Summary
The vascular endothelium is a hot spot in the response to radiation therapy for both tumors and normal tissues. To improve patient outcomes, interpretable systemic hypotheses are needed to help radiobiologists and radiation oncologists propose endothelial targets that could protect normal tissues from the adverse effects of radiation therapy and/or enhance its antitumor potential. To this end, we captured the kinetics of multi-omics layers—i.e. miRNome, targeted transcriptome, proteome, and metabolome—in irradiated primary human endothelial cells cultured in vitro. We then designed a strategy of deep learning as in convolutional graph networks that facilitates unsupervised high-level feature extraction of important omics data to learn how ionizing radiation-induced endothelial dysfunction may evolve over time. Last, we present experimental data showing that some of the features identified using our approach are involved in the alteration of angiogenesis by ionizing radiation.
Subject areas: Radiation biology, Systems biology, Omics, Proteomics, Metabolomics, Transcriptomics
Graphical abstract

Highlights
-
•
Irradiation promotes cell death, senescence, and impaired angiogenesis in HUVECs
-
•
Multi-omics analyses enable tracking pathways from early to late dysfunction
-
•
Dynamic modeling allows accurate learning of essential dysfunction checkpoints
-
•
Loss-/gain-of-function experiments identify key players of angiogenesis in vitro
Radiation biology; Systems biology; Omics; Proteomics; Metabolomics; Transcriptomics
Introduction
Radiation therapy, alone or in combination with other therapeutic strategies, is one of the most effective treatments for cancer. However, the radioresistance of tumors and the toxicity to healthy tissues present in the irradiation field are two major limitations regarding this treatment. Despite consistent improvements in ballistic configurations, there is still a point of no return at dose-volume thresholds that cannot be crossed without severely altering the patient's quality of life.
Profoundly modified by ionizing radiation, the vascular endothelium is in the spotlight to improve the overall benefit/risk ratio of radiation therapy (Guipaud et al., 2018; Wijerathne et al., 2021). New interdisciplinary approaches are useful to better understand radiation-induced biological effects and thus optimize radiation therapy outcomes for patients. These approaches can provide a comprehensive understanding of the response of the different cellular compartments involved in normal tissue and tumor responses and could therefore increase the likelihood of cancer cure and/or decrease toxicity to normal tissue. Molecular changes after exposure to ionizing radiation are numerous and of multiple types. They include molecular changes at the transcriptional, translational, and post-translational levels, phenotypic alterations, as well as deregulation of the microenvironment through the production and secretion of soluble factors such as reactive oxygen and nitrogen species, chemokines, cytokines, and growth factors. The cascade of molecular alterations triggered by irradiation may determine the phenotype of endothelial cells and alter the function of the vasculature, thereby contributing to the initiation and progression of radiation-induced damage in healthy organs and/or causing the recruitment of immune cells into tumors following radiation therapy (Guipaud et al., 2018; Korpela and Liu, 2014). In this regard, recent cancer therapies have successfully combined an effective immune response with radiation therapy (Zitvogel and Kroemer, 2015). On the other hand, strategies based on the endothelium for therapeutic gain in radiation oncology attempt to protect the vasculature of normal tissues from damage due to radiation, for instance by protecting endothelial cells from death, or by inhibiting vascular inflammation and endothelial cell activation. Because early alterations of endothelial cells by ionizing radiation are associated with late endothelial dysfunction (Rannou et al., 2015; Toullec et al., 2018), it is critical to better understand the continuum and dynamics of molecular changes that propagate over time and lead to vascular injury.
In this work, we propose a systemic analysis of ionizing radiation-induced dysfunction in vascular endothelial cells using topological data analysis (Zomorodian and Carlsson, 2004) and deep learning on graphs (Hamilton et al., 2017a, b). Both of these approaches are enhanced by systems biology methods (Heinonen et al., 2015, 2018; Herskind et al., 2016) that significantly improve the characterization of biological processes across the heterogeneous samples of omics layers assembled for this study. These datasets describe the proteome, targeted transcriptome, metabolome, and miRNome of primary human endothelial cells grown in vitro, and the results are aligned with experimental validation steps. Briefly, this interdisciplinary approach was carried out in three main steps. First, we defined two feature representations of the data, i.e. early and late cell dysfunction based on the proximity of the sample to the response of vascular endothelial cells to irradiation over time at an irradiation dose of 20 Gy. This distinction allowed us to identify the molecular signatures of vascular dysfunction on each dataset. To do this, we used persistent homology algorithms based on discrete Morse theory (Scoville, 2019). We then integrated each signature into a global gene regulatory network (OGRN) (Chen and Mar, 2018), which led to identification of the global molecular signature. Last, this network was dynamically modeled using semi-supervised learning of the two feature representations (Ding et al., 2018), providing the top five candidates per feature for experimental validation. Ultimately, this methodology improves interpretable molecular hypotheses, providing radiobiologists with a systems biology approach useful for proposing potential endothelial targets to protect normal tissues from radiation and/or increase tumor control with radiation therapy.
Results
In this work, we sought to exploit temporal molecular data from unirradiated and irradiated primary human endothelial cells (HUVECs) to identify key molecular players involved in the development of radiation-induced vascular dysfunction. To this end, we analyzed four levels of molecules, namely miRNAs, transcripts, proteins, and metabolites, to better understand the molecular response of HUVECs (Figure 1). The following results were derived from a systematic modeling approach encompassing multifaceted high-throughput screening, topological data analysis, homology, functional analysis, regulatory network reconstruction, and dynamic learning based on convolutional networks. We first present the recurrent temporal profiles of the data that were projected onto a 3D space where we found topologically important modules by homology. We then applied a network projection theorem capturing such homology in a practical way. Next, we analyzed the functional systems that the components of these modules deregulate. We subsequently considered the integration of molecular entities most likely to act as regulators in the molecular pathways of interest. This regulatory network helped us to understand the dynamics of the endothelial cell response to irradiation by constructing convolutional networks associated with each of the two characteristics considered, i.e. early and late responses. Last, we highlight an important biological process altered by radiation exposure, namely angiogenesis, and propose candidate genes to target in order to improve the effects of ionizing radiation.
Figure 1.
The main workflow leveraged in this manuscript
Time vectors were defined based on endothelial cell response measurements and omics data collection over time, i.e. miRNAs, targeted transcripts, proteins, and metabolites. Then, projection of omics layers in a lower space through t-SNE were used to seek recurrent homology between data by means of alpha complex building, enabling the construction of complex networks and determination of average data expression per homological module. Functional analysis of the identified signature and progression over time of the signature depending on the early or late response of the endothelial cells to irradiation were explored using deep learning modeling to propose targets of interest in connection with a cellular process altered by irradiation. Finally, candidates were studied experimentally using gain-of-function and loss-of-function strategies.
Behavior of HUVECs postirradiation
After irradiation at 20 Gy, the cell layer became progressively less dense, likely due to death-induced cell detachment (Figure 2A). The dose of 20 Gy led to the loss of about 66% of the cells at day 4, 80% at day 7, 86% at day 14, and 93% at day 21, as compared with the nonirradiated cells (Figure 2C). At the same time, the cells became larger and displayed a higher amount of proteins and RNA per cell (Figures 2B and 2C), suggesting that they were still metabolically active. At 21 days after 20 Gy, on average 85% (40% at day 7) of the remaining cells were β-gal positive (Figure 2D), suggesting that almost all cells were senescent at this time point after irradiation.
Figure 2.
Endothelial cell behavior over time after irradiation: confluence, shape, number, volume, RNA and protein contents, and senescence
(A) Representative images of HUVECs on the day of irradiation and 4, 7, 14, and 21 days after irradiation at 20 Gy compared with nonirradiated cells. Scale bar, 50 μm.
(B) Estimation of the cell volumes of HUVECs after irradiation at 0, 2, and 20 Gy. n = 12 technical replicates.
(C) Estimation of changes in cell volume, number, and RNA and protein contents of HUVECs at different time points after irradiation at at 20 Gy compared with nonirradiated cells.
(D) Estimation of the proportion of senescent cells 7 and 21 days after irradiation at 0, 2, and 20 Gy. At the indicated time following 0, 2, or 20 Gy, cells were stained using a β-Gal Staining Kit. N = 4 technical replicates. One-way ANOVA test with Tukey correction. ns, not significant, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. Representative images of HUVECs 21 days after irradiation are displayed on the right of the panel. Scale bar, 50 μm.
Dataset time profiles
We collected 4 omics datasets from unexposed HUVECS and HUVECS exposed to 20 Gy of ionizing radiation (12 h–21 days after irradiation, 8 time points) (Figure 1). For the miRNomics and the transcriptomics studies, we focused on microRNAs (miRNAs) and genes of interest using TaqMan array microfluidic cards to assess levels of expression using RT-qPCR. We thus generated expression matrices containing the 2−ΔCt values of 293 miRNAs (Data S1) and 308 gene transcripts (Data S2) over time. For the proteomics study, we performed quantitative iTRAQ LC-MS/MS analysis following isoelectric point-based peptide fractionation. This enabled the identification and quantification as a ratio of 4 exposed to 4 unexposed samples associated to a credible interval of about 1,700–2,800 proteins per time point (Data S3). For the metabolomics study, we achieved semi-quantitative ultra-high-performance LC-MS/MS. Using the LC-MS annotation with bank in-house tab enabled the quantification and identification of 115 metabolites for each technical replicate and each time point (Data S4).
From the 3D t-SNE projection of the clustered omics datasets (Figure S1A), we can visualize three differentiated time vectors depending on sample closeness to the response of HUVECs to irradiation. Thus, we defined T1 as the acute vector composed by the measures captured in the range between days 0 and 7 postirradiation, T2 as the mid-acute vector captured on day 14 postirradiation, and T3 as the late response captured on day 21 postirradiation. Data were then translated into sets of NIR (nonirradiated) and IR (irradiated) elements describing the number of samples captured at each time depending on the irradiation dose and the time vector, i.e. Tk k = 1, 2, or 3, in which they were placed.
Tracking the pathway from early and nonirradiated to late endothelial dysfunction
Next, evaluating the barcodes associated with each omics dataset allowed us to accurately assign an optimal number of persistent modules (Figure S1B), herein referred to as p-cubes, to each mapper graph (see Videos S1, S2–S5). This assignment was also confirmed by checking the landscape and silhouette distributions of each omics barcode (Figures S1C and S2). We could then locally characterize each mapper graph as NIR or early cell dysfunction versus IR or late cell dysfunction regions. This distinction is based on the homology score average displayed by the molecular entities that compose the p-cubes.
In particular, the proteomics mapper graph (Video S3) is determined by 7 p-cubes. This relatively high number of cubes mainly traces the full path going from early to late endothelial cell dysfunction following a dose of 20 Gy. In contrast, the other omics datasets showed a much lower number of p-cubes: the transcriptome by 3 p-cubes (Video S4), the miRNome, and the metabolome by 2 p-cubes (respectively Videos S2 and S5). From Videos S2–S5, we observe how the mapper graph is described by p-cubes tracking the path going from NIR to early IR. This trajectory is homologically relevant in terms of the time vectors Ts for a dose of 20 Gy. The miRNA mapper graph is characterized by NIR to mid-IR full time path, also associated with a dose of 20 Gy. Next, the transcriptomics mapper graph, mainly associated to 20 Gy irradiation, aptly describes the transition from NIR to late-IR with a temporary delayed pass by early IR cell dysfunction.
Global molecular signature of the irradiated endothelium
Molecules in each omics dataset subject to functional reprogramming were identified by considering overlapping entities in the p-cubes assembly performed by the mapper algorithm. Subsequently, a thorough examination of the mapper graphs described in the previous section identified 83 candidate checkpoints controlling the response of endothelial cells to irradiation (Table 1). We inferred miRNA targets from the TarBase database, one of the embedded databases in DIANA webtools, leading to the identification of the CDKN2A gene as a target of hsa-miR-10b-5p, which is strongly linked to radiation-induced senescence in vitro and in vivo (Lafargue et al., 2017; Soysouvanh et al., 2020).
Table 1.
Proposed checkpoints controlling the response of endothelial cells to irradiation in each omic layer
| miRNA | Description/alias | Transcript | Description/alias | Protein | Description/alias | Metaboliteb | Description/alias |
|---|---|---|---|---|---|---|---|
| hsa-miR-10ba | hsa-miR-10b-5p | ADAMTS1 | ADAM metallopeptidase with thrombospondin type 1 motif 1 | AHCY | Adenosylhomocysteinase | Alanine |
D-arabino-1,4-lactone |
| hsa-let-7b-5p | ALDOA | Aldolase, fructose-bisphosphate A | Aminoadipic acid | ||||
| hsa-let-7d-5p | CDCA3 | Cell-division-cycle-associated protein 3 | AMP | ||||
| hsa-miR-15b-5p | BHLHA15 | Basic helix-loop-helix family member a15 (also named BHLHB8) | DDX6 | DEAD-box helicase 6 | Aspartate | ||
| EIF4E | Eukaryotic translation initiation factor 4E | D-arabinonic acid | |||||
| hsa-miR-16-5p | ESD | Esterase D | |||||
| hsa-miR181a | |||||||
| hsa-miR181c | CD44 | CD44 molecule (Indian blood group) | FGFR1 | Fibroblast growth factor receptor 1 | Fumarate |
L-leucine |
|
| hsa-miR-191-5p | FN1 | Fibronectin 1 | Galactitol | ||||
| hsa-miR-19a-3p | EPHB4 | EPH receptor B4 | GANAB | Glucosidase II alpha subunit | Guanosine | ||
| hsa-miR-210-3p | FABP5 | Fatty acid binding protein 5 | GPX1 | Glutathione peroxidase 1 | Leucine | ||
| hsa-miR-218-5p | GRB2 | Growth factor receptor bound protein 2 | Maleic acid | ||||
| hsa-miR26a-11 | hsa-miR-26a-1- | IL18 | Interleukin 18 | HBA1 | Hemoglobin subunit alpha 1 | Methionine |
N-methylaspartate |
| 3p | LRRC2 | Leucine rich repeat containing 2 | HMGB1 | High-mobility group box 1 | N-acetylserine | ||
| hsa-miR-26b1 | hsa-miR-26b-3p | LRPPRC | Leucine-rich pentatricopeptide repeat-containing protein | NMDA | |||
| hsa-miR-320a-3p | Pantothenic acid | ||||||
| hsa-miR-365a-5p | MAPK14 | Mitogen-activated protein kinase 14 | MCTS1 | MCTS1 re-initiation and release factor | PPi | ||
| hsa-miR-374a-5p |
L-tryptophan |
||||||
| hsa-miR-532-3p | MCL1 | MCL1 apoptosis regulator, BCL2 family member | MMRN1 | Multimerin 1 | Tryptophan | ||
| hsa-miR-5582-3p | NUTF2 | Nuclear transport factor 2 | |||||
| hsa-miR-92a-3p | PABPC4 | Poly(A)-binding protein cytoplasmic 4 | |||||
| NFKBIA | NFKB inhibitor alpha | PLOD1 | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 | ||||
| PDE4B | Phosphodiesterase 4B | ||||||
| SELP | Selectin P | PSMB3 | Proteasome 20S subunit beta 3 | ||||
| SMAD3 | SMAD family member 3 | RPL13 | Ribosomal protein L13 | ||||
| TIMP3 | TIMP metallopeptidase inhibitor 3 | RPL24 | Ribosomal protein L24 | ||||
| RPL26 | Ribosomal protein L26 | ||||||
| TNFSF10 | TNF superfamily member 10 | RTN4 | Reticulon 4 | ||||
| SERPINE1 | Serpin family E member 1 | ||||||
| SPARC | Secreted protein acidic and cysteine rich | ||||||
| TAGLN2 | Transgelin 2 | ||||||
| TMEM263 | Transmembrane protein 263 | ||||||
| TPM1 | Tropomyosin 1 | ||||||
| TPM2 | Tropomyosin 2 | ||||||
| TUBB6 | Tubulin beta 6 class V | ||||||
| VDAC1 | Voltage dependent anion channel 1 | ||||||
| VIM | Vimentin |
Complementary arm of the “reference” miRNA; those are originated from the same hairpin structure (pri- and pre-miRNA). When the data are not sufficient to determine which sequence is the predominant one, the majority sense in DIANA database is considered and put inside brackets (see Supplemental Information text for further details on their gene targets).
See Supplemental Information text for further details on their associated enzymes.
Highly altered biological functions
We used the subnetwork enrichment analysis module of Pathway Studio software to search for enriched regulated processes for each of the four sets of checkpoint candidates learnt from dynamic modeling. The proteome graph mapping is largely characterized by key players implied in the earlier processes of vascular endothelium injuries caused by ionizing radiation. Mainly, there is an overabundance of entities related to cell death, cytotoxicity, ROS generation, and oxidative stress as well as senescence. Other more unspecific biological processes to describe vascular endothelium injuries such as endothelial cell proliferation, cell regeneration, and cell infiltration were also reported. Similarly, as for the proteome, general terms indicating cytotoxicity (cell damage), senescence (aging), endothelial cell proliferation, and cell infiltration (cell adhesion) are also screened in the transcriptome graph mapping. Groups of transcripts enriched in cell processes such as angiogenesis, inflammatory, and immune responses are reported. However, other processes mainly involved in neovascularization and proinflammatory phenotype are also highlighted. Similarly, the miRNA graph mapping records and displays an enrichment in entities related to inflammatory response as well as cell growth, cell death, cell differentiation, and angiogenesis. It also shows an alternative endothelial/epithelial-to-mesenchymal transition as typically occurs in fibrosis (Morilla et al., 2017). Further, terms describing the early stages of endothelial injury, such as cell proliferation, death, or senescence, seem to be characterized by cell-cycle dysregulation (G0/G1 and G1/S) and double-strand DNA repair. Last, the graph mapping associated with the metabolome contains metabolites enriched in the set of biological processes describing endothelial damage after irradiation. Supporting this, we found cellular processes associated with early stages of dysfunction, such as regulators of ROS generation, inflammatory response, and oxidative stress.
From the list of 83 candidate checkpoint entities obtained by dynamic modeling, we searched for clusters of molecular entities enriched in gene ontology (GO) annotations that ideally would be involved in common biological processes relevant to the radiation-induced response of endothelial cells. We have pinpointed the top 25 enriched GO cell process categories identified in this study by excluding overly general biological cell processes (Figure S3). An expected enrichment of some GO terms is related to death and programmed cell death (apoptosis), in agreement with previous studies, as well as terms related to a response to stress and to defense and inflammatory responses, in line with knowledge on endothelial cell response to DNA damage (Corre et al., 2010; Guipaud et al., 2018; Korpela and Liu, 2014). Interestingly, more than half of the top enriched cellular processes may be directly or indirectly related to angiogenesis, suggesting that a large portion of the identified entities are related to angiogenesis and that this process is strongly altered by ionizing radiation in our study.
Taking this further, we used the subnetwork enrichment analysis module in Pathway Studio software to search for protein regulators and protein targets, and then we searched for enriched regulated processes and cells using the 83 candidates and 200 most enriched regulated proteins (Figure 3). The network built using Pathway Studio and Cytoscape (Figure 3B) confirms the previous analysis of GO pathways/groups enriched with the 83 checkpoint candidate entities (cell death and apoptosis). In addition, it highlights other previously known processes involved in the response of endothelial cells to irradiation, i.e. aging and senescence, as well as a connection to the immune response through cell adhesion and relationships with leukocytes, especially macrophages. Last, it indicates angiogenesis as an important process in response to irradiation. Figure 4 shows an alternative representation of the network, keeping only angiogenesis as a cellular process. It indicates the level of expression of the candidates, showing different responses at early and late times.
Figure 3.
Common regulators and expanded cells, targets, and cell processes of selected candidates
Enrichment analyses using Pathway Studio software enable highlighting of essential processes potentially altered by exposure of endothelial cells to ionizing radiation.
(A) Strategy of enrichment: enrichment of common regulators and targets using the 83 candidates on the one hand, enrichment of cells and cell processes using candidates and targets on the other hand.
(B) Schematic view using Cytoscape of connections between candidates, common regulators, targets, cells, and cell processes. Angiogenesis appears as one of the cell processes potentially impaired by ionizing radiation.
Figure 4.
Interaction network of the 83 experimental candidate entities with their predicted regulators, targets, and the cell process angiogenesis
(A) Network overview obtained using Pathway Studio software. The 83 candidate checkpoints controlling the response of endothelial cells to irradiation in each omics layer are shown in the middle of the array, whereas the regulators and targets predicted by enrichment are placed above and below the candidates, respectively. Note that CDKN2A, an interesting miRNA target highlighted in our study, is also included in the network although not experimentally identified.
(B and C) Same network where candidate entities are pinpointed according to their level of expression compared with nonirradiated cells (red: increased expression, blue: decreased expression) in the early (0.5–7 days) (B) and late (14–21 days) (C) time points following irradiation.
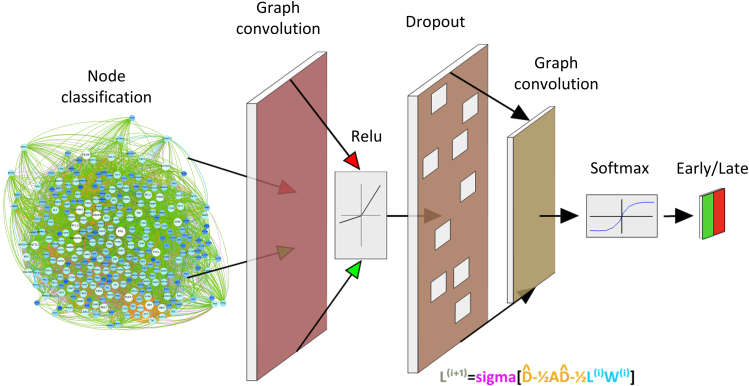
Dynamic modeling enables accurate learning of essential checkpoints of vascular endothelial cell dysfunction after irradiation
We built a hybrid dynamic model based on the semi-supervised learning of vascular endothelial cell dysfunction after irradiation (Figure 5). Visualization of the model's performance (Figure S4) was achieved by applying a spectral graph convolution to the integrated omics graph as shown in an mp4 file (Video S6). The weighted average of the first model is 0.56 supported by 111 and 176 samples to the early and late feature representations, respectively. This figure increases to 0.79 in the second learning model. These measures asymptotically reach a steady state of accuracy close to 0.77 with the progressive inclusion of many tested layers up to 1,024.
Figure 5.
Classification of candidates in early and late dysfunction after ionizing irradiation
The figure presents an overview of the graphical learning model calibrated in this work to predict early or late-stage molecular dysfunction after ionizing irradiation. We operated in layer starting from the regulatory gene network with all potential associations between the selected candidates at the multiscale level. Each interaction of a pair was integrated into a first convolutional layer according to its early or late initial stage (highlighted in green and red, respectively). At the entrance of the first layer, all this information was merged (in red and green) with the other interactions of the network, which is why we have colored in dark brown as a reference in this layer. Then, we sorted all these associations by means of a ReLU activation layer. Because this is a sort of diffusion process from the original interactions to the output of the model, we have drawn lighter shades respecting the colors of the original arrows. Then, a 50% exclusion layer followed to avoid overfitting. There, the interactions associated with the first convolutional layer were probabilistically clustered (the colors become even lighter) while waiting for the applications of the last layers. A final convolutional layer was then designed to contain all the information about the aggregations associated with an initial pair of genes. Finally, we used Softmax activation to transform the probability distributions of the last convolutional layer into a binary matrix of information that will be the output of the model. This computation ensured the final prediction of the initial genes in an early or late dysfunctional stage. The equation: this equation describes how the machine learning model recursively passed through the layers, namely: (1) we normalized the network structure (highlighted in orange); (2) after normalization, we multiplied its structure by that captured by a previous epoch (i- > i+1). We called this the gene properties (highlighted in blue). Finally, we applied sigma to the gene properties and model weights (colored in pink). Sigma is a nonlinear function based on the Adam optimizer that computes the minimization problem associated with the loss of information produced between the eventual output of the model in a particular epoch and its immediately preceding solution.
Dynamic modeling thus enabled shortening of the initial molecular signature by the best candidates learned to be closest to the feature representations, i.e. both hsa-miR-10b-5p and hsa-miR-5582-3p at a similar level, MCL1, hsa-miR-181c-5p, RTN4, and SELP regarding the early cellular dysfunction on the one hand, and fumarate, adenosine monophosphate, hsa-miR-181a, and CD44 for the late dysfunction on the other hand. In addition, the model identifies the existence of an intermediate pattern composed of molecules such as TAGLN2 or SERPINE1 that trace the path from early to late dysfunction but end up halfway. It is conceivable that their reprogramming could prevent the effect of radiation-induced late dysfunction. In Figure 6, we display the interaction network of top candidates, cell processes, and radiation as a treatment, established on a bibliographic basis using the text mining module of pathway studio. This figure sums up the many relations between top candidates and between top candidates and cell processes such as cell damage, cell death, apoptosis, senescence, cell adhesion, immune response, and angiogenesis.
Figure 6.
Interaction network of top candidates, cell processes, and radiation
(A) Protein network of the top candidates identified in this study, enriched cell processes, and the term “Radiation” as treatment was obtained using Pathway Studio software. The top 13 candidates include the top 6 entities for early dysfunction (has-miR-10b-5hashsa-miR-5582-3p, has1, hsa-miR-181c-5p, RTN4, and SELP), the top 4 entities for late dysfunction (fumarate, adenosine monophosphate, hsa-miR-181a, and CD44), and the top 2 entities for intermediate patterns (TAGLN2 and SERPINE1). CDKN2A, target of miR10B, is also included in the list of top candidates.
(B and C) Same network where candidate entities are identified according to their level of expression compared with nonirradiated cells (red: increased expression, blue: decreased expression) in the early (0.5–7 days) (B) and late (14–21 days) (C) time points following irradiation.
Angiogenesis as an essential cell process impaired by radiation exposure: experimental validation of some checkpoint candidates learned from dynamic modeling
We then focused on several targets highlighted by dynamic modeling to study their involvement in the alteration of tube formation after irradiation. We selected mir181c and CDKN2A (a target of miR10b) as features involved in the early dysfunction, SERPINE1 that traces the path from early to late dysfunction, and miR181a and CD44 for the late dysfunction. SERPINE1, CD44, and CDKN2A (at late time) were upregulated in radiation-exposed cells (Figures S5A and S5B). Surprisingly, miR181a and miR181c were hardly modulated after irradiation regarding the RT-qPCR results (Figure S5A), whereas dynamic modeling highlighted these 2 miRNAs as important features. This may possibly be caused by the closeness of these two miRNAs in expression to the homology generators observed during the complex creation. To obtain more information on their potential relevance in the response to irradiation, we measured their level of expression by RT-qPCR in the gut of mice exposed to ionizing radiation (Figure S6). The results show a statistically significant decrease in the expression of miRNA181a and miR181c on day 3 and day 7 after irradiation, confirming in vivo the result of the modeling. We also show that angiogenic genes ANGPTL1, interleukin-6 (IL-6), MMP2, and SERPINE1 were upregulated in the intestine exposed to irradiation (Figure S6B), suggesting a role of angiogenesis in response to tissue damage.
To establish the contribution of SERPINE1, CD44, CDKN2A, miR181a, and miR181c in angiogenesis in vitro, we used siRNAs and miRNA mimics in HUVECs and estimated tube formation on Matrigel after irradiation. The administration of siRNA against SERPINE1, CD44, or CDKN2A in unexposed or irradiated HUVECs showed 74%–88% inhibition of gene expression (Figure S7A). Regarding miRNAs, we verified that mimics enter durably into the cells (Figure S7B). Administration of miR181c mimic was deleterious for HUVECs at day 7 posttransfection (Figure S7C). Nevertheless, miRNA mimics led to an overexpression of both miR181a and miR181c at day 3 and day 7 posttransfection and following 20 Gy at day 5 postirradiation for the miR181a mimic (Figure S7D).
We first checked that angiogenesis capacities of HUVECs were impaired by ionizing radiation exposure. Looking at untreated and nontargeting control siRNA conditions, a 40%–50% decrease in the number of branching points of HUVEC tubes formed on Matrigel was observed 18 h after irradiation at 20 Gy compared with unexposed cells (Figure 7). The administration of siRNAs against SERPINE1, CD44, and CDKN2A was associated with a marked impairment of the number of branch points in the tubes formed by HUVECs (Figures 7A and 7B). Also, siRNAs against SERPINE1 and CD44 induced a decrease in gene expression of CD34 (Figure S8A), a key player in angiogenesis (Siemerink et al., 2012). The administration of miR181a and miR181c mimics was associated with an increased number of branch points in tube formed by HUVECs after irradiation compared with untreated cells (Figures 7C and 7D) and influenced the expression of the essential angiogenic genes CD44, PDGFRA, and TIMP3 in nonirradiated cells and IL-6 and VEGFA in both nonirradiated and irradiated cells (Figures S8B and S8C). These results suggest that the radiation-induced alteration of angiogenic capacity may be partly counterbalanced by overexpression of miR181a and to a lesser extent miR181c.
Figure 7.
Tube formation in irradiated HUVECs treated with siRNAs or miRNA mimics
(A and C) Representative images of HUVECs seeded on Matrigel 18 h postirradiation at 20 Gy after treatment by siRNA against SERPINE1, CD44, or CDKN2A (A) or by miR181a or miR181c mimics (C). Scale bars, 400 μm.
(B and D) Number of branch points per microscope field of view were quantified and plotted. Data are mean ± SEM of three independent experiments. One-way ANOVA test with Sidak correction. ns, not significant, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001.
Discussion
The vascular endothelium represents a promising target for improving the therapeutic potential of radiation therapy in the future (Guipaud et al., 2018; Wijerathne et al., 2021). In this study, we used high single doses of ionizing radiation, namely 20 Gy for HUVECs and 19 Gy for mouse intestine, which may seem far from conventional fractionated radiation therapy protocols that deliver daily dose fractions of 1.8–2 Gy.
The animal model we used was first established in rats to investigate the effects of localized irradiation in the long term (Hauer-Jensen et al., 1983) and then developed in mice (Zheng et al., 2000). The same team also developed a model in which a loop of small intestine was transposed into the left part of the scrotum in order to irradiate the intestine repeatedly (Hauer-Jensen et al., 1988, 1990). This model results in reproducible complications very similar to those observed in patients with acute or late radiation enteropathy and is considered a clinically relevant model (Hauer-Jensen et al., 2014). In addition, localized single-dose and fractionated irradiation to the small intestine caused in this model similar responses in the rats in terms of survival and radiation-induced pathology (Langberg et al., 1992). However, due to the small size of mice, this model can only be performed in rats. For all these reasons, it is considered that in mice, like in rats, a single dose of 19 Gy to a small part of the intestine induces late lesions such as those seen in patients treated with radiation therapy. Importantly, it is the only model that can generate radiation-induced intestinal fibrosis in mice. To better understand the adverse effects of radiation therapy, we therefore used this model of localized single-dose irradiation of the intestine as a model of radiation-induced late intestinal injury. We previously showed using this model of single-dose irradiation that endothelial cells played a critical role in the initiation and development of radiation-induced intestinal toxicity in mice (Rannou et al., 2015; Toullec et al., 2018). On the other hand, advances in treatment planning and delivery now allow delivery of one or more high-dose ionizing radiation fractions (40–60 Gy in 1–5 fractions) to tumors by stereotactic body radiation therapy (SBRT) (Lo et al., 2010; Qiu et al., 2020), which is increasingly used to treat patients (Brown et al., 2019; Simone et al., 2013). Thus, we assume that endothelial cells from normal tissues and tumors can be exposed to one or more high-dose radiation fractions such as that used in our in vitro model and in SBRT.
The endothelial cell model of radiation-induced dysfunction we used has been extensively characterized in our previous work (Ben Kacem et al., 2020; Heinonen et al., 2015, 2018; Jaillet et al., 2017; Paget et al., 2019). In this model, at a dose of 20 Gy, the cells stopped proliferating as early as one day after irradiation and did not proliferate at all by the third day (Ben Kacem et al., 2020; Paget et al., 2019). In addition, we estimated that at day 7 post-20 Gy only 2%–3% of cells had divided, and at most once, during the first 7 days after irradiation (Paget et al., 2019). Here, we show that a dose of 20 Gy led to the progressive loss of almost all cells (7% of irradiated cells remained 21 days after exposure), probably due to cell death, and impaired the angiogenesis function of HUVECs. Also, our results suggest that these cells were metabolically active, as they were larger and contain more RNA and proteins, and were probably all in senescence, as suggested by the result of β-gal staining. These results can be connected to cellular processes revealed by pathway enrichment analyses such as cell death, senescence, and angiogenesis, showing that our multi-omics approach can reveal processes observed experimentally at the cell culture level, which validates our method. Although this model is far from what endothelial cells undergo in vivo, several works suggest that irradiation leads to endothelial cell senescence, which would participate in radiation-induced late effects (Azimzadeh et al., 2015; Soysouvanh et al., 2020). In this sense, it could be hypothesized that endothelial cells remaining in irradiated tissues are senescent, dysfunctional, and contribute significantly to late radiation-induced damage. Considering all these data and our own results, this in vitro model could recapitulate the steps that lead irradiated endothelial cells to become senescent in vivo.
To decipher the molecular response of endothelial cells, we analyzed the miRNA, transcript, protein, and metabolite contents of primary vascular endothelial cells to model the continuum of radiation-induced injury processes between early and late vascular dysfunction in healthy tissue and tumors during radiation therapy. The great depth of omics datasets exploited is a major advantage of this analysis to fill the gap in our understanding between early and late endothelial dysfunction, which is thought to contribute significantly to the tissue effects of radiation therapy. This allowed us to propose molecular targets related to the radiation-induced alteration of angiogenesis, a fundamental function of endothelial cells.
Integration of multi-omics data identifies a molecular signature of radiation-induced endothelial dysfunction
Deep models of integrated multi-omics data enable learning the entire molecular process accurately (Eraslan et al., 2019). To build these models, we exhaustively interrogated a heterogeneous set of available kinetic samples using data shaping methods whose core comes from topological data analysis. The models successfully captured the complex molecular regulation between early endothelial cell response and late endothelial cell dysfunction, which likely occurs in radiation therapy and causes adverse effects despite dose fractionation (Christersdottir et al., 2019; Halle et al., 2010a, 2010b; Rannou et al., 2015; Toullec et al., 2018). After a thorough examination of the samples, we found that their temporal expression profiles showed three initial shapes that corresponded to the boundary points of the different experimental conditions associated with each measurement. In this way, this accumulation of samples suggests a real effect due to the time period considered after irradiation. Our analysis accounts for such an effect by translating each dataset into time expression matrices subject to these models. A regression of the datasets to their fold-change shapes allowed for proper interpretation of the results in the subsequent parts of the study. At some stages, a projection of the datasets preserving their original shape was useful to create the topological skeleton of each omics layer. In each of these discrete spaces, we were able to compare groups of elements in a homological way, converting these spaces into continuous omics spaces, conveniently represented by graphs (discrete Morse theory used in the mapper algorithm). This allowed us to identify topologically important molecular feature modules to perform continuous conversions between omics spaces, which goes beyond descriptive clustering. As the omics datasets are expressed in terms of the expression-time vectors introduced earlier, the molecules comprising these modules appear to be excellent candidates for forming the basis of the molecular signature describing the mechanisms from early to late endothelial cell dysfunction after irradiation.
From the molecular signature to a gene regulatory network
Because molecules do not function independently in the cell, we provide a global view of the exchanges produced in the response after ionizing radiation with the unsupervised integration of each molecular candidate as a gene regulatory network. In this respect, there is a high degree of homological similarity between the integrated omics graph and the graph resulting from the mapper algorithm of the identified persistent candidates. The former then relies on the topological representation of the data, which formalizes the notions of proximity, continuity, and robust denoising through an analytical study of spaces and maps. This perspective enables learning of global representations from local information, unlike more traditional methods that tend to rely on overly simplistic and less flexible assumptions (Munch, 2017). In particular, the global structure of our complex data can be rigorously quantified through those topological invariants. As invariant, those representations became ideal features to be used as the basis of the graph neural network (Tauzin et al., 2020), herein applied to dynamically learn molecular interactions and patterns (Knyazev et al., 2018; Rhee et al., 2018) occurring within cells after irradiation.
Systems biology and pathway analysis identify the most promising molecules
The use of systems biology methods has greatly improved the extraction of relevant biological information about the full range of processes underlying endothelial dysfunction. From the graph mapping, we deduced three behavioral patterns with respect to molecular functions that could lead to toxicity during radiation therapy. These graphs are closely related to the earlier and intermediate mechanisms of response to endothelium injuries triggered by DNA/cell damage, apoptosis, senescence, cell migration, and angiogenesis. They primarily account for immune and inflammatory responses characterized by cell adhesion, and, ultimately, fibrosis predicted by the miRNome. These three sets of graphs accurately mapped the deregulated pathways required to release cellular stress along the endothelial barrier disruption as well as the alteration of the angiogenesis process. Their dynamics were obtained naturally by applying spectral convolutional aggregation on the nodes of the omics integrated network. Although there was no particular reason for these aggregations to be robust, we eventually found that they were. Thus, this process made possible to any given early, intermediate, and late feature representations of cells dysfunction, the semi-supervised learning of the aggregation process leading to a list of the most promising molecules to be experimentally validated: MCL1, SELP, RTN4, and CDKN2A as a target of the selected miR10b, SERPINE1, CD44, TAGLN2, miR10b, mi181a, miR181c, miR5582, AMP, and fumarate.
Biological relevance of the most promising molecules
Some of the 13 selected entities using the supervised learning have clearly direct links with irradiation. Upregulated by IL-6, the BCL2 homolog MCL1 has an antiapoptotic effect in HUVECs following radiation exposure (Chou et al., 2009). The vascular P-selectin (SELP) is a cell adhesion molecule that is upregulated in response to ionizing radiation and participates in the recruitment of leukocytes (Molla et al., 1999). The cyclin-dependent kinase inhibitor 2A (CDKN2A), also known as p16INK4A, acts as a negative regulator of normal cell proliferation and is a marker for and an effector of senescence (Campisi and d'Adda di Fagagna, 2007). Also a marker and a mediator of senescence (Vaughan et al., 2017), the endothelial plasminogen activator inhibitor type-1 (SERPINE1), known as PAI-1, plays an essential role in radiation enteropathy (Milliat et al., 2008; Rannou et al., 2015). Involved in cell–cell interactions, cell adhesion, and migration, CD44 is upregulated in irradiated HUVECs, associated with radiation-induced senescence, and promotes cell adhesion in response to irradiation of endothelial cells (Kim et al., 2014; Lowe and Raj, 2014). Last, transgelin-2 (TAGLN2, also known as SM22-alpha, for smooth muscle 22-alpha) has been shown to be upregulated in endothelial cells that undergo endothelial-mesenchymal transition, a process that intersects with senescence (Mintet et al., 2015; Sasaki et al., 2020). On the other hand, the link between ionizing radiation and reticulon 4 (RTN4, also known as Nogo-B), which is a member of the reticulon protein family and an inducer of apoptosis in certain types of cancer cell (Kawaguchi et al., 2018; Li et al., 2001), is not documented.
Further, miRNAs are able to modulate the angiogenic properties of HUVECs (Poliseno et al., 2006) and are differentially expressed following radiation in endothelial cells (Wagner-Ecker et al., 2010). More generally, miRNAs are modulated in various cells, tissues, and fluids in response to ionizing radiation (Belli and Tabocchini, 2020; Metheetrairut and Slack, 2013). However, the roles of the 4 miRNAs we selected, i.e. miR10b, mi181a, miR181c, and miR5582, have never been established in endothelial cells in response to irradiation.
The local production of fumarate, defined as an oncometabolite, by fumarate hydratase has been characterized in cancer cells as a DNA repair factor required in nonhomologous end joining (Jiang et al., 2015; Yogev et al., 2010) and may have a direct role in checkpoint signaling (Johnson et al., 2018). Also, we identified AMP as an essential factor in the development of late dysfunction induced by irradiation. Cyclic adenosine monophosphate (cAMP) is the prototypical second messenger that is generated by adenylyl cyclase on stimulation of G protein–coupled receptors. cAMP exerts an inhibitory effect on DNA replication, leading to cell arrest in S phase (Naderi et al., 2005). The activation of cAMP signaling inhibits DNA damage-induced apoptosis through the DNA damage–induced accumulation of p53 (Naderi et al., 2009).
Angiogenesis, a function strongly altered by radiation, can be modified in vitro
Among the processes predicted by modeling and systems biology, angiogenesis is a key function that can be affected by ionizing radiation. Radiation therapy is considered to inhibit angiogenesis by damaging the endothelium (Fuks et al., 1992; Hatjikondi et al., 1996; Rose et al., 1999). However, there are also in vivo studies that show that ionizing radiation induces angiogenesis (Kaminski et al., 1978, 1983) and increases the expression of genes that favor it (Polytarchou et al., 2004), but no study explains these contradictory effects. Irradiation has been shown in vivo to influence the expression, deposition, and turnover of extracellular matrix proteins in a way that secondarily promotes normal and tumor angiogenesis (Giannopoulou et al., 2001). These data indicate that radiation first inhibits angiogenesis and then indirectly stimulates angiogenesis through the necessary healing process that results from radiation-induced damage. Similarly, in this study, we observed that ionizing radiation inhibits the expression of proangiogenic genes and the formation of tubes formed by HUVECs on Matrigel, whereas, on the other hand, we showed that proangiogenic genes were stimulated in mice locally exposed to radiation. We modulated in HUVECs the expression of 5 entities highlighted by dynamic modeling to study their involvement in the alteration of tube formation after irradiation. Inhibition of the expression of SERPINE1, CD44, and CDKN2A was found to inhibit angiogenesis, even more so when the cells had undergone irradiation. This antiangiogenic effect was associated with the decrease in the expression of CD34, an essential key player in angiogenesis (Siemerink et al., 2012). Conversely, administration of miR181a and miR181c limited or even slightly counteracted the irradiation-induced inhibition of angiogenesis and stimulated the expression of proangiogenic genes. These validations confirmed that the best candidates obtained by our machine learning model were functionally modified in vascular endothelial cells with respect to angiogenesis after irradiation in vitro.
Conclusions
The strategy we present here facilitates unsupervised high-level feature extraction of important omics data to learn how ionizing-radiation-induced endothelial dysfunction may evolve over time. As a proof of principle, the experimental validation of such a dysfunction, here angiogenesis, allows us to propose new targets to restore impaired angiogenesis or inhibit neovascularization in growing tumors.
Limitations of the study
This study focused on identifying molecular pathways and target molecules involved in the initiation and development of ionizing-radiation-induced vascular endothelial dysfunction using large-scale omics analyses. For the transcriptomic study, we chose to target the analysis on genes of interest using premade TaqMan low-density 384-well arrays (TLDA) for human angiogenesis, inflammation, apoptosis, immune response, and protein kinase panels, representing approximately 400 genes, whereas the number of genes expressed and detected in HUVECs can be expected to average 1,500–2,500 genes per cell in sc-RNAseq experiments (Zirkel et al., 2018). Although this strategy represents an obvious advantage, as gene expression is measured with the robust RT-PCR technique, assessment of unmeasured gene expression levels may provide other interesting information that we will need to investigate in the future.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Dulbecco's Phosphate Buffered Saline | Thermo Fisher Scientific | Cat#: 14190-094 |
| mirVana™ miRNA Isolation kit | Thermo Fisher Scientific | Cat#: AM1560 |
| RNeasy Mini Kit | Qiagen | Cat#: 74104 |
| EBM-2, Endothelial Cell Basal Medium-2 | Lonza | Cat#: CC-3156 |
| Trypsin-EDTA | Thermo Fisher Scientific | Cat#: 25300054 |
| Trypan blue dye | Bio-Rad | Cat#: 145-0013 |
| Dharmafect 1, Transfection Reagent | Thermo Fisher Scientific | Cat#: T-2001-02 |
| Nuclease-free water | Qiagen | Cat#: 129114 |
| Gibco™ Opti-MEM™ I Reduced Serum Medium | Thermo Fisher Scientific | Cat#: 31985-065 |
| BD Matrigel Basement Membrane Matrix | BD Biosciences | Cat#: 354234 |
| AErrane (Isoflurane) | Abbot GmbH | Cat#: DDG9623 |
| High-Capacity cDNA Reverse Transcription Kit | Thermo Fisher Scientific | Cat#: 4368814 |
| Ethanol absolute | VWR | Cat#: 20821.365 |
| TaqMan™ Fast Universal PCR Master Mix (2X), no AmpErase™ UNG | Thermo Fisher Scientific | Cat#: 4366072 |
| TaqMan™ Gene Expression Master Mix | Thermo Fisher Scientific | Cat#: 4370074 |
| Megaplex™ RT Primers, Human Pool Set v3.0 | Thermo Fisher Scientific | Cat#: 4444745 |
| TaqMan™ PreAmp Master Mix | Thermo Fisher Scientific | Cat#: 4391128 |
| Bicinchoninic Acid (BCA) Kit for Protein Determination | Sigma-Aldrich | Cat#: BCA1-1KT |
| Urea | Sigma-Aldrich | Cat#: 51456 |
| Triethylammonium bicarbonate buffer | Thermo Fisher Scientific | Cat#: 15215753 |
| cOmplete™, Mini, EDTA-free Protease Inhibitor Cocktail | Sigma-Aldrich | Cat#: 11836170001 |
| ProteaseMAX™ Surfactant, Trypsin Enhancer | Promega | Cat#: V2071 |
| Tris(2-carboxyethyl)phosphine hydrochloride | Sigma-Aldrich | Cat#: 75259 |
| Iodoaceatmide | Sigma-Aldrich | Cat#: I1149 |
| Sequencing Grade Modified Trypsin | Promega | Cat#: V5111 |
| iTRAQ® Reagents Multiplex Kit | SCIEX | Cat#: 4352135 |
| 3100 OFFGEL Fractionator High Res Kit, pH 3-10 | Agilent | Cat#: 5188-6424 |
| Acetonitrile | Sigma-Aldrich | Cat#: 34851 |
| Formic acid | Sigma-Aldrich | Cat#: 695076 |
| Methanol | Sigma-Aldrich | Cat#: 179337 |
| β-Gal Staining Kit | Thermo Fischer Scientific | Cat#: K146501 |
| Critical commercial assays | ||
| TaqMan™ Array Human Immune Panel | Thermo Fisher Scientific | Cat#: 4370499 |
| TaqMan™ Array Human Protein Kinase Panel | Thermo Fisher Scientific | Cat#: 4365299 |
| TaqMan™ Array Human Apoptosis Panel | Thermo Fisher Scientific | Cat#: 4378716 |
| TaqMan™ Array Human Inflammation Panel | Thermo Fisher Scientific | Cat#: 4378722 |
| TaqMan™ Array Human Angiogenesis Panel | Thermo Fisher Scientific | Cat#: 4378725 |
| TaqMan™ Array Human MicroRNA A+B Cards Set v3.0 | Thermo Fisher Scientific | Cat#: 4444913 |
| Custom TaqMan® Array Human MicroRNA Cards | Thermo Fisher Scientific | Cat#: 4342265 |
| Deposited data | ||
| Raw proteomics data | This paper | PRIDE: PXD030572 |
| Raw metabolomics data | This paper | MetaboLights: MTBLS3680 |
| SWISS-PROT protein sequence database (release 20170315) | Uniprot | https://www.uniprot.org/ |
| Mammal (Anatomy; CellEffect™; DiseaseFx®; GeneticVariant; Viruses) version 12.4.0.3 (Updated April 25, 2021) database from Elsevier | Elsevier | www.elsevier.com/pathway-studio |
| Human metabolome database (HMDB) | https://hmdb.ca/ | |
| Experimental models: Cell lines | ||
| HUVEC – Human Umbilical Vein Endothelial Cells | Lonza | Cat#: C2519A |
| Experimental models: Organisms/strains | ||
| C57BL/6J mouse | Charles River | Cat#: 000664 |
| Oligonucleotides | ||
| Primers for TaqMan® Human gene expression assay: SERPINE1 - Assay ID: Hs01126606_m1 | Thermo Fisher Scientific | Cat#: 4331182 |
| Primers for TaqMan® Human gene expression assay: CD44 - Assay ID: Hs01075861_m1 | Thermo Fisher Scientific | Cat#: 4331182 |
| Primers for TaqMan® Human gene expression assay: CDKN2A - Assay ID: Hs00924091_m1 | Thermo Fisher Scientific | Cat#: 4331182 |
| Primers for TaqMan® Human gene expression assay: CD34 - Assay ID: Hs00990732_m1 | Thermo Fisher Scientific | Cat#: 4331182 |
| Primers for TaqMan® Human gene expression assay: COL4A2 - Assay ID: Hs01098873_m1 | Thermo Fisher Scientific | Cat#: 4331182 |
| Primers for TaqMan® Human gene expression assay: IL6 - Assay ID: Hs00174131_m1 | Thermo Fisher Scientific | Cat#: 4331182 |
| Primers for TaqMan® Human gene expression assay: PDGFRA - Assay ID: Hs00998018_m1 | Thermo Fisher Scientific | Cat#: 4331182 |
| Primers for TaqMan® Human gene expression assay: TIMP3 - Assay ID: Hs00165949_m1 | Thermo Fisher Scientific | Cat#: 4331182 |
| Primers for TaqMan® Human gene expression assay: VEGFA - Assay ID: Hs00900055_m1 | Thermo Fisher Scientific | Cat#: 4331182 |
| Primers for TaqMan® Human gene expression assay: ACTB - Assay ID: Hs99999903_m1 | Thermo Fisher Scientific | Cat#: 4331182 |
| Primers for TaqMan® Human/Mouse miRNA expression assay: miR181a-5p - Assay ID: 000480 | Thermo Fisher Scientific | Cat#: 4427975 |
| Primers for TaqMan® Human/Mouse miRNA expression assay: miR181c-5p - Assay ID: 000482 | Thermo Fisher Scientific | Cat#: 4427975 |
| Primers for TaqMan® Human/Mouse miRNA expression assay: U6 snRNA - Assay ID: 001973 | Thermo Fisher Scientific | Cat#: 4427975 |
| Primers for TaqMan® Mouse gene expression assay: SERPINE1 - Assay ID: Mm00437306_m1 | Thermo Fisher Scientific | Cat#: 4331182 |
| Primers for TaqMan® Mouse gene expression assay: ANGPT1 - Assay ID: Mm00456503_m1 | Thermo Fisher Scientific | Cat#: 4331182 |
| Primers for TaqMan® Mouse gene expression assay: ANGPTL1 - Assay ID: Mm01291815_m1 | Thermo Fisher Scientific | Cat#: 4331182 |
| Primers for TaqMan® Mouse gene expression assay: IL6 - Assay ID: Hs00174131_m1 |
Thermo Fisher Scientific | Cat#: 4331182 |
| Primers for TaqMan® Mouse gene expression assay: MMP2 - Assay ID: Hs01548727_m1 | Thermo Fisher Scientific | Cat#: 4331182 |
| Non targeting siRNA | GE Healthcare | Cat#: D-001810-10-20 |
| siRNA against human SERPINE1 | GE Healthcare | Cat#: L-019376-01-0050 |
| siRNA against human CD44 | GE Healthcare | Cat#: L-009999-00-0020 |
| siRNA against human CDKN2A | GE Healthcare | Cat#: L-011007-00-0020 |
| Human miR181a-5p Mimic | Thermo Fisher Scientific | Cat#: MC10421 |
| Human miR181c-5p Mimic | Thermo Fisher Scientific | Cat#: MC10181 |
| miRIDIAN microRNA Mimic Transfection Control with Dy547 | Thermo Fisher Scientific | Cat#: CP-004500-01-05 |
| Software and algorithms | ||
| Spectrum Mill, Rev B.04.00.127 | Agilent Technologies | Cat#: G2721AA/G2733AA |
| R software | www.r-project.org | www.r-project.org |
| iQuantitator | Schwacke et al., 2009 | https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-10-342#Sec17 |
| Chromeleon 6.8 software | Thermo Fisher Scientific | Cat#: CHROMELEON6 |
| Xcalibur 3.0.63 | Thermo Fisher Scientific | Cat#: XCALI-97553 |
| ProteoWizard | Chambers et al., 2012 | http://proteowizard.sourceforge.net/ |
| van der Kloet algorithm | van der Kloet et al., 2009 | https://pubs.acs.org/doi/abs/10.1021/pr900499r |
| Workflow4Metabolomics | Giacomoni et al., 2015 | https://workflow4metabolomics.org |
| Pathway Studio Web Mammal version 12.4.0.3 | Nikitin et al., 2003 | www.elsevier.com/pathway-studio |
| DIANA Tools | Vlachos et al., 2012 | http://diana.imis.athena-innovation.gr/DianaTools/ |
| Topological Data Analysis version 1.6.9 | Fasy et al., 2019 | https://cran.r-project.org/web/packages/TDA/index.html |
| DataAssis software version 3.01 | Thermo Fisher Scientific | https://www.thermofisher.com/ |
| GraphPad Prism version 8.1.1 | GraphPad Software | www.graphpad.com |
| ImageJ version 1.52a software | National Institutes of Health | http://imagej.nih.gov/ij |
| Other | ||
| Gene Ontology resource | Ashburner et al., 2000 | http://geneontology.org/ |
| PANTHER classification resource version 14 | Mi et al., 2019 | http://pantherdb.org/ |
| g:Profiler | Raudvere et al., 2019 | https://biit.cs.ut.ee/gprofiler/ |
| GeneMANIA | Warde-Farley et al., 2010 | https://genemania.org/ |
| JavaPlex | Adams et al., 2014 | http://appliedtopology.github.io/javaplex/ |
| FFmpeg function | Korbel, 2012 | http://ffmpeg.org/ |
| R package mapmate | Leonawicz, 2017 | https://github.com/leonawicz/mapmate |
| Keplermapper | van Veen and Saul, 2019 | http://doi.org/10.5281/zenodo.1054444 |
| Angiogenesis Analyzer plugin for Image J | http://image.bio.methods.free.fr/ImageJ/?Angiogenesis-Analyzer-for-ImageJ | https://imagej.nih.gov/ij/macros/toolsets/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Olivier Guipaud (olivier.guipaud@irsn.fr).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Cells
Human umbilical vein endothelial cells (HUVECs, pooled donors, newborn, male/female mixed, product code C2519A, batch number 0000465419) from Lonza were grown at 37°C with 5% CO2 in EBM-2 MV medium containing 5% FBS (Lonza). Cells were authenticated by the suppliers and tested negative for mycoplasma, bacteria, yeast, and fungi, as guaranteed by the suppliers.
Animals
Male C57BL/6 mice came from Charles River Laboratories and were 10 weeks of age upon arrival. Animals were housed at the IRSN animal facilities authorized by the French Ministry of Agriculture for performing experiments on rodents. Animal experiments were performed in strict compliance with French and European guidelines and regulations on the protection of animals used for scientific purposes (EC Directive, 2010/63/EU and French Decree No. 2013–118). They were approved by IRSN's Animal Experimentation Ethics Committee, registered under number 81 and authorized by the French Ministry of Research under reference APAFIS#6209-2016072213325989 v2 (internal project number P15-04).
Method details
Irradiation procedure
Irradiation of HUVECs at a single dose of 20 Gy was performed using a 137Cs source (IBL 637, CisBio, 1 Gy.min−1). Non-irradiated cells (control samples) were left for the same amount of time on a benchtop near the irradiation room. All irradiations were carried out at 90–100% cell confluence and the same number of population doublings (passage 3, corresponding to 9 to 12 population doublings). The cells were collected at 0.5, 1, 2, 3, 4, 7, 14 and 21 days after irradiation or without having been irradiated (control samples) in order to prepare proteins (4 replicates per dose and per time point), mRNAs (3 replicates per dose and per time point), miRNA (3 replicates per dose and per time point) and metabolites (5 replicates per dose and per time point). For long-term experiments (14- and 21-days post-irradiation), the culture medium was changed every week.
Radiation enteropathy was induced by exposure of an intestinal segment to a single dose of 19 Gy (Milliat et al., 2008). Briefly, mice were anesthetized by spontaneous inhalation of isoflurane-N2O gas (Abbott GmbH) and, after laparotomy, a 3 cm-long intestinal segment was exteriorized and exposed to a single dose of 19 Gy using an Elekta Synergy Platform delivering 4 MV X-rays at 2.3 Gy.min−1. Sham-irradiation (sham-IR) was performed by maintaining the intestinal segment exteriorized without radiation exposure. After radiation exposure or sham-IR, the exposed segment was returned to the abdominal cavity and peritoneum/abdominal muscles and skin were separately closed with interrupted sutures.
Estimation of the number and volume of cells
For HUVEC counting and cell volume estimation experiments, cells were trypsinized (trypsin-EDTA, Life Technologies) and then counted by a TC20 Automated Cell Counter (Biorad) in the presence of trypan blue (Biorad) to estimate mortality. Cell volumes were calculated from the mean diameters determined by the TC20 Automated Cell Counter.
RNA extraction
For mRNA analysis, RNAs were prepared using the Rneasy Mini Kit (Qiagen). For miRNA analysis, total RNAs were prepared using the mirVana miRNA isolation kit (Thermo Fisher Scientific). RNA concentrations were quantified on a NanoDrop ND-1000 apparatus (NanoDrop Technologies), and miRNA quality was checked using an Agilent Bioanalyser. Experiments were performed in 3 replicates for each dose of irradiation and each time point of the kinetic.
mRNA expression analysis
Reverse transcription was performed using the High Capacity Reverse Transcription Kit (Thermo Fisher Scientific) according to the manufacturer's instructions. For targeted transcriptomics studies, gene expression assays were performed using pre-made TaqMan low-density 384-well arrays (TLDA) for human angiogenesis, inflammation, apoptosis, immune response and protein kinase panels (Thermo Fisher Scientific). cDNA (400 ng per sample) was loaded onto the port of each gene signature array card and PCR was performed using the ABI PRISM 7900 Sequence detection system (Thermo Fisher Scientific). Data Assist software (Thermo Fisher Scientific) was used to determine 2−ΔΔCt values, with the following criteria: a maximum allowable Ct value at 37 was set and maximum Ct values were not included in calculations. Normalization was performed using a global normalization method on a per sample basis (Mestdagh et al., 2009). For specific gene expression studies, pre-developed TaqMan Gene Expression Assays (Thermo Fisher Scientific) were used and PCR was performed with the ABI PRISM 7900 Sequence detection system. For in vitro experiments, PCR fluorescent signals were normalized to a PCR fluorescent signal obtained from the housekeeping ACTB gene (for mRNAs) or the U6 snRNA (for miRNAs). For in vivo experiments, normalization was performed using the global normalization method on a per sample basis as indicated above. For unsupervised hierarchical clustering analyses and heatmap creation using Data Assist software, distances between samples were calculated for hierarchical clustering based on the ΔCT values using Pearson's correlation, assay centric as map type, and average linkage as clustering method.
miRNA expression analysis
Reverse transcription and qPCR were performed using different MegaPlex Human Pools (Thermo Fisher Scientific). RT MegaPlex and preamplification were performed on an Eppendorf Mastercycler Ep Gradient according to manufacturer instructions. We first evaluated microRNAs expression in HUVECs using RT Megaplex pool primers and TaqMan Array MicroRNA A and B Cards (759 microRNAs) (Thermo Fisher Scientific) using quantitative PCRs with the ABI PRISM 7900 Sequence detection system (Thermo Fisher Scientific). We selected a list of 312 microRNAS expressed in control and irradiated HUVECs 4 days post-irradiation by applying a cut-off with a maximum allowable CT value at. We next designed 4 custom TaqMan Array MicroRNA Cards to assess the expression level of the 312 microRNAs. In each card, a panel of 4 housekeeping microRNAs was included (Thermo Fisher Scientific). These 312 miRNAs were measured in triplicate at 0.5, 1, 2, 3, 4, 7, 14 and 21 days after 0 and 20 Gy. The global matrix was analyzed using Data Assist software (Thermo Fisher Scientific) and a final list of 289 miRNAs was used for implementation in the computational analysis including all biological entities.
Protein expression analysis
Protein extraction
HUVECs were washed 3 times with PBS and lysed, strongly vortexed and sonicated for 5 min in 1 mL of protein extraction buffer containing 8 M urea (Sigma), 100 mM triethylammonium bicarbonate buffer pH 8.5 (Thermo Fischer Scientific) and cOmplete Mini EDTA-free Protease Inhibitor Cocktail (Sigma-Aldrich). After addition of 0.03% (v/v) of ProteaseMAX Surfactant, Trypsin Enhancer (Promega), samples were incubated for 45 min at room temperature, centrifuged at 20,000 × g for 1 h and the supernatants were collected. Protein concentration was determined using the BCA protein assay kit (Sigma-Aldrich). Experiments were performed in 4 replicates for each time point of the kinetic and each dose of irradiation.
Peptide fractionation
Protein samples were reduced with 5 mM Tris-(2-carboxyethy) phosphine (Sigma-Aldrich) at 37°C for 1 h and alkylated with 60 mM iodoacetamide (Sigma-Aldrich) at room temperature in dark for 30 min. Then, 200 μg of protein were digested overnight at 37°C with 3.5% (W/W) sequencing grade modified porcine trypsin (Promega). 100 μg of the digests were dried in a SpeedVac concentrator (Thermo Fisher Scientific) and labeled with iTRAQ in a 4-Plex experimental design (114 and 116 reagents for control samples, 116 and 117 reagents for treated cells) as per the standard protocol supplied by the manufacturer (SCIEX). After labeling, samples were pooled and dried with the SpeedVac concentrator prior to peptide fractionation by isoelectric focusing. To perform peptide fractionation, 3100 OFFGEL Fractionator and the high resolution OFFGEL Kit (linear pH 3–10 gradient, 24 fractions) from Agilent Technologies were used as per the user's guide protocol. After focusing, each fraction was concentrated with a SpeedVac concentrator prior to LC-MS/MS analysis.
Liquid chromatography–electrospray ionization MS and data analysis of proteins
Peptides were resuspended in 15–80 μL of 3% (v/v) acetonitrile/0.2% (v/v) formic acid depending on the initial volume of the fraction and then analyzed with a nano-LC1200 system coupled to a Q-TOF 6520 mass spectrometer equipped with a nanospray source and an HPLC-chip cube interface (Agilent Technologies). Briefly, peptides were enriched and desalted on a 160-nL RP-C18 trap column and separated on a Zorbax C18 column (30-nm pore size, 5-μm particle size, 150-mm long 75 μm inner diameter; Agilent Technologies) using a 55-min linear gradient (3–75% acetonitrile in 0.1% formic acid) at a flow rate of 400 nL/min. Full autoMS1 scans from 290 to 2,400 m/z and autoMS2 from 59 to 3,200 m/z were recorded. In each cycle, up to five precursors sorted by charge state (preferred 2 + ions and single-charge ions excluded) were isolated and fragmented in the collision cell. The collision cell energy was automatically adjusted according to the m/z ratio. Active exclusion of these precursors was enabled after 1 spectrum in 0.2 min, and the precursor selection threshold was set at 1,000 counts. Peptide and protein identification were performed using the Spectrum Mill MS Proteomic Workbench (Rev B.04.00.127; Agilent Technologies). The following parameters were used for data extraction: MH + mass range from 600 to 4,000, scan time range from 0 to 300 min, similarity merging of scan with same precursor (+/− 15 s and 0.05 m/z) and minimum MS s/n set to 25. The searches were performed with the following specific parameters: enzyme specificity, trypsin; two missed cleavage permitted; iTRAQ (N-term, K) fixed modifications; variable modifications, methionine oxidation, cysteine carbamidomethylation and Gln pyro-Glu (N-ter Q); maximum ambiguous precursor charge set to 3; mass tolerance for precursor ions, 20 ppm; mass tolerance for fragment ions, 50 ppm; ESI-QUAD-TOF as instrument; taxonomy, Human; database, SWISS-PROT release 20,170,315; 50% minimum scored peak intensity; calculate reversed database scores and dynamic peak thresholding. Identified proteins and peptides were auto-validated using default parameters. Validated peptides were then exported in an Excel.ssv file using the following filter parameters: score of peptides >6 and % of SPI >60. Last, differential protein expression of iTRAQ labeling data was performed using iQuantitator open source software and the eDesign default parameter value as described (Schwacke et al., 2009).
Metabolite expression analysis
Preparation of metabolites
HUVECs were washed with PBS, trypsinized, pelleted and stored at −80°C. Five million cells per experimental condition were vortexed for 2 min with cold (−80°C) 80% methanol for the precipitation of proteins. After 30 min at −80°C, the mixture was centrifuged for 10 min at 4°C, 14,000 × g. Supernatants were evaporated under a gentle stream of nitrogen and the dry extracts were resuspended in 70 μL of a 20% acetonitrile solution. Experiments were performed in 5 replicates for each time point of the kinetic and each dose of irradiation. 20 μL of each sample were gathered to obtain a pooled sample used as a quality control. A set of diluted pool samples was prepared making pools diluted 2, 4, 8, 16 and 32 times. The Mixture used for dissolution of dry extracts was also used as blank sample.
Ultra-high-performance liquid chromatography-high resolution mass spectrometry analysis of metabolites
Extracted metabolites were analyzed using reverse phase liquid chromatography hyphenated to high-resolution mass spectrometry. Chromatographic separation was carried out on a Dionex UltiMate 3,000 (Thermo Fisher Scientific) operated by Chromeleon 6.8 software. 5 μL of each sample was injected in a Hypersil Gold (100 mm × 2.1 mm x 1.9 μm) column (Thermo Fisher Scientific). The column was kept at 40°C and the flow rate was maintained at 400 μL/mL. Mobile phases were 0.1% formic acid solutions in water (A) and acetonitrile (B). For the first minute, a 0% of B was kept isocratically following by 10 min of linear gradient up to 100% B which was maintained in isocratic mode for 2 min. Initial conditions were reached in 1 min and the column equilibrated for 2 min. Along with chromatographic separation, mass spectrometry analysis was performed on the Q-Exactive Plus hybrid mass spectrometer (Thermo Fisher Scientific) with a Heated Electrospray Ionization (H-ESI II) probe working in the positive and negative ionization modes. Spray voltage was maintained to a ±3,500 V. Transfer capillary was heated to 320°C, sheath and auxiliary gas flow rates were maintained at 30 and 8 arbitrary units and the gas was heated to 310°C. S-lens RF was kept at 55 V in order to transfer ions of large m/z range toward the orbitrap mass analyzer. Mass spectra were acquired in the 80–1,000 m/z range. Each scan was acquired by collecting the ions into the C-trap during up to 250 ms in order to obtain spectra with 35,000 FWHM (Full Width Half Maximum) resolving power for the theoretical m/z 200. Instrument setup and LC-MS system control were achieved under Xcalibur 3.0.63 spectrometer software (Thermo Fisher Scientific) and control of the mass spectrometer was achieved using Tune Q Exactive Plus 2.5 application (Thermo Fisher Scientific). The samples were analyzed in the following order: blank samples were analyzed 5 times, then diluted pools were analyzed in triplicate from most diluted to least diluted, followed by experimental samples in random order and spaced every 5 samples by a pool sample. At the end, a pool sample was subjected to a High Collision Dissociation (HCD) experiment using Data Dependent Analysis in order to obtain structural information from the MS/MS spectra.
Pre-processing of metabolomics data
Raw data were converted into mzXML files using ProteoWizard (Chambers et al., 2012). Data extraction was achieved using XCMS library under the R environment for the positive and negative ion modes separately. Peak detection was performed using the centWave method. Chromatographic peak width was set from 2 to 15 s, signal to noise threshold at 3, while the noise was set to 10,000 and the m/z tolerance between two consecutive scans was set to 5 ppm. Pre-filtering of detected peaks was set to 4 consecutive scans with intensities higher than 50,000. Peak alignment between samples and peak grouping were achieved using the Obiwarp algorithm and density method, respectively. Extracted data matrices were then filtered in order to eliminate analytical background and correct analytical drift. Peaks coming from the blank samples and from instrument noise were removed from the matrix, together with any peaks that varied by more than 30% in all pool samples. Peaks in diluted samples that did not linearly increase in line with concentration were removed for all samples. Analytical batch drift was corrected using the van der Kloet algorithm (van der Kloet et al., 2009). The final matrices contained 661 ions in the positive ionization mode and 658 ions in the negative mode. The two-ionization matrices were used separately for the untargeted metabolomics analysis. For the semi-targeted approach, matrices were matched to our in-house database, containing retention time and m/z information on about 1,300 metabolites for the peak annotation. Matching was performed using Workflow4Metabolomics (Giacomoni et al., 2015), using the LC-MS annotation with bank in-house tab (Rochat, 2017). The final matrix contains 115 metabolites identified with different levels of identification. Prior to statistical analysis, metabolite intensity is normalized by row, i.e. dividing it by the addition of metabolite intensities in all the samples. Next, we “autoscale” the matrix by mean-centering each metabolite and then multiplying by the inverse of its standard deviation.
Omics data visualization and denoising
Before any analysis, the data were projected into a three-dimensional space using the t-distributed stochastic neighbor embedding (t-SNE) method (van der Maaten and Hinton, 2008). The aim of such translation was to explore the eventual presence of recurrent profiles, both in time and in expression.
Since only the protein dataset was expressed in fold-change terms, and with a view to completing a homogeneous comparison, we linearly regressed to their fold-change-like ratio transcripts, metabolites and miRNAs. This may be calculated by solving the following optimization problem (Morilla and Ranea, 2017):
where typically y is the vector of observed values, i.e. up- or downregulated relative to their prior measurement, Xß the product between the matrices X (dataset expression) and ß (variable to be calculated), and δ is the correlated random error which follows a normal distribution.
Additionally, those molecular entities with an associated confidence interval of Z-scores ranging [-1, 1] in multi-testing were considered as unchanged and consequently removed from the initial analysis of the omics models. These discarded entities were only recovered if they were functionally enriched in some GO (Ashburner et al., 2000) or Panther (Mi et al., 2019) terms in the following functional analysis step.
Construction of the irradiated complexes
In this step of the analysis, we intended to contract the discrete space determined by the projection of the data denoised in the previous section to a continuous space. The rationale underlying this transformation was to identify lasting modules. To this end, we constructed simplicial complexes making use of the Vietoris-Rips filter (Hausmann, 1995), which provides a suitable method for associating vector spaces using linear maps. This topological configuration (i.e. the nerve theorem (Borsuk, 1948)) would enhance by homology (Stillwell, 1993) those clusters relevant to successfully completing the entire conversion. Those clusters are referred to as persistence modules and within them, they contain precious information on the high dimensional structure of any natural dataset.
Filtered simplicial complexes of proteins, transcripts, metabolites and miRNAs were carried out by means of in-house scripts using Javaplex software (Adams et al., 2014), the R package Mapmate (Leonawicz, 2017) and the FFmpeg (Korbel, 2012) function in R. The homological assembly was performed in two steps. First, the Vietoris-Rips filter coordinates and simplicial alpha complex chains were calculated using Javaplex. both coordinates and chains information were then used as input in a Mapmate-based R script. This script captures the trace of each homology variance resulting during the conversion in individual frames. Last, we take the FFmpeg function to neatly glue each of these frames in a video tracking the whole conversion between spaces.
Summary of the topological invariants in the datasets: barcodes landscapes and silhouettes
Next, we summarized the structural information encapsulated in the persistence modules considering barcodes (Carlsson et al., 2005) at different dimensions, a notion based on the durability that two topological subspaces are approached as homologically independent within the same complex during the construction of the filtered simplicial complex. Given an interval IinR and a persistence module M = Mt we defined the indecomposable persistence modules as interval modules (Im)t equal to the filtration complex if tεI or 0 otherwise. Hence M is a direct sum of these interval modules [Gabriel, Zomorodian-Carlsson, Crawley-Boevey Theorem (Oudot, 2015)]. Last, we denominated barcodes to the random variable composed by the set of possible intervals [Ik] (Bubenik, 2015) agreeing with that theorem. other versions of the barcodes, such as landscapes (i.e. a continuous and stable barcode) (Bubenik, 2015) or silhouettes (smooth piecewise barcodes) (Chazal et al., 2015) were also explored during this step in the analysis.
The barcodes along with their related other versions summarizing the filtered simplicial complex were computed on each dataset by tailored R scripts based on TDA package, version 1.6.9 (Fasy et al., 2019).
Mapping of topological invariants
Another in-house script, based on the Mapper algorithm (Singh et al., 2007) and its python module Kepler mapper (Kmapper) (van Veen and Saul, 2019), was developed to perform the mapping between the persistence modules identified by their barcodes, landscapes or silhouettes and their projections in R2. This mapping gave us the simplicial complex in a graphical form which is very practical for our purposes: the mapper graph.
First, we automatically detected samples that are different from regular samples by means of anomaly detection algorithm (i.e. isolation random forest) generating a 1-D lens or filter function that is a lower dimensional representation of data (van Veen and Saul, 2019). Then, we built another 1-D lens with t-SNE projection and combine both lenses to create a 2-D lens. Next, we created the simplicial complex as indicated in the preceding sections. The existence of some cyclic temporary periods found in our datasets penalizes the use of the Hausdorff distance as the default measure applied in the filter. Instead, we used the more convenient Dynamic Time Warping (DTW) distance (Lauwers and De Moor, 2017):
where with the (l,m)−th element of the matrix determined by the time series yi. Since the time points were discriminated later at D14 and D21, the alignment of the curves was done earlier, between 12 h and D7. Then, the sum over the path WDTW is the DTW distance. Hence, we constructed the map induced by the nerve of the simplicial complex cover composed of 15 clusters, the so-called cubes, which may overlap by up to 50%, and used the K-means method (Lloyd, 1982) to group the cubes with the DTW distance. Last, an interactive html file was created to clearly visualize the graph.
Functional analysis of persistent candidates
At this stage, the mapping produced ad described in the previous section was functionally examined under the prism of GO annotations. We searched for enriched terms in the set of persistent candidates that might reveal relevant biological pathways and processes involved in the response to irradiation. In addition, we implemented a Python API to query the Panther database. Moreover, the Pathway Studio Web Mammal software, version 12.4, querying the Mammal (Anatomy; CellEffect; DiseaseFx; GeneticVariant; Viruses) version 12.4.0.3 (Updated April 25, 2021) database from Elsevier (www.elsevier.com/pathway-studio) (Nikitin et al., 2003) and the g:Profiler webtool (Raudvere et al., 2019) were also used in the functional analysis of the candidates.
Integrated omics-graph construction
The molecular identities we identified do not work independently. There is an intricate exchange of information at many levels that determines their global function in the endothelial cells. To untangle such a complex of wired regulatory interactions, we constructed an overall gene regulatory network (OGRN). The GeneMANIA webtool (Warde-Farley et al., 2010) worked as primary reference (Lees et al., 2015) to construct this OGRN. Thus, the persistent candidates obtained in the earlier sections were the bricks used to build the OGRN. In particular, we considered DIANA Tools (Vlachos et al., 2012) to detect the most likely experimental or predicted miRNA-target interaction. For the metabolite dataset, we used the metabolite-protein interactome publicly available in the Human Metabolome Database (HMDB) (Wishart et al., 2018). If no information could be retrieved from HMDB for a specific metabolite, we retrieved related terms instead.
Graph convolutional networks of integrated omics-graph
A deep learning model on the OGRN composed of the persistent candidates was presented in the last step of the analysis. This model somehow accurately predicts how differentiated molecular entities involved in relevant processes of radiation-induced dysfunction evolve in terms of feature representation, i.e. early or late irradiation response. To build the model, we equipped the OGRN with a convolutional design (i.e. neural networks) and a spectral rule of node aggregation similar to graph convolutional networks (GCNs) (Defferrard et al., 2016) in order to extract and exploit the complex information contained in its dynamic graphic structure. Two hybrid sequential models were combined to learn the endothelial cells progression after irradiation: (1) using the identity matrix I as features and the adjacency matrix A to build the model using the spectral rule:
where D the degree matrix, and (2) considering the distance of the shortest path to acute and late cell dysfunction as an additional feature. These models were designed in two layers with 512 units in the layers and a 2D transformation of the activation function tanh. Instead, the relu activation function was applied in the spectral rule at the beginning of the layer implementation. The number of epochs was set to 5,000 and 250, respectively. We applied the stochastic gradient descent (sgd) optimizer during the training with a learning rate and momentum regularization set to 0.001 and flagged true, respectively.
An in-house python script based on MXNet implementation (Cruchant, 2020) performed the semi-supervised classification (early or late response) of nodes in the OGRN graph (Kipf and Welling, 2016).
siRNA and miRNA mimic transfection
HUVECs were transfected at 50–60% confluence with 100 nM of non-targeting siRNA or siRNA targeting SERPINE1, CD44 or CDKN2A (Thermo Scientific), or with 50 nM of miR181a or miR181c mimics (Thermo Scientific) using Dharmafect (Thermo Scientific) as transfection reagent. Cells were lysed for RNA extraction as described above at the times indicated post-transfection. Knockdown efficiency was measured by RT-qPCR.
In vitro tube formation assay
HUVECs were trypsinized 48 h after transfection. BD Matrigel Basement Membrane Matrix (BD Biosciences) was added to 24-well plates and allowed to solidify at 37°C. Once the Matrigel solidified, HUVECs were re-seeded into Matrigel-coated wells (180,000 per well) and irradiated as described earlier. Angiogenic ability was assessed 18 h after re-seeding and irradiation by evaluating the total number of junctions after observation under a light microscope and the use of the Angiogenesis Analyzer plugin of ImageJ version 1.52a software (National Institutes of Health).
β-gal staining
At the indicated time following 0, 2 or 20 Gy, cells were stained using a β-Gal Staining Kit (Thermo Fischer Scientific) according to the supplier's instructions.
Quantification and statistical analysis
For the proteomics dataset, we used iQuantitator software which is based on a modeling approach, statistical methods and tools for estimating relative changes in protein expression under different treatments and experimental conditions (Schwacke et al., 2009). Parameter inference in iQuantitator uses computational methods developed using the Bayesian statistical framework. As a result, the normalized expression data are means ± credible intervals. A differential expression is statistically significant (noted with ∗ on the graphs) when the 95% credible interval does not contain 1 (the null effect). For other datasets, results are expressed as mean ± SEM. When the data meet the normality assumption (Shapiro-Wilk), we performed t-tests and one-way ANOVA followed by multiple comparisons tests (Tukey, Dunnett or Sidak). Otherwise, we used non-parametric statistical tests (Mann-Whitney Rank-Sum Test or Kruskal-Wallis one-way ANOVA on Ranks). All p values were corrected for multiple comparisons tests (Tukey or Dunn's). The type of test used is indicated in the figure legend for each dataset. A p value of <0.05 was considered statistically significant. ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001 for indicated comparisons, error bars represent standard error of means. The tests were performed using GraphPad Prism, version 8.1.1 for Windows, which was also used to build the graphs.
Acknowledgments
This work is supported by Electricité de France (Groupe Gestion Projet Radioprotection), the Institut de Radioprotection et de Sûreté Nucléaire (ROSIRIS program), INSERM, the University of Normandy-Rouen, the Region Normandie, and the European Union. Europe gets involved in Normandy with the European Regional Development Fund (ERDF). The funding agencies were not involved in any research plan relative to the study design, data collection and analysis, decision to publish, or drafting of the manuscript.
Author contributions
A.F., F.M., and O.G. conceived and designed the experiments. F.C., P.C., S. S., S.L., V.B., G.T., B.M., V.P., A.F., L.S., F.M., and O.G. performed experiments. I.M., G.T., and F.C. analyzed the miRNomic data. I.M., G.T., V.B., A.F., F.M., and O.G. analyzed the transcriptomic data. I.M., P.C., and D.V. analyzed the proteomic data. I.M., L.S., and J.C.M. analyzed the metabolomic data. I.M. performed formal analysis. I.M., P.C., L.S., J.C.M., and O.G. wrote the original manuscript. I.M., P.C., D.V., L.S., J.C.M., V.P., M.S, M.A.B., F.M., and O.G. reviewed and edited the manuscript. Funding acquisition was done by F.M. and O.G.; F.M. and O.G. supervised the project. All authors contributed to the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: January 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103685.
Contributor Information
Ian Morilla, Email: ian.morilla@gmail.com.
Olivier Guipaud, Email: olivier.guipaud@irsn.fr.
Supplemental information
Data S1 shows the 2-ΔCt and the Ct values of 293 miRNAs measured using RT-qPCR in unexposed HUVECs and HUVECs exposed to 20 Gy of ionizing radiation at different times post-irradiation
Data S2 shows the 2-ΔCt and the Ct values of 308 mRNAs measured using RT-qPCR in unexposed HUVECs and HUVECs exposed to 20 Gy of ionizing radiation at different times post-irradiation
Data S3 presents the mean expression ratios and credible intervals, as well as mass spectrometry data, of proteins quantified and identified using iTRAQ-LC-MS/MS in unexposed HUVECs and HUVECs exposed to 20 Gy of ionizing radiation at different times post-irradiation
Data S4 presents the intensity values and identifications names of metabolites quantified and identified using LC-MS/MS in unexposed HUVECs and HUVECs exposed to 20 Gy of ionizing radiation at different times post-irradiation
Chains of omics complexes are displayed in a user-friendly fashion as networks of homological measurements. To do this, we used interactive files in html format. In each file, nodes are colored according to the expression range and edges indicate the homological overlap between connected entities. It is also possible to hover over each node to numerically identify groups of entities with a similar lower or upper expression relative to the average. molecular entities are then selected depending on the number of barcodes considered in Figure S1 and their average profile expressions. MF 2, miRNome, MF 3, proteome, MF 4, transcriptome and MF 5, metabolome
Simulations of persistent homology per omics layer (miRNome, transcriptome, proteome and metabolome). This provides the trace of modules whose molecular entities topologically resemble each other therein. This is a recurrent homology observed between modules and throughout the whole transformation of projected data into continuous space process. Node and community colors amount to control or the different molecular response to irradiation
This learning model provides a better understanding of the inherent non-linear dynamic displayed by the expression of the selected signature in response to irradiation. The panels show the first (Epoch: 0) and the last (Epoch: 250) frames of the mp4 video. Node numbers are equivalent to each of those entities identified in the regulatory network (Figure S4) while they are colored according to their early (i.e. red) or late (i.e. green) response to irradiation
Data and code availability
-
•
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with accession number PRIDE: PXD030572. The mass spectrometry metabolomics data have been deposited at the MetaboLights database (Haug et al., 2020) with accession number MetaboLights: MTBLS3680. Both datasets are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Raw data from real-time quantitative PCR and microscopy data reported in this paper will be shared by the lead contact upon request. The miRNA, mRNA, proteome and metabolome processed data reported in this study are supplied as Data S1, S2, S3, and S4.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adams H., Vejdemo-Johansson M., Tausz A. In: Proceedings of ICMS. Hong H., Yap C., editors. Springer; 2014. JavaPlex: a research software package for persistent (co)-homology; pp. 129–136. [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh O., Sievert W., Sarioglu H., Merl-Pham J., Yentrapalli R., Bakshi M.V., Janik D., Ueffing M., Atkinson M.J., Multhoff G., et al. Integrative proteomics and targeted transcriptomics analyses in cardiac endothelial cells unravel mechanisms of long-term radiation-induced vascular dysfunction. J. Proteome Res. 2015;14:1203–1219. doi: 10.1021/pr501141b. [DOI] [PubMed] [Google Scholar]
- Belli M., Tabocchini M.A. Ionizing radiation-induced epigenetic modifications and their relevance to radiation protection. Int. J. Mol. Sci. 2020;21:5993. doi: 10.3390/ijms21175993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Kacem M., Benadjaoud M.A., Dos Santos M., Soysouvanh F., Buard V., Tarlet G., Le Guen B., Francois A., Guipaud O., Milliat F., et al. Variation of 4 MV X-ray dose rate strongly impacts biological response both in vitro and in vivo. Sci. Rep. 2020;10:7021. doi: 10.1038/s41598-020-64067-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsuk K. On the imbedding of systems of compacta in simplicial complexes. Fund. Math. 1948;35:217–234. [Google Scholar]
- Brown S., Banfill K., Aznar M.C., Whitehurst P., Faivre Finn C. The evolving role of radiotherapy in non-small cell lung cancer. Br. J. Radiol. 2019;92:20190524. doi: 10.1259/bjr.20190524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubenik P. Statistical topological data analysis using persistence landscapes. J. Mach. Learn. Res. 2015;16:77–102. [Google Scholar]
- Campisi J., d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- Carlsson G.E., Zomorodian A., Collins A., Guibas L. Persistence barcodes for shapes. Int. J. Shape Model. 2005;11:149–187. [Google Scholar]
- Chambers M.C., Maclean B., Burke R., Amodei D., Ruderman D.L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazal F., Fasy B.T., Lecc i.F., Rinaldo A., Wasserman L. Stochastic convergence of persistence landscapes and silhouettes. J. Comput. Geom. 2015;6:140–161. [Google Scholar]
- Chen S., Mar J.C. Evaluating methods of inferring gene regulatory networks highlights their lack of performance for single cell gene expression data. BMC Bioinf. 2018;19:232. doi: 10.1186/s12859-018-2217-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C.H., Chen S.U., Cheng J.C. Radiation-induced interleukin-6 expression through MAPK/p38/NF-kappaB signaling pathway and the resultant antiapoptotic effect on endothelial cells through Mcl-1 expression with sIL6-Ralpha. Int. J. Radiat. Oncol. Biol. Phys. 2009;75:1553–1561. doi: 10.1016/j.ijrobp.2009.08.034. [DOI] [PubMed] [Google Scholar]
- Christersdottir T., Pirault J., Gistera A., Bergman O., Gallina A.L., Baumgartner R., Lundberg A.M., Eriksson P., Yan Z.Q., Paulsson-Berne G., et al. Prevention of radiotherapy-induced arterial inflammation by interleukin-1 blockade. Eur. Heart J. 2019;40:2495–2503. doi: 10.1093/eurheartj/ehz206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corre I., Niaudet C., Paris F. Plasma membrane signaling induced by ionizing radiation. Mutat. Res. 2010;704:61–67. doi: 10.1016/j.mrrev.2010.01.014. [DOI] [PubMed] [Google Scholar]
- Cruchant O. The Apache Software Foundation; 2020. Faster, Cheaper, Leaner: Improving Real-Time ML Inference Using Apache MXNet.https://medium.com/apache-mxnet/faster-cheaper-leaner-improving-real-time-ml-inference-using-apache-mxnet-2ee245668b55 [Google Scholar]
- Defferrard M., Bresson X., Vandergheynst P. Convolutional neural networks on graphs with fast localised spectral filtering. Adv. Neural Inf. Process. Syst. 2016;29:3844–3852. [Google Scholar]
- Ding M., Tang J., Zhang J. CIKM '18: Proceedings of the 27th ACM International Conference on Information and Knowledge. ACM; ): 2018. Semi-supervised learning on graphs with generative adversarial nets; pp. 313–322. [Google Scholar]
- Eraslan G., Avsec Z., Gagneur J., Theis F.J. Deep learning: new computational modelling techniques for genomics. Nat. Rev. Genet. 2019;20:389–403. doi: 10.1038/s41576-019-0122-6. [DOI] [PubMed] [Google Scholar]
- Fasy B.T., Kim J., Lecci F., Maria C., Millman D.L., Rouvreau V. CRAN-R repository; 2019. TDA: Statistical Tools for Topological Data Analysis.https://cran.r-project.org/web/packages/TDA/index.html [Google Scholar]
- Fuks Z., Vlodavsky I., Andreeff M., McLoughlin M., Haimovitz-Friedman A. Effects of extracellular matrix on the response of endothelial cells to radiation in vitro. Eur. J. Cancer. 1992;28A:725–731. doi: 10.1016/0959-8049(92)90104-a. [DOI] [PubMed] [Google Scholar]
- Giacomoni F., Le Corguille G., Monsoor M., Landi M., Pericard P., Petera M., Duperier C., Tremblay-Franco M., Martin J.F., Jacob D., et al. Workflow4Metabolomics: a collaborative research infrastructure for computational metabolomics. Bioinformatics. 2015;31:1493–1495. doi: 10.1093/bioinformatics/btu813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannopoulou E., Katsoris P., Hatziapostolou M., Kardamakis D., Kotsaki E., Polytarchou C., Parthymou A., Papaioannou S., Papadimitriou E. X-rays modulate extracellular matrix in vivo. Int. J. Cancer. 2001;94:690–698. doi: 10.1002/ijc.1535. [DOI] [PubMed] [Google Scholar]
- Guipaud O., Jaillet C., Clement-Colmou K., Francois A., Supiot S., Milliat F. The importance of the vascular endothelial barrier in the immune-inflammatory response induced by radiotherapy. Br. J. Radiol. 2018;91:20170762. doi: 10.1259/bjr.20170762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halle M., Ekstrom M., Farnebo F., Tornvall P. Endothelial activation with prothrombotic response in irradiated microvascular recipient veins. J. Plast. Reconstr. Aesthet. Surg. 2010;63:1910–1916. doi: 10.1016/j.bjps.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Halle M., Gabrielsen A., Paulsson-Berne G., Gahm C., Agardh H.E., Farnebo F., Tornvall P. Sustained inflammation due to nuclear factor-kappa B activation in irradiated human arteries. J. Am. Coll. Cardiol. 2010;55:1227–1236. doi: 10.1016/j.jacc.2009.10.047. [DOI] [PubMed] [Google Scholar]
- Hamilton W.L., Ying R., Leskovec J. Inductive representation learning on large graphs. arXiv preprint. 2017 https://arxiv.org/abs/1609.02907v4 [Google Scholar]
- Hamilton W.L., Ying R., Leskovec J. Representation learning on graphs: methods and applications. arXiv preprint. 2017 https://arxiv.org/abs/1709.05584v3 [Google Scholar]
- Hatjikondi O., Ravazoula P., Kardamakis D., Dimopoulos J., Papaioannou S. In vivo experimental evidence that the nitric oxide pathway is involved in the X-ray-induced antiangiogenicity. Br. J. Cancer. 1996;74:1916–1923. doi: 10.1038/bjc.1996.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauer-Jensen M., Denham J.W., Andreyev H.J. Radiation enteropathy--pathogenesis, treatment and prevention. Nat. Rev. Gastroenterol. Hepatol. 2014;11:470–479. doi: 10.1038/nrgastro.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauer-Jensen M., Poulakos L., Osborne J.W. Effects of accelerated fractionation on radiation injury of the small intestine: a new rat model. Int. J. Radiat. Oncol. Biol. Phys. 1988;14:1205–1212. doi: 10.1016/0360-3016(88)90399-9. [DOI] [PubMed] [Google Scholar]
- Hauer-Jensen M., Poulakos L., Osborne J.W. Intestinal complications following accelerated fractionated x-irradiation. An experimental study in the rat. Acta Oncol. 1990;29:229–234. doi: 10.3109/02841869009126549. [DOI] [PubMed] [Google Scholar]
- Hauer-Jensen M., Sauer T., Devik F., Nygaard K. Late changes following single dose roentgen irradiation of rat small intestine. Acta Radiol. Oncol. 1983;22:299–303. doi: 10.3109/02841868309134045. [DOI] [PubMed] [Google Scholar]
- Haug K., Cochrane K., Nainala V.C., Williams M., Chang J., Jayaseelan K.V., O'Donovan C. MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020;48:D440–D444. doi: 10.1093/nar/gkz1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann J.C. On the Vietoris-Rips complexes and a cohomology theory for metric spaces. Ann. Math. Stud. 1995;138:175–188. [Google Scholar]
- Heinonen M., Guipaud O., Milliat F., Buard V., Micheau B., Tarlet G., Benderitter M., Zehraoui F., d'Alche-Buc F. Detecting time periods of differential gene expression using Gaussian processes: an application to endothelial cells exposed to radiotherapy dose fraction. Bioinformatics. 2015;31:728–735. doi: 10.1093/bioinformatics/btu699. [DOI] [PubMed] [Google Scholar]
- Heinonen M., Milliat F., Benadjaoud M.A., Francois A., Buard V., Tarlet G., d'Alche-Buc F., Guipaud O. Temporal clustering analysis of endothelial cell gene expression following exposure to a conventional radiotherapy dose fraction using Gaussian process clustering. PLoS One. 2018;13:e0204960. doi: 10.1371/journal.pone.0204960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskind C., Talbot C.J., Kerns S.L., Veldwijk M.R., Rosenstein B.S., West C.M. Radiogenomics: a systems biology approach to understanding genetic risk factors for radiotherapy toxicity? Cancer Lett. 2016;382:95–109. doi: 10.1016/j.canlet.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillet C., Morelle W., Slomianny M.C., Paget V., Tarlet G., Buard V., Selbonne S., Caffin F., Rannou E., Martinez P., et al. Radiation-induced changes in the glycome of endothelial cells with functional consequences. Sci. Rep. 2017;7:5290. doi: 10.1038/s41598-017-05563-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Qian X., Shen J., Wang Y., Li X., Liu R., Xia Y., Chen Q., Peng G., Lin S.Y., et al. Local generation of fumarate promotes DNA repair through inhibition of histone H3 demethylation. Nat. Cell Biol. 2015;17:1158–1168. doi: 10.1038/ncb3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T.I., Costa A.S.H., Ferguson A.N., Frezza C. Fumarate hydratase loss promotes mitotic entry in the presence of DNA damage after ionising radiation. Cell Death Dis. 2018;9:913. doi: 10.1038/s41419-018-0912-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski M., Majewski S., Kaminska G., Bem W., Szmurlo A. Enhancing effect of X-ray irradiation on a new blood vessel formation in mice tested by lymphocyte induced angiogenesis assay. Arch. Immunol. Ther. Exp. 1978;26:1075–1078. [PubMed] [Google Scholar]
- Kaminski M.J., Majewski S., Kaminska G., Bem W., Szmurlo A. Protease-mediated enhancement of lymphocyte-induced angiogenesis in X-ray irradiated mice. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1983;43:149–156. doi: 10.1080/09553008314550161. [DOI] [PubMed] [Google Scholar]
- Kawaguchi N., Tashiro K., Taniguchi K., Kawai M., Tanaka K., Okuda J., Hayashi M., Uchiyama K. Nogo-B (Reticulon-4B) functions as a negative regulator of the apoptotic pathway through the interaction with c-FLIP in colorectal cancer cells. Biochim. Biophys. Acta Mol. Basis Dis. 2018;1864:2600–2609. doi: 10.1016/j.bbadis.2018.04.018. [DOI] [PubMed] [Google Scholar]
- Kim K.S., Kim J.E., Choi K.J., Bae S., Kim D.H. Characterization of DNA damage-induced cellular senescence by ionizing radiation in endothelial cells. Int. J. Radiat. Biol. 2014;90:71–80. doi: 10.3109/09553002.2014.859763. [DOI] [PubMed] [Google Scholar]
- Kipf T.N., Welling M. Semi-supervised classification with graph convolutional networks. arXiv preprint. 2016 https://arxiv.org/abs/1609.02907v4 [Google Scholar]
- Knyazev B., Lin X., Amer M.R., Taylor G.W. Spectral multigraph networks for discovering and fusing relationships in molecules. arXiv preprint. 2018 https://arxiv.org/abs/1811.09595 [Google Scholar]
- Korbel F. CreateSpace Independent Publishing Platform); 2012. FFmpeg Basics: Multimedia Handling with a Fast Audio and Video Encoder. [Google Scholar]
- Korpela E., Liu S.K. Endothelial perturbations and therapeutic strategies in normal tissue radiation damage. Radiat. Oncol. 2014;9:266. doi: 10.1186/s13014-014-0266-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafargue A., Degorre C., Corre I., Alves-Guerra M.C., Gaugler M.H., Vallette F., Pecqueur C., Paris F. Ionizing radiation induces long-term senescence in endothelial cells through mitochondrial respiratory complex II dysfunction and superoxide generation. Free Radic. Biol. Med. 2017;108:750–759. doi: 10.1016/j.freeradbiomed.2017.04.019. [DOI] [PubMed] [Google Scholar]
- Langberg C.W., Sauer T., Reitan J.B., Hauer-Jensen M. Tolerance of rat small intestine to localized single dose and fractionated irradiation. Acta Oncol. 1992;31:781–787. doi: 10.3109/02841869209083871. [DOI] [PubMed] [Google Scholar]
- Lauwers O., De Moor B. A time series distance measure for efficient clustering of input/output signals by their underlying dynamics. IEEE Control Syst. Lett. 2017;1:286–291. [Google Scholar]
- Lees J.G., Heriche J.K., Morilla I., Fernandez J.M., Adler P., Krallinger M., Vilo J., Valencia A., Ellenberg J., Ranea J.A., et al. FUN-L: gene prioritization for RNAi screens. Bioinformatics. 2015;31:2052–2053. doi: 10.1093/bioinformatics/btv073. [DOI] [PubMed] [Google Scholar]
- Leonawicz M. 2017. Mapmate.https://github.com/leonawicz/mapmate [Google Scholar]
- Li Q., Qi B., Oka K., Shimakage M., Yoshioka N., Inoue H., Hakura A., Kodama K., Stanbridge E.J., Yutsudo M. Link of a new type of apoptosis-inducing gene ASY/Nogo-B to human cancer. Oncogene. 2001;20:3929–3936. doi: 10.1038/sj.onc.1204536. [DOI] [PubMed] [Google Scholar]
- Lloyd S.P. Least squares quantization in PCM. IEEE Trans. Inf. Theor. 1982;28:129–137. [Google Scholar]
- Lo S.S., Fakiris A.J., Chang E.L., Mayr N.A., Wang J.Z., Papiez L., Teh B.S., McGarry R.C., Cardenes H.R., Timmerman R.D. Stereotactic body radiation therapy: a novel treatment modality. Nat. Rev. Clin. Oncol. 2010;7:44–54. doi: 10.1038/nrclinonc.2009.188. [DOI] [PubMed] [Google Scholar]
- Lowe D., Raj K. Premature aging induced by radiation exhibits pro-atherosclerotic effects mediated by epigenetic activation of CD44 expression. Aging Cell. 2014;13:900–910. doi: 10.1111/acel.12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P., Van Vlierberghe P., De Weer A., Muth D., Westermann F., Speleman F., Vandesompele J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10:R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metheetrairut C., Slack F.J. MicroRNAs in the ionizing radiation response and in radiotherapy. Curr. Opin. Genet. Dev. 2013;23:12–19. doi: 10.1016/j.gde.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Ebert D., Huang X., Thomas P.D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47:D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milliat F., Sabourin J.C., Tarlet G., Holler V., Deutsch E., Buard V., Tamarat R., Atfi A., Benderitter M., Francois A. Essential role of plasminogen activator inhibitor type-1 in radiation enteropathy. Am. J. Pathol. 2008;172:691–701. doi: 10.2353/ajpath.2008.070930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintet E., Rannou E., Buard V., West G., Guipaud O., Tarlet G., Sabourin J.C., Benderitter M., Fiocchi C., Milliat F., et al. Identification of endothelial-to-mesenchymal transition as a potential participant in radiation proctitis. Am. J. Pathol. 2015;185:2550–2562. doi: 10.1016/j.ajpath.2015.04.028. [DOI] [PubMed] [Google Scholar]
- Molla M., Panes J., Casadevall M., Salas A., Conill C., Biete A., Anderson D.C., Granger D.N., Pique J.M. Influence of dose-rate on inflammatory damage and adhesion molecule expression after abdominal radiation in the rat. Int. J. Radiat. Oncol. Biol. Phys. 1999;45:1011–1018. doi: 10.1016/s0360-3016(99)00286-2. [DOI] [PubMed] [Google Scholar]
- Morilla I., Doblas S., Garteiser P., Zappa M., Ogier-Denis E. In: Bioinform Biomed Eng. Rojas I., Ortuño F., editors. Springer; 2017. Scores of intestinal fibrosis from wavelet-based magnetic resonance imaging models; pp. 569–578. [Google Scholar]
- Morilla I., Ranea J.A. Mathematical deconvolution uncovers the genetic regulatory signal of cancer cellular heterogeneity on resistance to paclitaxel. Mol. Genet. Genom. 2017;292:857–869. doi: 10.1007/s00438-017-1316-2. [DOI] [PubMed] [Google Scholar]
- Munch E. A user’s guide to topological data analysis. J. Learn. Analytics. 2017;4:47–61. [Google Scholar]
- Naderi E.H., Findley H.W., Ruud E., Blomhoff H.K., Naderi S. Activation of cAMP signaling inhibits DNA damage-induced apoptosis in BCP-ALL cells through abrogation of p53 accumulation. Blood. 2009;114:608–618. doi: 10.1182/blood-2009-02-204883. [DOI] [PubMed] [Google Scholar]
- Naderi S., Wang J.Y., Chen T.T., Gutzkow K.B., Blomhoff H.K. cAMP-mediated inhibition of DNA replication and S phase progression: involvement of Rb, p21Cip1, and PCNA. Mol. Biol. Cell. 2005;16:1527–1542. doi: 10.1091/mbc.E04-06-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitin A., Egorov S., Daraselia N., Mazo I. Pathway studio–the analysis and navigation of molecular networks. Bioinformatics. 2003;19:2155–2157. doi: 10.1093/bioinformatics/btg290. [DOI] [PubMed] [Google Scholar]
- Oudot S.Y. American Mathematical Society; 2015. Persistence Theory: From Quiver Representations to Data Analysis. [Google Scholar]
- Paget V., Ben Kacem M., Dos Santos M., Benadjaoud M.A., Soysouvanh F., Buard V., Georges T., Vaurijoux A., Gruel G., Francois A., et al. Multiparametric radiobiological assays show that variation of X-ray energy strongly impacts relative biological effectiveness: comparison between 220 kV and 4 MV. Sci. Rep. 2019;9:14328. doi: 10.1038/s41598-019-50908-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L., Tuccoli A., Mariani L., Evangelista M., Citti L., Woods K., Mercatanti A., Hammond S., Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- Polytarchou C., Gligoris T., Kardamakis D., Kotsaki E., Papadimitriou E. X-rays affect the expression of genes involved in angiogenesis. Anticancer Res. 2004;24:2941–2945. [PubMed] [Google Scholar]
- Qiu B., Aili A., Xue L., Jiang P., Wang J. Advances in radiobiology of stereotactic ablative radiotherapy. Front. Oncol. 2020;10:1165. doi: 10.3389/fonc.2020.01165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannou E., Francois A., Toullec A., Guipaud O., Buard V., Tarlet G., Mintet E., Jaillet C., Iruela-Arispe M.L., Benderitter M., et al. In vivo evidence for an endothelium-dependent mechanism in radiation-induced normal tissue injury. Sci. Rep. 2015;5:15738. doi: 10.1038/srep15738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., Vilo J. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S., Seo S., Kim S. Hybrid approach of relation network and localized graph convolutional filtering for breast cancer subtype classification. arXiv preprint. 2018 https://arxiv.org/abs/1711.05859 [Google Scholar]
- Rochat B. Proposed confidence scale and ID score in the identification of known-unknown compounds using high resolution MS data. J. Am. Soc. Mass Spectrom. 2017;28:709–723. doi: 10.1007/s13361-016-1556-0. [DOI] [PubMed] [Google Scholar]
- Rose R.W., Grant D.S., O'Hara M.D., Williamson S.K. The role of laminin-1 in the modulation of radiation damage in endothelial cells and differentiation. Radiat. Res. 1999;152:14–28. [PubMed] [Google Scholar]
- Sasaki N., Itakura Y., Toyoda M. Rapamycin promotes endothelial-mesenchymal transition during stress-induced premature senescence through the activation of autophagy. Cell Commun. Signal. 2020;18:43. doi: 10.1186/s12964-020-00533-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke J.H., Hill E.G., Krug E.L., Comte-Walters S., Schey K.L. iQuantitator: a tool for protein expression inference using iTRAQ. BMC Bioinf. 2009;10:342. doi: 10.1186/1471-2105-10-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoville N.A. American Mathematical Society; 2019. Discrete Morse Theory. [Google Scholar]
- Siemerink M.J., Klaassen I., Vogels I.M., Griffioen A.W., Van Noorden C.J., Schlingemann R.O. CD34 marks angiogenic tip cells in human vascular endothelial cell cultures. Angiogenesis. 2012;15:151–163. doi: 10.1007/s10456-011-9251-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone C.B., 2nd, Wildt B., Haas A.R., Pope G., Rengan R., Hahn S.M. Stereotactic body radiation therapy for lung cancer. Chest. 2013;143:1784–1790. doi: 10.1378/chest.12-2580. [DOI] [PubMed] [Google Scholar]
- Singh G., Memoli F., Carlsson G.E. In: Eurographics Symposium on Point-Based Graphics. Botsch M., Pajarola R., Chen B., Zwicker M., editors. The Eurographics Association; 2007. Topological methods for the analysis of high dimensional data sets and 3D object recognition; pp. 91–100. ()). [Google Scholar]
- Soysouvanh F., Benadjaoud M.A., Dos Santos M., Mondini M., Lavigne J., Bertho A., Buard V., Tarlet G., Adnot S., Deutsch E., et al. Stereotactic lung irradiation in mice promotes long-term senescence and lung injury. Int. J. Radiat. Oncol. Biol. Phys. 2020;106:1017–1027. doi: 10.1016/j.ijrobp.2019.12.039. [DOI] [PubMed] [Google Scholar]
- Stillwell J. Springer-Verlag New York; 1993. Classical Topology and Combinatorial Group Theory. [Google Scholar]
- Tauzin G., Lupo U., Tunstall L., Burella Pérez J., Caorsi M., Reise W., Medina-Mardones A., Dassatti A., Hess K. Giotto-Tda: a topological data analysis toolkit for machine learning and data exploration. arXiv preprint. 2020 https://arxiv.org/abs/2004.02551 [Google Scholar]
- Toullec A., Buard V., Rannou E., Tarlet G., Guipaud O., Robine S., Iruela-Arispe M.L., Francois A., Milliat F. HIF-1alpha deletion in the endothelium, but not in the epithelium, protects from radiation-induced enteritis. Cell. Mol. Gastroenterol. Hepatol. 2018;5:15–30. doi: 10.1016/j.jcmgh.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kloet F.M., Bobeldijk I., Verheij E.R., Jellema R.H. Analytical error reduction using single point calibration for accurate and precise metabolomic phenotyping. J. Proteome Res. 2009;8:5132–5141. doi: 10.1021/pr900499r. [DOI] [PubMed] [Google Scholar]
- van der Maaten L.J.P., Hinton G.E. Visualizing data using t-SNE. J. Mach Learn. Res. 2008;9:2579–2605. [Google Scholar]
- van Veen H.J., Saul N. 2019. KeplerMapper. [DOI] [Google Scholar]
- Vaughan D.E., Rai R., Khan S.S., Eren M., Ghosh A.K. Plasminogen activator inhibitor-1 is a marker and a mediator of senescence. Arterioscler. Thromb. Vasc. Biol. 2017;37:1446–1452. doi: 10.1161/ATVBAHA.117.309451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos I.S., Kostoulas N., Vergoulis T., Georgakilas G., Reczko M., Maragkakis M., Paraskevopoulou M.D., Prionidis K., Dalamagas T., Hatzigeorgiou A.G. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012;40:W498–W504. doi: 10.1093/nar/gks494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner-Ecker M., Schwager C., Wirkner U., Abdollahi A., Huber P.E. MicroRNA expression after ionizing radiation in human endothelial cells. Radiat. Oncol. 2010;5:25. doi: 10.1186/1748-717X-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C.T., et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijerathne H., Langston J.C., Yang Q., Sun S., Miyamoto C., Kilpatrick L.E., Kiani M.F. Mechanisms of radiation-induced endothelium damage: emerging models and technologies. Radiother. Oncol. 2021;158:21–32. doi: 10.1016/j.radonc.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S., Feunang Y.D., Marcu A., Guo A.C., Liang K., Vazquez-Fresno R., Sajed T., Johnson D., Li C., Karu N., et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46:D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev O., Singer E., Shaulian E., Goldberg M., Fox T.D., Pines O. Fumarase: a mitochondrial metabolic enzyme and a cytosolic/nuclear component of the DNA damage response. PLoS Biol. 2010;8:e1000328. doi: 10.1371/journal.pbio.1000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Wang J., Koteliansky V.E., Gotwals P.J., Hauer-Jensen M. Recombinant soluble transforming growth factor beta type II receptor ameliorates radiation enteropathy in mice. Gastroenterology. 2000;119:1286–1296. doi: 10.1053/gast.2000.19282. [DOI] [PubMed] [Google Scholar]
- Zirkel A., Nikolic M., Sofiadis K., Mallm J.P., Brackley C.A., Gothe H., Drechsel O., Becker C., Altmuller J., Josipovic N., et al. HMGB2 loss upon senescence entry disrupts genomic organization and induces CTCF clustering across cell types. Mol. Cell. 2018;70:730–744.e736. doi: 10.1016/j.molcel.2018.03.030. [DOI] [PubMed] [Google Scholar]
- Zitvogel L., Kroemer G. Subversion of anticancer immunosurveillance by radiotherapy. Nat. Immunol. 2015;16:1005–1007. doi: 10.1038/ni.3236. [DOI] [PubMed] [Google Scholar]
- Zomorodian A., Carlsson G.E. Computing persistent homology. Discrete Comput. Geom. 2004;33:249–274. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 shows the 2-ΔCt and the Ct values of 293 miRNAs measured using RT-qPCR in unexposed HUVECs and HUVECs exposed to 20 Gy of ionizing radiation at different times post-irradiation
Data S2 shows the 2-ΔCt and the Ct values of 308 mRNAs measured using RT-qPCR in unexposed HUVECs and HUVECs exposed to 20 Gy of ionizing radiation at different times post-irradiation
Data S3 presents the mean expression ratios and credible intervals, as well as mass spectrometry data, of proteins quantified and identified using iTRAQ-LC-MS/MS in unexposed HUVECs and HUVECs exposed to 20 Gy of ionizing radiation at different times post-irradiation
Data S4 presents the intensity values and identifications names of metabolites quantified and identified using LC-MS/MS in unexposed HUVECs and HUVECs exposed to 20 Gy of ionizing radiation at different times post-irradiation
Chains of omics complexes are displayed in a user-friendly fashion as networks of homological measurements. To do this, we used interactive files in html format. In each file, nodes are colored according to the expression range and edges indicate the homological overlap between connected entities. It is also possible to hover over each node to numerically identify groups of entities with a similar lower or upper expression relative to the average. molecular entities are then selected depending on the number of barcodes considered in Figure S1 and their average profile expressions. MF 2, miRNome, MF 3, proteome, MF 4, transcriptome and MF 5, metabolome
Simulations of persistent homology per omics layer (miRNome, transcriptome, proteome and metabolome). This provides the trace of modules whose molecular entities topologically resemble each other therein. This is a recurrent homology observed between modules and throughout the whole transformation of projected data into continuous space process. Node and community colors amount to control or the different molecular response to irradiation
This learning model provides a better understanding of the inherent non-linear dynamic displayed by the expression of the selected signature in response to irradiation. The panels show the first (Epoch: 0) and the last (Epoch: 250) frames of the mp4 video. Node numbers are equivalent to each of those entities identified in the regulatory network (Figure S4) while they are colored according to their early (i.e. red) or late (i.e. green) response to irradiation
Data Availability Statement
-
•
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with accession number PRIDE: PXD030572. The mass spectrometry metabolomics data have been deposited at the MetaboLights database (Haug et al., 2020) with accession number MetaboLights: MTBLS3680. Both datasets are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Raw data from real-time quantitative PCR and microscopy data reported in this paper will be shared by the lead contact upon request. The miRNA, mRNA, proteome and metabolome processed data reported in this study are supplied as Data S1, S2, S3, and S4.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.