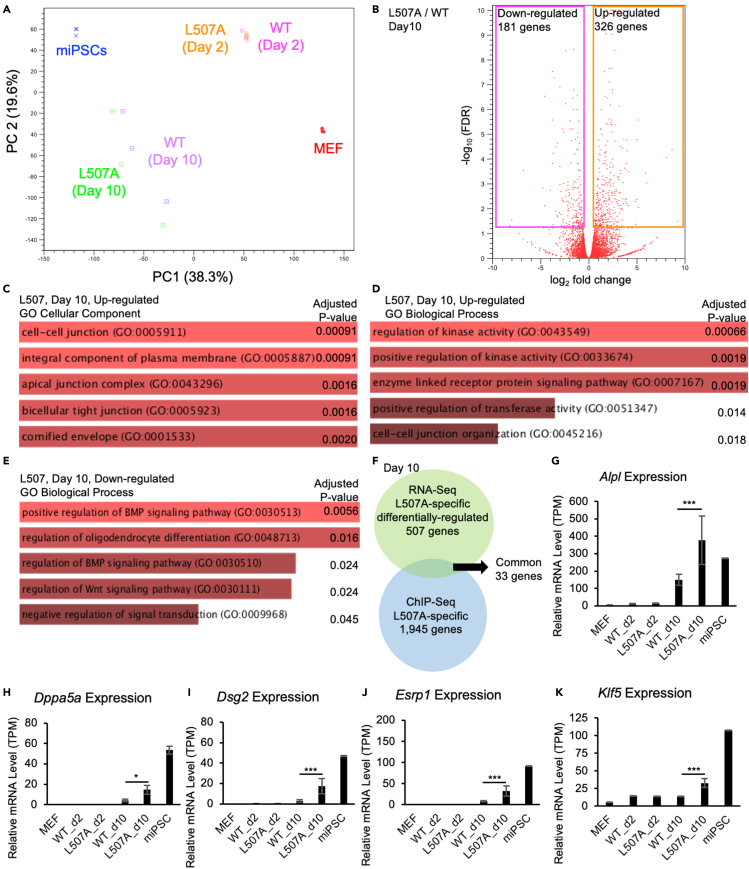
Figure 7.
Global expression similarity and differences between reprogramming cells between KLF4 WT and L507A
(A) PCA of global gene expression in MEF, miPSCs, and reprogramming MEF induced by KLF4 WT or L507A on day 2 or 10 after transduction of each SeVdp vector.
(B) A volcano plot shows log2 fold change as X axis and -log10 (FDR) as Y axis. All the genes were plotted in red circles. Significant genes with FDR<0.05 were rectangularly surrounded with pink (downregulated) and orange (upregulated).
(C-E) GO (Gene Ontology) enrichment analysis was performed on the gene sets, composed of the genes which differentially expressed between WT and L507A conditions on day 10 samples. GO Cellular Component enriched from upregulated genes in L507A samples were shown in (C); GO Biological Process enriched from upregulated genes in L507A samples were shown in (D); GO Biological Process enriched from down-regulated genes in L507A samples were shown in (E). The top 5 GO terms that are enriched with statistically significant are illustrated with adjusted p values.
(F) A Venn diagram of gene sets obtained from specifically regulated by L507A as shown above in RNA-seq data and ChIP-seq data. 33 genes, including Klf5, were extracted in common.
(G–K) As examples of the genes which are commonly identified in RNA-seq and ChIP-seq experiments in this study, the expression levels (i.e., TPM (Transcripts Per Million)) of Alpl, Dppa5a, Dsg2, Esrp1, and Klf5, are shown from RNA-seq data. Results are mean and SE, n = 3. p values calculated with FDR correction were shown based on the comparison between WT and L507A KLF4 conditions on day 10 samples.