Abstract
The key role of inflammation in COVID-19 induced many authors to study the cytokine storm, whereas the role of other inflammatory mediators such as oxylipins is still poorly understood.
IMPRECOVID was a monocentric retrospective observational pilot study with COVID-19 related pneumonia patients (n = 52) admitted to Pisa University Hospital between March and April 2020. Our MS-based analytical platform permitted the simultaneous determination of sixty plasma oxylipins in a single run at ppt levels for a comprehensive characterisation of the inflammatory cascade in COVID-19 patients. The datasets containing oxylipin and cytokine plasma levels were analysed by principal component analysis (PCA), computation of Fisher’s canonical variable, and a multivariate receiver operating characteristic (ROC) curve.
Differently from cytokines, the panel of oxylipins clearly differentiated samples collected in COVID-19 wards (n = 43) and Intensive Care Units (ICUs) (n = 27), as shown by the PCA and the multivariate ROC curve with a resulting AUC equal to 0.92. ICU patients showed lower (down to two orders of magnitude) plasma concentrations of anti-inflammatory and pro-resolving lipid mediators, suggesting an impaired inflammation response as part of a prolonged and unsolvable pro-inflammatory status. In conclusion, our targeted oxylipidomics platform helped shedding new light in this field. Targeting the lipid mediator class switching is extremely important for a timely picture of a patient’s ability to respond to the viral attack. A prediction model exploiting selected lipid mediators as biomarkers seems to have good chances to classify patients at risk of severe COVID-19.
Keywords: COVID-19, Oxylipins, Inflammation regulation, Severity predictors, Lipid mediator class switching, UHPLC-MS/MS
1. Introduction
Approximately 24 months after the onset of the COVID-19 pandemic, despite the unprecedented effort of the scientific community, there are still many open questions regarding the pathophysiology of the disease, the therapy, and the clinical management of patients.
Severe patients are characterized by a hyper immuno-activation leading generally to respiratory failure, systemic inflammation and multi-organ fibrosis. Markers associated with this acute immune-inflammatory response and severe COVID-19 symptoms could be found among chemicals involved in the cytokine and oxylipin storm [1].
The cytokine storm results in a detrimental dysregulation of T cell responses and in an uncontrolled overproduction of immune cells and cytokines, such as tumour necrosis factor-α (TNF-α), interferon-γ (IFN-γ), interleukins (IL-1β, IL-2, IL-6, IL-7, IL-8), and a large number of chemokines (CCL2, CCL3, CCL5, CCL9, CCL10) [2]. Besides cytokines, other important inflammatory mediators, e.g., oxylipins, sustain the inflammatory response observed in severe COVID-19 cases [[3], [4], [5], [6], [7]]. COVID-19 patients showed higher levels of these mediators compared to healthy subjects [4,5,7]. Bosio et al. suggested a lipid dysregulation from moderate to severe disease. However, neither clear lipidome dysregulation nor significant separation between the two groups was observed [6].
Oxylipins are bioactive lipids generated from both ω-3 and ω-6 polyunsaturated fatty acids (PUFAs) through enzymatic (e.g., prostanoids, epoxy, and hydroxy fatty acids) and non-enzymatic (e.g., isoprostanoids) oxidation reactions [8,9]. Because of their anti-inflammatory action, ω-3 PUFAs seem to limit the level and duration of the critical inflammatory phase [10], and reduce ICU admissions of COVID-19 patients [11]. At the same time, PUFAs act as precursors for pro-inflammatory, anti-inflammatory, and specialized pro-resolving lipid mediators (SPM) [12,13]. Firstly, pro-inflammatory mediators such as prostaglandins, thromboxanes, and leukotrienes are released into the body, leading to the classic signs of inflammation [14]. The production of oxylipins then undergoes lipid mediator class switching mainly because of CYP450-derived epoxy fatty acids, thus shifting from the lipoxygenase pathway to the specialized pro-resolving mediators [14]. SPMs mostly consist of lipoxins, resolvins, maresins, and protectins, which could lower the inflammatory response and even promote its resolution [15,16] without being immunosuppressive unlike classic anti-inflammatory drugs [17].
Consequently, a better understanding of the cytokine storm and its coupling with the oxylipin storm should provide new insights into the complex immuno-inflammatory cascade induced by SARS-CoV-2 and lead to novel strategies for managing patients. The levels of these bioactive mediators may mirror basic biological processes determining the evolution of the pathology and could provide clinicians with clear indications regarding the urgency of patient transfer to ICU.
In this paper, a powerful in-house oxylipidomics platform was successfully employed for the monitoring of the inflammatory response in COVID-19. In more detail, the plasma levels of 48 oxylipins and 5 cytokines in COVID-19 ward and ICU patients were compared. The results highlighted that the oxylipin plasma levels can be used to discriminate between the two groups by multivariate analysis. A biological interpretation linking the findings of the present study to a possible evolution of the inflammatory process are proposed, shedding new light on the pathophysiological mechanism of the COVID-19 disease.
2. Methods
2.1. Study design and participants
This monocentric retrospective observational study (IMPRE-COVID-19) was approved by the local Ethics Committee. Patients eligible for enrolment were aged 18 years or older and admitted to the University Hospital of Pisa (Pisa, Italy) in March-April 2020 with COVID-19 related pneumonia (wild type). SARS-CoV-2 infection was confirmed by polymerase chain reaction in a nasopharyngeal swab, while pneumonia was demonstrated by CT scan. Members of the Pisa COVID group collected all the clinical information, including co-morbidities, drug intake and routine laboratory data, which were de-identified and stored according to the recommendations of the Ethics Committee and made available to academic researchers. Residuals of plasma samples used for routine clinical measurements were de-identified and stored in the BMS Multispecialistic Biobank of Pisa University Hospital.
Oxylipin levels were analysed in a convenient sample of 52 patients randomly selected among those with a complete set of cytokine values evaluated for clinical purposes. In a few cases, plasma samples collected on different days from a single patient were available at the biobank and were analysed to understand the evolution over time of the oxylipin levels during hospitalization.
2.2. Procedures
2.2.1. Virological tests
Viral nucleic acids were manually extracted from 250 μL of plasma or serum samples using the EXTRA blood kit (ELITechGroup, Turin, Italy) according to the manufacturer’s instructions. After extraction, purified RNA samples were screened by RT-qPCR using the SARS-CoV-2 R-Gene assay (Biomerioux, Marcy-l’Etoile, France) on an ABI 7500 FAST thermocycler (Applied Biosystems). The real-time SARS-CoV-2 R-Gene assay is carried out by two triplex PCRs. The first PCR detects the N gene and the RdRp gene whereas the second PCR detects the E gene of the SARS-CoV-2 genome. The assay contains internal controls to check PCR processing, and a cellular control to check sampling for certain results.
2.2.2. Sample collection and processing
Peripheral blood samples were collected from COVID-19 ward and ICU patients with COVID-19 during their hospitalization, as part of the routine clinical activity of the Laboratory of Clinical Pathology of Pisa University Hospital. Whole blood was collected in EDTA tubes (BD Vacutainer®) and centrifuged at 1500 g for 10 min at 25 °C to separate blood cells and plasma. Plasma was removed and stored in aliquots at −80 °C until analysis. For the oxylipin analyses, the antioxidant butylhydroxytoluene (BHT, 15 mg/mL in methanol) was added before storage (BHT:sample volume ratio of 1:100) to preserve polyunsaturated fatty acids from in vitro lipid peroxidation. Before oxylipin analysis, plasma samples were checked to identify SARS-CoV-2 virus. Positive samples were excluded from the quantification of the oxylipin content for safety reasons.
2.2.3. Quantification of oxylipins
The MS-based targeted profiling of 60 oxylipins (e.g., prostaglandins, lipoxins, protectins, resolvins, hydroxy- and epoxy-fatty acids, F2-isoprostanes, F3-isoprostanes, F2-dihomo-isoprostanes, and F4-neuroprostane) [[18], [19], [20]] was performed using micro-extraction by packed sorbent (MEPS) ultra-high performances liquid chromatography tandem mass spectrometry (MEPS-UHPLC-MS/MS) platform [[21], [22], [23]]. Briefly, plasma proteins were precipitated by the sequential addition of salts (i.e., 250 μL of CuSO4·5H2O 10% w/v and 250 μL of Na2WO4·2H2O 12% w/v) and acetonitrile (500 μL) to the plasma sample (500 μL). The supernatant was then diluted (1:6 v/v) with water and loaded onto the MEPS C18 cartridge. The cartridge was activated by drawing and discharging 3 times 100 μL of methanol (3 × 100 μL), and then conditioned with 3 × 100 μL of water at 0.6 mL/min. The diluted supernatant (3000 μL) was loaded up and down twelve times at 0.3 mL/min by discarding it. The cartridge was then washed with 100 μL of a water:methanol mixture (95:5 v/v) at 0.6 mL/min to remove potential interferences. Analytes were eluted with 30 μL of methanol at 0.3 mL/min and then injected into the UHPLC-MS/MS instrument. We employed an Agilent 1290 Infinity II LC system coupled to a 6495 Triple Quadrupole mass spectrometer, which was equipped with a Jet Stream electrospray (ESI) ionization source (Agilent Technologies, USA). The chromatographic separation was achieved using a Polaris 3C18-A column (50 × 4.6 mm, 3 μm, Agilent Technologies, USA) and a gradient elution with a mobile phase consisting of 0.1% aqueous formic acid and 50:50 v/v methanol:acetonitrile. The mass spectrometer operated in ESI negative ionization mode and performed multiple reaction monitoring (MRM) with unit mass resolution. Detailed chromatographic parameters, ESI and MRM operating conditions are shown in the Supporting Information (Table S1). Compounds were quantified by calibration curves plotting the analyte to an internal standard peak area ratio (Quantifier transition) versus the corresponding concentration ratio. Table S2 lists the main analytical figures of merit of the MEPS-UHPLC-MS/MS platform. Fig. S1 shows the chromatographic profiles for the sixty oxylipins.
2.2.4. Quantification of cytokines
Plasma cytokines (i.e., IL-6, IL-1β, IL-10, TNF-α, and CCL2) and granulocyte-macrophage colony-stimulating factor (GM-CSF) were quantified by automated ELISA assays according to the manufacturer’s instructions (see supplementary information).
2.3. Statistical analysis
Datasets D1 and D2, which include the plasma levels of 48 oxylipins (Table S3) and 5 cytokines (IL-6, IL-1β, IL-10, TNF-α, and CCL2, Table S4), respectively, were obtained from the analyses of 70 samples collected from patients hospitalised in COVID-19 wards (patients = 32, samples = 43) and ICUs (patients = 24, samples = 27) (four patients were in both groups). Twelve out of sixty oxylipins (Table S2) were excluded from D1 as the concentrations were below the limit of quantification for more than 50% of samples. A decimal logarithmic transform was used to correct for asymmetry characterising all the variables [24]. Samples were randomly split into a training (n = 56) and a test set (n = 14): the former was used to build the models and the latter to independently estimate performances and consistency.
Data were analysed by a multivariate exploratory method (principal component analysis, PCA) [25,26].
In the present study, PCA was applied after column autoscaling to ensure the same importance a priori to be given to all variables, irrespectively of their magnitude [27].
Fisher’s canonical variable was then computed in the plane described by the two lowest-order PCs, as the direction that maximises the ratio between inter-class and intra-class variances [28]. This axis represents the most discriminant direction, and its loadings, multiplied by the loadings of the PCs considered, indicate the importance of the original input variables in the differentiation of the classes.
Finally, a multivariate receiver operating characteristic (ROC) curve was obtained by varying the confidence level of unequal class models and computing, at each step, sensitivity and specificity [[29], [30], [31]]. Multivariate data processing was performed by in-house Matlab routines (The MathWorks, Inc., Natick, USA, Version 2019b).
3. Results
Between 1 March and 30 April 2020, 52 patients were admitted to Pisa University Hospital, 28 in COVID-19 wards, 20 in ICUs and 4, initially hospitalised in COVID-19 wards, who were then transferred to the ICUs due to deteriorating health conditions. The clinical characteristics, comorbidities and outcome of patients are reported in Table 1 .
Table 1.
Demographic and clinical baseline characteristics of enrolled patients from COVID-19 wards (W) and intensive care units (ICU). Data are represented as median (first and third quartile). Statistics: Student’s t-test (difference between means) on log-transformed data and Fisher’s test to compare prevalence of comorbidities and drug intake between the two groups.
| W (n = 32) | ICU (n = 20) | p | |
|---|---|---|---|
| Comorbidities | |||
| Diabetes (n; %) | 1; 3 | 5; 25 | 0.0263 |
| Hypertension (n; %) | 10; 31 | 7; 35 | 0.7630 |
| COPD/asthma (n; %) | 3; 9 | 0; 0 | 0.2760 |
| Hypercholesterolemia (n; %) | 4; 13 | 4; 20 | 0.6949 |
| Heart disease (n; %) | 13; 41 | 3; 15 | 0.0679 |
| Drugs | |||
| Lopinavir plus ritonavir (n; %) | 29; 91 | 13; 65 | 0.0327 |
| Remdesivir (n; %) | 1; 3 | 3; 15 | 0.2855 |
| Coricosteroids (n; %) | 11; 34 | 12; 60 | 0.0901 |
| Tolicizumab (n; %) | 7; 22 | 1; 5 | 0.1324 |
| Baricitinib (n; %) | 5; 16 | 4; 20 | 0.7151 |
| Heparin (n; %) | 22; 69 | 16; 80 | 0.5237 |
| Clinical characteristics | |||
| Age, years | 60 (51 – 78) | 63 (57 – 73) | 0.7700 |
| P/F admission, mmHg | 313 (263 – 377) | 256 (207-327) | 0.0215 |
| P/F nadir, mmHg | 182 (95 – 288) | 96 (72 – 118) | 0.0055 |
| SOFA score | 2.0 (1.0 – 3.0) | 2.0 (1.8 – 3.3) | 0.3724 |
| Creatinine, mg/dL | 0.98 (0.80 – 1.27) | 1.13 (0.83 – 1.46) | 0.4274 |
| While blood cells, cells.103/μL | 5.9 (4.5 – 7.8) | 8.0 (6.9 – 11.9) | 0.0117 |
| Neutrophils, cells.103/μL | 3.8 (2.9 – 5.8) | 6.1 (3.7 – 8.3) | 0.0623 |
| Lymphocytes, cells.103/μL | 1.1 (0.7 – 1.3) | 0.6 (0.5 – 1.1) | 0.0309 |
| C-reactive protein, mg/dL | 5.7 (2.8 – 11.0) | 8.8 (3.1 – 13.6) | 0.4470 |
| D-Dimer, μg/mL | 0.35 (0.19 – 0.52) | 0.65 (0.27 – 1.26) | 0.0166 |
| Outcome | |||
| Survived (n; %) | 27; 84 | 17; 85 | |
| ETI (n; %) | 0; 0 | 12; 60 | |
Baseline characteristics were generally similar between the two groups, with the exception of diabetes prevalence, which was higher in ICU patients, and of the Horowitz index for lung function (P/F ratio) that identified the acute hypoxemic respiratory condition of ICU patients. These patients were also characterized by leucocytosis, lymphocytopenia and higher D-dimer levels, which were all related to the enhanced inflammatory response and more severe viral infection. Pharmacological therapy was similar in the two-study group except for the antiretroviral drugs that were more represented in the ward group.
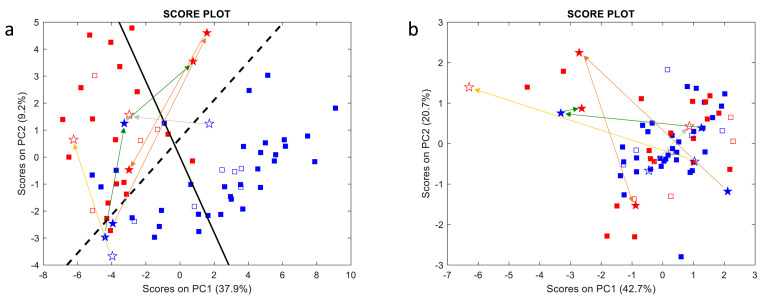
The full data-set concerning the plasma concentrations of oxylipins and cytokines are reported in Table S3 and Table S4, respectively. A clear separation of samples collected from COVID-19 wards (blue symbols) and ICUs (red symbols) is visible in the oxylipin score plot (Fig. 1 a), whereas it is not observed in the case of cytokines (Fig. 1b). This pattern is consistent for items of both the training set (full symbols), used to build the model and calculate PCs, and the test set (empty symbols), which were simply projected onto the PC plane [24] for validation purposes.
Fig. 1.
Oxylipin (a) and cytokine (b) score plots. Blue and red symbols represent samples collected from COVID-19 wards and ICUs, respectively, whereas full and empty symbols belong to the training and test sets, respectively. Samples collected from the same patient on different days are represented as stars connected by coloured arrows. The full black and dashed lines represent Fisher’s canonical variable, i.e., the direction of maximum discrimination between the two classes, and the delimiter separating the two classes, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Interestingly, many borderline samples were collected from COVID-19 ward patients who were subsequently transferred to the ICUs a few days after sample collection due to their deteriorating health conditions. Samples collected from the same patient on different days (stars) are connected by coloured arrows: shifts in the oxylipin score plot from the blue to the red zone seem to replicate the movements of patients from COVID-19 wards to the ICUs, however this is not reflected in the cytokine score plot.
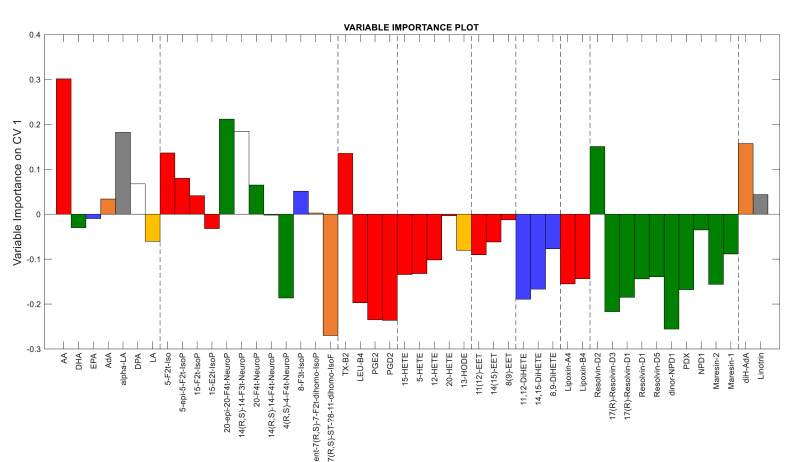
Fig. 1a shows that PC1 scores provide the main contribution (about 38% of the total variance) to the separation of the two sample classes, though PC2 also contributes for about 10%. To understand which oxylipins differentiate samples, the most discriminant direction was computed as Fisher’s canonical variable (full black line), whose loadings (more precisely their absolute values) are representative of oxylipin contributions to discrimination (Fig. 2 ). The variables with positive and negative weights in Fig. 2 were over-expressed in ICU and COVID-19 ward samples, respectively.
Fig. 2.
Loadings of Fisher’s canonical variable indicating the importance of the input variables in discriminating between the two classes of samples. Colours refer to different PUFAs as oxylipin precursors: red – arachidonic acid (AA); green –docosahexanoic acid (DHA); blue – eicosapentaenoic acid (EPA); orange – adrenic acid (AdA), grey – alpha-linolenic acid (alpha-LA), white – docosapentaenoic acid (DPA); yellow – linoleic acid (LA). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Samples from ICU patients were characterized by higher concentrations [median (first and third quartile)] of most PUFAs (e.g. arachidonic acid: ICU [208 μg/mL (90.1–473)] vs. W [19.5 μg/mL (8.27–27.1)], p = 5.90E-03) and almost comparable levels of isoprostanoids. The latter represents a group of oxylipins produced by the non-enzymatic peroxidation of membrane PUFAs, such as arachidonic acid (AA), docosahexaenoic acid (DHA), and eicosapentaenoic acid (EPA). AA-derived F2-IsoPs resulted significantly different (p = 0.004) between the two populations, whereas no distinction (p > 0.05) was observed between (ω-3)-derived and (ω-6)-derived isoprostanoids (Fig. S2). Interestingly, thromboxane B2 (TX-B2), which is representative of the TX-A2 synthesis through platelet COX-1 and endothelial COX-1, and COX-2, was over-expressed (ICU [0.324 ng/mL (0.039–1.65)] vs. W [0.283 ng/mL (0.174–0.595)], p = 9.93E-01) in these samples. On the other hand, COVID-19 ward samples showed higher concentrations of most oxylipins of enzymatic origin: prostanoids, hydroxy-eicosatetraenoic acid as HETEs and HODE (e.g. 15-HETE: ICU [0.861 ng/mL (0.562–4.51)] vs. W [24.7 ng/mL (2.56–82.8)], p = 1.37E-08), dihydroxyeicosatetraenoic acid (DiHETEs) epoxy-eicosatrienoic acid (EETs), lipoxins, and SPMs as resolvins, protectins, maresin (e.g. resolvin-D5: ICU [0.091 ng/mL (0.038–0.489)] vs. W [1.73 ng/mL (0.632–3.95)], p = 3.88E-08). Only the multivariate analysis of oxylipins provides such a clear separation between the two classes of samples, whereas individual variables exhibit a lower discriminant capability (see scatter column plots for individual oxylipins in Fig. S3).
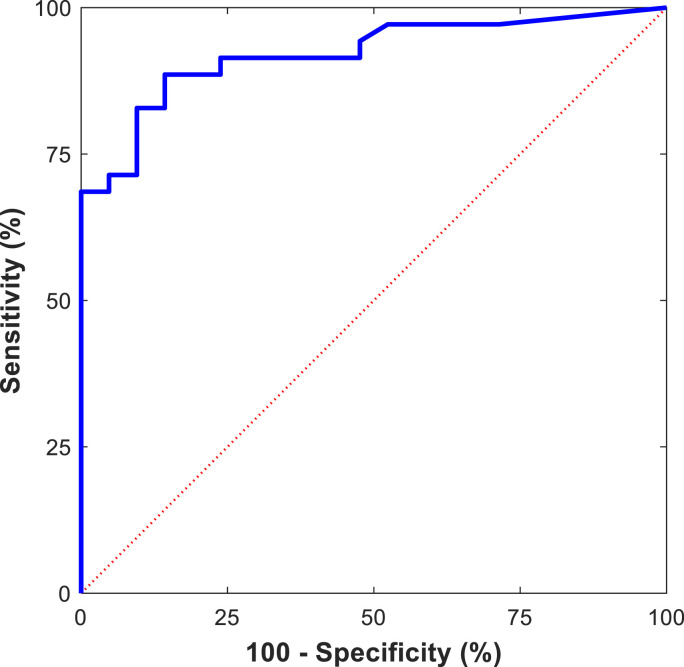
To further confirm the classification capacity of the full oxylipin set, a multivariate ROC curve was obtained by the UNEQ class-modelling strategy (Fig. 3 ). The multivariate model was computed using the four lowest-order PCs as the input variables and COVID-19 ward samples as the target class. The resulting area under the curve (AUC, 0.92) exceeded all the AUC outcomes obtained for individual oxylipins (selected univariate ROC curves are reported in Fig. S4).
Fig. 3.
Multivariate ROC curve obtained from the UNEQ class model computed for the COVID-19 ward class with the four lowest-order PCs (AUC = 0.92).
4. Discussion
Multivariate analysis of oxylipin plasma levels enabled the correct classification of COVID-19 ward and ICU samples, whereas separation was not achieved by processing cytokine values. The fact that many borderline samples were collected from COVID-19 ward patients that were about to be transferred to ICUs suggests that sample differences were not a trivial consequence of different treatments received from patients in different wards and ICUs. In addition, the analysis of the relative contributions of individual oxylipins to the classification of the two groups fits well with our present understanding of COVID-19 and may provide new insights into its pathophysiology.
Several papers have been published in the last year discussing factors related to the severity of COVID-19 and defining risk scores for the prediction of severity and outcome of the disease [[32], [33], [34], [35]]. Most of the proposed scores are based on demographic and anamnestic data as well as on clinical parameters easily measured or calculated from a routine blood test, e.g., age, comorbidities, oxygen saturation, lactate dehydrogenase or neutrophil-to-lymphocyte ratio. If from one side this makes these scores practical for clinical use, on the other it does not provide any improved understanding on the pathophysiological mechanism of the disease or information relevant to the evolution of the biological processes occurring in the patient.
In our case, ICU patients showed almost comparable levels of pro-inflammatory isoprostanoids produced from the non-enzymatic oxidation of PUFAs such as AA, DHA and EPA, and a pronounced and selective deficiency of oxylipins such as prostaglandins, leukotrienes, anti-inflammatory, and pro-resolving oxylipins originating from their enzymatic conversion. The isoprostanoid 4(R,S)-4-F4t-neuroprostane was down-regulated in ICU patients (ICU [0.026 ng/mL (0.020–0.074)] vs. W [0.238 ng/mL (0.092–0.594)], p = 1.39E-05). This lipid mediator prevents oxidation of the RyR ryanodine receptors, a family of Ca2+ release channels that controls the intracellular calcium exchange and whose oxidation may cause leaks of Ca2+ and lead to heart failure, pulmonary insufficiency and cognitive dysfunction in COVID-19 [36]. Furthermore, TX-B2 was the only prostanoid with an enzymatic origin over-expressed in ICU patients, which agrees with the need for heparin therapy in these patients to prevent microcirculatory damage.
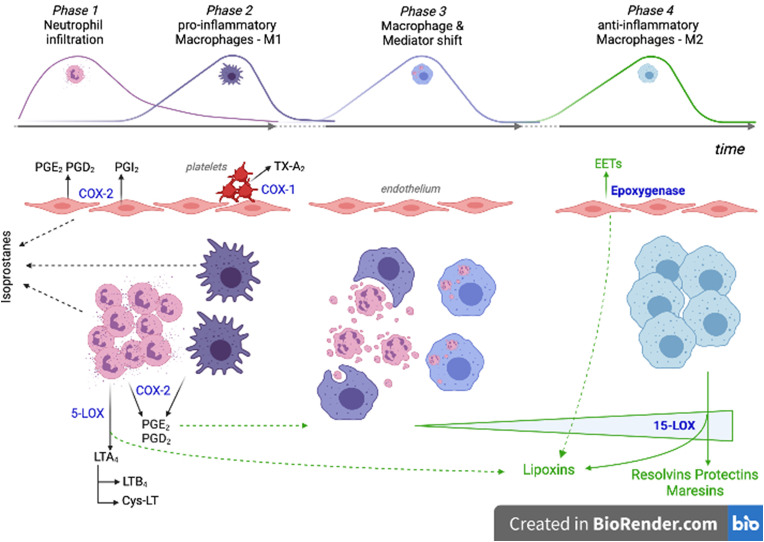
Inflammation is a highly coordinated transcriptional process whose evolution towards resolution or persistence depends on a dynamic balance between pro- and anti-inflammatory mediators. In a controlled inflammatory process, immune tissue-resident cells activate processes producing chemokines and cytokines. Endothelial cells then respond thereby facilitating the recruitment of neutrophils (first) and monocytes (at a later stage), which differentiate into pro-inflammatory M1 macrophages (Fig. 4 ) [37]. Pro-inflammatory cytokines activate the phospholipase A2 enzymes, which cause the release from membranes of AA, which is then converted into prostanoids or leukotrienes by cell-specific enzymatic activities. This phase is characterized by a prevalence of COX-2, both in neutrophils and M1-macrophages, and 5-LOX activity, above all in neutrophils. COX-2 and 5-LOX are responsible for the production of prostaglandins (PGD2 and PGE2) and leukotrienes (LTs), respectively [38].
Fig. 4.
Cellular and oxylipin interplay during the evolution of an inflammatory process. Phase 1 and Phase 2: rapid neutrophil and delayed monocyte extravasation in response to cytokines produced by activated immune tissue-resident cells. Pro-inflammatory oxylipins are produced mainly by neutrophils, M1-macrophages, activated endothelial cells and platelets. Phase 3: neutrophil apoptotic bodies and prostaglandins promote the macrophage shift towards a resolution-phase function. Phase 4: inflammation resolution is promoted by increasing production of the specialized pro-resolving mediators. PGE2: prostaglandin E2; PGD2: prostaglandin D2; PGI2: prostacyclin I2; TX-A2: thromboxane A2; LTA4: leukotriene A4; LTB4: leukotriene B4; Cys-LT: cysteine-leukotrienes; EETs: epoxyeicosatrienoic acids; COX-1: cyclooxygenase-1; COX-2: cyclooxygenase-1; 5-LOX: 5-lipoxygenase; 15-LOX: 15-lipoxygenase.
The switch of lipid mediators from prostanoids to lipoxins and SPMs (i.e., resolvins, protectins, and maresins) is critical for inflammation resolution [39]. In fact, PGE2 facilitates the transformation of pro-inflammatory M1 into anti-inflammatory M2-macrophages characterized by the up-regulation of the 15-LOX enzyme, which is primarily involved in the synthesis of SPMs [40]. PGE2 also helps to switch the pro-inflammatory (e.g., TNF-α, IL-1β, and IL-6) into the anti-inflammatory interleukins synthetized by M2-macrophages (e.g., IL-10) [41]. The intermediate products of 5-LOX and 15-LOX activities (5-HPTEs and 5-HETE; 15-HPTE and 15-HETE) are also precursors for the biosynthesis of lipoxins LXA4 and LXB4, which are important agonists of the resolution of inflammation since they inhibit neutrophil recruitment, stimulate vasodilation, and promote efferocytosis [41].
Together with prostanoids, neutrophils play a key role in the modulation of the macrophage function through the release of apoptotic bodies and microvesicles. In fact, the phagocytosis of the microvescicles by M1 macrophages is essential to trigger their functional reprogramming into M2-macrophages (Fig. 4) [42].
For these reasons, we speculate that the oxylipin pattern observed in ICU patients affected by severe COVID-19 mirrors an impaired inflammation response which is part of a prolonged and unsolvable pro-inflammatory status characterized by a relative lack of oxylipins produced from the enzymatic processing of PUFAs. The impaired production of anti-inflammatory and pro-resolving oxylipins is not a consequence of the reduced availability of PUFA precursors, which appear unchanged or even increased in ICU patients. The presence of soluble isoprostanoids in both classes of patients confirms that the availability of membrane PUFAs is not a limitation for their enzymatic processing [43].
Much remains to be understood regarding the pathogenetic mechanism of COVID-19, however defective innate and specific immune responses are likely to be critical features [44]. Schulte-Schrepping et al. [45] showed an increase in dysfunctional neutrophils and monocytes in severe COVID-19 patients that seems to be in agreement with our findings.
A massive endothelial dysfunction resulting from the cytokine storm and the infiltration of SARS-CoV-2 is a further characteristic of severe COVID-19 that determines the loss of vessel barrier, the promotion of leukocyte infiltration, and the activation of platelet aggregation and coagulation [44,46]. Interestingly, in ICU patients we found higher levels of TX-B2, the biological inactive catabolite of TX-A2 (a potent activator of platelet aggregation and thus of coagulation). This is in line with the diffused microthrombosis observed in COVID-19 [44,47] and with the lower levels of EETs, which are mainly produced by endothelial epoxygenase and are important mediators of all pro-resolving mechanisms [48], including the lipid mediator class switching [49,50].
In conclusion, our powerful in-house oxylipidomics platform revealed that the more severe disease in ICU patients is accompanied by an inefficient enzymatic synthesis of the anti-inflammatory oxylipins resulting in an ineffective resolution mechanism of inflammation, likely worsened by endothelial damage. This hypothesis, once supported by prospective studies, might provide a basis for the identification of early biomarkers of poor disease outcome. In addition, the possible imbalance between the production of anti-inflammatory and pro-resolving oxylipins in ICU patients might explain the poor outcome of therapies based on cytokine inhibitors, and opens up new perspectives for the therapy of severe COVID-19 and, in general, of lung diseases.
Data sharing
All de-identified data will be shared upon approval from the IMPRE-COVID-19 steering committee and a signed data access agreement. All requests should be sent to denise.biagini@dcci.unipi.it.
Author contributions
DB, FM, AP and FDF designed the study. DB, TL and SG developed the method for the analysis of oxylipins, LB, TD, CO and JMG synthesized 22 oxylipins needed for calibration that were not commercially available, DB, FV and AB measured oxylipin levels, MF measured cytokine levels whereas LM performed virological tests. PO performed data analysis, MF, AC and AP provided biological and clinical interpretation of data. DB, MF, PO and FDF drafted the paper, all the authors revised the paper.
Funding
Institutional funds from the University of Pisa supported the study.
Role of the funding source
The funders of the study had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. All the authors had full access to all data and shared the final responsibility for the decision to submit for publication.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgments
Members of the Pisa COVID group are gratefully acknowledged for collecting the clinical information, Dr. Simone Lapi and the BMS Multispecialistic Biobank of Pisa University Hospital are acknowledged for supplying the plasma samples. The study received financial support from institutional funds of the University of Pisa.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.freeradbiomed.2022.01.021.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
The Supporting Information section contains a detailed description of materials, Table S1 (list of quantified oxylipins and MRM transitions), Table S2 (analytical figures of merit for the oxylipin quantification), Table S3 (oxylipin concentration levels in COVID-19 ward and ICU samples), Table S4 (cytokine concentration levels in COVID-19 ward and ICU samples), and Figure S1 (ROC curves of the best performing oxylipins in the classification of samples).
References
- 1.Balta M.G., Papathanasiou E., Christopoulos P.F. Specialized pro-resolving mediators as potential regulators of inflammatory macrophage responses in COVID-19. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.632238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu B., Huang S., Yin L. The cytokine storm and COVID-19. J. Med. Virol. 2021;93:250–256. doi: 10.1002/jmv.26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Das U.N. Can bioactive lipids inactivate coronavirus (COVID-19)? Arch. Med. Res. 2020;51:282–286. doi: 10.1016/j.arcmed.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.M.M. Pérez, V.E. Pimentel, C.A. Fuzo, P. v da Silva-Neto, D.M. Toro, C.O. S Souza, T.F. C Fraga-Silva, L. Gustavo Gardinassi, S. de Carvalho, N.T. Neto, I. Carmona-Garcia, C.N. S Oliveira, C.M. Milanezi, V. Nardini Takahashi, T. Canassa, D. Leo, L.C. Rodrigues, C.F. S L Dias, R.S. Parra, J.J. R da Rocha, O. Feres, F.C. Vilar, G.G. Gaspar, R.C. da Silva, L.F. Constant, F.M. Ostini, A.P. de Amorim, A.M. Degiovani, D.P. da Silva, R.C. C Barbieri, I.K. F M Santos, S.R. C Maruyama, E.M. S Russo, A.L. Viana, A.P. M Fernandes, V.L. D Bonato, C.R. B Cardoso, C.A. Sorgi, M. Dias-Baruffi, M. Martínez Pérez, V. Eduardo Pimentel, B. Sc, P. Vieira da Silva-Neto, L. Helena Faccioli, Cholinergic and lipid mediators crosstalk in Covid-19 and the impact of glucocorticoid therapy, (n.d.). 10.1101/2021.01.07.20248970. [DOI]
- 5.McReynolds C.B., Cortes-Puch I., Ravindran R., Khan I.H., Hammock B.G., Shih P. an B., Hammock B.D., Yang J. Plasma linoleate diols are potential biomarkers for severe COVID-19 infections. Front. Physiol. 2021;12 doi: 10.3389/fphys.2021.663869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosio C.M., Ko A.I., IMPACT Team Y., dela Cruz Casanovas-Massana C.S., Minasyan M., Farhadian S., Bermejo S., Leighton I., Benjamin Schwarz A., Sharma L., Roberts L. Eicosanoid immune mediators lipidome, resulting in dysregulation of serum in humans is defined by a shift in the cutting edge: severe SARS-CoV-2 infection. 2021. [DOI] [PMC free article] [PubMed]
- 7.Archambault A.S., Zaid Y., Rakotoarivelo V., Turcotte C., Doré É., Dubuc I., Martin C., Flamand O., Amar Y., Cheikh A., Fares H., el Hassani A., Tijani Y., Côté A., Laviolette M., Boilard É., Flamand L., Flamand N. High levels of eicosanoids and docosanoids in the lungs of intubated COVID-19 patients. FASEB (Fed. Am. Soc. Exp. Biol.) J. 2021;35 doi: 10.1096/fj.202100540R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curtis-Prior P. 2004. The Eicosanoids. [Google Scholar]
- 9.Galano J.-M., Lee Y.Y., Oger C., Vigor C., Vercauteren J., Durand T., Giera M., Lee J.C.-Y. Isoprostanes, neuroprostanes and phytoprostanes: an overview of 25 years of research in chemistry and biology. Prog. Lipid Res. 2017;68:83–108. doi: 10.1016/j.plipres.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Weill P., Plissonneau C., Legrand P., Rioux V., Thibault R. May omega-3 fatty acid dietary supplementation help reduce severe complications in Covid-19 patients? Biochimie. 2020;179:275–280. doi: 10.1016/j.biochi.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogero M.M., Leão M. de C., Santana T.M., Pimentel M.V. de M.B., Carlini G.C.G., da Silveira T.F.F., Gonçalves R.C., Castro I.A. Potential benefits and risks of omega-3 fatty acids supplementation to patients with COVID-19. Free Radic. Biol. Med. 2020;156:190–199. doi: 10.1016/j.freeradbiomed.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnardottir H., Pawelzik S.C., Öhlund Wistbacka U., Artiach G., Hofmann R., Reinholdsson I., Braunschweig F., Tornvall P., Religa D., Bäck M. Stimulating the resolution of inflammation through omega-3 polyunsaturated fatty acids in COVID-19: rationale for the COVID-omega-F trial. Front. Physiol. 2021;11 doi: 10.3389/fphys.2020.624657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serhan C.N. Pro-resolving lipid mediators are leads for resolution physiology. Nature. 2014;510:92–101. doi: 10.1038/nature13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammock B.D., Wang W., Gilligan M.M., Panigrahy D. Eicosanoids: the overlooked storm in coronavirus disease 2019 (COVID-19)? Am. J. Pathol. 2020;190:1782–1788. doi: 10.1016/j.ajpath.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serhan C.N., Levy B.D. Resolvins in inflammation: emergence of the pro-resolving superfamily of mediators. J. Clin. Invest. 2018;128:2657–2669. doi: 10.1172/JCI97943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levy B.D., Clish C.B., Schmidt B., Gronert K., Serhan C.N. Lipid mediator class switching during acute inflammation: signals in resolution. 2001. http://immunol.nature.com [DOI] [PubMed]
- 17.Panigrahy D., Gilligan M.M., Huang S., Gartung A., Cortés-Puch I., Sime P.J., Phipps R.P., Serhan C.N., Hammock B.D. Inflammation resolution: a dual-pronged approach to averting cytokine storms in COVID-19? Cancer Metastasis Rev. 2020;39:337–340. doi: 10.1007/s10555-020-09889-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oger C., Brinkmann Y., Bouazzaoui S., Durand T., Galano J.-M. Stereocontrolled access to isoprostanes via a bicyclo [3.3. 0] octene framework. Org. Lett. 2008;10:5087–5090. doi: 10.1021/ol802104z. [DOI] [PubMed] [Google Scholar]
- 19.Balas L., Durand T. Dihydroxylated E, E, Z-docosatrienes. An overview of their synthesis and biological significance. Prog. Lipid Res. 2016;61:1–18. doi: 10.1016/j.plipres.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Balas L., Risé P., Gandrath D., Rovati G., Bolego C., Stellari F., Trenti A., Buccellati C., Durand T., Sala A. Rapid metabolization of protectin D1 by β-oxidation of its polar head chain. J. Med. Chem. 2019;62:9961–9975. doi: 10.1021/acs.jmedchem.9b01463. [DOI] [PubMed] [Google Scholar]
- 21.Biagini D., Antoni S., Lomonaco T., Ghimenti S., Salvo P., Bellagambi F.G., Scaramuzzo R.T., Ciantelli M., Cuttano A., Fuoco R., di Francesco F. Micro-extraction by packed sorbent combined with UHPLC-ESI-MS/MS for the determination of prostanoids and isoprostanoids in dried blood spots. Talanta. 2020;206 doi: 10.1016/j.talanta.2019.120236. [DOI] [PubMed] [Google Scholar]
- 22.Biagini D., Lomonaco T., Ghimenti S., Fusi J., Cerri E., de Angelis F., Bellagambi F.G., Oger C., Galano J.M., Bramanti E., Franzoni F., Fuoco R., di Francesco F. Saliva as a non-invasive tool for monitoring oxidative stress in swimmers athletes performing a VO2max cycle ergometer test. Talanta. 2020;216 doi: 10.1016/j.talanta.2020.120979. [DOI] [PubMed] [Google Scholar]
- 23.Ghimenti S., Lomonaco T., Bellagambi F.G., Biagini D., Salvo P., Trivella M.G., Scali M.C., Barletta V., Marzilli M., di Francesco F., Errachid A., Fuoco R. Salivary lactate and 8-isoprostaglandin F2α as potential non-invasive biomarkers for monitoring heart failure: a pilot study. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-64112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliveri P., Forina M. Chemical Analysis of Food: Techniques and Applications. 2012. Data analysis and chemometrics; pp. 25–57. [DOI] [Google Scholar]
- 25.Jolliffe I.T. Springer-Verlag; New York: 2002. Principal Component Analysis. [Google Scholar]
- 26.Oliveri P., Malegori C., Mustorgi E., Casale M. Qualitative pattern recognition in chemistry: theoretical background and practical guidelines. Microchem. J. 2021;162 doi: 10.1016/j.microc.2020.105725. [DOI] [Google Scholar]
- 27.Oliveri P., Malegori C., Simonetti R., Casale M. The impact of signal pre-processing on the final interpretation of analytical outcomes – a tutorial. Anal. Chim. Acta. 2019;1058:9–17. doi: 10.1016/J.ACA.2018.10.055. [DOI] [PubMed] [Google Scholar]
- 28.Oliveri P., Malegori C., Casale M. Chemometrics and statistics | multivariate classification techniques. 2019. [DOI]
- 29.Pirro V., Oliveri P., Sciutteri B., Salvo R., Salomone A., Lanteri S., Vincenti M. Multivariate strategies for screening evaluation of harmful drinking. Bioanalysis. 2013;5:687–699. doi: 10.4155/bio.13.12. [DOI] [PubMed] [Google Scholar]
- 30.Oliveri P. Class-modelling in food analytical chemistry: development, sampling, optimisation and validation issues – a tutorial. Anal. Chim. Acta. 2017;982:9–19. doi: 10.1016/j.aca.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 31.Derde M.P., Massart D.L. 1986. UNEQ: A DISJOINT MODELLING TECHNIQUE FOR PATTERN RECOGNITION BASED ON NORMAL DISTRIBUTION. [Google Scholar]
- 32.Dashti H., Roche E.C., Bates D.W., Mora S., Demler O. SARS2 simplified scores to estimate risk of hospitalization and death among patients with COVID-19. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-84603-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costa Dos Santos Junior G., Pereira C.M., Kelly Da Silva Fidalgo T., Valente A.P. Saliva NMR-based metabolomics in the war against COVID-19. Anal. Chem. 2020;92:15688–15692. doi: 10.1021/acs.analchem.0c04679. [DOI] [PubMed] [Google Scholar]
- 34.Banerjee A., Gokhale A., Bankar R., Palanivel V., Salkar A., Robinson H., Shastri J.S., Agrawal S., Hartel G., Hill M.M., Srivastava S. Rapid classification of COVID-19 severity by ATR-FTIR spectroscopy of plasma samples. Anal. Chem. 2021;93:10391–10396. doi: 10.1021/acs.analchem.1c00596. [DOI] [PubMed] [Google Scholar]
- 35.Delafiori J., Navarro L.C., Siciliano R.F., de Melo G.C., Busanello E.N.B., Nicolau J.C., Sales G.M., de Oliveira A.N., Val F.F.A., de Oliveira D.N., Eguti A., dos Santos L.A., Dalçóquio T.F., Bertolin A.J., Abreu-Netto R.L., Salsoso R., Baía-Da-Silva D., Marcondes-Braga F.G., Sampaio V.S., Judice C.C., Costa F.T.M., Durán N., Perroud M.W., Sabino E.C., Lacerda M.V.G., Reis L.O., Fávaro W.J., Monteiro W.M., Rocha A.R., Catharino R.R. Covid-19 automated diagnosis and risk assessment through metabolomics and machine learning. Anal. Chem. 2021;93:2471–2479. doi: 10.1021/acs.analchem.0c04497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reiken S., Dridi H., Sittenfeld L., Liu X., Marks A.R. BioRxiv : The Preprint Server for Biology; 2021. Alzheimer’s-like Remodeling of Neuronal Ryanodine Receptor in COVID-19. [DOI] [Google Scholar]
- 37.Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- 38.Martinez F.O., Gordon S., Locati M., Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 39.Levy B.D., Clish C.B., Schmidt B., Gronert K., Serhan C.N. Lipid mediator class switching during acute inflammation: signals in resolution. 2001. http://immunol.nature.com [DOI] [PubMed]
- 40.Elliott M.R., Koster K.M., Murphy P.S. Efferocytosis signaling in the regulation of macrophage inflammatory responses. J. Immunol. 2017;198:1387–1394. doi: 10.4049/jimmunol.1601520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serhan C.N., Chiang N., van Dyke T.E. Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat. Rev. Immunol. 2008;8:349–361. doi: 10.1038/nri2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugimoto M.A., Sousa L.P., Pinho V., Perretti M., Teixeira M.M. Resolution of inflammation: what controls its onset? Front. Immunol. 2016;7:160. doi: 10.3389/fimmu.2016.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Basu S., Helmersson J. Factors regulating isoprostane formation in vivo. Antioxidants Redox Signal. 2005;7:221–235. doi: 10.1089/ars.2005.7.221. [DOI] [PubMed] [Google Scholar]
- 44.Perico L., Benigni A., Casiraghi F., Ng L.F.P., Renia L., Remuzzi G. Immunity, endothelial injury and complement-induced coagulopathy in COVID-19. Nat. Rev. Nephrol. 2021;17:46–64. doi: 10.1038/s41581-020-00357-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., Bonaguro L. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin Y., Ji W., Yang H., Chen S., Zhang W., Duan G. Signal Transduction and Targeted Therapy. Vol. 5. 2020. Endothelial activation and dysfunction in COVID-19: from basic mechanisms to potential therapeutic approaches; pp. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Canzano P., Brambilla M., Porro B., Cosentino N., Tortorici E., Vicini S., Poggio P., Cascella A., Pengo M.F., Veglia F. Platelet and endothelial activation as potential mechanisms behind the thrombotic complications of COVID-19 patients. Basic Transl. Sci. 2021;6:202–218. doi: 10.1016/j.jacbts.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng Y., Edin M.L., Theken K.N., Schuck R.N., Flake G.P., Kannon M.A., DeGraff L.M., Lih F.B., Foley J., Bradbury J.A. Endothelial CYP epoxygenase overexpression and soluble epoxide hydrolase disruption attenuate acute vascular inflammatory responses in mice. Faseb. J. 2011;25:703–713. doi: 10.1096/fj.10-171488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ono E., Dutile S., Kazani S., Wechsler M.E., Yang J., Hammock B.D., Douda D.N., Tabet Y., Khaddaj-Mallat R., Sirois M. Lipoxin generation is related to soluble epoxide hydrolase activity in severe asthma. Am. J. Respir. Crit. Care Med. 2014;190:886–897. doi: 10.1164/rccm.201403-0544OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.S.M. Shoieb, M.A. El-Ghiaty, A.O.S. El-Kadi, Targeting arachidonic acid-related metabolites in COVID-19 patients: potential use of drug-loaded nanoparticles, (n.d.). 10.1007/s42247-020-00136-8/Published. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.