Abstract
Invasive mucinous adenocarcinoma (IMA) of the lung frequently presents with diffuse pneumonic-type features or multifocal lesions, which are regarded as a pattern of intrapulmonary metastases. However, the genomics of multifocal IMAs have not been well studied. We performed whole exome sequencing on samples taken from 2 to 5 regions in seven patients with synchronous multifocal IMAs of the lung (24 regions total). Early initiating driver events, such as KRAS, NKX2-1, TP53, or ARID1A mutations, are clonal mutations and were present in all multifocal IMAs in each patient. The tumor mutational burden of multifocal IMAs was low (mean: 1.13/mega base), but further analyses suggested intra-tumor heterogeneity. The mutational signature analysis found that IMAs were predominantly associated with endogenous mutational process (signature 1), APOBEC activity (signatures 2 and 13), and defective DNA mismatch repair (signature 6), but not related to smoking signature. IMAs synchronously located in the bilateral lower lobes of two patients with background usual interstitial pneumonia had different mutation types, suggesting that they were double primaries. In conclusion, genomic evidence found in this study indicated the clonal intrapulmonary spread of diffuse pneumonic-type or multifocal IMAs, although they can occur in multicentric origins in the background of usual interstitial pneumonia. IMAs exhibited a heterogeneous genomic landscape despite the low somatic mutation burden. Further studies are warranted to determine the clinical significance of the genomic characteristics of IMAs in expanded cohorts.
Subject terms: Non-small-cell lung cancer, Cancer genomics
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide. The most common subtype of lung cancer is adenocarcinoma [1]. Invasive mucinous adenocarcinoma (IMA) accounts for approximately 5% of all lung adenocarcinoma cases [2]. IMA is well known for its unique clinicopathologic characteristics [3]. Patients with IMA usually present with diffuse pneumonic patterns that manifest as multiple lung lesions [4]. Although recurrence in the lungs is frequently observed, extrathoracic or nodal metastasis can occur [5]. Histologically, IMA is characterized by goblet or columnar cells containing intracellular mucin with basally located nuclei [3]. Immunohistochemistry showed the loss of TTF-1/NKX2-1 protein expression, the lineage-specific protein, which is frequently used to distinguish between lung and other organ adenocarcinomas, in more than 80% of IMAs [6, 7]. KRAS mutations are the most common type of oncogenic driver mutations [5, 7–9]. KRAS G12D mutations are the most common subtype of KRAS mutations, followed by G12V, G12C, and G12A mutations [5, 9]. In addition to KRAS mutations, BRAF and ERBB2 mutations, and gene rearrangements of NRG1 have also been observed [5, 9–12].
Lung cancers with multiple lesions are challenging to classify. Generally, lung cancers involving multiple pulmonary sites occur in one of four patterns: (1) second primary cancers, (2) separate tumor nodules, (3) multifocal ground glass or lepidic nodules, and (4) diffuse pneumonic-type [4]. IMAs are usually classified as diffuse pneumonic-type cancers. In CT scans, they usually exhibit a diffuse consolidative pattern in the absence of proximal bronchial obstructions [4, 13, 14]. Histologically, IMAs are regarded as occurring through intrapulmonary metastasis and have a poorer prognosis than ground glass or lepidic nodules [4]. However, the genomic basis of this classification has not been well studied. While this study is in progress, only one study on comparative molecular profiling of spatially separate IMAs has been recently reported by Yang and colleagues [15]. The genomic structures of IMAs with multiple pulmonary sites of involvement are important because such knowledge can increase the accuracy of stage classifications, prognoses, and treatment plans. In this study, the genomics of spatially separated regions of multifocal IMAs were examined using whole exome sequencing (WES).
Methods and materials
Patients
Patients with IMAs who underwent surgical resection were enrolled from January 2014 to December 2018 at our institution. The institutional review board approved this retrospective study (No. 4-2019-0468). Clinical data including age, gender, smoking history, and radiologic findings were retrieved from electronic medical records.
Histological review
Surgical specimens were fixed in 10% neutral-buffered formalin and embedded in paraffin blocks. For each formalin-fixed, paraffin-embedded (FFPE) tissue block, 4 μm sections were cut and stained with hematoxylin and eosin. IMA was diagnosed based on the 2015 World Health Organization classification [2].
Whole exome sequencing
DNA was extracted from FFPE tissue using the QIAamp DNA FFPE Tissue kit according to the manufacturer’s protocols (Qiagen, Valencia, CA, USA). The concentration and quality of DNA were assessed using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). WES was performed using the SureSelect Human All Exon V6 kit (Agilent Technologies, Santa Clara, CA, USA) and processed on the HiSeq 2500 platform to obtain a mean depth of 200× (Illumina, San Diego, CA, USA). Sequence reads were aligned to the human reference genome hg19 using the Burrows−Wheeler Aligner-MEM algorithm [16]. Duplicate or low-quality reads were marked using sambamba [17] and base quality score recalibration was performed using a genome analysis tool kit (GATK) [18].
Somatic mutation calling
Somatic mutations were called using GATK MuTect2 using its default settings by comparing the sequences of tumor samples with those of matched normal samples [19]. Variant filtration functions in the GATK, such as FilterMutectCalls, CollectSequencingArtifactMetrics, and FilterByOrientationBias, were applied for the confident somatic calls. Somatic variants were annotated using the SnpEff and SnpSift functions [20]. To reduce the effect of false-positive variants, the following variants were excluded: (i) variants with a minor allele frequency of more than 1% in the genome aggregation [21], Exome Aggregation Consortium [22], and Korean population databases, (ii) variants with oxidized guanine to 8-oxoguanin (OxoG) artifacts, (iii) variants with mutated read counts less than 3, (iv) variants with total read depth less than 20, and (v) variants with a variant allele frequency (VAF) less than 3%. A manual review of KRAS codons 12, 13, and 61 for mutations below the VAF threshold for calling was performed using Integrated Genomics Viewer.
Phylogenetic tree analysis
Hamming distances between samples were calculated using the presence and absence of somatic variants across samples. For each patient, phylogenetic tree analysis was conducted using the neighbor-joining algorithm in the APE package [23].
Tumor mutational burden (TMB) analysis
TMB was defined as the total number of somatic nonsynonymous missense mutations per megabase (Mb) [24, 25]. Germline variants were subtracted with matched normal samples [26, 27].
Intra-tumor heterogeneity (ITH) analysis
The percentage of mutations that were branch and private mutations, which were not present in all sections of a tumor, was determined to evaluate the degree of ITH [28, 29].
Jaccard similarity coefficient
The Jaccard similarity coefficient was defined as the proportion of intersections between samples A and B divided by the proportion of their union [30–33]:
Mutational signature analysis
Mutational signatures were determined using Mutalisk in which mutations were decomposed by linear regression [34]. The somatic variants filtered with paired normal variants were pooled according to lung cancer signatures 1, 2, 4, 5, 6, 13, 15, and 17 in the Catalogue of Somatic Mutations in Cancer database. The mutational signatures were validated by signeR based on a non-negative matrix factorization algorithm [35, 36]. The mutational signature spectrum was compared and confirmed considering cosine similarity with same as above lung signatures.
Purity and copy number variation (CNV) analysis
Somatic CNVs were identified using a CNVkit with the default settings [37]. The circular binary segmentation method was used to integrate noisy copy number signals into the same copy number. Mutations’ normalized copy ratios and b allele frequencies were used to estimate tumor purity and ploidy using absCNseq [38]. This purity information was then used to identify the final CNVs for each sample using the CNVkit’s purity analysis function.
Results
Clinical characteristics
Out of 1,948 chemotherapy-naïve patients who underwent surgical resection for invasive adenocarcinoma of the lung, 132 (6.8%) were diagnosed with IMA. Eight patients with synchronous multifocal IMAs resected by surgery were enrolled in this study and designated as P01–P08, respectively. Patient characteristics are summarized in Table 1. The median age was 59 years and the ages ranged from 36 to 79 years. Two were male and six were female. Three patients were ex-smokers and five had never smoked. P01 and P06 had undergone lung transplantation due to clinically diffuse interstitial pneumonia. Subsequent pathologic examination of the explanted lungs revealed IMAs involving multiple lobes in the background of usual interstitial pneumonia and fibrosing non-specific interstitial pneumonia, respectively. In P07, pathological examination of non-neoplastic lung showed findings consistent with usual interstitial pneumonia. In the remaining patients, no evidence of fibrosing interstitial pneumonia was observed. P02 was excluded during sequencing due to the poor quality of extracted DNA.
Table 1.
Patient cohort.
| Patient No. | Age | Sex | Smoking history | Main tumor size (cm) | Stage | Location |
|---|---|---|---|---|---|---|
| P01 | 53 | M | Ex-smoker | >10 | IV | RUL, RML, RLL, LUL, LLL |
| P02 | 72 | F | Never-smoker | 8.6 | IV | RUL1, RUL2, RUL3, LUL |
| P03 | 79 | F | Never-smoker | 6.7 | IIIB | RLL1, RLL2, RLL3 |
| P04 | 65 | M | Ex-smoker | 3.5, 1.0 | IIIA | RLL, RUL1, RUL2 |
| P05 | 65 | F | Never-smoker | 5.8 | IIB | RLL1, RLL2, RLL3 |
| P06 | 36 | F | Ex-smoker | >10 | IV | RUL, RML, LUL, LN |
| P07 | 52 | F | Never-smoker | 5.6, 3.2 | IIB | LLL, RLL |
| P08 | 48 | F | Never-smoker | 5.5 | IIB | LLL1, LLL2, LLL3, LLL4 |
RUL right upper lobe, RML right middle lobe, RLL right lower lobe, LUL left upper lobe, LLL left lower lobe, LN lymph node metastasis.
Genomic landscape of IMAs
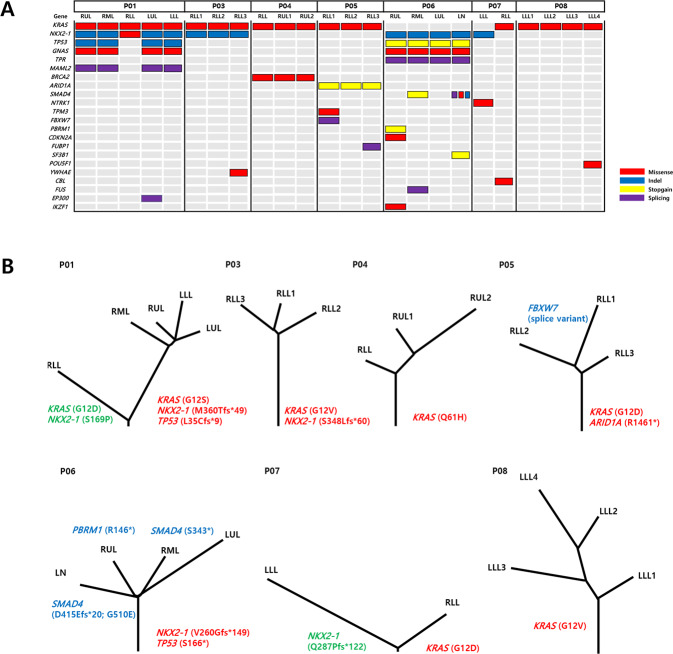
In total, 2–5 regions per patient were sequenced for a total of 24 sectors (Fig. 1). Sequencing occurred at a mean depth of 207×. A total of 4,059 somatic single nucleotide variants and small insertion or deletion mutations were identified with an average of 169.2 mutations/region. Mutation heat maps and phylogenetic trees were generated (Fig. 1).
Fig. 1. Landscape of clonal and subclonal cancer-related mutations in IMAs.
A OncoPrint heatmap of mutations in IMAs. B Phylogenetic trees generated for seven patients with multifocal IMAs (letters in red: trunk mutations, letters in green: mutation in contralateral lower lobes, letters in blue: branch mutations). (RUL right upper lobe, RML right middle lobe, RLL right lower lobe, LUL left upper lobe, LLL left lower lobe, LN lymph node metastasis, Indel insertion and deletion mutation).
All KRAS mutations, namely G12D, G12S, G12V, and Q61H, were confirmed to be truncal events as they were present in all sectors of the tumors, underscoring KRAS mutations’ role as an early driver of tumor initiation (Fig. 1 and Supplementary Table S1). However, KRAS mutations were different between synchronous tumors in the bilateral lower lobes of individuals (G12S vs. G12D in P01 and wild type vs. G12D in P07) (Fig. 1 and Supplementary Table S1). The second most frequently mutated gene was NKX2-1, found in four out of seven tumors (Fig. 1 and Supplementary Table S1) and was also a truncal event. NKX2-1 mutations differed between bilateral lower lobes in individuals (e.g., M360Tfs*49 vs. S169P in P01 and Q287Pfs*122 vs. wild type in P07) (Fig. 1). TP53 mutations in P01 and P06 were also truncal mutations (Fig. 1). In addition, ARID1A R1461* mutation was a truncal mutation in P05 (Fig. 1). The mutational status with variant allele frequencies for each patient is described in Supplementary Table S1.
TMB of IMAs
The mean TMB was 1.13/Mb and the TMB range was 0.33–1.63 Mb (Table 2). The TMB values of IMAs were considerably lower than those of non-small-cell lung cancer, which have been reported to be 3–8/Mb [39, 40].
Table 2.
Tumor mutational burden (TMB) of multifocal IMAs.
| Patient | Tumor location | TMB |
|---|---|---|
| P01 | RUL | 1.13 |
| RML | 0.92 | |
| RLL | 0.74 | |
| LUL | 1.04 | |
| LLL | 1.28 | |
| P03 | RLL1 | 0.86 |
| RLL2 | 0.89 | |
| RLL3 | 1.01 | |
| P04 | RLL | 0.77 |
| RUL1 | 1.10 | |
| RUL2 | 1.48 | |
| P05 | RLL1 | 1.39 |
| RLL2 | 1.63 | |
| RLL3 | 1.34 | |
| P06 | RUL | 1.04 |
| RML | 0.89 | |
| LUL | 1.13 | |
| LN | 1.25 | |
| P07 | LLL | 0.74 |
| RLL | 0.33 | |
| P08 | LLL1 | 0.56 |
| LLL2 | 0.86 | |
| LLL3 | 0.80 | |
| LLL4 | 0.98 |
RUL right upper lobe, RML right middle lobe, RLL right lower lobe, LUL left upper lobe, LLL left lower lobe, LN lymph node metastasis.
ITH index and Jaccard coefficient of IMAs
The tumors had a median ITH index of 68.1%, which was significantly higher than the previously reported 30% ITH index in lung adenocarcinoma (Table 3) [28, 29]. The median Jaccard similarity coefficient ranged from 0.16 to 0.58 except for P07 (Table 4). In P07, the Jaccard similarity coefficient was 0.01. P07’s synchronous tumors in the bilateral lower lobes had different truncal mutations (Fig. 1). The Jaccard similarity coefficient for the synchronous bilateral lower lobe tumors in P01 was 0.01, which also differed in terms of truncal events (Figs. 1 and 2).
Table 3.
Percentage of truncal and branch mutations in IMA patients.
| Branch mutations | Truncal mutations | Percentage of branch mutations (%) (branch mutations/total mutations) | |
|---|---|---|---|
| P01 | 37 | 37 | 50 |
| P03 | 30 | 31 | 49.1 |
| P04 | 112 | 24 | 82.3 |
| P05 | 68 | 36 | 65.4 |
| P06 | 78 | 25 | 75.7 |
| P07 | 42 | 0 | N/A |
| P08 | 41 | 17 | 70.7 |
Table 4.
Jaccard similarity coefficients based on exome sequencing.
| Patient | Tumor location | Median | |||||
|---|---|---|---|---|---|---|---|
| P01 | RUL | RML | RLL | LUL | LLL | ||
| RUL | 1.00 | 0.50 | |||||
| RML | 0.52 | 1.00 | |||||
| RLL | 0.01 | 0.01 | 1.00 | ||||
| LUL | 0.60 | 0.55 | 0.01 | 1.00 | |||
| LLL | 0.53 | 0.49 | 0.01 | 0.58 | 1.00 | ||
| P03 | RLL1 | RLL2 | RLL3 | ||||
| RLL1 | 1.00 | 0.52 | |||||
| RLL2 | 0.52 | 1.00 | |||||
| RLL3 | 0.59 | 0.48 | 1.00 | ||||
| P04 | RLL | RUL1 | RUL2 | ||||
| RLL | 1.00 | 0.26 | |||||
| RUL1 | 0.26 | 1.00 | |||||
| RUL2 | 0.21 | 0.38 | 1.00 | ||||
| P05 | RLL1 | RLL2 | RLL3 | ||||
| RLL1 | 1.00 | 0.29 | |||||
| RLL2 | 0.29 | 1.00 | |||||
| RLL3 | 0.30 | 0.27 | 1.00 | ||||
| P06 | RUL | RML | LUL | LN | |||
| RUL | 1.00 | 0.30 | |||||
| RML | 0.35 | 1.00 | |||||
| LUL | 0.27 | 0.26 | 1.00 | ||||
| LN | 0.34 | 0.32 | 0.23 | 1.00 | |||
| P07 | LLL | RLL | |||||
| LLL | 1.00 | 0.01 | |||||
| RLL | 0.01 | 1.00 | |||||
| P08 | LLL1 | LLL2 | LLL3 | LLL4 | |||
| LLL1 | 1.00 | 0.24 | |||||
| LLL2 | 0.20 | 1.00 | |||||
| LLL3 | 0.19 | 0.31 | 1.00 | ||||
| LLL4 | 0.16 | 0.47 | 0.28 | 1.00 | |||
RUL right upper lobe, RML right middle lobe, RLL right lower lobe, LUL left upper lobe, LLL left lower lobe, LN lymph node metastasis.
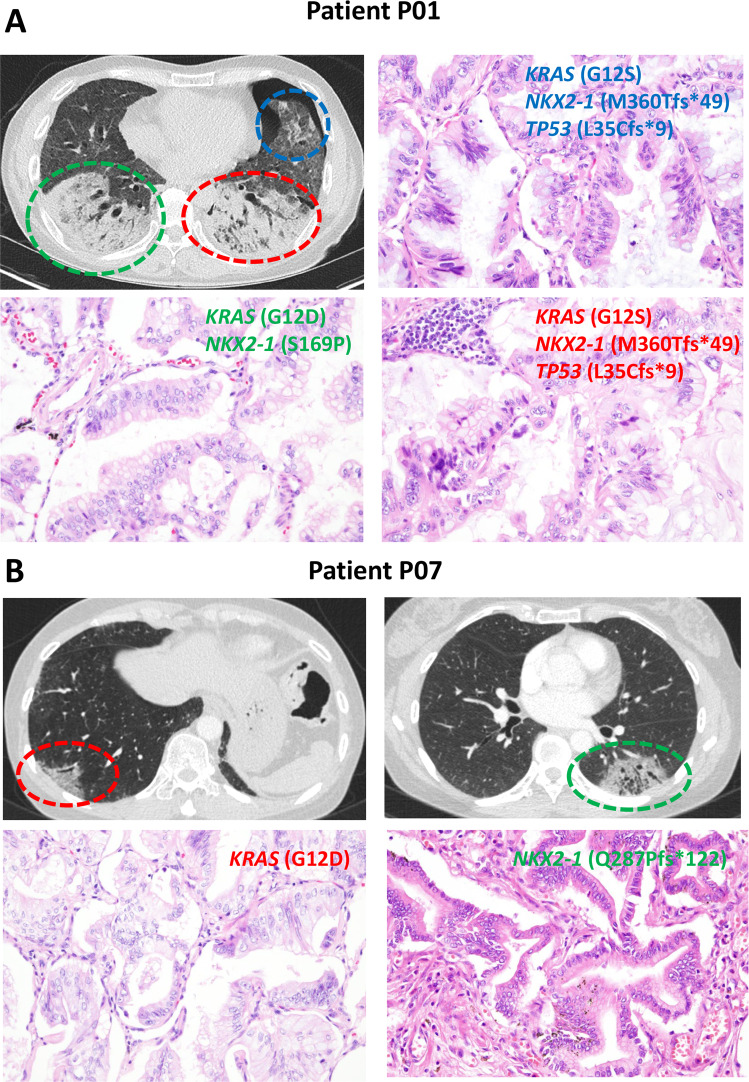
Fig. 2. Representative radiologic and histologic findings of IMAs in P01 and P07.
A The radiologic features of patient P01 show lobar consolidations with ground-glass opacities in bilateral lobes (right lower lobe-green color; left lower lobe-red color; left lingular segment-blue color). The tumors in the left lower lobe and left upper lobe share the same truncal mutations (KRAS G12S, NKX2-1 M360Tfs*49, and TP53 L35Cfs*9). The right lower lobe tumor is clonally distinct, harboring KRAS G12D and NKX2-1 S169P mutations. B In P07, a mass-like ground-glass opacity with traction bronchiolectasis and internal low attenuation is observed in the subpleural area of the left lower lobe (green color), and diffuse subpleural ground-glass opacity is observed in the subpleural areas of the right lower lobe (red color). These tumors are clonally unrelated. Original magnification of histologic sections, 20× objective.
Mutational signatures of IMAs
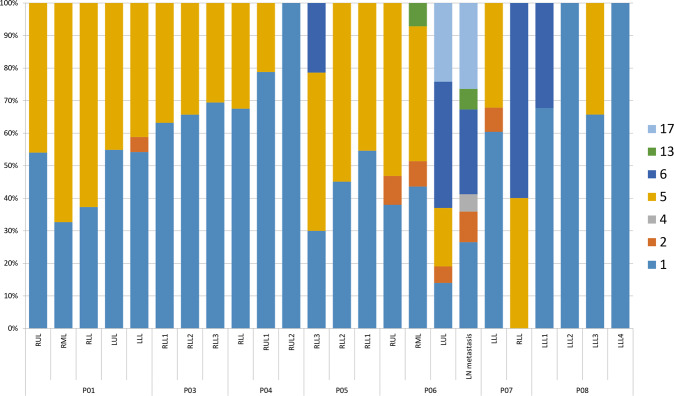
Among seven mutation signatures identified in our samples (Fig. 3), signature 1, the result of an endogenous mutational process, was found in almost all samples (95.8%, 23/24). Signatures 2 and 13, APOBEC-related signatures, were found in six samples (25.0%). Signature 6, associated with defective DNA mismatch repair (MMR), was found in five out of 24 samples (20.8%). Signature 4, a smoking-related signature, was observed in only one sample with a small portion.
Fig. 3. Mutational signatures of multifocal IMAs.
The right axis of the graph is arranged in accordance with the heatmap in Fig. 1. Signature 1 is the result of an endogenous mutational process. Signature 2 has been attributed to the activity of the AID/APOBEC family of cytidine deaminases. Signature 4 is associated with smoking. The etiology of signature 5 is unknown. Signature 6 is associated with defective DNA mismatch repair. Signature 13 has been attributed to the activity of the AID/APOBEC family of cytidine deaminases converting cytosine to uracil. The etiology of signature 17 remains unknown. Signature 1 is found in almost all samples (95.8%, 23/24). Signatures 2 and 13 were found in six samples (25.0%). Signature 6 was found in five out of 24 samples (20.8%).
CNVs of IMAs
Genome-wide CNVs showed distinct patterns between patients, and no recurrent CNVs were present in all sectors of the tumors (Supplementary Fig. S1, S2). CNVs observed in each patient provided the evidence of the clonal relationship in some cases. In P01, some CNVs (such as chromosome 12p duplication, and deletion of chromosome 17p and 19p) were identified in tumors of the left lower lobe, left upper lobe, and right upper lobe (Supplementary Fig. S2A). CNVs in chromosomes 9 and 14 were also present in all sectors of P05 (Supplementary Fig. S2B). In P06, loss of chromosome 9 and partial duplication of chromosomes 6, 8, and 17 were present in all tumor sectors (Supplementary Fig. S2C). We also found that amplifications of KRAS and ERBB2 gene were present only in the lymph node metastasis and right middle lobe of P06, respectively.
Discussion
We have characterized the genomic architecture of multifocal IMAs through multi-region whole exome sequencing. KRAS, NKX2-1, TP53, or ARID1A mutations were found in all multifocal IMAs, serving as genetic evidence for the occurrence of intrapulmonary metastases. KRAS mutations account for 60–70% of IMA driver mutations [5, 9, 41]. In this study’s cohort, various KRAS mutations (G12D, G12V, G12S, and Q61H) were identified, of which three were G12D and two were G12V. This number of instances was small, but their frequency was consistent with that found in previous studies [5, 9]. Recent studies on KRAS mutations have reported encouraging results [42]. IMA patients with KRAS G12D mutation responded exceptionally well to the proteasome inhibitor bortezomib [43]. Thus, it may present new therapeutic opportunities for patients with IMA.
Previous studies have reported NKX2-1 mutations in IMAs [6]. NKX2-1, also known as TTF- 1, plays role in lung morphogenesis and differentiation [44]. NKX2-1 is an organ-specific marker and is expressed in most lung primary adenocarcinomas, so its immunohistochemistry is commonly used to differentiate between primary lung tumors and metastatic neoplasm [45]. Recurrent NKX2-1 mutations have been observed in IMAs, which suggests that NKX2-1 plays a role in IMA carcinogenesis [6]. However, this study was the first to observe the truncal nature of NKX2-1 mutations. This study’s results suggest that NKX2-1 mutations may be involved in the early carcinogenesis of IMA. Of the five types of NKX2-1 mutations found in this study, four were frameshift mutations, which was consistent with the results of previous studies [6, 46]. Transgenic mice with inactivated Nkx2-1/Ttf-1 develop mucinous adenocarcinomas in the lungs that resemble human IMAs [47]. Taken together, these findings suggest that NKX2-1 mutations play a pivotal role in the development of IMA.
A nonsense mutation R1461* in ARID1A gene was found in all separate areas of P05, implying a truncal mutation. As Hung and colleagues reported, we thought that loss of function mutations of ARID1A is also clonal [48].
In this study, two patients presented with tumor involvement in their bilateral lower lobes. Unexpectedly, the genetic types of KRAS and NKX2-1 mutations differed in each lobe, suggesting that these tumors were clonally unrelated. Since no KRAS mutation was found in the left lower lobe tumor of patient P07, we went through the following confirmation process. First, we manually reviewed sequencing reads for the KRAS mutation, but no KRAS mutation was found. Second, we also performed pyrosequencing (Qiagen, Hilden, Germany) and PNA clamp-based real-time PCR (Panagene, Daejeon, Korea), but no KRAS mutation was found. Third, we further analyzed the mitochondrial mutations, and the results supported that the tumors in the lower lobes of P01 and P07 were clonally unrelated to each other (Supplementary Fig. S3). We have noticed that the two patients had background usual interstitial pneumonia. It is known that underlying usual interstitial pneumonia may predispose to the formation of multiple IMAs [49, 50]. This result indicates that synchronous multifocal IMAs can rarely occur in multicentric origins, especially in the background of fibrosing interstitial pneumonia. We recently experienced the same type of KRAS mutation (G12V) in a patient with metachronous multifocal IMAs involving the left lower lobe and then the right lower lobe (data not shown). Taking these findings together, caution is required when staging these tumors, and in some cases, mutational analysis can be helpful. We previously found that approximately 75% of IMA patients had primary masses in their lower lobes [41]. This result may indicate that IMAs, which spread to multiple lobes, including the lower lobes, are likely to originate from the lower lobe in probability. In this cohort, six out of seven patients (86%) had IMAs involving the lower lobe.
TP53 mutations were found in the IMAs of two patients who presented with the involvement of multiple lobes and were given poor prognoses. There is accumulating evidence that co-occurring mutations can affect the biological behavior of lung cancers [51]. Recent studies have found that treatment, patient prognosis, and responses to targeted therapy or immunotherapy may differ according to these subtypes [52, 53]. Although the number of cases is limited, the presence of TP53 mutations in IMA suggests a more aggressive biological behavior, as demonstrated in other lung cancers [54].
In this study, IMAs had a low somatic mutation burden, but they did have a heterogeneous genomic landscape, which was similar to Asian EGFR-mutant lung adenocarcinomas [54]. In contrast, Caucasian lung cancers have a high-mutation burden and low intra-tumor heterogeneity [28, 29]. The median proportion of branch mutation in smoker-dominated Caucasian lung adenocarcinomas was about 30%. In contrast, the proportion of branch mutation in Asian EGFR-mutant adenocarcinomas was 62.3%, which was similar to that of IMAs in our cohort. In addition, from TMB analysis, it appears even clonally related tumors can differ somewhat in TMB counts. Both intratumor genetic heterogeneity and tumor purity can affect the TMB counts [55]. In this study, since whole exome sequencing was performed on samples having a tumor purity of 20% or more, we thought that the main factor affecting TMB counts was intratumor genetic heterogeneity. However, it cannot be excluded that the low tumor purity may have affected the TMB counts in some cases.
The mutational signature analysis found that IMAs were predominantly associated with endogenous mutational process (signature 1), APOBEC activity (signatures 2 and 13), and MMR deficiency (signature 6), not related to smoking. These findings suggest that IMAs are distinguished from conventional adenocarcinomas or KRAS-mutant non-mucinous adenocarcinomas, which are closely related to smoking signatures [56]. In addition, the prevalence of APOBEC-related signatures can contribute to subclonal diversification and ITH of IMAs found in this study [30]. Interestingly, MMR deficiency signature was found in five of 24 areas. We performed immunohistochemistry for MMR proteins (MLH1, MSH2, MSH6, and PMS2) and microsatellite instability (MSI) RT-PCR test using U-TOP MSI Detection Kit (SEASUN Biomaterials, Daejeon, Korea) for those samples. No MSI-high tumors were found, and all MMR proteins were preserved. According to reports so far, the frequency of MSI-high in lung cancer is very low, ranging from 0.5 to 0.8% [57, 58]. On the other hand, the MMR deficiency signature has been reported in in situ or early lung adenocarcinoma, and lung adenocarcinomas with MMR deficiency signature were reported to have a better prognosis compared to tumors with APOBEC signature [59]. We think that it is necessary to distinguish between a tumor with MMR deficiency signature and an MSI-high tumor. Because these cases are not an MSI-high tumor, it is not considered to be a candidate for cancer immunotherapy. Further studies are necessary to elucidate the clinical and biological significance of MMR deficiency signature in lung cancers.
This study has several limitations. Due to the limited sample size, the clinical impact or significance of genomic findings in multifocal IMAs has not been investigated. Also, since low tumor purity is a well-known issue with the sequencing of IMAs, the possibility that low tumor purity may have affected the genomic results in some cases cannot be completely excluded.
In conclusion, genomic evidence found in this study indicated the clonal intrapulmonary spread of diffuse pneumonic-type or multifocal IMAs, although they can occur in multicentric origins in the background of usual interstitial pneumonia. IMAs exhibited a heterogeneous genomic landscape despite the low somatic mutation burden. Further studies are warranted to determine the clinical significance of the genomic characteristics of IMAs in expanded cohorts.
Supplementary information
Author contributions
HSS performed study concept and design; MK, JH, KAK, SH, HJL, and HSS performed development of methodology and writing; JYJ, JGL, YJC, and HSS provided samples and acquisition of data; all authors performed review and revision of the paper; all authors read and approved the final paper.
Funding
This research was supported by a faculty research grant of Yonsei University College of Medicine (6-2020-0105) and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF- 2018R1D1A1B07047811).
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Competing interests
The authors declare no competing interests.
Ethics approval
The institutional review board of Severance Hospital approved this retrospective study (No. 4-2019-0468). Written informed consent from the patients was waived due to the retrospective nature of the study. The study was performed in accordance with the Declaration of Helsinki.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: M. Kim, J. Hwang.
Supplementary information
The online version contains supplementary material available at 10.1038/s41379-021-00872-0.
References
- 1.Reck M, Rabe KF. Precision diagnosis and treatment for advanced non-small-cell lung cancer. N Engl J Med. 2017;377:849–61. doi: 10.1056/NEJMra1703413. [DOI] [PubMed] [Google Scholar]
- 2.Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, et al. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:1243–60. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 3.Cha YJ, Shim HS. Biology of invasive mucinous adenocarcinoma of the lung. Transl Lung Cancer Res. 2017;6:508–12. doi: 10.21037/tlcr.2017.06.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Detterbeck FC, Nicholson AG, Franklin WA, Marom EM, Travis WD, Girard N, et al. The IASLC lung cancer staging project: summary of proposals for revisions of the classification of lung cancers with multiple pulmonary sites of involvement in the forthcoming eighth edition of the TNM classification. J Thorac Oncol. 2016;11:639–50. doi: 10.1016/j.jtho.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 5.Shim HS, Kenudson M, Zheng Z, Liebers M, Cha YJ, Hoang Ho Q,, et al. Unique genetic and survival characteristics of invasive mucinous adenocarcinoma of the lung. J Thorac Oncol. 2015;10:1156–62. doi: 10.1097/JTO.0000000000000579. [DOI] [PubMed] [Google Scholar]
- 6.Hwang DH, Sholl LM, Rojas-Rudilla V, Hall DL, Shivdasani P, Garcia EP, et al. KRAS and NKX2-1 mutations in invasive mucinous adenocarcinoma of the lung. J Thorac Oncol. 2016;11:496–503. doi: 10.1016/j.jtho.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 7.Kishikawa S, Hayashi T, Saito T, Takamochi K, Kohsaka S, Sano K, et al. Diffuse expression of MUC6 defines a distinct clinicopathological subset of pulmonary invasive mucinous adenocarcinoma. Mod Pathol. 2021;34:786–97. doi: 10.1038/s41379-020-00690-w. [DOI] [PubMed] [Google Scholar]
- 8.Maeshima A, Sakamoto M, Hirohashi S. Mixed mucinous-type and non-mucinous-type adenocarcinoma of the lung: immunohistochemical examination and K-ras gene mutation. Virchows Arch. 2002;440:598–603. doi: 10.1007/s00428-002-0629-6. [DOI] [PubMed] [Google Scholar]
- 9.Chang JC, Offin M, Falcon C, Brown D, Houck-Loomis BR, Meng F, et al. Comprehensive molecular and clinicopathologic analysis of 200 pulmonary invasive mucinous adenocarcinomas identifies distinct characteristics of molecular subtypes. Clin Cancer Res. 2021;27:4066–76. [DOI] [PMC free article] [PubMed]
- 10.Murayama T, Nakaoku T, Enari M, Nishimura T, Tominaga K, Nakata A, et al. Oncogenic fusion gene CD74-NRG1 confers cancer stem cell-like properties in lung cancer through a IGF2 autocrine/paracrine circuit. Cancer Res. 2016;76:974–83. doi: 10.1158/0008-5472.CAN-15-2135. [DOI] [PubMed] [Google Scholar]
- 11.Duruisseaux M, McLeer-Florin A, Antoine M, Alavizadeh S, Poulot V, Lacave R, et al. NRG1 fusion in a French cohort of invasive mucinous lung adenocarcinoma. Cancer Med. 2016;5:3579–85. doi: 10.1002/cam4.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung Y, Yong S, Kim P, Lee HY, Jung Y, Keum J, et al. VAMP2-NRG1 fusion gene is a novel oncogenic driver of non-small-cell lung adenocarcinoma. J Thorac Oncol. 2015;10:1107–11. doi: 10.1097/JTO.0000000000000544. [DOI] [PubMed] [Google Scholar]
- 13.Lee HY, Lee KS, Han J, Kim BT, Cho YS, Shim YM, et al. Mucinous versus nonmucinous solitary pulmonary nodular bronchioloalveolar carcinoma: CT and FDG PET findings and pathologic comparisons. Lung Cancer. 2009;65:170–5. doi: 10.1016/j.lungcan.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Miyake H, Matsumoto A, Terada A, Yoshida S, Takaki H, Mori H. Mucin-producing tumor of the lung: CT findings. J Thorac Imaging. 1995;10:96–8. doi: 10.1097/00005382-199521000-00003. [DOI] [PubMed] [Google Scholar]
- 15.Yang SR, Chang JC, Leduc C, Tan KS, Dogan S, Benayed R, et al. Invasive mucinous adenocarcinomas with spatially separate lung lesions: analysis of clonal relationship by comparative molecular profiling. J Thorac Oncol. 2021;16:1188–99. doi: 10.1016/j.jtho.2021.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Durbin R. Fast and accurate short read alignment with Burrows−Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31:2032–4. doi: 10.1093/bioinformatics/btv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–43. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–8. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 24.Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16:2598–608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Büttner R, Longshore JW, López-Ríos F, Merkelbach-Bruse S, Normanno N, Rouleau E, et al. Implementing TMB measurement in clinical practice: considerations on assay requirements. ESMO open. 2019;4:e000442. doi: 10.1136/esmoopen-2018-000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–13. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang H, Sasson A, Srinivasan S, Golhar R, Greenawalt DM, Geese WJ, et al. Bioinformatic methods and bridging of assay results for reliable tumor mutational burden assessment in non-small-cell lung cancer. Mol Diagn Ther. 2019;23:507–20. doi: 10.1007/s40291-019-00408-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256–9. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–6. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roper N, Gao S, Maity TK, Banday AR, Zhang X, Venugopalan A, et al. APOBEC mutagenesis and copy-number alterations are drivers of proteogenomic tumor evolution and heterogeneity in metastatic thoracic tumors. Cell Rep. 2019;26:2651–2666.e2656. doi: 10.1016/j.celrep.2019.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi W, Ng CKY, Lim RS, Jiang T, Kumar S, Li X, et al. Reliability of whole-exome sequencing for assessing intratumor genetic heterogeneity. Cell Rep. 2018;25:1446–57. doi: 10.1016/j.celrep.2018.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makohon-Moore AP, Zhang M, Reiter JG, Bozic I, Allen B, Kundu D, et al. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat Genet. 2017;49:358–66. doi: 10.1038/ng.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levandowsky M, Winter D. Distance between sets. Nature. 1971;234:34–5. [Google Scholar]
- 34.Lee JJ, Park S, Park H, Kim S, Lee J, Lee J, et al. Tracing oncogene rearrangements in the mutational history of lung adenocarcinoma. Cell. 2019;177:1842–1857.e1821. doi: 10.1016/j.cell.2019.05.013. [DOI] [PubMed] [Google Scholar]
- 35.Rosales RA, Drummond RD, Valieris R, Dias-Neto E, da Silva IT. signeR: an empirical Bayesian approach to mutational signature discovery. Bioinformatics. 2017;33:8–16. doi: 10.1093/bioinformatics/btw572. [DOI] [PubMed] [Google Scholar]
- 36.Lee J, Lee AJ, Lee JK, Park J, Kwon Y, Park S, et al. Mutalisk: a web-based somatic MUTation AnaLyIS toolKit for genomic, transcriptional, and epigenomic signatures. Nucleic Acids Res. 2018;46:W102–w108. doi: 10.1093/nar/gky406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016;12:e1004873. doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bao L, Pu M, Messer K. AbsCN-seq: a statistical method to estimate tumor purity, ploidy, and absolute copy numbers from next-generation sequencing data. Bioinformatics. 2014;30:1056–63. doi: 10.1093/bioinformatics/btt759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berland L, Heeke S, Humbert O, Macocco A, Long-Mira E, Lassalle S, et al. Current views on tumor mutational burden in patients with non-small cell lung cancer treated by immune checkpoint inhibitors. J Thorac Dis. 2019;11:S71–s80. doi: 10.21037/jtd.2018.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alexander M, Galeas J, Cheng H. Tumor mutation burden in lung cancer: a new predictive biomarker for immunotherapy or too soon to tell? J Thorac Dis. 2018;10:S3994–s3998. doi: 10.21037/jtd.2018.09.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cha YJ, Kim HR, Lee HJ, Cho BC, Shim HS. Clinical course of stage IV invasive mucinous adenocarcinoma of the lung. Lung Cancer. 2016;102:82–8. doi: 10.1016/j.lungcan.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 42.Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217–23. doi: 10.1038/s41586-019-1694-1. [DOI] [PubMed] [Google Scholar]
- 43.Drilon A, Schoenfeld AJ, Arbour KC, Litvak A, Ni A, Montecalvo J, et al. Exceptional responders with invasive mucinous adenocarcinomas: a phase 2 trial of bortezomib in patients with KRAS G12D-mutant lung cancers. Cold Spring Harb Mol Case Stud. 2019;5:a003665. [DOI] [PMC free article] [PubMed]
- 44.Kimura S. Thyroid-specific enhancer-binding protein role in thyroid function and organogenesis. Trends Endocrinol Metab. 1996;7:247–52. doi: 10.1016/s1043-2760(96)00115-4. [DOI] [PubMed] [Google Scholar]
- 45.Moldvay J, Jackel M, Bogos K, Soltész I, Agócs L, Kovács G, et al. The role of TTF-1 in differentiating primary and metastatic lung adenocarcinomas. Pathol Oncol Res. 2004;10:85–8. doi: 10.1007/BF02893461. [DOI] [PubMed] [Google Scholar]
- 46.Matsubara D, Soda M, Yoshimoto T, Amano Y, Sakuma Y, Yamato A, et al. Inactivating mutations and hypermethylation of the NKX2-1/TTF-1 gene in non- terminal respiratory unit-type lung adenocarcinomas. Cancer Sci. 2017;108:1888–96. doi: 10.1111/cas.13313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snyder EL, Watanabe H, Magendantz M, Hoersch S, Chen TA, Wang DG, et al. Nkx2-1 represses a latent gastric differentiation program in lung adenocarcinoma. Mol Cell. 2013;50:185–99. doi: 10.1016/j.molcel.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hung YP, Redig A, Hornick JL, Sholl LM. ARID1A mutations and expression loss in non-small cell lung carcinomas: clinicopathologic and molecular analysis. Mod Pathol. 2020;33:2256–68. doi: 10.1038/s41379-020-0592-2. [DOI] [PubMed] [Google Scholar]
- 49.Masai K, Tsuta K, Motoi N, Shiraishi K, Furuta K, Suzuki S, et al. Clinicopathological, immunohistochemical, and genetic features of primary lung adenocarcinoma occurring in the setting of usual interstitial pneumonia pattern. J Thorac Oncol. 2016;11:2141–9. doi: 10.1016/j.jtho.2016.07.034. [DOI] [PubMed] [Google Scholar]
- 50.Honda T, Sakashita H, Masai K, Totsuka H, Motoi N, Kobayashi M, et al. Deleterious pulmonary surfactant system gene mutations in lung adenocarcinomas associated with usual interstitial pneumonia. JCO Precis Oncol. 2018;2:1–24. doi: 10.1200/PO.17.00301. [DOI] [PubMed] [Google Scholar]
- 51.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5:860–77. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer. 2019;19:495–509. doi: 10.1038/s41568-019-0179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arbour KC, Jordan E, Kim HR, Dienstag J, Yu HA, Sanchez-Vega F, et al. Effects of co-occurring genomic alterations on outcomes in patients with KRAS-mutant non-small cell lung cancer. Clin Cancer Res. 2018;24:334–40. doi: 10.1158/1078-0432.CCR-17-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nahar R, Zhai W, Zhang T, Takano A, Khng AJ, Lee YY, et al. Elucidating the genomic architecture of Asian EGFR-mutant lung adenocarcinoma through multi-region exome sequencing. Nat Commun. 2018;9:216. doi: 10.1038/s41467-017-02584-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kazdal D, Endris V, Allgäuer M, Kriegsmann M, Leichsenring J, Volckmar AL, et al. Spatial and temporal heterogeneity of panel-based tumor mutational burden in pulmonary adenocarcinoma: separating biology from technical artifacts. J Thorac Oncol. 2019;14:1935–47. doi: 10.1016/j.jtho.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 56.Alexandrov LB, Ju YS, Haase K, Van Loo P, Martincorena I, Nik-Zainal S, et al. Mutational signatures associated with tobacco smoking in human cancer. Science. 2016;354:618–22. doi: 10.1126/science.aag0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warth A, Körner S, Penzel R, Muley T, Dienemann H, Schirmacher P, et al. Microsatellite instability in pulmonary adenocarcinomas: a comprehensive study of 480 cases. Virchows Arch. 2016;468:313–9. doi: 10.1007/s00428-015-1892-7. [DOI] [PubMed] [Google Scholar]
- 58.Bonneville R, Krook MA, Kautto EA, Miya J, Wing MR, Chen HZ, et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol. 2017;1:PO.17.00073. [DOI] [PMC free article] [PubMed]
- 59.Qian J, Zhao S, Zou Y, Rahman SMJ, Senosain MF, Stricker T, et al. Genomic underpinnings of tumor behavior in in situ and early lung adenocarcinoma. Am J Respir Crit Care Med. 2020;201:697–706. doi: 10.1164/rccm.201902-0294OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.