Abstract
The Tol–Pal system of Gram-negative bacteria is necessary for maintaining outer membrane integrity. It is a multiprotein complex of five envelope proteins, TolQ, TolR, TolA, TolB, and Pal. These proteins were first investigated in E. coli, and subsequently been identified in many other bacterial genera. However, the function of the Tol–Pal system in Salmonella Choleraesuis pathogenesis is still unclear. Here, we reported the role of three of these proteins in the phenotype and biology of S. Choleraesuis. We found that mutations in tolA, tolB, and tolR caused severe damage to the cell wall, which was supported by observing the microstructure of spherical forms, long chains, flagella defects, and membrane blebbing. We confirmed that all the mutants significantly decreased S. Choleraesuis survival when exposed to sodium deoxycholate and exhibited a high sensitivity to vancomycin, which may be explained by the disruption of envelope integrity. In addition, tolA, tolB, and tolR mutants displayed attenuated virulence in a mouse infection model. This could be interpreted as a series of defective phenotypes in the mutants, such as severe defects in envelope integrity, growth, and motility. Further investigation showed that all the genes participate in outer membrane vesicles (OMVs) biogenesis. Interestingly, immunization with OMVs from ΔtolB efficiently enhanced murine viability in contrast to OMVs from the wild-type S. Choleraesuis, suggesting its potential use in vaccination strategies. Collectively, this study provides an insight into the biological role of the S. Choleraesuis Tol–Pal system.
Keywords: Tol–Pal system, tolA, tolB, tolR, Salmonella Choleraesuis, Outer membrane vesicles, Virulence
Introduction
Salmonella enterica serovar Choleraesuis (S. Choleraesuis), a Gram-negative bacterium, is an important swine pathogen that cause a series of severe diseases, including meningitis, hepatitis, pneumonia, and other systemic diseases (Reed et al. 1986). Moreover, it's a major zoonotic agent that could be occasionally isolated from humans and triggers huge economic damage in the porcine sector across the globe (Allison et al. 1969; Bangtrakulnonth et al. 2004; Gray et al. 1995). Thus far, the pathogenesis of S. Choleraesuis infections is still not fully understood (Chiu et al. 2004). Hence, it's imperative to elucidate the pathogenic mechanism of S. Choleraesuis.
The Tol–Pal system of Gram-negative bacteria is a multiprotein composite traversing the inner membrane, periplasm, and outer membrane (OM) (Hirakawa et al. 2020). It comprises five envelope proteins, corresponding to TolQ, TolR, TolA, TolB, and Pal (Henry et al. 2004). Three inner membrane proteins TolQ, TolA, and TolR exhibit interaction with each other through their trans-membraneous domains (Derouiche et al. 1995). Pal is an OM anchored protein interacting with the periplasmic protein TolB (Ray et al. 2000). The system plays numerous biologic functions in Gram-negative bacteria, including cell morphology, sensitivity to bile salts, and bacterial virulence (Dubuisson et al. 2005; Lahiri et al. 2011; Paterson et al. 2009). Furthermore, it has also been displayed that inactivation of the tol–pal genes negatively impacts the outer membrane integrity, resulting in increased formation of outer membrane vesicles (OMVs). OMVs primarily comprise phosphatides, periplasm and OM proteins in Gram-negative microbes, and the Tol–Pal system proteins are the essential components of the OMVs. The Tol–Pal system was 1st characterized in E. coli (Webster 1991), and subsequently been reported in many other bacterial genera, including Pseudomonas aeruginosa (Dennis et al. 1996), Vibrio cholerae (Heilpern and Waldor 2000), Pseudomonas putida (Llamas et al. 2000), Salmonella Typhimurium (Prouty et al. 2002), Erwinia chrysanthemi (Dubuisson et al. 2005), and Salmonella Typhi (Lahiri et al. 2011). Although some progress has been made in the research of other Salmonella enterica serovars, the function of the Tol–Pal system in S. Choleraesuis has not been documented.
Of particular note, data obtained from S. Typhimurium or S. Typhi (less from other bacteria) may not be inferred directly to S. Choleraesuis without experimental results (Nevermann et al. 2019; Urrutia et al. 2014). Compared with S. Typhimurium and S. Typhi, S. Choleraesuis behaves obvious differences in terms of disease progression and host range. They are differences in pathogenic mechanisms, probably due to the different molecular functions of some proteins. Lahiri et al. have demonstrated that there is a considerable difference in the sequence of tolA between S. Typhi and S. Typhimurium (Lahiri et al. 2011). Deletion of tolA of the two serovars exhibits entirely different phenotypes, including membrane organization, detergent resistance, and cell morphology. Nevermann et al. showed that the tolR mutation of S. Typhimurium and S. Typhi also presents fully differently regarding sensitivity to vancomycin, motility, and OMVs production (Nevermann et al. 2019). These results indicated that an experimental approach has to be implemented to better elucidate the function of Tol–Pal system of S. Choleraesuis.
In this study, and with the aim to explore the roles of tolA, tolB, and tolR genes of the Tol–Pal system in S. Choleraesuis, we constructed tolA, tolB, and tolR mutants via homologous recombination. Identifying these genes will help us better comprehend the additional roles of the Tol–Pal system that are difficult to observe in other bacterial genera. We found that all these genes are involved in cell morphology, membrane integrity, cell growth, motility, virulence, and OMVs biogenesis. In addition, we also described the immune responses and protective efficacy of S. Choleraesuis OMVs in a mouse model. In general, this study expanded our understanding of the biological role of the S. Choleraesuis Tol–Pal system.
Materials and methods
Plasmids, strains, and growth conditions
Plasmid pRE112 and E. coli strain χ7213 were kindly offered by Dr. Roy Curtiss III. S. Choleraesuis strain C78-3 (CVCC79103) were bought from China Institute of Veterinary Drugs Control. E. coli χ7213 and S. Choleraesuis strains were grown on LB agar plates or in LB broth (OXOID). When required, 25 µg/ml chloromycetin (Cm) or 50 µg/ml diaminopimelic acid was supplemented into the LB media. Plasmids and strains used in this study are presented in Table 1.
Table 1.
Characteristics of the bacterial strains and plasmids used in this study
| Strains or plasmids | General characteristicsa | Sources or references |
|---|---|---|
| Bacterial strains | ||
| C78-3 | Wild type, virulent, CVCC79103 | Ji et al. (2015) |
| ∆tolA | Isogenic tolA mutant of strain C78-3 | This study |
| ∆tolB | Isogenic tolB mutant of strain C78-3 | This study |
| ∆tolR | Isogenic tolR mutant of strain C78-3 | This study |
| χ7213 | thi‑1 thr‑1 leuB6 fhuA21 lacY1 glnV44 asdA4 recA1 RP4 2‑Tc::Mu pir | Roland et al. (1999) |
| Plasmids | ||
| pRE112 | sacB mobRP4 R6 K oriV oriT; suicide vector; Cmr | Edwards et al. (1998) |
| pRE112-tolA | Suicide vector for ∆tolA; pRE112 derivative; Cmr | This study |
| pRE112-tolB | Suicide vector for ∆tolB; pRE112 derivative; Cmr | This study |
| pRE112-tolR | Suicide vector for ∆tolR; pRE112 derivative; Cmr | This study |
aCmr: chloramphenicol resistance
Ethics statement
Female BALB/c mice (6-week-old) were bought from the Comparative Medicine Center of Yangzhou University. The entire murine studies were completed at Yangzhou University and approved by the Administrative Committee for Laboratory Animals of Jiangsu Province [permission number SCXK (SU) 2017-0007]. The process complied with the protocols of Jiangsu Laboratory Animal Welfare and Ethical guidelines, and all endeavors were performed for the purpose of minimizing the pain of the mice.
Construction of the tolA, tolB, and tolR mutants
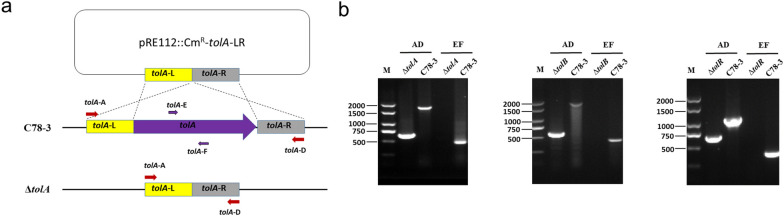
Three mutations ΔtolA, ΔtolB, and ΔtolR were applied for S. Choleraesuis strain C78-3 using correspondent suicide vectors. The primers used for PCR amplification of DNA fragments corresponding to the upstream and downstream flanking regions of the tolA, tolB, and tolR genes are listed in Table 2. In brief, the upstream flanking regions (L) and downstream flanking regions (R) of the target genes were fused as complete fragments (LR) via overlapping PCR, and then cloned into pRE112 via the SacI and KpnI restriction sites. The mutations were constructed in C78-3 by conjugating with χ7213 carrying suicide plasmids as previously reported (Curtiss et al. 2009; Roland et al. 1999). PCR confirmation of the deletions using two primer sets of flanking regions (A/D) and internal regions (E/F), and sequencing by Tsingke Biotechnology Co., Ltd. (Beijing, China).
Table 2.
Primers used for PCR amplification and detection
| Primers | Sequences (5′–3′)a | Function | Length (bp) | Restriction enzyme |
|---|---|---|---|---|
| tolA-A | CGCAGAGCTCATTATTGAGGTTTCCGGAGTA | Upstream flanking regions of tolA | 301 | SacI |
| tolA-B | TCTCGGTTCCCAAAAAACTGT | |||
| tolA-C | ACAGTTTTTTGGGAACCGAGAATACTTTTCTTTATGGAAGTT | Downstream flanking regions of tolA | 322 | |
| tolA-D | CGGGGTACCTACCGCTATTGCGTAAATCTG | KpnI | ||
| tolA-E | AGGAGCGGTTGAAACAACTTG | Internal regions of tolA | 562 | |
| tolA-F | CTGAGATCGCCAAGCAGATCG | |||
| tolB-A | CGCAGAGCTCAATGTGTCTTGCATATTAGCCTG | Upstream flanking regions of tolB | 305 | SacI |
| tolB-B | CATATCTCCCATACCTGGGCCTG | |||
| tolB-C | CAGGCCCAGGTATGGGAGATATGTAATAATTAATTGATTACTAA | Downstream flanking regions of tolB | 269 | |
| tolB-D | CGGGGTACCACTTGTCGAGATCGAAGTAAA | Kpn I | ||
| tolB-E | TGCGTTATGCAGGTCATACCG | Internal regions of tolB | 415 | |
| tolB-F | TGACCGGAGGCGAGATCCATA | |||
| tolR-A | CGCAGAGCTCCGTTTCTTGGCACGGTAGGCT | Upstream flanking regions of tolR | 314 | SacI |
| tolR-B | GGCTTACCCCTTGTTGCTTTC | |||
| tolR-C | GAAAGCAACAAGGGGTAAGCCAGTCTGCGTCCCGTTGGCTTG | Downstream flanking regions of tolR | 320 | |
| tolR-D | CGGGGTACCGTTGCAGCTTTTTACGCTCTT | KpnI | ||
| tolR-E | AGGTCGTCGCGAACTTAAGTC | Internal regions of tolR | 351 | |
| tolR-F | AGCGCTTTAATTATTTCATCG |
aBold nucleotides denote enzyme restriction sites
Transmission electron microscopy (TEM)
TEM analysis was completed to investigate the role of tolA, tolB, and tolR on the morphology of S. Choleraesuis as previously described with minor modification (Elhenawy et al. 2016). In brief, bacterial strains were cultivated in LB liquid medium with shaking at 37 °C and collected at the mid-exponential phase (OD600 = 0.9). Afterwards, the cells were allowed to absorb onto carbon-coated copper grids and negatively stained with 1% uranyl acetate. The samples were allowed to air dry and examined with a Tecnai T12 transmission electron microscope.
Analysis of resistance to sodium deoxycholate and vancomycin
Assays for resistance to deoxycholic acid were performed as previously described (Nevermann et al. 2019). Briefly, bacterial strains were grown in LB to an OD600 of 0.9 and harvested via centrifugating. Then, the cells were cleaned two times with PBS and subjected to resuspension in 0.5% sodium deoxycholate or PBS at 37 °C for 2 h. Microbial survival was counted following plating serial dilutions onto LB agar. The survival rate was computed as (CFU in deoxycholic acid/CFU in PBS) × 100%. The antibiotic sensitivity assay of vancomycin was performed as previously described (Li et al. 2019), using Kirby-Bauer disc diffusion technique. The vancomycin disks contained 30 µg of the antibiotic. Every experiment was finished in 3 independently performed biology duplicates.
Bacterial growth curve assays
The S. Choleraesuis C78-3 and its mutants (ΔtolA, ΔtolB, and ΔtolR) were cultivated in LB to an OD600 of 0.9 and added to 50 ml LB broth (1:200). The cultures were cultivated by vigorous shaking (200 rpm) at 37 °C for 12 h. The OD600 of C78-3 and three mutants were quantified at 60 min interval via a spectral photometer (Bio-Rad). Meanwhile, bacteria numbers were counted every hour following plating serial dilutions onto LB agar. Every experiment was finished in 3 independently conducted biology duplicates.
Motility assays
Motility assays were performed according to a previously described method with minor modification (Morgan et al. 2014). In short, bacterial strains were cultured in LB to an OD600 of 0.9, and then diluted 1:10 in fresh LB and 1 µl was inoculated on semi-solid (0.5%) LB agar plates containing 0.02% arabinose. The plates were cultivated for 5 h at 37 °C and cell motility was assessed via the diameter of growth halo (mm). Each assay was performed in triplicate and repeated in 3 independent replicates.
Assessment of LD50 via a mouse model
To investigate the effect of inactivating tolA, tolB, and tolR on the virulence of S. Choleraesuis, the LD50 of C78-3 and three mutants was tested by intraperitoneal challenges with a mouse model. Briefly, bacterial strains were cultivated in LB to an OD600 of 0.9 and washed twice with PBS. Four groups of BALB/c mice (n = 4) were subjected to injection with the doses of 3, 3 × 101, 3 × 102, and 3 × 103 CFU/mouse in 100 µl PBS of wild-type strain C78-3. Meanwhile, 12 groups of mice (n = 4) were subjected to injection with the doses of 5 × 103, 5 × 104, 5 × 105, and 5 × 106 CFU/mouse in 100 µl PBS of ΔtolA, ΔtolB, or ΔtolR, respectively. The animals with the corresponding infection were supervised daily for 30 days. LD50 was determined by the approach of Reed and Muench (1938).
Purification and quantification of OMVs
Outer membrane vesicles (OMVs) of S. Choleraesuis C78-3 and its mutants were isolated as previously described (Muralinath et al. 2011). In brief, bacterial strains were cultivated in LB liquid medium (300 ml) at 37 °C overnight (OD600 = 2.1) and harvested by centrifugation (12,000×g, 10 min). Subsequently, the supernatants were treated with filtration by 0.45 µm sterile filtering device (Millipore, USA). OMVs were collected from the supernatant by ultracentrifugation (150,000×g, 3 h, 4 °C) and washed once with PBS. OMVs were then purified by density gradient centrifugation (150,000×g, 12 h, 4 °C) on a discontinuous gradient from 20 to 45% of Optiprep (Axis-Shield). OMVs fractions were pooled and ultracentrifuged again. The vesicles were resuspended in 2 ml PBS and stored at − 80 °C. The obtained OMVs isolation was analyzed by TEM as previously described (Nevermann et al. 2019). The yield of OMVs from S. Choleraesuis C78-3 and its mutants was evaluated by the protein concentration in the OMVs. Quantification of the OMVs concentration was quantified via a BCA protein analysis kit. All OMVs samples from the strains were subjected to purification and quantification at least 3 times. Each OMVs sample (8 µl) was separated by 12% SDS-PAGE, and then the protein profiles of OMVs were visualized using Coomassie Brilliant Blue R-250.
Immunization and challenge of mice
Five groups of BALB/c mice (6-week-old, n = 5) were immunized with 100 µl PBS involving 10 µg OMVs via the intraperitoneal route. Intraperitoneal immunizations of 100 µl PBS was the negative controls. Booster immunizations were administered 3 weeks posterior to the primary immunization. Blood specimens were harvested 5 weeks posterior to the initial immunization via orbital sinus puncture. Serum IgG were evaluated by ELISA as previously described (Li et al. 2017). Two weeks after the booster immunizations, the animals were treated with 3 × 106 CFU (nearly 100 × LD50) of the wild-type C78-3 in 20 µl PBS via the oral route. The infected mice were supervised every day for 30 days. The protection assays were finished two times, and the results were integrated for analysis.
Statistical analysis
The numerical results were assayed via GraphPad Prism (GraphPad Prism 5, GraphPad Software, USA). Unpaired two-tailed Student’s t-test was employed to evaluate statistic significance. Differences were considered as significant at P < 0.05. All results were obtained from at least 3 independent replicates, and values were expressed as mean ± SEM.
Results
Construction and confirmation of the tolA, tolB, and tolR knockout mutants
To probe the roles of Tol–Pal system in S. Choleraesuis, three mutants of tolA, tolB, and tolR were constructed via homologous recombination. A schematic representation of the homologous recombination strategy is shown (Fig. 1a). The generation of the tolA, tolB, and tolR mutants using a mediator based on the suicide vector pRE112. The tolA, tolB, and tolR mutants were verified via integrated PCR analysis using two pairs of primers, as well as sequencing. As presented in Fig. 1b, there were no fragments of ΔtolA, ΔtolB, and ΔtolR using inner primers (E/F), while the flanking primers (A/D) amplified smaller fragments from the mutants in contrast to those amplified from the parental strain C78-3. These results showed that ΔtolA, ΔtolB, and ΔtolR were constructed successfully.
Fig. 1.
Construction and confirmation of the tolA, tolB, and tolR knockout mutants. a Schematic diagram for the generation of the tolA, tolB, and tolR mutants. b The tolA, tolB, and tolR mutants were verified through combined PCR. PCR confirmation of the deletions using two primer sets of flanking regions (A/D) and internal regions (E/F)
Morphological characterization of S. Choleraesuis C78-3 and three mutants
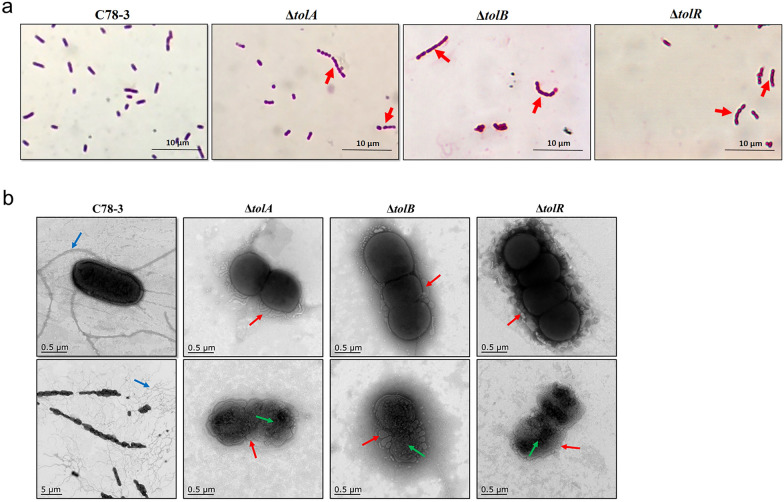
Considering that the Tol–Pal system of Gram-negative bacteria toward maintaining outer membrane stability, we speculated that deletion of tolA, tolB, and tolR might influence the phenotypes of S. Choleraesuis. Cell morphology of C78-3 and three mutants were examined by light microscopy after Gram staining. In LB broth, the wild-type S. Choleraesuis C78-3 grew as single rods (Fig. 2a). Under the same conditions, ΔtolA, ΔtolB, and ΔtolR grew in chains (3 to 10 cells) of coccobacilli (Fig. 2a) (red arrow). In order to further confirm the results, we evaluated the cell morphology by TEM. C78-3 was rod-shaped with the average size of 1.6 × 0.7 µm and presented long flagella (blue arrow). In contrast, ΔtolA, ΔtolB, and ΔtolR presented an altered morphology with spherical forms and long chains, which is consistent with the above observations (Fig. 2b). Of particular note, the cell morphology among the ΔtolA, ΔtolB, and ΔtolR strains are very similar. The three mutants formed vesicles at the cell surface (red arrow), but had no flagella. We also found that some mutants were severely damaged in their cell morphology (green arrow). Therefore, S. Choleraesuis tolA, tolB, and tolR genes participate in the maintenance of cell morphology.
Fig. 2.
Morphological characterization of S. Choleraesuis C78-3 and three mutants. a Gram staining of C78-3 and three mutants grown in LB were examined by light microscopy (×1000). The ΔtolA, ΔtolB, and ΔtolR strains grew in chains of coccobacilli (red arrow). b Morphology characteristics of C78-3 and three mutants were evaluated by TEM. C78-3 was rod-shaped and presented long flagella (blue arrow). The ΔtolA, ΔtolB, and ΔtolR strains showed an altered morphology with long chains and formed vesicles at the cell surface (red arrow), while some mutants were severely damaged in their cell morphology (green arrow)
Characterization of ΔtolA, ΔtolB, and ΔtolR regarding the envelope integrity
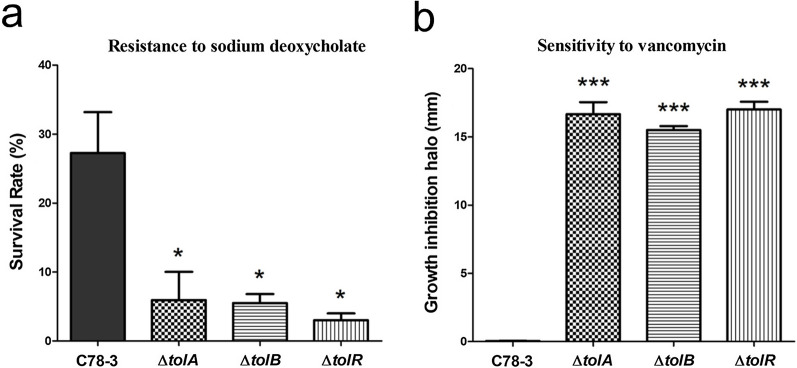
Previous studies found that the tol–pal genes of E. coli are important in maintaining outer membrane integrity (Lazzaroni et al. 1999). This phenomenon was also reported in S. Typhimurium (Paterson et al. 2009) and Erwinia chrysanthemi (Dubuisson et al. 2005). To determine whether deletion of tolA, tolB, and tolR affect the envelope integrity of S. Choleraesuis, assays for resistance to deoxycholic acid were performed. All mutants were more susceptible to 0.5% sodium deoxycholate in contrast to that of the parental strain (Fig. 3a). To further probe the envelope integrity, the sensitivity of ΔtolA, ΔtolB, and ΔtolR towards vancomycin were analyzed. In essence, Gram-negative bacteria exhibit resistance to vancomycin due to the limit of diffusible molecules through the microbial envelope (Pimenta et al. 1999). Therefore, the increase in sensitivity to vancomycin can be explained by the increase in permeability. Our results showed that wild-type S. Choleraesuis presented full resistance to vancomycin. In contrast, all the mutants revealed complete sensitivity to vancomycin (Fig. 3b). The tolA, tolB, and tolR mutants exhibited increased susceptibility to sodium deoxycholate and vancomycin, indicating that the envelope integrity might be damaged in these cases.
Fig. 3.
Resistance to sodium deoxycholate and vancomycin of C78-3 and three mutants. a Survival of C78-3, ΔtolA, ΔtolB, and ΔtolR in 0.5% sodium deoxycholate. Bacterial strains were cultivated in LB to an OD600 of 0.9. Then, the cells were resuspended in 0.5% sodium deoxycholate or PBS at 37 °C for 2 h. The survival rate was computed as (CFU in deoxycholic acid/CFU in PBS) × 100%. b Resistance to vancomycin of C78-3, ΔtolA, ΔtolB, and ΔtolR strains. The vancomycin disks contained 30 µg of the antibiotic. Each assay was completed in 3 independently conducted biology duplicates. *P < 0.05; **P < 0.01; ***P < 0.001
The roles of tolA, tolB, and tolR in cell growth, motility, and virulence of S. Choleraesuis
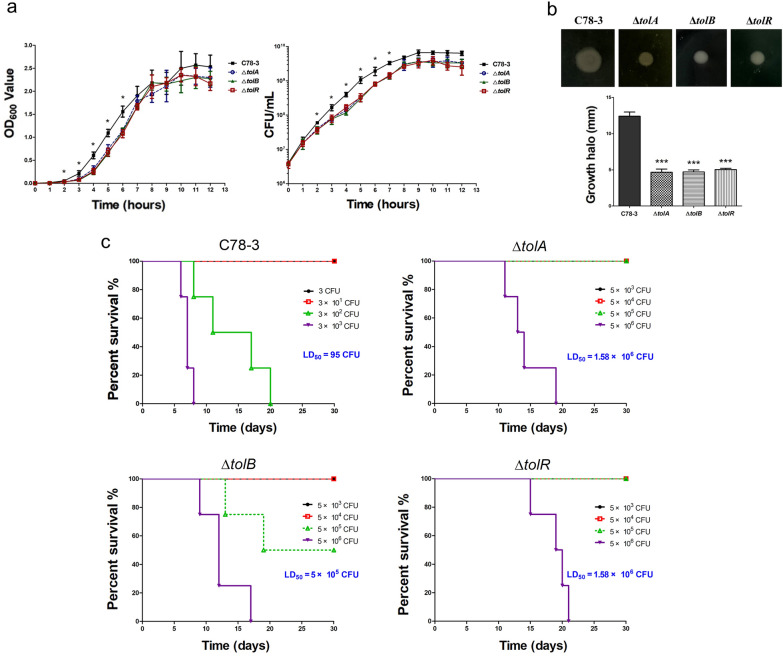
To investigate the biological roles of tolA, tolB, and tolR in S. Choleraesuis, the growth curve, motility and virulence of the wild type C78-3 and mutants were studied. The growth of the ΔtolA, ΔtolB, and ΔtolR strains was significantly slower than C78-3 in the exponential phase, while there were no observed growth differences among the three mutants (Fig. 4a). The swimming halo diameter of C78-3, ΔtolA, ΔtolB, and ΔtolR was 12.4 mm, 4.7 mm, 4.7 mm, or 5.0 mm, respectively. These results indicated that the motility of S. Choleraesuis ΔtolA, ΔtolB, and ΔtolR was markedly impaired compared with the wild-type strain (Fig. 4b). We then examined the virulence of C78-3 and three mutants in a mouse model through the intraperitoneal route, the LD50 of C78-3, ΔtolA, ΔtolB, and ΔtolR were 95 CFU, 1.58 × 106 CFU, 5 × 105 CFU, or 1.58 × 106 CFU, respectively (Fig. 4c). The LD50 value of three S. Choleraesuis mutants was significantly higher than that of C78-3. The results suggested that deletion of tolA, tolB, and tolR displayed an attenuated virulence of S. Choleraesuis in a mouse infection model.
Fig. 4.
The roles of tolA, tolB, and tolR in cell growth, motility, and virulence of S. Choleraesuis. a Growth curves of C78-3, ΔtolA, ΔtolB, and ΔtolR using OD600 measurements, and the bacterial suspensions were serially diluted and plated to determine CFU numbers per milliliter. b Motility was evaluated by measuring the growth halo of C78-3, ΔtolA, ΔtolB, and ΔtolR. Each assay was completed in 3 independently conducted biology duplicates. *P < 0.05; **P < 0.01; ***P < 0.001. c LD50 evaluation of C78-3, ΔtolA, ΔtolB, and ΔtolR with a murine model. The infected mice were supervised every day for 30 days
The involvement of tolA, tolB, and tolR in OMVs biogenesis of S. Choleraesuis
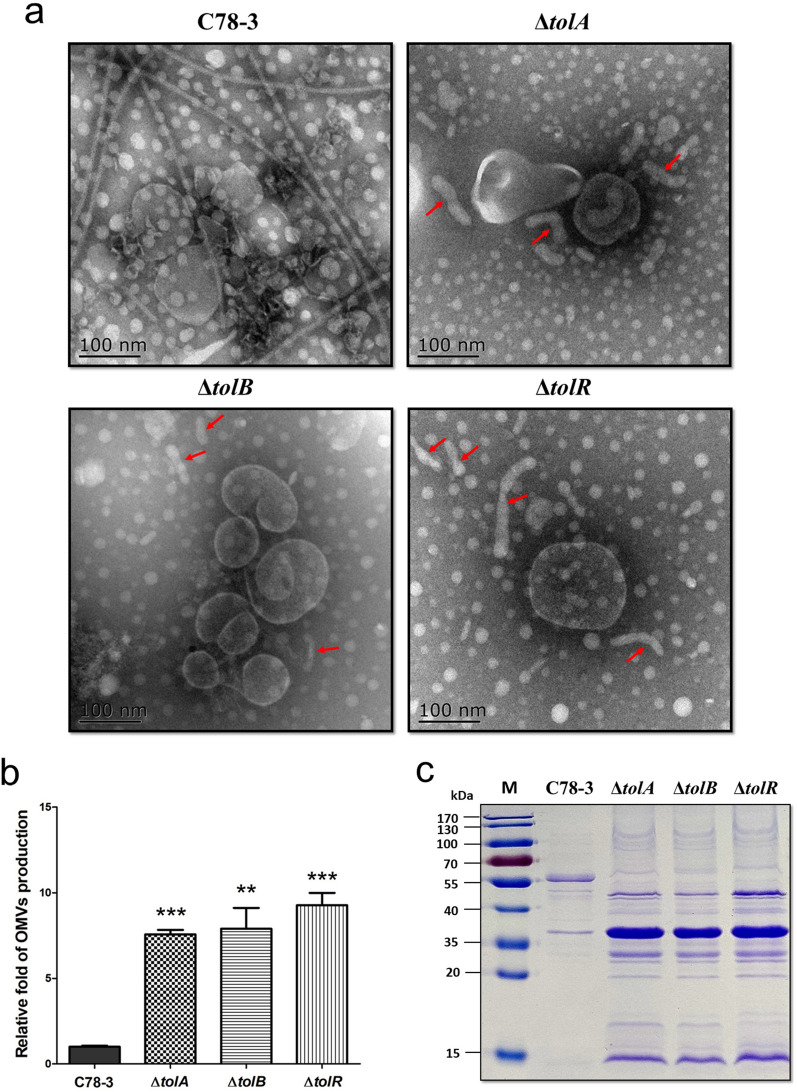
At this point, we speculated that S. Choleraesuis tolA, tolB, and tolR genes participate in OMVs biogenesis for the following reasons: (1) all S. Choleraesuis tolA, tolB, and tolR mutants showed an impaired envelope integrity compared with the wild-type strain (Fig. 3); (2) vesicles could be clearly observed at the surface of S. Choleraesuis tolA, tolB, and tolR mutants (Fig. 2b). (3) according to previous reports, tolA, tolB, and tolR genes contribute to the OMVs biogenesis in Salmonella enterica serovars, like tolA and tolB of S. Typhimurium (Deatherage et al. 2009), and tolR of S. Typhi (Nevermann et al. 2019). To determine whether disruption of tolA, tolB, and tolR influence the OMVs biogenesis of S. Choleraesuis, we isolated OMVs from the wild-type and three mutants and further examined by TEM. As shown in Fig. 5a, all S. Choleraesuis mutants produced morphologically diverse OMVs regarding their shape (red arrow) and component (with or without flagella) compared with the wild-type strain. The OMVs of wild-type C78-3 has a large number of flagella. In contrast, no obvious flagella were observed in the OMVs preparations of the three mutants, which is consistent with the expressed non motile phenotype observed. In addition, the amount of OMVs in the ΔtolA, ΔtolB, and ΔtolR strains was higher than that in C78-3. To further test the OMVs yield of S. Choleraesuis C78-3 and its mutants, we determined the protein concentration of OMVs as an abundance indicator normalizing with CFU/ml according to a previous study (Deatherage et al. 2009). Our results showed that the ΔtolA, ΔtolB, and ΔtolR strains obviously exhibited more proteins in the OMVs fraction than the wild-type strain (Fig. 5b). Collectively, these data suggested that tolA, tolB, and tolR in S. Choleraesuis participate in the OMVs biogenesis.
Fig. 5.
The involvement of tolA, tolB, and tolR in OMVs biogenesis of S. Choleraesuis. a OMVs derived from C78-3, ΔtolA, ΔtolB, and ΔtolR were evaluated by TEM. b OMVs production of C78-3 and its three mutants. The yield of OMVs was determined by quantitating the protein concentration. The assay was completed in 3 independently conducted biology duplicates. *P < 0.05; **P < 0.01; ***P < 0.001. c SDS-PAGE profile of OMVs derived from C78-3, ΔtolA, ΔtolB, and ΔtolR strains. Proteins were visualized using Coomassie Brilliant Blue R-250
A previous study demonstrated that OMVs cargo selection is closely related to the OMVs biogenesis (Schwechheimer and Kuehn 2015). Due to tolA, tolB, and tolR gene products are involved in the OMVs biogenesis, we speculated that OMVs cargo selection derived from S. Choleraesuis ΔtolA, ΔtolB, and ΔtolR strains should be influenced. To examine the differences in OMVs cargo selection, an SDS-PAGE was performed from the OMVs extracts. We observed that S. Choleraesuis wild-type OMVs presented few detectable proteins, with a major protein band at ~ 55 kDa (corresponding to flagellin) (Fig. 5c). The similar pattern of OMVs was also reported in a number of wild-type Salmonella enterica serovars, including S. Typhi (Nevermann et al. 2019), S. Typhimurium (Liu et al. 2016a), S. Choleraesuis (Liu et al. 2017b), and S. Enteritidis (Liu et al. 2017a). We found that the SDS-PAGE profiles of OMVs derived from S. Choleraesuis ΔtolA, ΔtolB, and ΔtolR strains (major protein bands at ~ 45 kDa, ~ 37 kDa, ~ 30 kDa, and ~ 15 kDa) were different from that of S. Choleraesuis wild-type OMVs, suggesting the involvement of tolA, tolB, and tolR genes in OMVs cargo selection.
Altogether, these results confirmed that tolA, tolB, and tolR genes participate in the OMVs biogenesis in S. Choleraesuis, increasing OMVs production, and affecting OMVs cargo selection.
Evaluation of immune responses and protection against S. Choleraesuis
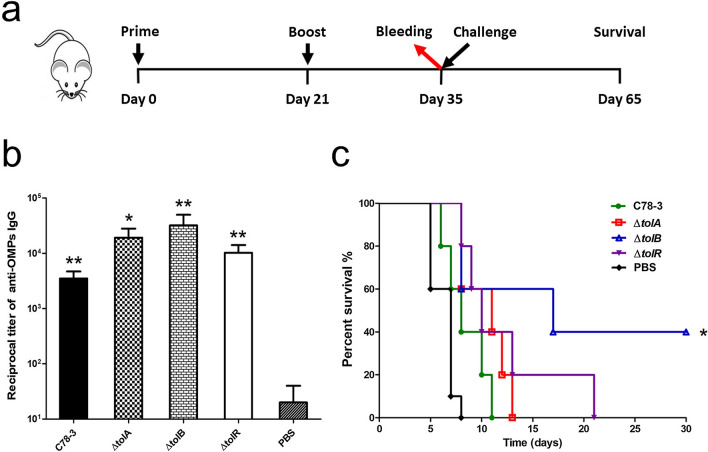
To investigate the immune responses induced by OMVs, serum IgG was measured by ELISA against outer membrane proteins (OMPs) that were derived from S. Choleraesuis. Five groups of BALB/c mice were subjected to immunization for two times with OMVs or PBS via the intraperitoneal route (Fig. 6a). As shown in Fig. 6b, IgG titers against OMPs from C78-3 and three mutants groups were remarkably higher in contrast to the PBS group. Remarkably, although IgG titers against OMPs from ΔtolA, ΔtolB, and ΔtolR groups were slightly higher than the wild-type group, they had no significant difference. Two weeks after the booster immunizations, all animals were challenged with wild-type C78-3 via the oral route. All of the PBS control mice died within 8 days after C78-3 challenge, while immunization with OMVs of C78-3, ΔtolA, and ΔtolR prolonged mice survival to 11 days, 13 days, or 21 days, respectively (Fig. 6c). Specifically, immunization with ΔtolB OMVs conferred 40% protection to mice (Fig. 6c), which is significantly higher than that of the wild-type S. Choleraesuis OMVs. These data revealed that ΔtolB OMVs was able to provide partial protection against the wild-type S. Choleraesuis.
Fig. 6.
Evaluation of immune responses and protection against S. Choleraesuis. a Scheme of immunization regimen. b Antibody responses induced by OMVs derived from C78-3, ΔtolA, ΔtolB, and ΔtolR strains. IgG titers against OMPs from C78-3 and three mutants groups were remarkably higher in contrast to the PBS group. c Survival of the mice subjected to immunization with OMVs obtained from C78-3, ΔtolA, ΔtolB, and ΔtolR strains. Intraperitoneal immunizations of PBS served as the negative controls. Two weeks posterior to the reinforce immunizations, all animals were challenged with the wild-type S. Choleraesuis. The infected mice were supervised every day for 30 days. *P < 0.05; **P < 0.01; ***P < 0.001
Discussion
The tol–pal genes are not fully characterized in S. Choleraesuis, but they may play important roles in this pathogen considering that the multiple functions of this system in other bacteria. The present study aimed to characterize the function of tolA, tolB, and tolR genes in S. Choleraesuis. Indeed, tol–pal mutants have been studied extensively in many Gram-negative bacteria (Dennis et al. 1996; Dubuisson et al. 2005; Heilpern and Waldor 2000; Lahiri et al. 2011; Llamas et al. 2000; Prouty et al. 2002; Webster 1991). Mutations in tol–pal genes are impaired in their envelope integrity, affected cell morphology, increased sensitivity to bile salts, promoted OMVs production, reduced the cell growth, motility, and bacterial virulence.
Our analysis confirmed that S. Choleraesuis ΔtolA, ΔtolB, and ΔtolR display an altered cell morphology, which was supported by observing the microstructure of spherical forms, long chains, flagella defects, and membrane blebbing. A phenotype of altered morphology was also reported in tol–pal mutants of S. Typhimurium (Masilamani et al. 2018), S. Typhi (Lahiri et al. 2011), and Erwinia chrysanthem (Dubuisson et al. 2005). This phenomenon is mainly due to the disruption of envelope integrity. As previously observed, the Tol–Pal system mediated phosphatidylglycerols trafficking might affect envelope homeostasis that alters cell morphology (Masilamani et al. 2018). Previous studies have demonstrated that the Tol–Pal system of Gram-negative microbes is involved in maintaining OM integrity (Lazzaroni et al. 1999; Masilamani et al. 2018). According to our results, ΔtolA, ΔtolB, and ΔtolR mutants exhibited increased susceptibility to sodium deoxycholate and vancomycin, revealing that the envelope integrity might be damaged in these cases.
In S. Choleraesuis, deletion of tolA, tolB, or tolR is detrimental to the bacteria, since mutants grow very slowly in LB broth. In support of this, the lack of tolA and tolB in Erwinia chrysanthemi reduces cell growth (Dubuisson et al. 2005). Our data showed that tolA, tolB, and tolR genes were involved in motility of S. Choleraesuis. This phenotype was also observed in tol–pal mutants of S. Typhimurium (Nevermann et al. 2019), E. coli (Morgan et al. 2014), and Erwinia chrysanthem (Dubuisson et al. 2005). Our TEM observations provide a valuable explanation for this point. The tolA, tolB, and tolR mutations lack flagella, while the wild-type had a large number of flagella around the cell surface (Fig. 2b). By contrast, S. Typhi ΔtolR did not affect the motility (Nevermann et al. 2019), indicating that mutants lacking tolR in different Salmonella enterica serovars are not entirely equivalent.
S. Choleraesuis tolA, tolB, and tolR mutants displayed an attenuated virulence in a mouse infection model. Reduced virulence was also reported in tol–pal mutants of other pathogens. In S. Typhimurium, the virulence of tolA, tolB, and tolR mutants were significantly attenuated (Masilamani et al. 2018). In E. coli, the virulence of ΔtolA was largely attenuated using a Galleria mellonella model (Morgan et al. 2014). In Erwinia chrysanthem, expression of TolA or TolB was necessary for the full virulence in a potato tuber model (Dubuisson et al. 2005). The impaired virulence of S. Choleraesuis tolA, tolB, and tolR mutants probably results from the defective phenotypes, such as serious defects in cell morphology, envelope integrity, growth, and motility.
Thus far, the knowledge on the biogenesis of OMVs is still limited and fragmentary in Salmonella serotypes (Deatherage et al. 2009), and significantly lacking in S. Choleraesuis. Evidence has revealed that cross-linking between peptidoglycan and bacterial envelope proteins is an important mechanism for OMV biogenesis (Schwechheimer and Kuehn 2015). The Tol–Pal system corresponds to five envelope proteins that participates in OMVs biogenesis by interacting with peptidoglycan. The yield of OMVs increases when the cross-linking decreases. Accordingly, S. Typhimurium tolA and tolB mutants contribute to the production of OMVs (Deatherage et al. 2009). Moreover, the tolR mutation of S. Typhi displayed an increased production of OMVs (Nevermann et al. 2019). In S. Choleraesuis, we found that tolA, tolB, and tolR genes participate in the OMVs biogenesis. This study confirmed that TolA, TolB, and TolR are critical cell envelope proteins essential for OMV biogenesis.
Many studies have shown that native OMVs obtained from pathogens were able to confer strong protection against the challenge of pathogenic bacteria, such as S. Typhimurium (Liu et al. 2016a, b), S. Enteritidis (Liu et al. 2017a), Shigella flexneri (Camacho et al. 2011), Acinetobacter baumannii (McConnell et al. 2011), Neisseria meningitidis (Serruto et al. 2012). Remarkably, immunization with OMVs isolated from S. Choleraesuis wild type via the intraperitoneal route failed to confer protection against S. Choleraesuis. This result is likely due to the non-essential immune responses and excessive pro-inflammatory responses (McSorley et al. 2000; Singh et al. 2003; Smith et al. 2003), resulting in the failure of immune protection induced by OMVs. In support of this, similar results has been reported in a previous study (Liu et al. 2017b). Specifically, immunization with OMVs isolated from ΔtolB conferred 40% protection to mice. The result suggested that deletion of tolB in S. Choleraesuis can significantly improve the immunogenicity of OMVs, which is an intriguing discovery for OMVs of S. Choleraesuis, and the mechanism needs to be further evaluated.
This study confirmed that deletion of S. Choleraesuis tolA, tolB, and tolR genes severely damaged cell morphology, impaired the envelope integrity, inhibited growth and motility ability, and reduced the bacterial virulence. Moreover, tolA, tolB, and tolR genes also participate in the OMVs biogenesis, promoting OMVs production, and affecting OMVs cargo selection. In summary, this study provides an insight into the biological role of the S. Choleraesuis Tol–Pal system.
Acknowledgements
We thank Dr. Roy Curtiss III for kindly providing χ7213 and pRE112.
Authors’ contributions
QL was responsible for implementation of the assays, interpreting the data and writing the first draft; ZL, XF, YT, and GZ was responsible for certain assays; SW was involved in the discussion and was responsible for the revision of the first draft; HS was involved in experiment design was responsible for the interpretation of the data, monitoring the exploration process. All authirs read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (Grant Numbers 32002301, 32172802, 31672516, 31172300, 30670079), the China Postdoctoral Science Foundation (Grant Number 2019M661953), the Natural Science Foundation of Jiangsu Province (Grant Number BK20190886), and A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Procedures involving the care and use of animals were approved by the Jiangsu Administrative Committee for Laboratory Animals (permission number SYXK-SU-2007-0005) and complied with the Jiangsu Laboratory Animal Welfare and Ethics guidelines of the Jiangsu Administrative Committee of Laboratory Animals.
Consent for publication
All authors gave their informed consent prior to their inclusion in the study.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Quan Li, Email: liquan2018@yzu.edu.cn.
Zheng Li, Email: 1156851868@qq.com.
Xia Fei, Email: 963367073@qq.com.
Yichen Tian, Email: 734855568@qq.com.
Guodong Zhou, Email: 1466732225@qq.com.
Yuhan Hu, Email: 924214484@qq.com.
Shifeng Wang, Email: shifengwang@ufl.edu.
Huoying Shi, Email: hyshi@yzu.edu.cn.
References
- Allison MJ, Dalton HP, Escobar MR, Martin CJ. Salmonella choleraesuis infections in man: a report of 19 cases and a critical literature review. South Med J. 1969;62:593–596. [PubMed] [Google Scholar]
- Bangtrakulnonth A, Pornreongwong S, Pulsrikarn C, Sawanpanyalert P, Hendriksen RS, Lo Fo Wong DM, Aarestrup FM. Salmonella serovars from humans and other sources in Thailand, 1993–2002. Emerg Infect Dis. 2004;10:131–136. doi: 10.3201/eid1001.02-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho AI, de Souza J, Sanchez-Gomez S, Pardo-Ros M, Irache JM, Gamazo C. Mucosal immunization with Shigella flexneri outer membrane vesicles induced protection in mice. Vaccine. 2011;29:8222–8229. doi: 10.1016/j.vaccine.2011.08.121. [DOI] [PubMed] [Google Scholar]
- Chiu CH, Su LH, Chu C. Salmonella enterica serotype Choleraesuis: epidemiology, pathogenesis, clinical disease, and treatment. Clin Microbiol Rev. 2004;17:311–322. doi: 10.1128/CMR.17.2.311-322.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtiss R, 3rd, Wanda SY, Gunn BM, Zhang X, Tinge SA, Ananthnarayan V, Mo H, Wang S, Kong W. Salmonella enterica serovar typhimurium strains with regulated delayed attenuation in vivo. Infect Immun. 2009;77:1071–1082. doi: 10.1128/IAI.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage BL, Lara JC, Bergsbaken T, Rassoulian Barrett SL, Lara S, Cookson BT. Biogenesis of bacterial membrane vesicles. Mol Microbiol. 2009;72:1395–1407. doi: 10.1111/j.1365-2958.2009.06731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis JJ, Lafontaine ER, Sokol PA. Identification and characterization of the tolQRA genes of Pseudomonas aeruginosa. J Bacteriol. 1996;178:7059–7068. doi: 10.1128/jb.178.24.7059-7068.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derouiche R, Benedetti H, Lazzaroni JC, Lazdunski C, Lloubes R. Protein complex within Escherichia coli inner membrane. TolA N-terminal domain interacts with TolQ and TolR proteins. J Biol Chem. 1995;270:11078–11084. doi: 10.1074/jbc.270.19.11078. [DOI] [PubMed] [Google Scholar]
- Dubuisson JF, Vianney A, Hugouvieux-Cotte-Pattat N, Lazzaroni JC. Tol–Pal proteins are critical cell envelope components of Erwinia chrysanthemi affecting cell morphology and virulence. Microbiology (Reading) 2005;151:3337–3347. doi: 10.1099/mic.0.28237-0. [DOI] [PubMed] [Google Scholar]
- Edwards RA, Keller LH, Schifferli DM. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene. 1998;207:149–157. doi: 10.1016/s0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- Elhenawy W, Bording-Jorgensen M, Valguarnera E, Haurat MF, Wine E, Feldman MF. LPS remodeling triggers formation of outer membrane vesicles in Salmonella. mBio. 2016;7:e00940-16. doi: 10.1128/mBio.00940-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JT, Fedorka-Cray PJ, Stabel TJ, Ackermann MR. Influence of inoculation route on the carrier state of Salmonella choleraesuis in swine. Vet Microbiol. 1995;47:43–59. doi: 10.1016/0378-1135(95)00060-n. [DOI] [PubMed] [Google Scholar]
- Heilpern AJ, Waldor MK. CTXphi infection of Vibrio cholerae requires the tolQRA gene products. J Bacteriol. 2000;182:1739–1747. doi: 10.1128/jb.182.6.1739-1747.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry T, Pommier S, Journet L, Bernadac A, Gorvel JP, Lloubes R. Improved methods for producing outer membrane vesicles in Gram-negative bacteria. Res Microbiol. 2004;155:437–446. doi: 10.1016/j.resmic.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Hirakawa H, Suzue K, Takita A, Awazu C, Kurushima J, Tomita H. Roles of the Tol–Pal system in the Type III secretion system and flagella-mediated virulence in enterohemorrhagic Escherichia coli. Sci Rep. 2020;10:15173. doi: 10.1038/s41598-020-72412-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Shang J, Li Y, Wang S, Shi H. Live attenuated Salmonella enterica serovar Choleraesuis vaccine vector displaying regulated delayed attenuation and regulated delayed antigen synthesis to confer protection against Streptococcus suis in mice. Vaccine. 2015;33:4858–4867. doi: 10.1016/j.vaccine.2015.07.063. [DOI] [PubMed] [Google Scholar]
- Lahiri A, Ananthalakshmi TK, Nagarajan AG, Ray S, Chakravortty D. TolA mediates the differential detergent resistance pattern between the Salmonella enterica subsp. enterica serovars Typhi and Typhimurium. Microbiology (Reading) 2011;157:1402–1415. doi: 10.1099/mic.0.046565-0. [DOI] [PubMed] [Google Scholar]
- Lazzaroni JC, Germon P, Ray MC, Vianney A. The Tol proteins of Escherichia coli and their involvement in the uptake of biomolecules and outer membrane stability. FEMS Microbiol Lett. 1999;177:191–197. doi: 10.1111/j.1574-6968.1999.tb13731.x. [DOI] [PubMed] [Google Scholar]
- Li YA, Ji Z, Wang X, Wang S, Shi H. Salmonella enterica serovar Choleraesuis vector delivering SaoA antigen confers protection against Streptococcus suis serotypes 2 and 7 in mice and pigs. Vet Res. 2017;48:89. doi: 10.1186/s13567-017-0494-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Yin J, Li Z, Li Z, Du Y, Guo W, Bellefleur M, Wang S, Shi H. Serotype distribution, antimicrobial susceptibility, antimicrobial resistance genes and virulence genes of Salmonella isolated from a pig slaughterhouse in Yangzhou, China. AMB Express. 2019;9:210. doi: 10.1186/s13568-019-0936-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Liu Q, Yi J, Liang K, Hu B, Zhang X, Curtiss R, 3rd, Kong Q. Outer membrane vesicles from flagellin-deficient Salmonella enterica serovar Typhimurium induce cross-reactive immunity and provide cross-protection against heterologous Salmonella challenge. Sci Rep. 2016;6:34776. doi: 10.1038/srep34776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Liu Q, Yi J, Liang K, Liu T, Roland KL, Jiang Y, Kong Q. Outer membrane vesicles derived from Salmonella Typhimurium mutants with truncated LPS induce cross-protective immune responses against infection of Salmonella enterica serovars in the mouse model. Int J Med Microbiol. 2016;306:697–706. doi: 10.1016/j.ijmm.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Yi J, Liang K, Zhang X, Liu Q. Outer membrane vesicles derived from Salmonella enteritidis protect against the virulent wild-type strain infection in a mouse model. J Microbiol Biotechnol. 2017;27:1519–1528. doi: 10.4014/jmb.1705.05028. [DOI] [PubMed] [Google Scholar]
- Liu Q, Yi J, Liang K, Zhang X, Liu Q. Salmonella choleraesuis outer membrane vesicles: proteomics and immunogenicity. J Basic Microbiol. 2017;57:852–861. doi: 10.1002/jobm.201700153. [DOI] [PubMed] [Google Scholar]
- Llamas MA, Ramos JL, Rodriguez-Herva JJ. Mutations in each of the tol genes of Pseudomonas putida reveal that they are critical for maintenance of outer membrane stability. J Bacteriol. 2000;182:4764–4772. doi: 10.1128/jb.182.17.4764-4772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masilamani R, Cian MB, Dalebroux ZD. Salmonella Tol–Pal reduces outer membrane glycerophospholipid levels for envelope homeostasis and survival during bacteremia. Infect Immun. 2018;86:e00173-18. doi: 10.1128/IAI.00173-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell MJ, Rumbo C, Bou G, Pachon J. Outer membrane vesicles as an acellular vaccine against Acinetobacter baumannii. Vaccine. 2011;29:5705–5710. doi: 10.1016/j.vaccine.2011.06.001. [DOI] [PubMed] [Google Scholar]
- McSorley SJ, Cookson BT, Jenkins MK. Characterization of CD4+ T cell responses during natural infection with Salmonella typhimurium. J Immunol. 2000;164:986–993. doi: 10.4049/jimmunol.164.2.986. [DOI] [PubMed] [Google Scholar]
- Morgan JK, Ortiz JA, Riordan JT. The role for TolA in enterohemorrhagic Escherichia coli pathogenesis and virulence gene transcription. Microb Pathog. 2014;77:42–52. doi: 10.1016/j.micpath.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Muralinath M, Kuehn MJ, Roland KL, Curtiss R., 3rd Immunization with Salmonella enterica serovar Typhimurium-derived outer membrane vesicles delivering the pneumococcal protein PspA confers protection against challenge with Streptococcus pneumoniae. Infect Immun. 2011;79:887–894. doi: 10.1128/IAI.00950-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevermann J, Silva A, Otero C, Oyarzun DP, Barrera B, Gil F, Calderon IL, Fuentes JA. Identification of genes involved in biogenesis of outer membrane vesicles (OMVs) in Salmonella enterica serovar Typhi. Front Microbiol. 2019;10:104. doi: 10.3389/fmicb.2019.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson GK, Northen H, Cone DB, Willers C, Peters SE, Maskell DJ. Deletion of tolA in Salmonella Typhimurium generates an attenuated strain with vaccine potential. Microbiology (Reading) 2009;155:220–228. doi: 10.1099/mic.0.021576-0. [DOI] [PubMed] [Google Scholar]
- Pimenta AL, Young J, Holland IB, Blight MA. Antibody analysis of the localisation, expression and stability of HlyD, the MFP component of the E. coli haemolysin translocator. Mol Gen Genet MGG. 1999;261:122–132. doi: 10.1007/s004380050949. [DOI] [PubMed] [Google Scholar]
- Prouty AM, Van Velkinburgh JC, Gunn JS. Salmonella enterica serovar typhimurium resistance to bile: identification and characterization of the tolQRA cluster. J Bacteriol. 2002;184:1270–1276. doi: 10.1128/JB.184.5.1270-1276.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray MC, Germon P, Vianney A, Portalier R, Lazzaroni JC. Identification by genetic suppression of Escherichia coli TolB residues important for TolB–Pal interaction. J Bacteriol. 2000;182:821–824. doi: 10.1128/jb.182.3.821-824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- Reed WM, Olander HJ, Thacker HL. Studies on the pathogenesis of Salmonella typhimurium and Salmonella choleraesuis var kunzendorf infection in weanling pigs. Am J Vet Res. 1986;47:75–83. [PubMed] [Google Scholar]
- Roland K, Curtiss R, 3rd, Sizemore D. Construction and evaluation of a delta cya delta crp Salmonella typhimurium strain expressing avian pathogenic Escherichia coli O78 LPS as a vaccine to prevent airsacculitis in chickens. Avian Dis. 1999;43:429–441. [PubMed] [Google Scholar]
- Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol. 2015;13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serruto D, Bottomley MJ, Ram S, Giuliani MM, Rappuoli R. The new multicomponent vaccine against meningococcal serogroup B, 4CMenB: immunological, functional and structural characterization of the antigens. Vaccine. 2012;30(Suppl 2):B87–97. doi: 10.1016/j.vaccine.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SP, Williams YU, Miller S, Nikaido H. The C-terminal domain of Salmonella enterica serovar typhimurium OmpA is an immunodominant antigen in mice but appears to be only partially exposed on the bacterial cell surface. Infect Immun. 2003;71:3937–3946. doi: 10.1128/IAI.71.7.3937-3946.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Andersen-Nissen E, Hayashi F, Strobe K, Bergman MA, Barrett SL, Cookson BT, Aderem A. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat Immunol. 2003;4:1247–1253. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- Urrutia IM, Fuentes JA, Valenzuela LM, Ortega AP, Hidalgo AA, Mora GC. Salmonella Typhi shdA: pseudogene or allelic variant? Infect Genet Evol. 2014;26:146–152. doi: 10.1016/j.meegid.2014.05.013. [DOI] [PubMed] [Google Scholar]
- Webster RE. The tol gene products and the import of macromolecules into Escherichia coli. Mol Microbiol. 1991;5:1005–1011. doi: 10.1111/j.1365-2958.1991.tb01873.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.