Abstract
The production of mannosidase activity by all currently recognized species of human viridans group streptococci was determined using an assay in which bacterial growth was dependent on the degradation of the high-mannose-type glycans of RNase B and subsequent utilization of released mannose. RNase B is an excellent substrate for the demonstration of mannosidase activity since it is a glycoprotein with a single glycosylation site which is occupied by high-mannose-type glycoforms containing five to nine mannose residues. Mannosidase activity was produced only by some members of the mitis group (Streptococcus mitis, Streptococcus oralis, Streptococcus gordonii, Streptococcus cristatus, Streptococcus infantis, Streptococcus parasanguinis, and Streptococcus pneumoniae) and Streptococcus intermedius of the anginosus group. None of the other species within the salivarius and mutans groups or Streptococcus peroris and Streptococcus sanguinis produced mannosidase activity. Using matrix-assisted laser desorption ionization time-of-flight mass spectrometry, it was demonstrated that the Man5 glycan alone was degraded while Man6 to Man9, which contain terminal α(1→2) mannose residues in addition to the α(1→3), α(1→6), and β(1→4) residues present in Man5, remained intact. Investigations on mannosidase production using synthetic (4-methylumbelliferone- or p-nitrophenol-linked) α- or β-mannosides as substrates indicated that there was no correlation between degradation of these substrates and degradation of the Man5 glycan of RNase B. No species degraded these α-linked mannosides, while degradation of the β-linked synthetic substrates was restricted to strains within the Streptococcus anginosus, S. gordonii, and S. intermedius species. The data generated using a native glycoprotein as the substrate demonstrate that mannosidase production within the viridans group streptococci is more widely distributed than had previously been considered.
The viridans group streptococci, which comprise a significant proportion of the normal flora of the oropharyngeal tract (8), form a highly heterogenous group of organisms (13, 37). Over the last decade, identification of these organisms at the species level has been facilitated by the development of a range of rapid phenotypic tests, some of which rely, at least in part, on the detection of preformed glycosidase activities, enzymes with the potential to cleave sugar residues from a range of glycoconjugates, including glycoproteins. A number of schemes for the identification of individual species of the viridans group streptococci have included phenotyping for the production of a range of glycosidases using synthetic substrates, glycosides with a fluorogenic (4-methylumbelliferone [4MU]) or chromogenic (p-nitrophenol [pNP]) leaving group, some of these being supplied as part of commercially available kits, such as the API-20STREP system (2, 5, 9, 18, 19, 20, 36, 39). These schemes for the classification of the viridans group streptococci have enabled associations between the isolation of a particular species and specific extra-oral diseases to be established (15, 16, 38), which is of major importance due to the emergence of these bacteria as significant pathogens of immunocompromised patients and neonates (1, 3, 29, 35). Little is known regarding the mechanisms by which these streptococci grow at sites removed from the oral cavity, but it has been suggested that glycosidase production may play a nutritional role by releasing carbohydrates which are essential for proliferation from host glycoproteins (1).
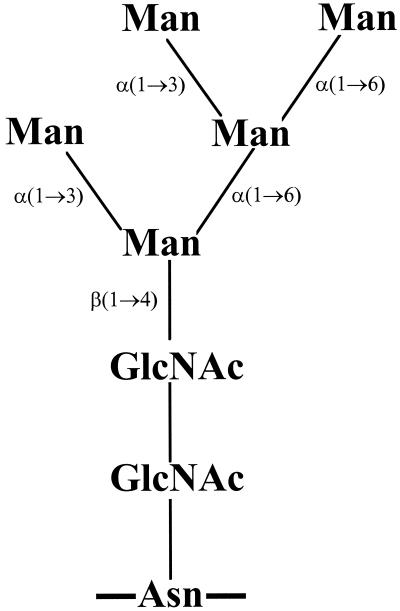
In a recent investigation it was demonstrated that the acute-phase human serum glycoprotein, α1-acid glycoprotein, supported in vitro growth of Streptococcus oralis (4), an important pathogen within the viridans group streptococci which is the cause of a large number of cases of infective endocarditis and septicemia in neutropenic patients (1, 6, 7). α1-acid glycoprotein, which comprises five fully sialylated complex-type oligosaccharide chains, was degraded to release all nonterminal sugar residues, including those mannose residues which, in addition to two N-acetylglucosamine residues, form part of the conserved pentasaccharide core (GlcNAc2Man5) of N-linked glycans. Thus, novel mannosidase activities were demonstrated in S. oralis and these enzymes were not detected using synthetic substrates. These observations were supported by demonstration of in vitro growth of S. oralis strain AR3 on RNase B (30). This glycoprotein contains a single glycosylation site, and posttranslational modification results in the addition of high-mannose-type glycans containing five to nine mannose residues attached to the chitobiose core (Man5 to Man9) (10, 24). Using a combination of high-pH anion-exchange chromatography with pulsed amperometric detection and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), it was demonstrated that S. oralis produced α-(1→3), α-(1→6), and β-(1→4) mannosidase activities which resulted in the degradation of the Man5 glycoform only (Fig. 1), leaving the remaining glycoforms (Man6 to Man9), which contain additional α-(1→2)-linked mannose residues, intact (30).
FIG. 1.
The oligosaccharide structure of the Man5 glycoform of RNase B. Asn represents the asparagine of the polypeptide backbone. N-Acetylglucosamine and mannose are represented by GlcNAc and Man, respectively, and numbers indicate the positions of the glycosidic linkages for mannose residues. The Man6 to Man9 glycoforms of RNase B are formed by the addition of further mannose units [all in the α-(1→2) configuration] to the outer three residues occurring in the Man5 glycoform.
In the present study, we have extended our studies on mannosidase production by S. oralis strain AR3 to all currently recognized human species of viridans group streptococci. We have used assays in which bacterial growth on the model glycoprotein RNase B is dependent on the production of mannosidase activity and utilization of liberated mannose and have monitored changes in RNase B glycoforms by MALDI-TOF MS.
MATERIALS AND METHODS
Bacterial strains and culture.
Representative strains, including type strains, of all currently recognized species of human viridans group streptococci were used (Table 1). Isolates were stored in vials containing cryopreservative (TSC Ltd., Heywood, Lancashire, United Kingdom) at −70°C and were routinely maintained by subculture onto Fastidious Anaerobe Agar (LabM, Bury, Lancashire, United Kingdom) supplemented with 5% (vol/vol) defibrinated horse blood (FAA). Cultures were incubated in an anaerobic cabinet (80% N2, 10% H2, 10% CO2; Don Whitley, Shipley, West Yorkshire, United Kingdom) for 24 h prior to use in growth studies.
TABLE 1.
Strains of viridans group streptococci included in this study
| Species identification | Strainsa |
|---|---|
| S. salivarius | ATCC 7073T, T267, H50, KPS1, NCTC 8606, A385 |
| S. vestibularis | ATCC 49124T, LV71, OP81, HV81, JW3 |
| S. mutans | ATCC 25175T, 4177, KPSK2, SE11, B48, 161 |
| S. sobrinus | ATCC 33478T, 279, OMZ65, TH62 |
| S. anginosus | ATCC 33397T, NCTC 8073, NCTC 11062, KR 687, PC4890, NMH 10 |
| S. constellatus | ATCC 27823T, AM699, NCTC 0714, NCTC 11063, NCTC 5389 |
| S. intermedius | ATCC 27335T, 415-87, NS35, NMH2 |
| S. mitis | ATCC 49456T, K208, PP53, NCTC 10712, HV51, 130411-14,b 130411-16,b 143511-6,b T6(2)-5,b T6(2)-10,b T6(2)-11,b T6(3)-12,b T6(2)-7b |
| S. oralis | ATCC 35037T, NCTC 7864, OPA1, PC1467, AR3, H362 |
| S. gordonii | ATCC 10558T, NCTC 7868, M5, GPF1, F90A |
| S. cristatus | ATCC 51100T, CC5A, AK1, CR3 |
| S. pneumoniaec | ATCC 33400T, 47213, 48626, 40726, BC76, 48764, 45419, 45309, 45422 |
| S. sanguinis | ATCC 10556T, P695, KPE2, NP506, SK96 |
| S. parasanguinis | ATCC 15912T, SS895, 85-81, SS-897, MGH143, FW213 |
| S. infantisd | 0-122T, 0-101, 1-134, 0-92, 0-103 |
| S. perorisd | 0-66T, 0-105, 0-91 |
Strains are human isolates characterized as described previously (14, 36) unless otherwise noted. T, type strain; ATCC, American Type Culture Collection, Manassas, Va.; NCTC, National Collection of Type Cultures, Central Public Health Laboratory, London, United Kingdom.
Strains were identified by sequencing of superoxide dismutase.
Strains (except for ATCC 33400T) were from blood culture isolates from the collection of the Department of Medical Microbiology, King's College Hospital, London, United Kingdom.
All strains were gifts from Y. Kawamura, Department of Microbiology, Gifu University School of Medicine, Gifu, Japan (18).
In order to determine preformed whole-cell glycosidase activities, cell growth was removed from FAA plates and put into 50 mM sodium phosphate buffer (pH 7.5). Aliquots of each suspension (200 μl) were dispensed into a flat-bottom 96-well microtiter tray, and the absorbance at 620 nm (A620) was recorded in a 96-well plate-reading spectrophotometer (Titertek Multiscan MCC 340; ICN-Flow, ICN Biomedicals Ltd., Thame, Oxfordshire, United Kingdom). A620 was adjusted to approximately 0.5, and cell suspensions were used in the assay of glycosidase activities using synthetic substrates, as described below.
Growth of bacteria in minimal media.
Bacterial colonies were removed from FAA plates into prereduced, filter-sterilized (0.2-μm-pore-size filters; Pall Gelman Sciences, Northampton, United Kingdom) 50 mM sodium phosphate buffer (pH 7.5) to yield a uniform suspension with an A620 of approximately 0.5. These suspensions were used as inocula for minimal media.
Minimal medium was prepared based on a modification of previously described methods (21, 32), with all sources of fermentable carbohydrates omitted. Preliminary investigations had demonstrated that growth of viridans group streptococci in this sugar-depleted medium was low unless supplemented with a source of fermentable carbohydrate. RNase B (from bovine pancreas; Sigma Chemical Co., Poole, Dorset, United Kingdom) was reconstituted and dialyzed against two changes of distilled water at 4°C. The glycoprotein was lyophilized and reconstituted to yield a concentration of 10 mg/ml. Basal medium was mixed with an equal volume of either 10 mg of RNase B/ml, 20 mM mannose, 20 mM glucose, 20 mM N-acetylglucosamine, or water and was filter sterilized (0.2-μm-pore-size membrane filtration unit). Media were dispensed into a sterile 96-well flat-bottom microtiter tray (Sterilin) in 200-μl aliquots, and the tray was prereduced for ca. 1 h in an anaerobic cabinet. An aliquot of bacterial suspension (10 μl) was added to each well, and 10 μl of sterile sodium phosphate buffer (pH 7.5) was added to control wells. Trays were sealed and incubated for up to 40 h at 37°C in a shaking-plate-reading spectrophotometer (iEMS; Labsystems, Life Sciences International, Hampshire, United Kingdom) with automated data acquisition. A620 was recorded every 30 min with a 10-s period of shaking (300 rpm) prior to each absorbance reading. For strains of viridans group streptococci which would not grow under these conditions, which were all members of the anginosus group (Streptococcus anginosus, Streptococcus constellatus, and Streptococcus intermedius), alternative growth conditions were employed. Supplemented basal media were dispensed into sterile bijou bottles (Sterilin) in 1-ml aliquots, prereduced under anaerobic conditions and inoculated with 50 μl of bacterial suspension prepared in sterile 50 mM sodium phosphate buffer. An aliquot of culture was removed for the determination of A620, and the remaining culture was incubated anaerobically. At the end of the growth period, the absorbance of the each culture was determined as a measure of growth.
At the end of the growth period, RNase B-supplemented cultures were transferred to microcentrifuge tubes with a nominal volume of 1.5 ml and centrifuged at 11,600 × g for 3 min at ambient temperature to pellet bacterial cells. The cell-free culture supernatants were transferred to fresh microcentrifuge tubes and stored at −20°C for a period of up to 2 weeks prior to analysis by MALDI-TOF MS.
Assay for glycosidase activities using synthetic substrates.
Glycosidase activities of bacterial suspensions from FAA plates were assayed using fluorogenic (4MU-linked) or chromogenic (pNP-linked) substrates. Fluorogenic assays were set up in 96-well microtiter trays and each were comprised as follows: 50 μl of 0.2 mM 4MU–α-d-mannopyranoside, 4MU–β-d-mannopyranoside, or 4MU–β-d-N-acetylglucosaminide (Sigma), 50 μl of 0.2 M sodium phosphate buffer (pH 7.0), an appropriate volume of cell suspension (up to 50 μl), and distilled water to a total volume of 200 μl. Trays were incubated at 37°C for up to 24 h and fluorescence values were recorded in a fluorimeter fitted with a 96-well plate reader (Fluoroscan Ascent FL; Labsystems) at excitation and emission wavelengths of 355 and 460 nm, respectively. Released 4MU was quantified by comparison with standard curves of authentic 4MU (Sigma) obtained under the same conditions. Chromogenic assays were set up in 96-well microtiter trays and each was comprised as follows: 25 μl of 0.2 mM pNP–α-d-mannopyranoside or pNP–β-d-mannopyranoside (Sigma), 25 μl of 0.2 M sodium phosphate buffer (pH 7.0), an appropriate volume of cell suspension (up to 25 μl), and distilled water to a total volume of 100 μl. Trays were incubated at 37°C for up to 24 h, and then the reaction was terminated and the pH of each assay was elevated by the addition of 100 μl of 0.1 M sodium carbonate buffer (pH 10.2). Absorbances at 450 nm were recorded by a 96-well plate-reading spectrophotometer (Titertek Multiscan MCC340), and released pNP was quantified by comparison with standard curves of authentic pNP (Sigma) obtained under the same conditions. Whole-cell protein in bacterial suspensions was determined using the bicinchoninic acid assay described by Smith et al. (28; reagents were purchased from Sigma) and was quantified by comparison with bovine serum albumin as a standard. Where appropriate, specific activities of enzymes are given as nanomoles/hour/milligram of whole-cell protein. Assays were carried out on at least two separate occasions, and mean data are presented.
MALDI-TOF-MS analysis of RNase B.
RNase B-supplemented culture supernatants were diluted 10-fold with 0.1% trifluoroacetic acid (Sigma), and 0.5 μl of each was applied to a gold-coated target plate. 3,5-Dimethoxy-4-hydroxycinnaminic acid (sinapinic acid; Aldrich) was used as matrix. This was freshly prepared to a concentration of 10 mg/ml in 50% acetonitrile in 0.1% trifluoroacetic acid, and 0.5 μl was applied to each sample. Spectra were acquired using a PerSeptives Voyager MALDI-TOF MS system operating in linear mode with delayed extraction and a nitrogen laser giving 337-nm output. Scans (typically 256) were acquired and averaged, and mass values were calibrated by reference to intact RNase B as an external standard or RNase A, which forms a minor component of all Sigma preparations of RNase B (10) as an internal standard.
RESULTS
Growth on RNase B as a measure of mannosidase production.
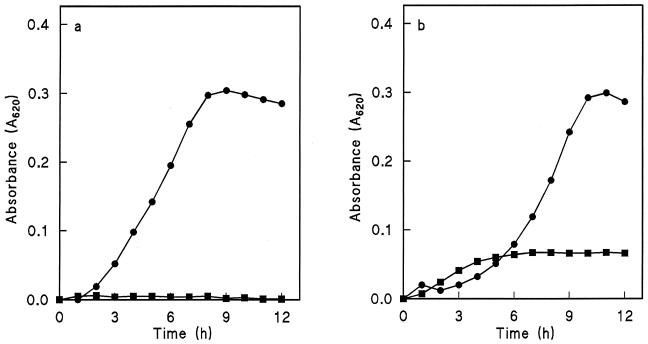
There was a linear relationship between the concentration of mannose supplied to S. oralis ATCC 35037 and Streptococcus mutans ATCC 25175 and the maximum increase in absorbance of cultures over the concentration range of 0 to 10 mM (r > 0.9 for both strains; data not shown), demonstrating the dependence of growth on the presence of a source of fermentable carbohydrate as the supplement to minimal medium. Both strains grew on 10 mM mannose with a maximum increase in A620 of 0.299 and 0.304, respectively (Fig. 2). RNase B supported growth of S. oralis cells, with a maximum increase in A620 of 0.067 (Fig. 2b), but S. mutans cells did not utilize the glycans of RNase B, with no significant increase in absorbance of the culture when compared with unsupplemented minimal media.
FIG. 2.
Growth of S. mutans and S. oralis cells on mannose and RNase B. S. mutans ATCC 25175T (a) and S. oralis ATCC 35037T (b) were cultured in minimal media supplemented with mannose (●) or RNase B (■), and growth was measured by monitoring A620.
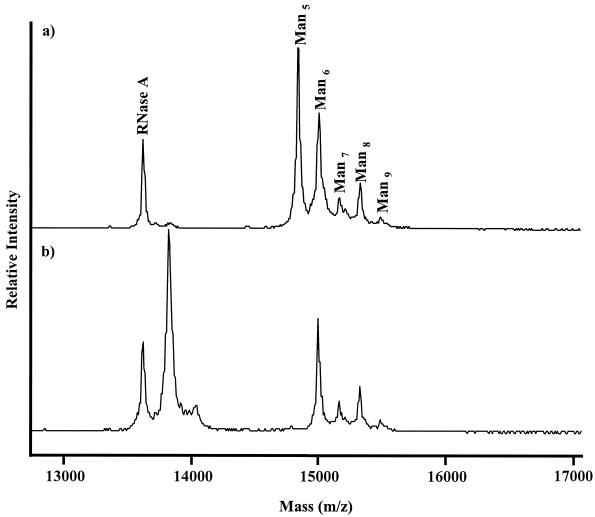
MALDI-TOF mass spectra of the supernatant derived from the spent S. mutans RNase B-supplemented culture demonstrated that Man5 to Man9 glycoforms of the glycoprotein were present, as indicated by the presence of species with m/z ratios of 14,899, 15,061, 15,224, 15,386, and 15,548, respectively (Fig. 3a), and produced a spectrum identical to that of control (uninoculated) minimal medium supplemented with RNase B (data not shown). At the end of the growth period, S. oralis cells had cleaved all constituent mannose residues of RNase B Man5 and further had cleaved the chitobiose core to leave a single N-acetylglucosamine residue attached to the polypeptide backbone, yielding a new product with an m/z of 13,885 (Fig. 3b). The Man6 to Man9 glycoforms remained intact, with a less than 5% change in the peak intensity of these species relative to that of the control.
FIG. 3.
Effect of S. mutans and S. oralis cells on RNase B glycoforms. Spent supernatants from RNase B-supplemented cultures of S. mutans ATCC 25175T (a) and S. oralis ATCC 35037T (b) were analyzed by MALDI-TOF MS with sinapic acid as the matrix. Man5, Man6, Man7, Man8, and Man9 indicate the Man5 to Man9 glycoforms of RNase B with m/z ratios of 14,899, 15,061, 15,224, 15,386, and 15,548, respectively. RNase A is the nonglycosylated form of the protein (m/z, 13,682).
Growth of bacteria on RNase B and mannosidase production monitored by MALDI-TOF MS.
Growth on RNase B was a reliable indicator of the production of mannosidase activity and thus the ability to degrade the Man5 glycoform of the glycoprotein, releasing fermentable carbohydrate (Table 2). For isolates which lacked this ability, the maximum increase in A620 for RNase B-supplemented minimal media was always less than 0.010 when compared with the control cultures without an added carbohydrate source, and no degradation of the RNase B glycoforms (Man5 to Man9) could be detected when culture supernatants were analyzed by MALDI-TOF MS. All isolates grew in glucose-supplemented minimal media (Table 2) and virtually all strains utilized mannose as efficiently as glucose, as judged by the maximum increase in A620 over a 24-h period and the doubling time of the cultures (data not shown). One notable exception to this was the low rate of growth of the Streptococcus peroris isolates on mannose, suggesting that mannose was utilized less efficiently than glucose.
TABLE 2.
Utilization of the Man5 glycoform of RNase B by viridans group streptococci and ability to hydrolyze synthetic mannosidase substrates
| Species groupa | Species (no. of strains) | A620 in basal media with 10 mM glucoseb | A620 in basal media with 5 mg of RNase B/mlb | Degradation of Man5 glycoformc | β-d-Mannosidase productiond | α-d-Mannosidase productiond | β-d-N-acetyl- Glucosaminidase productiond |
|---|---|---|---|---|---|---|---|
| Salivarius | S. salivarius (6) | 0.406 | NDe | −f | − | − | − |
| S. vestibularis (5) | 0.382 | ND | − | − | − | − | |
| Mutans | S. mutans (6) | 0.324 | ND | − | − | − | − |
| S. sobrinus (4) | 0.305 | ND | − | − | − | − | |
| Anginosus | S. anginosus (6) | 0.227 | ND | − | + | − | − |
| S. constellatus (5) | 0.280 | ND | − | − | − | − | |
| S. intermedius (4) | 0.257 | 0.043 | + | + | − | + | |
| Mitis | S. mitis (13) | 0.312 | 0.040 (n = 9) | + | − | − | − |
| ND (n = 4) | − | − | − | − | |||
| S. oralis (6) | 0.326 | 0.074 | + | − | − | + | |
| S. gordonii (5) | 0.323 | 0.051 | + | + | − | + | |
| S. cristatus (4) | 0.230 | 0.042 | + | − | − | + | |
| S. pneumoniae (9) | 0.232 | 0.050 (n = 7) | + | − | − | + | |
| ND (n = 2) | − | − | − | + | |||
| S. sanguinis (5) | 0.292 | ND | − | − | − | + | |
| S. parasanguinis (9) | 0.273 | 0.055 | + | − | − | + | |
| S. infantis (5) | 0.326 | 0.053 (n = 3) | + | − | − | + | |
| ND (n = 2) | − | − | − | − | |||
| S. peroris (3) | 0.372 | ND | − | − | − | − |
Species groups were assigned on the basis of 16S rRNA sequence data (17).
Growth is recorded as the mean maximum increase in A620 for all strains within the group.
Degradation of the Man5 glycoform was monitored by MALDI-TOF MS. Utilization of the Man5 glycoform indicates production of functional α-(1→3), α-(1→6), and β-(1→4) mannosidase and β-d-N-acetylglucosaminidase activities.
Preformed glycosidase activities were monitored using pNP- and MU-linked glycosides at 2 mM.
ND, not detected.
Reactions were given by all strains within the group.
The production of mannosidase activities was restricted to strains within the anginosus and mitis groups of streptococci (Table 2). Among the anginosus group, S. intermedius alone degraded the Man5 glycoform of RNase B. All S. oralis, Streptococcus gordonii, Streptococcus cristatus and Streptococcus parasanguinis strains produced mannosidases, as demonstrated by the A620 for RNase B-supplemented media and the loss of the Man5 glycoform. Production of mannosidase by Streptococcus mitis, Streptococcus pneumoniae, and Streptococcus infantis was variable, with 9 of 13, 7 of 9, and 3 of 5 isolates, respectively, producing mannosidase activity. In all cases, the Man5 glycoform alone was modified and a new peak corresponding to the protein with a single N-acetylglucosamine residue attached was observed, suggesting that, in addition to α-(1→3), α-(1→6), and β-(1→4) mannosidase activities, these isolates also produced N-acetylglucosaminidase activity. Among the mitis group, Streptococcus sanguinis and S. peroris were the only species which did not exhibit Man5-degrading activity (Table 2). For strains degrading the Man5 glycan of RNase B, there was no evidence for proteolytic cleavage of the RNase B polypeptide backbone; no new species with an m/z ratio of less than 13,682 were detected in spent culture supernatants.
Glycosidase activities measured using synthetic substrates.
Measurement of mannosidase activities using chromogenic or fluorogenic substrates revealed a different pattern of production by viridans group streptococci, and there was no correlation between the production of these enzymic activities and the ability to degrade RNase B (Table 2). None of the isolates investigated degraded 4MU–α-d-mannopyranoside or pNP–α-d-mannopyranoside, all strains yielding specific activities of less than 0.5 nmol/h/mg of bacterial protein. 4MU–β-d-mannopyranoside and pNP–β-d-mannopyranoside degradation was limited to isolates of S. gordonii, Streptococcus anginosus, and S. intermedius, with all strains degrading these substrates. Specific activities for the degradation of 4MU–β-d-mannopyranoside within these isolates ranged from 1.1 to 9.8 nmol/h/mg of bacterial protein (data not shown). All strains which degraded the Man5 glycan produced N-acetylglucosaminidase activity, which was detectable using either the respective chromogenic or fluorogenic substrates, with the notable exception of S. mitis strains, which did not hydrolyze this substrate. All isolates which degraded the Man5 glycoform grew when N-acetylglucosamine was provided as the supplement to minimal media (data not shown).
DISCUSSION
In our earlier studies with S. oralis strain AR3, we demonstrated the presence of α- and β-mannosidase activities which cleaved mannose residues from the core pentasaccharide of the complex N-glycans of human serum α1-acid glycoprotein (4). Subsequently, we used bovine pancreatic RNase B to show that of all the high-mannose glycans (Man5 to Man9), the Man5 glycoform alone was degraded during in vitro growth of S. oralis cells in what appeared to be a single-step reaction involving these α- and β-mannosidases (30), and the mannosidase activities were undetectable with synthetic substrates. We now extend these observations to all currently recognized species of human viridans group streptococci by using RNase B as a model. The ability to utilize the high-mannose glycans of RNase B as sole carbohydrate sources during in vitro growth was restricted to the mitis group (S. oralis, S. mitis, Streptococcus parasanguinis, S. gordonii, S. cristatus, S. infantis, and S. pneumoniae) and one species within the anginosus group, namely S. intermedius. Analysis of the spent culture supernatants from these organisms by MALDI-TOF MS of RNase B demonstrated that only the Man5 glycoform was degraded, yielding a product corresponding to the polypeptide backbone with a single N-acetylglucosamine residue attached. This degradation may be accounted for by the production of α-(1→3), α-(1→6), and β-(1→4) mannosidase activities and a β-N-acetylglucosaminidase activity cleaving the chitobiose core. The lack of ability to degrade glycoforms higher than Man5 has been ascribed to the lack of an α-(1→2) mannosidase in S. oralis (30), and our data indicate that all viridans group streptococci degrading RNase B do so via a mechanism similar to that observed for S. oralis. In addition, the degradation of the Man5 glycoform of RNase B is brought about without production of a detectable endoglycosidase activity (30), thus differing from the mechanism by which these glycans are utilized by Enterococcus faecalis (23), formerly part of the genus Streptococcus.
These mannosidase activities were not detected using synthetic substrates. β-Mannosidase activity was detected in only a limited number of species of the viridans group streptococci (S. gordonii, S. intermedius, and S. anginosus) when assayed using synthetic 4MU-linked or pNP-linked substrates, and these data were consistent irrespective of the leaving group or the concentration of substrate presented to bacterial cell suspensions. This extends the observations of Kilian et al. (20), who reported the activity in S. gordonii and S. anginosus only when using a chromogenic substrate. In addition, we have used the corresponding α-linked mannosides as substrates and demonstrated that none of the viridans group streptococci had the capacity to degrade these substrates. α-Mannosidases from yeast or mammalian sources which cleave α-(1→2) mannose residues from native glycoproteins or free glycans have previously been shown to lack activity when pNP–α-d-mannoside or 4MU–α-d-mannoside was presented as a substrate (26) and that synthetic substrates may underrepresent the incidence of this enzyme activity. Given the failure of synthetic substrates to detect mannosidases, a number of strategies have been adopted for the measurement of the these enzymes, including monitoring the release of mannose from free oligosaccharides and resolving enzyme-treated glycans using high-performance liquid chromatography methodologies (26, 34). In the present study, we have utilized MALDI-TOF MS to monitor RNase B degradation. MALDI-TOF MS is particularly applicable to the rapid analysis of biological samples due to its ability to tolerate high concentrations of salts and buffers as encountered in biological systems and is relatively rapid. This methodology has recently been shown to be applicable to the detection of mannosidases acting on RNase B (31) and has been applied here to investigate the presence of mannosidase activities within the viridans group streptococci.
The recent availability of genomic sequence data for a number of bacterial species, including S. pneumoniae (http://www.tigr.org/tdb, The Institute for Genomic Research) has facilitated investigations linking genome organization with the production of functional proteins. Interrogation of these data using the WIT suite of software (WIT; http://wit.integratedgenomics.com/IGwit) indicates the presence of an open reading frame in S. pneumoniae coding for a protein with homology to an hypothetical α-mannosidase present in E. faecalis and Streptococcus pyogenes (P score = 1.17 × 10−115 and 6.86 × 10−111, respectively). These in silico data are in accordance with our demonstration of functional α-mannosidase in this organism and other closely related species. There is no evidence for a β-mannosidase in this genomic data, but this may form one of a number of genes for which no function can be ascribed at present. Further biochemical characterization of mannosidase of viridans group streptococci and the substrate specificity of the enzyme(s) will be required to elucidate the mechanism by which this degradation is achieved.
The function of mannosidases produced by viridans group streptococci is unclear. High-mannose-type glycans are frequent constituents of host tissues and are found as components of the extracellular matrix, e.g., laminin, being involved in a wide range of cellular functions (11, 25, 27). In addition, the major platelet integrin, GP IIb/IIIa, contains high-mannose-type oligosaccharides, predominantly the Man5 and Man6 glycoforms (33). Degradation of the Man5 glycoform of human glycoproteins in vivo may not only modulate their function but also release carbohydrates and play a role in the persistence of viridans group streptococci at infection sites. We have demonstrated that mannosidase activities are produced by major pathogens within the viridans group of streptococci, including S. oralis and S. mitis, which are associated with infective endocarditis and infections in immunocompromised patients, S. pneumoniae, which is the cause of pneumonia, otitis media, and meningitis, and S. intermedius, which is associated with purulent abscesses of the liver and brain (3, 12, 13, 22, 37). The effect of mannosidase activities on host tissues and high-mannose-type glycoproteins remains to be established. The ability of viridans group streptococci to grow on RNase B, however, may be a useful test to assist in the identification of this complex group of microorganisms.
ACKNOWLEDGMENTS
This work was supported in part by grant PG/97015 from the British Heart Foundation.
Y. Kawamura (Department of Microbiology, Gifu University School of Medicine, Gifu, Japan) is thanked for providing S. infantis and S. peroris strains.
REFERENCES
- 1.Beighton D, Carr A D, Oppenheim B A. Identification of viridans streptococci associated with bacteraemia in neutropenic cancer patients. J Med Microbiol. 1994;40:202–204. doi: 10.1099/00222615-40-3-202. [DOI] [PubMed] [Google Scholar]
- 2.Beighton D, Hardie J M, Whiley R A. A scheme for the identification of viridans streptococci. J Med Microbiol. 1991;35:367–372. doi: 10.1099/00222615-35-6-367. [DOI] [PubMed] [Google Scholar]
- 3.Bochud P-Y, Calandra T, Francioli P. Bacteremia due to viridans streptococci in neutropenic patients: a review. Am J Med. 1994;97:256–264. doi: 10.1016/0002-9343(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 4.Byers H L, Tarelli E, Homer K A, Beighton D. Sequential deglycosylation and utilization of the N-linked, complex-type glycans of human α1-acid glycoprotein mediates growth of Streptococcus oralis. Glycobiology. 1999;9:469–479. doi: 10.1093/glycob/9.5.469. [DOI] [PubMed] [Google Scholar]
- 5.Coykendall A L. Classification and identification of the viridans streptococci. Clin Microbiol Rev. 1989;2:275–286. doi: 10.1128/cmr.2.3.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douglas C W I. Pathogenic mechanisms in infective endocarditis. Rev Med Microbiol. 1993;4:130–137. [Google Scholar]
- 7.Douglas C W I, Heath J, Hampton K K, Preston F E. Identity of viridans streptococci isolated from cases of infective endocarditis. J Med Microbiol. 1993;39:179–182. doi: 10.1099/00222615-39-3-179. [DOI] [PubMed] [Google Scholar]
- 8.Frandsen E V, Pedrazzoli V, Kilian M. Ecology of viridans streptococci in the oral cavity and pharynx. Oral Microbiol Immunol. 1991;6:129–133. doi: 10.1111/j.1399-302x.1991.tb00466.x. [DOI] [PubMed] [Google Scholar]
- 9.French G L, Talsania H, Charlton J R H, Phillips I. A physiological classification of viridans streptococci by use of the API-20STREP system. J Med Microbiol. 1989;28:275–286. doi: 10.1099/00222615-28-4-275. [DOI] [PubMed] [Google Scholar]
- 10.Fu D T, Chen L, O'Neill R A. A detailed structural characterization of ribonuclease-B oligosaccharides by H1-NMR spectroscopy and mass spectrometry. Carbohydr Res. 1994;261:173–186. doi: 10.1016/0008-6215(94)84015-6. [DOI] [PubMed] [Google Scholar]
- 11.Fujiwara S, Shinkai H, Deutzmann R, Paulsson M, Timpl R. Structure and distribution of N-linked oligosaccharide chains on various domains of mouse tumor laminin. Biochem J. 1988;252:453–461. doi: 10.1042/bj2520453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardie J M, Whiley R A. Recent developments in streptococcal taxonomy: their relation to infection. Rev Med Microbiol. 1994;5:151–162. [Google Scholar]
- 13.Hardie J M, Whiley R A. Classification and overview of the genera Streptococcus and Enterococcus. Soc Appl Bacteriol Symp Ser. 1997;26:1S–11S. [PubMed] [Google Scholar]
- 14.Hogg S D, Whiley R A, de Soet J J. Occurrence of lipoteichoic acid in oral streptococci. Int J Syst Bacteriol. 1997;47:62–66. doi: 10.1099/00207713-47-1-62. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs J A, Schouten H C, Stobberingh E E, Soeters P B. Viridans streptococci isolated from the bloodstream. Relevance of species identification. Diagn Microbiol Infect Dis. 1995;22:267–273. doi: 10.1016/0732-8893(95)00137-y. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs J A, Stobberingh E E. Hydrolytic enzymes of Streptococcus anginosus, Streptococcus constellatus and Streptococcus intermedius in relation to infection. Eur J Clin Microbiol Infect Dis. 1995;14:818–820. doi: 10.1007/BF01691002. [DOI] [PubMed] [Google Scholar]
- 17.Kawamura Y, Hou X G, Sultana F, Miura H, Ezaki T. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J Syst Bacteriol. 1995;45:406–408. doi: 10.1099/00207713-45-2-406. [DOI] [PubMed] [Google Scholar]
- 18.Kawamura Y, Hou X G, Todome Y, Sultana F, Hirose K, Shu S E, Ezaki T, Ohkuni H. Streptococcus peroris sp. nov. and Streptococcus infantis sp. nov., new members of the Streptococcus mitis group, isolated from human clinical specimens. Microbiology. 1998;48:921–927. doi: 10.1099/00207713-48-3-921. [DOI] [PubMed] [Google Scholar]
- 19.Kikuchi K, Enari T, Totsuka K, Shimizu K. Comparison of phenotypic characteristics, DNA-DNA hybridization results, and results with a commercial rapid biochemical and enzymatic reaction system for identification of viridans group streptococci. J Clin Microbiol. 1995;33:1215–1222. doi: 10.1128/jcm.33.5.1215-1222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kilian M, Mikkelsen L, Henrichsen J. Taxonomic study of viridans streṕtococci: description of Streptococcus gordonii sp. nov. and emended descriptions of Streptococcus sanguis (White and Niven 1946), Streptococcus oralis (Bridge and Sneath 1982), and Streptococcus mitis (Andrewes and Horder 1906) Int J Syst Bacteriol. 1989;39:471–484. [Google Scholar]
- 21.Lacks S, Hotchkiss R D. A study of the genetic material determining an enzyme activity in the pneumococcus. Biochim Biophys Acta. 1960;39:508–518. doi: 10.1016/0006-3002(60)90205-5. [DOI] [PubMed] [Google Scholar]
- 22.Richard P, Amador Del Valle G, Moreau P, Milpied N, Felice M P, Daeschler T, Harousseau J L, Richet H. Viridans streptococcal bacteraemia in patients with neutropenia. Lancet. 1995;345:1607–1609. doi: 10.1016/s0140-6736(95)90117-5. [DOI] [PubMed] [Google Scholar]
- 23.Roberts G, Tarelli E, Homer K A, Philpott-Howard J, Beighton D. Production of an endo-β-N-acetylglucosaminidase activity mediates growth of Enterococcus faecalis on a high-mannose-type glycoprotein. J Bacteriol. 2000;182:882–890. doi: 10.1128/jb.182.4.882-890.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rudd P M, Scragg I G, Coghill E, Dwek R A. Separation and analysis of the glycoform populations of ribonuclease B using capillary electrophoresis. Glycoconj J. 1992;9:86–91. doi: 10.1007/BF00731704. [DOI] [PubMed] [Google Scholar]
- 25.Sathyamoorthy N, Decker J M, Sherblom A P, Muchmore A. Evidence that specific high mannose structures directly regulate multiple cellular activities. Mol Cell Biochem. 1991;102:139–147. doi: 10.1007/BF00234571. [DOI] [PubMed] [Google Scholar]
- 26.Scaman C H, Lipari F, Herscovics A. A spectrophotometric assay for alpha-mannosidase activity. Glycobiology. 1996;6:265–270. doi: 10.1093/glycob/6.3.265. [DOI] [PubMed] [Google Scholar]
- 27.Sherblom A P, Smagula R M. High-mannose chains of mammalian glycoproteins. Methods Mol Biol. 1993;14:143–149. doi: 10.1385/0-89603-226-4:143. [DOI] [PubMed] [Google Scholar]
- 28.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 29.Sriskandan S, Soto A, Evans T J, Cohen J. Viridans streptococcal bacteraemia: a clinical survey. Q J Med. 1995;88:415–420. [PubMed] [Google Scholar]
- 30.Tarelli E, Byers H L, Homer K A, Beighton D. Evidence for mannosidase activities in Streptococcus oralis when grown on glycoproteins as carbohydrate source. Carbohydr Res. 1998;312:159–164. doi: 10.1016/s0008-6215(98)00246-8. [DOI] [PubMed] [Google Scholar]
- 31.Tarelli E, Byers H L, Wilson M, Roberts G, Homer K A, Beighton D. Detecting mannosidase activities using ribonuclease B and matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Anal Biochem. 2000;282:165–172. doi: 10.1006/abio.2000.4606. [DOI] [PubMed] [Google Scholar]
- 32.Tomasz A, Hotchkiss R D. Regulation of the transformability of pneumococcal cultures by macromolecular cell products. Proc Natl Acad Sci USA. 1964;51:480–487. doi: 10.1073/pnas.51.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuji T, Osawa T. Structures of the carbohydrate chains of membrane glycoproteins IIb and IIIa of human platelets. J Biochem. 1986;100:1387–1398. doi: 10.1093/oxfordjournals.jbchem.a121845. [DOI] [PubMed] [Google Scholar]
- 34.Tyagarajan K, Forte J G, Townsend R R. Exoglycosidase purity and linkage specificity: assessment using oligosaccharide substrates and high-pH anion-exchange chromatography with pulsed amperometric detection. Glycobiology. 1996;6:83–93. doi: 10.1093/glycob/6.1.83. [DOI] [PubMed] [Google Scholar]
- 35.West P W J, Al-Sawan R, Foster H A, Electricwala Q, Alex A, Panigrahi B. Speciation of presumptive viridans streptococci from early onset neonatal sepsis. J Med Microbiol. 1998;47:923–928. doi: 10.1099/00222615-47-10-923. [DOI] [PubMed] [Google Scholar]
- 36.Whiley R A, Beighton D. Emended descriptions and recognition of Streptococcus constellatus, Streptococcus intermedius, and Streptococcus anginosus as distinct species. Int J Syst Bacteriol. 1991;41:1–5. doi: 10.1099/00207713-41-1-1. [DOI] [PubMed] [Google Scholar]
- 37.Whiley R A, Beighton D. Current classification of the oral streptococci. Oral Microbiol Immunol. 1998;13:195–216. doi: 10.1111/j.1399-302x.1998.tb00698.x. [DOI] [PubMed] [Google Scholar]
- 38.Whiley R A, Beighton D, Winstanley T G, Fraser H Y, Hardie J M. Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus (the Streptococcus milleri group): association with different body sites and clinical infections. J Clin Microbiol. 1992;30:243–244. doi: 10.1128/jcm.30.1.243-244.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whiley R A, Fraser H, Hardie J M, Beighton D. Phenotypic differentiation of Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus strains within the “Streptococcus milleri group.”. J Clin Microbiol. 1990;28:1497–1501. doi: 10.1128/jcm.28.7.1497-1501.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]