Abstract
Multiplex electrochemical biosensors have been used for eliminating the matrix effect in complex bodily fluids or enabling the detection of two or more bioanalytes, overall resulting in more sensitive assays and accurate diagnostics. Many electrochemical biosensors lack reliable and low-cost multiplexing to meet the requirements of point-of-care detection due to either limited functional biosensors for multi-electrode detection or incompatible readout systems. We developed a new dual electrochemical biosensing unit accompanied by a customized potentiostat to address the unmet need for point-of-care multi-electrode electrochemical biosensing. The two-working electrode system was developed using screen-printing of a carboxyl-rich nanomaterial containing ink, with both working electrodes offering active sites for recognition of bioanalytes. The low-cost bi-potentiostat system (∼$80) was developed and customized specifically to the bi-electrode design and used for rapid, repeatable, and accurate measurement of electrochemical impedance spectroscopy signals from the dual biosensor. This binary electrochemical data acquisition (Bi-ECDAQ) system accurately and selectively detected SARS-CoV-2 Nucleocapsid protein (N-protein) in both spiked samples and clinical nasopharyngeal swab samples of COVID-19 patients within 30 min. The two working electrodes offered the limit of detection of 116 fg/mL and 150 fg/mL, respectively, with the dynamic detection range of 1–10,000 pg/mL and the sensitivity range of 2744–2936 Ω mL/pg.mm2 for the detection of N-protein. The potentiostat performed comparable or better than commercial Autolab potentiostats while it is significantly lower cost. The open-source Bi-ECDAQ presents a customizable and flexible approach towards addressing the need for rapid and accurate point-of-care electrochemical biosensors for the rapid detection of various diseases.
Keywords: Dual working screen-printed electrode, Electrochemical immunosensing, Bi-potentiostat readout system, SARS-CoV-2 nucleocapsid protein, COVID-19, All-in-one data acquisition platform
Graphical abstract
1. Introduction
The accuracy, selectivity, and diagnostic value of conventional analytical diagnostic technologies focusing on the detection of a single analyte are limited for many disease conditions because they provide limited sample information for disease diagnosis (He et al., 2017; Jones et al., 2019; Kadimisetty et al., 2015). Tests that can be used at the point of care/in the field/in the home (POCT) that are rapid, reliable, adequately sensitive/specific, cheap, use low sample volume, and can simultaneously detect multiple analytes using a single testing assay, commonly known as multiplexed detection, are required for a number of use cases (Dai et al., 2021; Huang et al., 2020; Timilsina et al., 2021).
For infectious diseases of clinical, agricultural, environmental, and food importance, accurate and low-cost multiplex testing techniques and assays yet to be developed to enhance the ability in responding to various public health emergencies (Fyfe et al., 2007; Gao et al., 2017; Ryu et al., 2013; Sood et al., 2020; Yang et al., 2006a; Zhang et al., 2021b). Multiplexing, for example for SARS-CoV-2, can provide time-dependent information about the dynamic of infection in the individual and population scales (Lin et al., 2020; Parihar et al., 2020; Rosado et al., 2021; Zhu et al., 2020). Antigen markers of the virus are present in different biofluids at the onset of the infection, making them attractive for early diagnosis of viral infectious (Schildgen et al., 2021). Simultaneous detection of viral antigens in a low-cost and accurate detection assay can be used to reliably detect the viral infection, independent from knowing the onset of the infection (Shan et al., 2021; Veyrenche et al., 2021; Zonneveld et al., 2021). Longitudinal monitoring of these biomarkers using point-of-care kits can exclusively provide personalized information about seroconversion of the infected individuals regardless of being symptomatic or asymptomatic (Norman et al., 2020; Ogata et al., 2020). Moreover, multiplex detection has also been used for detecting one biomarker but to further i) suppress the matrix effect on sensing signals in complex biofluids, ii) using similar or different detection methods for performing self-validation (Roda et al., 2021; Sun and Liu, 2018; Zhang et al., 2021a), and iii) increasing the sensitivity of the testing biosensors.
Numerous efforts have been devoted to realizing simultaneous detection of multi-analytes using a range of multiplex testing technologies, such as chromatography, mass-spectrometry, multiplex polymerase chain reaction, thermal cycling and isothermal amplification devices, and metagenomic next-generation sequencing (Gomaa and Boye, 2015; Gu et al., 2021; Huang et al., 2012; Neubert et al., 2020; Radmard et al., 2019; Reboud et al., 2019; Wang et al. 2021, 2022). These analytical methods significantly increase detection efficiency primarily in an accredited laboratory and clinical settings. However, most are too sophisticated and time- and reagent-consuming to be used as POCT devices. Some multiplex PCR assays are available for POCT but require instrumentation costing thousands of dollars (e.g. Xpert, Biofire) and are not designed to be portable, which limits widespread deployment at the point of clinical care, in the home or the field. In infectious diseases such as SARS-CoV-2 POCT that are not multiplex, PCRs detect only one or two analytes plus control and are limited by poor sensitivity (Döhla et al., 2020). Multiplex testing immunoassays have also been extensively used for the detection of more than one analyte using microsphere immunoassays, label-free array technologies, and fluorescent protein microarrays but their implementation deals with several manual processes which make them complicated for the POCT use (Schultz et al., 2021; Wong et al., 2017).
Biosensing systems are of great interest for multiplex point-of-care diagnostics due to their low-cost, high sensitivity, and ease of use (Liu et al., 2020; Van Nguyen et al., 2019). The use of immuno-biosensors has gained competitive advantages for point of care (POC) detection of single or multiple analytes (Khetani et al., 2021; Xue et al., 2020). Whereas single-plex optical systems are widely available in the market for the detection of various biomarkers, there are currently a few manufacturers that offer multiplex immunoassays for rapid disease diagnostics (Davis et al., 2020; Furuya et al., 2019; Harmon et al., 2017; Marquette et al., 2012). Optical-based detection of multi-analytes mainly utilizes multiple labels such as fluorescent materials and metal compounds to tag capture molecules or the corresponding analytes (Zanchetta et al., 2021). These methods mostly suffer from a limited number of labels, signal overlapping, need for large readout instruments, and differing optimal reaction conditions of the labels, constantly challenging their wide clinical application in multiplex analysis (Asghari et al., 2021; Joshi et al., 2021). Label-free optical technologies such as surface plasmon resonance have been developed with the aim of addressing the POC multiplexing needs of biosensors, however, because of their limited specificity, sensitivity and selectivity, the label-free mode has not been widely employed in clinical diagnosis (Choi et al., 2020; Hausler et al., 2019; Jebelli et al., 2020; Patil et al., 2019).
Alternatively, label-free, accurate, quantitative, and low-cost electrochemical biosensors are appealing for multiplex POC biomarker detection and have been practiced by several groups for the detection of multiple biomarkers in cancer (Akhavan et al., 2014; Pimalai et al., 2021), infectious diseases (Wu et al., 2018), and brain injuries (Khetani et al., 2021). However, a majority of low-cost electrochemical biosensors can only detect a single analyte or have limited accuracy and repeatability (Abdullah et al., 2019; Bianchi et al., 2019; Hoilett et al., 2020; Pruna et al., 2018; Wu et al., 2018). New multiplex electrode designs, electrochemical biosensors, and hand-held potentiostats with the capability of low-cost point-of-care detecting of multiple bioanalytes in complex clinical samples are needed to address the unmet need of multiplex and/or self-validating needs of point-of-care biosensing.
The design of electrodes has become an important aspect of the reliable and stable performance of biosensors. Multi-electrode detection needs can be met on the basis of the parallel assembly of three-electrode configuration screen printed electrodes (SPEs) (Gao et al., 2017; Sin et al., 2013). This strategy is relatively simple, with careful design and optimization, preventing cross-contamination and mutual interference among different analytes. However, increased reagent and sample volumes and sample addition times are typically required to complete the analysis (Arduini et al., 2016). The multi-electrode designs with multiple isolated working electrodes (WEs), known as spatial multiplexing, while using the common reference and counter electrodes in one single electrochemical sensing unit is an appealing strategy to increase the simultaneous detection capacity of the biosensors (Rafi and Zestos, 2021). To address this need, dual or multiple electrode designs including designs with circled, centered, or elliptic WEs have been developed and tested for multiplex analyte detection (Eissa and Zourob, 2020; Liu et al., 2018; Viswanathan et al., 2012) (Eissa and Zourob, 2020; Neves et al., 2013; Viswanathan et al., 2012). Despite the success of multiplex electrochemical biosensors for detection of small molecules and metabolites (e.g. maltose and glucose) in different biofluids, their utility for detecting low abundance concentrations of protein analytes in complex biofluids has shown to be limited (Jothimuthu et al., 2011; Neves et al., 2013).
Highly sensitive detection of protein requires coating of WEs with conductive nanomaterials followed by functionalizing the electrodes but due to the small size of electrodes and multistep preparation and functionalization steps needed for multiplex electrode designs, the coating and functionalization of WEs is a cumbersome process and often deal with a limited reproducibility (Parlak et al., 2017; Prakash et al., 2013; Salahandish et al. 2018a, 2018b, 2019a, 2019b; Yang et al., 2006b). To eliminate these challenges, single-step nanomaterial coating protocols and approaches have been exploited to eliminate complex nanomaterial coating and improve sensing reproducibility while preserving the sensitivity needed for ultrasensitive and quantitative detection of biomarkers (Khetani et al., 2019; Shu et al., 2015; Sun et al., 2021). However, their utility for multiplex biosensing has yet to be explored.
On the other hand, several groups have developed portable multichannel potentiostats but they are still expensive to be used for low-cost point-of-care sensing in settings like detection of COVID-19 outside of a laboratory (Alam et al., 2021; Ramfos et al., 2013; Wu et al., 2018). Also, the curve-shaped output readings of these potentiostats are not universally understood, require further data processing and interpretations, and complicate the practical implementation of multiplex electrochemical immuno-biosensors for POCT applications. We recently developed a multiplex μDrop potentiostat readout that enables simultaneous detection of up to eight protein biomarkers and demonstrated its equivalent performance to the commercial potentiostats (Khetani et al., 2021). However, this multiplex potentiostat is still expensive (<$1500) and bulky. Also, to record accurate signals from each of the sensing electrodes, the signals need to be recorded at each testing frequency for all WEs before switching to the subsequent testing frequency. Further miniaturization, parallelization, and lower cost multiplex potentiostats (e.g., <$100) are needed to meet the needs of multiplex potentiostats for PoC applications.
In the present work, we developed a binary electrochemical data acquisition (Bi-ECDAQ) system with novel electrode designs offering multiple specific sensing areas, accompanied by a customized potentiostat to address the unmet need of POC multiplexing for electrochemical biosensors. A two-working electrode design was printed using a novel carboxyl-rich nanomaterial containing ink, with both working electrodes offering active and independent sites for accurate recognition of bioanalytes without the need for any functionalization step. Also, a bi-potentiostat system (∼$80) was developed and customized specifically to the bi-electrode design and aimed to perform rapid, repeatable, and accurate simultaneous measurement of electrochemical impedance spectroscopy signals recorded from both WEs. The combined dual biosensors and potentiostats was then used for measuring SARS-CoV-2 Nucleocapsid protein (N-protein) in both spiked samples and clinical nasopharyngeal swab samples from SARS-CoV-2 infected patients (An alpha variant of concern). This Bi-ECDAQ system presents a breakthrough, customizable, and flexible approach towards addressing the need for multiplex point-of-care electrochemical biosensors.
2. Materials and methods
2.1. Electrode design and fabrication
All technical drawings for the creation of the two-working electrode design were sketched using AutoCAD 2021 Version R.47.0.0. The electrode dimensions and lead distances were selected according to the commercial connector (Molex, 0472861001), screen-printing manufacturing constraints, and the commercially available sensing strips in the market. A Graphene@PEDOT:PSS hybrid ink dispersed in Dimethylformamide (DMF) (Sigma, USA) was mixed with the carbon ink (7102, Dupont, USA) and subject to standard mechanical stirring with 200 rpm rotational speed. Micro Flatbed Printer (A.W.T. World Trade, Inc., USA) was used for printing the electrodes using a polyester mesh. The printed electrodes were then cured and dried by an InfraRed curing machine (Customized Natgraph Air Force Dryer, Natgraph Ltd, UK). Double-sided pressure-sensitive adhesive sheets (ARcare 90106NB, Adhesive Research Inc., USA) were cut using Speedy 360 flex CO2 laser engraver machine (Trotec, USA) to produce the insulator masks, acting as a dielectric layer for covering the conductive areas of the electrodes, leaving only the desired parts in contact with the redox system.
2.2. Bi-ECDAQ definition and parameters
The potentiostat unit of the Bi-ECDAQ is a high precision, programmable, auto-calibrated readout system composed of Impedance converter (AD5933), Analog Front End, Memory, Programmable clock, and Microcontroller Unit breakout board (SparkFun Pro nRF52840). The proposed potentiostat reads the impedance values of the two integrated but independent immuno-biosensors, and transfer the corresponding digital data to the computer or smartphone through wired or wireless communication protocols such as Universal Serial Bus (USB) and Bluetooth, respectively. The full details of the design and implantation of the custom-made potentiostat of the Bi-ECDAQ are shown in the supplementary (Fig. S8 to Fig. S24; Table S3 to Table S7). The microcontroller receives from the GUI, all the commands to initialize, calibrate the system, and request the impedance converter to generate the signals to apply to the immuno-biosensors. The impedance recorded changes proportion to the concentration of the target analyte. To expedite the sensing without compromising the accuracy, the complete impedance spectrum is obtained from 7 logarithmically spaced points over the frequency range from 1400 Hz to 1 Hz for both working electrodes (WE1 and WE2).
2.3. Numerical simulation of electrochemical biosensing
The electrochemical module of COMSOL Multiphysics 5.6 was used for 3D simulation of the primary current distribution and the potential contour of the four-electrode design. The domain and boundaries of the simulated system, the surface of the electrodes, and the boundaries of the electrolyte were defined. The primary current distribution interface in the electrochemistry module specifies the transport of charged ions in an electrolyte of homogeneous composition and current conduction on the electrode by Ohm's law and the charge balance principle. (COMSOL user guide module). Ohm's law is considered in charge transfer inside the electrolyte. This modeling considers homogeneous electrolyte and equilibrium value of the potential of the electrolyte–electrode interface without any significant effect of convection on current density (Stevenson et al., 2018). The counter, reference, the two WEs, and the surrounding electrolyte were used to form the geometric model of the biosensor. According to the open circuit potential (E ocp), 170 and 0 mV potential were provided to the working and counter electrodes, respectively, to apply the physics of interest. On the other hand, electrical insulation was applied to the reference via a von Neumann boundary condition. The governing equations for this simulation in COMSOL Multiphysics were provided in the supplementary (SI-A).
2.4. Characterization of electrodes and biosensors
Scanning Electron Microscopy (SEM, JSM-7001FA) was used to characterize the morphology of the WEs’ surface. Combining the SME data with the simultaneous elemental analysis using the same apparatus, through the mapping module, characterized the presence and distribution of chemical elements on WEs. The microscopy images were acquired under high voltage (7.5 kV) and high vacuum (∼10−3 Pa) conditions without any need for pre-coating of the electrode. PGSTAT 204 Potentiostat/Galvanostat (Metrohm, USA) was utilized as the commercial potentiostat for comparing the performance of the portable Bi-ECDAQ potentiostat. Electrochemical Impedimetric Spectroscopy (EIS) measurements were performed with the signal amplitude of 10 mV, the single sinusoidal wave type, and the frequency range of 1400 Hz–1 Hz, with 7 or 50 points as the number of frequencies tested. The E ocp was selected to be 170 mV.
2.5. Immuno-biosensor preparation and testing
SARS-CoV-2 Nucleocapsid Antibody (GenScript Inc. USA, HC2003, #A02039) was diluted from the stock solution to the concentration of 50 μg/mL in phosphate buffer saline (PBS) (1X, Growcells, USA, pH = 7.4, #MRGF-6230) and drop cast on the surface of both working electrodes using a manual pipette. Electrodes were then incubated for 1 h at room temperature followed by overnight incubation at 4 °C. Then, the immobilized electrodes were rinsed using Milli-Q water three times. As for the blocking agent (the passivation step), 0.005% (w/v) bovine serum albumin (BSA; ChemCruz, #sc-2323, USA), prepared in PBS, was cast coated on WEs and incubated at room temperature for 30 min. Various concentrations of SARS-CoV-2 N-protein antigen (GenScript Inc. USA, #Z03488), ranging from 100 fg/mL to 10 ng/mL were prepared by serial dilution of the spiked protein in PBS. Nasopharyngeal samples positive or negative for SARS-CoV-2 were obtained from the Alberta Public Health Laboratory (Alberta Precision Laboratories, Calgary, AB). Samples were initially collected in universal/viral transport media for clinical diagnosis of COVID-19 and frozen at −20 °C after testing. All positive SARS-CoV-2 samples were identified as the Alpha variant of concern (B.1.1.7 lineage). The clinical sample testing was performed in a Biocontainment Level Class II laboratory at the University of Calgary (Ethics ID: REB20-1032), without any prior heat treatment, deactivation, or other sample preparation steps. BSA, IgG from human serum (Sigma, USA, #I4506), and SARS-CoV-2 Spike protein (S1) (Genscript Inc., USA, #T80202) were used to assess the specificity of the N-protein biosensors. All electrochemical measurements were conducted using a PBS solution containing 2.5 mM of potassium hexacyanoferrate (II) trihydrate (#33358, Alfa Aesar, Thermo Fisher Scientific., USA) and 2.5 mM Potassium ferricyanide (III) (#702587, Sigma, USA) as the redox couple. Panbio COVID-19 N-protein Ag Rapid Test devices were purchased from Abbott, USA.
3. Results and discussion
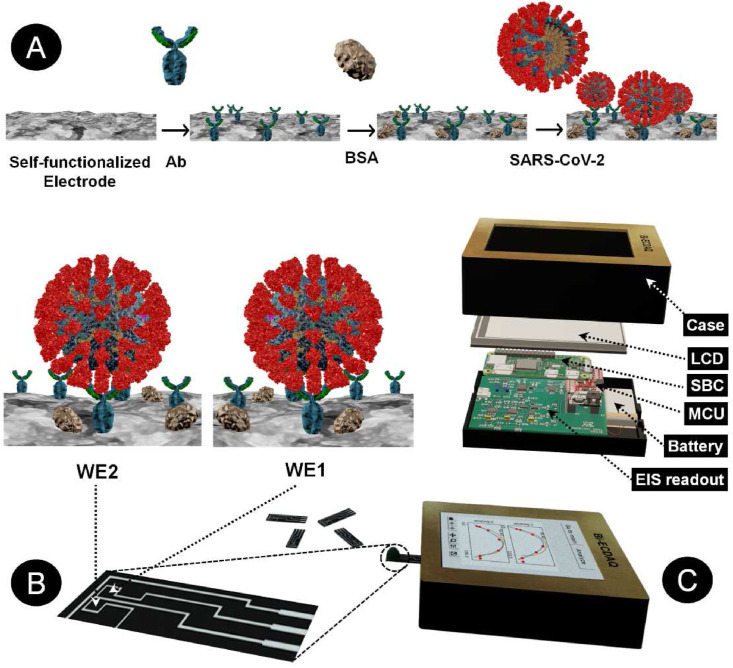
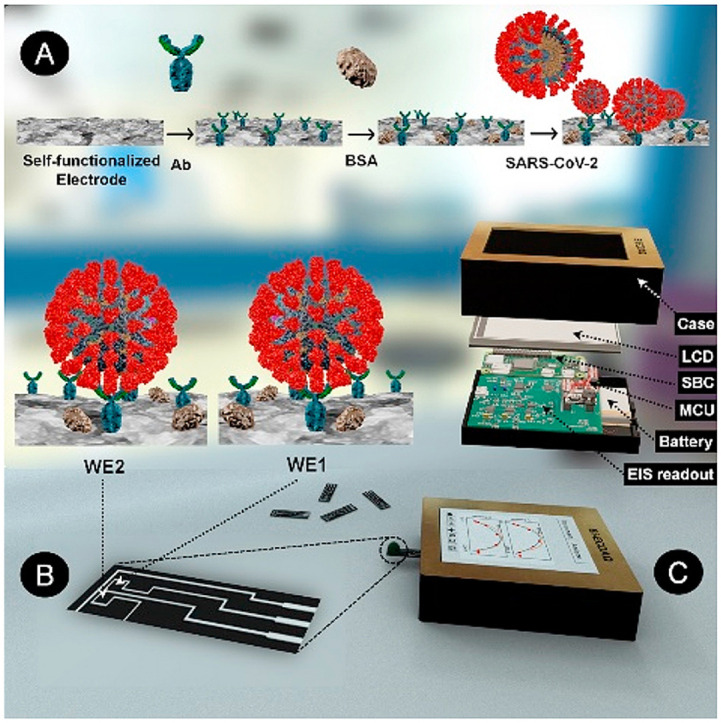
An electrochemical screen-printed electrode was designed in accordance with the principles of three-working electrode configurations, with adding an additional working electrode enabling either the detection of two different analytes or recognizing one analyte and its matrix effect (Scheme 1 ). The working electrodes are located at the center of the design, surrounded by a continuous counter electrode, providing the path for proper current transfer between the two electrode types (Scheme 1A). The electrode designs, customized for screen-printing using the Carbon/Graphene@PEDOT:PSS hybrid ink, ensure the consistent gap of 0.3 mm between the electrodes, printed on a flexible polyethylene-terephthalate (PET) layer. Scheme 1B depicts the step-by-step modification of the surface of the homemade 2 WEs strip to have a specific sensor for the detecting N-protein of SARS-CoV-2. The EIS custom-made readout set-up is also shown in Scheme 1C, containing the EIS readout, battery, MicroController Unit (MCU), Single Board Computer (SBC), Liquid Crystal Display (LCD), and the case. The electrode strip is 6.65 mm × 28.2 mm in size, with each working electrode's surface area to be 2.76 mm2. To avoid the shielding effect, the reference electrode is located at an optimal distance from WEs, preserving the electrical potential between the counter and WEs (Fig. 1 A–C). Also, the successful production of the intended design, in terms of final feature size and less than 0.5 mm gap between the electrodes, shows that the inks were suitably mixed to form a uniform dispersion with no clots and possess suitable viscosity for printing and curing. Numerical electro-analysis was performed to validate the performance of the design in terms of the acceptable distribution of potential and current streamlines.
Scheme 1.
A representation of various components of the Binary Elechtrochemcial Data Acquisition (Bi-ECDAQ) biosensing platform. A) and B) The strip component of the Bi-ECDAQ platform is composed of a two-working electrode (WE) design screen-printed with inks containing adherent carboxyl functional groups for covalent bonding with the antibody probe. The surface of both WEs is blocked with bovine serum albumin (BSA), preventing non-specific bindings upon the incubation with nasopharyngeal swab samples containing SARS-CoV-2 virus particles. C) The custom-made bi-potentiostat readout of the Bi-ECDAQ extracts the Electrochemical Impedance Spectroscopy (EIS) response (Nyquist plots and Charge transfer resistance) of the plugged biosensor strip. The extracted data are processed through the embedded MicroController Unit (MCU) and Single Board Computer (SBC), while represented to the user via an in-place Liquid Crystal Display (LCD).
Fig. 1.
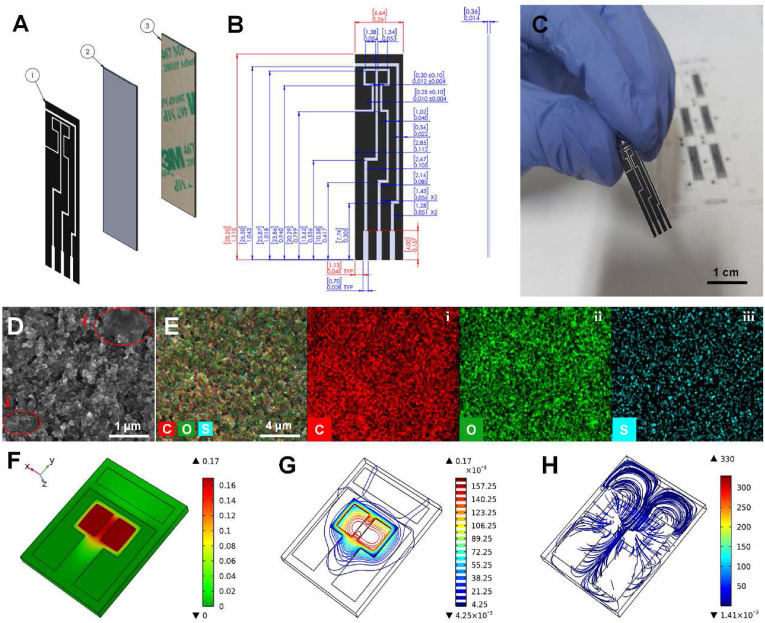
Technical dimensions and physical characterizations of the dual working electrode design. A) A representation of different layers of the electrode including 1) printed hybrid ink (Carbon/Graphene@PEDOT:PSS), 2) screen-printing polyethylene terephthalate (PET) substrate, and 3) The lamination layer to reach the desired thickness of the strip. B) Technical drawing of the electrode representing the dimensions and tolerances for screen-printing production. C) Real image of the printed electrode. D) Scanning Electron Microscopy (SEM) images provided in 7.5 kV and 10-3 Pa (scale bar: 1 μm), with showing semi-transparent graphene sheets (i) and E) Elemental analysis (mapping) showing the (i) carbon, (ii) oxygen, and (iii) sulfur contents of the printed electrodes (scale bar: 4 μm). The simulation results for F) surface plot of the electrolyte potential, G) contour plot of the electrolyte potential, show the isopotential surfaces within the electrolyte. H) Vector plot showing the distribution of current density.
3.1. Characterization of the screen-printed electrodes and finite element electroanalysis
Morphological characteristics of the WEs were characterized using SEM (Fig. 1D). To provide functional groups needed for biosensing over WEs, the base carbon ink was intermixed with Graphene@PEDOT:PSS hybrid ink and screen printed on the PET substrate to create the continuous and uniform electrode design. The SEM images depict the uniform distribution of the base carbon particles and the ink binders. The highlighted area in Fig. 1D, i represents the graphene characterized in the form of semi-transparent sheets on top of the underlying carbon features. The image also indicates a semi-porous surface with fragmented structures uniformly attached by the binders.
The elemental analysis performed for the area of interest in the SEM image shows the uniform distribution of the chemical elements present in the carbon base ink and the added Graphene@PEDOT:PSS hybrid ink. The absence of cracks in the printed electrodes, which is considered an essential factor for ensuring the integrity of the ink and the cured electrode, confirms that the properties of the final ink mixture were compatible with the needs of screen printing processes. Carbon is present continuously throughout the imaging zone, represented with red color, the high intensity of which can be associated with the carbon-based accounting for 80% of the final printing mixture (Fig. 1E, i). The high intensity of the oxygen, with green color, is attributed to the abundant presence of oxygen in the PEDOT:PSS chemical structure. The pattern of the un-merged mapping images confirms the presence of sulfur present in the PEDOT:PSS polymeric chain, illustrated with blue color.
Finite element analysis using Electrochemistry Module (Primary Current Distribution) in COMSOL Multiphysics was conducted to simulate the electrode response in the presence of the electrolyte and optimize the electrode design for the improved performance (Fig. S1A). Simulations were run to characterize electric field distribution on the electrodes and determine the quality of multi-sensing. To assess the surface-electrolyte potential difference and charge distribution at the electrode-electrolyte interface, a very fine mesh was used to simulate surface current density and electrolyte potential. The tetrahedral mesh with the high resolution near the WEs and the counter electrode was used to divide the model into about 106 finite elements (Fig. S1B). The electrolyte potential distribution was simulated at both surfaces of the electrodes and the electrolyte volume, through the heat map and contour representations, respectively. The results show that electrolyte potential distribution is very strong near the WE1 and WE2 and steadily diminished toward zero at the counter electrode (CE) while the contours are symmetric in the region of the WEs (Fig. 1F and G). Moreover, the vectors of electrolyte current density are shown to be perpendicular to the boundaries of the reference electrode (RE), showing that there is no current passing through the insulated region of the RE. Additionally, current vectors are symmetric due to the position of the electrodes in the symmetric design.
The potential and current density distributions of the electrolyte along the cut line (Fig. S1A) were investigated to assess the symmetric behavior of the two WEs. The simulation result shows that along the cut line, the output potential and current varied from 170 mV to 0 mV and 260 A/m2 to 0 A/m2, respectively (Figs. S1C and S1D). The potential of 170 mV and the charge density of 20 A/m2 were obtained between the two WEs. The magnitude of the current density between the two WEs was 91% less than the maximum current density at the region between the working and counter electrodes which implies an enhanced electrochemical response for the electrode design.
3.2. Electrochemical performance of Bi-ECDAQ
The Bi-ECDAQ potentiostat is a hand-held, portable, and low-cost solution to detect and quantify multiple biological molecules based on impedance spectroscopy technique. Against commercial potentiostats, the custom-made Bi-ECDAQ was programmed to measure the impedance in particular frequencies instead of scanning in a range of frequencies that are routinely used in the benchtop EIS spectroscopies. With this capability, the proposed Bi-ECDAQ not only offers the advantages of a low-cost and handheld impedimetric readout system but also tunes the number of measurement frequency points to achieve high accuracy without extending the response time. In other words, the programmable, handheld, and low-cost Bi-ECDAQ potentiostat offers the advantages of higher accuracy in the short response time. The total price of this reader is about $80.25 which is significantly cheaper than the multiplex Autolab instrument potentiostats (>$6000). A list of elements used in the Bi-ECDAQ potentiostat reader along with their prices is shown in the supplementary (Table S5). The price would be much less in mass production of this handheld device. Additionally, the dimensions of the hand-held Bi-ECDAQ is 8.3 × 6 × 2.4 cm3 which is smaller than the desktop-based Autolab instrument (15 × 26 × 20 cm3). The functional tests performed with different concentrations of the redox solutions showed the same trend of high impedance values for low redox concentrations as well as low impedance for high redox concentrations, given that the ions in the electrolyte favor the transfer of charge.
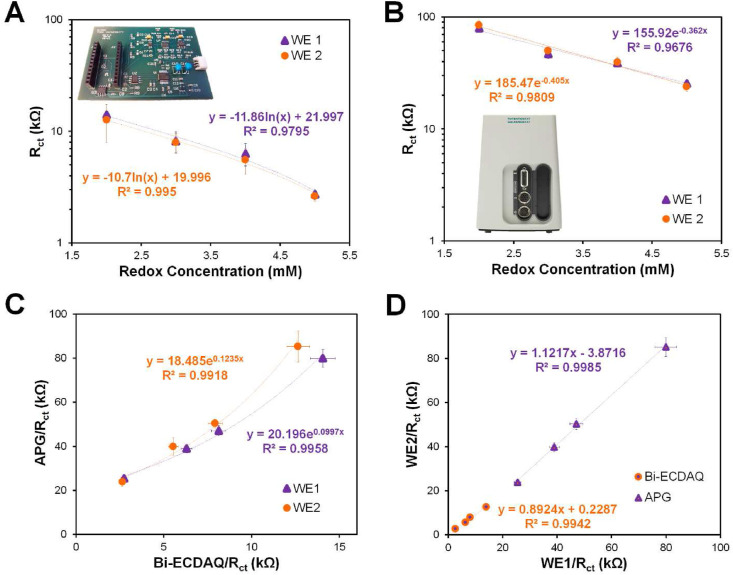
The electrochemical functionality of the Bi-ECDAQ potentiostat was compared to the available commercial Autolab Potentiostat/Galvanostat (APG) (PGSTAT, Metrohm Autolab) by conducting EIS measurement within the customized frequency range (1400 Hz–1 Hz) for 7-point data (Fig. 2 ; Figs. S20 and S22). This 7-point data was shown to be the minimum number of frequency points that the potentiostat can reliably detect the impedance without compromising the accuracy. The signals of both WEs were extracted by drop-casting the redox solution, [Fe(CN)6]3-/4- prepared in 1X PBS, with various concentrations of 2–5 mM on top of the four-electrode system to form the electrochemical cell. Both WEs were functional within the range of redox probe concentrations tested using the Bi-ECDAQ potentiostat, providing a logarithmic correlation between the redox concentration and charge transfer resistance signals (Rct), with R2 of 0.97 for WE1 and 0.99 for WE2 (Fig. 2A). Also, the results showed an exponential relationship between Rct values measured by APG potentiostat for WE1 and WE2 for all redox concentrations (Fig. 2B). The EIS signals for each working electrode measured by the Bi-ECDAQ potentiostat were fitted in an exponential correlation with the signals obtained using the APG for all redox concentrations in the redox range of 2 mM–5 mM, with R2 of 0.99 for both WE1 and WE2 (Fig. 2C). To evaluate the identical performance of the two WEs and the potential difference in the signals obtained, Rct measured with both APG and Bi-ECDAQ potentiostats for different redox concentrations were plotted for WE1 versus WE2 (Fig. 2D). The curves for the Bi-ECDAQ (orange) and APG (purple) were fitted linearly with R2 of 0.99, and with 10.76% and 12.17% in relative error percentage deviation from y = x line, respectively. For all measurements, the utilized sensors were randomly selected from various batches of manufactured electrodes, which were maintained in the laboratory environment for over six months. The associated error bars in Fig. 2 are attributed to three replicates of these electrodes which depict the high stability of the printed sensors over the mentioned time slot.
Fig. 2.
Electrochemical comparison of the homemade Bi-ECDAQ and commercial Autolab potentiostats by measuring EIS signals for both working electrode (WE1) and WE2 drop-casted with different concentrations of [Fe(CN)6]3-/4- redox probe. Measurements for both WEs performed using A) Bi-ECDAQ system and B) Autolab Potentiostat/Galvanostat (APG). C) Exponential correlation of the signals measured by Bi-ECDAQ and APG for both WE1 and WE2. D) Evaluating the identical performance of WE1 and WE2 in different redox concentrations measured by Bi-ECDAQ and APG potentiostats.
3.3. Electrochemical detection of SARS-CoV-2 nucleocapsid (N-) protein using Bi-ECDAQ potentiostat
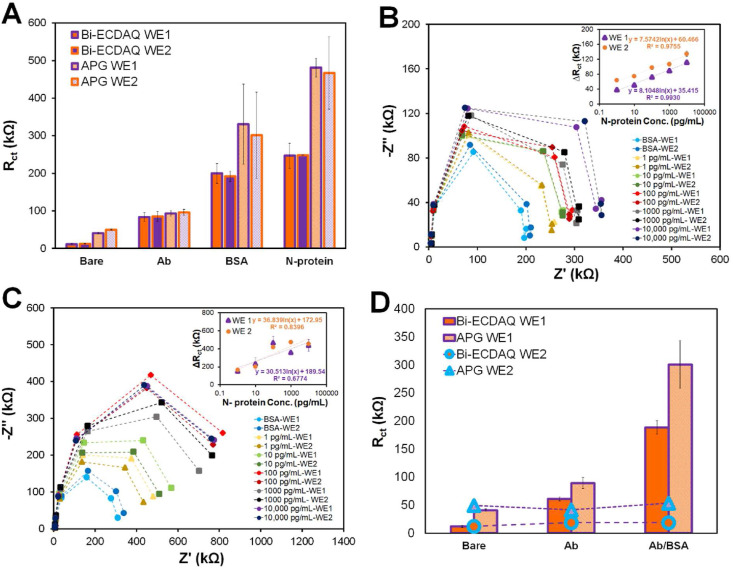
Standard concentrations of SARS-CoV-2 N-protein, ranging from 0.1 pg/mL to 10,000 pg/mL, were prepared by serial dilution of the spiked protein in PBS. To assess the performance of the immuno-biosensors for detecting SARS-CoV-2 N-protein, it was prepared by immobilizing the antibody on both WEs and passivating it with BSA (details in section 2.3). The spiked samples containing the defined concentrations of the prepared antigen solution were incubated on the biosensor for 30 min at room temperature. EIS testing was carried out in 1X PBS solution containing 2.5 mM [Fe(CN)6]3-/4- redox probe. Both WEs were measured simultaneously, using the Bi-ECDAQ reader, with an average measurement time of 120 s per test. Fig. 3 A shows the corresponding bar chart of Rct variations according to each step of surface modification, from the bare electrode to the signal recorded for detecting 1 pg/mL N-protein antigen. The addition of antibodies to the surface of both WEs results in a rise in their Rct, reaching 76.7 kΩ for WE1 and 76.5 kΩ for WE2, confirming the successful immobilization of the bio-capture antibody on WEs. Similarly, incubating the antibody-immobilized WEs with BSA increased Rct to 120.4 kΩ and 132.4 kΩ for WE1 and WE2, respectively. These signals were also obtained using the APG potentiostat where both WEs were successful in detecting the deposited layers. The corresponding Nyquist plots were also represented for the stepwise surface modification (Fig. S2). Fig. 3B represents the Nyquist plot and the corresponding calibration curve for the Bi-ECDAQ measurements for different concentrations of N-protein in PBS. The concentration range is correlated with the Rct signal based on the logarithmic equation y = 8.1ln(x) + 35.4 for WE1 and y = 7.6ln(x) + 60.4 for WE2. Increasing the concentration of the protein increases the charge transfer, confirming the formation of a protein layer on the electrode surface which hinders the transfer of electrons accordingly. In a similar manner, the Nyquist plot and the associated calibration curve for the measurements performed by APG is presented in Fig. 3C. Inconsistencies can be observed for higher concentrations of the protein (1000 pg/mL to 10,000 pg/mL) for this system, resulting in a lower R2 of 0.67. As shown by the calibration curves, the linear detection range of the biosensor recorded by the Bi-ECDAQ potentiostat is determined to be 1 pg/mL to 10,000 pg/mL for detecting SARS-CoV-2 N-protein in PBS, with the limit of detection (LoD) of 115 fg/mL for WE1 and 150 fg/mL for WE2, confirming the reproducible response of the biosensor and its credential for detecting N-proteins in clinical samples at the early stage of the infection (Norman et al., 2020), as well as the capacity of electrochemical biosensors for yielding ultra-low limit of detections (Akhavan et al., 2012; Rahmati et al., 2021).
Fig. 3.
Analytical performance of the Bi-ECDAQ potentiostat reader for measuring spiked concentrations of SARS-CoV-2 N-protein in PBS using the N-protein immuno-biosensor. A) Electrochemical characterization (in terms of Rct response) of the stepwise surface modification of the immuno-biosensors on both WEs measured by Bi-ECDAD and APG. B) Nyquist plot representation and calibration curves of the EIS measurements performed by Bi-ECDAQ for various concentrations of N-protein analyte. C) Nyquist plot representation and calibration curves of EIS response measured by Autolab potentiostat for various concentrations of N-protein spiked in PBS. D) Rct values representing the independent response of WE1 from WE2, when only WE1 is modified with antibody and BSA while WE2 remained bare. The measurements were performed using both Bi-ECDAC and APG readers.
To assess the risk of cross-reactivity between the two working electrodes, the Rct signals were obtained for a condition where WE2 is Ab and BSA-free (bare) while WE1 is coated with the antibody and BSA. The signals recorded by both Bi-ECDAQ and APG readers measured from WE2 show no significant difference among the three states of I; one-way ANOVA) uncoated WE1 and uncoated WE2, ii) coated WE1with Ab and uncoated WE2, and iii) coated WE1 with Ab/blocked BSA and uncoated WE2, wherein in all these three states, WE2 shows the same signal as the bare WE2 (Fig. 3D), confirming the independent performance of each WEs in the system.
To evaluate whether the Rct signals measured by the 7-point frequency scan carried out by the Bi-ECDAQ reader have any dependency on the number of frequency points, the same immuno-biosensors underwent the EIS measurements with similar conditions (i.e., the frequency range of 1400 Hz–1 Hz and E ocp of 170 mV) but recorded at two different numbers of frequency points (7-point frequency and 50-point frequency) using the APG potentiostat. Comparing the Rct values measured show that there exists a linear correlation between the Rct signals obtained using 7-point and 50-point frequency scan numbers for each of the WEs (Fig. S3). The linear curve of the 7-point frequency scan versus 50-point frequency scan is fitted with the slope of 0.90 and 0.85 (R2 of 0.99), as compared to the ideal slope of 1 for WE1 and WE2, respectively. The analytical performance of the presented biosensor in this work in terms of LoD, dynamic range, and potential for multi-sensing was compared to the other electrochemical biosensors developed for the detection of one or multiple SARS-CoV-2 biomarkers (Table S1).
3.4. Selectivity assessment of the immuno-biosensor measured by Bi-ECDAQ
The selective response of the immuno-biosensor to the SARS-CoV-2 N-protein analyte was evaluated by exposing the biosensor to other proteins that may have interaction with the selective antibody, such as Immunoglobulin G (IgG) and the SARS-CoV-2 Spike protein (Crescenzo-Chaigne et al., 2021; Lee et al., 2021). These proteins are abundantly found in NP swab samples of COVID-19 infected patients (Strömer et al., 2020). The electrode immobilized with 50 μg/mL SARS-CoV-2 Nucleocapsid antibodies were incubated with 1 pg/mL of each of the non-specific proteins (in PBS for 30 min). The results obtained by one-way ANOVA analysis show no significant difference among Rct signals of the biosensor for both WEs in the absence and presence of IgG or S-protein. The selective response of the immuno-biosensor to N-protein is confirmed with the increase in the EIS signals of the biosensor for both WEs incubated with samples containing N-protein. The difference is also not significant in the signals collected from the biosensors for samples spiked with both N- and S-proteins compared to the samples spiked only with N-protein. The difference is significant in the signal of the samples with and without N-protein, confirming high selectivity of the immunosensor for detection of N-protein compared to other proteins (P value < 0.05) (Fig. S4).
3.5. Detection of SARS-CoV-2 in NP swab samples from infected patients using the point-of-care Bi-ECDAQ system
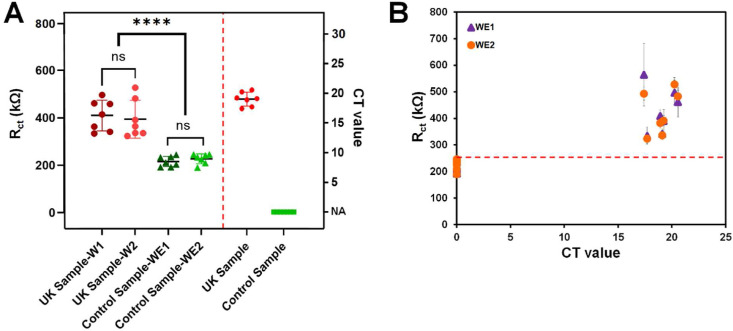
The performance of the immuno-biosensor and the Bi-ECDAQ reader was evaluated by detecting seven SARA-CoV-2 positive NP swab samples collected in Universal/Viral Transport Medium (UTM) and seven negative controls. These samples were previously detected by real-time polymerase chain reaction (RT-PCR) and determined to be the Alpha variant (lineage B.1.1.7) SARS-CoV-2 samples. EIS was shown to be effective for detecting this variant (Torres et al., 2021). The clinical samples were dispensed on both WEs and incubated at room temperature for 30 min. The electrodes were then rinsed 3 times and plugged into the Bi-ECDAQ system, where the EIS measurements were conducted using the 2.5 mM [Fe(CN)6]3-/4- redox solution. Fig. 4 A shows Rct for both WEs for clinical samples. The cycle threshold (Ct) values of the RT-PCR test for both patients and controls were compared to the EIS results. The biosensor data show that Rct values measured for the UK variant patient samples are significantly different from the values obtained for the control groups (P value = 0.043), with the cut-off value of 247 kΩ. The results of EIS signals measured for the paired WEs (both detecting the target N-protein in the same clinical sample; Fig. 4B) show the success of both WEs in detecting COVID-19 patients, resulting in the highly similar Rct measured by the paired WEs and enabling the self-validation of COVID-19 testing using this system. These paired Rct data of SARS-CoV-2 positive NP swabs were also compared to the Rct of SARS-CoV-2 negative controls where the Ct values of each sample obtained from RT-PCR tests falling within the range of 17–21 is presented. The Rct of WEs measured upon exposure to the positive samples containing the target virus increased significantly, indicating the interaction of N-proteins with the immobilized N-protein antibodies on the surface of the immuno-biosensor. The biosensor and the readout system were able to successfully differentiate all positive samples from negative controls. The results from the clinical performance of the Bi-ECDAQ system were compared to the point-of-care Panbio COVID-19 N-protein Ag Rapid Tests performed as per the manufacturer's instructions. The Bi-ECDAQ platform was 100% in positive and negative agreement with the Panbio tests, demonstrating the clinical utility of the Bi-ECDAQ platform for rapid and point-of-care detection of SARS-CoV-2. In addition to the detection of SARS-CoV-2, the Bi-ECDAQ system presents quantitatively and digitally the viral load of the clinical samples, something that is not possible to report by the Panbio testing kits. The quantification of viral load is clinically important to assess disease severity and prognosis, delineate the stage of disease and monitor some COVID-19 patients (e.g severely immunocompromised) to assess their response to therapeutics and to help determine the length of isolation (Pujadas et al., 2020; Song et al., 2021). Further analytical and clinical validations with a larger number of samples are needed to determine the capability of the Bi-ECDAQ system to the quantitive measurement of SARS-CoV-2 viral loads. To evaluate the stability of the immobilized sensors at room temperature during the 5-h preparation and measurement time, the Rct signals of the immunosensors associated with EIS measurements for eight control samples (four samples in each day) were indicated in Fig. S5. The measurements were performed on two different days, with each sample tested on the immobilized sensors maintained in room temperature and humidity conditions after 3–5 h of placing the immobilized electrodes outside of the 4 °C refrigerator. Each measurement was conducted in a 30-min timespan for each of the tested clinical control samples. Three different electrodes, from different manufacturing batches, were used for replications represented in the error bars. The signals show that the immunosensors maintain their stability at room temperature, with no significant difference between the results of the measurements for each of the control samples even after 5 h of remaining at room temperature.
Fig. 4.
The performance of the Bi-ECDAQ sensing platform (combined immuno-biosensor and the potentiostat readout) for electrochemical detection of SARS-CoV-2 Alpha variant real-time polymerase chain reaction (RT-PCR) positive and negative nasopharyngeal (NP) swab samples. A) Charge transfer resistance signals measured for each of WEs for each sample and control groups and their comparison with the corresponding cycle threshold (Ct) values of RT-PCR. B) The paired WEs generated by the Bi-ECDAQ reader, resulting in the self-validation of COVID-19 detection performed by the Bi-ECDAQ.
3.6. Intra-reproducibility assessment of Bi-ECDAQ reader
Three different Bi-ECDAQ readers were used to measure the signals of the bare electrodes for evaluating the intra-reproducibility of this readout system. The EIS measurements were conducted within the frequency range of 1400 Hz–1 Hz and in the presence of 2.5 mM of redox probe solution. The mean value of Rct for each of the readers tested is 12.2 kΩ for WE1 and 11.2 kΩ for WE2, with Coefficient of Variations (CoV) of 6.9% and 8.0%, respectively (Table S2). The accuracy of Bi-ECDAQ can be improved in the future, by increasing the number of resistors shown in the impedance range extender block (See Fig. S2). This block serves as a self-calibration system.
3.7. System assembly, software development, and remote communication
To enable the use of the Bi-ECDAQ system for quantitative and digital point-of-care detection of SARS-CoV-2, we integrated three components: i) the reader's electronic board (EIS module), ii) 3.5-inch liquid crystal display (LCD), and iii) a low-power and low-cost Linux-based single-board computer (1x ARM core at 1 GHz). The single-board computer had the built-in wireless capability for communication with the Cloud through the internet or the devices in the local area network, enabling data gathering, storage, and analysis independently and fully automated (Fig. S6). The data gathered was monitored via the administrator or user panel of the designed telehealth web application. The application and all the analysis modules could be updated remotely. One charging module and one 18500 battery were also integrated to operate it for longer than 8 h. The standard serial port was used to develop the communication between the EIS board and the single-board computer.
The steps of GUI interface are illustrated in Fig. S7 while the operation procedure of the graphical user interfaces GUI is detailed in Figs. S18 and S19. First, the user logs into the device when the device is turned on (Fig. S7A). The user ID and a unique device ID (MAC address) are used to save data on the Cloud. The hashed MAC address is used for the key transfer between the web server and the device to increase communication security. Then the sample number is asked for the data processing and future data retrieving (Fig. S7B). After inserting the sample-incubated biosensor into the device, the start button initiates the EIS measurement of the biosensor for the sample tested (Fig. S7C). The sample number and its time and date are saved into a table in the database. After collecting the EIS data from the readers, the Nyquist plot of the complex and the fitted curve are used to find the charge transfer resistance Rct (Fig. S7D). In the Nyquist plot, the axis is defined as Imaginary (Z″) versus Real (Z′) with the frequency as a parameter, where Z is in kΩ. The semicircle is fitted with a high-frequency intercept on the real axis of Solution Resistance (Rs) and the diameter of Rct. NumPy and matplotlib libraries in python were used to extract the curve fitting algorithm, draw the graph, and develop the computational GUI tools. The extracted Rct was used in the exponential relationship to find the N-protein concentration in pg/mL (Fig. S7E). The results and raw data were also saved in the device and could be manipulated using the provided essential tools (Fig. S7F). This system has an option to send data to the Cloud (the web-based designed application) or a health center for telediagnosis or healthcare data collection or processing.
4. Conclusion
In this study, the technological advancements for the realization of fast and accurate multiplex electrochemical biosensing using a new Bi-ECDAQ biosensing system were explored. For this purpose, new electrode designs and biosensing strips were developed in accordance with the governing electrochemical principles, offering two functional working electrodes that enable the detection of the Nucleocapsid protein of SARS-CoV-2. The electrode design and its carboxyl-rich carbon-based ink (Carbon/Graphene@PEDOT:PSS) material, along with the precise screen-printing manufacturing technique, ensure high reproducibility of the strips, low electrical noise interference, and optimal distribution of the current density and electrical potential, as demonstrated by the numerical simulation and experimental testing. Also, to address the challenge of reading signals from bi-electrode designs using a low-cost but accurate reader, a new bi-potentiostat readout system (∼$80) was developed to record simultaneously the EIS signals from the two adjacent working electrodes of the strip in less than 2 min. The high reproducibility of the Bi-ECDAQ bi-potentiostat, along with the repeatable response of the fabricated N-protein antigen immuno-biosensors, offers a reliable biosensing platform for rapid detection of SARS-CoV-2, Nucleocapsid proteins, and quantification of viral loads. Both working electrodes functioned independently, depicted a low detection limit down to hundreds of fg/mL, and performed in a wide linear detection range of 1 pg/mL to 10 ng/mL. The Bi-ECDAQ bi-potentiostat performed equivalent or outperformed the commercial Autolab potentiostat, in all steps of immunosensing and redox concentration optimization. Each subsystem of the Bi-ECDAQ platform was integrated into an all-in-one system, including the strips, Bi-ECDAC reader, and data processing and transfer unit, therefore it can illustrate the COVID-19 test results to the user. The Bi-ECDAQ platform could successfully detect SARS-CoV-2 in patient samples and perform comparably to the Panbio Antigen Rapid Test device. In conclusion, the Bi-ECDAC biosensing platform addresses the unmet needs of electrochemical biosensing for clinical diagnostics with a high potential for multiplex biosensing. It can be further used for rapid and accurate quantification of various protein bioanalytes, showcasing promising potentials for electrochemical multiplexing.
Notes
The authors declare no competing financial interests.
CRediT authorship contribution statement
Razieh Salahandish: Concept and design, Experimentation & Data acquisition, Data analysis and/or interpretation, Numerical analysis, Clinical sample analysis, Drafting and revising the manuscript. Fatemeh Haghayegh: Experimentation & Data acquisition, Data analysis and/or interpretation, Numerical analysis, Drafting and revising the manuscript. Giancarlo Ayala-Charca: Concept and design, Experimentation & Data acquisition, Data analysis and/or interpretation, Drafting and revising the manuscript. Jae Eun Hyun: Experimentation & Data acquisition, Clinical sample analysis. Mahmood Khalghollah: Experimentation & Data acquisition, Drafting and revising the manuscript. Azam Zare: Numerical analysis, Drafting and revising the manuscript. Behrouz Far: Drafting and revising the manuscript. Byron M. Berenger: Clinical sample analysis, Drafting and revising the manuscript. Yan Dong Niu: Data analysis and/or interpretation, Clinical sample analysis, Drafting and revising the manuscript. Ebrahim Ghafar-Zadeh: Concept and design, Data analysis and/or interpretation. Amir Sanati-Nezhad: Data analysis and/or interpretation, Clinical sample analysis, Drafting and revising the manuscript, Drafting and revising the manuscript, Concept and design.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors acknowledge the Canadian Institutes of Health Research (CIHR) and Natural Sciences and Engineering Research of Canada (NSERC) for their Rapid COVID-19 Response funding. The authors also acknowledge the Canada Research Chair, NSERC CREATE Wearable Technology Research and Collaboration (We-TRAC) Training Program (Project No. CREATE/511166-2018), University of Calgary, and CMC – Microsystems, Canada for supporting this research. The authors would also like to thank the Alberta Public Health Laboratory in Calgary and their research staff members LeeAnn Turnbull and Kanti Pabbaraju for their support in providing access to clinical samples.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2022.114018.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
SI file contains the following:
Governing equations for numerical analysis, electrochemical characterization of BiECDAQ and APG, comparison between 7-point and 50-point frequency signals, evaluation of the immunosensor's selectivity in the presence of different proteins, comparing the performance of the biosensor, reproducibility testing of home-made bi-potentiostat, the stability test of biosensor, the functionality of the Bi-ECDAQ, electronics, and the list of different components of the readout system, are listed in the supporting information file.
References
- Abdullah S., Tonello S., Borghetti M., Sardini E., Serpelloni M. Journal of Sensors. 2019:2019. [Google Scholar]
- Akhavan O., Ghaderi E., Rahighi R. ACS Nano. 2012;6(4):2904–2916. doi: 10.1021/nn300261t. [DOI] [PubMed] [Google Scholar]
- Akhavan O., Ghaderi E., Rahighi R., Abdolahad M. Carbon. 2014;79:654–663. [Google Scholar]
- Alam F., Hasan M.M., Siddiquee M.R., Forouzanfar S., Jalal A.H., Pala N. Miniaturized, wireless multi-channel potentiostat platform for wearable sensing and monitoring applications. Smart Biomed. Physiol. Sens. Technol. 2021;XVIII (International Society for Optics and Photonics) [Google Scholar]
- Arduini F., Micheli L., Moscone D., Palleschi G., Piermarini S., Ricci F., Volpe G. Trac. Trends Anal. Chem. 2016;79:114–126. [Google Scholar]
- Asghari A., Wang C., Yoo K.M., Rostamian A., Xu X., Shin J.-D., Dalir H., Chen R.T. Appl. Phys. Rev. 2021;8(3) doi: 10.1063/5.0022211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi V., Boni A., Fortunati S., Giannetto M., Careri M., De Munari I. IEEE Trans. Instrum. Meas. 2019;69(6):3232–3240. [Google Scholar]
- Choi J.-H., Lee J.-H., Son J., Choi J.-W. Sensors. 2020;20(4):1003. [Google Scholar]
- Crescenzo-Chaigne B., Behillil S., Enouf V., Escriou N., Petres S., Ungeheuer M.N., Ghosn J., Tubiana S., Bouadma L., van der Werf S. 2021. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai B., Yin C., Wu J., Li W., Zheng L., Lin F., Han X., Fu Y., Zhang D., Zhuang S. Lab Chip. 2021;21(1):143–153. doi: 10.1039/d0lc00944j. [DOI] [PubMed] [Google Scholar]
- Davis J., Raisis A.L., Miller D.W., Rossi G. J. Vet. Diagn. Invest. 2020;32(5):656–663. doi: 10.1177/1040638720939520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhla M., Boesecke C., Schulte B., Diegmann C., Sib E., Richter E., Eschbach-Bludau M., Aldabbagh S., Marx B., Eis-Hübinger A.-M. Publ. Health. 2020;182:170–172. doi: 10.1016/j.puhe.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissa S., Zourob M. Microchim. Acta. 2020;187(9):1–11. doi: 10.1007/s00604-020-04423-3. [DOI] [PubMed] [Google Scholar]
- Furuya H., Pagano I., Chee K., Kobayashi T., Wong R.S., Lee R., Rosser C.J. Diagnostics. 2019;9(4):166. doi: 10.3390/diagnostics9040166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyfe J.A., Lavender C.J., Johnson P.D., Globan M., Sievers A., Azuolas J., Stinear T.P. Appl. Environ. Microbiol. 2007;73(15):4733–4740. doi: 10.1128/AEM.02971-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Jeffries L., Mach K.E., Craft D.W., Thomas N.J., Gau V., Liao J.C., Wong P.K. SLAS Technol.: Transl. Life Sci. Innovat. 2017;22(4):466–474. doi: 10.1177/2211068216651232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomaa A., Boye J. Food Chem. 2015;175:585–592. doi: 10.1016/j.foodchem.2014.12.017. [DOI] [PubMed] [Google Scholar]
- Gu W., Deng X., Lee M., Sucu Y.D., Arevalo S., Stryke D., Federman S., Gopez A., Reyes K., Zorn K. Nat. Med. 2021;27(1):115–124. doi: 10.1038/s41591-020-1105-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon J.R., Nichol S.T., Spiropoulou C.F., McElroy A.K. Viral Immunol. 2017;30(9):671–674. doi: 10.1089/vim.2017.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausler P., Roth C., Vitzthumecker T., Bierl R. Springer; 2019. Miniaturized Surface Plasmon Resonance Based Sensor Systems—Opportunities and Challenges. Optics, Photonics and Laser Technology 2018; pp. 169–195. [Google Scholar]
- He X.-P., Hu X.-L., James T.D., Yoon J., Tian H. Chem. Soc. Rev. 2017;46(22):6687–6696. doi: 10.1039/c6cs00778c. [DOI] [PubMed] [Google Scholar]
- Hoilett O.S., Walker J.F., Balash B.M., Jaras N.J., Boppana S., Linnes J.C. Sensors. 2020;20(8):2407. doi: 10.3390/s20082407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Sun J., Chen Y., Song Y., Dong L., Zhan Q., Zhang R., Abliz Z. Anal. Chim. Acta. 2012;711:60–68. doi: 10.1016/j.aca.2011.10.058. [DOI] [PubMed] [Google Scholar]
- Huang L., Tian S., Zhao W., Liu K., Ma X., Guo J. Analyst. 2020;145(8):2828–2840. doi: 10.1039/c9an02485a. [DOI] [PubMed] [Google Scholar]
- Jebelli A., Oroojalian F., Fathi F., Mokhtarzadeh A., de la Guardia M. Biosens. Bioelectron. 2020;169 doi: 10.1016/j.bios.2020.112599. [DOI] [PubMed] [Google Scholar]
- Jones A., Dhanapala L., Kankanamage R.N., Kumar C.V., Rusling J.F. Anal. Chem. 2019;92(1):345–362. doi: 10.1021/acs.analchem.9b05080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A., Vishnu A.G., Sakorikar T., Kamal A.M., Vaidya J.S., Pandya H.J. Nanoscale Adv. 2021;3:5542–5564. doi: 10.1039/d1na00453k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothimuthu P., Wilson R.A., Herren J., Haynes E.N., Heineman W.R., Papautsky I. Biomed. Microdevices. 2011;13(4):695–703. doi: 10.1007/s10544-011-9539-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadimisetty K., Malla S., Sardesai N.P., Joshi A.A., Faria R.C., Lee N.H., Rusling J.F. Anal. Chem. 2015;87(8):4472–4478. doi: 10.1021/acs.analchem.5b00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetani S., Kollath V.O., Eastick E., Debert C., Sen A., Karan K., Sanati-Nezhad A. Biosens. Bioelectron. 2019;145 doi: 10.1016/j.bios.2019.111715. [DOI] [PubMed] [Google Scholar]
- Khetani S., Singh A., Besler B., Butterworth S., Lijnse T., Loughery K., Smith K., Hosseini E., Narang R., Karan K. Biosens. Bioelectron. 2021;178 doi: 10.1016/j.bios.2021.113033. [DOI] [PubMed] [Google Scholar]
- Lee J.-H., Choi M., Jung Y., Lee S.K., Lee C.-S., Kim J., Kim J., Kim N.H., Kim B.-T., Kim H.G. Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Wen D., Wu J., Liu L., Wu W., Fang X., Kong J. Anal. Chem. 2020;92(14):9454–9458. doi: 10.1021/acs.analchem.0c01635. [DOI] [PubMed] [Google Scholar]
- Liu D., Wang J., Wu L., Huang Y., Zhang Y., Zhu M., Wang Y., Zhu Z., Yang C. Trac. Trends Anal. Chem. 2020;122 [Google Scholar]
- Liu F., Ni L., Zhe J. Biomicrofluidics. 2018;12(2) doi: 10.1063/1.5022168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquette C.A., Corgier B.P., Blum L.J. Bioanalysis. 2012;4(8):927–936. doi: 10.4155/bio.12.56. [DOI] [PubMed] [Google Scholar]
- Neubert H., Shuford C.M., Olah T.V., Garofolo F., Schultz G.A., Jones B.R., Amaravadi L., Laterza O.F., Xu K., Ackermann B.L. Clin. Chem. 2020;66(2):282–301. doi: 10.1093/clinchem/hvz022. [DOI] [PubMed] [Google Scholar]
- Neves M.M., González-García M.B., Delerue-Matos C., Costa-García A. Sensor. Actuator. B Chem. 2013;187:33–39. [Google Scholar]
- Norman M., Gilboa T., Ogata A.F., Maley A.M., Cohen L., Busch E.L., Lazarovits R., Mao C.-P., Cai Y., Zhang J. Nat. Biomed. Eng. 2020;4(12):1180–1187. doi: 10.1038/s41551-020-00611-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata A.F., Maley A.M., Wu C., Gilboa T., Norman M., Lazarovits R., Mao C.-P., Newton G., Chang M., Nguyen K. Clin. Chem. 2020;66(12):1562–1572. doi: 10.1093/clinchem/hvaa213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parihar A., Ranjan P., Sanghi S.K., Srivastava A.K., Khan R. ACS Appl. Bio Mater. 2020;3(11):7326–7343. doi: 10.1021/acsabm.0c01083. [DOI] [PubMed] [Google Scholar]
- Parlak O., İncel A., Uzun L., Turner A.P., Tiwari A. Biosens. Bioelectron. 2017;89:545–550. doi: 10.1016/j.bios.2016.03.024. [DOI] [PubMed] [Google Scholar]
- Patil P.O., Pandey G.R., Patil A.G., Borse V.B., Deshmukh P.K., Patil D.R., Tade R.S., Nangare S.N., Khan Z.G., Patil A.M. Biosens. Bioelectron. 2019;139 doi: 10.1016/j.bios.2019.111324. [DOI] [PubMed] [Google Scholar]
- Pimalai D., Putnin T., Waiwinya W., Chotsuwan C., Aroonyadet N., Japrung D. Microchim. Acta. 2021;188(10):1–10. doi: 10.1007/s00604-021-04995-8. [DOI] [PubMed] [Google Scholar]
- Prakash S., Chakrabarty T., Singh A.K., Shahi V.K. Biosens. Bioelectron. 2013;41:43–53. doi: 10.1016/j.bios.2012.09.031. [DOI] [PubMed] [Google Scholar]
- Pruna R., Palacio F., Baraket A., Zine N., Streklas A., Bausells J., Errachid A., López M. Biosens. Bioelectron. 2018;100:533–540. doi: 10.1016/j.bios.2017.09.049. [DOI] [PubMed] [Google Scholar]
- Pujadas E., Chaudhry F., McBride R., Richter F., Zhao S., Wajnberg A., Nadkarni G., Glicksberg B.S., Houldsworth J., Cordon-Cardo C. The lancet. Respir. Med. 2020;8(9) doi: 10.1016/S2213-2600(20)30354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radmard S., Reid S., Ciryam P., Boubour A., Ho N., Zucker J., Sayre D., Greendyke W.G., Miko B.A., Pereira M.R. Front. Neurol. 2019;10:281. doi: 10.3389/fneur.2019.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafi H., Zestos A.G. Anal. Bioanal. Chem. 2021:1–12. doi: 10.1007/s00216-021-03526-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmati Z., Roushani M., Hosseini H., Choobin H. Microchim. Acta. 2021;188(3):1–9. doi: 10.1007/s00604-021-04762-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramfos I., Vassiliadis N., Blionas S., Efstathiou K., Fragoso A., O'Sullivan C.K., Birbas A. Biosens. Bioelectron. 2013;47:482–489. doi: 10.1016/j.bios.2013.03.068. [DOI] [PubMed] [Google Scholar]
- Reboud J., Xu G., Garrett A., Adriko M., Yang Z., Tukahebwa E.M., Rowell C., Cooper J.M. Proc. Natl. Acad. Sci. Unit. States Am. 2019;116(11):4834–4842. doi: 10.1073/pnas.1812296116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roda A., Cavalera S., Di Nardo F., Calabria D., Rosati S., Simoni P., Colitti B., Baggiani C., Roda M., Anfossi L. Biosens. Bioelectron. 2021;172 doi: 10.1016/j.bios.2020.112765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado J., Pelleau S., Cockram C., Merkling S.H., Nekkab N., Demeret C., Meola A., Kerneis S., Terrier B., Fafi-Kremer S. The Lancet Microbe. 2021;2(2):e60–e69. doi: 10.1016/S2666-5247(20)30197-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J., Park S.H., Yeom Y.S., Shrivastav A., Lee S.-H., Kim Y.-R., Kim H.-Y. Food Control. 2013;32(2):659–664. [Google Scholar]
- Salahandish R., Ghaffarinejad A., Naghib S.M., Majidzadeh-A K., Zargartalebi H., Sanati-Nezhad A. Biosens. Bioelectron. 2018;117:104–111. doi: 10.1016/j.bios.2018.05.043. [DOI] [PubMed] [Google Scholar]
- Salahandish R., Ghaffarinejad A., Naghib S.M., Niyazi A., Majidzadeh-A K., Janmaleki M., Sanati-Nezhad A. Sci. Rep. 2019;9(1):1–11. doi: 10.1038/s41598-018-37573-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salahandish R., Ghaffarinejad A., Omidinia E., Zargartalebi H., Majidzadeh-A K., Naghib S.M., Sanati-Nezhad A. Biosens. Bioelectron. 2018;120:129–136. doi: 10.1016/j.bios.2018.08.025. [DOI] [PubMed] [Google Scholar]
- Salahandish R., Zargartalebi H., Janmaleki M., Khetani S., Azarmanesh M., Ashani M.M., Aburashed R., Vatani M., Ghaffarinejad A., Sanati‐Nezhad A. Adv. Mater. Technol. 2019;4(11) [Google Scholar]
- Schildgen V., Demuth S., Lüsebrink J., Schildgen O. Pathogens. 2021;10(1):38. doi: 10.3390/pathogens10010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J.S., McCarthy M.K., Rester C., Sabourin K.R., Annen K., DomBourian M., Eisenmesser E., Frazer-Abel A., Knight V., Jaenisch T. J. Clin. Microbiol. 2021;59(6) doi: 10.1128/JCM.00290-21. 00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan D., Johnson J.M., Fernandes S.C., Suib H., Hwang S., Wuelfing D., Mendes M., Holdridge M., Burke E.M., Beauregard K. Nat. Commun. 2021;12(1):1–8. doi: 10.1038/s41467-021-22072-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu H., Chang G., Su J., Cao L., Huang Q., Zhang Y., Xia T., He Y. Sensor. Actuator. B Chem. 2015;220:331–339. [Google Scholar]
- Sin M.L.Y., Gau V., Liao J.C., Wong P.K. J. Microelectromech. Sys. 2013;22(5):1126–1132. doi: 10.1109/JMEMS.2013.2253545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K.-H., Kim D.-M., Lee H., Ham S.Y., Oh S.-M., Jeong H., Jung J., Kang C.K., Park J.Y., Kang Y.M. Kor. J. Intern. Med. 2021;36(1):11. doi: 10.3904/kjim.2020.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood A., Sui Y., McDonough E., Santamaría-Pang A., Al-Kofahi Y., Pang Z., Jahrling P.B., Kuhn J.H., Ginty F. Viruses. 2020;12(8):787. doi: 10.3390/v12080787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson H., Radha Shanmugam N., Paneer Selvam A., Prasad S. SLAS Technol.: Transl. Life Sci. Innovat. 2018;23(1):5–15. doi: 10.1177/2472630317738700. [DOI] [PubMed] [Google Scholar]
- Strömer A., Rose R., Grobe O., Neumann F., Fickenscher H., Lorentz T., Krumbholz A. Microorganisms. 2020;8(10):1572. doi: 10.3390/microorganisms8101572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Liu Y. Micromachines. 2018;9(4):142. doi: 10.3390/mi9040142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Liu X., Zhang X., Zuo M., Zou X., Niu Z., Pan H., Li J., Gao Y. Sensor. Actuator. B Chem. 2021;338 [Google Scholar]
- Timilsina S.S., Jolly P., Durr N., Yafia M., Ingber D.E. Acc. Chem. Res. 2021:1143–1149. doi: 10.1021/acs.accounts.1c00382. [DOI] [PubMed] [Google Scholar]
- Torres M.D., de Araujo W.R., de Lima L.F., Ferreira A.L., de la Fuente-Nunez C. Matter. 2021 [Google Scholar]
- Van Nguyen H., Nguyen V.D., Lee E.Y., Seo T.S. Biosens. Bioelectron. 2019;136:132–139. doi: 10.1016/j.bios.2019.04.035. [DOI] [PubMed] [Google Scholar]
- Veyrenche N., Bolloré K., Pisoni A., Bedin A.S., Mondain A.M., Ducos J., Segondy M., Montes B., Pastor P., Morquin D. J. Med. Virol. 2021;93(5):3069–3076. doi: 10.1002/jmv.26855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan S., Rani C., Ho J.-a.A. Talanta. 2012;94:315–319. doi: 10.1016/j.talanta.2012.03.049. [DOI] [PubMed] [Google Scholar]
- Wang Y., Li K., Xu G., Chen C., Song G., Dong Z., Lin L., Wang Y., Xu Z., Yu M. Research. 2021:2021. doi: 10.34133/2021/2813643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wang Y., Lin L., Wu T., Zhao Z., Ying B., Chang L. Biosens. Bioelectron. 2022;195 doi: 10.1016/j.bios.2021.113663. [DOI] [PubMed] [Google Scholar]
- Wong S.J., Furuya A., Zou J., Xie X., Dupuis A.P., II, Kramer L.D., Shi P.-Y. EBioMedicine. 2017;16:136–140. doi: 10.1016/j.ebiom.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Rios-Aguirre D., Chounlakone M., Camacho-Leon S., Voldman J. Biosens. Bioelectron. 2018;117:522–529. doi: 10.1016/j.bios.2018.06.049. [DOI] [PubMed] [Google Scholar]
- Xue W., Tan X., Oo M.K.K., Kulkarni G., Ilgen M.A., Fan X. Analyst. 2020;145(4):1346–1354. doi: 10.1039/c9an02498k. [DOI] [PubMed] [Google Scholar]
- Yang B., Song X.L., Huang J., Shi C.Y., Liu Q.H., Liu L. J. Fish. Dis. 2006;29(5):301–305. doi: 10.1111/j.1365-2761.2006.00713.x. [DOI] [PubMed] [Google Scholar]
- Yang M., Yang Y., Liu Y., Shen G., Yu R. Biosens. Bioelectron. 2006;21(7):1125–1131. doi: 10.1016/j.bios.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Zanchetta G., Carzaniga T., Vanjur L., Casiraghi L., Tagliabue G., Morasso C., Bellini T., Buscaglia M. Biosens. Bioelectron. 2021;172 doi: 10.1016/j.bios.2020.112751. [DOI] [PubMed] [Google Scholar]
- Zhang G., Liu Z., Fan L., Han Y., Guo Y. Biosens. Bioelectron. 2021;173 doi: 10.1016/j.bios.2020.112785. [DOI] [PubMed] [Google Scholar]
- Zhang H., Klose A.M., Miller B.L. Bioconjugate Chem. 2021;32(3):533–540. doi: 10.1021/acs.bioconjchem.0c00718. [DOI] [PubMed] [Google Scholar]
- Zhu X., Wang X., Han L., Chen T., Wang L., Li H., Li S., He L., Fu X., Chen S. Biosens. Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonneveld R., Jurriaans S., van Gool T., Hofstra J.J., Hekker T.A., Defoer P., Broekhuizen-van Haaften P.E., Wentink-Bonnema E.M., Boonkamp L., Teunissen C.E. J. Clin. Virol. 2021;139 doi: 10.1016/j.jcv.2021.104821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.