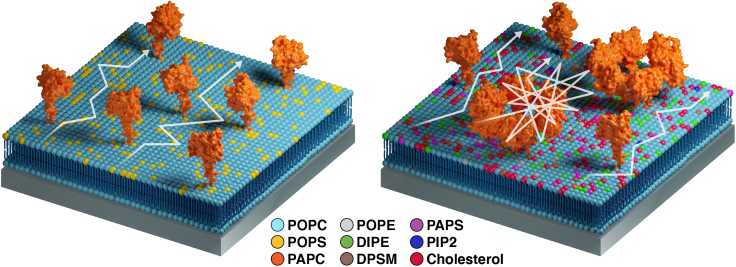
Summary
Understanding the spatiotemporal distribution and dynamics of RAS on the plasma membrane (PM) is the key for elucidating the molecular mechanisms of the RAS signaling pathway. Single particle tracking (SPT) experiments show that in cells, KRAS diffuses in at least three interchanging states on the cellular PM; however, KRAS remains monomeric and always shows homogeneous diffusion on artificial membranes. Here, we show for the first time on a supported lipid bilayer composed of heterogeneous lipid components that we can recapitulate the three-state diffusion of KRAS seen in cells. The use of a biologically relevant eight-lipid system opens a new frontier in the biophysical studies of RAS and other membrane associated proteins on a biomimetic system that recapitulates the complexity of a cellular PM.
Subject areas: Molecular biology, Membrane architecture, Biomimetics, Biotechnology, Biophysics
Graphical abstract
Highlights
-
•
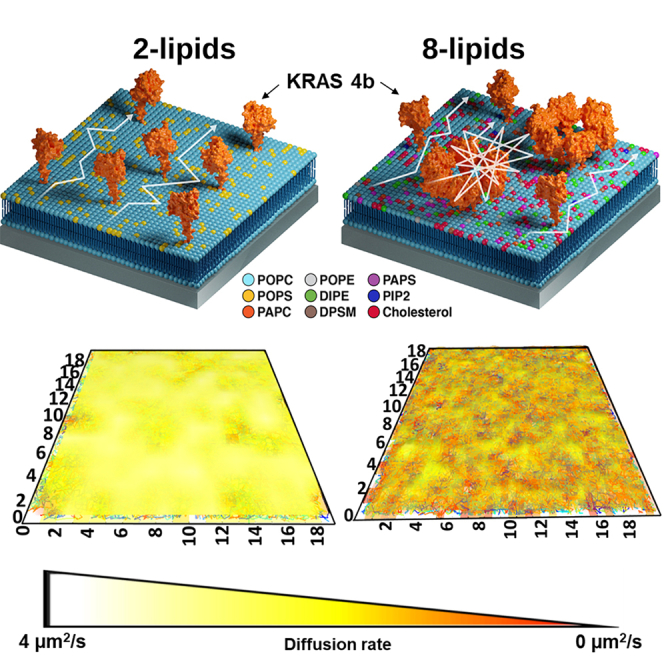
KRAS4b shows homogeneous diffusion on simple 2-lipids bilayer
-
•
KRAS4b shows a cell-like, three-state diffusion on a complex 8-lipid bilayer
-
•
Phase separation in lipids favors the multi-state diffusion of KRAS4b
-
•
The complex lipid composition favors RAS nanoclustering irrespective of nucleotide state
Molecular biology; Membrane architecture; Biomimetics; Biotechnology; Biophysics
Introduction
RAS proteins are membrane-localized molecules that initiate signaling pathways such as the mitogen-activated protein kinase (MAPK) pathway that are essential for cell survival, proliferation, and migration (Stephen et al., 2014). Oncogenic mutations in RAS constitutively turn on core pathways, leading to uncontrolled cell proliferation, and eventually oncogenesis. RAS proteins are small GTPases that toggle between the GDP-loaded inactive, or "off" state, and the GTP-loaded active, or "on" state. RAS is only capable of activating its effectors when it is tethered to the membrane via its post-translational modified (prenylated) C-terminus. When it is in the on-state it binds to its effectors, such as RAF1, and activates downstream signaling cascades. The precise mechanism of the activation step is not fully understood, but it is thought to involve dynamic conformational changes in the effector binding G-domain of RAS (Iversen et al., 2014). The cancer research community has been targeting this "undruggable" protein for over 30 years, and it was not until recently, with the advent of KRASG12C covalent inhibitors, that targeting RAS seemed possible (Lito et al., 2016). However, the G12C inhibitors target only some of the cancer-causing mutant alleles of KRAS and understanding the complex and dynamic relationship between RAS and membrane could open new frontiers in targeting RAS. The four major isoforms of RAS: HRAS, NRAS, and two splice variants KRAS4a and KRAS4b share 95% homology in their G-domain but vary in the membrane associated C-terminal hypervariable region (HVR). The HVR contains isoform specific posttranslational modifications which targets the proteins to distinct membrane microdomains leading to different signaling outputs (Hancock, 2003).
Unlike other RAS isoforms, KRAS4b association with the membrane is mediated by two features in the HVR: (i) farnesylation of cysteine 185 and (ii) six positively charged lysine residues in the HVR (Banerjee et al., 2016; Gillette et al., 2015). The positively charged lysines are suggested to form a combinatorial code to recognize or recruit dynamically, negatively charged phospholipids such as phosphatidylserines (PS) (Weise et al., 2011; Zhou et al., 2017). Studies suggest that lipid-specific interactions drive KRAS4b organization into higher-order oligomers and nanoclusters on the membrane, which act as signaling hubs for effector recruitment (Zhou et al., 2014). In human cancers, KRAS4b is more frequently mutated than any other RAS isoforms; 15-20% in all human cancers and 85% in RAS related cancer (Zhou et al., 2018); therefore, characterizing and understanding KRAS4b membrane interactions and dynamics is a critical area of study.
Cellular plasma membranes (PM) are complex and heterogeneous entities that dynamically evolve across various lengths and time scales (Cooper, 2000). The PM is composed of thousands of phospholipid species with varying polar head groups, acyl chains with different length and saturation, fatty acids, sterols, and proteins that are embedded within or anchored at the membrane surface via electrostatic and hydrophobic interactions. Together, they orchestrate highly dynamic and transitory substructures, such as lipid rafts, protein and lipid complexes and nanoclusters. The actin cytoskeleton is thought to further compartmentalize lipid domains existing within the PM creating an organizational platform for protein localization, effector binding, and signal transduction (Kusumi et al., 2010, 2012). Therefore, it is imperative to learn how signaling proteins are regulated by these transient membrane domains ranging from nanoscale to mesoscale, in order to gain mechanistic insights on protein-membrane biology and to determine whether it can be exploited for therapeutic purposes.
The lipid composition of the inner leaflet PM is highly complex and closely regulates the spatial and temporal organization of KRAS4b on the membrane (Zhou and Hancock, 2015). Single-particle tracking (SPT) experiments revealed that KRAS4b predominantly exists as freely diffusing molecules on the PM and, upon activation, assembles into slower moving nanoclusters of signaling complexes linking membrane mobility to activation (Lommerse et al., 2006; Murakoshi et al., 2004). This model was recently improved upon by Goswami et al. (Goswami et al., 2020) and Nan et al. (Lee et al., 2019) when they identified three diffusion states for KRAS4b-a fast, intermediate, and slow state. However, in a study by Chung et al., KRAS4b remained monomeric and showed homogeneous diffusion over a wide range of surface densities and membrane compositions, including over 20% negatively charged phosphatidylserine (PS) (Chung et al., 2018). Although the in vivo and in vitro studies suggest a more significant role of the membrane heterogeneity in RAS organization and recruitment into signaling complexes, the exact molecular mechanism is yet to be elucidated.
In this work, we captured a cellular PM-like environment on a glass coverslip and investigated how heterogeneous lipid composition regulates the spatiotemporal organization of KRAS4b on model membranes. We implemented a bottom-up approach where we built up the membrane complexity starting from a simple two lipid, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS) bilayer and compared this system to a complex eight-lipid bilayer, including the most abundantly found lipid species in the inner leaflet of the PM (Cooper, 2000; Ingolfsson et al., 2021). We performed total internal reflection fluorescence (TIRF)-SPT experiments to track single molecules of purified full-length KRAS4b, both farnesylated and methylated, as it diffuses along the 2D plane of the supported lipid bilayer, and utilized atomic force microscopy (AFM) to characterize the lipid bilayers. We provide the first direct evidence of a cell-like three-state diffusion of KRAS4b on any reconstituted membrane system. We characterize a fast state composed of freely diffusing KRAS4b, an intermediate state containing membrane mediated KRAS4b multimers, and a slow state representing large nanoclusters. Using atomic force microscopy, we show that the eight different lipids altogether lead to a heterogeneous membrane structure which collectively regulates the spatial and temporal distribution of KRAS4b on the membrane and is independent of the nucleotide state. We hypothesize that membrane microdomains containing slow moving KRAS4b molecules provide an initial platform for effector engagement. Together, our data suggests that modulation of RAS spatiotemporal dynamics on the PM by perturbations in membrane heterogeneity could be an alternative approach to targeting RAS therapeutically.
Results
KRAS shows free diffusion on a simple two-lipid bilayer
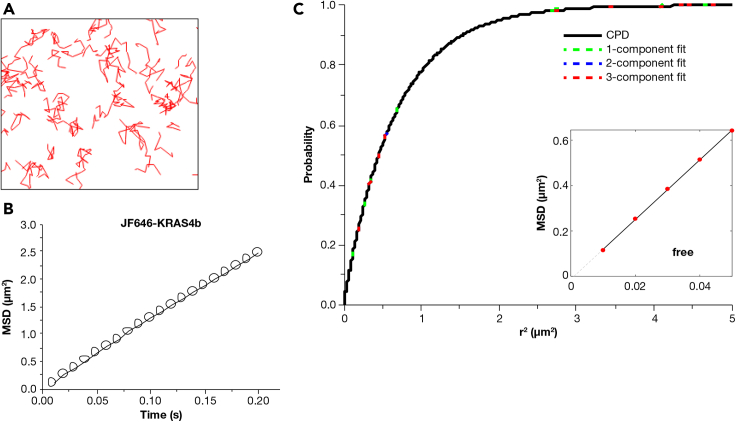
We performed single-particle tracking experiments of JF646 (Grimm et al., 2015) labeled KRAS4b on a simple two-lipid bilayer composed of 87% POPC and 13% POPS to validate our experimental and data analysis methods. The negatively charged head group of PS is essential for the localization and nanoscale spatial organization of KRAS4b on the plasma membrane (Goswami et al., 2020; Zhou et al., 2017). A few examples of the representative tracks are displayed in Figure 1A. To extract statistical information on the diffusion behavior of KRAS4b, we first performed a mean square displacement (MSD) analysis of the thousands of trajectories obtained for KRAS4b. MSD analysis is an ensemble calculation that provides information on the average mode of diffusion that a particle has undergone and is evaluated by the degree of bend in the MSD curve (Matysik and Kraut, 2014; Michalet, 2010). A straight line indicates free diffusion, whereas a bent line represents confinement. In our homogeneous two-lipid supported lipid bilayer, the MSD plot yielded a straight line—evidence of simple Brownian-like free diffusion for KRAS4b (Figure 1B). We next calculated the cumulative probability distribution (CPD) curve and a single component yielded the best fit. No further improvement was seen upon increasing the number of components in the fit model, as shown in Figure 1C.
Figure 1.
KRAS4b diffusion on a simple POPC/POPS (87:13) bilayer (Video S1)
(A–C) (A) Single molecule tracks, (B) Ensemble mean square displacement (MSD) plot, and (C) Cumulative probability distribution plots of the tracks obtained for full-length KRAS4b labeled with JF646 reconstituted on a simple POPC/POPS bilayer. The inset in (C) represents the MSD plot of the one-component identified by CDP analysis. Data are represented as mean +/− standard deviation.
Furthermore, the MSD plot obtained for the single component fit in the CPD analysis also displayed a straight line (Figure 1C inset). This further substantiates the finding that KRAS4b undergoes free diffusion on a simple two lipid bilayer with an average diffusion coefficient of ∼3.2 μm2/s. Our data is consistent with previous diffusion studies on KRAS4b that were performed on a planar supported lipid bilayer using fluorescence based techniques such as fluorescence recovery after photobleaching, fluorescence correlation spectroscopy (Chung et al., 2018), and single-particle tracking (Chung et al., 2018). However, we do not observe the cell-like three-state diffusion on this simple two lipid bilayer. Therefore we hypothesize that the widely used membrane mimetic systems composed of either two or three lipids lack the membrane complexity and thus the RAS dynamics were observed on the cellular PM (Goswami et al., 2020; Lee et al., 2019).
KRAS4b shows confined, 3-state diffusion on a complex eight-lipid bilayer
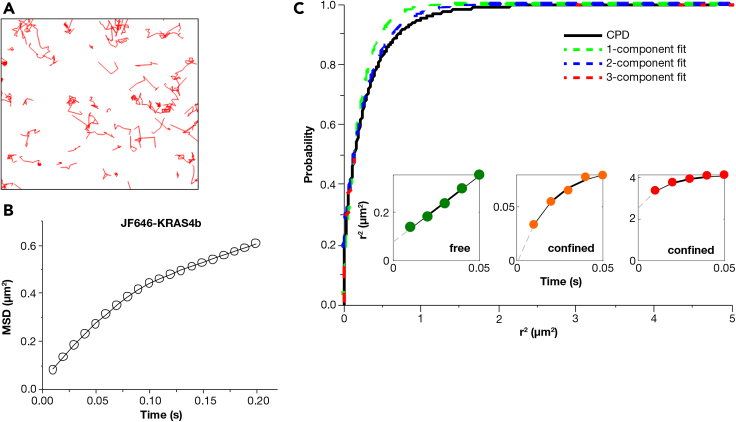
To recapitulate the complexity of a biological plasma membrane, we introduced additional components into our reconstituted membrane to generate cell-like heterogeneity. Recently, a joint partnership between the US Department of Energy and National Cancer Institute launched the JDACS4C and Pilot 2 program with the goal to advance exascale development through cancer research. The campaign primarily focused on understanding RAS behavior on the membrane at microscale length and nanosecond timescales. In a collaboration involving five different national labs, scientists performed an extensive automated multi-scale simulation of KRAS4b nestled on a biologically relevant PM model (Ingolfsson et al., 2021). The in silico PM model was composed of an asymmetric eight-lipid mixture of cholesterol, phosphatidylcholines (PC), phosphatidylethanolamines (PE), phosphatidylserine (PS), phosphatidylinositol bisphosphate (PIP2), and sphingomyelin (SM). The lipid classes represent the most abundant lipid species in the inner leaflet of the PM. Each type of lipid has a different head group, acyl chain length, and saturation. A detailed characterization of the eight-lipid bilayer including diffusion properties of individual lipid types can be found in (Ingolfsson et al., 2021). This computational simulation revealed a dynamic rearrangement of lipid composition, especially increased PIP2 and decreased cholesterol, creating a local lipid fingerprint that induced lateral segregation of KRAS4b into multimers and suggested a greater role for lipid patterning in effector recruitment and downstream signal transduction. In this study, we utilized the same eight-lipid composition to prepare supported lipid bilayer and perform single particle tracking studies of laterally-diffusing, fluorescently labeled KRAS4b. The molecular structure and mole percent of each lipid type are described in detail in Table S1. To isolate the effect of membrane complexity from the electrostatic effect, we maintained the overall charge of the lipid composition at −13 to match the negative charge of the previously described POPC/POPS bilayer. Figure 2A shows examples of the representative tracks. Here, unlike the simple two-lipid bilayer, the tracks are non-uniform and display occasional confinement. The ensemble MSD plot obtained from KRAS4b tracks is bent, indicating that average KRAS4b diffusion is more confined in the more complex system, as shown in Figure 2B. The lipids displayed a brownian like free diffusion as indicated by a straight line MSD plot in Figure S1. CPD analysis of the square displacements also showed a three-component best fit, unlike the one-component best fit in the simple two lipid bilayer (Figure 2C). The corresponding MSD curves of the three components (Figure 2C inset) showed that KRAS underwent free, confined, and confined diffusion in each state, respectively.
Figure 2.
KRAS4b diffusion on a complex eight-lipid bilayer, explained in Table S1, Video S2
KRAS4b diffusion on a complex eight-lipid bilayer, explained in Table S1.
(A–C) (A) Single molecule tracks, (B) Ensemble mean square displacement (MSD) plot, and (C) Cumulative probability distribution plots of the tracks obtained for full length KRAS4b labeled with JF646 reconstituted on an eight-lipid bilayer. The inset represents the MSD plots of the three-components identified by CPD analysis. Data are represented as mean +/− standard deviation.
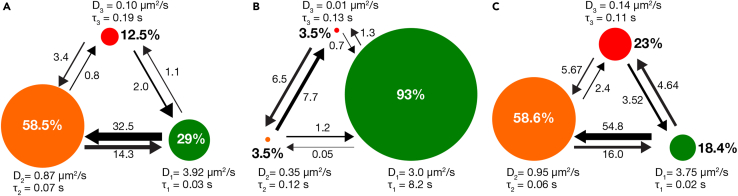
Although CPD analysis correctly predicts the nature of the underlying diffusion states, it cannot precisely calculate the diffusion parameters. For example, CPD analysis only fits the linear region of the MSD plot and incorrectly defines the diffusion coefficient for the non-linear or bent curves (Matysik and Kraut, 2014). To overcome this mathematical limitation, we next employed Hidden Markov Modeling (HMM) analysis using Variational Bayes for Single Particle Tracking (vbSPT) software to extract quantitative information on diffusion parameters, such as diffusion coefficients, state occupancy percentages, dwell times in individual states, and the probability that a molecule will transition from one state to another (Persson et al., 2013). HMM analysis corroborated our CPD analysis results, and yielded three diffusion states for KRAS4b; fast, intermediate, and slow with diffusion coefficients of D1 = 3.9 μm2/s, D2 = 0.87 μm2/s and D3 = 0.10 μm2/s, respectively (represented by green, orange, and red circles in Figure 3A) (Table S2). The fast diffusion resembled the diffusion of the freely moving KRAS4b on simple artificial lipid bilayer reported earlier in this paper and in various other in vitro experiments (Chung et al., 2018). The intermediate and the slow diffusion rates were closer to the diffusion rates obtained on cellular PM (Goswami et al., 2020; Lee et al., 2019; Murakoshi et al., 2004).
Figure 3.
The three-state diffusion model for KRAS4b on supported lipid bilayer yielded by the Hidden Markov Modeling (HMM) analysis using vbSPT software
(A) KRAS4b loaded with GDP on eight-lipid bilayer.
(B and C) (B) KRAS4b loaded with GppNHp on simple POPC/POPS (87/13) bilayer (C)) KRAS4b loaded with GppNHp on eight-lipid bilayer. D1 (green), D2 (orange), and D3 (red) represent the fast, intermediate, and slow state, respectively. The circle represents the fractional occupancy of each state, τi represents the dwell time in each state and the arrows represent transition between states. The diameter of the circle and the width of the arrows are drawn in proportion to their relative values.
A further advantage of HMM analysis is that it characterizes the dynamics of the states by yielding the percent occupancy in each state, the time a molecule spends in an individual state and the probability that molecules will transition between states. KRAS4b showed highest occupancy in the intermediate state, ∼ 58%, followed by the fast, ∼29%, and the slow state, ∼13%. The transition probabilities between states indicate that KRAS4b is most likely to transition from the fast state to the intermediate state and vice versa and rarely transitions to other states. Similarly, in terms of dwell time in the individual states, the fast state is very short lived (t = 0.03s), followed by the intermediate state (t = 0.07s), and longest for the slow state (t = 0.14s). Taken together, the results suggest an order of assembly that once KRAS is bound to the membrane, it diffuses freely in the fast state and then quickly transitions into the intermediate state where molecules slow down and occasionally assemble into a relatively stable slow-moving state. This diffusion behavior of KRAS4b is different from a single component Brownian diffusion observed on simple POPC/POPS bilayer and resembles the complex three-state diffusion on the PM of living cells.
KRAS4b dynamics is independent of nucleotide state
RAS activation from GDP-loaded “off” state to GTP-loaded state “on” by guanine exchange factors (GEFs) is a crucial step in effector recruitment and signal transduction. Understanding how the on and off states of KRAS4b interact with the membrane could yield insights into how KRAS4b binds to the effector partners including the RAF protein. Here, we investigated if KRAS4b membrane dynamics are dependent on its nucleotide state. For imaging purposes, we utilized Alexa 647-labeled KRAS4b loaded with GppNHp, a non-hydrolysable GTP analog and allowed it to laterally diffuse on the supported lipid bilayer. The diffusion behavior of GTP-loaded KRAS4b was similar to the diffusion of GDP-loaded KRAS4b on both the POPC/POPS and eight-lipid bilayers. HMM analysis of the single molecule tracks yielded mostly fast diffusion (∼93%) for KRAS4b on the simple POPC/POPS bilayer with a diffusion coefficient of 3.0 μm2/s (Figure 3B), whereas a distinct three-state diffusion was observed on the eight-lipid bilayer (Figure 3C) (Table S2). The intermediate state had the highest fractional occupancy of ∼59%, consistent with GDP-loaded KRAS4b (Figure 3A), followed by the slow and then fast state.
Phase separation of the membrane components promotes the intermediate state and slow state
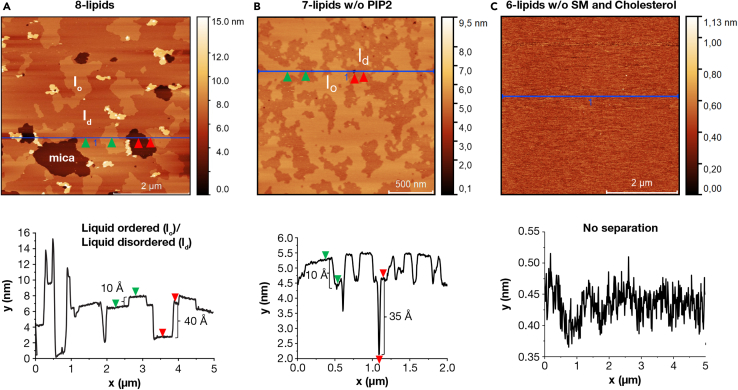
To further characterize what biophysical properties of the eight-lipid bilayer facilitate the three-state diffusion of KRAS4b, we performed atomic force microscopy (AFM) experiments on supported lipid bilayer prepared on a freshly cleaved mica surface. AFM is a label-free microscopy technique with sub-atomic resolution that provides information on the topography of the sample surface (Frederix et al., 2009). AFM has been widely used in membrane biology and biophysics to visualize and characterize the phase separation process in model membranes, as well as cellular plasma membranes (Connell and Smith, 2006; Johnston, 2007). In this study, we imaged lipid bilayers made by collapsing both eight-lipid and two-lipid vesicles on mica. AFM images of the eight-lipid bilayer, shown in Figure 4A, exhibited phase separation into two different domains with a height difference of ∼10 angstroms, whereas no phase separation was visible in a simple POPC/POPS bilayer (Figure S2).
Figure 4.
Biophysical characterization of the complex lipid bilayer
(A–C) Atomic force microscopy topography images of (A) eight-lipids, (B) 7-lipids without PIP2, and (C) 6-lipids without cholesterol and sphingomyelin deposited on mica sheets. The height information is represented with different shades of brown. The dark brown represents the exposed mica and the lighter shades represent the height of the lipid bilayer. The line graph at the bottom corresponds to the height information along the blue line drawn in their corresponding images above. The green arrows represent the difference between different domains and the red arrows represent the difference between bare mica and lipid bilayer.
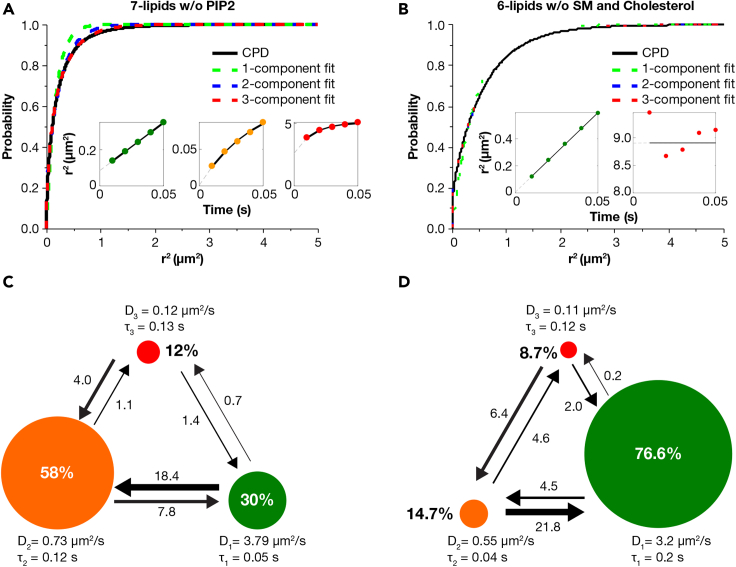
We know from previous biophysical studies that different molecular affinities and immiscibility among different classes of lipids create raft domains in the plasma membrane that are predominantly enriched in cholesterol and sphingolipids (Connell and Smith, 2006; Kusumi et al., 2012). Similarly, highly charged phosphoinositides, such as PIP2, are distinguished by their propensity to form electrostatic-based clusters on model and biological membranes (Bilkova et al., 2017; Wen et al., 2018). To identify the molecular mechanism of this phase separation phenomenon in our experiments, we modified the lipid composition in two different ways: (1) 7-lipids without PIP2 and (2) 6-lipids without sphingomyelin and cholesterol, while maintaining the total negative charge at 13% in both cases. AFM results, shown in Figures 4B and 4C show that the deletion of PIP2 did not have any effect on the domain formation, whereas eliminating cholesterol and sphingomyelin completely abrogated the phase separation process, respectively.
SPT studies on these two modified lipid bilayers revealed striking differences in the lateral mobility patterns of KRAS4b. The results are shown in Figure 5 for 7-lipid without PIP2 (A and C) and for 6-lipid bilayer without sphingomyelin and cholesterol (B and D). Careful analysis of the tracks revealed that excluding PIP2 from the original eight-lipid composition had minimal effect on the diffusion. For example, both CPD and HMM analysis predicted a three-state model with similar properties to that of the eight-lipid bilayer, i.e., three-state diffusion with the intermediate state as the most prominent state. But when cholesterol and sphingomyelin were removed from the lipid composition, the CPD analysis of KRAS4b tracks on the 6-lipid bilayer showed only a 2-component system. The MSD plots of the two components suggested that the majority of KRAS4b underwent free diffusion, and only a minor population showed anomalous diffusion. In addition, HMM analysis showed that most KRAS4b exhibited fast diffusion (76% occupancy), resembling diffusion on a POPC/POPS bilayer.
Figure 5.
KRAS4b diffusion on the modified eight-lipid bilayer
(A–D) Cumulative probability distribution (CPD) and Hidden Markov Modeling (HMM) analysis of KRAS4b tracks obtained on (A and C) 7-lipids without PIP2 and (B and D) 6-lipids without cholesterol and sphingomyelin. The insets represent the mean-square displacement (MSD) plots of the individual components identified by CPD analysis.
Artificial multimerization of KRAS4b slows diffusion and enhances intermediate and slow diffusion states
Next, we interrogated the correlation between KRAS4b multimers and the slower diffusion states. Previous FCS experiments indicated that the diffusion coefficient of artificially induced KRAS4b dimers fused to a Leucine zipper dimer molecule on a simple POPC/POPS bilayer is approximately 2.3 μm2/s (Chung et al., 2018). The intermediate and the slow diffusion coefficients measured in our experiments are lower than the reported dimer diffusion coefficient and hint toward large scale multimerization. However, in a two-dimensional (2D) diffusion experiment, one cannot infer a proportional relationship between particle size and lateral mobility based on the Stokes-Einstein diffusion model. The 2-D diffusion coefficient is much more complex, and the drag coefficient is convoluted with the effects of protein structure, lipid environment, and protein-lipid interaction (Ziemba and Falke, 2013). Hence, it is more conclusive to extract the diffusion parameters based on experimental work, rather than using theoretical calculations using the Stokes-Einstein diffusion equation.
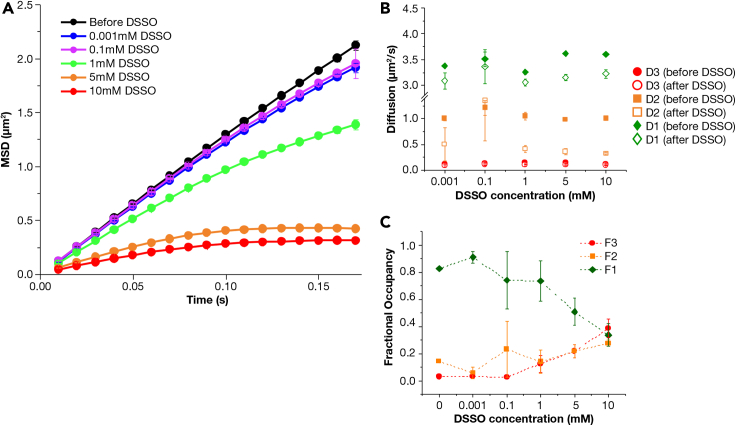
To examine the effect of large scale multimerization on the mobility states of KRAS4b, we introduced artificial multimers of KRAS4b into our system using the crosslinker molecule disuccinimidyl sulfoxide (DSSO). DSSO is capable of forming crosslinks between primary amines that are within 20 angstroms of each other and should impact lateral diffusion either by slowing down or by enhancing molecule clustering (Kao et al., 2011). For this study, we used a simple POPC/POPS lipid bilayer and investigated how increasing concentrations of DSSO affect KRAS4b dynamics. We calculated the ensemble MSD plots and fitted the tracks to three diffusion states using vbSPT HMM analysis. The MSD plots in Figure 6A clearly show systematic increase in confined diffusion with increasing concentrations of DSSO. The diffusion rates of all three states decreased after addition of DSSO at all concentrations as shown in Figure 6B. In case of fractional occupancy, the fast state gradually declined while the intermediate and slow states increased steadily (Figure 6C) with higher DSSO concentration. Most intriguingly, at the highest concentration of 10 mM DSSO, the fractional occupancy of the slow state surpassed both the fast and the intermediate states. The steady increase in the fractional occupancy of the slow state with increasing DSSO concentration confirms a direct correlation between the cluster size and the slow diffusion state. Hence, this result corroborates our hypothesis that the slow diffusion state is comprised of KRAS4b clusters that are mediated by the interaction between complex lipids and RAS on an eight-lipid bilayer.
Figure 6.
Effect of DSSO on KRAS diffusion on POPC/POPS bilayer
(A) Mean-square displacement (MSD) analysis of KRAS4b tracks before DSSO and after addition of DSSO at concentrations ranging from 1 μM up to 10 mM.
(B) Diffusion coefficients of fast (D1, green), intermediate (D2, orange), and slow (D3, red) calculated from HMM analysis before (filled) and after (unfilled) addition of DSSO.
(C) Fractional occupancy of fast (D1, green), intermediate (D2, orange), and slow (D3, red) calculated from HMM analysis in presence of DSSO. Data are represented as mean +/− standard deviation.
Discussion
KRAS4b traverses across a multi-tiered, hierarchical membrane environment that leads to multi-component, highly transitory, and dynamic sub-diffusion states. The fact that RAS engagement to the membrane is necessary for the MAPK signaling and the finding that RAS molecules slow down upon EGFR activation, make this an important area of research. Recently, using high spatial and temporal resolution techniques, Goswami et al. and Nan et al. demonstrated a unique, isoform specific three-state model of KRAS4b diffusion on the plasma membrane of live cells, and related it to KRAS4b signaling and membrane recycling (Goswami et al., 2020; Lee et al., 2019). However, because of the limitations of the techniques and the complex cellular environment, the exact molecular mechanism behind the three-state diffusion model of KRAS4b is unclear. Here, we aimed to unravel the molecular mechanism behind the diffusion of membrane tethered KRAS4b by comparing its mobility on simple two lipid bilayer to one composed of eight lipid mixture that closely mimics the most abundant lipid classes in the inner leaflet of the plasma membrane. In the simple POPC/POPS bilayer, we observed free diffusion that was best fit by a single component (Figure 7). In the eight-lipid bilayer, however, KRAS4b showed complex diffusion with three components and three rates: fast, intermediate, and slow (Figure 7). Using CPD analysis, we found that the fast state showed Brownian type free diffusion, whereas RAS molecules in the intermediate and slow states show anomalous and confined diffusion, respectively. We hypothesize and later deduce that first, the fast diffusing KRAS4b molecules are monomeric, primarily based on their Brownian motion and the diffusion coefficient values previously reported for monomeric KRAS4b on artificial membranes (Chung et al., 2018); second, the intermediate diffusion is from KRAS4b molecules that are intermittently interacting with heterogeneous membrane structures (such as phase separation); and third, the slow diffusion comes from relatively stable KRAS4b nanoclusters supported by the crosslinking experiment.
Figure 7.
Schematic representation of KRAS4b diffusion on POPC/POPS lipid bilayer (left) and eight-lipid bilayer (right).
KRAS4b shows a single component free diffusion on a two-lipid bilayer but a 3-state diffusion with confined behavior on an eight-lipid bilayer.
Although the diffusion model presented in this work resembles the diffusion model observed in live cells, there are differences in the diffusion parameters. For example, the diffusion coefficients (D1 = 3.9 μm2/s, and D2 = 0.87 μm2/s) retrieved for the fast and intermediate state on the supported lipid bilayer is faster than diffusion constants measured on live cells (D1 = 0.9 μm2/s and D2 = 0.35 μm2/s, respectively (Goswami et al., 2020; Lee et al., 2019)). Our model membrane still lacks biological moieties such as transmembrane proteins, ion channels, and sterols that make the plasma membrane more viscous and present barriers to free diffusion. This may explain why we observe faster diffusion on supported lipid bilayer compared to the cellular plasma membrane.
Similarly, in the in vivo experiments, the state occupancy of the fast state was highest followed by the intermediate and the slow states, whereas in our in vitro experiments, the intermediate state was the most populated state despite the eight-lipid composition. To explain their cellular results, Nan et al. proposed a non-equilibrium steady state (NESS) model in which KRAS4b is continuously depleted from the plasma membrane via the slow state and replenished back into the membrane from the cytosol via the fast state (Lee et al., 2019). This cellular recycling mechanism helps maintain the highest fractional occupancy of KRAS4b in the fast state. In our artificial system, there is no active recycling or internalization of KRAS4b to and from the membrane. However, the system still maintains a steady-state equilibrium where KRAS4b can freely transition between different diffusion states. As suggested by HMM analysis, KRAS4b instantaneously transitions into the intermediate state once its membrane is bound and thus accumulates the most in the intermediate state. This suggests an alternative interpretation of why the fractional occupancy of the fast state in the reconstituted bilayer is smaller compared to the live cells.
Upon membrane binding, as previously shown by electron microscopy, KRAS4b assembles into transient, non-random, and functional nanoclusters to build a platform for the construction of signaling complexes (Weise et al., 2011; Zhou et al., 2018). The exact molecular mechanism behind the formation of RAS nanoclusters is unclear. Researchers have proposed the dimerization of RAS as an underlying step in forming a functional nanocluster (Chen et al., 2016; Prior et al., 2003; Zhou and Hancock, 2015). However, RAS dimerization is exceptionally transient, and to date, we lack direct experimental evidence of a stable protein-protein interaction interface. Most experiments using model membranes used artificially forced RAS dimers (Güldenhaupt et al., 2012; Inouye et al., 2000) and some were biased by the resolution of the experimental techniques (Lee et al., 2020; Nan et al., 2015). For example, in a supported lipid bilayer-based diffusion study, Chung et al. created controlled artificial KRAS4b dimers by binding it to RAS binding domain (RBD) of C-Raf fused to a leucine zipper molecule, which forms a constitutive dimer (Chung et al., 2018; Lin et al., 2014). Using FCS, they measured the diffusion of this leucine zipper dimer complex and found it to be slower, ∼ 2 μm2/s, than the diffusion of KRAS4b monomer, ∼ 4 μm2/s. In our experiments, even though we observed multiple-state diffusion for KRAS4b on a complex membrane, none of the diffusion coefficients matched with the apparent diffusion coefficient of KRAS4b dimer. According to our results, KRAS4b either exists as a monomer, indicated by fast state diffusion of ∼4 μm2/s, or complex larger than dimer, indicated by the slower diffusion rate of ∼0.87 μm2/s and ∼0.1 μm2/s (Chung et al., 2018; Li et al., 2018; Weise et al., 2011). Also, in a separate experiment, we crosslinked KRAS4b molecules using DSSO, and under these conditions observed multiple state diffusion of KRAS4b on a simple two lipid system. The amount of DSSO added in the experiments correlated to the fractional occupancy of the slow and intermediate state. This provided evidence that RAS nanoclusters, enhanced by higher concentration of DSSO, are indeed responsible for the slower diffusion rates. Nevertheless, it is quite possible that transient dimerization occurs at a timescale beyond the time resolution of SPT experiments and hence are only detectable by an ultra-sensitive technique, such as paramagnetic relaxation enhancement (PRE) NMR (Lee et al., 2020). Similarly, intermediate-sized oligomers including trimers, tetramers and pentamers have also been estimated in live cells expressing GFP-tagged wild-type and mutant KRAS4b determined by molecular brightness analysis (N&B analysis) on the fluorescent intensity images obtained through raster image correlation spectroscopy (RICS) and supported by MD simulation (Sarkar-Banerjee et al., 2017). Still, the diffusion coefficient of these intermediate-sized oligomers are yet faster (2.19 μm2/s) than the diffusion obtained in our intermediate state. We also believe that these discrepancies come from the differences in the limit of resolution of different fluorescence techniques employed to measure diffusion and the field can benefit from an improved image analysis pipeline that facilitates better particle localization and particle tracking if one hopes to directly link diffusion to particle size.
RAS activation by GEFs is believed to be at the apex of the signaling cascade in the MAPK pathway. Previously, researchers have used diffusion as a tool to understand the order of events during a signal transduction process (Murakoshi et al., 2004; Nussinov et al., 2019). Particularly, KRAS4b has been shown to substantially slow down or immobilize on the plasma membrane upon activation with growth factor stimulation in cells (Murakoshi et al., 2004). These results suggest the cooperative formation of large, activated RAS-signaling complexes, which are transient and membrane microdomain specific. Here, we evaluated the role of SOS- mediated GTP loading of KRAS4b on the spatiotemporal distribution of KRAS4b by tracking the “on” state KRAS4b that are loaded with GppNHp, a non-hydrolyzable GTP analog, on both simple and complex membrane systems. In the absence of downstream effector molecules, KRAS4b did not show any significant difference in diffusion behavior between the GDP-loaded "inactive" state versus the GppNHp-loaded "active" state. Our results confirm that the “on” and “off” states defined solely by the nucleotide state does not affect the mobility of KRAS4b. Hence, the nucleotide exchange step in the signal transduction pipeline does not initiate clustering and slows down KRAS4b, but possibly orients KRAS4b in the correct conformation suitable for effector binding (Fang et al., 2018).
Lastly, this work highlights the importance of reconstituting a physiologically relevant lipid environment in model membranes when studying membrane-protein interactions. Biological membranes are a crowded 3D mixture of membrane proteins and lipids and their geometrical (membrane thickness, curvature, and protein size), mechanical (elastic moduli, viscosity), and chemical properties (electrostatic, chemical composition) affect the lateral mobility of membrane proteins. Most model membranes used in biophysical studies are oversimplified, and hence, the results are often inconsistent with live-cell studies. We created an artificial lipid bilayer with complex lipid composition that recapitulated the intrinsic heterogeneity of the plasma membrane. Using this novel eight-lipid membrane mimetic, we were able to recreate cell-like diffusion of KRAS4b on reconstituted membrane and gain insight about the molecular mechanisms of the early steps in the signaling pathway. For example, earlier studies have suggested that it is GTP-loading that leads to RAS immobilization and assembly (Murakoshi et al., 2004). However, here we showed that KRAS4b diffusion is indifferent to its nucleotide state and it is in fact the lipid-protein interaction that leads to RAS partitioning into slow moving and clustered states. This study successfully demonstrates the importance of KRAS4b-membrane interaction in the assembly of a signaling platform and emphasizes lipid composition as a therapeutic target. Our future studies include the usage of eight-lipid composition in biophysical assays targeting RAS nanoclusters, as well as examining RAS interactions with its downstream effectors including RAF.
Limitations of the study
Although single particle tracking studies allow visualization of single molecules of proteins, the size of the point spread function is diffraction limited; therefore, the size of the bright spots cannot be directly correlated to the size of the protein. In addition, to visualize single molecules, the number of labeled proteins is kept at minimum and so many proteins are invisible (dark matter). To overcome this limitation, we collect thousands of trajectories, yet we do not fully capture all the protein-interactions. In our study, we observed that, as in cells, KRAS4b diffuses in three different diffusion states on complex lipid membranes. We propose and later deduce from the crosslinker experiment that the slower diffusing states come from KRAS4b nanoclusters. However, it is difficult to establish the size of the cluster. We are working toward an improved image analysis pipeline that will improve the localization precision and identify merge and split events. By keeping track of how many molecules come together before merge and split events, we hope to gain insight into the cluster size. Lastly, we acknowledge that our complex 8-lipid mixture captured the complexity of the lipid environment in the plasma membrane; however, we still lack many other cellular components including actin filaments, transmembrane proteins, and other complex sterols that are vital to structure, function, and dynamics of membrane proteins including RAS. We continue to pursue our goal to recreate a cell-like complex membrane environment on the bench-top to share with the scientific community.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine | Avanti Polar Lipids Inc. | Cat#850457C |
| 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine | Avanti Polar Lipids Inc. | Cat#850459C |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (sodium salt) | Avanti Polar Lipids Inc. | Cat#840034C |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine | Avanti Polar Lipids Inc. | Cat#850757C |
| 1,2-dilinoleoyl-sn-glycero-3-phosphoethanolamine | Avanti Polar Lipids Inc. | Cat#850755C |
| N-palmitoyl-D-erythro-sphingosylphosphorylcholine | Avanti Polar Lipids Inc. | Cat#860584P |
| 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phospho-L-serine (sodium salt) | Avanti Polar Lipids Inc. | Cat#840061C |
| 1-stearoyl-2-arachidonoyl-sn-glycero-3-phospho-(1'-myo-inositol-4',5'-bisphosphate) (ammonium salt) | Avanti Polar Lipids Inc. | Cat#850165P |
| Cholesterol | Avanti Polar Lipids Inc. | Cat#700100P |
| Atto550 DOPE | Atto-tec Gmbh | Cat#AD 550-161 |
| 2-Mercaptoethanol | Sigma-Aldrich | CAS:60-24-2 |
| Gppnhp-tetralithium salt | Jena Bioscience | Cat#NU-401-50 |
| Janelia Fluor 646, Maleimide | Tocris | Cat#6590 |
| Alexa Fluor 647 C2 Maleimide | ThermoFisher Scientific | Cat#A20347 |
| alkaline phosphatase beads | Millipore Sigma | Cat# P0762 |
| beta-cyclodextrin | Millipore Sigma | Cat# C4767 |
| KRAS (Hs.KRAS4b (Ac-2-185-FMe) | Messing et al., 2021 | https://doi.org/10.1007/978-1-0716-1190-6_6 |
| GG-Hs.KRAS4b(2-185-FMe) S106C/C118S | This paper | N/A |
| GG-Hs.KRAS4b(2-185-FMe) C118S/S171C | This paper | N/A |
| Deposited data | ||
| Single particle tracking movies | Mendeley data | https://doi.org/10.17632/4x6hy98krb.1 |
| Experimental models: Cell lines | ||
| Trichoplusia ni Tni-FNL insect cell line | Talsania et al. | https://doi.org/10.3390/genes10020079 |
| Software and algorithms | ||
| Image J | Schneider et al., 2012 | https://imagej.net/software/fiji/ |
| Localizer | Dedecker et al., 2012 | |
| Graph Pad Prism | https://www.graphpad.com/ | |
| Gwyddion | Nečas and Klapetek, 2012 | http://gwyddion.net/ |
| NIS Element Viewer | N/A | https://www.microscope.healthcare.nikon.com/products/software/nis-elements/viewer |
| Track Art | Matysik and Kraut, 2014 | https://sourceforge.net/projects/trackart/ |
| vbSPT HMM analysis | Persson et al., 2013 | https://sourceforge.net/projects/vbspt/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Rebika Shrestha (rebika.shrestha@nih.gov).
Materials availability
-
•
This study did not generate unique reagents.
Experimental model and subject details
Materials
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS), 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine (PAPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1,2- dilinoleoyl-sn-glycero-3-phosphoethanolamine (DIPE), N-stearoyl-D-erythro-sphingosylphosphorylcholine (DPSM), 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphatidylserine (PAPS), L-α-phosphatidylinositol-4,5-bisphosphate (Brain PI(4,5)P2) and Cholesterol were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL) and used without further purification. Janelia Fluor 646 Maleimide was purchased from Tocris Bioscience, a Bio-techne brand (Minneapolis, MN). Alexa647 Maleimide and DSSO were purchased from Thermo Fisher Scientific.
Vesicle preparation
Small unilamellar vesicle (SUV)s were prepared using the sonication technique. Briefly, desired amounts of lipid species were aliquoted in a clean glass vial. The choloroform was evaporated under a gentle stream of Argon gas and further dried in a freeze vacuum overnight. The dried lipids were resuspended in 1 mL of 20 mM Hepes, 200 mM NaCl, pH 7.4 at room temperature for 1 hour, vortexed at maximum strength for 5 minutes and then freeze-thawed for a minimum of ten cycles. The mixture was then sonicated in a bath sonicator at the highest power for at least 1 hour or until the mixture turned clear from a milky solution. All vesicle samples were stored at room temperature and centrifuged at 15000 rpm for 15 minutes prior to use to remove any large aggregates or residuals.
Supported lipid bilayer preparation
The supported lipid bilayers (SLB) were prepared using the vesicle fusion technique (Cremer and Boxer, 1999) in a Bioptech’s FCS2 flow cell chamber. The implement used #1.5, 40 mm borosilicate glass coverslips which were cleaned rigorously in 1% (v/v) Hellmanex III solution followed by the plasma cleaner. 2 μL of the prespun vesicle samples were deposited on a clean glass coverslip, incubated for 30 minutes at room temperature and washed with 10 mL of 20 mM Hepes, 200 mM NaCl, pH 7.4. For RAS imaging experiments, 700 μL of 500 nM wild type KRAS4b plus 50 nM of KRAS4b S106C /JF646 or Alexa647 was added to the flow cell chamber using a 1 mL syringe, incubated for 30 minutes and buffer exchanged with 10 mL of imaging buffer containing 20 mM Hepes, 300 mM NaCl, 5 mM beta-mercaptoethanol (BME) at pH 7.4. BME was necessary to prevent any crosslinking of proteins via photosensitized oxidation (Chung et al., 2016). For DSSO experiments, 400 μL of desired concentration of DSSO prepared in Hepes buffer was flowed through the SLBs containing RAS. No difference was observed in images taken before and after washing.
Protein preparation
Wild type, native N-terminus (minus 1Met, acetylated on 2Thr), prenylated KRAS (Hs.KRAS4b (Ac-2-185-FMe)) was cloned, expressed, and purified following previously published protocols (Messing et al., 2021). Mutated prenylated KRAS proteins, GG-Hs.KRAS4b(2-185-FMe) S106C/C118S and GG-Hs.KRAS4b(2-185-FMe) C118S/S171C were cloned, expressed, and purified following previously published protocols (Gillette et al., 2019).
Nucleotide exchange
The following was added, in order, to 4.9 mL of 130 μM KRAS4b (2-185) S106C/C118S FMe: 128 mL of 50 mM GppNHp (dissolved in 0.5 mM HEPES, pH 7.4), 0.366 mL of buffer (20 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM TCEP), 6 μL of 100 mM ZnCl2, 600 μL of 2 M ammonium sulfate. The ammonium sulfate is added last and mixed quickly to avoid localized areas of high concentration. Final reaction conditions were 107 μM protein, 1.07 mM GppNHp, 0.1 mM ZnCl2, and 200 mM ammonium sulfate. To the reaction was added 55 μL alkaline phosphatase beads (Millipore Sigma, cat# P0762) and the reaction rotated slowly for 2 hr at room temperature (∼21°C). The reaction tube was centrifuged at 1000 × g for 1 minute, the supernatant removed to a fresh tube, the reaction adjusted to 5 mM MgCl2, additional GppNHp added (128 μL of 50 mM GppNHp dissolved in 0.5 mM HEPES, pH 7.4), and the reaction incubated at room temperature for 2 hr then overnight at 4°C. To prevent substantial loss of prenylated KRAS4b to the desalting column used to buffer exchange the protein, non-exchanged KRAS4b(2-185) S106C/C118S FMe was used to precondition the desalting column. This was achieved by passing 1.6 mg of protein over the desalting column, collecting the eluted protein and repeating the process twice. The column was washed with 20 column volumes of 20 mM HEPES, pH 7.4, 150 mM NaCl, 2 mM MgCl2, 1 mM TCEP. To further reduce protein loss to the column, the exchanged protein was amended with 1 mM beta-cyclodextrin. The conditioned column was then used as per the manufacturer’s protocol to buffer exchange the GppNHp-exchanged protein into a final buffer of 20 mM HEPES, pH 7.4, 150 mM NaCl, 2 mM MgCl2, 1 mM TCEP. The collected protein was then dispensed into 0.25 mL aliquots, snap frozen in liquid nitrogen, and stored at -80°C. HPLC analysis (not shown) indicated the protein was 99.5% exchanged to GppNHp.
For imaging experiments, the mutant proteins were labeled with Janelia Fluor 646 (JF646) maleimide fluorophore. Briefly, the protein was buffer exchanged to the labelling buffer, 20mM Hepes, 150 mM NaCl, 5 mM MgCl2 and 1 mM TCEP, pH 7.4. The labelling reaction was initiated by adding 30 μL of 10 mM JF646 dye and ran for 2 hours at 4°C protected from light. The reaction was quenched by adding 10 μL of BME. The free fluorophores were separated from the protein-bound fluororphores by passing through a Superdex S-75 column (GE healthcare). The proteins were concentrated to ∼1.6 mg/mL concentration, flash-frozen in liquid nitrogen and stored at -80°C. The labelling was confirmed using native mass spectrometry. For labelling active KRAS4b, the wild type GDP loaded KRAS4b was first nucleotide exchanged to GppNHp, a non-hydrolyzable GTP analog, and then labelled with Alexa647 maleimide dye following the above described procedure.
Method details
SPT experiments
Single particle tracking experiments were carried out on a Nikon NStorm Ti-81 inverted microscope. The samples were loaded onto a stage and imaged with a 100x 1.49 N.A. oil immersion TIRF objective (Nikon, Japan) and a Andor iX EMCCD camera with 16 μm pixel size (Andor technologies, USA). Prior to recording, the sample was first photobleached with high intensity laser power to isolate any immobile fluorophores stuck to the glass surface. For imaging the lipids, 561 nm laser from an Agilent laser module was used at 10% power and for KRAS4b, and the 647nm laser also from the Agilent laser module was used at 20% power setting. The Alexa647-labeled, GppNHp loaded KRAS4b was imaged with 647 nm laser using 10% laser power. The samples were illuminated under TIRF mode by changing the illumination angle to 2980 through the Nikon TIRF box controlled by the NIS-Element AR 4.4 by Nikon. Time-lapse images were acquired under continuous illumination with 10 ms exposure time and zero-time delay between two subsequent frames. For each experiment, a minimum of three samples with 15 TIRF movies per sample and 5000 frames per movie were acquired.
AFM experiments
Atomic force microscopy (AFM) experiments were performed on an Asylum Cypher VRS Video Rate AFM (Oxford Instruments Asylum Research, Santa Barbara, CA). The SLB samples were prepared by depositing ∼50 μL of SUVs on a freshly cleaved mica surface, incubated for 60 min followed by careful rinsing with ∼2 mL of 20 mM Hepes, 200 mM NaCl, pH 7.4 buffer. The samples were maintained under aqueous conditions throughout the preparation and experimental procedures. AFM images were obtained under the tapping- mode in Hepes buffer using a Biolever mini silicon nitride tip (BL-AC40TS) (Oxford Instruments Asylum Research) with a spring constant of 0.09 N/m and tip-sample force of <100 pN. The topography images were analyzed using an open source software, Gwyddion (Nečas and Klapetek, 2012).
Data processing
Single particle tracking movies collected in NIS-Element AR 4.4 were first converted into .tif files using Image J (Schneider et al., 2012) and then processed using the Localizer plug-in available in the Igor Pro software (Dedecker et al., 2012). Single particles in each frame were localized as spots based on the eight-way adjacency particle detection algorithm with generalized likelihood ratio test (GLRT) sensitivity of 10, point spread function (PSF) of 1.3 pixels and smooth factor of 5. A symmetric 2D Gaussian fit function was used to estimate the position of PSF in each frame. Any localized particles persistent for more than 6 frames were then linked between the subsequent frames into tracks with a set of criteria. The particles were allowed a maximum jump distance of 5 pixels and at least one frame blinking.
Mean square displacement (MSD) and Cumulative probability distribution (CPD) analysis of the trajectories were performed on a Matlab based TrackArt software (Matysik and Kraut, 2014). Single molecule tracks from several movies were organized and combined into a single matlab file before input into the TrackArt and vbSPT for HMM analysis. The iteration steps and bootstrapping were fixed at 250 for HMM analysis.
Quantification and statistical analysis
Single particle tracks were collected from at least 15 movies composed of 5000 frames per movie for each experiment. 3 replicates of each experiments were performed. Figures 1, 2, 3, 4, and 5 represent results from one representative experiment. Table S2 shows the mean and the standard deviation for all experiment.
Acknowledgments
We would like to thank Matt Drew, Dom Esposito, William Gillette, Vijaya Gowda, Jen Mehalko, Simon Messing, Shelley Perkins, Kelly Snead, and Vanessa Wall from the Protein Expression Laboratory at the NCI RAS Initiative, Frederick National Lab for Cancer Research (FNLCR) for protein cloning, expression, and purification. We are grateful to Timothy Waybright (FNLCR), Will Heinz at the Optical Microscopy and Analysis Laboratory (FNLCR), Joe Meyer at the Scientific Publications, Graphics and Media at FNLCR, and Arvind Ramanathan at Argonne National Laboratory for creating the KRAS PyMOL model. We would also like to acknowledge Andrew G. Stephen, Tim Tran, Constance Agamasu, and JDACS4C and the Pilot 2 team for helpful discussions. We thank Frank McCormick, University of California San Francisco for scientific conversations. This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Author contributions
R.S. and T.J.T conceptualized the study. R.S. performed the single-particle tracking experiments. P.F. expressed and purified the proteins. R.S., D.C., and P.F. analyzed the data. R.S. prepared the manuscript. All authors contributed to the reviewing and editing of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: January 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103608.
Supplemental information
Data and code availability
-
•
Single particle tracking data have been deposited at Mendeley Data and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
No original code was created as a part of this work.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Banerjee A., Jang H., Nussinov R., Gaponenko V. The disordered hypervariable region and the folded catalytic domain of oncogenic K-Ras4B partner in phospholipid binding. Curr. Opin. Struct. Biol. 2016;36:10–17. doi: 10.1016/j.sbi.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilkova E., Pleskot R., Rissanen S., Sun S., Czogalla A., Cwiklik L., Róg T., Vattulainen I., Cremer P.S., Jungwirth P., et al. Calcium directly regulates phosphatidylinositol 4,5-bisphosphate headgroup conformation and recognition. J. Am. Chem. Soc. 2017;139:4019–4024. doi: 10.1021/jacs.6b11760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Peters A., Huang T., Nan X. Ras dimer formation as a new signaling mechanism and potential cancer therapeutic target. Mini Rev. Med. Chem. 2016;16:391–403. doi: 10.2174/1389557515666151001152212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J.K., Lee Y.K., Lam H.Y.M., Groves J.T. Covalent ras dimerization on membrane surfaces through photosensitized oxidation. J. Am. Chem. Soc. 2016;138:1800–1803. doi: 10.1021/jacs.5b12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J.K., Lee Y.K., Denson J.-P., Gillette W.K., Alvarez S., Stephen A.G., Groves J.T. K-Ras4B remains monomeric on membranes over a wide range of surface densities and lipid compositions. Biophys. J. 2018;114:137–145. doi: 10.1016/j.bpj.2017.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell S.D., Smith D.A. The atomic force microscope as a tool for studying phase separation in lipid membranes. Mol. Membr. Biol. 2006;23:17–28. doi: 10.1080/09687860500501158. [DOI] [PubMed] [Google Scholar]
- Cooper G.M. The Cell: A Molecular Approach. Second edition. Sinauer Associates; 2000. Structure of the plasma membrane. [Google Scholar]
- Cremer P.S., Boxer S.G. Formation and spreading of lipid bilayers on planar glass supports. J. Phys. Chem. B. 1999;103:2554–2559. [Google Scholar]
- Dedecker P., Duwé S., Neely R.K., Zhang J. Localizer: fast, accurate, open-source, and modular software package for superresolution microscopy. J. Biomed. Opt. 2012;17:126008. doi: 10.1117/1.JBO.17.12.126008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z., Marshall C.B., Nishikawa T., Gossert A.D., Jansen J.M., Jahnke W., Ikura M. Inhibition of K-RAS4B by a unique mechanism of action: stabilizing membrane-dependent occlusion of the effector-binding site. Cell Chem. Biol. 2018;25:1327–1336.e4. doi: 10.1016/j.chembiol.2018.07.009. [DOI] [PubMed] [Google Scholar]
- Frederix P.L.T.M., Bosshart P.D., Engel A. Atomic force microscopy of biological membranes. Biophys. J. 2009;96:329–338. doi: 10.1016/j.bpj.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette W., Frank P., Perkins S., Drew M., Grose C., Esposito D. In: Protein Lipidation: Methods and Protocols. Linder M.E., editor. Springer; 2019. Production of farnesylated and methylated proteins in an engineered insect cell system; pp. 259–277. [DOI] [PubMed] [Google Scholar]
- Gillette W.K., Esposito D., Abreu Blanco M., Alexander P., Bindu L., Bittner C., Chertov O., Frank P.H., Grose C., Jones J.E., et al. Farnesylated and methylated KRAS4b: high yield production of protein suitable for biophysical studies of prenylated protein-lipid interactions. Sci. Rep. 2015;5:15916. doi: 10.1038/srep15916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami D., Chen D., Yang Y., Gudla P.R., Columbus J., Worthy K., Rigby M., Wheeler M., Mukhopadhyay S., Powell K., et al. Membrane interactions of the globular domain and the hypervariable region of KRAS4b define its unique diffusion behavior. ELife. 2020;9:e47654. doi: 10.7554/eLife.47654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm J.B., English B.P., Chen J., Slaughter J.P., Zhang Z., Revyakin A., Patel R., Macklin J.J., Normanno D., Singer R.H., et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods. 2015;12:244–250. doi: 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güldenhaupt J., Rudack T., Bachler P., Mann D., Triola G., Waldmann H., Kötting C., Gerwert K. N-Ras forms dimers at POPC membranes. Biophys. J. 2012;103:1585–1593. doi: 10.1016/j.bpj.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock J.F. Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell Biol. 2003;4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- Ingolfsson H., Neale C., Carpenter T., Shrestha R., Lopez C., Tran T., Oppelstrup T., Bhatia H., Stanton L., Zhang X., et al. Machine learning-driven multiscale modeling reveals lipid-dependent dynamics of RAS signaling proteins. PNAS. 2021 doi: 10.1073/pnas.2113297119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye K., Mizutani S., Koide H., Kaziro Y. Formation of the Ras dimer is essential for Raf-1 activation. J. Biol. Chem. 2000;275:3737–3740. doi: 10.1074/jbc.275.6.3737. [DOI] [PubMed] [Google Scholar]
- Iversen L., Tu H.-L., Lin W.-C., Christensen S.M., Abel S.M., Iwig J., Wu H.-J., Gureasko J., Rhodes C., Petit R.S., et al. Molecular kinetics. Ras activation by SOS: allosteric regulation by altered fluctuation dynamics. Science. 2014;345:50–54. doi: 10.1126/science.1250373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L.J. Nanoscale imaging of domains in supported lipid membranes. Langmuir. 2007;23:5886–5895. doi: 10.1021/la070108t. [DOI] [PubMed] [Google Scholar]
- Kao A., Chiu C., Vellucci D., Yang Y., Patel V.R., Guan S., Randall A., Baldi P., Rychnovsky S.D., Huang L. Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.002212. M110.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi A., Shirai Y.M., Koyama-Honda I., Suzuki K.G.N., Fujiwara T.K. Hierarchical organization of the plasma membrane: investigations by single-molecule tracking vs. fluorescence correlation spectroscopy. FEBS Lett. 2010;584:1814–1823. doi: 10.1016/j.febslet.2010.02.047. [DOI] [PubMed] [Google Scholar]
- Kusumi A., Fujiwara T.K., Chadda R., Xie M., Tsunoyama T.A., Kalay Z., Kasai R.S., Suzuki K.G.N. Dynamic organizing principles of the plasma membrane that regulate signal transduction: commemorating the fortieth anniversary of singer and Nicolson’s fluid-mosaic model. Annu. Rev. Cell Dev. Biol. 2012;28:215–250. doi: 10.1146/annurev-cellbio-100809-151736. [DOI] [PubMed] [Google Scholar]
- Lee K.-Y., Fang Z., Enomoto M., Gasmi-Seabrook G., Zheng L., Koide S., Ikura M., Marshall C.B. Two distinct structures of membrane-associated homodimers of GTP- and GDP-bound KRAS4B revealed by paramagnetic relaxation enhancement. Angew. Chem. Int. Ed. Engl. 2020;59:11037–11045. doi: 10.1002/anie.202001758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Phelps C., Huang T., Mostofian B., Wu L., Zhang Y., Tao K., Chang Y.H., Stork P.J., Gray J.W., et al. High-throughput, single-particle tracking reveals nested membrane domains that dictate KRasG12D diffusion and trafficking. ELife. 2019;8:e46393. doi: 10.7554/eLife.46393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Dwivedi M., Erwin N., Möbitz S., Nussbaumer P., Winter R. Interaction of KRas4B protein with C6-ceramide containing lipid model membranes. Biochim. Biophys. Acta Biomembr. 2018;1860:1008–1014. doi: 10.1016/j.bbamem.2018.01.016. [DOI] [PubMed] [Google Scholar]
- Lin W.-C., Iversen L., Tu H.-L., Rhodes C., Christensen S.M., Iwig J.S., Hansen S.D., Huang W.Y.C., Groves J.T. H-Ras forms dimers on membrane surfaces via a protein-protein interface. Proc. Natl. Acad. Sci. U S A. 2014;111:2996–3001. doi: 10.1073/pnas.1321155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lito P., Solomon M., Li L.-S., Hansen R., Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science. 2016;351:604–608. doi: 10.1126/science.aad6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommerse P.H.M., Vastenhoud K., Pirinen N.J., Magee A.I., Spaink H.P., Schmidt T. Single-molecule diffusion reveals similar mobility for the Lck, H-ras, and K-ras membrane anchors. Biophys. J. 2006;91:1090–1097. doi: 10.1529/biophysj.105.079053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matysik A., Kraut R.S. TrackArt: the user friendly interface for single molecule tracking data analysis and simulation applied to complex diffusion in mica supported lipid bilayers. BMC Res. Notes. 2014;7:274. doi: 10.1186/1756-0500-7-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing S., Agamasu C., Drew M., DeHart C.J., Stephen A.G., Gillette W.K. In: Ras Activity and Signaling: Methods and Protocols. Rubio I., Prior I., editors. Springer US; 2021. Production and membrane binding of N-terminally acetylated, C-terminally farnesylated and carboxymethylated KRAS4b; pp. 105–116. [DOI] [PubMed] [Google Scholar]
- Michalet X. Mean square displacement analysis of single-particle trajectories with localization error: brownian motion in isotropic medium. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2010;82:041914. doi: 10.1103/PhysRevE.82.041914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakoshi H., Iino R., Kobayashi T., Fujiwara T., Ohshima C., Yoshimura A., Kusumi A. Single-molecule imaging analysis of Ras activation in living cells. Proc. Natl. Acad. Sci. U S A. 2004;101:7317–7322. doi: 10.1073/pnas.0401354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X., Tamgüney T.M., Collisson E.A., Lin L.-J., Pitt C., Galeas J., Lewis S., Gray J.W., McCormick F., Chu S. Ras-GTP dimers activate the mitogen-activated protein kinase (MAPK) pathway. Proc. Natl. Acad. Sci. U S A. 2015;112:7996–8001. doi: 10.1073/pnas.1509123112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nečas D., Klapetek P. Gwyddion: an open-source software for SPM data analysis. Cent. Eur. J. Phys. 2012;10:181–188. [Google Scholar]
- Nussinov R., Tsai C.-J., Jang H. Oncogenic KRas mobility in the membrane and signaling response. Semin. Cancer Biol. 2019;54:109–113. doi: 10.1016/j.semcancer.2018.02.009. [DOI] [PubMed] [Google Scholar]
- Persson F., Lindén M., Unoson C., Elf J. Extracting intracellular diffusive states and transition rates from single-molecule tracking data. Nat. Methods. 2013;10:265–269. doi: 10.1038/nmeth.2367. [DOI] [PubMed] [Google Scholar]
- Prior I.A., Muncke C., Parton R.G., Hancock J.F. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 2003;160:165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar-Banerjee S., Sayyed-Ahmad A., Prakash P., Cho K.-J., Waxham M.N., Hancock J.F., Gorfe A.A. Spatiotemporal analysis of K-Ras plasma membrane interactions reveals multiple high order homo-oligomeric complexes. J. Am. Chem. Soc. 2017;139:13466–13475. doi: 10.1021/jacs.7b06292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen A.G., Esposito D., Bagni R.K., McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25:272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- Weise K., Kapoor S., Denter C., Nikolaus J., Opitz N., Koch S., Triola G., Herrmann A., Waldmann H., Winter R. Membrane-mediated induction and sorting of K-Ras microdomain signaling platforms. J. Am. Chem. Soc. 2011;133:880–887. doi: 10.1021/ja107532q. [DOI] [PubMed] [Google Scholar]
- Wen Y., Vogt V.M., Feigenson G.W. Multivalent cation-bridged PI(4,5)P2 clusters form at very low concentrations. Biophys. J. 2018;114:2630–2639. doi: 10.1016/j.bpj.2018.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Hancock J.F. Ras nanoclusters: versatile lipid-based signaling platforms. Biochim. Biophys. Acta. 2015;1853:841–849. doi: 10.1016/j.bbamcr.2014.09.008. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Liang H., Rodkey T., Ariotti N., Parton R.G., Hancock J.F. Signal integration by lipid-mediated spatial cross talk between ras nanoclusters. Mol. Cell. Biol. 2014;34:862. doi: 10.1128/MCB.01227-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Prakash P., Liang H., Cho K.-J., Gorfe A.A., Hancock J.F. Lipid-Sorting specificity encoded in K-Ras membrane anchor regulates signal output. Cell. 2017;168:239–251.e16. doi: 10.1016/j.cell.2016.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Prakash P., Gorfe A.A., Hancock J.F. Ras and the plasma membrane: a complicated relationship. Cold Spring Harb. Perspect. Med. 2018;8:a031831. doi: 10.1101/cshperspect.a031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemba B.P., Falke J.J. Lateral diffusion of peripheral membrane proteins on supported lipid bilayers is controlled by the additive frictional drags of (1) bound lipids and (2) protein domains penetrating into the bilayer hydrocarbon core. Chem. Phys. Lipids. 2013;172–173:67–77. doi: 10.1016/j.chemphyslip.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The tracked molecules are JF646 labeled KRAS4b captured at 10 ms/ frame and played at 25 frames/s.
The tracked molecules are JF646 labeled KRAS4b captured at 10 ms/ frame and played at 25 frames/s.
Data Availability Statement
-
•
Single particle tracking data have been deposited at Mendeley Data and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
No original code was created as a part of this work.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.