repeats B/C follows Xist coating over the Xi and is initially observed at intergenic regions [8,10,27]. Subsequent spreading to active genes is only observed following transcriptional silencing [27]. The dynamics of accumulation of PcG marks over active genes could be attributed to a direct role of Repeat A in PcG recruitment to genes [13] or simply be a secondary event to the transcriptional silencing imposed by the Repeat A/SPEN pathway. We favor this second hypothesis given that the transcriptional silencing of CG-rich promoters, such as the ones for many X-linked genes, creates a chromatin environment permissive for PRC2 recruitment [38–40]. In this respect, mapping H3K27me3/H2AK119ub enrichment over the Xi in Spen SPOC mutants will be important to disentangle between a direct or a passive role for Repeat A in PcG recruitment to X-linked genes undergoing silencing.
Whereas Repeat A seems to be the main actor on the initial steps of XCI, Repeats B and C appear to take the center stage once XCI has been set in motion. Accordingly, X-linked gene silencing is initiated but is partially defective in Xist ΔB/C mutants [8–10,41] (Table 1). Likewise, removal of PCGF3/PCGF5–PRC1, recruited by Repeats B/C, also causes reduced Xist-mediated gene repression [10,32]. This important role of Repeats B/C in the maintenance of XCI, may depend on the involvement of PcG proteins on Xist spreading over the Xi [9], but also on several redundant mechanisms of repression enabled by PcG protein recruitment. For instance, Repeats B/C are important for the deposition of the H4K20me1 heterochromatin mark, presumably secondary to PRC2 recruitment [37,42] (Figure 1C). The B/C module also allows the recruitment of the structural protein SMCHD1 (Structural Maintenance of Chromosomes flexible Hinge Domain containing 1), downstream of the PCGF3/PCGF5–PRC1 complex [43] (Figure 1C). SMCHD1 is fundamental for promoting the characteristic folding of the Xi in two mega domains largely depleted of Topologically Associating Domains (TADs) [44,45]. A recent study also implicated Repeat B in this module in Xi compaction [41]. Whether other hallmarks of the Xi, such as recruitment of the macroH2A histone variant, DNA methylation of gene promoters are dependent on Repeats B/C remain to be tested. In summary, by recruiting PcG complexes, Repeats B/C play a key role in heterochromatin formation and topological reconfiguration of the X chromosome, being essential for long-lasting transcriptional silencing along the entire Xi.
Repeat E — The hub for Xist localization to the inactive X territory
Repeat E is localized at the beginning of exon 7 and maps to nucleotides ∼10 275–11 400 of mouse Xist transcript. It is composed of two types of tandem repeats: around 35 C/U/G-rich motifs of ∼16–27 bp at the 5′end and ∼25 C/U-rich motifs of 6–19 bp towards its 3′end. Different Xist ΔE deletions invariably result in delocalization of Xist from the Xi territory that aggravates as cells enter the maintenance stage of XCI [11,46–48] (Table 1). Repeat E is not necessary for initiating X-linked gene silencing or PcG recruitment, but the inability of Xist ΔE to coat the X chromosome ultimately results in gene reactivation and loss of PcG marks at the maintenance stage of XCI [11].
Several proteins were shown to bind Repeat E and to be important for tethering Xist to the Xi territory. One of the first interactors identified was CIZ1 (CDKN1A-Interacting Zinc finger protein 1). CIZ1 is a component of the nuclear matrix, a filamentous insoluble protein network that appears to participate in restricting Xist RNA molecules to the Xi [46,47] (Figure 1D). More recently, several RBPs, such as PTBP1, MATR3, CELF1, and TDP-43 were shown to directly interact with Repeat E [11] (Figure 1D). They form a distinct functional complex that assembles at Repeat E independently of CIZ1, being important to stabilize Xist coating after the initial wave of transcriptional silencing and PcG recruitment by Repeat A and Repeats B/C, respectively [11]. These proteins, namely PTBP1 and MATR3 have the capacity to form liquid droplets in vitro [49,50]. In particular, PTBP1 was shown to form phase-separated droplets upon binding to the Repeat E module [11]. This observation suggests that Repeat E may be important for the assembly of a specialized phase-separated sub-nuclear compartment necessary for the efficient maintenance of XCI [11,51]. This is an attractive idea to explain the remarkably stable epigenetic silencing of the Xi once XCI is established. Whether a Xi phase-separated condensate is formed and whether this biophysical compartment plays a role in the maintenance of XCI still awaits experimental confirmation. Overall, Repeat E interacts with several RBPs, which are essential for anchoring Xist RNA molecules to the Xi territory (Figure 1D), playing a critical role in the maintenance of XCI.
Other relevant repeat domains in Xist
The other two repetitive modules, Repeats F and D have attracted less attention. Repeat F is the shortest of all repeats and is made of two copies of a 10 bp motif (UGGCGGGCUU) mapping to nucleotides ∼1540–1575 in the mouse Xist RNA. Studies addressing the role of Repeat F resorted so far to larger deletions that extended beyond this module [9,10,52,53] (Table 1). The deletion of this region, also known as LBS, disrupts several binding sites for YY1 (Yin and Yang 1), a transcription factor important for Xist expression [54,55] and interferes with the binding of LBR (Lamin B receptor) in the vicinity of Repeat F, which consequences for XCI remain conflicting [10,52]. It is important to consider that such deletions can interfere with important DNA regulatory elements for XCI, besides having a direct influence on Xist RNA, a phenomenon that has previously been documented for some ΔA mutants [14,16] (Table 1). To disentangle between these, a study using inducible XIST from its endogenous locus in male ESCs shows that deletion of this region has a major impact on transcriptional silencing due to loss of tethering of the Xi to Lamin B at the nuclear periphery [52]. In contrast, a recent report using a similar inducible system in female ESCs shows that deletion of the LBS region only has marginal effects on gene silencing [10]. A precise deletion of Repeat F will be necessary to unveil its function in XCI.
The Repeat D is the least characterized repetitive module of mouse Xist. Originally, a single ∼200 bp core motif was identified in mouse Xist [56], but up to 10 truncated Repeat D motifs, located at nucleotides ∼5200–7900 towards the 3′ end of exon 1, was later described [6] (Figure 1A). The only ΔD deletion described, that includes the core motif, shows no noticeable phenotypes in terms of Xist coating and PcG recruitment in Mouse Embryonic Fibroblats (MEFs) [9] (Table 1). This does not exclude a role for Repeat D in the initial stages of XCI or when the full 3 kb region is deleted. It is however important to reiterate that Repeat D in rodents is heavily degenerated in contrast with other mammals, suggesting that deletion analysis of the mouse Repeat D might not be relevant for other mammalian species.
Mouse versus human Xist
Most of our understanding about the role of Xist in XCI comes from the mouse model. However, an increasing body of evidence has defied the murine-centric view of XCI, revealing that many of the developmental and molecular features observed in mice are not conserved in humans. Three major differences stand out between mouse and human XCI (reviewed in [57,58]): in the mouse preimplantation embryo, an initial form of paternally imprinted XCI takes place, which is maintained in extraembryonic lineages but reversed in the embryo proper; in human blastocysts, XIST is expressed from both active X chromosomes, prior the onset of random XCI; the number of genes escaping silencing in humans is remarkably higher than in mice. Whether these differences reflect a distinct species-specific modular organization of the Xist molecule is only starting to be dissected.
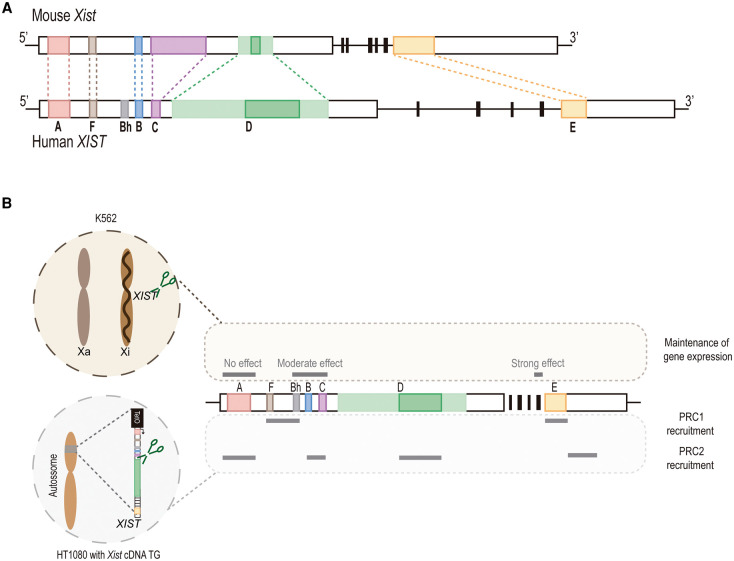
Overall, the gene structure of Xist is conserved between mouse and humans (Figure 2A). Repeats are amongst the most conserved blocks, in particular Repeats A, E and F, but some important differences in the number of repetitive motifs are seen for the other repeats. For instance, Repeat B is duplicated in human XIST, with an extra B-like repeat, known as Bh, located between Repeat F and B. Repeats C and D are the ones that differ the most between mouse and human Xist. While Repeat C is considerably expanded in the mouse, a feature that seems to be rodent-specific, it is reduced to a single motif in humans. In contrast, Repeat D is considerably expanded in human XIST, while it is mostly made of truncated motifs in rodent lineages [6,7] (Figure 2A).
Figure 2. Similarities and differences between mouse and human Xist.
(A) Comparison between mouse and human Xist gene structure and tandem repeats. Core Repeat D and its extended truncated motifs are marked in dark and light green, respectively. (B) Outline of the putative role of human XIST repeats for XCI maintenance assessed in female K562 lymphoblast cell line and for XCI initiation in male HT1080 fibrosarcoma cell line with a Xist cDNA autosomal transgene; see main text for details.
Despite these sequence differences, the higher-order folding of Xist RNA was shown to be conserved between mouse and human, being partitioned in five major structural modules: Repeats A, F, B/C/D, E and exon 6/7 [12,29]. Moreover, mouse and human Xist also share an extensive fraction of their protein interactome [20,59]. Proteomic screening of XIST interactors in human B lymphocytes by ChIRP-MS (Comprehensive Identification of RNA-binding Proteins by Mass Spectrometry) identified factors known to interact with mouse Xist, such as SPEN, RBM15/RBM15B, HNRNPK or CIZ1 as well as novel functional interactors, like TRIM28 (TRIpartite Motif-containing 28, commonly known as KAP1) [59]. Whether TRIM28 is a species- or cell type-specific XIST interactor is not yet known. Besides a shared set of protein interactors, these interactors distribute in a similar fashion along the mouse and human Xist molecules as revealed by enhanced CLIP-seq (UV Cross-Linking and ImmunoPrecipitation coupled with high-throughput sequencing) [60]. Small differences, namely on the interaction of HNRNPK with the expanded human Repeat D, have been described [29]. This raises the intriguing possibility that the larger Repeat D might replace the function of Repeat C in humans, which is reduced to one incomplete motif. It also suggests that the distribution of RBPs between different repetitive modules might confer slightly different functions to Xist in different species.
Recent studies, resorting to segmental deletions of human XIST in cancer cell lines, have started to explore the role of the repeats in the initiation and maintenance stages of human XCI, allowing to further uncover similarities and differences with mouse Xist [61,62]. In K562 erythromyeloblastoid leukemia cell line, where XCI has already occurred, a homozygous deletion of the entire XIST gene results in a global up-regulation of X-linked genes [62]. Several deletions along human XIST were also made in K562, many of which result in multiple defects at the transcriptional and splicing levels and/or affect XIST coating [62]. For instance, ΔD mutants show decreased RNA levels and splicing abnormalities that result in moderate X-linked reactivation, as previously observed in 293FT cell line [63]. Concentrating on XIST mutants with no such defects in K562, it was revealed that exon 5, which does not contain any repeat module, has an important role in XCI maintenance that has not yet been mechanistically explored. In contrast, deletions of Repeat A and Repeat B/C show no or moderate effects on XCI maintenance, respectively, in K562 cells [62] (Figure 2B).
A crucial role for human XIST in XCI maintenance in K562 contrasts with the situation in the mouse, where no visible effect on gene silencing is observed when Xist or its repeats are deleted in MEFs [9,64]. However, these results must be taken with caution, as this cancer cell line presents several (epi)genetic abnormalities that hinder the robustness of this cellular model to study XCI maintenance. For example, the Xi shows an atypical XCI status, being devoid of PcG marks [62]. As such, it might be over-simplistic to assume that human XIST, but not mouse Xist, plays a role in the maintenance of XCI. As a matter of fact, the emerging picture is that the role of Xist in XCI maintenance is complex and cell-type specific, whether in mouse or human [59,65–68].
A more recent study investigated the initiation phase of human XCI, using inducible autosomal cDNA transgenes in the HT1080 male fibrosarcoma cell line [61]. Hallmarks of facultative heterochromatin of the Xi are recapitulated in this system and were used to identify the regions of human XIST essential for H3K27me3 and H2AK119ub accumulation on the Xi. In contrast to mouse ESCs, PRC1 and PRC2 recruitment in HT1080 cells, rely on different XIST modules (Figure 2B): Repeats F/Bh and E are important for the accumulation of the PRC2 mark, H3K27me3, while enrichment of H2AK119ub by PRC1 is dependent on Repeats A, B/C, D and the 3′ non-repetitive part of XIST. The independent recruitment of PRC1 and PRC2 was further confirmed using chemical inhibition of the catalytic domains of the two PcG complexes. Overall, these results differ remarkably from mouse cells, where PRC2 recruitment is mostly downstream of the Xist-mediated recruitment of PCGF3/5-PRC1 through HNRNPK [9,10,32]. However, direct comparison between studies on mouse ESCs and human cancer cells should be taken cautiously, especially when XIST anchoring to the Xi differs in primary versus transformed cells [69]. A more appropriate comparison would be to look into XIST mutants in human ESCs. Unfortunately, human ESCs are mostly found in a post-XCI or in a culture-induced eroded XCI state (reviewed in [70]). Continuing progress in naïve culture conditions, which should reset human ESCs to a pre-XCI state [71], will be fundamental to reveal similarities and differences between human and mouse Xist and their role in the initiation of XCI.
Concluding remarks
Since its discovery, in the early nineties, Xist has been under the spotlight of lncRNA research. Although a myriad of intergenic, intronic and antisense lncRNAs have now been identified, the biological relevance of these molecules has remained challenging to define. In no small measure, our understanding of Xist has been central in promoting and inspiring the study of other nuclear-located lncRNAs which, despite being poorly conserved compared with protein-coding genes, share several functions and mechanisms of action. In this sense, the idea that lncRNAs can organize in structural modules, similarly to Xist, is a tantalizing hypothesis to explain how lncRNAs develop their tissue- and species-specific roles. Indeed, it was recently shown that Rsx (RNA-on-the-silent X), the lncRNA responsible for XCI in marsupials but with no homology to Xist, presents multiple tandem repetitive modules reminiscent of Xist repeats [72]. Just like Xist, Rsx might resort to different repetitive modules as functional entities to regulate XCI in marsupials. This example illustrates how further research on Xist RNA might have important implications for our understanding of other lncRNAs.
Although the knowledge that has been acquired about Xist in the past decades is undeniable, many open questions still remain. For example, the crosstalk between the different RNA modules and their protein interactors still needs to be mechanistically explored. Furthermore, while most studies have focused on dissecting Xist function during the establishment phase of XCI, a maintenance role for Xist has just started to be unveiled with cell-type-specific characteristics [59,68]. This is particularly relevant in the context of diseases predominantly affecting women for which incorrect maintenance of X-linked gene dosage may play a role, such as auto-immune diseases like systemic lupus erythematosus or multiple sclerosis (reviewed in [73]). A possible link between XIST and human disease will certainly be one of the areas to explore in the future.
Perspectives
Xist lncRNA is the master regulator of X-chromosome inactivation. Discovered thirty years ago, the molecular mechanisms through which Xist renders an entire chromosome into a stable inactive state remain enigmatic.
A series of deletions in mouse cells revealed that Xist possesses several modules of functional specialization mapping to the conserved Repeats A-to-F. These functional modules execute specific tasks through interaction with various RNA-binding proteins in a coordinated manner during XCI. This modular organization and multitasking nature of Xist is conserved in humans, although the functional specialization of some repeats might have evolved novel functions.
It still remains to be deciphered the molecular mechanisms underlying the interplay between Xist repeats for the control of XCI. Also, the conservation of these events in humans is unknown and will need to be investigated in more appropriate cellular models. Finally, the translation of these findings to other lncRNAs might reveal new molecular insights in the function of these molecules in a vast plethora of biological systems.
Acknowledgements
We thank Evguenia P. Bekman and Bruno Silva for critical reading of the manuscript.
Abbreviations
CBX, Chromobox homolog; CHIRP-MS, comprehensive identification of RNA-binding protein by mass spectrometry; CIZ1, CDKN1A-interacting zinc finger protein 1; CLIP-seq, UV cross-linking and immunoprecipitation coupled with high-throughput sequencing; ESCs, embryonic stem cells; HDAC3, Histone deacetylase 3; HNRNPK, Heterogeneous nuclear ribonucleoprotein K; lncRNA, long non-coding RNA; m6A, N6-methyladenosine; MEFs, mouse embryonic fibroblasts; NCoR/SMRT, Nuclear receptor co-repressor/Silencing mediator of retinoic acid and thyroid hormone receptor; NuRD, Nucleosome remodeling and deacelyase; PcG, Polycomb group; PRC1, Polycomb Repressive Complex 1; PRC2, Polycomb Repressive Complex 2; PolII, RNA polymerase II; LBR, lamin B receptor; RBPs, RNA binding proteins; RBM15, RNA binding motif protein 15; Rsx, RNA-on-the-silent X; RRM, RNA recognition motifs; SMCHD1, Structural maintenance of chromosomes flexible hinge domain containing 1; SPEN, Split Ends; SPOC, SPEN paralogue/orthologue C-terminal; TADs, Topologically associating domains; TRIM28, Tripatite motif-containing 28; XCI, X-chromosome Inactivation; Xi, inactive X chromosome; Xist, X-inactive-specific transcript; YTHDC1, YTH domain containing 1; YY1, Yin yang 1
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by Fundação para a Ciência e Tecnologia (FCT)/Ministério da Ciência, Tecnologia e Ensino Superior (MCTES) through the project grants PTDC/BIA-MOL/29320/2017 (S.T.d.R. and A.C.R.), and PTDC/BTM-TEC/28534/2017 (M.C.). S.T.d.R. and A.-V. G. are supported by assistant research contracts from FCT/MCTES (CEECIND/01234/2017 and CEECIND/02085/2018, respectively; A.C.R. is supported by a PhD fellowship from FCT/MCTES (SFRH/BD/137099/2018).
Author Contributions
A.C.R., M.C., A.-V. G. and S.T.d.R. wrote the manuscript. A.C.R. developed the Table and the Figures under the guidance of M.C., A.V. G. and S.T.d.R.
References
- 1.Marahrens, Y., Panning, B., Dausman, J., Strauss, W. and Jaenisch, R. (1997) Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 11, 156–166 10.1101/gad.11.2.156 [DOI] [PubMed] [Google Scholar]
- 2.Penny, G.D., Kay, G.F., Sheardown, S.A., Rastan, S. and Brockdorff, N. (1996) Requirement for Xist in X chromosome inactivation. Nature 379, 131–137 10.1038/379131a0 [DOI] [PubMed] [Google Scholar]
- 3.Mutzel, V. and Schulz, E.G. (2020) Dosage sensing, threshold responses, and epigenetic memory: a systems biology perspective on random X-chromosome inactivation. Bioessays 42, e1900163 10.1002/bies.201900163 [DOI] [PubMed] [Google Scholar]
- 4.da Rocha, S.T. and Heard, E. (2017) Novel players in X inactivation: insights into Xist-mediated gene silencing and chromosome conformation. Nat. Struct. Mol. Biol. 24, 197–204 10.1038/nsmb.3370 [DOI] [PubMed] [Google Scholar]
- 5.Dossin, F. and Heard, E. (2021) The molecular and nuclear dynamics of X-chromosome inactivation. Cold Spring Harb. Perspect. Biol. Online ahead of print 10.1101/cshperspect.a040196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nesterova, T.B., Slobodyanyuk, S.Y., Elisaphenko, E.A., Shevchenko, A.I., Johnston, C., Pavlova, M.E.et al. (2001) Characterization of the genomic Xist locus in rodents reveals conservation of overall gene structure and tandem repeats but rapid evolution of unique sequence. Genome Res. 11, 833–849 10.1101/gr.174901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yen, Z.C., Meyer, I.M., Karalic, S. and Brown, C.J. (2007) A cross-species comparison of X-chromosome inactivation in Eutheria. Genomics 90, 453–463 10.1016/j.ygeno.2007.07.002 [DOI] [PubMed] [Google Scholar]
- 8.Bousard, A., Raposo, A.C., Zylicz, J.J., Picard, C., Pires, V.B., Qi, Y.et al. (2019) The role of Xist-mediated Polycomb recruitment in the initiation of X-chromosome inactivation. EMBO Rep. 20, e48019 10.15252/embr.201948019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colognori, D., Sunwoo, H., Kriz, A.J., Wang, C.Y. and Lee, J.T. (2019) Xist deletional analysis reveals an interdependency between Xist RNA and polycomb complexes for spreading along the inactive X. Mol. Cell 74, 101–17 e10 10.1016/j.molcel.2019.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nesterova, T.B., Wei, G., Coker, H., Pintacuda, G., Bowness, J.S., Zhang, T.et al. (2019) Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nat. Commun. 10, 3129 10.1038/s41467-019-11171-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pandya-Jones, A., Markaki, Y., Serizay, J., Chitiashvili, T., Mancia Leon, W.R., Damianov, A.et al. (2020) A protein assembly mediates Xist localization and gene silencing. Nature 587, 145–151 10.1038/s41586-020-2703-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu, Z., Zhang, Q.C., Lee, B., Flynn, R.A., Smith, M.A., Robinson, J.T.et al. (2016) RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165, 1267–1279 10.1016/j.cell.2016.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colognori, D., Sunwoo, H., Wang, D., Wang, C.Y. and Lee, J.T. (2020) Xist repeats A and B account for two distinct phases of X inactivation establishment. Dev. Cell 54, 21–32 e5 10.1016/j.devcel.2020.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoki, Y., Kimura, N., Kanbayashi, M., Amakawa, Y., Ohhata, T., Sasaki, H.et al. (2009) A proximal conserved repeat in the Xist gene is essential as a genomic element for X-inactivation in mouse. Development 136, 139–146 10.1242/dev.026427 [DOI] [PubMed] [Google Scholar]
- 15.Sakata, Y., Nagao, K., Hoki, Y., Sasaki, H., Obuse, C. and Sado, T. (2017) Defects in dosage compensation impact global gene regulation in the mouse trophoblast. Development 144, 2784–2797 10.1242/dev.149138 [DOI] [PubMed] [Google Scholar]
- 16.Wang, Y., Zhong, Y., Zhou, Y., Tanaseichuk, O., Li, Z. and Zhao, J.C. (2019) Identification of a Xist silencing domain by tiling CRISPR. Sci. Rep. 9, 2408 10.1038/s41598-018-36750-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wutz, A., Rasmussen, T.P. and Jaenisch, R. (2002) Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 30, 167–174 10.1038/ng820 [DOI] [PubMed] [Google Scholar]
- 18.Zhao, J., Sun, B.K., Erwin, J.A., Song, J.J. and Lee, J.T. (2008) Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322, 750–756 10.1126/science.1163045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pintacuda, G., Wei, G., Roustan, C., Kirmizitas, B.A., Solcan, N., Cerase, A.et al. (2017) hnRNPK recruits PCGF3/5-PRC1 to the Xist RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol. Cell 68, 955–69 e10 10.1016/j.molcel.2017.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu, C., Zhang, Q.C., da Rocha, S.T., Flynn, R.A., Bharadwaj, M., Calabrese, J.M.et al. (2015) Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416 10.1016/j.cell.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patil, D.P., Chen, C.K., Pickering, B.F., Chow, A., Jackson, C., Guttman, M.et al. (2016) . m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373 10.1038/nature19342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dossin, F., Pinheiro, I., Zylicz, J.J., Roensch, J., Collombet, S., Le Saux, A.et al. (2020) SPEN integrates transcriptional and epigenetic control of X-inactivation. Nature 578, 455–460 10.1038/s41586-020-1974-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McHugh, C.A., Chen, C.K., Chow, A., Surka, C.F., Tran, C., McDonel, P.et al. (2015) The Xist lncRNA interacts directly with SHARP o silence transcription through HDAC3. Nature 521, 232–236 10.1038/nature14443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moindrot, B., Cerase, A., Coker, H., Masui, O., Grijzenhout, A., Pintacuda, G.et al. (2015) A pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for xist RNA-mediated silencing. Cell Rep. 12, 562–572 10.1016/j.celrep.2015.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monfort, A., Di Minin, G., Postlmayr, A., Freimann, R., Arieti, F., Thore, S.et al. (2015) Identification of Spen as a crucial factor for Xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep. 12, 554–561 10.1016/j.celrep.2015.06.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodermund, L., Coker, H., Oldenkamp, R., Wei, G., Bowness, J., Rajkumar, B.et al. (2021) Time-resolved structured illumination microscopy reveals key principles of Xist RNA spreading. Science 372, eabe7500 10.1126/science.abe7500 [DOI] [PubMed] [Google Scholar]
- 27.Zylicz, J.J., Bousard, A., Zumer, K., Dossin, F., Mohammad, E., da Rocha, S.T.et al. (2019) The implication of early chromatin changes in X chromosome inactivation. Cell 176, 182–97 e23 10.1016/j.cell.2018.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang, X., Liu, B., Nie, Z., Duan, L., Xiong, Q., Jin, Z.et al. (2021) The role of m6A modification in the biological functions and diseases. Signal. Transduct. Target. Ther. 6, 74 10.1038/s41392-020-00450-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu, Z., Guo, J.K., Wei, Y., Dou, D.R., Zarnegar, B., Ma, Q.et al. (2020) Structural modularity of the XIST ribonucleoprotein complex. Nat. Commun. 11, 6163 10.1038/s41467-020-20040-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coker, H., Wei, G., Moindrot, B., Mohammed, S., Nesterova, T. and Brockdorff, N. (2020) The role of the Xist 5′ m6A region and RBM15 in X chromosome inactivation. Wellcome Open Res. 5, 31 10.12688/wellcomeopenres.15711.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.da Rocha, S.T., Boeva, V., Escamilla-Del-Arenal, M., Ancelin, K., Granier, C., Matias, N.R.et al. (2014) Jarid2 is implicated in the initial Xist-induced targeting of PRC2 to the inactive X chromosome. Mol. Cell 53, 301–316 10.1016/j.molcel.2014.01.002 [DOI] [PubMed] [Google Scholar]
- 32.Almeida, M., Pintacuda, G., Masui, O., Koseki, Y., Gdula, M., Cerase, A.et al. (2017) PCGF3/5-PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science 356, 1081–1084 10.1126/science.aal2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cirillo, D., Blanco, M., Armaos, A., Buness, A., Avner, P., Guttman, M.et al. (2016) Quantitative predictions of protein interactions with long noncoding RNAs. Nat. Methods 14, 5–6 10.1038/nmeth.4100 [DOI] [PubMed] [Google Scholar]
- 34.Nakamoto, M.Y., Lammer, N.C., Batey, R.T. and Wuttke, D.S. (2020) hnRNPK recognition of the B motif of Xist and other biological RNAs. Nucleic Acids Res. 48, 9320–9335 10.1093/nar/gkaa677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper, S., Grijzenhout, A., Underwood, E., Ancelin, K., Zhang, T., Nesterova, T.B.et al. (2016) Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat. Commun. 7, 13661 10.1038/ncomms13661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kasinath, V., Beck, C., Sauer, P., Poepsel, S., Kosmatka, J., Faini, M.et al. (2021) JARID2 and AEBP2 regulate PRC2 in the presence of H2AK119ub1 and other histone modifications. Science 371, eabc3393 10.1126/science.abc3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schoeftner, S., Sengupta, A.K., Kubicek, S., Mechtler, K., Spahn, L., Koseki, H.et al. (2006) Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 25, 3110–3122 10.1038/sj.emboj.7601187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blackledge, N.P. and Klose, R.J. (2021) The molecular principles of gene regulation by Polycomb repressive complexes. Nat. Rev. Mol. Cell Biol. 22, 815–833 10.1038/s41580-021-00398-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mendenhall, E.M., Koche, R.P., Truong, T., Zhou, V.W., Issac, B., Chi, A.S.et al. (2010) GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 6, e1001244 10.1371/journal.pgen.1001244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riising, E.M., Comet, I., Leblanc, B., Wu, X., Johansen, J.V. and Helin, K. (2014) Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol. Cell 55, 347–360 10.1016/j.molcel.2014.06.005 [DOI] [PubMed] [Google Scholar]
- 41.Markaki, Y., Chong, J.C., Wang, Y., Jacobson, E.C., Luong, C., Tan, S.Y.X.et al. (2021) Xist nucleates local protein gradients to propagate silencing across the X chromosome. Cell 184, 1–18 10.1016/j.cell.2021.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tjalsma, S.J.D., Hori, M., Sato, Y., Bousard, A., Ohi, A., Raposo, A.C.et al. (2021) H4k20me1 and H3K27me3 are concurrently loaded onto the inactive X chromosome but dispensable for inducing gene silencing. EMBO Rep. 22, e51989 10.15252/embr.202051989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jansz, N., Nesterova, T., Keniry, A., Iminitoff, M., Hickey, P.F., Pintacuda, G.et al. (2018) Smchd1 targeting to the inactive X is dependent on the Xist-HnrnpK-PRC1 pathway. Cell Rep. 25, 1912–23 e9 10.1016/j.celrep.2018.10.044 [DOI] [PubMed] [Google Scholar]
- 44.Jansz, N., Keniry, A., Trussart, M., Bildsoe, H., Beck, T., Tonks, I.D.et al. (2018) Smchd1 regulates long-range chromatin interactions on the inactive X chromosome and at Hox clusters. Nat. Struct. Mol. Biol. 25, 766–777 10.1038/s41594-018-0111-z [DOI] [PubMed] [Google Scholar]
- 45.Wang, C.Y., Colognori, D., Sunwoo, H., Wang, D. and Lee, J.T. (2019) PRC1 collaborates with SMCHD1 to fold the X-chromosome and spread Xist RNA between chromosome compartments. Nat. Commun. 10, 2950 10.1038/s41467-019-10755-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ridings-Figueroa, R., Stewart, E.R., Nesterova, T.B., Coker, H., Pintacuda, G., Godwin, J.et al. (2017) The nuclear matrix protein CIZ1 facilitates localization of Xist RNA to the inactive X-chromosome territory. Genes Dev. 31, 876–888 10.1101/gad.295907.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sunwoo, H., Colognori, D., Froberg, J.E., Jeon, Y. and Lee, J.T. (2017) Repeat E anchors Xist RNA to the inactive X chromosomal compartment through CDKN1A-interacting protein (CIZ1). Proc. Natl Acad. Sci. U.S.A. 114, 10654–9 10.1073/pnas.1711206114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yue, M., Ogawa, A., Yamada, N., Charles Richard, J.L., Barski, A. and Ogawa, Y. (2017) Xist RNA repeat E is essential for ASH2L recruitment to the inactive X and regulates histone modifications and escape gene expression. PLoS Genet. 13, e1006890 10.1371/journal.pgen.1006890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gallego-Iradi, M.C., Strunk, H., Crown, A.M., Davila, R., Brown, H., Rodriguez-Lebron, E.et al. (2019) N-terminal sequences in matrin 3 mediate phase separation into droplet-like structures that recruit TDP43 variants lacking RNA binding elements. Lab. Invest. 99, 1030–1040 10.1038/s41374-019-0260-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li, P., Banjade, S., Cheng, H.C., Kim, S., Chen, B., Guo, L.et al. (2012) Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cerase, A., Armaos, A., Neumayer, C., Avner, P., Guttman, M. and Tartaglia, G.G. (2019) Phase separation drives X-chromosome inactivation: a hypothesis. Nat. Struct. Mol. Biol. 26, 331–334 10.1038/s41594-019-0223-0 [DOI] [PubMed] [Google Scholar]
- 52.Chen, C.K., Blanco, M., Jackson, C., Aznauryan, E., Ollikainen, N., Surka, C.et al. (2016) Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354, 468–472 10.1126/science.aae0047 [DOI] [PubMed] [Google Scholar]
- 53.Jeon, Y. and Lee, J.T. (2011) YY1 tethers xist RNA to the inactive X nucleation center. Cell 146, 119–133 10.1016/j.cell.2011.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chapman, A.G., Cotton, A.M., Kelsey, A.D. and Brown, C.J. (2014) Differentially methylated CpG island within human XIST mediates alternative P2 transcription and YY1 binding. BMC Genet. 15, 89 10.1186/s12863-014-0089-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Makhlouf, M., Ouimette, J.F., Oldfield, A., Navarro, P., Neuillet, D. and Rougeulle, C. (2014) A prominent and conserved role for YY1 in Xist transcriptional activation. Nat. Commun. 5, 4878 10.1038/ncomms5878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown, C.J., Hendrich, B.D., Rupert, J.L., Lafreniere, R.G., Xing, Y., Lawrence, J.et al. (1992) The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71, 527–542 10.1016/0092-8674(92)90520-M [DOI] [PubMed] [Google Scholar]
- 57.Patrat, C., Ouimette, J.F. and Rougeulle, C. (2020) X chromosome inactivation in human development. Development 147, dev183095 10.1242/dev.183095 [DOI] [PubMed] [Google Scholar]
- 58.Posynick, B.J. and Brown, C.J. (2019) Escape from X-chromosome inactivation: an evolutionary perspective. Front. Cell Dev. Biol. 7, 241 10.3389/fcell.2019.00241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu, B., Qi, Y., Li, R., Shi, Q., Satpathy, A.T. and Chang, H.Y. (2021) B cell-specific XIST complex enforces X-inactivation and restrains atypical B cells. Cell 184, 1790–803 e17 10.1016/j.cell.2021.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Nostrand, E.L., Pratt, G.A., Shishkin, A.A., Gelboin-Burkhart, C., Fang, M.Y., Sundararaman, B.et al. (2016) Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 13, 508–514 10.1038/nmeth.3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dixon-McDougall, T. and Brown, C.J. (2021) Independent domains for recruitment of PRC1 and PRC2 by human XIST. PLoS Genet. 17, e1009123 10.1371/journal.pgen.1009123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee, H.J., Gopalappa, R., Sunwoo, H., Choi, S.W., Ramakrishna, S., Lee, J.T.et al. (2019) En bloc and segmental deletions of human XIST reveal X chromosome inactivation-involving RNA elements. Nucleic Acids Res. 47, 3875–3887 10.1093/nar/gkz109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lv, Q., Yuan, L., Song, Y., Sui, T., Li, Z. and Lai, L. (2016) D-repeat in the XIST gene is required for X chromosome inactivation. RNA Biol. 13, 172–176 10.1080/15476286.2015.1137420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Csankovszki, G., Panning, B., Bates, B., Pehrson, J.R. and Jaenisch, R. (1999) Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat. Genet. 22, 323–324 10.1038/11887 [DOI] [PubMed] [Google Scholar]
- 65.Brown, C.J. and Willard, H.F. (1994) The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature 368, 154–156 10.1038/368154a0 [DOI] [PubMed] [Google Scholar]
- 66.Cantone, I., Bagci, H., Dormann, D., Dharmalingam, G., Nesterova, T., Brockdorff, N.et al. (2016) Ordered chromatin changes and human X chromosome reactivation by cell fusion-mediated pluripotent reprogramming. Nat. Commun. 7, 12354 10.1038/ncomms12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Westervelt, N., Yoest, A., Sayed, S., Von Zimmerman, M., Kaps, K. and Chadwick, B.P. (2021) Deletion of the XIST promoter from the human inactive X chromosome compromises polycomb heterochromatin maintenance. Chromosoma 130, 177–197 10.1007/s00412-021-00754-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang, L., Yildirim, E., Kirby, J.E., Press, W. and Lee, J.T. (2020) Widespread organ tolerance to Xist loss and X reactivation except under chronic stress in the gut. Proc. Natl Acad. Sci. U.S.A. 117, 4262–4272 10.1073/pnas.1917203117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kolpa, H.J., Fackelmayer, F.O. and Lawrence, J.B. (2016) SAF-A requirement in anchoring XIST RNA to chromatin varies in transformed and primary cells. Dev. Cell 39, 9–10 10.1016/j.devcel.2016.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sahakyan, A., Plath, K. and Rougeulle, C. (2017) Regulation of X-chromosome dosage compensation in human: mechanisms and model systems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160363 10.1098/rstb.2016.0363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.An, C., Feng, G., Zhang, J., Cao, S., Wang, Y., Wang, N.et al. (2020) Overcoming autocrine FGF signaling-induced heterogeneity in naive human ESCs enables modeling of random X chromosome inactivation. Cell Stem Cell 27, 482–97 e4 10.1016/j.stem.2020.06.002 [DOI] [PubMed] [Google Scholar]
- 72.Sprague, D., Waters, S.A., Kirk, J.M., Wang, J.R., Samollow, P.B., Waters, P.D.et al. (2019) Nonlinear sequence similarity between the Xist and Rsx long noncoding RNAs suggests shared functions of tandem repeat domains. RNA 25, 1004–1019 10.1261/rna.069815.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Youness, A., Miquel, C.H. and Guery, J.C. (2021) Escape from X chromosome inactivation and the female predominance in autoimmune diseases. Int. J. Mol. Sci. 22, 1114 10.3390/ijms22031114 [DOI] [PMC free article] [PubMed] [Google Scholar]