Abstract
Aim
Accurate preoperative diagnosis of lateral lymph node metastasis (LLNM) from lower rectal cancer is important to identify patients who require lateral lymph node dissection (LLND). We aimed to create an effective prediction model for LLNM using machine learning by combining preoperative information.
Methods
We retrospectively examined patients who underwent primary rectal cancer surgery with unilateral or bilateral LLND between April 2010 and March 2020 at a single institution. Using the machine learning software “Prediction One” (Sony Network Communications), we developed a prediction model in the training cohort that included 267 consecutive patients (500 sides) from April 2010. Clinicopathological data obtained from the preoperative examinations were used as the learning items. In the validation cohort that included subsequent patients until March 2020, we compared the discriminating powers of the prediction model and the conventional method using the short‐axis diameter of the largest lateral lymph node, as detected on magnetic resonance imaging.
Results
The area under the receiver operating characteristic curve (AUC) of the prediction model was 0.903 in the validation cohort comprising 56 patients (107 sides). This indicated significantly higher predictive power than that of the conventional method (AUC = 0.754; P = .022). Using the cutoff values defined in the training cohort, the accuracy, sensitivity, and specificity of the prediction model were 80.4%, 90.0%, and 79.4%, respectively. The model was able to correctly predict four of five sides comprising LLNM with the short‐axis diameters ≤4 mm.
Conclusion
Machine learning contributed to the creation of an effective prediction model for LLNM.
Keywords: lateral lymph node dissection, lateral lymph node metastasis, machine learning, preoperative diagnosis, rectal cancer
Accurate preoperative diagnosis of lateral lymph node metastasis from lower rectal cancer is important to identify patients who require lateral lymph node dissection. Machine learning based on deep learning contributed to the creation of an effective prediction model for lateral lymph node metastasis.
1. INTRODUCTION
During the evolution of surgical treatment for rectal cancer, Ernest Miles investigated the precise pathological nature of the lymphatic spread of lower rectal cancer. Lymphatic spread from rectal cancer could occur in three directions, namely, upwards, downwards, and laterally. 1 Numerous additional studies have revealed the presence of lateral lymph node metastasis (LLNM) from lower rectal cancer. Retrospective studies from Japan reported that the incidence of pathological LLNM in patients with T3 or T4 lower rectal cancer was approximately 15%, and reliable treatment strategies are required to prevent local recurrences caused by LLNM. 2
Preoperative chemoradiotherapy (CRT) combined with total mesorectal excision (TME) is the standard therapy for locally advanced rectal cancer in Western countries. 3 Recently, the effectiveness of lateral lymph node dissection (LLND) for the local control of lower rectal cancer has gained interest 4 because preoperative CRT with only TME for patients with enlarged lateral lymph node (LLN) is insufficient to prevent a lateral pelvic local recurrence. 5 , 6 Considering the high efficacy of LLND for patients with LLNM, 7 an accurate preoperative diagnosis of LLNM is important to identify those who require LLND.
Magnetic resonance imaging (MRI) is one of the most useful tools for preoperative diagnosis. 2 , 3 Measuring the size of LLN using MRI is a common and conventional method for staging rectal cancer. However, it is difficult to diagnose LLNM, particularly in patients with small LLN or no enlarged LLN. This necessitates improved predictive methods that diagnose with high sensitivity regardless of the status of node enlargement in the era of selective LLND.
In recent years, the effectiveness of artificial intelligence (AI) has been investigated in various medical fields, such as risk factors, diagnosis, and prognosis prediction. 8 , 9 Machine learning, as a type of AI, supposedly has better flexibility and scalability than conventional biostatistical methods. Factors associated with LLNM include not only the status of node enlargement but also multiple clinicopathological factors. 10 Thus, machine learning, which can use these parameters comprehensively for diagnosis, may facilitate accurate diagnosis. To the best of our knowledge, there have been no reports of using machine learning to create a prediction model for LLNM from lower rectal cancer.
Herein, we aimed to create a prediction model for LLNM using machine learning by combining preoperative images and clinicopathological factors. Moreover, we intended to determine if the prediction model could help distinguish between patients with lower rectal cancer, with and without LLNM, and to compare the discriminating power of the prediction model with that of the conventional method using MRI.
2. PATIENTS AND METHODS
2.1. Patients
We retrospectively examined the patients who underwent primary rectal cancer surgery with LLND between April 2010 and March 2020 at the Shizuoka Cancer Center Hospital, using a prospectively collected database. We included patients who underwent unilateral or bilateral LLND dissecting the internal iliac node and the obturator node. Patients who underwent preoperative CRT, were diagnosed with cStage IV, underwent total pelvic exenteration, and did not have preoperative MRI images were excluded. We analyzed the study patients on each side. In other words, a patient with bilateral LLND was dealt as two sides, and one with unilateral LLND was dealt as one side. All study protocols were approved by the Institutional Review Board of the Shizuoka Cancer Center (Institutional code: J2020‐57‐2020‐1–3).
2.2. Preoperative diagnosis
Preoperative tumor staging was performed by a digital examination, barium enema, colonoscopy, computed tomography (CT), and MRI. High‐resolution MRI was performed using a 3.0‐T system (Achieva 3.0T dStream; Royal Philips Healthcare). We primarily used T2‐weighted images with a slice thickness of 5 mm to diagnose the depth of tumor invasion and lymph node metastasis. All lymph nodes detected on MRI were measured by colorectal surgeons before the surgery, of which lymph nodes with the short‐axis diameters ≥6 mm were considered as metastases. Patients were staged using the tumor node metastasis classification (Union for International Cancer Control, eighth edition). The multidisciplinary team consisting of surgeons and physicians specialized in colorectal cancer eventually confirmed their preoperative diagnosis and treatment strategies.
2.3. Treatment strategies for locally advanced lower rectal cancer
Based on the Japan guidelines, 2 our indications for LLND were either lower rectal cancer with cT3‐4anyN or cT1‐2 rectal cancer with LLNM on preoperative images. The standard treatment for locally advanced lower rectal cancer was TME with bilateral LLND, which included the complete lymph node dissection for three parts (the common iliac node, internal iliac node, and obturator node). However, for patients older than 75 years or those with severe comorbid conditions, we omitted LLND or considered only unilateral LLND for the metastatic site. Neoadjuvant CRT (50.4 Gy in 28 fractions for 6 weeks with systemic capecitabine chemotherapy) was indicated only for patients who required tumor shrinkage to either obtain a clear resection margin, preserve the anus, or avoid urinary diversion at our institution.
2.4. Machine learning
We used the machine‐learning software “Prediction One” (Sony Network Communications, https://predictionone.sony.biz/). This software uses “Neural Network Libraries” (https://nnabla.org/), which is an open‐source software of deep learning. Based on the learning items, it automatically performs machine‐learning analysis, such as neural networks and gradient boosting trees, and creates a prediction model easily and rapidly by cross‐validation. The prediction model calculates the predicted value by entering the items of each patient. Despite no information on the details of its analysis, this software, which had been created originally for nonmedical applications, is likely to be applied in various medical fields. 11 Moreover, it can be used without connecting to the Internet to analyze anonymized information. Thus, there is no concern about the leakage of patient information.
2.5. Learning items
To develop the prediction model for LLNM from lower rectal cancer, we used the clinicopathological data obtained from only the preoperative examinations as the learning items. The learning items included the age, sex, body mass index, serum carcinoembryonic antigen, and carbohydrate antigen 19‐9 levels. In addition, the items comprised tumor characteristics, including the distance from the anal verge, macroscopic type determined by colonoscopy, tumor diameter, the circumferential rate of lumen, tumor localization of the rectal wall (anterior, posterior, right, and left side, and entire circumference), histological type proven by endoscopic biopsy, and cT stage.
Furthermore, the short‐axis diameter of the largest perirectal lymph node (PLN) and LLN, as detected on T2‐weighted axial images of MRI, were included in the learning items. Two researchers, S.K., who is a doctor with 6 years of experience specialized in colorectal surgery, and A.S., who is an expert colorectal surgeon with over 20 years of experience in colorectal cancer, unaware of the presence of lymph node metastasis or the prognosis of the patients, measured and matched the maximum lymph node diameter of each patient. Moreover, the measured lymph nodes did not always correspond one‐to‐one with the pathological results.
2.6. An evaluation of the prediction model and statistical analyses
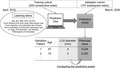
Figure 1 depicts the development of a prediction model for LLNM from lower rectal cancer and the steps to evaluate its predictive power. Using the hold‐out method, the training cohort included 500 consecutive sides from April 2010, while the validation cohort included 107 subsequent sides until March 2020. First, “Prediction One” created a model from the aforementioned learning items of the training cohort, considering the information about pathological LLNM. Next, the model predicted the LLNM for the validation cohort, and we checked the success or failure of the predictions based on the information about pathological LLNM. We measured the receiver operating characteristic (ROC) curves for the discriminating power of the prediction model. Furthermore, we measured the area under ROC curves (AUC) to compare the model and the conventional method using the short‐axis diameter of the largest LLN, as detected on MRI. The Delong test was used to compare the ROC curves. In addition, we determined the predicted cutoff values for LLNM and examined the accuracy, sensitivity, and specificity of the validation cohort using the Youden index, 12 defined as the maximum of (sensitivity + specificity – 1) in the ROC analysis of the training cohort.
FIGURE 1.
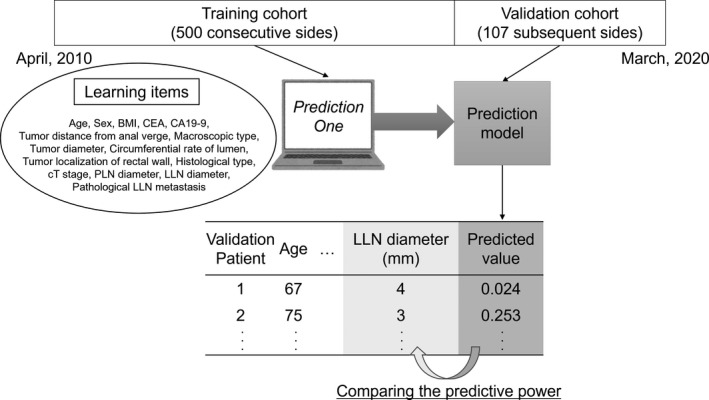
The development of the prediction model. “Prediction One (Sony Network Communications)” has created a model based on the learning items of the training cohort. The prediction model calculates the value of each patient in the validation cohort. The predictive power of this model has been compared to that of the conventional method using the LLN diameter. BMI, body mass index; CEA, carcinoembryonic antigen; CA19‐9, carbohydrate antigen 19‐9; LLN, lateral lymph node; PLN, perirectal lymph node
While we conducted Fisher's exact tests to analyze the categorical variables, the Mann–Whitney U‐test was performed to compare continuous variables between the groups. All P‐values were two‐sided, and values <.05 were considered statistically significant. We performed all statistical analyses, including the evaluation of the machine learning, with EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan), a graphical user interface for R (R Foundation for Statistical Computing, Vienna, Austria, v. 2.13.0). 13 More precisely, it is a modified version of R commander (v. 1.6‐3), designed to add frequently used statistical functions in biostatistics.
3. RESULTS
We evaluated 323 patients, comprising 267 consecutive patients (500 sides) and 56 subsequent patients (107 sides) in the training and validation cohort, respectively (Figure 2). Table 1 presents the characteristics of patients in both cohorts. Despite some significant differences in the learning items between the cohorts, there was no significant difference in the rates of LLNM.
FIGURE 2.
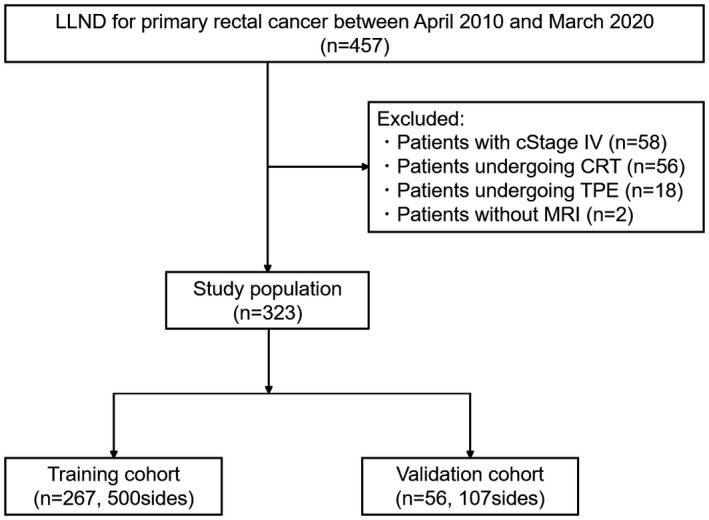
Patient selection process. CRT, chemoradiotherapy; LLND, lateral lymph node dissection; MRI, magnetic response imaging; TPE, total pelvic exenteration
TABLE 1.
Characteristics of patients in the training and validation cohort
| Training cohort (500 sides) | Validation cohort (107 sides) | P‐value | |
|---|---|---|---|
| Age (years) | 63 [26–84] | 67 [40–79] | .017 |
| Sex male/female | 367/133 | 61/46 | .001 |
| Side right/left | 251/249 | 53/54 | .916 |
| BMI (kg/m2) | 22.8 [15.7–37.6] | 22.5 [14.3–31.2] | .058 |
| CEA (ng/mL) | 5.7 [0.5–252.6] | 2.7 [0.7–61.7] | <.001 |
| CA19‐9 (U/mL) | 11 [2–2145] | 9 [2–120] | .466 |
| Tumor distance from anal verge (cm) | 5 [0–11] | 5 [1–10] | .029 |
| Macroscopic type 0/1/2/3/4/5 | 3/16/468/11/0/2 | 0/0/105/2/0/0 | .332 |
| Tumor diameter (cm) | 5 [1.5–11] | 4.5 [2–9] | .012 |
| Circumferential rate of lumen (%) | 50 [20–100] | 50 [20–100] | .469 |
| Tumor localization of rectal wall ant/post/same/opposite/circ | 135/128/80/72/85 | 38/28/13/12/16 | .445 |
| Histological type pap/well/mod/por/muc | 0/202/276/10/12 | 4/51/42/3/7 | <.001 |
| cT 1/2/3/4 | 2/9/408/81 | 0/3/89/15 | .721 |
| PLN diameter (mm) | 6 [2–19] | 6 [3–17] | .003 |
| LLN diameter (mm) | 3 [2–15] | 3 [2–11] | .228 |
| Operative approach open/laparoscopic/robotic | 128/46/326 | 2/0/105 | <.001 |
| Operative procedure LAR/ISR/APR/Hartmann | 287/109/102/2 | 69/16/22/0 | .369 |
| Pathological LLN metastasis presence/absence | 46/454 | 10/97 | >.999 |
Abbreviations: ant, anterior side; BMI, body mass index; CA19‐9, carbohydrate antigen 19‐9; CEA, carcinoembryonic antigen; circ, entire circumference; APR, abdominoperineal resection; opposite, opposite side as the side dissected; ISR, intersphincteric resection; LAR, low anterior resection; LLN, lateral lymph node; mod, moderately differentiated tubular adenocarcinoma; muc, mucinous adenocarcinoma; pap, papillary adenocarcinoma; PLN, perirectal lymph node; por, poorly differentiated adenocarcinoma; post, posterior side; same, same side as the side dissected; well, well differentiated tubular adenocarcinoma.
“Prediction One” created the model for LLNM from lower rectal cancer, based on the learning items of the training cohort. Although the details of the analysis could not be obtained from the software, LLN diameter, carcinoembryonic antigen, body mass index, carbohydrate antigen 19‐9, and PLN diameter were identified as influential factors in this order in predicting LLNM. In the training cohort, the AUC of the prediction model was 0.969 (95% confidence interval (CI), 0.944–0.965), which indicated significantly higher predictive power than the conventional method using the short‐axis diameters of LLN, as detected on MRI (AUC = 0.855; 95% CI, 0.792–0.917; P < .001) (Figure 3). Figure 4 displays the ROC curves of the prediction model and the conventional method in the validation cohort. In the validation cohort, the AUC of the prediction model was 0.903 (95% CI, 0.832–0.974), which indicated significantly higher predictive power than the conventional method (AUC = 0.754; 95% CI, 0.578–0.930; P = .022).
FIGURE 3.
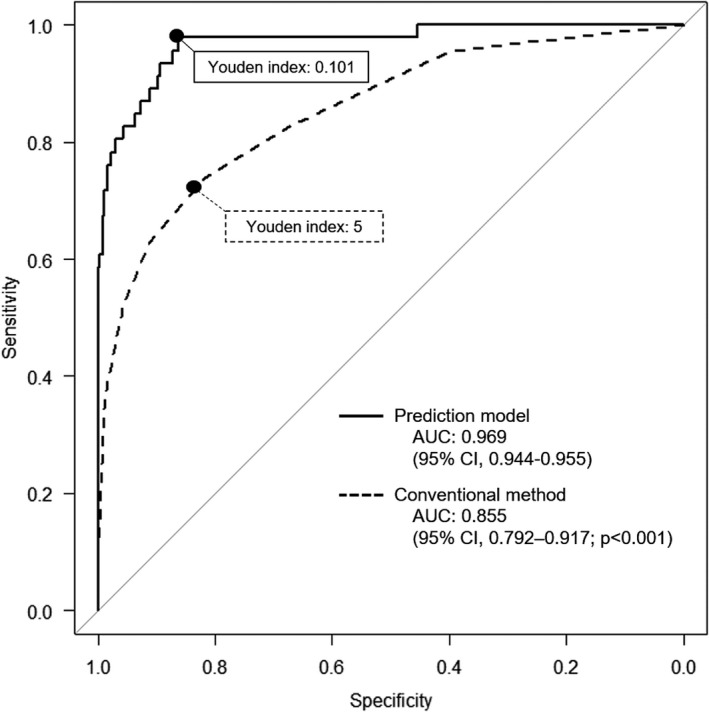
ROC curves of the prediction model and the conventional diagnosis method in the training cohort. The predictive power of the prediction model was compared to that of the conventional method using the short‐axis diameter of the largest LLN, as detected on MRI in the training cohort. The predicted cutoff values for LLN metastasis determined using the Youden index were ≥0.101 (sensitivity, 97.8%; specificity, 86.3%), while the conventional cutoff values of short‐axis diameters were ≥5 mm (sensitivity, 73.9%; specificity, 81.7%). AUC, area under ROC curves; CI, confidence interval; LLN, lateral lymph node; MRI, magnetic resonance imaging; ROC curves, receiver operating characteristic curves
FIGURE 4.
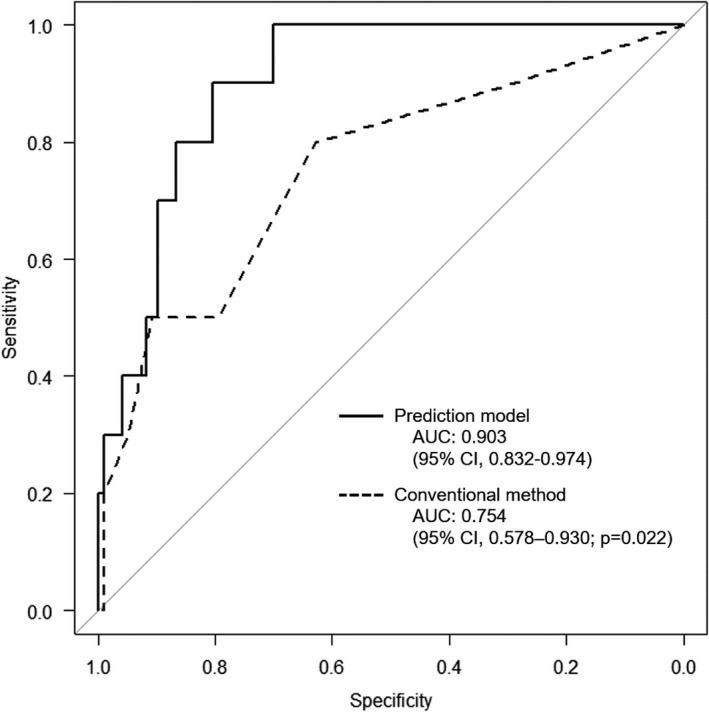
ROC curves of the prediction model and the conventional diagnosis method in the validation cohort. The predictive power of the prediction model was compared to that of the conventional method using the short‐axis diameter of the largest LLN, as detected on MRI in the validation cohort. AUC, area under ROC curves; CI, confidence interval; LLN, lateral lymph node; MRI, magnetic resonance imaging; ROC curves, receiver operating characteristic curves
The ROC analysis of the training cohort and the Youden index demonstrated predicted cutoff values ≥0.101 for LLNM from lower rectal cancer (Figure 3). This cutoff value in the validation cohort generated an accuracy, sensitivity, and specificity of 80.4%, 90.0%, and 79.4%, respectively. In contrast, the conventional cutoff values of short‐axis diameters ≥5 mm on MRI in the validation cohort produced an accuracy, sensitivity, and specificity of 76.6%, 50.0%, and 79.4%, respectively (Table 2). Table 3 presents the patients with LLNM in the validation cohort. The model was able to correctly predict four of five sides comprising LLNM with the short‐axis diameters ≤4 mm. However, only one side could not be predicted correctly by the prediction model.
TABLE 2.
Sensitivity, specificity, and accuracy of each method in the validation cohort
| Method | Cutoff | Sensitivity | Specificity | Accuracy | Positive predictive value | Negative predictive value |
|---|---|---|---|---|---|---|
| Conventional method | >4 mm | 80.0% (8/10) | 62.9% (61/97) | 64.5% (69/107) | 18.2% (8/44) | 96.8% (61/63) |
| Conventional method | >5 mm | 50.0% (5/10) | 79.4% (77/97) | 76.6% (82/107) | 20.0% (5/25) | 93.9% (77/82) |
| Conventional method | >6 mm | 50.0% (5/10) | 90.7% (88/97) | 86.9% (93/107) | 35.7% (5/14) | 94.6% (88/93) |
| Prediction model | >0.101 | 90.0% (9/10) | 79.4% (77/97) | 80.4% (86/107) | 31.0% (9/29) | 98.7% (77/78) |
The cutoff value of the prediction model was determined by the ROC analysis of the training cohort. The prediction model was compared to the conventional method using the short‐axis diameter of the largest LLN, as detected on MRI.
Abbreviations: LLN, lateral lymph node; MRI, magnetic resonance imaging; ROC curves, receiver operating characteristic curves.
TABLE 3.
Patients with LLN metastasis in the validation cohort
| Age (y) | Sex | Side | BMI (kg/m2) | CEA (ng/mL) | CA19‐9 (U/mL) | Tumor distance from anal verge (cm) | cT | Macroscopic Type | Tumor Diameter (cm) | Circumferential rate of lumen (%) | Tumor localization of rectal wall | Histological type | PLN diameter (mm) | LLN diameter (mm) | Predicted value | Predicted Result |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 76 | Male | Left | 24.4 | 10.4 | 31 | 3 | 2 | 2 | 2.5 | 33 | Post | Por | 6 | 6 | 0.144 | Positive |
| 69 | Male | Right | 18.5 | 1.2 | 26 | 5 | 3 | 2 | 6 | 67 | Same | Mod | 5 | 4 | 0.238 | Positive |
| 69 | Male | Left | 18.5 | 1.2 | 26 | 5 | 3 | 2 | 6 | 67 | Opposite | Mod | 5 | 3 | 0.138 | Positive |
| 79 | Male | Right | 26.2 | 4.5 | 25 | 4 | 3 | 2 | 5 | 50 | Post | Mod | 4 | 7 | 0.648 | Positive |
| 73 | Male | Left | 21.9 | 2.7 | 35 | 3 | 3 | 2 | 4 | 50 | Post | Muc | 5 | 9 | 0.713 | Positive |
| 49 | Female | Right | 14.3 | 24.5 | 39 | 5 | 4 | 2 | 4.5 | 100 | Circ | Well | 8 | 4 | 0.108 | Positive |
| 49 | Female | Left | 14.3 | 24.5 | 39 | 5 | 4 | 2 | 4.5 | 100 | Circ | Well | 8 | 2 | 0.082 | Negative |
| 50 | Female | Right | 25.6 | 0.8 | 7 | 7 | 3 | 2 | 5 | 67 | Post | Mod | 6 | 9 | 0.529 | Positive |
| 76 | Male | Right | 18.5 | 10.8 | 11 | 4 | 3 | 2 | 6 | 100 | Circ | Well | 11 | 6 | 0.405 | Positive |
| 64 | Female | Right | 17.3 | 10.1 | 6 | 3.5 | 3 | 2 | 2 | 33 | Same | Well | 12 | 4 | 0.125 | Positive |
Ten sides of the patients in the validation cohort had rectal cancer with pathological LLN metastasis. Using the predicted cutoff values determined by the training cohort (>0.101), only one side of the patients could not be predicted correctly.
Abbreviations: BMI, body mass index; CEA, carcinoembryonic antigen; CA19‐9, carbohydrate antigen 19‐9; ant, anterior side; post, posterior side; same, same side as the side dissected, opposite, opposite side as the side dissected; circ, entire circumference; pap, papillary adenocarcinoma; well, well differentiated tubular adenocarcinoma; mod, moderately differentiated tubular adenocarcinoma; por, poorly differentiated adenocarcinoma; muc, mucinous adenocarcinoma; PLN, perirectal lymph node; and LLN, lateral lymph node.
4. DISCUSSION
The prediction model for LLNM from lower rectal cancer using “Prediction One” had higher predictive power than the conventional diagnosis method using the short‐axis diameter, as detected on MRI. In addition, our model could provide high diagnostic sensitivity for small lymph nodes. Considering that LLND should be performed for patients with LLNM, 2 the prediction model could diagnose even small LLNM with high sensitivity and would be more useful than other conventional methods. This study is a novel attempt to demonstrate the effectiveness of machine learning based on deep learning in predicting LLNM, and these findings have never been reported before.
In the 1950s, Bacon and Deddish first reported that LLND, also called “aortoileopelvic lymph node dissection,” for rectal cancer could reduce the rate of local recurrence and improve survival. Both groups reported 8% and 5% improvement in 5‐year survival rates for patients who underwent LLND and historical controls who underwent conventional rectal resection, respectively. However, the difference was statistically insignificant. 14 , 15 Nevertheless, there was severe urinary dysfunction in the LLND group, and almost all the male patients became impotent. Moreover, Glass et al compared patients who underwent LLND to those who underwent conventional surgery. 16 There was no difference in the complication rate and survival rate between the groups. Since then, LLND has been rarely performed for rectal cancer in Western countries, owing to persistent concerns such as operative morbidity and urogenital dysfunction. 17
Lateral lymph node dissection requires a significantly longer operation time and greater blood loss than TME alone, 18 and is technically demanding, particularly in Western countries, comprising numerous patients with obesity. However, the improvement of surgical techniques and the development of new modalities, such as laparoscopic surgery and robotic surgery, could lead to acceptable perioperative morbidity even after CRT. 19 , 20 Furthermore, TME with only CRT is insufficient to prevent lateral local recurrence, particularly for patients with enlarged LLN. 5 , 6 Therefore, there is a growing need to reassess the significance of LLND in Western countries. A review of the treatment of locally advanced lower rectal cancer proposed that LLND should be performed for patients with nonresponsive LLN after CRT. 4
A Japanese multicenter randomized study suggested that LLND had oncological merit in patients with cStage III rather than in those with cStage II. 21 Thus, patients who require LLND will be stratified in the near future. LLND was considered effective for patients with LLNM, and LLNM could also be present in unenlarged LLN. 7 Hence, precisely diagnosing LLNM regardless of the size of LLN may facilitate the selection of patients who substantially benefit from LLND. Colorectal surgeons should determine the indication for LLND, which could contribute to local control, considering the surgical risks and tumor progression. This necessitates precise preoperative diagnosis for LLNM to determine the treatment strategies.
Lymph node metastasis of rectal cancer has been predominantly diagnosed by CT and MRI. Studies have not only measured the size of lymph nodes but also reported on various methods to improve the diagnostic power. However, a meta‐analysis evaluating the ability to diagnose PLN metastasis revealed that the sensitivity and specificity were 79% and 76% for CT and 77% and 76% for MRI, respectively. 22 Brown et al 23 reported on the effectiveness of the border contour and signal intensity characteristics of PLN instead of the size criteria using MRI. However, this method was difficult to evaluate in small lymph nodes and required the advanced ability to interpret images. In addition, diffusion‐weighted MRI and positron emission tomography / CT were reportedly effective in diagnosing lymph node metastasis. 24 , 25 However, these examinations were also unsuitable for evaluating small lymph nodes and often provided false‐positives results, reflecting inflammation around. Furthermore, ultrasmall superparamagnetic iron oxide‐enhanced MRI, which was expected to be extremely sensitive and specific in the detection of lymph node metastasis for various tumors, was uncommon in daily clinical practice. 26 It was difficult to identify patients with LLNM, despite using these diagnostic methods. This necessitates the establishment of precise diagnostic methods that are simpler and more sensitive than the conventional methods.
Recently, an attempt was made to create a logistic model combining multiple risk factors for LLNM to improve the diagnostic power. 10 This logistic model, including risk factors for the short‐axis diameter of the largest LLN as detected on MRI, histopathological grade, and pathological PLN metastasis, had significantly better prediction performance for LLNM than a model based on MRI findings alone. However, the postoperative information about pathological PLN metastasis was not useful to preoperatively determine if LLND should be performed. In addition, the sensitivity of the model was roughly 60%, and high rates of false‐negative results resulted in the exclusion of patients requiring LLND. In contrast, the prediction model in the present study was developed using only preoperative factors and had high sensitivity. Therefore, it could presumably contribute to preoperatively identifying patients requiring LLND.
The development of nomograms based on a complicated multivariate analysis was considered useful, while creating a better prediction model by combining several factors. In recent years, however, the development of prediction models using machine learning has been attempted in various medical fields. 8 , 9 The effectiveness of machine learning was reported in areas that required an accurate prediction in daily clinical practice, such as predicting diagnosis based on examinations and recurrence based on risk factors. Moreover, further applications of the method are expected. The establishment of machine learning can facilitate the easy handling of huge amounts of data and the development of prediction models. Moreover, the accumulation of big data with clinical meaning would enable the creation of better prediction models. In contrast, the machine‐learning software used in the present study could easily and quickly create prediction models, with high predictive power even with a small number of cases. 11 In addition, the model for LLNM had high predictive power, despite including only single‐center cases.
Our study has a future perspective. Deep‐learning methods for reading images, one of the advantages of AI, have been established in recent years. According to several reports, AI identifies lymph node metastasis. 27 Furthermore, there have been increasing reports that an imaging analysis called “radiomics,” which allows the integration of multiple imaging features, is useful for diagnosing lymph node metastasis. 28 Prediction models created by combining radiomics and multiple clinicopathological factors are likely to be more useful diagnostic tools than conventional methods. 29 In the present study, one of the learning items was the short‐axis diameter of LLN, which was as an imaging factor measured by colorectal surgeons. Nonetheless, better prediction models will be developed in the future by combining software that can read images, while interpreting the characteristics of lymph nodes.
Our study has some limitations. First, this retrospective cohort study was conducted at a single institution. The number of eligible patients was limited, and there were only 10 sides with LLNM in the validation cohort. There was some selection bias between the training and validation cohorts, with significant differences observed in several learning items. One of the advantages of machine learning is its ability to easily handle huge amounts of data. The higher the number of patients, the more accurate and universal will the prediction model be. This necessitates developing a better prediction model using additional patient data from different centers and confirming if the model has more precise predictive power. Second, we excluded patients undergoing preoperative CRT, the standard treatment for lower rectal cancer in Western countries. The efficacy of LLND for patients with LLNM after CRT 30 necessitates developing another prediction model based on the information before and after CRT. Third, the method of machine learning used had room for consideration. We used “Prediction One” as the machine‐learning software and selected almost all preoperative patient data as learning items. Nonetheless, we had to consider the possibility of including the unnecessary learning items and excluding other important factors involved in LLNM. We should continue to search for better machine‐learning software and clinicopathological factors involved in LLNM, in order to create more precise prediction models.
In conclusion, machine learning could contribute to the creation of an effective prediction model for LLNM from lower rectal cancer. The prediction model was developed using preoperatively diagnosable factors, and had better predictive power than the conventional method using MRI findings alone. In addition, it would help determine the treatment of locally advanced lower rectal cancer. Further studies, such as the utilization of big data and the development of novel machine‐learning methods, will lead to the creation of excellent prediction models for LLNM in daily clinical practice.
DISCLOSURE
Conflict of interest: The authors declare no conflicts of interest for this article.
Ethical approval: All study protocols were approved by the Institutional Review Board of the Shizuoka Cancer Center (Institutional code: J2020‐57‐2020‐1–3).
ACKNOWLEDGMENTS
None.
Kasai S, Shiomi A, Kagawa H, Hino H, Manabe S, Yamaoka Y, et al. The Effectiveness of Machine Learning in Predicting Lateral Lymph Node Metastasis From Lower Rectal Cancer: A Single Center Development and Validation Study. Ann Gastroenterol Surg.2022;6:92–100. 10.1002/ags3.12504
REFERENCES
- 1. Miles WE. A method of performing abdominoperineal excision for carcinoma of the rectum and of the terminal portion of the pelvic colon. Lancet. 1908;172(4451):1812–3. [Google Scholar]
- 2. Hashiguchi Y, Muro K, Saito Y, Ito Y, Ajioka Y, Hamaguchi T, et al. Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines 2019 for the treatment of colorectal cancer. Int J Clin Oncol. 2020;25(1):1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Glynne‐Jones R, Wyrwicz L, Tiret E, Brown G, Rödel C, Cervantes A, et al. Rectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow‐up. Ann Oncol. 2017;28:iv22‐iv40. [DOI] [PubMed] [Google Scholar]
- 4. Williamson JS, Quyn AJ, Sagar PM. Rectal cancer lateral pelvic sidewall lymph nodes: a review of controversies and management. Br J Surg. 2020;107(12):1562–9. [DOI] [PubMed] [Google Scholar]
- 5. Kusters M, Slater A, Muirhead R, Hompes R, Guy RJ, Jones OM, et al. What to do with lateral nodal disease in low locally advanced rectal cancer? A call for further reflection and research. Dis Colon Rectum. 2017;60(6):577–85. [DOI] [PubMed] [Google Scholar]
- 6. Ogura A, Konishi T, Cunningham C, Garcia‐Aguilar J, Iversen H, Toda S, et al. Neoadjuvant (chemo)radiotherapy with total mesorectal excision only is not sufficient to prevent lateral local recurrence in enlarged nodes: results of the multicenter lateral node study of patients with low cT3/4 rectal cancer. J Clin Oncol. 2019;37(1):33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ueno H, Mochizuki H, Hashiguchi Y, Ishiguro M, Miyoshi M, Kajiwara Y, et al. Potential prognostic benefit of lateral pelvic node dissection for rectal cancer located below the peritoneal reflection. Ann Surg. 2007;245(1):80–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ngiam KY, Khor IW. Big data and machine learning algorithms for health‐care delivery. Lancet Oncol. 2019;20(5):e262–e73. [DOI] [PubMed] [Google Scholar]
- 9. Hashimoto DA, Rosman G, Rus D, Meireles OR. Artificial intelligence in surgery: promises and perils. Ann Surg. 2018l;268(1):70–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ogawa S, Hida JI, Ike H, Kinugasa T, Ota M, Shinto E, et al. Prediction of lateral pelvic lymph node metastasis from lower rectal cancer using magnetic resonance imaging and risk factors for metastasis: multicenter study of the lymph node committee of the Japanese society for cancer of the colon and rectum. Int J Colorectal Dis. 2017;32(10):1479–87. [DOI] [PubMed] [Google Scholar]
- 11. Katsuki M, Kakizawa Y, Nishikawa A, Yamamoto Y, Uchiyama T. Easily created prediction model using deep learning software (Prediction One, Sony Network Communications Inc.) for subarachnoid hemorrhage outcomes from small dataset at admission. Surg Neurol Int. 2020;6(11):374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fluss R, Faraggi D, Reiser B. Estimation of the Youden Index and its associated cutoff point. Biom J. 2005;47(4):458–72. [DOI] [PubMed] [Google Scholar]
- 13. Kanda Y. Investigation of the freely available easy‐to‐use software 'EZR' for medical statistics. Bone Marrow Transplant. 2013;48(3):452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bacon HE. Cancer of the colon, rectum and anal canal: surgical approach with rates of five‐ and ten‐year survival. Am J Surg. 1957;1(94):567–72. [DOI] [PubMed] [Google Scholar]
- 15. Stearns MW Jr, Deddish MR. Five‐year results of abdominopelvic lymph node dissection for carcinoma of the rectum. Dis Colon Rectum. 1959;1(2):169–72. [DOI] [PubMed] [Google Scholar]
- 16. Glass RE, Ritchie JK, Thompson HR, Mann CV. The results of surgical treatment of cancer of the rectum by radical resection and extended abdomino‐iliac lymphadenectomy. Br J Surg. 1985;72(8):599–601. [DOI] [PubMed] [Google Scholar]
- 17. Georgiou P, Tan E, Gouvas N, Antoniou A, Brown G, Nicholls RJ, et al. Extended lymphadenectomy versus conventional surgery for rectal cancer: a meta‐analysis. Lancet Oncol. 2009;10(11):1053–62. [DOI] [PubMed] [Google Scholar]
- 18. Fujita S, Akasu T, Mizusawa J, Saito N, Kinugasa Y, Kanemitsu Y, et al. Postoperative morbidity and mortality after mesorectal excision with and without lateral lymph node dissection for clinical stage II or stage III lower rectal cancer (JCOG0212): results from a multicentre, randomised controlled, non‐inferiority trial. Lancet Oncol. 2012;13(6):616–21. [DOI] [PubMed] [Google Scholar]
- 19. Perez RO, Sao Juliao GP, Vailati BB, Fernandez LM, Mattacheo AE, Konishi T. Lateral node dissection in rectal cancer in the era of minimally invasive surgery: a step‐by‐step description for the surgeon unacquainted with this complex procedure with the use of the laparoscopic approach. Dis Colon Rectum. 2018;61(10):1237–40. [DOI] [PubMed] [Google Scholar]
- 20. Peacock O, Limvorapitak T, Bednarski BK, Kaur H, Taggart MW, Dasari A, et al. Robotic lateral pelvic lymph node dissection after chemoradiation for rectal cancer: a Western perspective. Colorectal Dis. 2020;22(12):2049–56. [DOI] [PubMed] [Google Scholar]
- 21. Tsukamoto S, Fujita S, Ota M, Mizusawa J, Shida D, Kanemitsu Y, et al. Long‐term follow‐up of the randomized trial of mesorectal excision with or without lateral lymph node dissection in rectal cancer (JCOG0212). Br J Surg. 2020;107(5):586–94. [DOI] [PubMed] [Google Scholar]
- 22. Li XT, Sun YS, Tang L, Cao K, Zhang XY. Evaluating local lymph node metastasis with magnetic resonance imaging, endoluminal ultrasound and computed tomography in rectal cancer: a meta‐analysis. Colorectal Dis. 2015;17(6):O129–35. [DOI] [PubMed] [Google Scholar]
- 23. Brown G, Richards CJ, Bourne MW, Newcombe RG, Radcliffe AG, Dallimore NS, et al. Morphologic predictors of lymph node status in rectal cancer with use of high‐spatial‐resolution MR imaging with histopathologic comparison. Radiology. 2003;227(2):371–7. [DOI] [PubMed] [Google Scholar]
- 24. Mizukami Y, Ueda S, Mizumoto A, Sasada T, Okumura R, Kohno S et al. Diffusion‐weighted magnetic resonance imaging for detecting lymph node metastasis of rectal cancer. World J Surg. 2011;35(4):895–9. [DOI] [PubMed] [Google Scholar]
- 25. Tateishi U, Maeda T, Morimoto T, Miyake M, Arai Y, Kim EE. Non‐enhanced CT versus contrast‐enhanced CT in integrated PET/CT studies for nodal staging of rectal cancer. Eur J Nucl Med Mol Imaging. 2007;34(10):1627–34. [DOI] [PubMed] [Google Scholar]
- 26. Lahaye MJ, Engelen SM, Kessels AG, de Bruïne AP, von Meyenfeldt MF, van Engelshoven JMA, et al. USPIO‐enhanced MR imaging for nodal staging in patients with primary rectal cancer: predictive criteria. Radiology. 2008;246(3):804–11. [DOI] [PubMed] [Google Scholar]
- 27. Lu Y, Yu Q, Gao Y, Zhou Y, Liu G, Dong Q, et al. Identification of metastatic lymph nodes in MR Imaging with faster region‐based convolutional neural networks. Cancer Res. 2018;78(17):5135–43. [DOI] [PubMed] [Google Scholar]
- 28. Nakanishi R, Akiyoshi T, Toda S, Murakami Y, Taguchi S, Oba K, et al. Radiomics approach outperforms diameter criteria for predicting pathological lateral lymph node metastasis after neoadjuvant (chemo)radiotherapy in advanced low rectal cancer. Ann Surg Oncol. 2020;27(11):4273–83. [DOI] [PubMed] [Google Scholar]
- 29. Huang YQ, Liang CH, He L, Tian J, Liang C‐S, Chen X, et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol. 2016;34(18):2157–64. [DOI] [PubMed] [Google Scholar]
- 30. Akiyoshi T, Matsueda K, Hiratsuka M, Unno T, Nagata J, Nagasaki T, et al. Indications for lateral pelvic lymph node dissection based on magnetic resonance imaging before and after preoperative chemoradiotherapy in patients with advanced low‐rectal cancer. Ann Surg Oncol. 2015;22(3):S614–20. [DOI] [PubMed] [Google Scholar]


