Summary
Non-genetically modified somatic cells can only be inefficiently and stochastically reprogrammed to pluripotency by exogenous expression of reprogramming factors. Low competence of natural reprogramming factors may prevent the majority of cells to successfully and synchronously reprogram. Here we screened DNA-interacting amino acid residues in the zinc-finger domain of KLF4 for enhanced reprogramming efficiency using alanine-substitution scanning methods. Identified KLF4 L507A mutant accelerated and stabilized reprogramming to pluripotency in both mouse and human somatic cells. By testing all the variants of L507 position, variants with smaller amino acid residues in the KLF4 L507 position showed higher reprogramming efficiency. L507A bound more to promoters or enhancers of pluripotency genes, such as KLF5, and drove gene expression of these genes during reprogramming. Molecular dynamics simulations predicted that L507A formed additional interactions with DNA. Our study demonstrates how modifications in amino acid residues of DNA-binding domains enable next-generation reprogramming technology with engineered reprogramming factors.
Subject areas: Stem cell plasticity, Molecular biology
Graphical abstract
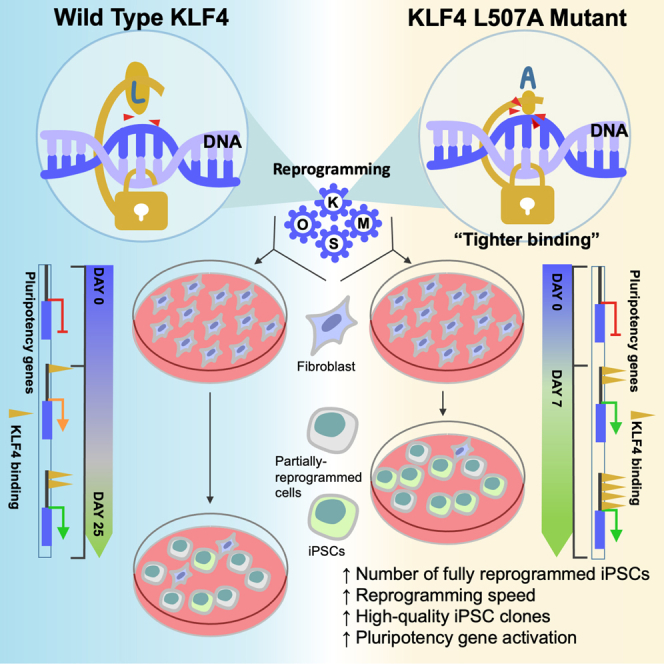
Highlights
-
•
KLF4 L507A variant accelerates and stabilizes reprogramming to pluripotency
-
•
KLF4 L507A has distinctive features of transcriptional binding and activation
-
•
KLF4 L507A may acquire a unique conformation with additional DNA interaction
-
•
Smaller amino acid residues in L507 position cause higher reprogramming efficiency
Stem cell plasticity; Molecular biology
Introduction
Reprogramming technology provides unprecedented opportunities for life sciences, drug discovery, and regenerative medicine (Gurdon and Melton, 2008; Yamanaka and Blau, 2010). Advances of reprogramming technology enable one type of somatic cells to turn into other desired cell types by ectopic expression of defined factors, which are mostly transcription factors (Adachi and Scholer, 2012; Gurdon, 2016; Takahashi and Yamanaka, 2016). These transcription factors cooperatively bind silenced or closed parts of DNA and initiate active transcriptional events as “pioneer factors” to regulate the expression of downstream genes and the modification of epigenetic status toward reprogrammed cell states (Guo and Morris, 2017; Iwafuchi-Doi and Zaret, 2014; Ninkovic and Gotz, 2018). Once a reprogramming method to generate a cell type from another has been developed, many studies have focused on searching additional or substitutive factors, epigenetic modifiers, culture conditions, and delivery methods of the reprogramming factors to improve the reprogramming results (Gonzalez et al., 2011; Hanna et al., 2010; Xu et al., 2015). However, the current stagnation of reprogramming technology still accounts for such drawbacks as a poor and low yield of high-quality reprogrammed cells from primary somatic cells, which results in delays of medical applications. On the other hand, the functional alteration of reprogramming factors remains disregarded or unexploited overall. Only a handful of studies have demonstrated that modification or addition of transcriptional activity in reprogramming factors enhanced or altered reprogramming results (Jauch, 2018; Tsang et al., 2014; Velychko et al., 2019). Generally, natural transcription factors are still used as reprogramming factors.
Among these reprogramming factors, KLF4 (Krüppel-like factor 4; as known as EZF or GKLF) is a ZnF (Zinc-Finger) protein with both transcriptional activation and repression domains and regulates downstream gene expression to control cellular states in a context-dependent manner (Garrett-Sinha et al., 1996; Rowland et al., 2005; Shields et al., 1996). Although KLF4 regulates the development and homeostasis of various tissues including cancers in natural biological contexts (Mahatan et al., 1999; Segre et al., 1999; Shie et al., 2000; Zhang et al., 2000), it plays a central role in the reprogramming of somatic cells into iPSCs (induced pluripotent stem cells) and other types of cells (Adachi and Scholer, 2012; Hayashi et al., 2014; Hiramatsu et al., 2011; Kitazawa et al., 2019; Takahashi and Yamanaka, 2006; Thier et al., 2012). In iPSC reprogramming, KLF4 regulates genome-wide epigenetic status and chromatin reorganization as a pioneer factor as well as its direct downstream transcription (Di Giammartino et al., 2019; Soufi et al., 2012, 2015). Although the functionality of KLF4 in reprogramming has been understood well as above, the structural basis of KLF4 that enables its unique reprogramming activity remains elusive.
Therefore in this study, we took a structure-function approach to answer if rational engineering of reprogramming factors might improve reprogramming. We focused on the ZnF DNA-binding domain of KLF4 to delineate its unique contribution to iPSC generation. We scanned alanine-substituted mutants of DNA-interacting amino acid residues in the KLF4 ZnF domain in iPSC generation. Although many of these mutants lost or decreased their reprogramming activity, one mutant, L507A, increased the speed and efficiency of iPSC generation. Human iPSC clones generated with this mutant showed relatively low heterogeneity in the expression of NANOG gene, low expression of exogenous vector RNA from SeVdp (Sendai Virus vector), and low expression of differentiation-defective iPSC markers (i.e., HERV-H and lincRNA-RoR (Ohnuki et al., 2014). By testing all the variants of L507 position, smaller amino acid residues of L507 position in KLF4 showed higher reprogramming efficiency. ChIP-Seq (chromatin immunoprecipitation – sequencing) and RNA-Seq experiments revealed that this L507A mutant bound more to promoters or enhancers of some pluripotency genes, such as KLF5, and drove the gene expression of them during reprogramming. Finally, MD (molecular dynamics) simulation analysis predicted that this mutant may additionally acquire a unique structural conformation in the protein-DNA complex by forming additional hydrogen bonds between several other amino acid residues and DNA. Our study demonstrates that only one amino acid modulation in a reprogramming factor can markedly improve natural transcription factor function to achieve faster and effective reprogramming, which leads to cost-effective advantages in future scientific and medical applications.
Results
Design of alanine scanning KLF4 ZnF mutants based on in silico identification of putative KLF4 DNA-interacting amino acid residues
First, we identified amino acid residues in the KLF4 ZnF domain directly interacting with target DNA by using PDB (protein data bank) ePISA program (Krissinel and Henrick, 2007) (Table S1). In this program, structural data of mouse Klf4 ZnF-DNA complex from two studies that solved its X-ray crystal structures were analyzed (Liu et al., 2014; Schuetz et al., 2011). The sequences of amino acid residues in the KLF4 ZnF domain are perfectly conserved between humans and mice. Next, we selected conserved amino acid residue among KLF2, KLF4, and KLF5, which were reported to show similar functions in pluripotency and reprogramming (Jeon et al., 2016; Jiang et al., 2008; Nakagawa et al., 2008; Yamane et al., 2018). As a result, 20 amino acid residues were identified as evolutionarily conserved and directly interacted with DNA (Table S1). Then, we generated 19 mutants of human KLF4, substituting wild-type residue with alanine (A) by site-directed mutagenesis in retroviral vectors except for an original alanine amino acid residue (Figure 1A). Retroviral expression and the transduced protein expression of these 19 KLF4 ZnF mutants were examined in human fibroblasts by Western blot analysis (Figures S1A–S1C). Although many of the mutant proteins were expressed comparable to WT (wild-type) KLF4, protein levels of K443A, H446A, H450A, F471A, R479A, F499A, R501A, and R504A were substantially lower than WT.
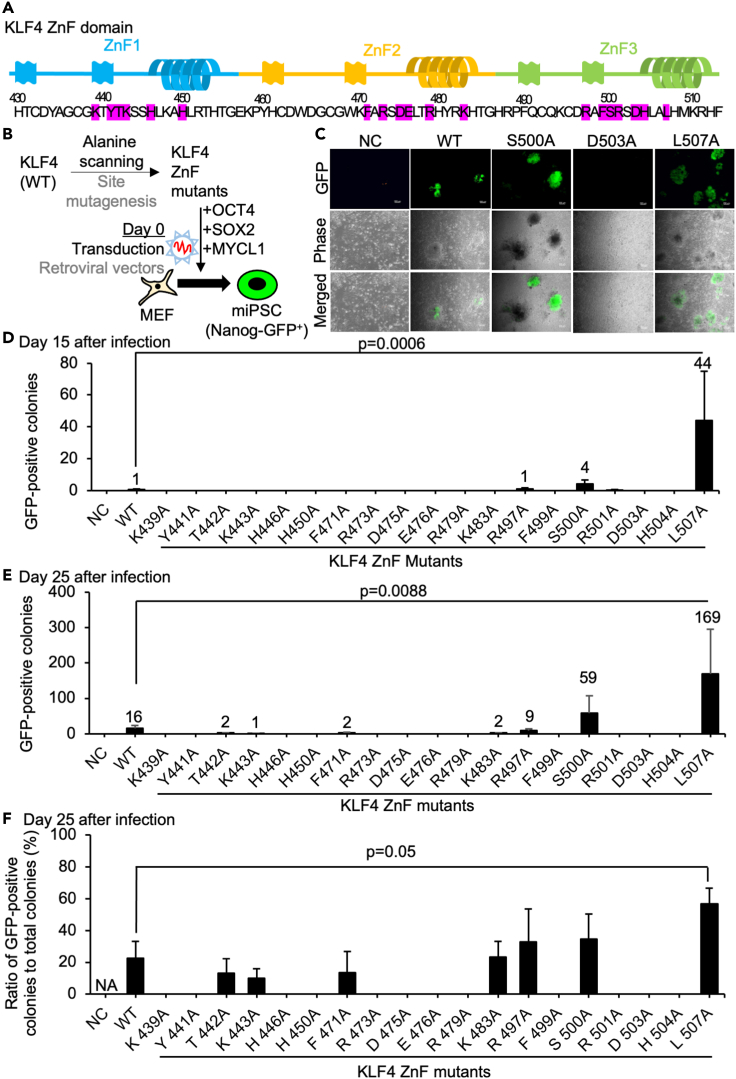
Figure 1.
The reprogramming efficiency of Nanog-GFP MEF using KLF4 ZnF alanine-scanned mutants
(A) Schematic of the protein sequence of the KLF4 ZnF domain. The amino acid residues highlighted in red are targeted by alanine scanning site mutagenesis in this study.
(B) Schematic of the generation of iPSCs using alanine-scanned KLF4 ZnF mutants made by site mutagenesis of KLF4 wild-type (WT) retroviral vector together with OCT4, SOX2, and MYCL1 retroviral vectors.
(C) The typical morphologies of generated iPSC colonies in NC (negative control with mRFP1 vector), WT (KLF4 wild-type), R497A mutant, D503A mutant, and L507A mutant. The images were shown in GFP (green channel fluorescence), Phase (phase contrast), and merged images. Scale bars, 200 μm.
(D) The number of Nanog-GFP-positive iPSC colonies were counted 15 days after infection of retroviral vectors to Nanog-GFP MEF from 10,000 cells. Results are mean and SE, n = 4. p value was calculated from Dunnett's test between WT and L507A conditions.
(E) The number of Nanog-GFP-positive colonies were counted 25 days after infection of retroviral vectors to Nanog-GFP MEF from 10,000 cells. Results are mean and SE, n = 4. p value was calculated from Dunnett's test between WT and L507A conditions.
(F) The ratio of Nanog-GFP-positive colonies was calculated 25 days after infection of retroviral vectors. The total number of Nanog-GFP-positives was counted from whole wells. Results are mean and SE, n = 4. p value was calculated from Dunnett's test between WT and L507A conditions.
iPSC generation from Nanog-GFP MEF using KLF4 ZnF mutants
To examine the role of these DNA-interacting residues in iPSC reprogramming, we generated iPSCs from MEF (mouse embryonic fibroblasts), which carry Nanog-GFP (Nanog-promoter-driven green fluorescent protein) transgene (Okita et al., 2007), using KLF4 ZnF alanine substitution mutants (Figure 1B). Retroviral vectors carrying KLF4 ZnF alanine substitution mutants were transduced together with OCT4, SOX2, and MYCL1 to generate iPSCs from these MEF (Nakagawa et al., 2010; Takahashi and Yamanaka, 2006). Nanog-GFP-positive colonies were observed and counted by a fluorescent microscope (Figure 1C). As early as day 15 after transduction, GFP-positive cells can be detected by fluorescent microscopy. On days 15 and 25 after transduction, more GFP-positive iPSC colonies were found in L507A mutant conditions than WT conditions (Figures 1D and E). Fewer or comparable numbers of GFP-positive iPSC colonies were found in the conditions using T442A, K443A, F471A, K483A, R497A, S500A mutants, and no or few GFP-positive iPSC colonies were found in the conditions using K439A, Y441A, H446A, H450A, R473A, D475A, E476A, R479A, F499A, R501A, D503A, and H504A mutants. The ratio of Nanog-GFP-positive iPSC colonies to total colonies was also calculated on day 25 after transduction to evaluate the heterogeneity of iPSC generation. L507A mutant conditions resulted in Nanog-GFP-positive colonies at more than 60%, whereas WT or all the other mutant conditions resulted in Nanog-GFP-positive colonies at around 30% (Figure 1F). These results indicated that L507A mutant improved both the efficiency and the speed of iPSC reprogramming from MEF and that targeted amino acid residues contained many critical sequences in reprogramming activity.
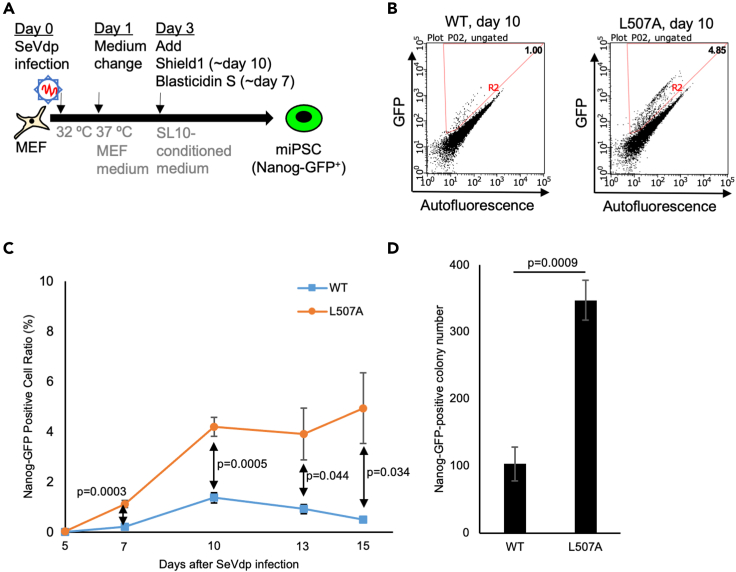
To test the effect of L507A mutant on iPSC generation in another vector system, we developed rapid and efficient iPSC generation from primary MEF using SeVdp (replication-defective and persistent Sendai virus) vectors in feeder-free conditions (Figure 2A). iPSC generation system using SeVdp vectors, which remains persistently in the cytoplasm without integrating into the host genome, has been developed considering safety and efficacy (Nishimura et al., 2011). In this study, we constructed L507A KLF4 mutant harboring SeVdp vector from SeVdp vector expressing WT KLF4 (Nishimura et al., 2014, 2017). Also in this system, because KLF4 is fused with a ligand-dependent destabilization domain (DD) at the N-terminus, we treated Shield-1 to stabilize exogenous KLF4 protein during reprogramming. In this culture condition, we analyzed the ratio of Nanog-GFP-positive cells using flow cytometry on day 5 - 15 after the infection of these SeVdp vectors and counted the Nanog-GFP-positive colony numbers on day 15. From day 7, Nanog-GFP-positive cells appeared, and the ratio of them was significantly higher in the L507A condition throughout these days (Figures 2B and 2C). The number of Nanog-GFP-positive colonies was also significantly higher in the L507A condition (Figure 2D). Compared with the retroviral vector system, these results suggested that SeVdp vector system carrying KLF4 L507A produced mouse iPSCs (miPSCs) from somatic cells more efficiently and faster.
Figure 2.
Efficient generation system of Nanog-GFP-positive miPSCs using SeVdp in feeder-free conditions
(A) Schematic of miPSC generation from Nanog-GFP MEF using SeVdp infection. On day 3 after infection, the medium was changed to serum replacement (SR)-based conditioned medium which was collected from the culture of the SL10 feeder cell line. In addition, Shield1 to stabilize KLF4 protein in the ProteoTuner Shield system and Blasticidin S to select the transduced cells were added on day 3 after infection until day 10 and day 7, respectively.
(B) A typical example of flow cytometric analysis of reprogrammed iPSCs in the conditioned medium or MEF medium using SeVdp on day 10 after infection.
(C) Flow cytometric analysis of the percentage of Nanog-GFP-positive cells after the infection of SeVdp carrying WT or L507A KLF4. Flow cytometric experiments were performed on days 5, 7, 10, 13, and 15 after the infection. Results are mean and SE, n = 5. p values calculated with t test were shown based on the comparison between WT and L507A KLF4 conditions.
(D) The number of Nanog-GFP-positive iPSC colonies were counted 13 days after infection of SeVdp to Nanog-GFP MEF from 10,000 cells. Results are mean and SE, n = 4. The p value calculated with the t test was shown based on the comparison between WT and L507A KLF4 conditions.
L507A mutant accelerated and stabilized human iPSC generation
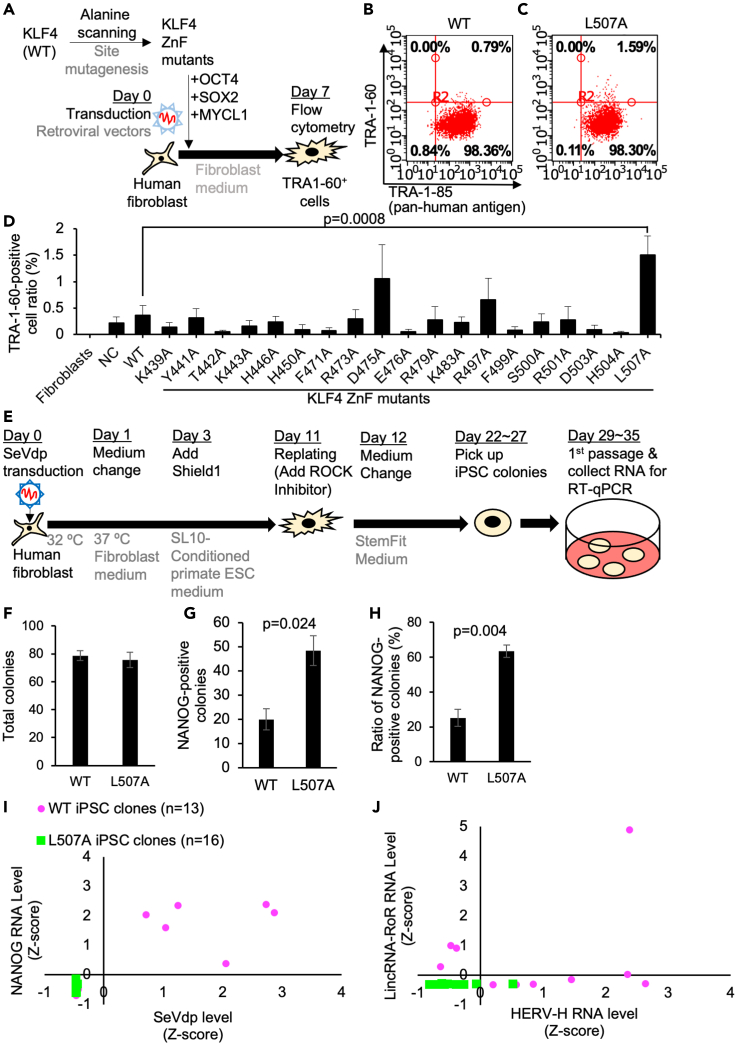
To determine whether KLF4 ZnF mutants affect the efficiency of iPSC reprogramming in human cells, we transduced KLF4 ZnF mutants together with OCT4, SOX2, and MYCL1 to human neonatal fibroblasts (Figure 3A). TRA (Tumor-Related Antigen)-1-60 was used to detect faithfully-reprogramming cells by flow cytometry (Chan et al., 2009). On day 7 after transduction, a higher ratio of TRA-1-60-positive cells was produced by L507A compared to WT (Figures 3B–3D). These results indicated that L507A enhanced the reprogramming process in human somatic cells. Consistent with earlier induction of Nanog-GFP-positive colonies in MEF on day 15 after transduction (Figure 1D), the effect of L507A on iPSC reprogramming may be common in mammals.
Figure 3.
The reprogramming efficiency and heterogeneity of human somatic cells using KLF4 ZnF alanine-scanned mutants
(A) Schematic of the reprogramming of human fibroblasts using alanine-scanned KLF4 ZnF mutants made by site mutagenesis of KLF4 wild-type (WT) retroviral vector together with OCT4, SOX2, and MYCL1 retroviral vectors.
(B and C) Typical data of flow cytometric analysis of the percentage of TRA1-60-positive cells in KLF4 alanine-scanned mutants 7 days after infection of reprogramming retrovirus vectors carrying KLF4 ZnF alanine-substituted mutants.
(D) Flow cytometric analysis of the percentage of TRA1-60-positive cells in KLF4 alanine-scanned mutants 7 days after infection of reprogramming retrovirus vectors carrying KLF4 ZnF alanine-substituted mutants. Results are mean and SE, n = 6. The p value was calculated with Dunnett's test between WT and L507A conditions.
(E) Schematic of the generation of human iPSCs using SeVdp carrying KLF4 or L507A mutant in feeder-free conditions. On day 3 after infection, the medium was changed to primate ESC medium-based conditioned medium which was collected from the culture of a feeder cell line. Because KLF4 is fused with a ligand-dependent destabilization domain (DD) at the N-terminus in this SeVdp, Shield1 to stabilize KLF4 protein in the ProteoTuner Shield system was added into one group until picking up iPSC colonies. Blasticidin S was added on day 3 after infection until day 7.
(F) Numbers of total colonies in each condition were counted by colony morphology on day 20. Results are mean and SE, n = 3.
(G) Numbers of NANOG-positive colonies in each condition were counted by immunocytochemistry. Results are mean and SE, n = 3. The p value calculated with the t test was shown based on the comparison between WT and L507A KLF4 conditions.
(H) The ratio of NANOG-positive colonies in each condition was calculated from numbers of total colonies and NANOG-positive colonies. Results are mean and SE, n = 3. The p value calculated with the t test was shown based on the comparison between WT and L507A KLF4 conditions.
(I and J) Variations of NANOG expression and residual SeVdp NP RNA amount (G) and HERV-H and lincRNA-RoR (H) in human iPSC clones generated with SeVdp carrying WT or L507A KLF4 were shown in scatter plots. Numbers (N) of the clones used in each condition are shown. Results are shown as Z-scores of all the samples.
Next, we generated human iPSCs with these vectors from human neonatal fibroblasts using SeVdp vector system in feeder-free conditions (Figure 2E). Comparable numbers of reprogrammed iPS-like colonies were observed based only on their morphologies (Figure 3F); however, the number and the ratio of NANOG-positive colonies were significantly higher in L507A condition (Figures 3F–3H). These results suggested that fully-reprogrammed iPSCs were generated more efficiently in the L507A condition. To evaluate the quality of iPSC clones generated with SeVdp vectors carrying KLF4 WT or L507A, the gene expression of NANOG, residual Sendai virus NP (Nucleocapsid Protein) RNA, and markers for differentiation-defective iPSCs (i.e., HERV-H and lincRNA-RoR) (Ohnuki et al., 2014) was examined by RT-qPCR (reverse transcription-quantitative polymerase chain reaction). RNA samples at first passage of more than 10 iPSC clones generated with WT or L507A conditions were collected. These iPSC clones generated with L507A mutant (n = 16) showed relatively homogeneous NANOG expression within a 2-fold range to a normal human iPSC line (Figure 3I). On the other hand, these iPSC clones generated with WT (n = 13) showed a huge variation in NANOG expression. Some of them showed below half of the NANOG expression than a normal human iPSC line. In addition, 6 out of 13 clones generated with WT, which carried higher NP RNA amounts at more than 10,000 times than a normal hiPSC line, showed aberrantly higher NANOG expression (p = 0.0029 by Fisher's exact test in comparison of 0 out of 17 clones generated with L507A). There were no iPSC clones generated with L507A which showed high expression of HERV-H and/or lincRNA-RoR although some iPSC clones generated with WT showed higher expression of them (Figure 3J). These results suggested that SeVdp vector carrying L507A mutant stably produced more homogeneous high-quality human iPSC clones without residual SeVdp vectors or the aberrantly high expression of differentiation-defective markers.
Screening all the natural amino acid residue variants at L507 position in reprogramming
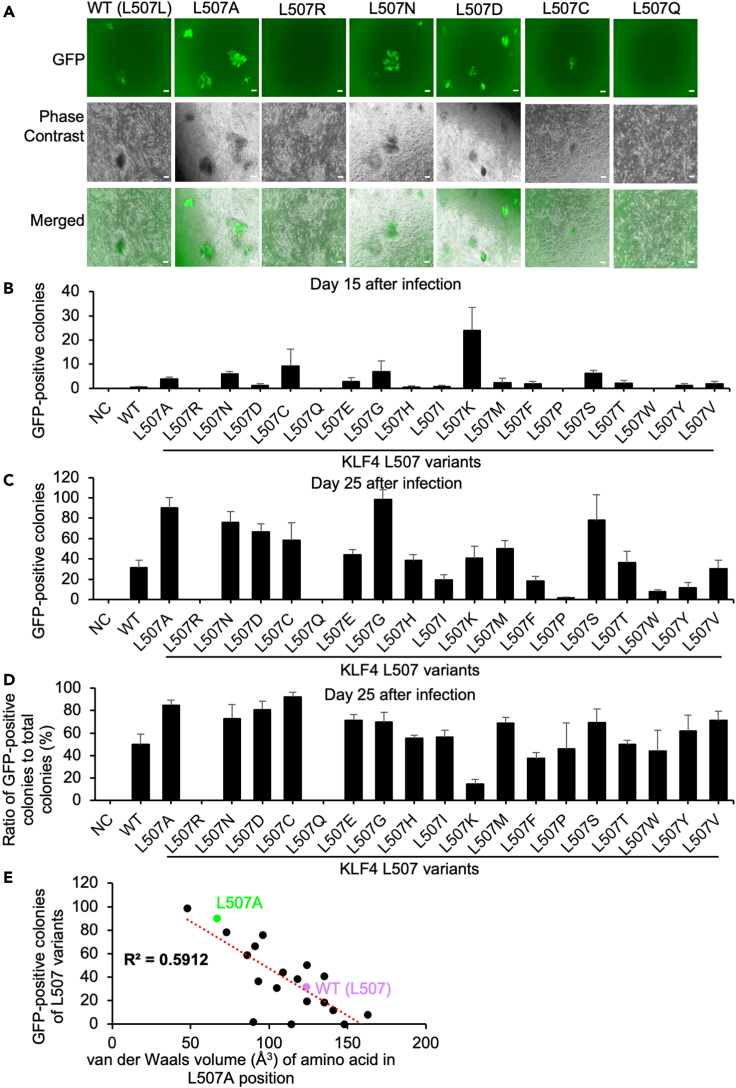
To determine how each type of amino acid residue in L507 position affected iPSC generation efficiency, we generated a series of retroviral vectors carrying every natural amino acid residue variant at L507 position. First, we checked the protein expression of every KLF4 L507 mutant. Although most of the variants were expressed comparably to WT, L507R had reduced expression (Figures S2A and S2B). Retroviral vectors carrying KLF4 L507 variants together with OCT4, SOX2, and MYCL1 were transduced to Nanog-GFP MEF to generate iPSCs (Takahashi and Yamanaka, 2006) (Figures 4A and S2C). On day 15 after transduction, more GFP-positive iPSC colonies were observed in the conditions of L507A, L507N, L507C, L507G, L507K, and L507S than in WT condition (Figure 4B). On day 25 after transduction, more GFP-positive iPSC colonies were found in L507A, L507N, L507D, L507C, L507G, and L507S than in WT (Figure 4C). No or few GFP-positive iPSC colonies were found in the conditions using L507R, L507Q, L507P, L507W, and L507Y. The ratio of Nanog-GFP-positive iPSC colonies to total colonies was also calculated on day 25 after transduction to evaluate the heterogeneity of iPSC generation. L507A, L507N, L507D, L507C, L507E, L507G, L507M, L507S, and L507V variants produced Nanog-GFP-positive colonies at a higher ratio than WT, whereas L507K variant produced Nanog-GFP-positive colonies at a lower ratio than WT (Figure 4D). These results indicated that L507A, L507N, L507D, L507C, L507G, and L507S variants improved both the efficiency and speed of iPSC reprogramming from MEF. Interestingly, a negative correlation (R2 = 0.5912) was identified between the numbers of Nanog-GFP-positive colonies of each KLF4 L507 variant on day 25 and the van der Waals volume of the amino acid residue of L507 position in each L507 variant (Figure 4E). These results indicated that KLF4 variants with smaller amino acid residues in the L507 position acquired higher reprogramming activity.
Figure 4.
The reprogramming efficiency of Nanog-GFP MEF using every KLF4 L507 amino acid residue variant
(A) Typical morphology of reprogrammed colonies Nanog-GFP MEF using retroviral vectors carrying every KLF4 L507 amino acid residue variant on day 25. Scale bars, 200 μm.
(B) The number of Nanog-GFP-positive iPSC colonies were counted on day 15 after infection of retroviral vectors to Nanog-GFP MEF from 10,000 cells. Results are mean and SE, n = 4.
(C) The number of Nanog-GFP-positive colonies was counted on day 25 after infection of retroviral vectors to Nanog-GFP MEF from 10,000 cells. Results are mean and SE, n = 4.
(D) The ratio of Nanog-GFP-positive colonies was calculated on day 25 after infection of retroviral vectors. The total number of Nanog-GFP-positives was counted from whole wells. Results are mean and SE, n = 4.
(E) Scatterplot shows a possible linear correlation between van der Waals volume (Å3) of the variants' amino acid residue and the mean numbers of Nanog-GFP-positive colonies on day 25. The red dot line and the value of R2 show the best-fitted trendline and the coefficient of determination (the square of the correlation coefficient), respectively.
Expression levels and protein stability of KLF4 L507A mutant are not causes of the effect on iPSC reprogramming
To examine the molecular mechanisms of the enhancing effect of KLF4 L507A on iPSC reprogramming, we first asked whether KLF4 L507A might hold different expression levels and/or stability from WT or not. Previous studies focused on characterizing other domains of KLF4 for its posttranslational modifications and protein stability in pluripotency and reprogramming (Dhaliwal et al., 2019; Kim et al., 2012, 2015; Reinhardt et al., 2020; Ye et al., 2018). KLF4 protein showed variable stability with a half-life of less than 2 h to more than 12 h depending on the different cellular status, which was regulated by protein-protein interaction, post-translational modification, and DNA-binding (Dhaliwal et al., 2019). Changes in KLF4 protein stability by the modulation of KLF4 N-terminal region regulated the efficiency of iPSC generation (Kim et al., 2015; Reinhardt et al., 2020). Thus, we examined the protein expression levels and stability of L507A in comparison with WT. First, L507A showed a similar protein expression level to WT transduced by retroviral vectors (Figures S1A and S1B). Next, we evaluated changes in protein stability by in silico analysis using an X-ray crystal structure of KLF4 ZnF and DNA complex. The L507A mutation was predicted to increase protein stability only slightly; however, some other mutations, which resulted in decreasing iPSC generation, were predicted to increase protein stability more (Figures S3A and S3B). Next, we evaluated the protein half-life of WT and L507A by cycloheximide chase assays. L507A protein showed similar stability to WT protein with a half-life of approximately 14.5 h (Figures S3C and S3D). Last, we evaluated the thermal stability of WT and L507A in the transduced cells by cellular thermal shift assays (Martinez Molina et al., 2013). KLF4 L507A protein showed similar thermal stability to WT protein (Figures S3E and S3F). Together, these results indicated that the expression levels, the protein lifetime, and the protein thermal stability of KLF4 L507A mutant were similar to those of WT. Thus, the change in iPSC reprogramming activity caused by L507A mutant should not be because of the change of its quantitative traits.
iPSCs generation with pro-reprogramming factors dissects independent or shared functions with KLF4 L507A
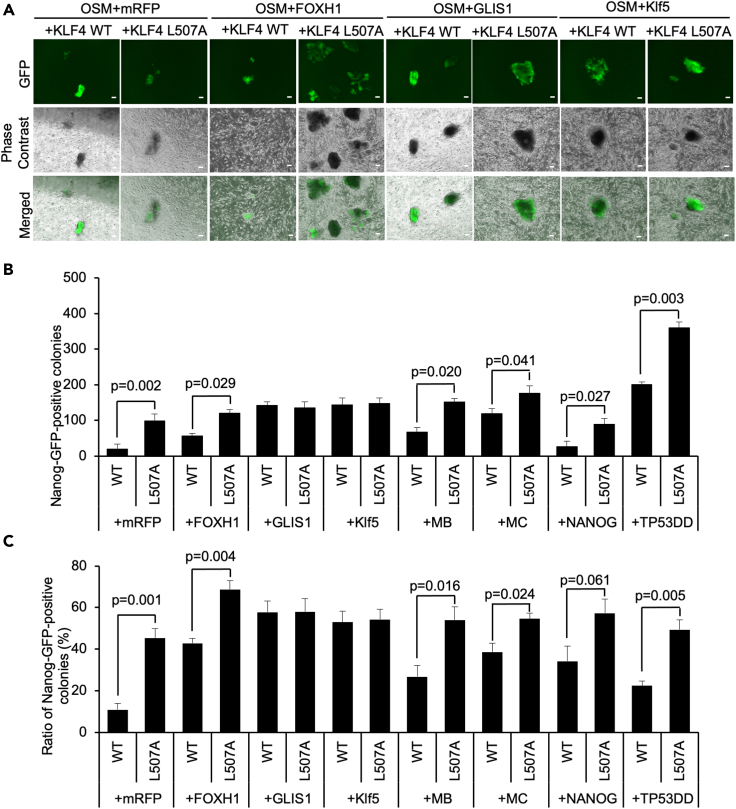
Then, we asked whether transcription traits of the KLF4 L507A mutant might be different from WT. First, to identify which known major pathways are related to enhanced reprogramming by KLF4 L507A, we used six putative pro-reprogramming factors capable of promoting iPSCs reprogramming when added to the conventional mixture of OSKM four reprogramming factors. Pro-reprogramming factors used in this assay were as follows: FOXH1 (Forkhead box H1) (Takahashi et al., 2014), GLIS1 (Glis Family Zinc Finger 1) (Maekawa et al., 2011; Worringer et al., 2014), Klf5 (Nakagawa et al., 2008), MB (microRNA 106a-363) (Liao et al., 2011), MC (microRNA Cluster 302–367) (Liao et al., 2011), NANOG (Yu et al., 2007), and TP53DD (p53 dominant-negative form) (Hong et al., 2009). In addition, mRFP1 (monomeric Red Fluorescent Protein 1) was used as a negative control (Hotta et al., 2009). We compared the efficiency of iPSCs generation between OSKM or OSK[L507A]M with each pro-reprogramming factor using the retroviral vector system in MEF. Overall, these pro-programming factors significantly increased the number and the ratio of Nanog-GFP-positive colonies in both WT and L507A conditions compared to mRFP conditions (Figures 5A–5C and S4). Only in the conditions that GLIS1 or Klf5 was added, there were no detectable changes between WT and L507A. These results suggested that the effect of L507A in reprogramming was epistatic to GLIS1 and Klf5 and independent from other pro-programming factors. Considering that Klf5 and GLIS1, which are both ZnF transcription factors, are identified as one of the factors which can be a substitute for KLF4 in iPSC reprogramming (Maekawa et al., 2011), these results suggested that the enhancing effect of L507A might be partially shared common functions and/or mechanisms with GLIS1 and Klf5.
Figure 5.
The reprogramming efficiency of Nanog-GFP MEF using retrovirus vector-based L507A mutants with pro-reprogramming factors
(A) Typical morphology of Nanog-GFP colonies in the reprogramming Nanog-GFP MEF on day 25 of reprogramming in KLF4 wildtype (WT) or L507A mutant with pro-programming factors. Scale bars, 200 μm.
(B) The number of Nanog-GFP colonies in the reprogramming Nanog-GFP MEF calculated on day 25 of reprogramming in KLF4 WT or L507A mutant with pro-programming factors. Results are mean and SE, n = 4. p values calculated with the t test were shown based on the comparison between WT and L507A KLF4 conditions.
(C) The ratio of Nanog-GFP colonies to the total colonies in the reprogramming Nanog-GFP MEF was calculated on day 25 of reprogramming in KLF4 WT or L507A mutant with pro-programming factors. Results are mean and SE, n = 4. p values calculated with the t test were shown based on the comparison between WT and L507A KLF4 conditions.
Distinctive features of DNA binding patterns of KLF4 L507A in reprogramming
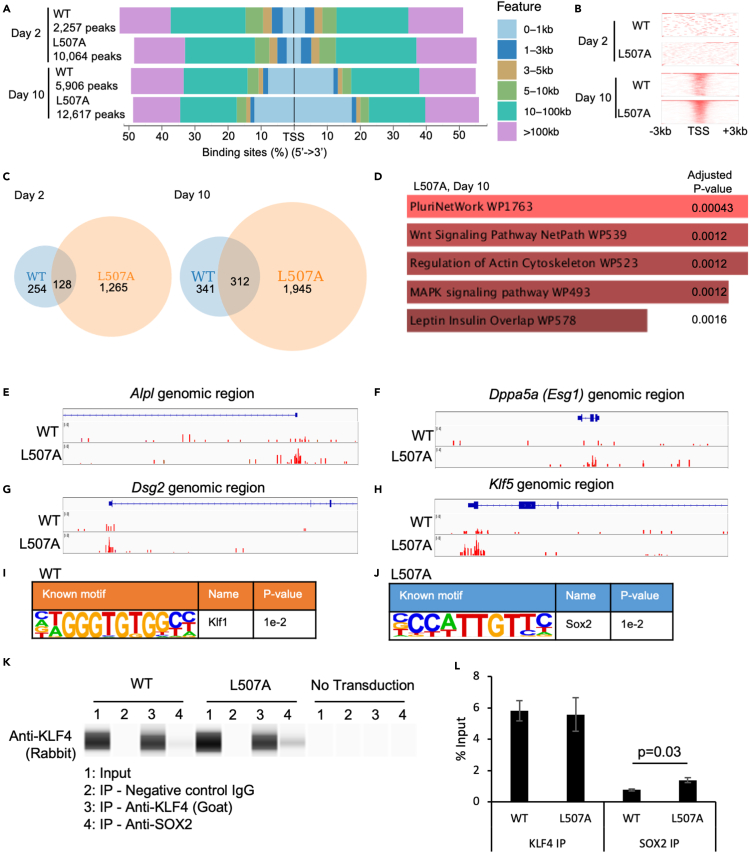
Next, we examined the genome-wide DNA binding patterns of KLF4 L507A by ChIP-Seq. To obtain clear DNA-binding patterns of KLF4 during reprogramming, we used rapid and efficient iPSC generation from primary MEF using SeVdp in feeder-free conditions (Figure 2A). On days 2 and 10 after transduction in this induction method, ChIP DNA samples were collected from reprogramming MEF transduced with SeVdp carrying L507A or WT. ChIP-Seq was performed using an anti-human KLF4 antibody. The numbers of total peaks in ChIP-seq data were 2,257 in WT 2d sample, 10,064 in L507A 2d sample, 5,906 in WT 10d sample, and 12,617 in L507A 10d sample (determined as FDR <0.05). Because these differences of the peaks were not reflected by the number of mapped reads (17 million in WT 2d sample, 8 million in L507A 2d sample, 18 million in WT 10d sample, and 18 million in L507A 10d sample), these results indicated that L507A had a higher affinity to specific regions of DNA/chromatin during reprogramming. The distribution of all peaks from these samples showed that binding sites were focused around TSS (transcription start site) more in day 10 samples than day 2 samples (Figures 6A and 6B). After extracting gene sets with specific binding peaks within 5 kilobase pairs from TSS from these ChIP-seq data, we compared these gene sets between KLF4 WT and L507A conditions (Figure 6C and Table S4). We found that 128 genes and 312 genes (33% and 48% of total genes in the WT sample) overlapped between the L507A and WT samples. To evaluate which biological pathways were involved in L507A-specific gene sets, pathway enrichment analysis was performed using the WikiPathway database (Kelder et al., 2009; Pico et al., 2008; Slenter et al., 2018). From a gene set composed of all the genes identified in the L507A sample, a category, “PuriNetWork (WP1763)”, was significantly ranked first (Figure 6D) although no pathways were significantly enriched from other conditions with the same analysis methods (Adjusted p values < 0.05); Figures S5A–S5C). From these pluripotency-related genes, mapped ChIP peaks around TSS of Alpl (Alkaline phosphatase), Dppa5a (Esg1), Dsg2 (Desmoglein-2), and Klf5, showed that L507A bound to the promoter or enhancer region of these genes more frequently than WT did (Figures 6E–6H). To identify what DNA sequence motifs are specifically bound by WT and L507A KLF4, motif discovery analysis was performed. Motif discovery program identified Klf1, a close family gene of Klf4, in the WT sample, confirming that WT sample preferentially bound to the consensus sequence of conventional KLF family proteins; however, Sox2, which is another reprogramming factor and pluripotency marker, was identified in the L507A sample (Figures 6I and 6J).
Figure 6.
KLF4 L507A mutant preferentially binds promoter/enhancer of pluripotency-related genes in reprogramming cells
(A) Heat maps of peak distribution and the numbers of peaks from the ChIP-seq data of KLF4.
(B) The distribution of ChIP-seq reads between -3kb and +3kb around TSS.
(C) A Venn diagram of gene sets obtained from the comparison of ChIP-seq data of KLF4 among reprogramming MEF induced by KLF4 WT or L507A on day 2 or 10. Genes that have specific ChIP-seq peaks within 5kb from TSS are categorized in each set. The numbers of the genes in each set are shown.
(D) Wikipathway enrichment analysis was performed on the gene set derived from the sample of L507A on day 10. The top 5 pathways that are enriched with statistically significant are illustrated with the adjusted p values.
(E–H) Mapped ChIP-seq peaks on pluripotency-related genes, Alpl (E), Dppa5 (F), Dsg2 (G), and Klf5 (H). These genes are extracted in the combination with RNA-seq data. Red bars indicate ChIP-seq peaks in each genomic region.
(I and J) Motif discovery analysis was performed on ChIP-seq data of KLF4 in reprogramming MEF induced by KLF4 WT or L507A. Discovered known motifs from WT (I) and L507A (J) were shown with the name of transcription factor and p value.
(K) A typical blot image of capillary Western blot assays on samples co-immunoprecipitated with anti-KLF4 antibody. Reprogramming MEFs infected with SeVdp carrying KLF4 WT or L507A or no transduction for 3 days were used.
(L) Protein abundance was calculated from the data of co-IP assays. Results are mean and SE, n = 3. The p value calculated with a t test was shown based on the comparison between WT and L507A KLF4 conditions.
Then we asked whether KLF4 L507A had stronger DNA binding activity to SOX2 protein than KLF4 WT did. By co-immunoprecipitation, SOX2 protein was more abundantly precipitated with KLF4 L507A protein than WT protein in the reprogramming cells (Figures 6L and 6K). These results suggested that KLF4 L507A protein existed as a complex with SOX2 protein more frequently and that KLF4 L507A might show apparently distinct patterns of DNA binding, which were closely related to the binding patterns of another reprogramming factor, SOX2, in iPSC generation.
Distinctive features of transcriptional changes driven by KLF4 L507A in reprogramming
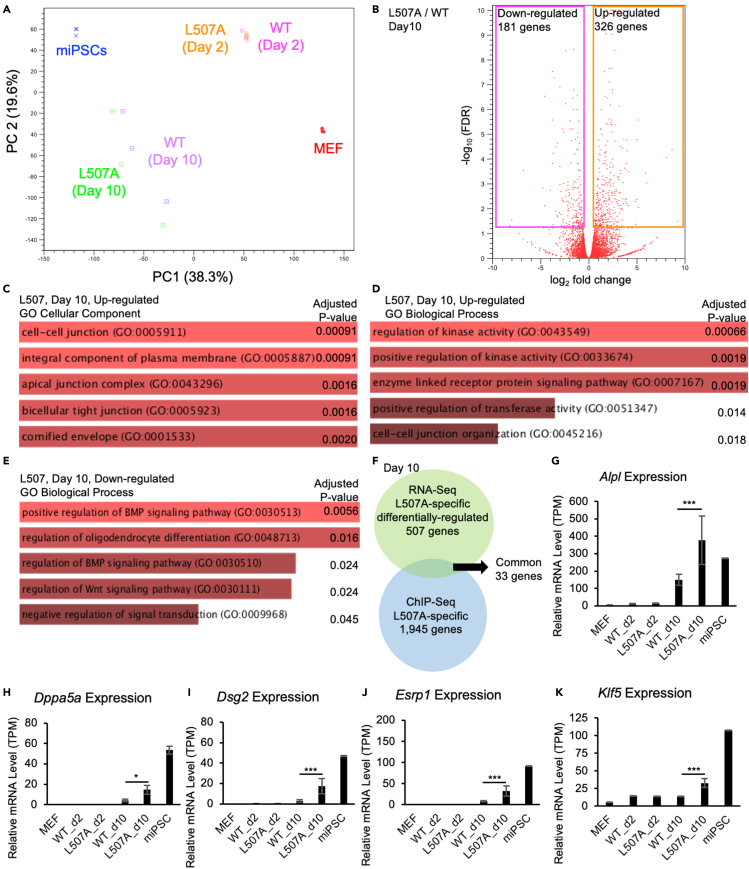
To evaluate global gene expression pattern changes between KLF4 L507A and WT during reprogramming, RNA-Seq was performed on the samples collected from the same reprogramming conditions as ChIP-Seq. Original MEF and resulting miPSCs were also used as samples. To visualize global gene expression patterns among these samples, we performed PCA (principal component analysis) (Figure 7A). KLF4 L507A and WT samples on day 2 were situated closely in the two-dimensional map composed of two major principal components, and there were no genes which significantly changed their expression levels. These results suggested that L507A had no or little specific effects on gene expression patterns at the initial phase of reprogramming. Among day 10 samples, L507A samples in each experimental time were situated slightly closer to miPSCs on the PC1 axis. In the comparison of Day 10 samples between WT and L507A, 507 genes, of which 181 genes were downregulated and 326 genes were upregulated (Figure 7B). To evaluate which biological pathways were involved in these L507A-specific gene sets, gene ontology (GO) enrichment analysis was performed. From a gene set composed of up-regulated genes in L507A samples, “cell-cell junction” and “regulation of kinase activity” were ranked first as GO cellular component and GO biological process, respectively (Figures 7C and 7D). In addition, from a gene set composed of down-regulated genes in L507A samples, “positive regulation of BMP signaling pathway” was ranked first as GO biological process, and no significant GO terms in GO cellular components (Figure 7E). Of note, in the “cell-cell junction” category, epithelial marker genes, such as Cdh1, Dsg2, and CLDN4, were included. These results suggested that L507A specifically enhanced mesenchymal-to-epithelial transition during reprogramming. To identify direct target genes which were specifically regulated by KLF4 L507A, we combined the gene sets from ChIP-Seq and RNA-Seq data. Thirty-three genes were overlapped, and Alpl, Dppa5a, Dsg2, Esrp1, and Klf5 were included in the overlapped genes (Figure 7F). The expression levels of these genes were low or undetectable in MEF and were gradually up-regulated toward miPSCs (Figures 7G–7K). Because we have shown that KLF4 L507A was functionally epistatic to the addition of Klf5 in iPSC generation (Figures 5B and 5C), Klf5 should be one of the direct and specific target genes of L507A during reprogramming. These results suggested that L507A mutant might enhance iPSC generation partly through amplifying Klf activity as a whole.
Figure 7.
Global expression similarity and differences between reprogramming cells between KLF4 WT and L507A
(A) PCA of global gene expression in MEF, miPSCs, and reprogramming MEF induced by KLF4 WT or L507A on day 2 or 10 after transduction of each SeVdp vector.
(B) A volcano plot shows log2 fold change as X axis and -log10 (FDR) as Y axis. All the genes were plotted in red circles. Significant genes with FDR<0.05 were rectangularly surrounded with pink (downregulated) and orange (upregulated).
(C-E) GO (Gene Ontology) enrichment analysis was performed on the gene sets, composed of the genes which differentially expressed between WT and L507A conditions on day 10 samples. GO Cellular Component enriched from upregulated genes in L507A samples were shown in (C); GO Biological Process enriched from upregulated genes in L507A samples were shown in (D); GO Biological Process enriched from down-regulated genes in L507A samples were shown in (E). The top 5 GO terms that are enriched with statistically significant are illustrated with adjusted p values.
(F) A Venn diagram of gene sets obtained from specifically regulated by L507A as shown above in RNA-seq data and ChIP-seq data. 33 genes, including Klf5, were extracted in common.
(G–K) As examples of the genes which are commonly identified in RNA-seq and ChIP-seq experiments in this study, the expression levels (i.e., TPM (Transcripts Per Million)) of Alpl, Dppa5a, Dsg2, Esrp1, and Klf5, are shown from RNA-seq data. Results are mean and SE, n = 3. p values calculated with FDR correction were shown based on the comparison between WT and L507A KLF4 conditions on day 10 samples.
L507A mutant was predicted to acquire a unique conformation of protein-DNA complex with additional DNA interaction
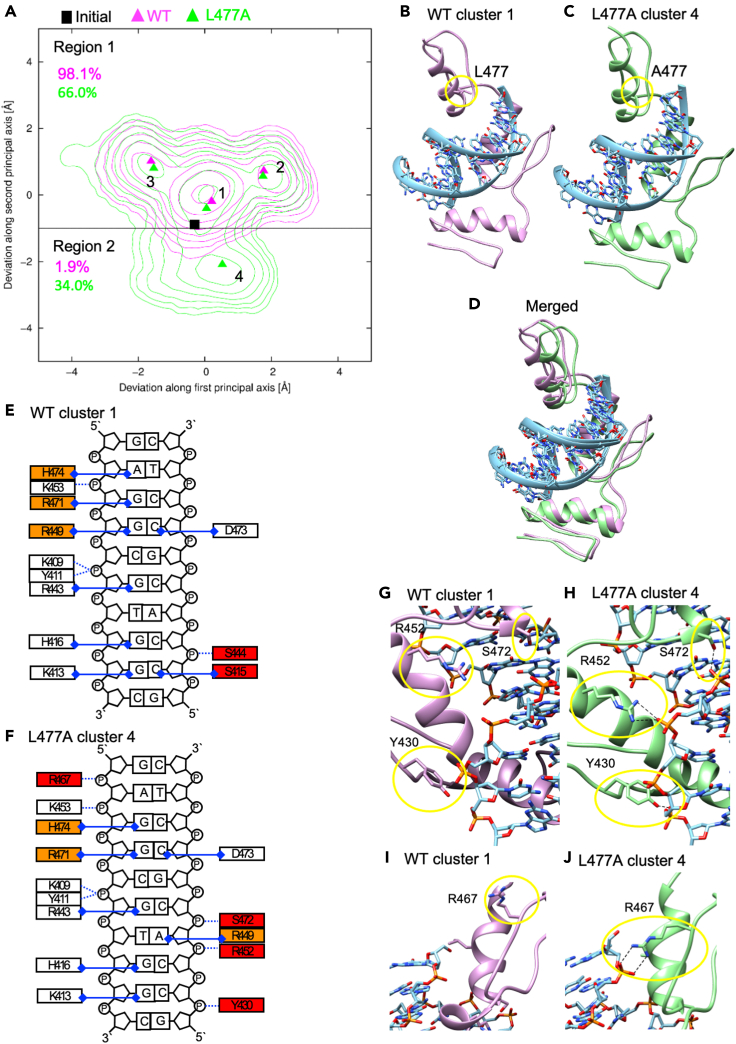
So far, we have revealed the functional changes in the reprogramming ability induced by the KLF4 L507A mutant. To examine the structural basis of these functional changes, we performed MD simulation analysis. The coordinates of the X-ray crystal structure of mouse Klf4 (Schuetz et al., 2011) were obtained from the PDB database (PDB ID: 2WBU). The structure of the Klf4 L477A (identical to human L507A) mutant was generated by removing the Cγ and Cδ atoms of L477. 2-μs MD simulations were performed three times for both the Klf4(WT)-DNA and Klf4(L477A)-DNA complexes in aqueous environments with different initial velocities. The complex structures were stably maintained during the MD simulations in both systems. The root-mean-square deviations (RMSDs) from the initial structures calculated after aligning the protein Cα atoms of the MD structures to those of the initial structure were 1.71 ± 0.30 Å and 2.78 ± 0.67 Å for the protein Cα atoms and the DNA P atoms of the Klf4(WT)-DNA complex and 1.65 ± 0.35 Å and 2.56 ± 0.55 Å for the protein Cα atoms and the DNA P atoms of the Klf4(L477A)-DNA complex. PCA was performed on the MD trajectories to visualize the conformational distributions of the complexes. The conformational distribution of the Klf4(WT)-DNA complex has three peaks and is well overlapped with that of the Klf4(L477A)-DNA complex (Figure 8A). In contrast, the conformational distribution of the Klf4(L477A)-DNA complex has an additional peak, which represents a conformation unique to the Klf4(L477A)-DNA complex. With the same methods of MD simulations on Klf4(L477R), Klf4(L477W), or Klf4(L477Y), which showed less or no iPSC generation activity (Figures 5A–5C), this additional peak was not be generated (Figures S6A–S6C). The population of the unique conformation (in the region below the black line; region 2) was about 33%. In this unique conformation, L477A triggered entire structural changes extended to three ZnFs (Figures 8B–8D). The mode of protein-DNA interaction was also changed (Figures 5E and F). Specifically, R449, R471, and H474 changed their DNA target base. Although S415 and S444 lost or decreased their interaction to DNA in the unique conformation, Y430, R452, R467, and S472 formed additional hydrogen bonds to the phosphate groups of DNA (Figures 8G–8J). These additional hydrogen bonds were less observed in the MD simulations on Klf4(L477R), Klf4(L477W), or Klf4(L477Y) (Figure S6D). These results suggest that the KLF4 L507A mutant should acquire an additional structural conformation that is stabilized by additional hydrogen bonds.
Figure 8.
MD simulation analysis of the effect of the mouse Klf4 L477A mutation (identical to human KLF4 L507A) on the structure of the Klf4 ZnF-DNA complex
(A) Conformational distribution of the mouse Klf4 (WT)-DNA complex (magenta contours) and the Klf4(L477A)-DNA complex (green contours). The level of the n-th contour is 2n−1 × 10−4. The positions of the initial structure and the representative structures obtained from the cluster analysis of the MD trajectories of the Klf4(WT)-DNA and the Klf4(L477A)-DNA complexes are indicated by a black rectangle, magenta triangles, green triangles, respectively. The black line separates conventional conformation distribution, Region 1, and unique conformation distribution of the Klf4(L477A)-DNA complex as Region 2.
(B–D) Comparison of the representative structure from cluster 1 of the Klf4(WT)-DNA complex (B) and that from cluster 4 of the Klf4(L477A)-DNA complex (C). A superposed view is also shown (D). L477 or A477 are highlighted in yellow circles.
(E–F) Schematic representation of protein-DNA interactions of WT cluster 1(E) and L477A cluster 4 (F). The interaction between an amino acid residue and a DNA base or DNA phosphate group is shown in a blue dotted line or a blue solid line with rhombus, respectively. Amino acid residues highlighted in tangerine and red indicate the ones that changed target DNA bases and that formed interactions to DNA in either WT cluster 1 or L477A cluster 4, respectively. Hydrogen-bond occupancy was calculated for each possible atom pair between the protein and the DNA of each cluster. The occupancies of the atom pairs within a pair of a residue and the base or the phosphate group of a nucleotide were summed. The pairs with the sum of the occupancies >0.7 are shown.
(G–J) Conformational changes and additional hydrogen bonds to phosphate groups of DNA in the representative structure from cluster 4 of the Klf4(L477A)-DNA complex. Y430, R452, R467, and S472 are highlighted in yellow circles. All the molecular graphic images were produced using UCSF Chimera (Pettersen et al., 2004).
Discussion
KLF4 L507A variant accelerates and stabilizes reprogramming to pluripotency
In this study, we identified that KLF4 L507A variant, which was discovered by alanine scanning mutagenesis methods on DNA-interacting amino acid residues in the KLF4 ZnF domain, improved iPSC generation. This KLF4 variant increased the number and the speed of iPSC generation in mouse and human somatic cells and enriched the fully-reprogrammed iPSCs among the transduced cell population. The benefits of using this variant in iPSC generation for regenerative medicine should be shortening the iPSC generation time and decreasing the required iPSC clones to be selected. While maintaining KLF4 function as a reprogramming factor, L507A variant additionally showed distinct DNA-binding patterns and downstream gene regulation from WT. Especially, we identified that KLF5 may be one of the direct transcriptional targets of KLF4 L507A specifically by both ChIP-Seq and RNA-Seq analysis. As KLF5 has similar functions in pluripotency and reprogramming of pluripotent stem cells (Aksoy et al., 2014; Azami et al., 2018; Nakagawa et al., 2008; Parisi et al., 2010; Yamane et al., 2018), KLF4 L507A may amplify KLF activity as a whole during reprogramming. In addition, because the effects of L507A on DNA-binding pattern and transcriptome are evident in Day 10, but not in Day 2 samples of SeVdp reprogramming, L507A may promote reprogramming in the transition from partially-reprogrammed cells to fully-reprogrammed cells by up-regulating Dppa5a, Dsg2, Esrp1, and Klf5 transcription directly. This notion is supported by the previous findings that higher expression of Dppa5a in reprogramming cells results in fully-reprogrammed fate (Guo et al., 2019; Qian et al., 2016).
We also found that the enhancing effect of KLF4 L507A on iPSC reprogramming was partially masked by the additional GLIS1. GLIS1 was identified as one of the transcription factors which can induce Nanog-GFP-positive iPSCs without KLF4 although the efficiency was much lower than KLF4 (Maekawa et al., 2011). GLIS1 also promoted iPSC generation together with other reprogramming factors as retroviral vectors or synthetic replicative RNA formats (Lee et al., 2017; Wang et al., 2019; Yoshioka et al., 2013). Like KLF4, GLIS1 is a Kruppel-like C2H2 ZnF transcription factor (Kim et al., 2002) and should not be a downstream factor of KLF4 L507A variant as the expression of GLIS1 was low in reprogramming MEF induced by either WT or L507A variant KLF4 as well as in mESCs and miPSCs (Maekawa et al., 2011). KLF4 L507A variant shares the functional effects with GLIS1 in reprogramming a part at least, thereby may render GLIS1 unnecessary to be added as a pro-reprogramming factor.
KLF4 L507 position represents a “molecular bump” to regulate structural stability of the interaction between ZnF domain and DNA complex
In iPSC generation experiments with all the amino acid residue variants of L507, we found that smaller amino acid residues tended to have higher iPSC reprogramming activity. In addition, in MD simulation analysis, L507A mutant is predicted to acquire a unique conformation of DNA-binding complex, which had additional hydrogen bonds between three other amino acid residues and phosphate groups of DNA. These results suggest that the L507 position represents a “molecular bump” in the KLF4 ZnF domain. Natural L507 seems to be tuned for mild interaction in this position. Our L507A and L507G variants may be tuned strongly for accomplishing herculean reprogramming tasks. Because L507 is conserved among three KLF family homologous members, we assume that the equivalents of L507 mutants in other KLF family genes possess improved reprogramming activity. It might be interesting to test how the same mutation will affect other KLF members in reprogramming and other biological contexts. This does not necessarily mean that this mutant simply might have a higher DNA-binding affinity overall, because we observed different DNA-binding patterns between L507A and WT. It might be interesting to test how this mutant could affect DNA binding affinity using biochemical analysis.
Coincidently, our previous study identified that an alanine-substituted mutant from leucine, L122A, in NANOG DNA-binding homeodomain showed enhanced properties to promote self-renewal and reprogramming of naive pluripotent stem cells (Hayashi et al., 2015). Although ZnF domain and homeodomain do not share the structural basis in DNA recognition and interaction, there might be some common features of these leucine residues in these DNA-binding domains.
Identification of critical amino acid residues of KLF4 ZnF domain in reprogramming
In addition, our results of alanine scanning experiments in the KLF4 ZnF domain identified functionally important residues in iPSC reprogramming. Specifically, K439, Y441, R473, E476, and D503 seemed critical because their alanine-substituted mutants were defective in iPSC reprogramming assays although these mutants were considerably expressed. These residues were reported to form critical interactions and/or to be located in the important places in the ZnF-DNA protein structure (Hashimoto et al., 2016; Liu et al., 2014; Schuetz et al., 2011). Thus, our study demonstrated the functional meaning of KLF4 ZnF domain and its DNA-binding interaction with regard to its pluripotency induction.
In summary, we have demonstrated the first generation of KLF4 ZnF variants with enhanced reprogramming activity. Only one amino acid modulation in a reprogramming factor can markedly improve natural transcription factor function to achieve faster and effective reprogramming, leading to cost-effective advantages in future scientific and medical applications. Moreover, as the protein bioengineering technology continues to proceed as ZnF proteins are used as versatile tools to bind any desired DNA sequences (Pabo et al., 2001), our study shares new insights regarding molecular mechanisms of transcription factors' DNA-binding with possible implications in designing highly engineered factors.
Limitations of the study
Our study aimed to focus on the improvement of reprogramming activity using KLF4 ZnF mutants. Although we determined the effects of KLF4 ZnF mutants on iPSC reprogramming in this study, we cannot assure that these effects can be reproducible in other biological contexts. Because KLF4 has pleiotropic effects in a context-dependent manner, it might be interesting to investigate the function and the effect of these variants in other biological contexts in the future. It was especially difficult to express some of the mutants as we have seen in Figures S1 and S2. There are two possible reasons for the reduction. One may be a biological reason that mutant proteins had defects in their expression and stability. Another may be a technical reason that the vectors of the mutants had problems. Although we verified the sequence of the whole length of the KLF4 mutants, we could not rule out that these vectors of the mutant had defects in the sequence outside KLF4. In addition, although we have addressed the positive effects of the L507A on iPSC generation in this study, there might be potential negative side effects of this mutant because of a shifting in binding affinities and target sequences. We could develop better mutants that avoid such negative side effects by aiming to prolong protein half-life and/or improve expression levels in the future.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat Anti-Human KLF4 affinity-purified polyclonal antibody | R&D | Cat#AF3640, RRID:AB_2130224 |
| RNA pol II mouse monoclonal antibody | Active Motif | Cat#39097, RRID:AB_2732926 |
| Alexa Fluor 488 Mouse anti-Human TRA-1-60 Antigen | BD Pharmingen | Cat#560173, RRID:AB_1645379 |
| Human TRA-1-85/CD147 APC-conjugated Antibody | R&D | Cat#FAB3195A, RRID:AB_663789 |
| Monoclonal ANTI-FLAG M2 antibody produced in mouse | Sigma | Cat#F1804, RRID:AB_262044 |
| Mouse Anti-Human/Mouse/Rat GAPDH/G3PDH Monoclonal Antibody | R&D | Cat#MAB5718, RRID:AB_10892505 |
| Anti-KLF4 polyclonal Rabbit Ig Antibodies | MBL | Cat#PM057, RRID:AB_10598180 |
| Ubiquitin (P4D1) Mouse mAb | Cell Signaling Technology | Cat#3936, RRID:AB_331292 |
| Anti-SOX2 antibody, Polyclonal Goat IgG | R&D systems | Cat#AF2018, RRID:AB_355110 |
| Normal Goat IgG Control | R&D systems | Cat#AB-108-C, RRID:AB_354267 |
| Chemicals, peptides, and recombinant proteins | ||
| ChIP-IT Protein G Magnetic Beads | Active Motif | Cat#53014 |
| Polybrene Solution | Nacalai Tesque | Cat#12996-81 |
| Blasticidin S Hydrochloride | Wako | Cat#029-18701 |
| FuGENE 6 Transfection Reagent | Promega | Cat#E2692 |
| Shield1 | Takara Bio | Cat#632189 |
| Fibroblast Growth Factor (basic) | Wako | Cat#060-04543 |
| CultureSure Y-27632 | Wako | Cat#034-24024 |
| iMatrix-511, silk (Laminin-511) | Nippi | Cat# 892021 |
| STEM-CELLBANKER GMP Grade | Takara Bio | Cat# CB045 |
| FuGENE6 Transfection Reagent | Promega | Cat#E2691 |
| SDS-PAGE Sample Buffer Solution without 2-ME(2x) for SDS-PAGE | Nacalai Tesque | Cat#30567-12 |
| 1mol/l-Dithiothreitol Solution (DTT) | Nacalai Tesque | Cat#14130-41 |
| Critical commercial assays | ||
| PrimeSTAR Mutagenesis Basal Kit | Takara Bio | Cat#R046A |
| ChIP-IT Express | Active Motif | Cat# 53008 |
| Chromatin IP DNA Purification Kit | Active Motif | Cat# 58002 |
| ChIP-IT Control qPCR Kit – Mouse | Active Motif | Cat# 53027 |
| Universal Magnetic Co-IP Kit | Active Motif | Cat# 54002 |
| Anti-Mouse Detection Module for Jess, Wes, Peggy Sue or Sally Sue | ProteinSimple | Cat#DM-002 |
| Monarch Total RNA Miniprep Kit | New England BioLabs | Cat#T2010S |
| FastGene Premium RNA extractioni Kit | Nippon Genetics Co. Ltd. | Cat#FG-81250 |
| Retrovirus Titer Set (for Real Time PCR) | Takara Bio | Cat#6166 |
| One Step TB Green PrimeScript RT-PCR Kit (Perfect Real Time) | Takara Bio | Cat#RR066A |
| Retro-X Concentrator | Takara Bio | Cat#Z1455N |
| Recombinant DNase I (RNase-free) | Takara Bio | Cat#2270A |
| Recombinant RNase Inhibitor | Takara Bio | Cat#2313A |
| 12-230 kDa Jess or Wes Separation Module, 8 x 25 capillary cartridges | Protein Simple | Cat#SM-W004 |
| Anti-Mouse Detection Module for Jess, Wes, Peggy Sue or Sally Sue | Protein Simple | Cat#DM-002 |
| Anti-Goat Detection Module for Jess, Wes, Peggy Sue or Sally Sue | Protein Simple | Cat#DM-006 |
| Deposited data | ||
| Crystal Data (KLF4 zinc finger) | www.rcsb.org | PDB ID: 2WBU |
| Crystal Data (Klf4 zinc finger) | www.rcsb.org | PDB ID: 4M9E |
| Raw and analyzed data of ChIP-seq | This paper | GEO: GSE157762 |
| Raw and analyzed data of RNA-seq | This paper | GEO: GSE184449 |
| Experimental models: Cell lines | ||
| Mouse embryonic fibroblasts isolated from Tg(Nanog-GFP,Puro)1Yam mice featuring transgenic GFP driven by Nanog promoter | This study | NA |
| Platinum-E (Plat-E) Retroviral Packaging Cell Line | Cell Biolabs, Inc. | Cat#RV-101 |
| Platinum-GP (Plat-GP) Retroviral Packaging Cell Line | Cell Biolabs, Inc. | Cat#RV-103 |
| Normal human skin fibroblast (NB1RGB) | RIKEN BRC Cell Bank | Cat#RCB0222 |
| SL10 feeder cells | Reprocell | Cat#RCHEFC001 |
| Experimental models: Organisms/strains | ||
| Mouse: STOCK Tg(Nanog-GFP,Puro)1Yam | RIKEN BRC (Okita et al., 2007) | RBRC 02290 |
| Oligonucleotides | ||
| Primers for plasmid construction, see Table S2 (KLF4 alanine mutants) | This paper | |
| Recombinant DNA | ||
| Plasmids made in this study, see Table S3 | This paper | |
| pMXs-hKLF4 | Laboratory of Plath (Lowry et al., 2008) | Addgene plasmid Cat#17967 |
| pMXs-hOCT3/4 | Laboratory of S. Yamanaka (Takahashi et al., 2007) | Addgene plasmid Cat#17217 |
| pMXs-hSOX2 | Laboratory of S. Yamanaka (Takahashi et al., 2007) | Addgene plasmid Cat#17218 |
| pMXs-Hu-L-Myc | Laboratory of S. Yamanaka (Nakagawa et al., 2008) | Addgene plasmid #26022 |
| pMXs-mRFP1 | Laboratory of J. Ellis (Hotta et al., 2009) | Addgene plasmid Cat#21315 |
| pMXs-p53DD | Laboratory of S. Yamanaka (Hong et al., 2009) | Addgene plasmid Cat#22729 |
| pCMV-VSV-G | Laboratory of H. Miyoshi, RIKEN BRC DNA Bank | Cat#RDB04392 |
| pMXs-hFOXH1 | Laboratory of S. Yamanaka (Takahashi et al., 2014) | Addgene plasmid Cat#59183 |
| pMXs-GLIS1 | Laboratory of S. Yamanaka (Maekawa et al., 2011; Worringer et al., 2014) | Addgene plasmid Cat#30166 |
| pMXs-ms-Klf5 | Laboratory of S. Yamanaka (Nakagawa et al., 2008) | Addgene plasmid Cat#50787 |
| pMXs-MC | Laboratory of D. Pei (Liao et al., 2011) | Addgene plasmid Cat#31704 |
| pMXs-MB | Laboratory of D. Pei (Liao et al., 2011) | Addgene plasmid Cat#31703 |
| pMXs-hNANOG (Plath) | Laboratory of K. Plath (Lowry et al., 2008) | Addgene plasmid Cat#18115 |
| SeVdp(fKgOSMaB) | This paper | NA |
| SeVdp(fK[507]gOSMaB) | This paper | NA |
| Software and algorithms | ||
| Bowtie2 | (Langmead and Salzberg, 2012) | http://bowtie-bio.sourceforge.net/index.shtml |
| MACS2 | (Zhang et al., 2008) | https://github.com/taoliu/MACS |
| CLC genomics workbench | Qiagen | https://digitalinsights.qiagen.com/ja/qiagen-clc-genomics-workbench/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yohei Hayashi (yohei.hayashi@riken.jp).
Materials availability
Plasmids generated in this study have been deposited to RIKEN BRC DNA bank. The details of the plasmids were in Table S3.
Experimental model and subject details
RIKEN Animal Research Committee has approved our animal research protocols. Nanog-GFP transgenic mice (i.e., Tg(Nanog-GFP,Puro)1Yam strain, RBRC 02290 in RIKEN BioResource Center) (Okita et al., 2007) was crossed to derive embryos. Male embryos on d.p.c.13.5 were isolated to derive mouse embryonic fibroblasts (MEFs). After genotyping by PCR on genomic DNA samples, MEFs harboring the Nanog-GFP construct were used for reprogramming experiments, ChIP- seq, Co-IP, and RNA-seq assays.
Method details
Plasmid construction
For generating human KLF4 mutant library, pMXs-hKLF4 (Addgene plasmid #17967) (Kitamura et al., 2003) was used as a template in site-directed mutagenesis to create pMXs-3XFLAG-KLF4-XXX (XXX indicating wild-type residue-polypeptide position-substituted residue). A 3XFLAG tag was fused to the N-terminus for detecting expressed mutant protein prior to mutagenesis. KLF4 alanine mutants were generated on pMXs-3XFLAG-hKLF4 using PrimeSTAR Mutagenesis Basal Kit (Takara) with PrimeSTAR Max DNA Polymerase following manufacturer protocols on PCR Thermal cycler (Applied Biosystems SimpliAmp). Specific primers were designed with 15 bp overlapping region containing substitution site. 3-step PCR Cycle conditions are 98°C for 10 s, 55°C (annealing step) for 15 s, 72°C (elongation step) for 30 s. Verification of the mutations was done by sequencing using two primers pMX-s1811 (FW) and pMX-as3205 (RV). All primers used for generating alanine mutants are listed in Table S2.
Primer sequences for adding N-terminal 3XFLAG tag are
(FW) 5-CATGACATCGACTACAAGGATGACGATGACAAGGCTGTCA GCGACGCGCTGCTCCCATCT-3′ and (RV) 5-ATCTTTATAATCACCGTCATGGTCTTTGTAGTCCATGAAT TCCCGTACCACCACACTGGG-3′. All the plasmid vectors generated in this study have been deposited in RIKEN DNA Bank (see Table S3).
To construct cDNA for SeVdp(fKOSMaB), we amplified the sequence of 2A peptide and blasticidin-resistant gene by PCR and inserted them after the c-Myc gene in SeVdp(fK-OSM) (Nishimura et al., 2014) using In-Fusion HD Cloning Kit (TaKaRa Bio). Similar mutagenesis as described above was applied to a construction of cDNA for SeVdp(fK[L507A]gOSMaB) from SeVdp(fKOSMaB).
Cell culture conditions and reprogramming experiments
Plat-E cells, Plat-GP cells, Nanog-GFP mouse embryonic fibroblasts, SL10 feeder cells (Reprocell), and NB1RGB human fibroblast line (Riken Cell Bank, RCB0222) were maintained in DMEM (4.5 g/L Glucose) with L-Gln and Sodium Pyruvate (Nacalai Tesque), supplemented with 10% fetal bovine serum (Biosera Cat#FB-1003/500), 1x penicillin/streptomycin (Nacalai Tesque). Mouse iPSCs were maintained in StemSure DMEM (High Glucose) with Phenol Red and Sodium Pyruvate (Wako), supplemented with 15% fetal bovine serum (Biosera), 1% StemSure 50 mmol/L monothioglycerol solution (Wako), 1x penicillin/streptomycin (Nacalai Tesque), L-Alanyl-L-Glutamine solution (Nacalai Tesque), 1% MEM non-essential amino acids (Nacalai Tesque), and 1,000 units of leukemia inhibitory factor (LIF) (Wako) from reprogramming day 8 on SL10 feeder cells. Experiments under feeder-free conditions (ChIP-seq and RNA-seq experiments) were performed using conditioned knockout serum replacement (KSR)-based mouse embryonic stem cell medium, formulated with StemSure DMEM (High Glucose) with Phenol Red and Sodium Pyruvate supplemented with 15% Stemsure Serum Replacement (Wako), 1% StemSure 50 mM monothioglycerol solution (Wako), 1x penicillin/streptomycin solution (Nacalai Tesque), L-Alanyl-L-Glutamine solution (Nacalai Tesque), 1% MEM non-essential amino acids (Nacalai Tesque), and 1,000 units/ml of leukemia inhibitory factor (LIF) (Wako). After conditioning by culturing SL10 feeder cells overnight, the medium was collected with filtering through a PTFE 0.45 μm syringe filter (AS ONE)
Retrovirus-based generation of iPSCs from Nanog-GFP mouse embryonic fibroblasts
Mouse iPSCs were generated from MEFs isolated from transgenic mice carrying the Nanog-GFP-IRES-Puro reporter construct (Okita et al., 2007) (provided by the RIKEN BioResource Research Center) (Nanog-GFP MEFs) by infection with an indicated retrovirus medium for 24 h supplemented with polybrene 4 μg/mL (Nacalai Tesque).
For retroviral production, Plat-E cells (Cell Biolabs) were transfected with the indicated retroviral vectors using Opti-MEM (Thermofisher Scientific) and FuGENE 6 (Promega). The viral supernatants were harvested after 48 h and 72 h after and filtered through PTFE 0.45 μm syringe filter (AS ONE). Reprogramming was performed according to a published procedure (Nakagawa et al., 2010) with minor changes. Briefly, retroviral supernatants containing the four reprogramming factors (OCT4, SOX2, KLF4 WT or a mutant, and L-MYC; OSKM) were mixed before infection at equal ratio. Nanog-GFP MEFs (10,000 cells) at early passages were prepared in a well of 6-well plate for the infection. Infected cells (10,000 cells) were reseeded to SL10 feeder cells (300,000 cells) on day 7 after transduction and maintained for 25 days. Nanog-GFP iPSC colonies were counted on day 15 or 25 after transduction using fluorescence microscope (BZ-X800, Keyence). For iPSC generation with pro-reprogramming factors, Nanog-GFP MEFs were infected with equal ratios of OSKM and indicated pro-reprogramming factor retroviral supernatants.
Retrovirus-based generation of iPSCs from normal human fibroblasts
Transduction of retroviruses into human fibroblasts was performed using Plat-GP cells with pCMV-vesicular stomatitis virus(VSV)-fusiogenic envelope G glycoprotein(G) vector. Viral supernatants were made by the transfection into Plat-GP cells using Opti-MEM (Thermo Fisher Scientific) and FuGENE 6 (Promega) with pMX vectors of OCT4 (#17217), SOX2 (#17218), L-MYC (#26022), and KLF4 or KLF4 mutants in combination with pCMV-VSV-G viral envelope vector at 3:1 ratio. The cells were infected with an indicated retrovirus medium for 24 h supplemented with polybrene 4 μg/mL (Nacalai Tesque). The medium was changed to the normal fibroblast medium described above next day and every other day. On day 7, the cells were collected and analyzed.
SeVdp-based generation of iPSCs from Nanog-GFP mouse embryonic fibroblasts
SeVdp vectors were produced as described previously (Nishimura et al., 2011). Mouse iPSCs were generated from Nanog-GFP MEFs (100,000 cells) by infection with an indicated SeVdp vectors (Nishimura et al., 2014) (MOI = 5) at 32°C for 24 h in a well of 12-well plate. On day 1, the medium was changed to normal MEF medium. On day 3, the medium was changed to conditioned medium with SL10 cells in feeder-free conditions. Then, the medium was changed every other day. The cells were cultured in the presence of 100 nM Shield1 (Takara Bio) with 1 μg/mL Blasticidin S (Wako) on days 3–7 after infection for selection of retroviral vector-infected cells and collected on specific days for the downstream analyses.
SeVdp-based generation of iPSCs from normal human fibroblasts
Human iPSCs were generated from normal human skin fibroblasts (NB1RGB) (100,000 cells) by infection with the indicated SeVdp vectors (MOI = 5) at 32°C for 24 h in a well of 12-well plate. The infected cells were grown in SL10-conditioned medium formulated with Primate ES cell medium (Reprocell), 1x penicillin/streptomycin (Nacalai Tesque), 10 ng/mL basic FGF (Wako) and in the presence of 100 nM Shield1 (Takara Bio). Cells were cultured with 1 μg/mL Blasticidin S (Wako) throughout days 3-7 for the selection and reseeded at day 11 at a 60 mm dish. On day 22, individual iPSC colonies were picked up and were cultured in StemFit AK02N medium (Ajinomoto) supplemented with 10 μM Y-27632 (Wako), 0.25 μg/cm2 iMatrix-511 (Nippi) in a well of 24-well plate , and then expanded to a 60 mm dish on day 32. Stocks prepared with STEM-CELLBANKER (Takara Bio) were stored until further gDNA and mRNA analyses.
Flow cytometry analysis
For the detection of TRA-1-60 and TRA-1-85 in the reprogramming human fibroblasts transduced with retroviral vectors, flow cytometry was performed on day 7 after infection. Briefly, the cells were dissociated with Accutase (Nacalai Tesque), resuspended in a home-prepared flow cytometry buffer (0.5% EDTA in PBS supplemented with 1% FBS) and incubated for one hour with antibodies: Alexa Fluor 488 Mouse anti-Human TRA-1-60 antibody (BD Pharmingen) and human TRA-1-85/CD147 APC-conjugated antibody (R&D systems). Cells were washed with the flow cytometry buffer and cell suspensions were evaluated on Guava easyCyte Flow Cytometer (Luminex).
For the detection of Nanog-GFP-positive cells in the reprogramming MEF transduced with SeVdp vectors, flow cytometry was performed on day specific days after infection. Briefly, the cells were dissociated with Accutase (Nacalai Tesque), resuspended in the flow cytometry buffer. Cell suspensions were evaluated on Guava easyCyte Flow Cytometer (Luminex).
Western Blot analysis
Protein expression assay
NB1RGB cells were seeded into 6-well plates at a density of 100,000 cells/well and infected the next day with the indicated retroviral supernatants (OSKM) for 24 h. On day 5 after infection, total proteins were collected and subjected to Western blot for KLF4 and GAPDH. Automatic capillary Western Blot analysis was performed using Wes device (Protein Simple) with Anti-Mouse Detection Module kit (Protein Simple). Antibodies used: anti-FLAG monoclonal mouse antibody (Sigma), mouse anti-Human/Mouse/Rat GAPDH/G3PDH Monoclonal Antibody (R&D).
Cycloheximide (CHX) chase assay
NB1RGB cells were seeded into 6-well plates at a density of 100,000 cells/well and infected next day with the indicated retroviral supernatants (OSKM) for 24 h. Cells were treated with 100 μg/mL CHX on day 5 after infection for varying lengths of time (0h, 8h, 24h, 48h, 56h, and 72h). Culture medium was changed next day after CHX treatment. Total proteins were collected and subjected to Western blot for KLF4, GAPDH, Ubiquitin. Briefly, the cells were harvested at indicated time point with 0.2 mL of Laemmli sample buffer and boiled at 95°C for 5 min. Automatic capillary Western Blot analysis was performed with Wes device (Protein Simple) with Anti-Mouse Detection Module kit (Protein Simple). Antibodies used: Anti-KLF4 polyclonal Rabbit Ig Antibodies at 1:50 dilution (MBL), Mouse Anti-Human/Mouse/Rat GAPDH/G3PDH Monoclonal Antibody 1:2000 dilution (R&D), Ubiquitin (P4D1) Mouse mAb 1:50 dilution (Cell Signaling Technology).
Co-immunoprecipitation (Co-IP)
Nanog-GFP MEF were seeded at 3 x 106 cells/100 mm dish samples. On the next day, the cells were infected with SeVdp (MOI = 5) carrying KLF4 WT or L507A or no infection. On day 3 after infection, co-IP assay was performed using Universal Magnetic Co-IP Kit (Active Motif, CA, USA) following the manufacturer's instruction. Total proteins were collected and subjected to immunoprecipitation with normal Goat IgG control (R&D systems, 2.5 μg/sample), anti-KLF4 antibody (Goat, R&D systems, 2.5 μg/sample), or anti-SOX2 antibody (Goat, R&D systems, 2.5 μg/sample) for overnight at 4°C with rotation. After immunoprecipitation with magnetic protein G beads and washing, automatic capillary Western blot was performed with Wes device (Protein Simple) with Anti-Rabbit Detection Module kit (Protein Simple) and Anti-KLF4 polyclonal Rabbit Ig Antibodies at 1:50 dilution (MBL). The amount of KLF4 protein was quantified from the peak area of the correspondence bands and calculated from protein input samples.
Quantitative RT-PCR
Total RNA was extracted using Monarch Total RNA Miniprep Kit (New England BioLabs) or FastGene RNA premium kit (Nippon Genetics) from cultured cells. Reverse transcription were performed using the ReverTra Ace qPCR RT kit (Toyobo), and qPCR was performed with THUNDERBIRD SYBR qPCR Mix (Toyobo) or THUNDERBIRD Probe qPCR Mix (Toyobo) on QuantStudio 3 Real-Time PCR System (Applied Biosystems). Taqman probes for GAPDH (Hs02786624_g1) and NANOG (Hs02387400_g1) were used. Primer sequence for SeV NP RNA are FW (ACCAACAGGCGGTGGTGCAA) and RV (TCCACCCCAACCCCTAGCGT).
Primer sequence for HERV-H are HERVH-S (TGG TGC CGT GAC TCG GAT) and HERVH-AS (GCT GAG TCC GAA AAG AGA GTC). Primer sequence for linRNA-RoR are lincROR-S558 (GAG GGG ACA TTT TCC ATC CT) and lincROR-AS700 (TCC AGT GGC TGT GCT AGA TG).
All quantitative PCR (qPCR) analyses were performed in technical triplicates.
Chromatin immunoprecipitation
All ChIP experiments were performed with ChIP-IT Express Chromatin Immunoprecipitation kit (Active Motif), following the kit protocol with minor modifications. Briefly, the reprogramming cells (seeding density is 6.0 x 105/cm2) in a 60-mm dish were crosslinked with 0.09 mL of 37% formaldehyde to 3.3 mL of growth medium. 1.5 mL of glycine (20X) was added to quench the unreacted formaldehyde. Cells in each 60-mm plate were collected and resuspended in 0.2 mL of lysis buffer. Genomic DNA was sheared to a targeted length of 150-200 bp on Covaris S220 Focused-ultrasonicator with optimized conditions. Sheared chromatin was subjected to cross-link reversal, treated with Proteinase K and RNase A, then extracted and analyzed for the shearing efficiency on 1% agarose gel electrophoresis. For KLF4 ChIP, 5 μg of anti-human KLF4 antibody (goat, R&D systems, AF3640) were used. Finally, DNA fragments were purified using Chromatin IP DNA Purification kit (Active Motif) following the manufacturer's protocol and used for real time qPCR and ChIP-seq library preparation.
ChIP-seq analysis
ChIP-seq library was generated using KAPA Hyper Plus kit (Roche) by Lab Droid “Maholo” (Robotic Biology Institute) automatically. Paired-end sequencing of the libraries was performed using Novaseq 6000 Sequencing system using SP 100 cycle kit (Illumina). ChIP-seq reads were mapped to the mouse genome (mm10 assembly) using BWA-MEM version 0.7.17 (Li and Durbin, 2010). To identify regions of ChIP-seq of TF enrichment over background, peak callings were performed by Bowtie 2 (Langmead and Salzberg, 2012) and MACS2 version 2.2.7 (Zhang et al., 2008). Peak regions were filtered for false discovery rate values <0.05. To visualize ChIP-seq tag counts in the IGV (Robinson et al., 2011), mapped reads were extended and converted into the bigWig format using bedGraphToBigWig. To link site accessibility to regulation of gene expression, we associated each peak to its nearest gene in the mm10 genome using ChIPpeakAnno version 3.22.2 (Zhu et al., 2010). Motif discovery was performed on the 100 bp vicinity of the peak summits using findMotifsGenome.pl from HOMER version 4.11 (Heinz et al., 2010). Pathway enrichment analysis was performed using Enrichr (https://amp.pharm.mssm.edu/Enrichr/) software (Chen et al., 2013). The p values are computed using a standard statistical method used by most enrichment analysis tools: Fisher's exact test or the hypergeometric test. This is a binomial proportion test that assumes a binomial distribution and independence for probability of any gene belonging to any set. Then, the adjusted p values were calculated with the Benjamini-Hochberg method for correction for multiple hypotheses testing.
RNA-seq analysis
RNA-seq library was generated using NEBNext Ultra Directional RNA Library Prep Kit (New England Biolabs) and NEBNext rRNA Delepletion Kit (New England Biolabs) by Lab Droid “Maholo” (Robotic Biology Institute) automatically. Paired-end sequencing of the libraries was performed using Novaseq 6000 SP Sequencing system using SP 100 cycle kit (Illumina). Reads from RNA-seq experiments were mapped to the mouse genome (GRCm38 or mm10) and analyzed for differential gene expression and PCA (principal component analysis) using CLC Genomics Workbench (Qiagen) software. Gene ontology and pathway enrichment analysis was performed using Enrichr (https://amp.pharm.mssm.edu/Enrichr/) software (Chen et al., 2013). The p values are computed using a standard statistical method used by most enrichment analysis tools: Fisher's exact test or the hypergeometric test. This is a binomial proportion test that assumes a binomial distribution and independence for probability of any gene belonging to any set. Then, the adjusted p values were calculated with the Benjamini-Hochberg method for correction for multiple hypotheses testing.
Molecular dynamics (MD) simulation
The coordinates of mouse Klf4 in complex with cognate DNA were obtained from Protein Data Bank (PDB) public database (PDB ID: 2WBU). The N-terminal of Klf4 was capped with an acetyl group. The structure of the Klf4 L477A mutant was generated by removing the Cγ and Cδ atoms of L477. Wild-type (WT) and mutant Klf4 models were solvated in a cubic water box with an edge length of about 80 Å and potassium ions were placed around the protein to neutralize the system. Amber ff14SB force field parameter (Maier et al., 2015) was used for the proteins, OL15 parameters (Krepl et al., 2012; Zgarbova et al., 2013, 2015) was used for DNA, and the TIP3P model (Jorgensen et al., 1983) was used for water. After energy minimization, each system was equilibrated at 300 K and 1.0 × 105 Pa with a 1-ns MD simulation. Position and distance restrain were imposed on the non-hydrogen atoms of the protein and the DNA and the atom pairs that form intermolecular hydrogen bonds between the protein and the DNA in the crystal structure, respectively. The position restraining force was gradually weakened during the simulation. Subsequently, a one hundred-ns MD simulation was performed with the distance restraints, of which force constant was gradually reduced during the simulation. Finally, a 2-μs MD simulation was performed without restraints. This series of the MD simulations were repeated three times for each of the Klf4(WT)-DNA complex and for the Klf4(L477A)-DNA complex systems with different initial velocities. In all MD simulations, the temperature was controlled by the velocity-rescaling method (Bussi et al., 2007) and the pressure was controlled by the Berendsen weak coupling method (Berendsen et al., 1984). Bond lengths involving hydrogen atoms were constrained using the LINCS algorithm (Hess, 2008; Hess et al., 1997) to allow the use of a large time step (2 fs). Electrostatic interactions were calculated with the particle mesh Ewald method (Darden et al., 1993; Essmann et al., 1995). MD simulations were performed with Gromacs 2019 (Hess et al., 2008), with coordinates recorded every 10 ps.
Principal component analysis (PCA) was performed to visualize conformational distribution of the complex in the MD simulations by using the method described in Terada and Kidera (2012) (Terada and Kidera, 2012). Briefly, each snapshot structure in the MD trajectories obtained from the six runs (three for the Klf4(WT)-DNA complex and three for the Klf4(L477A)-DNA complex) was aligned to the initial structure by minimizing the root-mean-square deviation (RMSD) calculated for the DNA P atoms between the snapshot structure and the initial structure. Then, the average structure was calculated from the aligned snapshot structures. The variance-covariance matrix was calculated from the deviations of the protein Cα and the DNA P atoms from those of the average structure. The matrix was diagonalized and the eigenvectors corresponding to the two largest eigenvalues were used to analyze the conformational distribution. In addition, the snapshot structures of each complex system were classified into clusters as described in Terada et al. (2008) (Terada et al., 2008), by using the RMSD values calculated for the protein Cα and the DNA P atoms as distance measure. The snapshot structure closest to the cluster center was used as the representative structure of the cluster.
Quantification and statistical analysis
In this manuscript, data are shown as mean ± SE in all the graphs generated with Microsoft Excel for Mac software (Microsoft, version 16.48). For comparison of two samples, p values were determined by unpaired two-tailed Student's t-test using Microsoft Excel for Mac software (Microsoft, version 16.48). For comparison of multiple samples more than two, p values were determined by Dunnett's test using Graphpad Prism 9 for macOS (version 9.2.0). Venn diagrams were drawn using "Venn Diagram Maker Online" (https://www.meta-chart.com/venn).
Acknowledgments
We would like to thank Ms. Kumiko Omori for her administrative support and all our colleagues and collaborators. This research was supported in part by Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED under Grant Number JP21am0101107 (support number 1907), the grants from a JSPS KAKENHI Grant-in-Aid for Research Activity start-up (JP16H06662) to Y.H., for Young Scientists (A) (JP17H05063) to Y.H., for scientific research (B) (JP17H04036 and JP19H03203) to Y.H., K.N., and K.H., “Nanotechnology Platform Project” operated by the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan (No. JPMXP09 S20NM0001), Takeda Science Foundation to Y.H., Uehara Memorial Foundation to Y.H., the Tokyo Biochemical Research Foundation to Y.H., and 2018 iPS Academia Japan Grant to Y.H. The Otsuka Toshimi Scholarship Foundation provided financial support during this study to E.B. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
Conceptualization, Y.H.; Methodology, E.B., K.N., T.T., K.A., E.S., T.S., and Y.H.; Investigation, E.B., K.N., M.T., J.L., Y.A., D.S., M.M.T., D.L., S.A., F.Y., T.T., K.H., and Y.H; Resources E.B., K.N., M.T., J.L., Y.A., D.S., M.M.T., S.A., and Y. H; Writing – Original Draft, E.B. and Y.H.; Writing – Review & Editing, E.B. K.N., E.S., M.M.T., T.T., K.H. and Y.H.; Visualization, E.B., K.N., T.T., and Y.H; Supervision, Y.H., K.N., Y.F., and K.H.; Project Administration, Y.H., Funding Acquisition, Y.H., K.N., and K.H.
Declaration of interests
E.B., K.N., F.Y., K.H., and Y.H. filed a patent related to this work.
Published: December 14, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103525.
Supplemental information
Data and code availability
The accession numbers for the ChIP-seq and RNA-seq datasets reported in this manuscript are GSE157762 and GSE184449 in NCBI GEO (gene expression omnibus), respectively.
References
- Adachi K., Scholer H.R. Directing reprogramming to pluripotency by transcription factors. Curr. Opin. Genet. Dev. 2012;22:416–422. doi: 10.1016/j.gde.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Aksoy I., Giudice V., Delahaye E., Wianny F., Aubry M., Mure M., Chen J., Jauch R., Bogu G.K., Nolden T., et al. Klf4 and Klf5 differentially inhibit mesoderm and endoderm differentiation in embryonic stem cells. Nat. Commun. 2014;5:3719. doi: 10.1038/ncomms4719. [DOI] [PubMed] [Google Scholar]
- Azami T., Matsumoto K., Jeon H., Waku T., Muratani M., Niwa H., Takahashi S., Ema M. Klf5 suppresses ERK signaling in mouse pluripotent stem cells. PLoS One. 2018;13:e0207321. doi: 10.1371/journal.pone.0207321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen H.J.C., Postma J.P.M., Vangunsteren W.F., Dinola A., Haak J.R. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Chan E.M., Ratanasirintrawoot S., Park I.H., Manos P.D., Loh Y.H., Huo H., Miller J.D., Hartung O., Rho J., Ince T.A., et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat. Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T., York D., Pedersen L. Particle mesh ewald - an N.Log(N) method for ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- Dhaliwal N.K., Abatti L.E., Mitchell J.A. KLF4 protein stability regulated by interaction with pluripotency transcription factors overrides transcriptional control. Genes Dev. 2019;33:1069–1082. doi: 10.1101/gad.324319.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino D.C., Kloetgen A., Polyzos A., Liu Y., Kim D., Murphy D., Abuhashem A., Cavaliere P., Aronson B., Shah V., et al. KLF4 is involved in the organization and regulation of pluripotency-associated three-dimensional enhancer networks. Nat. Cell Biol. 2019;21:1179–1190. doi: 10.1038/s41556-019-0390-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- Garrett-Sinha L.A., Eberspaecher H., Seldin M.F., de Crombrugghe B. A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J. Biol. Chem. 1996;271:31384–31390. doi: 10.1074/jbc.271.49.31384. [DOI] [PubMed] [Google Scholar]
- Gonzalez F., Boue S., Izpisua Belmonte J.C. Methods for making induced pluripotent stem cells: reprogramming a la carte. Nat. Rev. Genet. 2011;12:231–242. doi: 10.1038/nrg2937. [DOI] [PubMed] [Google Scholar]
- Guo L., Lin L., Wang X., Gao M., Cao S., Mai Y., Wu F., Kuang J., Liu H., Yang J., et al. Resolving cell fate decisions during somatic cell reprogramming by single-cell RNA-Seq. Mol. Cell. 2019;73:815–829. doi: 10.1016/j.molcel.2019.01.042. [DOI] [PubMed] [Google Scholar]
- Guo C., Morris S.A. Engineering cell identity: establishing new gene regulatory and chromatin landscapes. Curr. Opin. Genet. Dev. 2017;46:50–57. doi: 10.1016/j.gde.2017.06.011. [DOI] [PubMed] [Google Scholar]
- Gurdon J.B. Cell fate determination by transcription factors. Curr. Top. Dev. Biol. 2016;116:445–454. doi: 10.1016/bs.ctdb.2015.10.005. [DOI] [PubMed] [Google Scholar]
- Gurdon J.B., Melton D.A. Nuclear reprogramming in cells. Science. 2008;322:1811–1815. doi: 10.1126/science.1160810. [DOI] [PubMed] [Google Scholar]
- Hanna J.H., Saha K., Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H., Wang D., Steves A.N., Jin P., Blumenthal R.M., Zhang X., Cheng X. Distinctive Klf4 mutants determine preference for DNA methylation status. Nucleic Acids Res. 2016;44:10177–10185. doi: 10.1093/nar/gkw774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Sasamura H., Nakamura M., Azegami T., Oguchi H., Sakamaki Y., Itoh H. KLF4-dependent epigenetic remodeling modulates podocyte phenotypes and attenuates proteinuria. J. Clin. Invest. 2014;124:2523–2537. doi: 10.1172/JCI69557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y., Caboni L., Das D., Yumoto F., Clayton T., Deller M.C., Nguyen P., Farr C.L., Chiu H.J., Miller M.D., et al. Structure-based discovery of NANOG variant with enhanced properties to promote self-renewal and reprogramming of pluripotent stem cells. Proc. Natl. Acad. Sci. U S A. 2015;112:4666–4671. doi: 10.1073/pnas.1502855112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B. P-LINCS: A parallel linear constraint solver for molecular simulation. J. Chem. Theor. Comput. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- Hess B., Kutzner C., van der Spoel D., Lindahl E. Gromacs 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theor. Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Hiramatsu K., Sasagawa S., Outani H., Nakagawa K., Yoshikawa H., Tsumaki N. Generation of hyaline cartilaginous tissue from mouse adult dermal fibroblast culture by defined factors. J. Clin. Invest. 2011;121:640–657. doi: 10.1172/JCI44605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H., Takahashi K., Ichisaka T., Aoi T., Kanagawa O., Nakagawa M., Okita K., Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta A., Cheung A.Y., Farra N., Vijayaragavan K., Seguin C.A., Draper J.S., Pasceri P., Maksakova I.A., Mager D.L., Rossant J., et al. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nat. Methods. 2009;6:370–376. doi: 10.1038/nmeth.1325. [DOI] [PubMed] [Google Scholar]
- Iwafuchi-Doi M., Zaret K.S. Pioneer transcription factors in cell reprogramming. Genes Dev. 2014;28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauch R. Cell fate reprogramming through engineering of native transcription factors. Curr. Opin. Genet. Dev. 2018;52:109–116. doi: 10.1016/j.gde.2018.05.013. [DOI] [PubMed] [Google Scholar]
- Jeon H., Waku T., Azami T., Khoa le T.P., Yanagisawa J., Takahashi S., Ema M. Comprehensive identification of kruppel-like factor family members contributing to the self-renewal of mouse embryonic stem cells and cellular reprogramming. PLoS One. 2016;11:e0150715. doi: 10.1371/journal.pone.0150715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Chan Y.S., Loh Y.H., Cai J., Tong G.Q., Lim C.A., Robson P., Zhong S., Ng H.H. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- Kelder T., Pico A.R., Hanspers K., van Iersel M.P., Evelo C., Conklin B.R. Mining biological pathways using Wiki Pathways web services. PLoS One. 2009;4:e6447. doi: 10.1371/journal.pone.0006447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.O., Kim S.H., Cho Y.Y., Nadas J., Jeong C.H., Yao K., Kim D.J., Yu D.H., Keum Y.S., Lee K.Y., et al. ERK1 and ERK2 regulate embryonic stem cell self-renewal through phosphorylation of Klf4. Nat. Struct. Mol. Biol. 2012;19:283–290. doi: 10.1038/nsmb.2217. [DOI] [PubMed] [Google Scholar]
- Kim S.I., Oceguera-Yanez F., Hirohata R., Linker S., Okita K., Yamada Y., Yamamoto T., Yamanaka S., Woltjen K. KLF4 N-terminal variance modulates induced reprogramming to pluripotency. Stem Cell Rep. 2015;4:727–743. doi: 10.1016/j.stemcr.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.S., Lewandoski M., Perantoni A.O., Kurebayashi S., Nakanishi G., Jetten A.M. Identification of Glis1, a novel Gli-related, Kruppel-like zinc finger protein containing transactivation and repressor functions. J. Biol. Chem. 2002;277:30901–30913. doi: 10.1074/jbc.M203563200. [DOI] [PubMed] [Google Scholar]
- Kitamura T., Koshino Y., Shibata F., Oki T., Nakajima H., Nosaka T., Kumagai H. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 2003;31:1007–1014. [PubMed] [Google Scholar]
- Kitazawa K., Hikichi T., Nakamura T., Nakamura M., Sotozono C., Masui S., Kinoshita S. Direct reprogramming into corneal epithelial cells using a transcriptional network comprising PAX6, OVOL2, and KLF4. Cornea. 2019;38:S34–S41. doi: 10.1097/ICO.0000000000002074. [DOI] [PubMed] [Google Scholar]
- Krepl M., Zgarbova M., Stadlbauer P., Otyepka M., Banas P., Koca J., Cheatham T.E., 3rd, Jurecka P., Sponer J. Reference simulations of noncanonical nucleic acids with different chi variants of the AMBER force field: quadruplex DNA, quadruplex RNA and Z-DNA. J. Chem. Theor. Comput. 2012;8:2506–2520. doi: 10.1021/ct300275s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.Y., Noh H.B., Kim H.T., Lee K.I., Hwang D.Y. Glis family proteins are differentially implicated in the cellular reprogramming of human somatic cells. Oncotarget. 2017;8:77041–77049. doi: 10.18632/oncotarget.20334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B., Bao X., Liu L., Feng S., Zovoilis A., Liu W., Xue Y., Cai J., Guo X., Qin B., et al. MicroRNA cluster 302-367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J. Biol. Chem. 2011;286:17359–17364. doi: 10.1074/jbc.C111.235960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Olanrewaju Y.O., Zheng Y., Hashimoto H., Blumenthal R.M., Zhang X., Cheng X. Structural basis for Klf4 recognition of methylated DNA. Nucleic Acids Res. 2014;42:4859–4867. doi: 10.1093/nar/gku134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry W.E., Richter L., Yachechko R., Pyle A.D., Chieu J., Sridharan R., Clark A.T., Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa M., Yamaguchi K., Nakamura T., Shibukawa R., Kodanaka I., Ichisaka T., Kawamura Y., Mochizuki H., Goshima N., Yamanaka S. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature. 2011;474:225–229. doi: 10.1038/nature10106. [DOI] [PubMed] [Google Scholar]
- Mahatan C.S., Kaestner K.H., Geiman D.E., Yang V.W. Characterization of the structure and regulation of the murine gene encoding gut-enriched Kruppel-like factor (Kruppel-like factor 4) Nucleic Acids Res. 1999;27:4562–4569. doi: 10.1093/nar/27.23.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theor. Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Molina D., Jafari R., Ignatushchenko M., Seki T., Larsson E.A., Dan C., Sreekumar L., Cao Y., Nordlund P. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Takizawa N., Narita M., Ichisaka T., Yamanaka S. Promotion of direct reprogramming by transformation-deficient Myc. Proc. Natl. Acad. Sci. U S A. 2010;107:14152–14157. doi: 10.1073/pnas.1009374107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninkovic J., Gotz M. Understanding direct neuronal reprogramming-from pioneer factors to 3D chromatin. Curr. Opin. Genet. Dev. 2018;52:65–69. doi: 10.1016/j.gde.2018.05.011. [DOI] [PubMed] [Google Scholar]
- Nishimura K., Aizawa S., Nugroho F.L., Shiomitsu E., Tran Y.T.H., Bui P.L., Borisova E., Sakuragi Y., Takada H., Kurisaki A., et al. A role for KLF4 in promoting the metabolic shift via TCL1 during induced pluripotent stem cell generation. Stem Cell Rep. 2017;8:787–801. doi: 10.1016/j.stemcr.2017.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., Kato T., Chen C., Oinam L., Shiomitsu E., Ayakawa D., Ohtaka M., Fukuda A., Nakanishi M., Hisatake K. Manipulation of KLF4 expression generates iPSCs paused at successive stages of reprogramming. Stem Cell Rep. 2014;3:915–929. doi: 10.1016/j.stemcr.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., Sano M., Ohtaka M., Furuta B., Umemura Y., Nakajima Y., Ikehara Y., Kobayashi T., Segawa H., Takayasu S., et al. Development of defective and persistent Sendai virus vector: a unique gene delivery/expression system ideal for cell reprogramming. J. Biol. Chem. 2011;286:4760–4771. doi: 10.1074/jbc.M110.183780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuki M., Tanabe K., Sutou K., Teramoto I., Sawamura Y., Narita M., Nakamura M., Tokunaga Y., Nakamura M., Watanabe A., et al. Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential. Proc. Natl. Acad. Sci. U S A. 2014;111:12426–12431. doi: 10.1073/pnas.1413299111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Pabo C.O., Peisach E., Grant R.A. Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- Parisi S., Cozzuto L., Tarantino C., Passaro F., Ciriello S., Aloia L., Antonini D., De Simone V., Pastore L., Russo T. Direct targets of Klf5 transcription factor contribute to the maintenance of mouse embryonic stem cell undifferentiated state. BMC Biol. 2010;8:128. doi: 10.1186/1741-7007-8-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pico A.R., Kelder T., van Iersel M.P., Hanspers K., Conklin B.R., Evelo C. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6:e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Kim J.K., Tong W., Villa-Diaz L.G., Krebsbach P.H. DPPA5 supports pluripotency and reprogramming by regulating NANOG turnover. Stem Cells. 2016;34:588–600. doi: 10.1002/stem.2252. [DOI] [PubMed] [Google Scholar]
- Reinhardt A., Kagawa H., Woltjen K. N-terminal amino acids determine KLF4 protein stability in 2A peptide-linked polycistronic reprogramming constructs. Stem Cell Rep. 2020;14:520–527. doi: 10.1016/j.stemcr.2020.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland B.D., Bernards R., Peeper D.S. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat. Cell Biol. 2005;7:1074–1082. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- Schuetz A., Nana D., Rose C., Zocher G., Milanovic M., Koenigsmann J., Blasig R., Heinemann U., Carstanjen D. The structure of the Klf4 DNA-binding domain links to self-renewal and macrophage differentiation. Cell. Mol. Life Sci. 2011;68:3121–3131. doi: 10.1007/s00018-010-0618-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre J.A., Bauer C., Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Shie J.L., Chen Z.Y., Fu M., Pestell R.G., Tseng C.C. Gut-enriched Kruppel-like factor represses cyclin D1 promoter activity through Sp1 motif. Nucleic Acids Res. 2000;28:2969–2976. doi: 10.1093/nar/28.15.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields J.M., Christy R.J., Yang V.W. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J. Biol. Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slenter D.N., Kutmon M., Hanspers K., Riutta A., Windsor J., Nunes N., Melius J., Cirillo E., Coort S.L., Digles D., et al. WikiPathways: A multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018;46:D661–D667. doi: 10.1093/nar/gkx1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A., Donahue G., Zaret K.S. Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A., Garcia M.F., Jaroszewicz A., Osman N., Pellegrini M., Zaret K.S. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161:555–568. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Sasaki A., Yamamoto M., Nakamura M., Sutou K., Osafune K., Yamanaka S. Induction of pluripotency in human somatic cells via a transient state resembling primitive streak-like mesendoderm. Nat. Commun. 2014;5:3678. doi: 10.1038/ncomms4678. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 2016;17:183–193. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- Terada T., Kidera A. Comparative molecular dynamics simulation study of crystal environment effect on protein structure. J. Phys. Chem. B. 2012;116:6810–6818. doi: 10.1021/jp2125558. [DOI] [PubMed] [Google Scholar]
- Terada T., Satoh D., Mikawa T., Ito Y., Shimizu K. Understanding the roles of amino acid residues in tertiary structure formation of chignolin by using molecular dynamics simulation. Proteins. 2008;73:621–631. doi: 10.1002/prot.22100. [DOI] [PubMed] [Google Scholar]
- Thier M., Worsdorfer P., Lakes Y.B., Gorris R., Herms S., Opitz T., Seiferling D., Quandel T., Hoffmann P., Nothen M.M., et al. Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell. 2012;10:473–479. doi: 10.1016/j.stem.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Tsang J.C., Gao X., Lu L., Liu P. Cellular reprogramming by transcription factor engineering. Curr. Opin. Genet. Dev. 2014;28:1–9. doi: 10.1016/j.gde.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Velychko S., Kang K., Kim S.M., Kwak T.H., Kim K.P., Park C., Hong K., Chung C., Hyun J.K., MacCarthy C.M., et al. Fusion of reprogramming factors alters the trajectory of somatic lineage conversion. Cell Rep. 2019;27:30–39 e34. doi: 10.1016/j.celrep.2019.03.023. [DOI] [PubMed] [Google Scholar]
- Wang L., Su Y., Huang C., Yin Y., Chu A., Knupp A., Tang Y. NANOG and LIN28 dramatically improve human cell reprogramming by modulating LIN41 and canonical WNT activities. Biol. Open. 2019;8:bio047225. doi: 10.1242/bio.047225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worringer K.A., Rand T.A., Hayashi Y., Sami S., Takahashi K., Tanabe K., Narita M., Srivastava D., Yamanaka S. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell. 2014;14:40–52. doi: 10.1016/j.stem.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Du Y., Deng H. Direct lineage reprogramming: strategies, mechanisms, and applications. Cell Stem Cell. 2015;16:119–134. doi: 10.1016/j.stem.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Yamanaka S., Blau H.M. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane M., Ohtsuka S., Matsuura K., Nakamura A., Niwa H. Overlapping functions of Kruppel-like factor family members: targeting multiple transcription factors to maintain the naive pluripotency of mouse embryonic stem cells. Development. 2018;145 doi: 10.1242/dev.162404. dev162404. [DOI] [PubMed] [Google Scholar]
- Ye B., Liu B., Hao L., Zhu X., Yang L., Wang S., Xia P., Du Y., Meng S., Huang G., et al. Klf4 glutamylation is required for cell reprogramming and early embryonic development in mice. Nat. Commun. 2018;9:1261. doi: 10.1038/s41467-018-03008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka N., Gros E., Li H.R., Kumar S., Deacon D.C., Maron C., Muotri A.R., Chi N.C., Fu X.D., Yu B.D., et al. Efficient generation of human iPSCs by a synthetic self-replicative RNA. Cell Stem Cell. 2013;13:246–254. doi: 10.1016/j.stem.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zgarbova M., Luque F.J., Sponer J., Cheatham T.E., 3rd, Otyepka M., Jurecka P. Toward improved description of DNA backbone: Revisiting epsilon and zeta torsion force field parameters. J. Chem. Theor. Comput. 2013;9:2339–2354. doi: 10.1021/ct400154j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgarbova M., Sponer J., Otyepka M., Cheatham T.E., 3rd, Galindo-Murillo R., Jurecka P. Refinement of the sugar-phosphate backbone torsion beta for AMBER force fields improves the description of Z- and B-DNA. J. Chem. Theor. Comput. 2015;11:5723–5736. doi: 10.1021/acs.jctc.5b00716. [DOI] [PubMed] [Google Scholar]
- Zhang W., Geiman D.E., Shields J.M., Dang D.T., Mahatan C.S., Kaestner K.H., Biggs J.R., Kraft A.S., Yang V.W. The gut-enriched Kruppel-like factor (Kruppel-like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J. Biol. Chem. 2000;275:18391–18398. doi: 10.1074/jbc.C000062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L.J., Gazin C., Lawson N.D., Pages H., Lin S.M., Lapointe D.S., Green M.R. ChIPpeakAnno: A bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinf. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for the ChIP-seq and RNA-seq datasets reported in this manuscript are GSE157762 and GSE184449 in NCBI GEO (gene expression omnibus), respectively.