Abstract
Here, based on the design of rational primers and copper nanoclusters (CuNCs), we present a method for the accurate detection of the SARS-CoV-2 Delta variant, which is capable of distinguishing the Delta variant with its single nucleotide polymorphism from the ‘wild type’ coronavirus (NC_045512.2), and realizing visualization signal out. Specifically, we show that dual priming oligonucleotide (DPO) primers and AT primers can be used to distinguish between wild types and mutations of this virus by polymerase chain reaction (PCR) analysis and that visualization can be achieved via the red fluorescence of CuNCs in ultraviolet radiation. Among the results, it was found that the R-1-down (DPO)-6I and F-1-30 AT, with the single nucleotide deletion site designed at the 3′ end of the downstream primer, showed the best specificity towards the Delta variant. Moreover, the use of AT primers increased the AT contents of the PCR products, thus meeting the template requirements generated by the CuNCs. It was also found that the AT primers could assist with improving detection specificity. Finally, we demonstrate that the visualization of the CuNCs-based detection assay exhibited a linear relationship in 0.5 pg μL−1–50 ng μL−1, with a limit of quantitation (LOQ) of 0.5 pg μL−1.
Keywords: SARS-CoV-2, Delta variant, Design of rational primers, Copper nanocluster
Graphical abstract
1. Introduction
Data shared by the World Health Organization (WHO) indicates that by October 1, 2021 more than 235 million people worldwide had tested positive for the SARS-CoV-2 (COVID-19) infection, with over 4.8 million confirmed related deaths. SARS-CoV-2 belongs to the genus β-coronavirus family of positive-sense single-stranded RNA (+ssRNA) viruses [1,2]. To optimize their survival, RNA viruses frequently adopt mechanisms of genetic variation through mutation, thereby increasing the rates of infectivity and mortality [3,4]. According to the Global initiative on sharing all influenza data database, to date four variant categories have been reported, namely the Alpha (United Kingdom, B.1.1.7), Beta (South African, B.1.351), Gamma (Brazilian, P.1) and Delta (Indian Double Mutant, B.1.617.2) variants (http://www.bioticainternational.com). Of these, the Delta coronavirus has been found to exhibit comparatively stronger heat resistance, transmission and viral load, and may even have immune escape capabilities, all providing this variant with strong pathogenicity [5]. Given the high human-to-human transmission potential of SARS-CoV-2, including from asymptomatic carriers, rapid and accurate diagnosis is critical for timely treatment and control of the outbreak [6,7]. However, it remains unclear how best to use the existing conditions and methods to achieve the accurate detection of SARS-CoV-2 variant strains and this issue requires urgent resolution.
At present, the detection of SARS-CoV-2 includes immune response tests, such as antigen and antibody detection [8] and virology tests, including nucleocapsid protein (NP) and nucleotide detection assays. Although an immune response test is generally immediate and inexpensive, it presents the challenge of false-negative diagnoses due to the variations in people's individual immune systems. As an antigen, NP is released into mainly the blood, saliva, nasopharynx, feces and urine in the early stage of coronavirus infection, and the expression of NP in SARS-CoV-2 is lower than in SARS-CoV or MERS-CoV. Moreover, especially in the receptor-binding domain and NP detection of SARS-CoV-2, its amino acid sequence is very similar to that of SARS-CoV, making it prone to misjudgments. The most common method of nucleotide detection is that used to determine the RNA sequence of the virus by reverse transcription-polymerase chain reaction (RT-PCR), which is also recognized as the current ‘gold standard’ [9,10]. More than 30 types of RT-PCR test kits have been approved by the United States Food and Drug Administration (FDA), however, their positive detection rates are reportedly only 30–60% accurate (https://www.fda.gov/MedicalDevices/Safety/EmergencySituations/ucm161496. htm). Dong et al. [4] found that, in 196 clinical samples, the RT-digital PCR method was comparatively more accurate and sensitive in the diagnosis of SARS-CoV-2 than RT-qPCR. While the above mature detection methods can realize the rapid and accurate detection of SARS-CoV-2, the visual distinction of the SARS-CoV-2 Delta variant, especially those with single nucleotide deletion, has become a problem that requires an urgent solution.
The characteristic of copper nanoclusters (CuNCs) to produce red fluorescence under ultraviolet (UV) radiation has attracted wide attention and applications of scientists in biosensor design [11,12]. Ligands such as nucleic acids [13], cysteine [14,15], and bovine serum albumin [16,17] can generate CuNCs in the presence of reducing agents. Among T-rich single-stranded sequences [18] and AT-rich double-stranded sequences [19] also can be used as specificity templates for the generation of CuNCs with visualization. In addition, Gorthi et al. also reported that the combination of sodium ascorbate and Taq buffer can also enhance the fluorescence of CuNCs [20,21]. Therefore, we associated with PCR products as the nucleic acid templates of CuNCs to realize the visualization detection.
Under the existing conditions, in order to improve the specificity of detection single nucleotide deletion, dual priming oligonucleotide (DPO) primers have been adopted. A DPO primer is comprised of two separate priming regions joined by a polydeoxyinosine linker [22]. Since its discovery in 2007, it has been widely used in the detection of multiple viruses, including those with single nucleotide polymorphism (SNP), with high specificity and sensitivity [23,24].
In this study, DPO primers were used to realize the highly sensitive detection of single nucleotide deletion by existing experiment conditions and equipment. As well as, AT primers increased the AT contents of the PCR products, thus meeting the template requirements generated by the CuNCs. Then, a method based on the design of rational primers and utilization of CuNCs for the accurate detection of the SARS-CoV-2 Delta variant is presented, which can distinguish between the single nucleotide deletion virus and wildtype coronavirus, and can also realize visualization with specificity.
2. Materials and methods
2.1. Materials and primers
TransStart® Green qPCR SuperMix was purchased from the TransGen Biotech Co. Ltd, (Beijing, China). The primers based on target sequences were designed using the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov). All oligonucleotides were synthesized by Sangon Biotech (Shanghai) Co. Ltd. (Shanghai, China), and purified using high performance liquid chromatography (HPLC). The sequences used in this work are shown in Table 1 . Ultrapure 3-(N-morpholino) propanesulfonic acid (MOPS), sodium chloride (NaCl), copper sulfate (CuSO4) and sodium ascorbate (SA) were of analytical grade and commercially obtained from the Sigma-Aldrich Chemical Company (Merck Sigma-Aldrich, St. Louis, MO, USA). Deionized water was prepared using a Millipore Milli-Q system (Merck Millipore, USA).
Table 1.
Sequences of primers in experiments.
| Name | Sequences (5′-3′) | Length (nt) | Tm |
|---|---|---|---|
| F-1 | CTTGTGGACAACAGCAGACAAC | 22 | 56.6 °C |
| R-1 | TTCATACTGACAGGTGGTGCT | 21 | 56 °C |
| R-1-down (DPO) | CACAAGTAAATGTACCATGCTTAAGIIIIIACTGACAG | 38 | 64.4 °C |
| R-1-down (PT) | CACAAGTAAATGTACCATGCTTAAGTTCATACTGACAG | 38 | 59.9 °C |
| F-1-15 AT | ATATATATATATATACTTGTGGACAACAGCAGACAAC | 37 | 56.3 °C |
| F-1-20 AT | ATATATATATATATATATATCTTGTGGACAACAGCAGACAAC | 42 | 56.3 °C |
| F-1-25 AT | ATATATATATATATATATATATATACTTGTGGACAACAGCAGACAAC | 47 | 56.4 °C |
| F-1-30 AT | ATATATATATATATATATATATATATATATCTTGTGGACAACAGCAGACAAC | 52 | 56.7 °C |
| R-1-down (DPO)-3′1 | CACAAGTAAATGTACCATGCTTAAGIIIIIACTGAC | 36 | 63.5 °C |
| R-1-down (DPO)-3′2 | CACAAGTAAATGTACCATGCTTAAGIIIIIACTGACA | 37 | 64.3 °C |
| R-1-down (DPO)-3′3 | CACAAGTAAATGTACCATGCTTAAGIIIIIACTGACAG | 38 | 64.4 °C |
| R-1-down (DPO)-poly6I | CACAAGTAAATGTACCATGCTTAAGIIIIIIACTGACAG | 39 | 65.3 °C |
| R-1-down (DPO)-poly4I | CACAAGTAAATGTACCATGCTTAAGIIIIACTGACAG | 37 | 63.4 °C |
| R-1-down (DPO)-poly3I | CACAAGTAAATGTACCATGCTTAAGIIIACTGACAG | 36 | 62.3 °C |
| R-1-down (DPO)-6I | CACAAGTAAATGTACCATGCTTAAIIIIIIACTGACAG | 38 | 64.3 °C |
| R-1-down (DPO)-4I | CACAAGTAAATGTACCATGCTTAAGTIIIIACTGACAG | 38 | 63.3 °C |
| R-1-down (DPO)-3I | CACAAGTAAATGTACCATGCTTAAGTTIIIACTGACAG | 38 | 63.3 °C |
| F-2 | GTCAGCACCACCTGTCAGTA | 20 | 56.7 °C |
| R-2 | AGTAAAGCACCGTCTATGCAA | 21 | 54.3 °C |
| F-2-up(DPO) | AGGAGTCACCTTTTGTTATGATGTCAGIIIIICCTGTCAG | 40 | 68.1 °C |
| F-2-up(PT) | AGGAGTCACCTTTTGTTATGATGTCAGCACCACCTGTCAG | 40 | 66 °C |
2.2. Contrived sample preparation
To facilitate this study, an artificial plasmid model was constructed to validate the methodology. Genes fragments (wild-SARS-CoV-2, Mut-SARS-CoV-2, FluA, MERS, rhinovirus, and SARS-CoV-1) were cloned into TOPO vectors with a T7 primer, using a TOPO PCR cloning kit (Beijing, China) (Table S1) [25]. The vectors were obtained via a TIANpure Midi Plasmid Kit (Tiangen Biotech Co, Ltd.). DNA concentrations were measured using NanoDrop 2000 and all the DNA samples were stored at 4 °C.
2.3. qPCR system
The PCR reaction mixture (25 μL) contained 1 × SuperMix, 600 nM each of the primers, and double distilled water (ddH2O). The real-time PCR assay was performed using a PCR amplifier (Gene 9000, Hangzhou Bioer Technology Co. Ltd, China) for 3 min at 93 °C, followed by 30 cycles of denaturation for 30 s at 95 °C, annealing for 30 s at 65 °C, and extension for 30 s at 72 °C, while fluorescence intensity was measured during the annealing phase at 65 °C. To enhance the sensitivity of the PCR, the annealing temperature and the sequences of the DPO primers and AT primers were optimized. All samples in this experiment are three parallels, and the results were shown as mean ± standard error of the mean.
2.4. Copper nanoclusters formed by PCR products
Next, 10 μL of the PCR products was mixed with 10 μL of 1 mM CuSO4 60 μL of MOPS buffer (10 mM MOPS, 150 mM NaCl, pH 7.6) and 20 μL of 10 mM SA, and the mixture was shaken immediately, according to Yu's experimental method [26,27], with partial modifications. Fluorescence measurements were performed at room temperature using a Cary Eclipse BioMelt Bundle (G9802AA, Agilent Technologies, USA) at an excitation wavelength of 340 nm. The fluorescence emissions of the reaction were recorded from 550 nm to 650 nm and, by directly using UV light, the colors of the sample could also be observed by the naked eye.
2.5. Sensitivity and specificity to SARS-CoV-2 Delta variant
In order to verify the performance of our method, 10-fold gradient dilution templates which is in vitro transcribed RNA samples (in Supporting information) were selected for a sensitivity assay by real-time RT-PCR and a visualization CuNCs-based detection assay. Thereafter, four pathogens of respiratory diseases, namely FluA, MERS, rhinovirus, and SARS-CoV-1, were used for specificity detection. The concentration of the SARS-CoV-2 Delta variant was 5 ng μl−1, while the other templates were 100 ng μl−1.
3. Results
3.1. Design principle
This study involves the application of rational primers design, including DPO primers and AT primers, for PCR amplification to realize the highly sensitive detection of the SARS-CoV-2 Delta variant. DPO primers contain two separate priming regions joined by a polydeoxyinosine linker, which can accurately identify SNP; and AT primers artificially add AT sequences at the 5′ end of primers, which can not only improve the AT contents of PCR products and be conducive to the generation of CuNCs, but also provide the unexpected function of improving detection specificity. In this novel process, shown in Fig. 1 A, the cDNA of the virus target is obtained through reverse transcription, whereafter conventional PCR amplification is carried out through the rational designed DPO and AT primers, and finally the PCR amplification products are generated into CuNCs to realize the visual detection of the SARS-CoV-2 Delta variant. It is especially important to note that the Delta variant sequence used in this experiment was only that of the single nucleotide deletion from the wild-SARS-CoV-2 sequences, as shown on the right side of Fig. 1A. Compared with the SARA-COV-2 sequence (Genebank: NC_045512.2), the SARS-CoV-2 Delta variant (B.1.617.2, Genebank: OK091006.1) sequence has many mutations and SNP site. The mutation site selected in this study is located at the 5700th base deletion on ORF1ab, which intensively affects the infectivity and pathogenicity of Delta variant. The PCR process of rational primers design in this study is consistent with that of traditional PCR, which is dependent on PCR amplification and procedure (Fig. 1B). Final results were evaluated according to the Ct value of the amplification curves.
Fig. 1.
Schematic of SARS-CoV-2 Delta variant detection: (A) complete detection process; (B) real-time PCR process.
3.2. Feasibility analysis
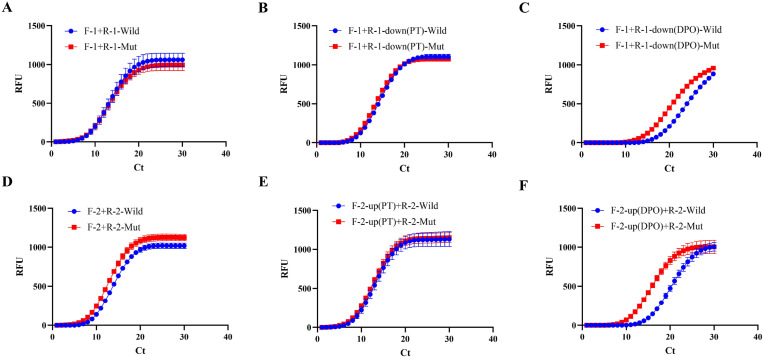
The effects of the primer design were assessed using real-time PCR. First, two pairs of DPO primers were designed as experiment groups, namely F-1 and R-1-down (DPO), and F-2-up (DPO) and R-2. Furthermore, F-1 and R-1, F-1 and R-1-down (PT), F-2 and R-2, and F-2-up (PT) and R-2 were selected as the control groups. Compared with the control groups, a gap was distinguished between the DPO primers’ amplification curves in the mutation and wild-type coronaviruses by real-time PCR (Fig. 2 A–F). In contrast, different type coronaviruses could not be distinguished by the DPO primers in agarose gel electrophoresis (Fig. S1). Thus, it is evident that the SNP of the single nucleotide deletion posed a significant challenge to the specificity capability of the DPO primers.
Fig. 2.
Real-time PCR amplification curves of experiment groups and control groups: (A) F-1 and R-1; (B) F-1 and R-1-down (PT); (C) F-1 and R-1-down (DPO); (D) F-2 and R-2; (E) F-2-up (PT) and R-2; (F) F-2-up (DPO) and R-2. Red lines indicate the Delta mutation type virus and blue lines indicate the wild-type SARS-CoV-2. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Annealing temperature optimization
The annealing temperature of the two pairs of primers was optimized to ensure the specificity of detection of the single base deletion. As can be seen in the graphs in Fig. 3 A-F, the PCR amplification curves gap between the mutation and wild-type SARS-CoV-2 by the F-1 and R-1-down (DPO) became increasingly obvious as the temperature increased. Similar results were also found in the amplification curves of the F-2-up (DPO) and R-2 (Fig. S2 A-F). These results indicated that the annealing temperature of 65 °C was best for the discrimination of the Delta variant. In order to intuitively analyze the experimental results, GraphPad Prism software was used to process the Ct values of the mutant and wild type coronavirus, ΔCt = Ctwild-Ctmut, the results of which are shown in Fig. 3 G-H. The ΔCt values of the R-1-down (DPO) and F-2-up (DPO) reached 7.1 and 9.3 respectively, 5.6 and 7 times higher, respectively, then those at 55 °C and with greater specificity in the detection. However, subsequent studies showed that the AT primers could also increase ΔCt, and the results of the downstream primers containing polyxanthine were better than those of the upstream primers. Therefore, F-1 and R-1-down (DPO) primers were optimized for subsequent investigations. When F-1 and R-1-down (DPO) were 0.6 μM in the system, the amplification effect was observed to be optimal, therefore, 0.6 μM was selected as the basis for subsequent optimization (Fig. S3).
Fig. 3.
Optimization of annealing temperature: (A–F) different templates' amplification curves of F-1 and R-1-down (DPO) primer at 55 °C, 56.8 °C, 59.1 °C, 61.5 °C, 63.9 °C, and 65 °C; (G) ΔCt values of F-1 and R-1-down (DPO) at different annealing temperatures; (H) ΔCt values of F-2-up (DPO) and R-2 in different annealing temperature. The blue line is the amplification curves of the “wild type”, and the red line is the amplification curves of the SARS-CoV-2 Delta variant. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. DPO primer optimization
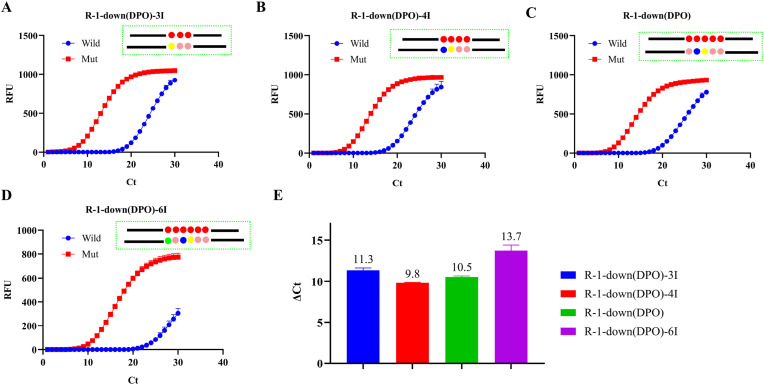
The rigorous challenge of single nucleotide deletion necessitated careful design of the base arrangement of the DPO primers. Previous results showed that the length of the 5' end sequence of DPO primers was 18–25 base, which was determined by the 3′ end sequence [22]. Consequently, this paper optimized mainly the site of the DPO primers’ 3′ end deletion bases and the number of deoxyinosine bases. Fig. S4 shows that R-1-down (DPO) that is 8 bases at the 3′ end and 50% guanine-cytosine (GC) content were the most suitable for these assays, with the deletion base located in middle site. The DPO primers recognized the SNP, relying mainly on polydeoxyinosine to interact with weak bases complementary to A, C and U through hydrogen bonds, thereafter achieving single nucleotide mutation [28]. It is evident from Fig. 4 A-D that the value ΔCt increased with the escalation of polydeoxyinosine. The ΔCt of R-1-down (DPO)-6I was 9.2 (Fig. 4 E), which proved that the specific recognition effect of R-1-down (DPO)-6I is better for Delta variant strains. With the increase of the number of hypoxanthine bases, it can not only improve the specificity of detection, but also have a certain impact on the sensitivity, so this study did not continue to increase the number of hypoxanthine bases. Therefore, R-1-down (DPO)-6I was selected as the best primer. At the same time, the primers that changed the number of polyhypoxanthine bases without changing the sequences of the 5′ and 3′ ends of DPO primers were also optimized (Fig. S5). It can be seen from the results that hypoxanthine bases can better identify Delta variant strains even if they are not complementary to template bases, thus indicating that the design of DPO primers in this method exhibited high tolerance and were more suitable for the popularization of SNP detection.
Fig. 4.
Optimization of DPO primers: (A) F-1 and R-1-down (DPO)-3I; (B) F-1 and R-1-down (DPO)-4I; (C) F-1 and R-1-down (DPO); (D) F-1 and R-1-down (DPO)-6I; (E) Data processing results of all four pairs of primers.
3.5. AT primer optimization
Although the novel DPO primers’ design principle was found to effectively distinguish between the wild-type SARS-CoV-2 and the Delta variant, precision real-time detection equipment is required to realize the discrimination effect. Here, a novel visualization method for the detection of the Delta variant was developed based on CuNCs under UV. As the aforementioned PCR products could not meet the requirements of CuNCs for the template AT content, the AT base sequence was artificially added at the 5′ end of the specific primers, with reference to the method of Chen et al. [29]. Unexpectedly, when the AT contents gradually increased, so too did the ΔCt, indicating that the specificity of detection became increasingly more effective (Fig. 5 ). In addition, the PCR products produced by different AT primers were used to generate CuNCs, the results of which are shown in (Fig. S6). The higher the content of AT primers, the higher the fluorescence of CuNCs, which is consistent with previously reported results [13]. The visualization effect of the CuNCs under UV irradiation can be seen in Fig. S6, and F-1-30 AT was clearly visible to the naked eye.
Fig. 5.
Optimization of AT primers: (A) F-1 and R-1-down (DPO)-6I; (B) F-1-15 AT and R-1-down (DPO)-6I; (C) F-1-20 AT and R-1-down (DPO)-6I; (D) F-1-25 AT and R-1-down (DPO)-6I; (E) F-1-30 AT and R-1-down (DPO)-6I; (F) Data processing results of all five pairs of primers.
3.6. Conditions for the optimization of CuNC generation
To analyze the visualization of the detection methods, we measured the fluorescence of CuNCs using UV. As the CuNCs were generated from precursors and the reducing agent, the concentrations of CuSO4 and SA were crucial for the reaction. As shown in Fig. 6 A, C, the CuNCs were maximally generated when the concentration of CuSO4 was 1 mM, however, no CuNCs were generated when the concentration of CuSO4 was reduced to 0.1 mM. Similarly, when the concentration of SA was reduced to 1 mM, Cu2+ could not be effectively reduced to form nanoclusters. Since the reduction effects were seen to be similar between 5 mM and 100 mM, 10 mM SA was selected as the subsequent reduction concentration (Fig. 6 B, D). In addition, this reaction was found to be rapid, with a stable fluorescence signal generated within 2 min (Fig. S7), thus enabling the rapid visualization of PCR products and providing the possibility for in situ detection.
Fig. 6.
Optimization of PCRs-CuNCs generation conditions: (A) Fluorescence amplification curve optimized for CuSO4 concentrations; (B) Fluorescence amplification curve optimized for SA concentrations; (C) Optimized result of CuSO4 concentrations; (D) Optimized result of SA concentrations.
3.7. Sensitivity and specificity assay of SARS-CoV-2 Delta variant
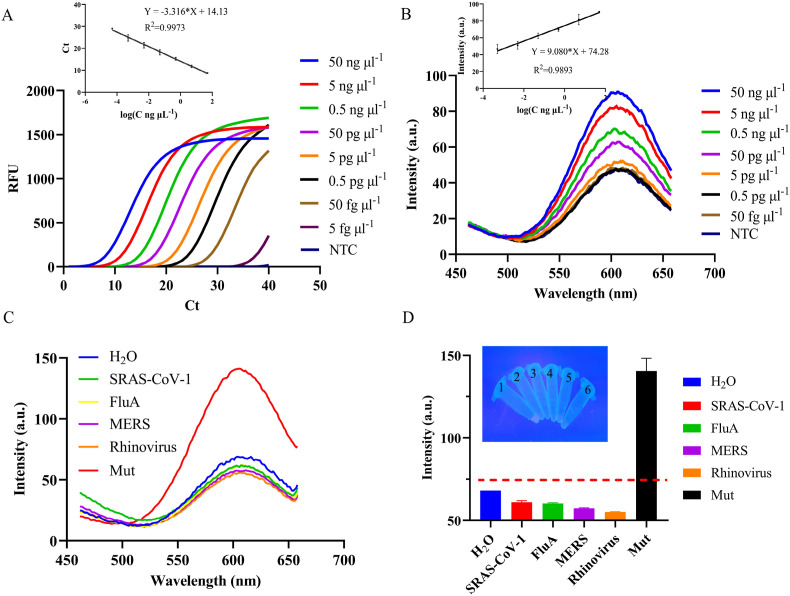
Sensitivity is an important metric used to assess the performance of a diagnostic assay. In this study, different concentrations of the SARS-CoV-2 Delta variant were detected via this novel method, under optimal conditions. As can be seen in Fig. 7 , the detection range of the fluorescence quantitative PCR instrument was 50 ng μl−1-50 fg μl−1, and the limit of quantitation (LOQ) was 50 fg μl−1 and limit of detection (LOD) was 5 fg μl−1 (Fig. 7 A). Furthermore, 110.9% amplification efficiency indicated that the linear relationship between Y = −3.316*X+14.13, R2 = 0.9973 was reliable. The signal output was achieved by the visualization CuNCs-based detection assay, the linear relationship was Y = 9.080*X+74.28, R2 = 0.9893, and the detection results were 50 ng μl−1-0.5 pg μl−1 with the LOQ of 0.5 pg μl−1 and LOD of 0.1 pg μl−1 (3σ) (Fig. 7 B), which is consistent with resolution of agarose gel electrophoresis (Fig. S8). At the same time, the sensitivity of this study was compared with that of other SARS-COV-2 detection, and the results are shown in Table S2.
Fig. 7.
Sensitivity and specificity of detection method. (A) Amplification result of real time-PCR and the linear relationship; (B) Fuorescence of PCR-CuNCs and the linear relationship; (C) Fluorescence curve generated by PCR products; (D) Fluorescence value at 602 nm, in which the red dotted line is the set detection standard, and the illustration shows the UV illumination diagram corresponding to D, with 1–6 indicating Mut (SARS-CoV-2 Delta variant), Rhinovirus, MERS, FluA, SARS-CoV-1, and H2O, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In order to evaluate the specificity of this assay, four other coronaviruses were selected for specificity experiment. It can be seen from results that only the SARS-CoV-2 Delta variant was amplified in real-time qPCR with amplification curves (Fig. S9), generating red fluorescence of the CuNCs (Fig. 7C–D). These results indicate that DPO primers and AT primers can effectively achieve distinction from cross-reactivity with other coronaviruses and realize on-site detection with rapid visualization.
4. Discussion and conclusion
In this study, visual rapid testing for the SARS-CoV-2 Delta coronavirus, in which single nucleotide deletion distinguishes it from wild-type SARS-Cov-2, was developed. To clarify the effect of DPO primers in SNP detection, we performed real-time PCR for the optimization of DPO-primers sequences and realized high specificity of the Delta variant. In addition, the function of AT primers was verified by copper nanoclusters, thereby satisfying need for rapid visual detection on site. Collectively, our results showed that the Delta variant was detected and distinguished from wild-type SARS-CoV-2 via rational primers design and the visualization of CuNCs with high sensitivity and specificity.
Primers in which polydeoxyinosine joins two separate priming regions have been widely reported for SNP assays of single nucleotide replacement, while conventional primers have a wider range of annealing temperature and higher priming specificity [30]. However, DPO primers have not previously been used for an SNP assay of single-base deletions, especially not for the novel coronavirus. It is speculated that the main reason for this may be that the detection of single nucleotide deletion is more difficult than single nucleotide replacement, and more stringent primers design is needed. As can be seen from Fig. 3, the DPO primers in this paper had a narrow annealing temperature range compared to those previously reported. It is well known that double-stranded DNA can generate CuNCs in the presence of reducing agents and CuSO4, and that the AT sequence is most favorable to CuNCs production [19,26]. Moreover, the application of universal primers shows that it can be realized to increase the universal sequence in PCR products, thus effectively avoiding amplification disparity [31,32]. Therefore, in this study AT-rich sequences were used as universal primers only to increase the AT content of PCR products. However, unexpectedly, the use of AT primers not only increased the AT content of the PCR products, but also improved the specificity of detection (Fig. 5 and Fig. S6). In the condition for CuNCs generation, the optimization concentration of CuSO4 was 1 mM, which was also the median (Fig. 6A). This result is consistent with the trend observed by Chen et al. [29]. This is mainly because a concentration of Cu2+ may generate hydroxyl radicals, causing the destruction of PCR products, thus reducing the template of CuNCs and inhibiting the formation of CuNCs [13]. Moreover, while the system of real-time qPCR contained SYBR Green dye, but practical results showed that it had no effect on the CuNCs fluorescence (Fig. 7B). Thus, the difficulties of single base diversity detection were solved by DPO primers and AT primers, while PCR products’ dependence on precision equipment was settled by the CuNCs fluorescence signal output.
In conclusion, this novel approach achieves the sensitive detection of SARS-CoV-2 Delta variants with visualization, as well as the first detection of single nucleotide deletion of the SARS-CoV-2 Delta variants. Notably, our assay could not only distinguish single nucleotide deletion in the target sites and maintain high specificity for the prevention and control of the spread of an epidemic, but it was also able to distinguish SARS-CoV-2 from other coronaviruses reliably to trace the source. Furthermore, the linear range of this method was shown to be wide, varying within 106. However, this method also has some limitations, 1) when the single nucleotide deletion site changes, the DPO-primers must be redesigned; 2) there is a lack of detection of actual samples which is limited by experiment conditions.
Author statement
Zaihui Du: Conceptualization, Methodology, Writing – original draft preparation. Longjiao Zhu: Validation, Supervision, Writing- Reviewing and Editing. Wentao Xu: Resources, Visualization, Validation, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was supported by HeBei Provincical key research and development program(CN) (21372801D), the National Natural Science Foundation of China (No.31871875),and the 2115 Talent Development Program of China Agricultural University (No. 00109016).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.talanta.2022.123266.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Feng W., Newbigging A.M., Le C., Pang B., Peng H., Cao Y., Wu J., Abbas G., Song J., Wang D.B., Cui M., Tao J., Tyrrell D.L., Zhang X.E., Zhang H., Le X.C. Molecular diagnosis of COVID-19: challenges and research needs. Anal. Chem. 2020;92(15):10196–10209. doi: 10.1021/acs.analchem.0c02060. [DOI] [PubMed] [Google Scholar]
- 2.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz M., Chen J., Janda M., Sullivan M., den Boon J., Ahlquist P. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell. 2002;9(3):505–514. doi: 10.1016/s1097-2765(02)00474-4. [DOI] [PubMed] [Google Scholar]
- 4.Dong L., Zhou J., Niu C., Wang Q., Pan Y., Sheng S., Wang X., Zhang Y., Yang J., Liu M., Zhao Y., Zhang X., Zhu T., Peng T., Xie J., Gao Y., Wang D., Dai X., Fang X. Highly accurate and sensitive diagnostic detection of SARS-CoV-2 by digital PCR. Talanta. 2021;224:121726. doi: 10.1016/j.talanta.2020.121726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saha I., Ghosh N., Maity D., Sharma N., Sarkar J.P., Mitra K. Genome-wide analysis of Indian SARS-CoV-2 genomes for the identification of genetic mutation and SNP. Infect. Genet. Evol. 2020;85:104457. doi: 10.1016/j.meegid.2020.104457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma P., Meng Q., Sun B., Zhao B., Dang L., Zhong M., Liu S., Xu H., Mei H., Liu J., Chi T., Yang G., Liu M., Huang X., Wang X. MeCas12a, a highly sensitive and specific system for COVID-19 detection. Adv. Sci. 2020:2001300. doi: 10.1002/advs.202001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaibun T., Puenpa J., Ngamdee T., Boonapatcharoen N., Athamanolap P., O'Mullane A.P., Vongpunsawad S., Poovorawan Y., Lee S.Y., Lertanantawong B. Rapid electrochemical detection of coronavirus SARS-CoV-2. Nat. Commun. 2021;12(1):802. doi: 10.1038/s41467-021-21121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji T., Liu Z., Wang G., Guo X., Akbar Khan S., Lai C., Chen H., Huang S., Xia S., Chen B., Jia H., Chen Y., Zhou Q. Detection of COVID-19: a review of the current literature and future perspectives. Biosens. Bioelectron. 2020;166:112455. doi: 10.1016/j.bios.2020.112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan J.F.-W., Yip C.C.-Y., To K.K.-W., Tang T.H.-C., Wong S.C.-Y., Leung K.-H., Fung A.Y.-F., Ng A.C.-K., Zou Z., Tsoi H.-W. Improved molecular diagnosis of COVID-19 by the Novel,Highly sensitive and specific COVID-19-RdRp/hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020;58(5):e00310–e00320. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ooi K.H., Liu M.M., Tay J.W.D., Teo S.Y., Kaewsapsak P., Jin S., Lee C.K., Hou J., Maurer-Stroh S., Lin W., Yan B., Yan G., Gao Y.G., Tan M.H. An engineered CRISPR-Cas12a variant and DNA-RNA hybrid guides enable robust and rapid COVID-19 testing. Nat. Commun. 2021;12(1):1739. doi: 10.1038/s41467-021-21996-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bu X., Fu Y., Jiang X., Jin H., Gui R. Self-assembly of DNA-templated copper nanoclusters and carbon dots for ratiometric fluorometric and visual determination of arginine and acetaminophen with a logic-gate operation. Mikrochim. Acta. 2020;187(3):154. doi: 10.1007/s00604-020-4146-6. [DOI] [PubMed] [Google Scholar]
- 12.Cui J., Han H., Piao J., Shi H., Zhou D., Gong X., Chang J. Construction of a novel biosensor based on the self-assembly of dual-enzyme cascade amplification-induced copper nanoparticles for ultrasensitive detection of MicroRNA153. ACS Appl. Mater. Interfaces. 2020;12(30):34130–34136. doi: 10.1021/acsami.0c06032. [DOI] [PubMed] [Google Scholar]
- 13.Rotaru A., Dutta S., Jentzsch E., Gothelf K., Mokhir A. Selective dsDNA-templated formation of copper nanoparticles in solution. Angew. Chem. Int. Ed. 2010;49(33):5665–5667. doi: 10.1002/anie.200907256. [DOI] [PubMed] [Google Scholar]
- 14.Borghei Y.S., Hosseini M., Khoobi M., Ganjali M.R. Novel fluorometric assay for detection of cysteine as a reducing agent and template in formation of copper nanoclusters. J. Fluoresc. 2017;27(2):529–536. doi: 10.1007/s10895-016-1980-3. [DOI] [PubMed] [Google Scholar]
- 15.Jiao M., Li Y., Jia Y., Xu L., Xu G., Guo Y., Luo X. Ligand-modulated aqueous synthesis of color-tunable copper nanoclusters for the photoluminescent assay of Hg(II) Mikrochim. Acta. 2020;187(10):545. doi: 10.1007/s00604-020-04539-6. [DOI] [PubMed] [Google Scholar]
- 16.Hu L., Yuan Y., Zhang L., Zhao J., Majeed S., Xu G. Copper nanoclusters as peroxidase mimetics and their applications to H2O2 and glucose detection. Anal. Chim. Acta. 2013;762:83–86. doi: 10.1016/j.aca.2012.11.056. [DOI] [PubMed] [Google Scholar]
- 17.Wang B., Gui R., Jin H., He W., Wang Z. Red-emitting BSA-stabilized copper nanoclusters acted as a sensitive probe for fluorescence sensing and visual imaging detection of rutin. Talanta. 2018;178:1006–1010. doi: 10.1016/j.talanta.2017.08.102. [DOI] [PubMed] [Google Scholar]
- 18.Qing Z., He X., He D., Wang K., Xu F., Qing T., Yang X. Poly(thymine)-templated selective formation of fluorescent copper nanoparticles. Angew. Chem. Int. Ed. 2013;52(37):9719–9722. doi: 10.1002/anie.201304631. [DOI] [PubMed] [Google Scholar]
- 19.Song Q., Shi Y., He D., Xu S., Ouyang J. Sequence-dependent dsDNA-templated formation of fluorescent copper nanoparticles. Chem. Eur J. 2015;21(6):2417–2422. doi: 10.1002/chem.201405726. [DOI] [PubMed] [Google Scholar]
- 20.N K.R., Gorthi S.S. Enhancement of the fluorescence properties of double stranded DNA templated copper nanoparticles. Mater. Sci. Eng. C-Mater. Biol. Appl. 2019;98:1034–1042. doi: 10.1016/j.msec.2019.01.042. [DOI] [PubMed] [Google Scholar]
- 21.Zhu X., Shi H., Shen Y., Zhang B., Zhao J., Li G. A green method of staining DNA in polyacrylamide gel electrophoresis based on fluorescent copper nanoclusters synthesized in situ. Nano Res. 2015;8(8):2714–2720. [Google Scholar]
- 22.Chun J.Y., Kim K.J., Hwang I.T., Kim Y.J., Lee D.H., Lee I.K., Kim J.K. Dual priming oligonucleotide system for the multiplex detection of respiratory viruses and SNP genotyping of CYP2C19 gene. Nucleic Acids Res. 2007;35(6):e40. doi: 10.1093/nar/gkm051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins O., Smith T.J. 3' tth endonuclease cleavage polymerase chain reaction (3TEC-PCR) Technology for single-base-specific multiplex pathogen detection using a two-oligonucleotide system. Int. J. Mol. Sci. 2021;22(11):6061. doi: 10.3390/ijms22116061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li D.D., Hao C.B., Liu Z.M., Wang S.J., Wang Y., Chao Z., Gao S.Y., Chen S. Development of a novel dual priming oligonucleotide system‐based PCR assay for specific detection of Salmonella from food samples. J. Food Saf. 2020;40(3) [Google Scholar]
- 25.Yang K., Chaput J.C. REVEALR: a multicomponent XNAzyme-based nucleic acid detection system for SARS-CoV-2. J. Am. Chem. Soc. 2021;143(24):8957–8961. doi: 10.1021/jacs.1c02664. [DOI] [PubMed] [Google Scholar]
- 26.Wang H.-B., Zhang H.-D., Chen Y., Huang K.-J., Liu Y.-M. A label-free and ultrasensitive fluorescent sensor for dopamine detection based on double-stranded DNA templated copper nanoparticles. Sensor. Actuator. B Chem. 2015;220:146–153. [Google Scholar]
- 27.Xiangyuan Ouyang M.W., Guo Linjie, Cui Chengjun, Liu Ting, Ren Yongan, Zhao Yan, Ge Zhilei, Guo Xiniu, Xie Gang, Jiang Li, Fan Chunhai, Wang Lihua. DNA Nanoribbon-Templated Self-Assembly of Ultrasmall Fluorescent copper nanoclusters with enhanced luminescnece. Angew. Chem. Int. Ed. 2020;132:11934–11942. doi: 10.1002/anie.202003905. [DOI] [PubMed] [Google Scholar]
- 28.Lee C.S., Kang B.K., Lee D.H., Lyou S.H., Park B.K., Ann S.K., Jung K., Song D.S. One-step multiplex RT-PCR for detection and subtyping of swine influenza H1, H3, N1, N2 viruses in clinical samples using a dual priming oligonucleotide (DPO) system. J. Virol. Methods. 2008;151(1):30–34. doi: 10.1016/j.jviromet.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Chen C.A., Wang C.C., Jong Y.J., Wu S.M. Label-Free fluorescent copper nanoclusters for genotyping of deletion and duplication of duchenne muscular dystrophy. Anal. Chem. 2015;87(12):6228–6232. doi: 10.1021/acs.analchem.5b00918. [DOI] [PubMed] [Google Scholar]
- 30.Jia S., Feng B., Wang Z., Ma Y., Gao X., Jiang Y., Cui W., Qiao X., Tang L., Li Y., Wang L., Xu Y. Dual priming oligonucleotide (DPO)-based real-time RT-PCR assay for accurate differentiation of four major viruses causing porcine viral diarrhea. Mol. Cell. Probes. 2019;47:101435. doi: 10.1016/j.mcp.2019.101435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shang Y., Xu Y., Huang K., Luo Y., Xu W. Multiplex pyrosequencing quantitative detection combined with universal primer-multiplex-PCR for genetically modified organisms. Food Chem. 2020;320:126634. doi: 10.1016/j.foodchem.2020.126634. [DOI] [PubMed] [Google Scholar]
- 32.Luo F., Li Z., Dai G., Lu Y., He P., Wang Q. Simultaneous detection of different bacteria by microchip electrophoresis combined with universal primer-duplex polymerase chain reaction. J. Chromatogr. A. 2020;1615:460734. doi: 10.1016/j.chroma.2019.460734. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.