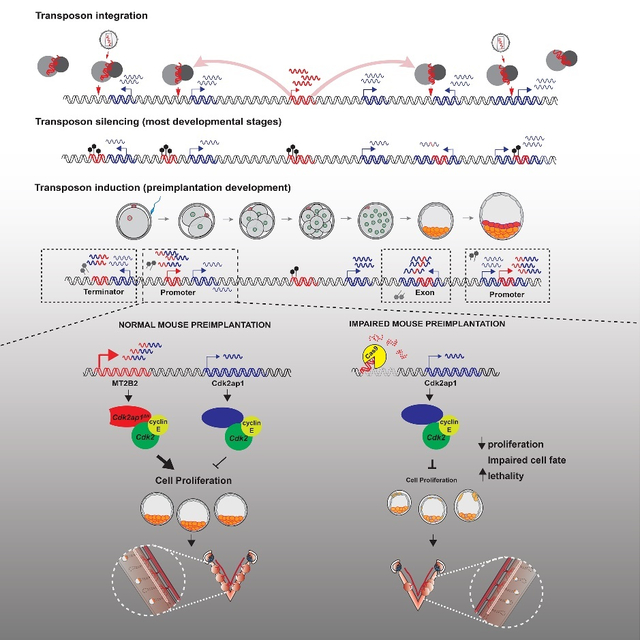
Summary
Retrotransposons mediate gene regulation in important developmental and pathological processes. Here, we characterized the transient retrotransposon induction during preimplantation development of eight mammals. Induced retrotransposons exhibit similar preimplantation profiles across species, conferring gene regulatory activities, particularly through LTR retrotransposon promoters. A mouse-specific MT2B2 retrotransposon promoter generates an N-terminally truncated Cdk2ap1ΔN that peaks in preimplantation embryos and promotes proliferation. In contrast, the canonical Cdk2ap1 peaks in mid-gestation and represses cell proliferation. This MT2B2 promoter is developmentally essential, whose deletion abolishes Cdk2ap1ΔN production, reduces cell proliferation and impairs embryo implantation. Intriguingly, Cdk2ap1ΔN is evolutionarily conserved in sequence and function, yet is driven by different promoters across mammals. The distinct preimplantation Cdk2ap1ΔN expression in each mammalian species correlates with the durations of its preimplantation development. Hence, species-specific transposon promoters can yield evolutionarily conserved, alternative protein isoforms, bestowing them with new functions and species-specific expression to govern essential biological divergence.
eTOC:
A transient retrotransposon induction in mammalian preimplantation embryos yields numerous gene regulatory events. Deletion of an MT2B2 retrotransposon promoter abolishes a Cdk2ap1 isoform (Cdk2ap1ΔN), impairing cell proliferation and causing embryonic lethality. Cdk2ap1ΔN is evolutionarily conserved, generated by species-specific promoters, including transposon-derived promoters, to yield divergent expression patterns.
Graphical Abstract
Introduction
Transposable elements constitute ~40% of mammalian genomes, due to their efficient propagation in the host genomes (Lanciano and Cristofari, 2020; Wells and Feschotte, 2020). The mammalian mobilome is derived from three classes of retrotransposons; Long Terminal Repeat (LTR) retrotransposons, Long Interspersed Nuclear Elements (LINEs) and Short Interspersed Nuclear Elements (SINEs), all propagating in host genomes using a “copy and paste” mechanism via RNA intermediates (Göke and Ng, 2016; Goodier, 2016). Once regarded as parasitic or “junk” DNAs, some retrotransposons are integral functional components of their host genomes (Cosby et al., 2019; Garcia-Perez et al., 2016; Kim et al., 2012). Specific retrotransposon encoded proteins have been co-opted for developmental functions in the host, regulating placental cytotrophoblast fusion in mammals, telomere maintenance in flies, and intracellular RNA transport across neurons (Dupressoir et al., 2009; ED et al., 2018; Levis et al., 1993; Ono et al., 2006; Sekita et al., 2008). More prevalently, retrotransposon exaptation provide numerous cis-regulatory elements for proximal host genes (Batut et al., 2013; Choudhary et al., 2020; Chuong et al., 2013; Rebollo et al., 2012; Sundaram et al., 2017; Wang et al., 2007; Xie et al., 2013). In particular, a subset of LTR retrotransposons, originated from ancient, exogenous retroviruses, still contain intact LTR elements, harboring intrinsic promoter and enhancer activities and splicing donor/acceptor sequences, greatly expanding gene regulation and transcript diversity (Choi et al., 2017; Flemr et al., 2013; Hackett et al., 2017; Macfarlan et al., 2012; Miao et al., 2020; Peaston et al., 2004; Sundaram et al., 2014). Due to their unique evolutionary history, retrotransposon-mediated gene regulation is often considered non-essential and species-specific (Ding et al., 2016; Flemr et al., 2013), and its functional importance in vivo remains largely obscure.
Most retrotransposon integrations are deleterious to genome integrity, necessitating inactivation through degenerative mutation or epigenetic silencing (Imbeault et al., 2017). However, a subset of retrotransposons are strongly induced and tightly regulated under specific developmental, physiological and pathological contexts, including preimplantation development (Boroviak et al., 2018; Gerdes et al., 2016; Gifford et al., 2013; Peaston et al., 2004), germ cell development (Inoue et al., 2017; Molaro et al., 2014; Pasquesi et al., 2020), immune response (Chuong et al., 2016; Grandi and Tramontano, 2018; Saleh et al., 2019), aging (Bravo et al., 2020; De Cecco et al., 2013; Sturm et al., 2015) and cancer (Burns, 2017; Chung et al., 2019; Jang et al., 2019; Kong et al., 2019). Hence, certain retrotransposons are likely exploited by their host for developmental and physiological functions.
A hallmark of the mammalian preimplantation embryo is the transient and robust retrotransposon induction, likely resulted from extensive epigenetic reprogramming (Tang et al., 2015). Here, we comprehensively analyzed retrotransposon expression and retrotransposon mediated gene regulation in preimplantation embryos from 8 mammalian species. We identified numerous alternative gene promoters derived from LTR retrotransposons, and characterized the gene structures of the retrotransposon-dependent gene isoforms. Importantly, we functionally characterized a mouse-specific MT2B2 retrotransposon promoter, which drives an N-terminally truncated, preimplantation-specific Cdk2ap1ΔN isoform. The canonical Cdk2ap1 negatively regulates cell proliferation, yet the MT2B2 driven Cdk2ap1ΔN strongly promotes cell proliferation in preimplantation embryos, rendering this MT2B2 promoter essential for mouse preimplantation development. The distinct expression patterns of Cdk2ap1ΔN and Cdk2ap1 govern their essential functions at different embryonic stages. Intriguingly, the Cdk2ap1ΔN protein is evolutionarily conserved in sequence and function, yet different mammalian species employ divergent regulatory mechanisms to confer species-specific, Cdk2ap1ΔN expression. This gives rise to a spectrum of Cdk2ap1ΔN abundance that inversely correlates with preimplantation duration across mammals. Altogether, species-specific transposon promoters can yield evolutionarily conserved protein isoforms with an alternative ORF, a distinct biological function, and a species-specific expression pattern to generate phenotypical divergence among species.
Results
Retrotransposons are strongly induced in mammalian preimplantation embryos
To comprehensively profile the retrotransposon landscape in mammalian preimplantation development, we analyzed published single-cell RNA-seq datasets from multiple eutherian mammals (human, rhesus monkey, marmoset, mouse, goat, cow, pig) and the metatherian opossum (Table S1). RNA-seq reads were mapped to their corresponding genomes with retrotransposon expression aggregated at the subfamily level (Figures 1A, S1A and Table S1). Retrotransposon expression was quantified either using uniquely mapped reads, or using both uniquely and multiply mapped reads by TEtranscripts (Jin et al., 2015) (Figures 1A, S1A). Both methods capture similar, global retrotransposon expression profiles (Figures 1A, S1A), yet TEtranscripts yields a higher estimation on the percentage of transcriptome derived from retrotransposon loci (Figure S1A). Retrotransposons collectively constitute one of the most abundant non-coding transcript species in preimplantation embryos, accounting for 9% to 38% transcriptome at peak expression across species (Figure 1A, Table S1). Although retrotransposon sequences and integration sites are highly divergent among species, primate, cow, pig, goat, mouse and opossum preimplantation embryos all exhibit a similar global retrotransposon profile, with a major switch at zygotic genome activation (ZGA) (Figure 1A).
Figure 1. Retrotransposons mediate gene regulation in mammalian preimplantation development.
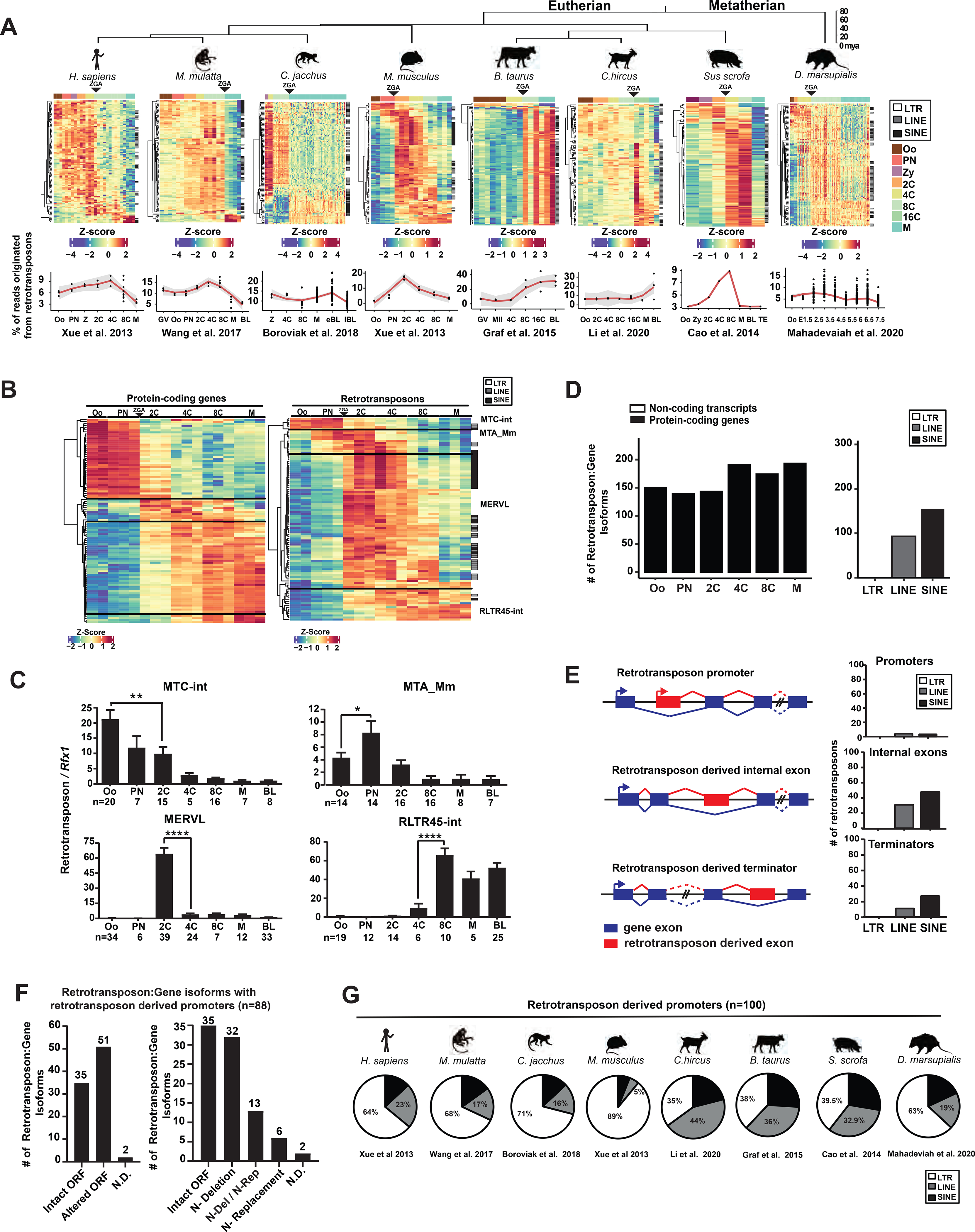
A. Retrotransposons are highly and dynamically expressed in preimplantation embryos across mammals. RNA-seq data from each species were subjected to TEtranscripts analyses to quantify the number of mappable RNA-seq reads at protein-coding genes, non-coding transcripts and retrotransposons. For each species, a heatmap exhibits the preimplantation profile of the top 100 most highly and differentially expressed retrotransposon subfamilies, and line graphs show the percentage of transcriptome from retrotransposon loci. B. TEtranscripts analyses revealed similar profiles of retrotransposon and protein-coding gene in mouse preimplantation embryos, as shown by the heatmap of the top 100 most highly and differentially expressed protein-coding genes (left) and retrotransposon subfamilies (right). Four distinct patterns emerged. A, B. Z-score, the number of standard deviations from the expression mean of a retrotransposon subfamily or a protein-coding gene. Oo, oocyte; Zy, zygotes; PN, pronucleus; 2C, two cell embryo; 4C, four cell embryo; 8C, eight cell embryo; 16C, sixteen cell embryos; M, morula; BL, blastocysts. C. Single embryo real time PCR analyses confirm the dynamic expression patterns of four representative retrotransposon subfamilies. Error bars, ± s.e.m. P values were calculated using unpaired, two-tailed Student’s t test. (MTC-int, Oo vs. 2C, **P = 0.009, t=2.8, df=33 MTA_Mm, Oo vs. PN, *P = 0.04 t=2.1, df=26; MERVL, 2C vs. 4C, ****P < 0.0001 t=7.4, df=62; RLTR45-int, 4C vs 8C, ****P < 0.0001 t=5.2, df=16). D. Preimplantation-specific, retrotransposon:gene splicing junctions preferentially associate with protein-coding genes in preimplantation embryos. Retrotransposon:gene isoforms of GENCODE annotated protein-coding genes (black) and non-coding transcripts (white) are shown as bar plots for all preimplantation stages (left). Retrotransposon:gene isoforms containing LTR, LINE or SINE retrotransposons are each quantified (right). Only highly expressed retrotransposon:gene splicing junctions (an average of ≥ 30 reads across preimplantation stages) are included in these analyses. E. Retrotransposons mediate gene regulation as alternative promoters, internal exons and terminators for proximal gene isoforms (left). The top 250 most highly and differentially expressed retrotransposons that yield gene promoters (TSS within retrotransposon), internal exons and terminators were classified by LTRs, LINEs and SINEs (right). F. Retrotransposon promoters frequently drive gene isoforms with N-terminally altered ORFs. Among the 250 most highly and differentially expressed retrotransposon:gene isoforms in mouse preimplantation embryos, 88 are driven by retrotransposon promoters. Manual curation predicts frequent ORFs alterations caused by retrotransposon promoters (left), which are further classified based on the mechanisms of ORF alteration (right). N-Deletion, predicted N-terminal truncation; N-Replacement, predicted sequence replacement of the protein N-terminus; N-Del/N-Rep, predicted as either N-terminal deletions or N-terminal sequence replacements, due to uncertainty in ATG prediction; N.D., not determined. G. Retrotransposon promoters in mammalian preimplantation embryos are enriched for LTR retrotransposons. The proportion of LTR, LINE or SINE retrotransposons was determined for the top 100 most highly and dynamically expressed retrotransposon promoters in preimplantation embryos of 8 mammalian species. RNA-seq data for 1B, 1D, 1E and 1F analyses were obtained from Xue et al. 2013. All P values were calculated using unpaired, two-tailed Student’s t test. n.s., not significant. See also Figure S1 and Tables S1–S5.
The global, dynamic retrotransposon expression profile in preimplantation embryos closely resembles that of protein-coding genes in each species (Figures 1B, S1B, Table S1, S2). Although a subset of retrotransposons are located within protein-coding gene introns (Figure S1C), these intronic retrotransposons do not confound the global expression profile of retrotransposons. Removing reads derived from intronic retrotransposons has no effect on the similar preimplantation expression patterns between retrotransposons and protein-coding genes (Figure S1D), implying that retrotransposons and protein-coding genes could be under similar transcription regulation.
In mouse embryos, retrotransposons exhibit four distinct expression patterns (Figures 1B), represented by MTC-int (peaks in oocytes and decreases upon ZGA), MTA_Mm (transiently peaks at pronuclear and 2C embryos), MERVL, ORR1A1 and IAPez-int (transiently peak in 2C-8C embryos), and RLTR45 and ERVB4_2-I_Mm (peak in morulae/blastocysts following an 8C induction) (Figures 1C, S1E). A subset of retrotransposon subfamilies that share the same expression pattern are related in sequence and classification (Figure S1F, Table S1).
Retrotransposons mediate gene regulation in mammalian preimplantation development
Hundreds of preimplantation-specific splicing events are detected between a transcribed retrotransposon element and a proximal gene exon in mouse preimplantation embryos (Figure S1G, Table S3). Retrotransposon-gene splicing events are significantly biased towards expressed protein-coding genes, rather than non-coding transcripts (Figures 1D, S1H). We ranked retrotransposon:gene splicing events based on extent of their differential expression and peak expression level, and then analyzed the impact of the top retrotransposons on host gene structure (Table S3, methods). Among the top 250 retrotransposon:gene transcripts, retrotransposons provide alternative promoters (37%), internal exons (46%) and terminators (17%) to proximal host genes (Figures 1E, S1I). Using 5’ and 3’ RACE and real time PCR, we experimentally validated the gene structure and preimplantation-specific expression patterns of 27 predicted retrotransposon:gene isoforms, with retrotransposons acting as alternative promoters (n=15), internal exons (n=4) or terminators (n=8) (Table S4). Interestingly, highly dynamic retrotransposon:gene isoforms differ from the corresponding canonical isoforms in gene structure, expression regulation, and frequently, open reading frames (ORFs) (Figures 1F, S1J, Tables S5). The retrotransposon:gene isoforms encoding an alternative ORF often harbor truncations, insertions or sequence replacement of the canonical protein sequences (Figures 1F, S1J), but rarely frame shift or non-sense mutations (Table S5).
Retrotransposon promoters in mouse preimplantation embryos are particularly enriched for LTR retrotransposons, but not LINEs or SINEs (Figure 1E). LTR retrotransposons exist either as full proviral sequences with two identical long terminal repeats (LTRs) flanking the internal region, or more frequently, as solo-LTRs. The LTR retrotransposon promoters confer new transcriptional regulation to the proximal host genes, contributing to alternative 5’UTRs and/or ORFs (Figure 1F). Among the 250 most highly and differentially expressed retrotransposon:gene isoforms in mouse embryos, 88 are driven by retrotransposon promoters. Manual curation of these 88 gene isoforms revealed that 58% were predicted to yield N-terminally altered ORFs (Figure 1F, Table S5). Our findings suggest that retrotransposon promoters frequently yield new gene isoform with an alternative ORF, and possibly, an alternative biological function.
The prevalence of retrotransposon promoters is not unique to mouse, as human, rhesus monkey, marmoset, cow, goat, pig and opossum all employ retrotransposon promoters in preimplantation embryos to generate alternative gene isoforms. In most cases, different mammals have different retrotransposon promoters (Figure S1K), which regulate host genes in a species-specific manner. In all species examined, LTR retrotransposons are enriched for retrotransposon-derived promoters in preimplantation embryos (Figures 1G, S1L, Tables S3, S5).
An MT2B2 retrotransposon promoter induces an N-terminally truncated Cdk2ap1 isoform
The frequency of retrotransposon initiated preimplantation gene isoforms prompted us to explore their functional importance in vivo. In mouse preimplantation embryos, one of the most highly and dynamically expressed gene isoforms driven by a retrotransposon promoter is the MT2B2 driven Cdk2ap1 (Cyclin dependent kinase associated protein 1) isoform (Figures 2A, S2A). The MT2B2 promoter, 8.2 kb upstream of Cdk2ap1, generates an N-terminally truncated Cdk2ap1ΔN (MT2B2) isoform (Figures 2A, S2B).
Figure 2. Canonical Cdk2ap1 and MT2B2 driven Cdk2ap1ΔN (MT2B2) differ in function.
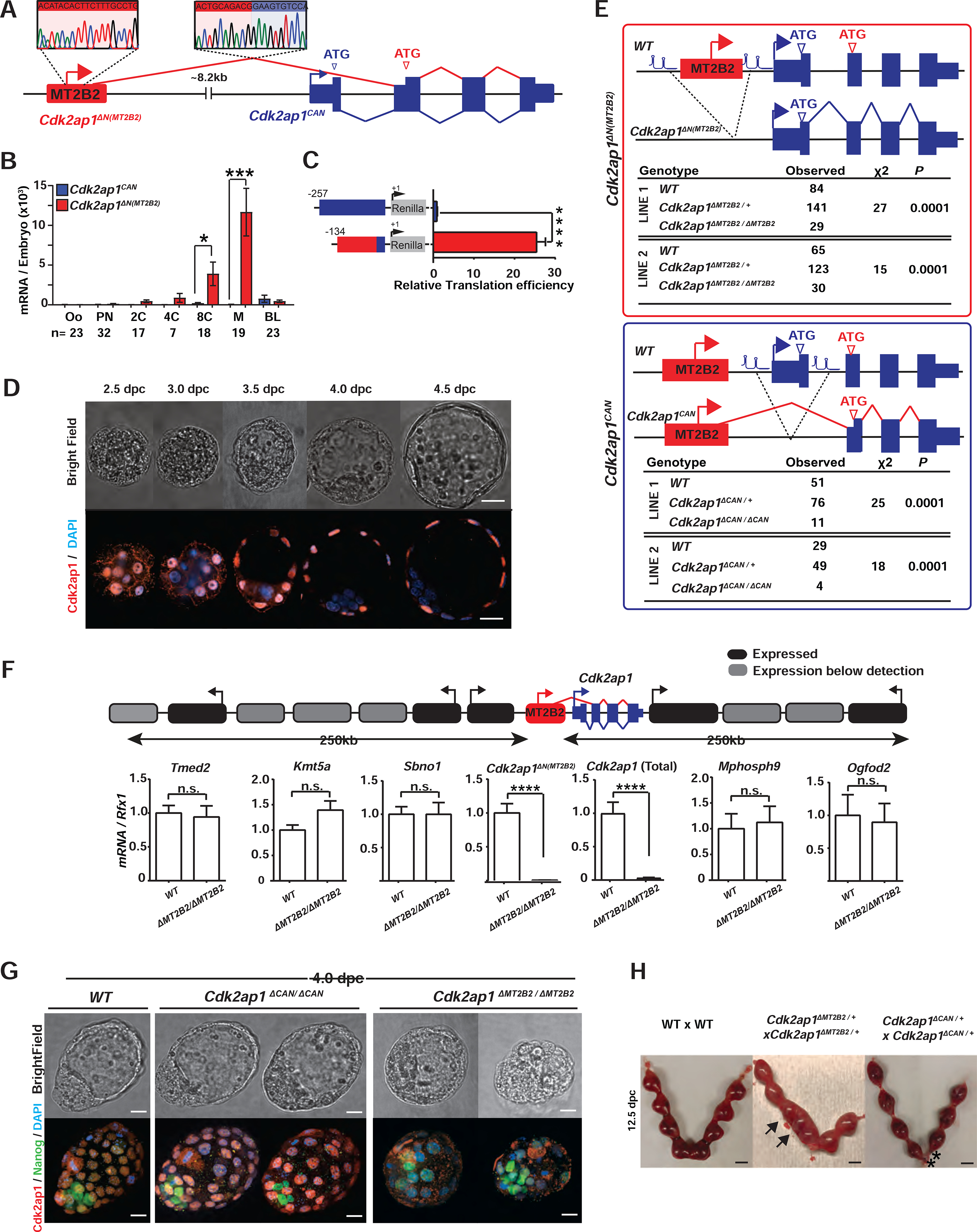
A. Diagram illustrates the gene structure of canonical Cdk2ap1CAN (blue) and Cdk2ap1ΔN (MT2B2) (red) isoforms. 5’ RACE confirms TSS within the MT2B2 element; RT-PCR confirms splicing between MT2B2 and Cdk2ap1 exon 2. B. Absolute real-time PCR quantification of single embryos compares the level of Cdk2ap1CAN and Cdk2ap1ΔN (MT2B2). Error bars, s.e.m. Cdk2ap1CAN vs. Cdk2ap1ΔN (MT2B2) at 8C, n=17, *P = 0.02, t=2.5, df=34; Cdk2ap1CAN vs. Cdk2ap1ΔN (MT2B2) at morula, n=19, ***P = 0.0004, t=3.9, df=36. C. MT2B2 derived 5’UTR enhances the translation efficiency of Cdk2ap1ΔN (MT2B2). The 5’UTR of Cdk2ap1CAN or Cdk2ap1ΔN (MT2B2) were each cloned 5’ to a Renilla luciferase reporter to measure its impact on translation in HEK293 cells. The MT2B2 derived 5’UTR was associated with a higher translation efficiency. Three independent experiments were performed in triplicate per condition. Error bars, s.e.m; **** P < 0.0001, t=20.44, df=4. D. Mouse preimplantation embryos between 2.5 dpc to 4.5 dpc were immunostained for Cdk2ap1. Cdk2ap1 protein expresses in the outer cells of morulae and the TE cells in blastocysts. Confocal images are representative of 4 or more embryos per stage. Scale bar, 20μm. E. Diagrams illustrate CRISPR genome engineering strategy for targeted deletion of Cdk2ap1ΔN (MT2B2) (top) and Cdk2ap1CAN (bottom). Mendelian ratios of progenies from Cdk2ap1ΔMT2B2/+ × Cdk2ap1ΔMT2B2/+ crosses (top) or Cdk2ap1ΔCAN/+ × Cdk2ap1ΔCAN/+ crosses (bottom) were documented at postnatal day 10 (p10), demonstrating a significant reduction of viability in both genotypes. Two independent Cdk2ap1ΔMT2B2/ΔMT2B2 and Cdk2ap1ΔCAN/ΔCAN lines were analyzed. F. The MT2B2 deletion specifically abolishes Cdk2ap1ΔN (MT2B2) expression, without impacting any neighboring genes. Age matched wildtype (n=9) and Cdk2ap1ΔMT2B2/ΔMT2B2 (n=7) morula embryos were collected from two independent WT × WT and Cdk2ap1ΔMT2B2/ΔMT2B2 × Cdk2ap1ΔMT2B2/ΔMT2B2 crosses, respectively, and were subjected to single embryo real-time PCR analyses to measure the expression of Cdk2ap1ΔN (MT2B2), total Cdk2ap1 and all neighboring genes with 250 kb of the deletion. Black, expressed genes; grey, genes below detection; error bars, s.e.m. Cdk2ap1 (Total), wildtype (n=3) vs. Cdk2ap1ΔMT2B2/ΔMT2B2 (n=3), ****P < 0.0001, t=16.8, df=4; Cdk2ap1ΔN (MT2B2), wildtype (n=9) vs. Cdk2ap1ΔMT2B2/ΔMT2B2 (n=7), ***P = 0.0002, t=4.9, df=14. G. Cdk2ap1ΔMT2B2/ΔMT2B2 embryos, but not Cdk2ap1ΔCAN/ΔCAN embryos, exhibit defective Cdk2ap1 protein expression in TE and impaired blastocyst formation. Representative confocal images for Cdk2ap1 and Nanog immunostaining are shown for wildtype (n=11), Cdk2ap1ΔCAN/ΔCAN (n=5) and Cdk2ap1ΔMT2B2/ΔMT2B2 (n=6) embryos. Scale bar, 25 μm. H. Deletion of Cdk2ap1ΔN (MT2B2), but not Cdk2ap1CAN, is associated with embryo implantation spacing defects. At E12.5, embryo crowding is evident in uteri from the Cdk2ap1ΔN (MT2B2)/+ × Cdk2ap1ΔN (MT2B2)/+ crosses (n=34), while resorption of correctly spaced embryos is evident in uteri from the Cdk2ap1ΔCAN/+ × Cdk2ap1ΔCAN/+ crosses (n=7). Black arrows, embryo crowding; *, resorbed embryos. Scale bars, 0.5 cm. All P values were calculated using unpaired, two-tailed Student’s t test. n.s., not significant. See also Figure S2 and Table S4.
The canonical Cdk2ap1 (Cdk2ap1CAN) is reported as a suppressor of cell proliferation, at least in part, by promoting Cdk2 degradation and repressing its kinase activity (Hu et al., 2004; Shintani et al., 2000; Wong et al., 2012). For Cdk2ap1CAN, both transcription start site (TSS) and the ATG start codon are within its exon 1 (Figures 2A, S2A). The MT2B2 driven Cdk2ap1ΔN (MT2B2) isoform is alternatively spliced to skip exon 1, utilizing a downstream ATG in exon 2 to generate an N-terminal truncation of 27 amino acids (Figures 2A, S2B). The MT2B2 element not only promotes strong Cdk2ap1ΔN (MT2B2) induction in 8C to morula embryos (Figure 2B), but also contributes to a hybrid 5’UTR with enhanced translation efficiency (Figure 2C).
Cdk2ap1CAN and Cdk2ap1ΔN (MT2B2) exhibit distinct expression patterns. Cdk2ap1CAN remained at a low level throughout preimplantation development, yet later peaked around 10.5 days post coitum (dpc) (Figures 2B, S2C). Cdk2ap1ΔN (MT2B2) is the predominant, preimplantation-specific isoform, whose expression peaks in 8C and morula embryos (Figure 2B). Cdk2ap1 protein expression was first detected in the nuclei of compacted morula blastomeres, and subsequently in the trophectoderm (TE) of blastocysts (Figure 2D). This is consistent with the Cdk2ap1 mRNA enrichment in the TE by the blastocysts stage (Figure S2D).
To determine which Cdk2ap1 protein isoform is expressed in preimplantation embryos, we engineered isoform-specific, V5 tagging at the N-terminus of endogenous Cdk2ap1CAN, the N-terminus of Cdk2ap1ΔN (MT2B2), and the C-terminus of all Cdk2ap1 isoforms. V5 Immunostaining revealed that most, if not all, Cdk2ap1 protein in preimplantation embryos is generated from the Cdk2ap1ΔN (MT2B2) isoform (Figure S2E).
The MT2B2 promoter for Cdk2ap1ΔN (MT2B2) is essential in preimplantation development
We next investigated the functional importance of the MT2B2 promoter. We employed CRISPR-EZ, a highly efficient CRISPR technology for mouse genome engineering (Chen et al., 2016; Modzelewski et al., 2018). We deleted the MT2B2 element or the Cdk2ap1 canonical exon 1, generating C57BL/6J mice deficient for either Cdk2ap1ΔN (MT2B2) or Cdk2ap1CAN, respectively (designated as Cdk2ap1ΔMT2B2/ΔMT2B2 and Cdk2ap1ΔCAN/ΔCAN mice, Figures 2E, S2F). The MT2B2 deletion specifically abolished Cdk2ap1ΔN (MT2B2), and significantly reduced total Cdk2ap1 mRNA in preimplantation embryos without impacting flanking genes (Figure 2F). While both Cdk2ap1ΔMT2B2/ΔMT2B2 and Cdk2ap1ΔCAN/ΔCAN mice exhibited significantly reduced viability by P10 (Figure 2E), only Cdk2ap1ΔMT2B2/ΔMT2B2 mice exhibited defective preimplantation development and embryo implantation (Figures 2G, 2H).
Two independent Cdk2ap1ΔMT2B2/ΔMT2B2 mouse lines exhibited 50–55% penetrance for lethality (Figure 2E); those that survive into adulthood appeared grossly normal and fertile. Cdk2ap1ΔMT2B2/ΔMT2B2 4.0 dpc embryos were recovered at the expected Mendelian ratio, yet 71% exhibited abnormal morphology, characterized by reduced cell number, aberrant cell organization and impaired blastocoel cavities. During post-implantation, the Cdk2ap1ΔMT2B2/ΔMT2B2 defect manifest as an embryo crowding event (Figure 2H), and nearly half of the embryos that survived implantation displayed developmental delays (Figure S2G). In comparison, Cdk2ap1ΔCAN/ΔCAN embryos were intact throughout preimplantation development (Figure 2G), but often displayed a higher frequency of resorption events in post-implantation development (Figure 2H). Hence, the different expression patterns of Cdk2ap1ΔN (MT2B2) and Cdk2ap1CAN underlie their distinct developmental functions.
Deficiency of Cdk2ap1ΔN (MT2B2), but not Cdk2ap1CAN, reduced cell proliferation in preimplantation embryos, as demonstrated by reduced total cell number and BrdU incorporation in Cdk2ap1ΔMT2B2/ΔMT2B2 morulae and blastocysts (Figures 3A–3C, S3A, S3B), particularly in the TE compartment. Aberrant Nanog and Cdx2 double-positive cells were frequently identified in 4.0 dpc Cdk2ap1ΔMT2B2/ΔMT2B2 blastocysts, mostly impacting TE blastomeres due to a delayed/impaired cell fate specification (Figures 3D, S3C). Consistently, Wnt5a, a temporal marker associated with maternal-fetal attachment at peri-implantation was impaired at the implantation sites (Figure S3D). Reduced TE cell number and impaired TE cell fate specification in Cdk2ap1ΔMT2B2/ΔMT2B2 blastocysts likely contribute to a decreased implantation rate, aberrant embryo spacing in uterus, and increased embryo lethality (Figure 3E). The blastocyst defects in Cdk2ap1ΔMT2B2/ΔMT2B2 embryos are consistent with the preimplantation lethality caused by targeted disruption of all Cdk2ap1 isoforms (Kim et al., 2009).
Figure 3. An MT2B2 promoter drives a Cdk2ap1ΔN (MT2B2) isoform to promote cell proliferation.
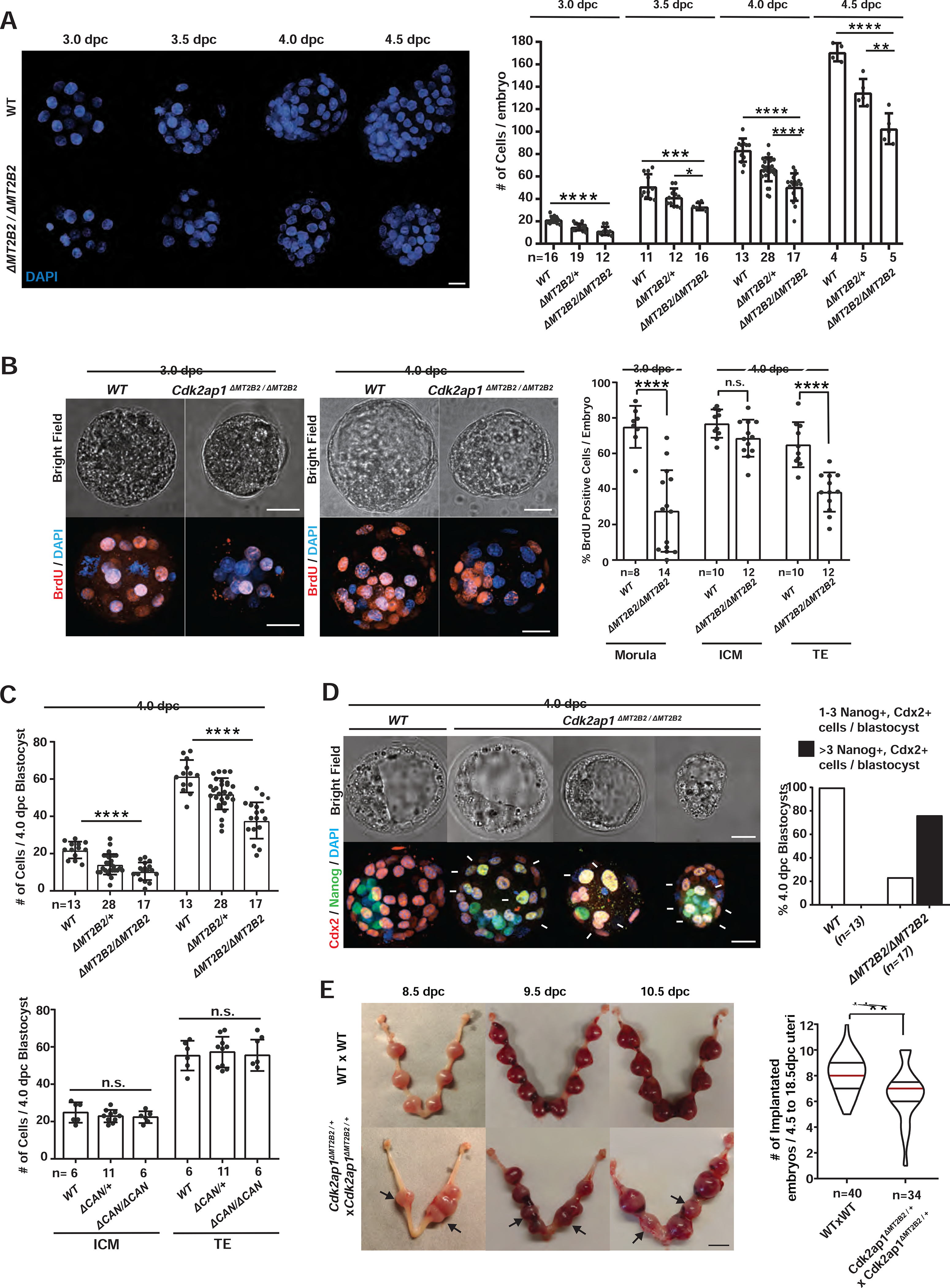
A. Cdk2ap1ΔMT2B2/ΔMT2B2 preimplantation embryos exhibited reduced cell number. Littermate-controlled wildtype (n=44), Cdk2ap1ΔMT2B2/+ (n= 64) and Cdk2ap1ΔMT2B2/ΔMT2B2 (n=50) embryos were collected at 3.0 dpc, 3.5 dpc, 4.0 dpc and 4.5 dpc from 29 Cdk2ap1ΔMT2B2/+ to Cdk2ap1ΔMT2B2/+ mating. Representative images of DAPI staining (left) and cell number quantitation (right) are shown for each stage. Scale bar, 25 μm; error bars, s.d.. Wildtype vs. Cdk2ap1ΔMT2B2/ΔMT2B2: 3.0 dpc, **** P < 0.0001, t=8.2, df=26; 3.5 dpc, *** P = 0.0007, t=4.2, df=15; 4.0 dpc, **** P < 0.0001, t=7.8, df=28; 4.5 dpc, **** P < 0.0001, t=8.7, df=7. Cdk2ap1ΔMT2B2/+vs Cdk2ap1ΔMT2B2/ΔMT2B2, 3.5 dpc, * P = 0.03, t=2.4, df=16; 4.0 dpc, **** P < 0.0001, t=4.5, df=43; 4.5 dpc, ** P = 0.005, t=3.9, df=8. B. Cdk2ap1ΔMT2B2/ΔMT2B2 embryos exhibit decreased BrdU incorporation. Representative confocal images (left) and quantitation (right) of BrdU staining are shown for embryos at 3.0 and 4.0 dpc. Age matched wildtype (n=18) and Cdk2ap1ΔMT2B2/ΔMT2B2 (n=26) morulae and blastocysts were collected from wildtype × wildtype and Cdk2ap1ΔMT2B2/ΔMT2B2 × Cdk2ap1ΔMT2B2/ΔMT2B2 mating, respectively. Scale bars, 20 μm; error bars, s.d.. Wildtype vs. Cdk2ap1ΔMT2B2/ΔMT2B2: morula, **** P < 0.0001, t=7.9, df=20; TE, **** P < 0.0001, t=5.3, df=20. C. Cdk2ap1ΔMT2B2/ΔMT2B2, but not Cdk2ap1ΔCAN/ΔCAN blastocysts, exhibit decreased cell number in ICM and TE. Blastocysts (n=58) from Cdk2ap1ΔMT2B2/+ × Cdk2ap1ΔMT2B2/+ crosses, and blastocysts (n=23) from Cdk2ap1ΔCAN/+ × Cdk2ap1ΔCAN/+ crosses were immunostained for Nanog and Cdx2 to quantify ICM and TE cell numbers, respectively. Scale bars, 25 μm; error bars, s.d.. Wildtype vs Cdk2ap1ΔMT2B2/ΔMT2B2: ICM, **** P < 0.0001, t=6.7, df=28; TE, **** P < 0.0001, t=6.9, df=28. D. The MT2B2 deletion impairs cell fate specification in blastocysts. Littermate controlled wildtype (n=13) and Cdk2ap1ΔMT2B2/ΔMT2B2 (n=17) blastocysts were immunostained for Nanog and Cdx2 at 4.0 dpc. Representative confocal images (left) and quantitation (right) are shown for Nanog and Cdx2 staining in wildtype and Cdk2ap1ΔMT2B2/ΔMT2B2 embryos. The presence of ≥ 3 Nanog and Cdx2 double positive cells in any blastocysts indicates impaired cell fate specification. White arrows, Nanog and Cdx2 double positive cells. Scale bar, 0.5 cm. E. The deletion of the MT2B2 element caused aberrant embryo spacing and impaired implantation. Representative images are shown for embryo implantation at 8.5, 9.5 and 10.5 dpc in wildtype × wildtype and Cdk2ap1ΔMT2B2/+ × Cdk2ap1ΔMT2B2/+ crosses (left). Black arrows, Cdk2ap1ΔMT2B2/ΔMT2B2 embryos; scale bar, 0.5 cm. Quantitation of implanted embryos from 4.5 to 18.5 dpc per uterus is shown for wildtype × wildtype (n=40), Cdk2ap1ΔMT2B2/+ × Cdk2ap1ΔMT2B2/+ (n=34), with median (red line) as well as lower (25%) and upper (75%) quartiles (black lines). Wildtype × wildtype vs. Cdk2ap1ΔMT2B2/+, ** P = 0.002, t=3.2, df=72. All P values were calculated using unpaired, two-tailed Student’s t test. n.s., not significant. See also Figure S3.
Previous studies have characterized the knockout phenotype of retrotransposon promoters in Drosophila and mice, yet those defects affect non-essential developmental processes, such as mating behavior and female fertility (Ding et al., 2016; Flemr et al., 2013). To our knowledge, MT2B2 is the first example of a retrotransposon promoter with an essential function in normal mammalian development.
Canonical Cdk2ap1 and MT2B2 driven Cdk2ap1ΔN (MT2B2) differ in developmental functions
In contrast to Cdk2ap1ΔMT2B2/ΔMT2B2 embryos, Cdk2ap1ΔCAN/ΔCAN blastocysts were morphologically intact, with no defects in cell number, cell proliferation, or cell fate specification (Figures 2G, 3C). Nevertheless, two independent Cdk2ap1ΔCAN/ΔCAN lines exhibited reduced viability at P10, with a 58–67% penetrance for lethality (Figure 2E). The lethality of Cdk2ap1ΔCAN/ΔCAN mice is likely attributed to impaired mid-gestation development, as the expression of Cdk2ap1CAN peaks on 10.5dpc (Figure S2B) and increased embryo resorption occurs during mid-gestation stage from the Cdk2ap1ΔCAN//+ × Cdk2ap1ΔCAN//+ mating (Figure 2H). In contrast, impaired implantation spacing is the major defect from the Cdk2ap1ΔMT2B2/+× Cdk2ap1ΔMT2B2/+ mating.
Cdk2ap1ΔN (MT2B2) and the canonical Cdk2ap1 have opposite effects on cell proliferation
The effect of Cdk2ap1ΔN (MT2B2) on cell proliferation is opposite from the anti-proliferative function of Cdk2ap1CAN (Figueiredo et al., 2006; Kim et al., 2005; Shintani et al., 2000). The decreased blastomere count in Cdk2ap1ΔMT2B2/ΔMT2B2 embryos supports a role for Cdk2ap1ΔN (MT2B2) in promoting proliferation (Figure 3A–3C). We compared Cdk2ap1ΔN (MT2B2) and Cdk2ap1CAN overexpression phenotype in preimplantation embryos. We optimized an electroporation-based method for mRNA delivery into mouse zygotes and achieved robust delivery efficiency (Figures 4A, S4A). Wildtype zygotes overexpressing Cdk2ap1ΔN (MT2B2) exhibited greater BrdU incorporation and increased total cell number (Figures S4B, S4C); those overexpressing Cdk2ap1CAN displayed reduced BrdU incorporation and decreased total cell number (Figures S4B, S4C). Importantly, ectopic Cdk2ap1ΔN (MT2B2) expression rescued cell proliferation defects of Cdk2ap1ΔMT2B2/ΔMT2B2 embryos, restoring BrdU incorporation and total cell number to wildtype levels (Figures 4B, 4C, S4D), and mitigating Nanog and Cdx2 double positivity in blastocysts (Figures 4D). In comparison, Cdk2ap1CAN overexpression in Cdk2ap1ΔMT2B2/ΔMT2B2 embryos exacerbated cell proliferation and cell fate defects (Figures 4B–4D, S4D). Hence, Cdk2ap1ΔN (MT2B2) and Cdk2ap1CAN exhibit opposite effects on cell proliferation in preimplantation embryos, but functional antagonism unlikely occurs in normal development due to their non-overlapping expression patterns.
Figure 4. Cdk2ap1ΔN (MT2B2) and Cdk2ap1CAN have opposite effects in cell proliferation.
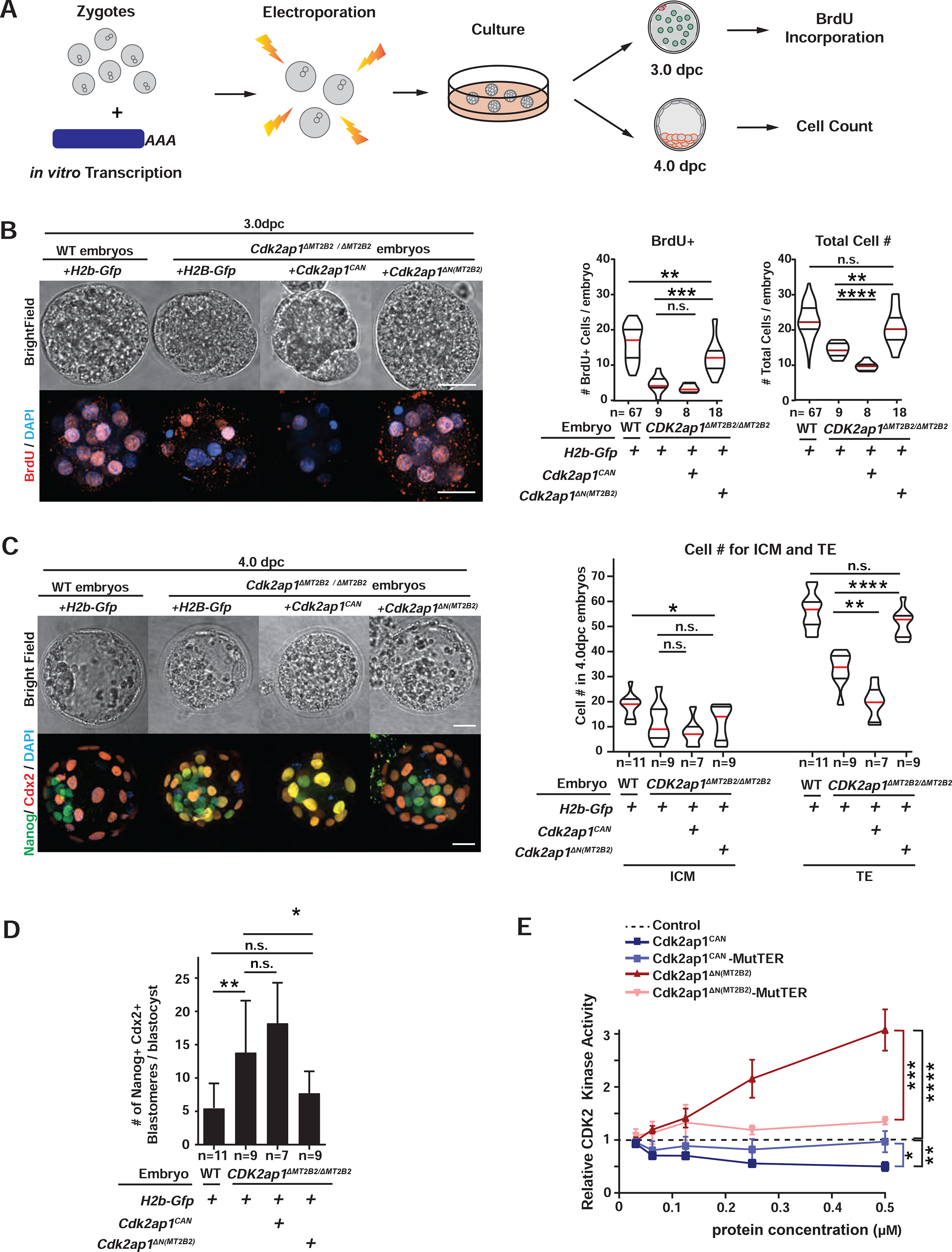
A. Diagram illustrates the experimental scheme for mRNA electroporation into zygotes. B, C. Cdk2ap1CAN and Cdk2ap1ΔN (MT2B2) have opposite effects on S-Phase entry and cell proliferation. H2b-Gfp, Cdk2ap1CAN or Cdk2ap1ΔN (MT2B2) mRNAs were each electroporated into Cdk2ap1ΔMT2B2/ΔMT2B2 zygotes, and (B) resulted morula were compared for BrdU incorporation at 3.0 dpc. Ectopic expression of Cdk2ap1ΔN (MT2B2) restores S-Phase entry and cell proliferation in Cdk2ap1ΔMT2B2/ΔMT2B2 embryos (B). Representative images (left) and quantitation of BrdU positive and total cell number (right) are shown. Violin plots are shown with median (red), as well as lower (25%) and upper (75%) quartiles (black). Scale bars, 20 μm. H2b-Gfp vs Cdk2ap1CAN in Cdk2ap1ΔMT2B2/ΔMT2B2 embryos: BrdU, n.s.; total cell number, **** P < 0.0001, t=5.7, df=15. H2b-Gfp vs Cdk2ap1ΔN (MT2B2) in Cdk2ap1ΔMT2B2/ΔMT2B2 embryos: BrdU, *** P =0.0002, t=4.5, df=25; total cell number, ** P =0.002, t=3.4, df=25. C, D. Ectopic expression of Cdk2ap1ΔN (MT2B2) rescues cell proliferation and cell fate specification defects in Cdk2ap1ΔMT2B2/ΔMT2B2 embryos. D. Representative confocal image of Cdx2 and Nanog immunostaining (left) and quantitation of ICM and TE cell number (right) are shown for 4.0 dpc Cdk2ap1ΔMT2B2/ΔMT2B2 embryos with overexpression of Cdk2ap1CAN or Cdk2ap1ΔN (MT2B2). Scale bars, 20 μm; White arrows, Nanog and Cdx2 double positive cells. H2b-Gfp vs Cdk2ap1CAN, TE, ** P =0.002, t=3.9, df=14; H2b-Gfp vs Cdk2ap1ΔN (MT2B2, TE, **** P < 0.0001, t=6.1, df=16. D. Quantitation of Nanog and Cdx2 double positive cells is shown for Cdk2ap1ΔMT2B2/ΔMT2B2 embryos overexpressing Cdk2ap1CAN or Cdk2ap1ΔN (MT2B2). H2b-Gfp-overexpressing wildtype vs. Cdk2ap1ΔMT2B2/ΔMT2B2 embryos, ** P = 0.007, t=3.1, df=17; H2b-Gfp vs Cdk2ap1ΔN (MT2B2) in Cdk2ap1ΔMT2B2/ΔMT2B2) embryos, * P =0.04, t=2.2, df=16. E. Cdk2ap1CAN and Cdk2ap1ΔN (MT2B2) have opposite effects on Cdk2 kinase activity. Recombinant Cdk2ap1CAN, Cdk2ap1CAN-MutTER, Cdk2ap1ΔN (MT2B2) or Cdk2ap1ΔN (MT2B2)-MutTER protein was incubated with recombinant CDK2, CYCLIN E, and HISTONE H1 in vitro to assay their effects on CDK2 activity at different concentrations. Three independent experiments were performed. Dashed line, baseline CDK2 kinase activity with elution buffer as the “control” input. Error bars, s.e.m. Control vs Cdk2ap1CAN, **P = 0.001, t=8.4, df=4. Cdk2ap1CAN vs Cdk2ap1CAN-MutTER, *P = 0.02, t=3.8, df=4. Control vs Cdk2ap1ΔN (MT2B2), ****P < 0.0001, t=10.5, df=6; Cdk2ap1ΔN (MT2B2) vs Cdk2ap1ΔN (MT2B2)-MutTER, ***P = 0.0003, t=8.9, df=5. All P values were calculated using unpaired, two-tailed Student’s t test. n.s., not significant. See also Figure S4.
We next explored the molecular basis for the opposite proliferation effects of Cdk2ap1ΔN (MT2B2) and Cdk2ap1CAN. Previous studies described Cdk2ap1CAN as a potent, negative cell cycle regulator that directly binds to Cdk2 via a three amino acid “TER motif” to reduce its abundance and inhibit its kinase activity (Shintani et al., 2000). Both Cdk2ap1CAN and Cdk2ap1ΔN(MT2B2) contain the TER motif and directly associate with Cdk2 in co-immunoprecipitation experiments (Figure S4E). In an in vitro CDK2 Kinase assay, immuno-precipitated Cdk2ap1 lysate from HEK293T cells overexpressing Cdk2ap1CAN or Cdk2ap1ΔN (MT2B2) were incubated with recombinant CDK2/CYCLIN E1 complex and substrate HISTONE H1 to quantify their effects on CDK2 kinase activity (Figure S4F). Similarly, purified recombinant Cdk2ap1CAN and Cdk2ap1ΔN (MT2B2) proteins were tested for their effects on CDK2 kinase activity (Figure 4E). In both experiments, Cdk2ap1CAN and Cdk2ap1ΔN (MT2B2) significantly inhibited and enhanced CDK2 kinase activity, respectively (Figures 4F, S4F), in line with their opposite effects on cell proliferation in vivo. Mutation of the TER motif in Cdk2ap1CAN or Cdk2ap1ΔN (MT2B2) abolished their effects on CDK2 kinase activity (Figures 4F, S4F), demonstrating the importance of direct Cdk2ap1-CDK2 binding for this regulation. It is possible that Cdk2ap1ΔN and Cdk2ap1CAN also regulate additional cell proliferation pathways (Alsayegh et al., 2018; Spruijt et al., 2010; Wong et al., 2012), because Cdk2 knockout alone is not sufficient to render any preimplantation defects (Singh et al., 2019).
The N-terminally truncated Cdk2ap1ΔN is evolutionarily conserved in human
The canonical Cdk2ap1 gene structure is highly conserved between mouse and human (Figure 5A), yet the mouse Cdk2ap1ΔN (MT2B2) isoform has a human orthologue CDK2AP1ΔN, generated from an alternative, human-specific upstream promoter that directly splices into exon2 (Figure 5A). Human CDK2AP1ΔN and mouse Cdk2ap1ΔN (MT2B2) both utilize the ATG start codon in exon 2 to initiate translation, and the N-terminally truncated Cdk2ap1 proteins from both species share 97% sequence identity (Figures 5A, 5B). Upon overexpression in mouse Cdk2ap1ΔMT2B2/ΔMT2B2 embryos, the human CDK2AP1ΔN isoform functionally resembled the mouse Cdk2ap1ΔN (MT2B2) isoform, restoring cell proliferation and cell fate specification to wildtype levels (Figures 5C–5E). In contrast, the canonical human CDK2AP1CAN isoform functionally resembled the mouse Cdk2ap1CAN isoform, as its overexpression reduced BrdU incorporation and total cell number in Cdk2ap1ΔMT2B2/ΔMT2B2 embryos, particularly in the TE compartment (Figures 5C, 5D). Hence, opposite functions of Cdk2ap1CAN and Cdk2ap1ΔN is evolutionarily conserved.
Figure 5. The MT2B2-driven Cdk2ap1ΔN isoform is evolutionarily conserved in human.
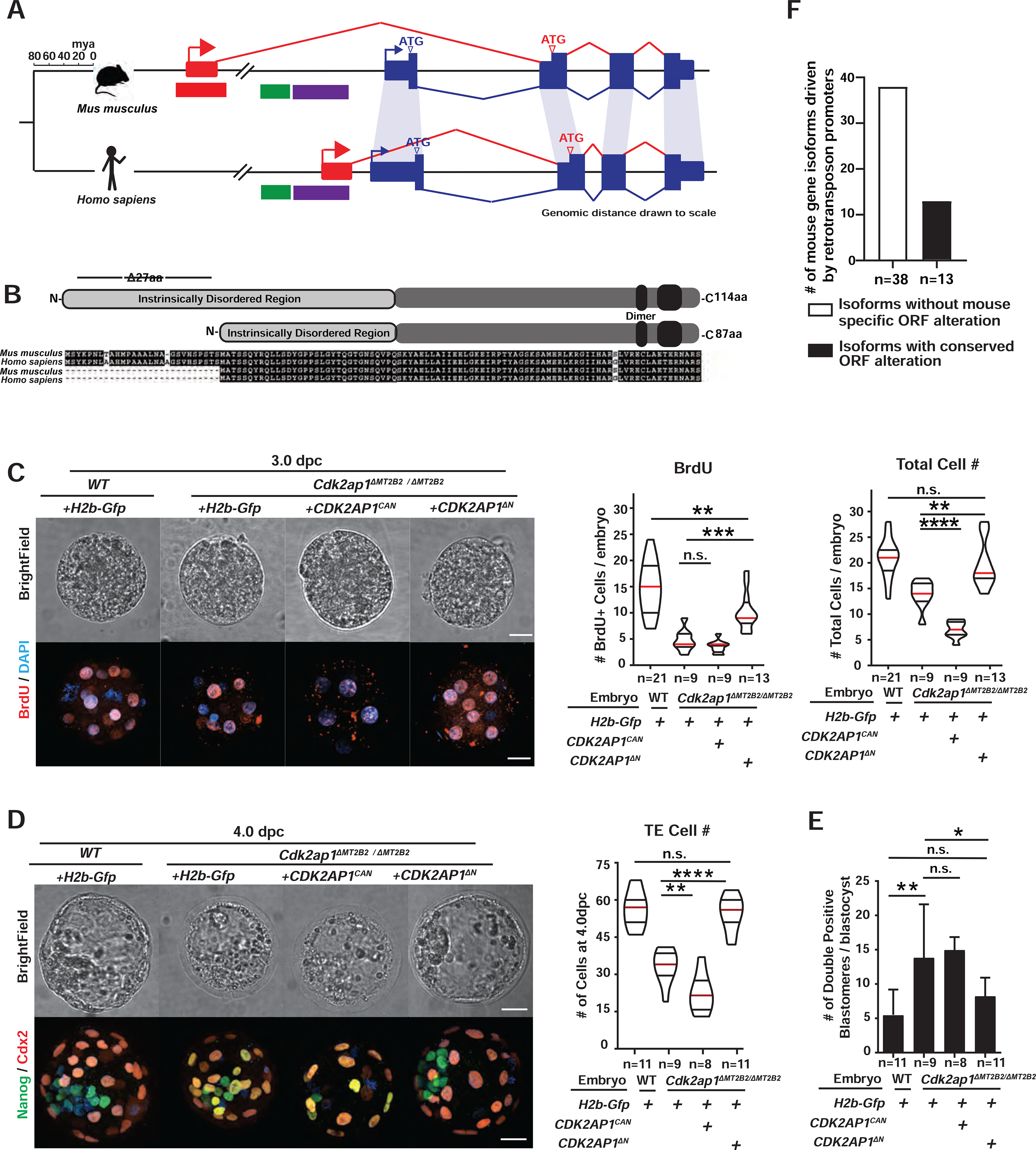
A, B. Preimplantation-specific Cdk2ap1ΔN isoforms are derived from species-specific promoters (A), but exhibit evolutionary conservation in protein sequences (B). A. Mouse Cdk2ap1ΔN originates from the MT2B2 promoter; human CDK2AP1ΔN originates from a promoter region containing an L2a and a Charlie4z hAT transposon element. Blue, canonical exons; red, alternative exons. B. Canonical Cdk2ap1 and Cdk2ap1ΔN isoforms are 97.4% and 98.8% identical, respectively, between mouse and human. C, D. Ectopic expression of CDK2AP1ΔN, but not CDK2AP1CAN, rescues defective cell proliferation in Cdk2ap1ΔMT2B2/ΔMT2B2 morulae (C) and blastocysts (D), as demonstrated by BrdU incorporation and total cell number. C. Representative confocal images of BrdU staining (left), quantification of BrdU incorporation (middle) and total cell number (right) are shown for 3.0 dpc embryos. D. Representative confocal images of Nanog and Cdx2 staining (left) and quantification of TE cell numbers (right) are shown for 4.0 dpc embryos. Scale bars, 20 μm. Quantitation is shown as violin plots with median (red), lower (25%) and upper (75%) quartiles (black). C. WT H2b-Gfp vs CDK2AP1ΔN (BrdU), ** P =0.0029, t=3.2, df=32; H2b-Gfp vs CDK2AP1ΔN (BrdU), *** P =0.0005, t=4.1, df=20; H2b-Gfp vs CDK2AP1CAN (total cell number), ****P < 0.0001, t=8.4, df=16; H2b-Gfp vs CDK2AP1ΔN (total cell number), **P = 0.0031, t=3.4, df=20. D. Cdk2ap1ΔMT2B2/ΔMT2B2 H2b-Gfp vs CDK2AP1ΔN (TE Cell number), ****P < 0.0001, t=6.9, df=18; Cdk2ap1ΔMT2B2/ΔMT2B2 H2b-Gfp vs CDK2AP1CAN (TE Cell number), **P = 0.007, t=3.1, df=15. E. Quantitation of Nanog and Cdx2 double positive cells in Cdk2ap1ΔMT2B2/ΔMT2B2 embryos overexpressing CDK2AP1ΔN or CDK2AP1CAN. H2b-Gfp-overexpressing wildtype vs. Cdk2ap1ΔMT2B2/ΔMT2B2 embryos, ** P = 0.007, t=3.1, df=17; H2b-Gfp vs CDK2AP1ΔN overexpression in Cdk2ap1ΔMT2B2/ΔMT2B2) embryos, * P =0.04, t=2.3, df=18. F. A subset of mouse-specific retrotransposon promoters drive gene isoforms harboring the evolutionarily conserved, N-terminal ORF alterations. Manual curation of the top 88 highly and differentially expressed mouse retrotransposon promoters reveals 51 that yield gene isoforms with altered ORFs. Among these, 13 (26%) correspond to Refseq annotated human isoforms that encode the same ORF alternation. See also Figure S5 and Table S5.
Cdk2ap1 is not an isolated case of species-specific retrotransposon promoters yielding evolutionarily conserved, N-terminally altered protein isoforms. Among the top 88 most highly and differentially expressed mouse retrotransposon promoters, 51 yield alternative gene isoforms with predicted alteration of the ORF (Figure 1F). Among these, 25% have RefSeq/Ensemble annotated human gene isoforms that carry a similar N-terminal ORF alteration (Figures 5F, S5A–S5C, Table S5). Interestingly, mouse and human often employ different mechanisms to generate alternative gene isoforms with a conserved ORF. This conservation spans ~85 million years of human-mouse divergence, indicating an evolutionary preservation of functionally important, alternative gene isoforms (Figures S5A–S5C). Hence, the intricate interaction between retrotransposon promoters and host genome may contribute to species-specific gene regulation of evolutionarily conserved gene isoforms, generating distinct expression patterns, important developmental functions and diverse phenotype among species.
Transposon-derived promoters yield species-specific Cdk2ap1ΔN expression in mammals
The canonical Cdk2ap1 proteins are highly conserved in sequences across mammals (Figure 6A). The predicted Cdk2ap1 ORFs from mouse, human, rhesus monkey, marmoset, cow, goat, pig and opossum genomes exhibit 86.1% sequence identity, all utilizing a conserved ATG start codon within exon 1 (Figure 6B). Although the MT2B2 promoter only exists in mice (Figure 6B), all examined mammalian species, with the exception of opossum, have annotated, species-specific gene isoforms that encode a conserved Cdk2ap1ΔN protein (Figure 6B). Annotated Cdk2ap1ΔN proteins in mammals are generated by isoforms driven by species-specific promoters; they all utilize the conserved ATG start codon within exon 2 and harbor an N-terminal truncation of 26–27 amino acids (Figures 6A, 6B).
Figure 6. Transposon promoters yield species-specific expression of evolutionarily conserved Cdk2ap1ΔN isoform.
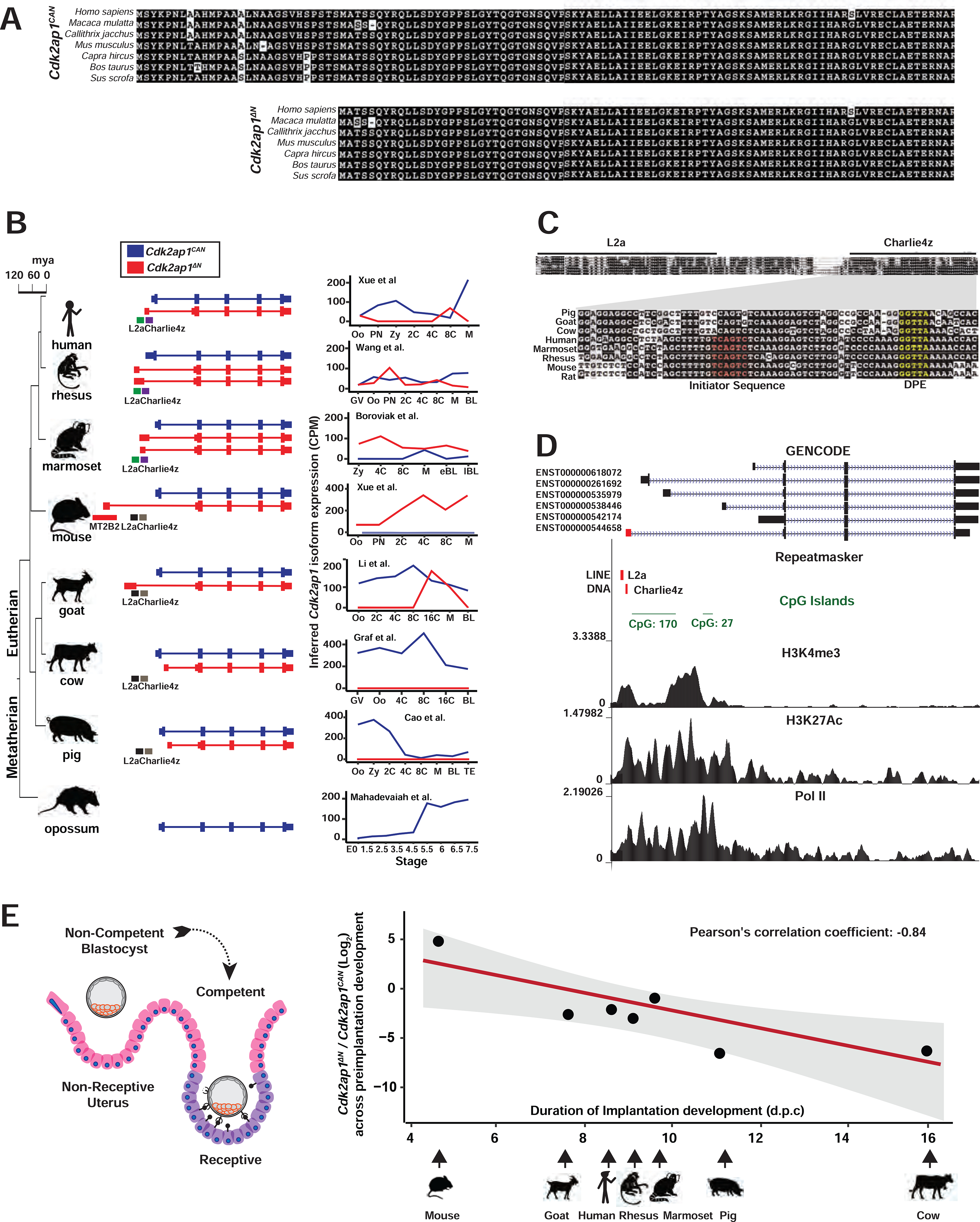
A. Alignment of Cdk2ap1CAN and Cdk2ap1ΔN isoforms across 8 mammals reveals strong evolutionary conservation in their protein sequences. B. Canonical Cdk2ap1 and Cdk2ap1ΔN exhibit species-specific differential expression in mammalian preimplantation embryos. Isoform specific expression of Cdk2ap1 in each species was determined by the total Cdk2ap1 expression and the ratio between isoform specific splicing junctions. C. In 8 mammals examined, the genomic regions containing the L2a/Charlie4z elements exhibit sequence conservation. The region between L2a and Charlie4z is the least conserved, with goat, pig and cattle harboring a small deletion, and rodents and primates exhibiting sequence variance. The Charlie4z element contains a predicted initiator sequence (red) and a DPE (Downstream Promoter Element, yellow), both implicating promoter functionality. D. The L2a/Charlie4z region acts as a bona fide CDK2AP1 promoter in human ESCs (Encode Consortium, 2012). Signatures of an active promoter (H3K4me3, H3K27Ac, and Pol II) in human ESCs are illustrated with ChIP-seq data from ENCODE and Roadmap Epigenomics project. E. The Cdk2ap1ΔN to Cdk2ap1CAN ratio is inversely correlated with the duration of preimplantation development in multiple mammals. The log2 ratio of Cdk2ap1ΔN to Cdk2ap1CAN, calculated based on the sum of normalized RNA-seq reads across isoform-specific junctions during preimplantation stages, is plotted against the duration of preimplantation development for each species. Pearson’s correlation coefficient between log2 (Cdk2ap1ΔN/Cdk2ap1CAN) and duration of preimplantation development equals to −0.84, ** P = 0.018, t = −3.5, df = 5; the P value was calculated as part of the Pearson’s product-moment correlation. See also Figure S6 and Tables S6 and S7.
In human preimplantation embryos, the predominant CDK2AP1ΔN isoform is driven by a putative promoter that contains an annotated L2a retrotransposon and a Charlie4z DNA transposon (Figure 6B). In human, rhesus monkey, marmoset and mouse genomes, the L2a/Charlie4z region is highly conserved (Figure 6C), containing predicted core promoter motifs, including an initiator motif near the TSS and a downstream DPE motif (downstream promoter element) (Burke and Kadonaga, 1997; Lo and Smale, 1996). Published ChIP-seq data in human ESCs support bona fide promoter activity at the L2a/Charlie4z region, as it exhibited enrichment for H3K4Me3 (Davis et al., 2018), H3K27Ac (Ernst et al., 2011) and RNA polymerase II association (Song et al., 2011) (Figures 6D). Consistently, published RNA-seq data support the transcription of the CDK2AP1ΔN isoform from the L2a/Charlie4z promoter region in human ESCs (Encode Consortium, 2012) (Figure S6A). Hence, the L2a/Charlie4z region likely possesses promoter activity to drive CDK2AP1ΔN in multiple species.
The Cdk2ap1ΔN isoform exhibits species-specific expression profiles in mammalian preimplantation embryos (Figure 6B). Mouse preimplantation embryos are characterized by the predominant and strong expression of Cdk2ap1ΔN (Figures 6B, 6E and S6B). Conversely, human, rhesus monkey, marmoset and goat preimplantation embryos express both Cdk2ap1CAN and Cdk2ap1ΔN isoforms, with Cdk2ap1ΔN peaking at different developmental stages in different species (Figures 6B, 6E and S6B). Pig and cow only express the canonical Cdk2ap1 in preimplantation embryos, with no detectable Cdk2ap1ΔN expression (Figure 6B, Table S6). Yet their genomes contain RefSeq annotated, alternative Cdk2ap1 isoforms predicted to encode Cdk2ap1ΔN, possibly in other tissue types. Opossum has no annotated Cdk2ap1ΔN isoform. Cdk2ap1ΔN regulation is likely achieved by species-specific promoter activity. The L2a/Charlie4z region is present in all 7 eutherian mammals examined (Figure 6B). In mouse, a strong, retrotransposon derived MT2B2 promoter drives the potent induction of Cdk2ap1ΔN, making it the predominant Cdk2ap1 isoform in preimplantation embryos; in human, and possibly other primates, the putative L2a/Charlie4z promoter drives a modest Cdk2ap1ΔN induction, which co-exists with Cdk2ap1CAN in preimplantation embryos; in pig and cow, the L2a/Charlie4z region lacks promoter activity, and the Cdk2ap1ΔN isoforms are likely produced from a different promoter that is inactive in preimplantation embryos (Figures 6B, 6C).
Rodents, cows, pig, goats, and primates exhibit considerable phenotypical differences in the duration of preimplantation development, with 4.5 days for mouse and ≥10 days for cow and pigs (Figures 6E, S6B, Table S7). Mammalian blastocysts consist of 100–200 cells, and their competency for implantation roughly correlates with the absolute number of blastomeres during uterine apposition (Kong et al., 2016), thus, cell proliferation rate in preimplantation development. Intriguingly, the ratio of Cdk2ap1ΔN to Cdk2ap1CAN in preimplantation embryos is inversely correlated with the duration of preimplantation development across all 7 eutherian mammals examined (Figure 6E). Given the importance of Cdk2ap1ΔN and Cdk2ap1CAN in promoting and repressing cell proliferation, respectively, we speculate that a high abundance of Cdk2ap1ΔN in mice could serve to promote cell proliferation to reach competency for implantation sooner, and that a low abundance of Cdk2ap1ΔN in pig and cow could serve to slow down cell proliferation to prolong preimplantation development. Altogether, a retrotransposon promoter can yield species-specific gene regulation of an alternative gene isoform, ultimately generating phenotypical diversity among species.
Discussion
Colonization of transposons pose considerable threats to genome integrity (Ardeljan et al., 2017; Beck et al., 2011), due to an increased risk of insertional mutagenesis (Gagnier et al., 2019; Kazazian et al., 1988), non-homologous recombination (Hancks and Kazazian, 2016), and genome instability (Ayarpadikannan and Kim, 2014; Maxwell et al., 2011). Yet a subset of transposons also provide abundant genetic material for gene regulatory sequences, substantially increasing the complexity of species-specific gene regulation (Cosby et al., 2019; Sundaram et al., 2014).
To date, the best characterized retrotransposon promoters are those that drive species-specific gene isoforms with a non-essential function (Ding et al., 2016; Flemr et al., 2013). For instance, a mouse intronic MTC promoter drives an N-terminally truncated, oocyte specific DicerO isoform, whose enhanced Dicer activity safeguard meiotic spindle formation in mice (Flemr et al., 2013). However, the MT2B2 promoter for Cdk2ap1ΔN is, surprisingly, essential. It is unclear when the MT2B2 element became essential during the evolutionary history of mouse. We favor the hypothesis that MT2B2 was not essential immediately upon its integration, yet its strong induction of Cdk2ap1ΔN may trigger additional events that render the MT2B2 element indispensable for preimplantation development. Our findings suggest that transposons can orchestrate species-specific gene expression and developmental functions and may eventually evolve to be essential (Figure 7).
Figure 7.
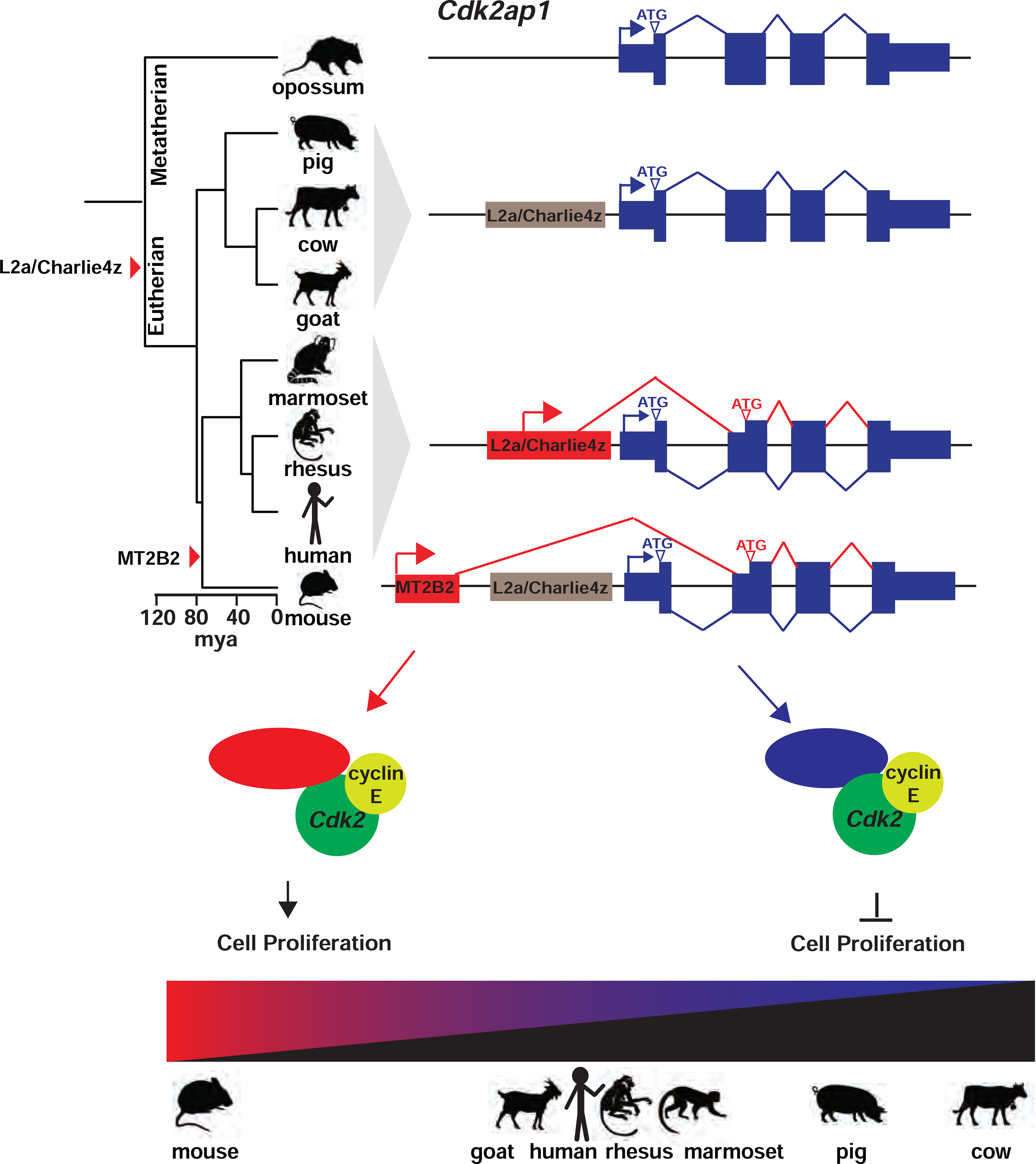
A model on the transposon-dependent gene regulation of Cdk2ap1 in mammalian preimplantation embryos.
In mouse embryos, the MT2B2 and the canonical Cdk2ap1 promoters yield two isoforms with distinct expression regulation and opposite biological functions. The alternative Cdk2ap1ΔN isoform is conserved in sequence and function across mammals, yet its gene regulation is divergent. The strong Cdk2ap1ΔN induction in mouse preimplantation embryos is driven the by MT2B2 promoter; the modest Cdk2ap1ΔN induction in human, and possibly rhesus monkey, marmoset and goat preimplantation embryos, is driven by a promoter containing an ancient L2a and Charlie4z integration; the lack of Cdk2ap1ΔN induction in pig and cow preimplantation embryos suggest the loss of promoter activity in the L2a/Charlize4 element. This leads to a diverse spectrum of Cdk2ap1ΔN abundance that is inversely correlated with duration of preimplantation development in each examined species (Figure 6E).
Transposon-derived sequences constitute an important mechanism for species-specific regulation on gene structure and gene expression. In some scenarios, transposon promoters generate a species-specific gene isoform with an alternative protein function for a unique biology of that species. In mouse oocytes, an MTC promoter drives an N-terminally truncated Dicer isoform to enhance the RNAi mechanism to safeguard meiotic spindle formation. In other scenarios, transposon-dependent gene regulation can generate evolutionarily conserved gene isoforms with species-specific expression patterns (Figure 6B). In the case of Cdk2ap1, the MT2B2 promoter in mouse, the L2a/Charlie4z promoters in primate, and the pig or cow specific promoters all drive Cdk2ap1 isoform transcripts that are alternatively spliced into exon 2, which employ the alternative ATG start codon within exon 2 to initiate translation. Hence, divergent promoters among different species yield an evolutionarily conserved, N-terminal truncated Cdk2ap1ΔN isoform, bestowing them with species-specific transcriptional and translational regulation via different promoter activity and 5’UTR sequences. Taken together, retrotransposons are important building blocks for evolutionary “tinkering”, promoting species-specific gene innovation and gene regulation, and possessing the capacity to generate either species-specific or evolutionarily conserved protein isoforms. Retrotransposon mediated gene regulation could contribute to species-specific gene regulation, and ultimately, phenotypical variance among species.
Limitations of the study
There are several limitations in our studies. First, retrotransposon expression and retrotransposon:gene splicing could be underestimated due to the short-read RNA-seq data and the repetitive nature of retrotransposons, particularly in the datasets with a strong 3’ signal bias. Second, reconstructing full-length transcripts using short-read RNA-seq data is challenging, hence ORF alterations in retrotransposon:gene isoforms were predicted using local sequence information. Third, our RNA-seq analyses across different mammals were performed using datasets generated by independent studies. While we included multiple dataset for each species whenever possible, we cannot rule out the possibility that our results are influenced by batch effects due to sample collection, library construction and sequencing. Fourth, the relative abundance of Cdk2ap1ΔN, inferred from the RNA-seq data, inversely correlates with the duration of preimplantation development in different mammals, yet experimental validation of this finding was not performed due to the difficulty to obtain the biological samples. Fifth, we characterized the opposing functions of Cdk2ap1 and Cdk2ap1ΔN, yet the function of the truncated N-terminal 27 amino acids remains unclear. Finally, we observed incomplete penetrance for the lethality phenotype of Cdk2ap1ΔMT2B2/ΔMT2B2 mice, and the underlying mechanism remains elusive. There could be compensatory production of Cdk2ap1ΔN via an alternative promoter.
STAR METHODS:
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Lin He (lhe@berkeley.edu)
Materials Availability
Both the Cdk2ap1ΔMT2B2/ΔMT2B2 and Cdk2ap1ΔCAN/ΔCAN mouse lines generated in this study will be deposited to Jackson Labs. Plasmids will be made available and deposited to AddGene.
Data and code availability
Our studies have employed published RNA-seq data, which are available from the Gene Expression Omnibus, ArrayExpress, or Short Read Archive, at accessions GSE44183, GSE45719, GSE36552, E-MTAB-7078, GSE86938, GSE143850, GSE25415, GSE129742, SRA076823, GSE139512, E-MTAB-7515, GSM733657, GSM646336, GSM748532, GSE23316.
Customized scripts used for quantifying retrotransposon:gene junction reads is publicly available as https://epigenome.wustl.edu/TE_Transcript_Assembly/tool.html.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental Models and subject details
Animals
Three-to-five week old C57BL/6J female mice and three-to-eight month old C57BL/6J male mice (stock 000664) were purchased from The Jackson Laboratory (Bar Harbor, Maine). Two-to-three month old CD-1 female mice (code 022) were purchased from Charles River (Wilmington, MA). The Cdk2ap1ΔMT2B2 allele was generated by deleting a 1.2kb region surrounding the 0.7kb MT2B2 locus upstream of Cdk2ap1, and the Cdk2ap1ΔCAN allele was generated by deleting the exon1 of Cdk2ap1 (0.7kb). Both knockout mouse lines were generated using CRISPR-EZ, a highly efficient mouse genome engineering technology (described in greater detail below). The Cdk2ap1ΔMT2B2/ΔMT2B2 and Cdk2ap1ΔCAN/ΔCAN mice were generated and maintained on an isogenic C57BL/6J background and housed in a non-barrier animal facility at UC-Berkeley. Wildtype or edited males used for breeding or timed mating are three-to-eight-month-old. Wildtype female mice used for superovulation are three-to-five-week-old. For phenotypical characterization of knockout embryos, we use two-to-four month old edited animals to generate embryos with desired genotype.
All mouse studies have appropriate authorizations acquired from institutional and/or federal regulatory bodies prior to performing these protocols, specifically our animal care and use protocol (AUP-2015–04-7485–1) has been reviewed and approved by our IACUC for this project. All mouse usage including but not limited to housing, breeding, production, sample collection for genotyping, and euthanasia, is in accordance with the Animal Welfare Act, the AVMA Guidelines on Euthanasia and are in compliance with the ILAR Guide for Care and Use of Laboratory Animals, and the UC Berkeley Institutional Animal Care and Use Committee (IACUC) guidelines and policies.
Method Details
RNA-seq data processing
RNA-seq raw sequencing files for mammalian preimplantation embryos were downloaded from NCBI Sequence Read Archive and EMBL-EBI ArrayExpress (Table S1). After trimming off adapter sequences with cutadapt (v. 2.10) (Martin, 2011), RNA-seq reads were mapped to the reference genomes using STAR (v. 2.7.1a) (Dobin et al., 2013). To increase the detection sensitivity of spliced RNA-seq reads, we applied the two-pass alignment strategy (Veeneman et al., 2015). For the first pass alignment, we aligned RNA-seq reads using STAR genome index files generated with the gene annotations provided by RefSeq (Table S1). Subsequently, we collected all the detected splice sites for each mammalian species, and updated the STAR genome index files by incorporating previously unannotated splice sites. To ensure the accuracy of the updated STAR index, we only considered splice sites that were confirmed by at least 3 mapped reads and were characterized by STAR-defined canonical intron motifs. These updated STAR genome index files were then employed for the second round of sequence alignment. To further reduce the number of spurious junctions, we only kept reads containing junctions that were included in the SJ.out.tab files (STAR option: --outFilterType BySJout). All the raw RNA-seq sequencing data used in this study are available from the Gene Expression Omnibus, ArrayExpress, or Short Read Archive, at accessions GSE44183, GSE45719, GSE36552, GSE86938, E-MTAB-7078, SRA076823, GSE139512, GSE143850, GSE52415, GSE129742, E-MTAB-7515.
Annotation of retrotransposon:gene junctions
We first performed transcript assembly using StringTie2 to identify novel exon structures that were absent from Refseq annotation (Kovaka et al., 2019). We then extracted split RNA-seq reads from aligned BAM files and only kept reads that had at least 6 nucleotides mapped to the genome at both ends. Only reads with splicing junctions between 50 and 100,000 bp in length in the genome were retained. A read was considered as a retrotransposon:gene junction read when it fulfilled the following two criteria: 1) both ends of the read were mapped to exons (assembled exons from RNA-seq data or annotated exons from RefSeq); 2) one end of the read was mapped to annotated protein-coding gene exons and the other end was mapped to an annotated retrotransposon. We then counted the number of retrotransposon:gene junction reads for each unique splicing junction. Due to the repetitive nature of retrotransposon sequences, this procedure may not be entirely accurate, especially in the presence of gene families and/or pseudogenes. Hence, only junctions with at least 10 reads in at least one samples were retained for downstream differential expression analysis.
Manual annotation of retrotransposon:gene isoforms
Following the bioinformatic identification of mouse retrotransposon:gene isoforms using published RNA-seq data (Xue et al., 2013) we performed manual annotation on the highest ranked retrotransposon:gene junction reads to predict the structure of the resulted retrotransposon:gene isoforms. Retrotransposon: gene junction reads were filtered for FDR < 0.05 then ranked based on averaged expression value during the developmental stage with peak expression. We predicted the mouse retrotransposon:gene isoforms that likely alter canonical ORFs, and explored if such ORF alternations are conserved in human. The top 100 or top 250 unique retrotransposon:gene junctions were manually curated with the following procedures.
RNA-seq reads across the retrotransposon-regulated genes were visualized using the Integrated Genomics Viewer (IGV, v2.9.4).
- Retrotransposon:gene junction reads were analyzed with regards to their splicing patterns. The position of the retrotransposon element with respect to the predicted retrotransposon:gene isoforms were classified as 5’ (the retrotransposon elements act as putative promoters), internal (the retrotransposon elements contribute to putative internal exons) and 3’ (the retrotransposon elements contribute to putative terminator exons). This classification is based on the following criteria:
- Retrotransposon positions are classified as 5’ when splicing only occurs between the retrotransposon and a downstream canonical gene exon. In most cases, the RNA-seq data support the existence of the transcription start site (TSS) within the retrotransposon element. Yet occasionally (n=5), transcription starts upstream of the annotated retrotransposon element and transcription continues through the retrotransposon and its downstream gene exon. Among the top 250 most highly and differentially expressed retrotransposon:gene junctions, 88 are 5’ retrotransposon promoter cases with evidence of a TSS.
- The retrotransposon positions were classified as “internal” exons. Our manual curation considers two scenarios for retrotransposon-derived internal exons. First, the retrotransposon-derived exon splices into both an upstream and a downstream host gene exon. Second, one splicing event splices into the annotated retrotransposon, and the other splicing event occurs immediately outside the retrotransposon. In our analyses, 65% retrotransposons that contribute to putative internal exons harbor only one splicing event, leaving the other splicing event occurring in its vicinity. In such cases, RNA-seq data support a continued transcription between the splicing site and the retrotransposons.
- The retrotransposon positions were classified as 3’ when splicing occurs exclusively between the retrotransposon and an upstream host gene exon.
-
We then determined if and how the ORFs encoded by the retrotransposon:gene isoforms could be altered compared to the canonical ORFs. This analysis was performed for all retrotransposon promoter cases in the top 250 retrotransposon:gene isoforms (n=88), and for all of the top 100 retrotransposon:gene isoforms. For each gene exon that harbors a splicing event with a proximal retrotransposon, we quantified the retrotransposon:exon splicing reads and all alternative, exon:exon splicing junction reads. All exons of the host genes were defined by RefSeq annotation. (a) In a subset of cases, only one exon:exon splicing event is alternative to the retrotransposon:exon splicing. We employed this exon:exon junction to predict the canonical amino acid and/or UTR sequences encoded by these two exons, and determined if the retrotransposon:gene splicing alter the canonical ORFs. (b) In a subset of cases, no splicing events are alternative to the retrotransposon:exon splicing in preimplantation embryos. In cases where a prominent Ref-seq annotation depicts the retrotransposon-independent gene isoform expressed in other tissues, we will define it as the putative canonical isoform. In cases where the retrotransposons have been exapted in the mouse genome as a bona fide gene exon, we predicted if the retrotransposon element contributed to ORF in this retrotransposon:gene isoform. (c) In a subset of cases, multiple exon:exon splicing events are alternative to the retrotransposon:exon splicing. We then selected the most highly expressed exon:exon splicing junction as the splicing event in the canonical isoform. We predicted if the retrotransposon:exon splicing could alter the amino acid and/or UTR sequences encoded by the two canonical exons.
We observed the following scenarios for the predicted ORFs encoded by the retrotransposon:gene isoforms: a) “Deletion”, the splicing between retrotransposons and gene exons are predicted to truncate N- or C-terminus of the canonical ORFs; b) “Replacement”, the retrotransposon:gene splicing events are predicted to truncate canonical ORFs, while the retrotransposon-derived sequence encode additional amino acids (Note: such retrotransposon:gene isoforms are supported by RefSeq annotation and are listed in Table S5), c) “insertion”, the retrotransposon derived exons are predicted to add additional amino acids to the canonical ORF, supported by RefSeq annotations, d) “Exaption”, the retrotransposons likely represent ancient integrations; they are fixed in the mouse genome and serve as bona fide protein-coding exons. Conserved gene isoforms in human are supported by RefSeq annotations (Table S5); e) “N-Del / N-Rep”, retrotransposon-derived exons are predicted to either cause N-terminal deletions or N-terminal replacements of canonical ORF, due to uncertainty in ATG prediction; f) “Intact ORF”, the retrotransposon:gene splicing events have no predicted impact on the canonical ORFs; g) “N.D.”, the ORFs of a small number of retrotransposon:gene isoforms could not be manually reconstructed due to low sequencing quality. It is important to note that we did not manually annotate the entire retrotransposon:gene isoform transcripts, and the ORF alterations were only predicted based on the exon structure and sequences proximal to the retrotransposon elements.
For the mouse retrotransposon:gene isoforms that exhibited an altered ORF (n=51 for retrotransposon promoter cases in the top 250, n=74 in the top 100), we examined all available Refseq and Ensembl annotated isoforms of the corresponding human genes. We identified annotated human gene isoforms with the identical or nearly identical ORF modifications as those generated by mouse retrotransposon:gene splicing events. It is important to note that our approach is only able to identify local ORF alterations, hence the conserved ORF modification between mouse and human isoforms was tested only locally and may or may not extend to the entirety of the gene isoform.
Cdk2ap1 promoter analysis with public data
The wiggle files for H3K4me3 ChIP-seq data (ENCODE Consortium2012) (GSM733657), H3K27Ac ChIP-seq data (Ernst et al., 2011) (GSM646336), and PolII ChIP-seq data (Song et al., 2011) (GSM748532) were obtained from Cistrome database (Mei et al., 2017) and displayed using UCSC genome browser (Kent et al., 2002). The human H1-ESC RNA-seq data were downloaded from NCBI GEO database (Edgar et al., 2002), GSE23316 (ENCODE Consortium 2012), and Kallisto (Bray et al., 2016) was used to quantify the isoform expression levels with GENCODE annotation (GRCh38 ver. 26) (Frankish et al., 2019). Four RNA-seq replicates with insert length of 200bp were used (Myers_H1-hESC_cell_2×75_200_1 through 4).
Phylogenetic analysis.
Genomic Phylogeny of various placental mammal taxa were generated by first organizing a selection of animal of interest in terms of their binomial nomenclature in Latin. This list is then imputed into TimeTree.org (Kumar et al., 2017), which generates timescales and species divergence nodes as a Newick file. This file is imported and modified using FigTree v1.4.4 for presentation.
Sequence Alignment
Current Sequence alignment was performed using clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) with default parameters. Alignment files were used as input for alignment shading (BoxShade v3.21 https://embnet.vital-it.ch/software/BOX_form.html). We aligned predicted Cdk2ap1 isoform amino acid sequences from each species using the NCBI GenPept entries; we also aligned the genomic sequence of the L2a/Charlie4z regions from each species. Zoomed in alignment for Charlie4z was manually adjusted and annotated for core promoter elements.
Mouse Embryo Isolation and Culture
3 to 5 weekold C57BL/6J female mice (Jackson Laboratory, 000664) were superovulated by intraperitoneal (IP) injection of 5 IU of Pregnant Mare Serum Gonadotropin (PMSG, Calbiochem, 367222), and 46–48 hours later, 5 IU of Human Chorion Gonadotropin (hCG, Calbiochem, 230734). Superovulated females were each housed at a 1:1 ratio with a 3- to 8-month-old C57BL/6J stud male to generate 1-cell zygotes at 0.5 dpc. Using forceps under a stereomicroscope (Nikon SMZ-U), the ampulla of oviduct was nicked, releasing fertilized zygotes associated with surrounding cumulus cells into 50 μl M2 + BSA media (M2 media (Millipore, MR-015-D) supplemented with 4 mg/mL bovine serum albumin (BSA, Sigma, A3311)). Using a handheld pipette set to 50 μl, we dissociate zygotes from cumulus cells, after the cumulus oocyte complexes were incubated in a 200 μl droplet of 1X Hyaluronidase in M2 solution (Millipore, MR-051-F) for 2 min, followed by five washes in the M2+BSA media to remove cumulus cells. From this point on, embryos were manipulated using a mouth-controlled assembly consisting of a glass needle pulled from glass capillary tubes (Sigma, pack of 250: P0674) over an open flame attached to a 15-inch aspirator tube (Sigma, pack of 5, A5177). Detailed instructions described previously (Modzelewski et al., 2018). Embryos were then transferred to KSOM + BSA media (KCl-enriched simplex optimization medium with amino acid supplement (Zenith Biotech, ZEKS-050), supplemented with 1 mg/ml BSA), which was equilibrated in an incubator to final embryo culture condition at least 3–4 hours prior to incubation to reach optimal temperature, CO2 and pH conditions. Embryos were cultured in 30 μl droplets of KSOM + BSA, overlaid with mineral oil (Millipore, ES-005-C) in 35 × 10 mm culture dishes (CellStar Greiner Bio-One, 627160) in a water-jacketed CO2 incubator under hypoxic conditions (5% O2, 5% CO2, 37 °C and 95% humidity).
Single-Embryo Quantitative RT-PCR
All single-embryo cDNA was prepared using a modified protocol of the Single Cell-to-Ct qRT-PCR kit (Life-Technologies, 4458236). Whole embryos were isolated at a desired developmental stage, and passed through three PBS washes. With a hand-held pipette set to 1 μL, a single embryo was collected in PBS and transferred to one tube of an 8 well PCR strip, and the successful transfer of each embryo was visually confirmed under microscope. To account for the larger volume of an embryo compared to a somatic cell, we modified the manufacturer’s protocol slightly, briefly: we incubated each embryo in 20 μl “Lysis/DNAse” reagent at room temperature (25°C) for 15 minutes, then added 2 μl of “Stop Solution” for a 2 min incubation at room temperature. Half reaction was stored at −80°C as a technical replicate, and the remaining sample (11 μl) continued through the Single Cell-to-Ct protocol per manufacturer’s recommendation. For each experiment, a single embryo was collected and reserved as a “-RT” control, 1 μl of PBS was collected as a “No Template Control”. All qRT-PCR analyses were performed on the StepOnePlus Real Time PCR system (Thermo, 437660). All real-time qPCR analyses were performed using SYBR FAST qPCR Master Mix (Kapa Biosystems, KK4604) following manufacturer’s protocol. Real time PCR analyses on retrotransposons detect their expression at the subfamily level, using primers designed from the retrotransposon consensus sequences. To detect retrotransposon gene isoform expression, primers were designed against the predicted isoform and to span the unique retrotransposon:gene splicing junctions, with one primer located within the retrotransposon sequence and the other located within the proximal gene exon. Rfx1 was used as a reference for both mRNA and retrotransposon quantitation in real time PCR analyses using preimplantation embryos. All real time PCR primers used in our studies are listed in Table S8.
Validation of Retrotransposon Gene Junction
Upon completion of qRT-PCR analysis, the amplification samples were mixed at a 1-to-1 ratio with non-processive TAQ-polymerase supplied as a 2x Master Mix (Promega, M7123) and incubated at 72°C for 10 min in order to append a single deoxyadenosine to the 3’ ends of the amplicon. The amplified fragments that captured retrotransposon:gene junction reads were purified through gel extraction (BioBasic, BS654) before TA cloned into pGEM-T easy vector (Promega, A1360). The plasmids were sequenced by Sanger Sequencing at the UC Berkeley DNA Sequencing Facility, and the retrotransposon:gene junctions were analyzed and visualized using SnapGene (version 2.3.2).
Rapid Extension of cDNA Ends (RACE)
All RACE experiments were conducted following manufacturer’s instructions (Clontech, 634858) with the following modifications. Input RNA was provided by pooling approximately 50 morula stages mouse embryos followed by trizol RNA extraction per manufacturer’s instruction (Life Technologies, 15596). A list of primers used in this experiment is listed in Table S8.
Luciferase Assay for translation efficiency
To analyze the impact of retrotransposon-derived 5’UTR on translation efficiency, we constructed luciferase reporters for translational assay using psiCheck2 luciferase reporter vector (Promega, C8021). The 5’UTRs of mouse canonical Cdk2ap1 and Cdk2ap1ΔΝ isoforms were cloned immediately upstream of the Renilla Luciferase ORF; the T7-promoter-FireFly luciferase reporter cassette from the siCheck2 vector was cloned as a control. All reporters were in vitro transcribed, 5’ capped and polyadenylated (HiScribe, NEB, e2060s). Renilla Luciferase and FireFly luciferase reporter mRNAs were co-transfected into HEK293T cells (600 ng Renilla Luciferase mRNA and 2200 ng FireFly luciferase mRNA per well of a 12-well plate), using Lipofectamine 2000 (Life Technologies, 11668027). Approximately 8 hours later, samples were assayed for luciferase activity by Dual-Luciferase® Reporter Assay System (Promega, E1910) as per manufacturer’s instructions using a Glomax 20/20 Luminometer (Promega).
Mouse genome engineering by CRISPR-EZ
Embryos were edited following the published CRISPR-EZ protocol (Chen et al., 2016, 2019; Modzelewski et al., 2018). Briefly, super ovulated C57BL/6J female mice were used to generate pronuclear stage embryos. Pronuclear stage embryos were dissociated from cumulus cells using Hyaluronidase (Millipore, MR-051-F), the zona was weakened with acid Tyrode’s solution (Sigma, T1788), and the embryos were subsequently washed in M2 buffer. For the MT2B2 deletion or the Cdk2ap1 exon 1 deletion, Cas9/sgRNA RNP complexes were assembled in vitro in a total of 10 μL by combining Cas9 protein (8 μM final concentration, MacroLab QB3, Berkeley CA) with two sgRNAs (2 μg per sgRNA) flanking the desired deletion. Assembled RNPs were then mixed with 50–75 zygotes in 10 μL OptiMEM media (Thermo, 31985062), and this 20 μL mixture was subjected to electroporation for Cas9/sgRNA RNP delivery (BioRad Genepulser XL, 1652660). Electroporation conditions were 30V, 6 Pulses, 3ms pulse length and 100ms Pulse interval. Electroporated embryos were immediately transferred into the oviduct of pseudo-pregnant CD-1 recipient females to generate genetically engineered mice. The Cdk2ap1ΔMT2B2/ΔMT2B2 and Cdk2ap1ΔCAN/ΔCAN mice were generated and maintained on an isogenic C57BL/6J background and housed in a non-barrier animal facility at UC-Berkeley.
For endogenous V5 tagging to specific Cdk2ap1 isoforms, a synthesized single stranded DNA donor oligo (IDT) for Homology Directed Repair (HDR) was added to the Cas9/sgRNA RNP Complex mixture at a final concentration of 20 μM in our CRISPR-EZ experiments (Chen et al., 2016; Modzelewski et al., 2018). Electroporated embryos were then cultured to appropriate developmental stages, fixed and processed for immunofluorescence staining using anti-V5 antibody (Gift from Dr. Robert Tjian and see below).
Correctly engineered mouse embryos or adult mice were confirmed by genotyping analyses. To extract DNA from embryos, embryos were washed twice with PBS, and 1 μl of PBS solution containing a single embryo was transferred into 10 μL of embryo lysis buffer containing 50 mM KCl (Fisher, catalog no. P217–3), 10 mM Tris-HCl, pH 8.5 (Fisher, BP1531), 2.5 mM MgCl2 (Fisher, M33–500), 0.1 mg/ml gelatin (Fisher, G7–500), 0.45% Nonidet P-40 (Fluka, 74385), 0.45% Tween 20 (Sigma, P7949–500), and 0.2 mg/ml proteinase K (Fisher, BP1700–100)). Lysis was performed in a thermocycler with the following conditions: 55 °C for 4 h, 95 °C for 10 min, and 10 °C hold. Due to the low success rate of embryo genotyping, 3–4 μl of the 11 μl of lysed material were used directly in a standard PCR reaction for genotyping, to allow for multiple attempts. To extract DNA from mouse tails, we used a standard Proteinase K extraction protocol. All genotyping primers are listed in Table S8.
mRNA Electroporation into mouse zygotes
Conditions for mRNA electroporation were identical to the parameters described by the CRISPR-EZ protocol (Chen et al., 2016; Modzelewski et al., 2018), except that mRNA was electroporated in place of RNP complexes. Prior to electroporation, H2b-Gfp control and Cdk2ap1 mRNAs were prepared by in vitro transcription (IVT) using the Hiscribe T7 ARCA w/Tailing kit, following manufacturer’s instructions (NEB, e2060). For each electroporation, 200 ng of control H2b-Gfp and 2000ng of experimental mRNA was mixed with 20 μl of Opti-MEM and combined with 25–75 mouse zygotes. Following electroporation, embryos were recovered and washed with M2 media, and cultured under mineral oil in KSOM+BSA until the appropriate developmental stage for subsequent analyses. A list of IVT templates and primers was summarized in Table S8.
Preimplantation embryos immunofluorescence
Embryos were fixed in 4% paraformaldehyde (Electron Microscopy Sciences, 19202) for 15 min at room temperature, and then transferred to wash buffer (PBS containing 0.1% bovine serum albumin, Sigma, A3311). Embryos were permeabilized with PBS containing 0.1% Triton X-100 and 0.1% BSA for 5 min at room temperature, blocked in blocking solution for 1 hour at room temperature in PBS containing 10% goat serum (Fisher 31872) and 0.1% BSA, then incubated with appropriate primary antibody in blocking solution at 4 °C overnight. The primary antibodies include antibodies against Cdx2 (1:100, Abcam, ab157524), Nanog (1:100, CosmoBio, REC-RCAB0002PF), Cdk2ap1 (1:50, Santa Cruz sc-390283), V5 (1:100, a gift from the Tjian Lab), BrdU (1:100, Thermo Fisher, 17–5071-41). On the following day, embryos were washed (PBS containing 0.1% bovine serum albumin, Sigma, A3311) twice before being incubated with appropriate secondary antibodies diluted in blocking solution at 4 °C overnight. The secondary antibodies used in our studies include goat anti-mouse IgG Alexa Fluor 594 (1:400, ThermoFisher, A11005), goat anti-rabbit IgG Alexa Fluor 594 (1:400, Thermo Fisher, A11037), goat anti-mouse IgG Alexa Fluor 488 (1:400, Thermo Fisher, A11001) and goat anti-rabbit IgG Alexa Fluor 488 (1:400, Thermo Fisher, A11034). Finally, embryos were stained with (4′,6-diamidino-2-phenylindole) (DAPI at 300 nM in PBS, Sigma, D9564) and subjected to imaging analyses using spinning disk scanning confocal microscopy (Nikon Eclipse TE200-E). Raw images were processed using ImageJ (Schneider et al., 2012). In order to match embryo genotypes to immunofluorescent images, after imaging, embryos were collected in the order they were imaged, lysed with temperature induced reverse-crosslinking and subjected to PCR based genotyping analysis. Lysis was performed in a thermocycler with the following conditions: 55 °C for 4 h, 95 °C for 10 min, and 10 °C hold.
Phenotypical analyses of embryo implantation
Uteri were collected at specific developmental stages after timed mating to analyze embryo implantation. Collected uterus was cleared of attached fat tissue, photographed with ruler, and placed in 10% PFA overnight at room temperature for standard paraffin embedding and tissue processing. After overnight RT incubation of uterus in 10% PFA, uterus was washed three times using PBS before long term storage in 4°C 70% EtOH, for up to 6 months. For embedding, uterine tissue was dehydrated by sequential exchange in higher concentration of EtOH and then clarified in 50% Histoclear-EtOH solution and 100% Histoclear (National Diagnostics, HS-200). Clarified tissue was embedded in paraffin (Fisher Histoplast, 22900700) by placing in 50% paraffin-Histoclear solution and then 100% paraffin in an embedding machine. Before final embedding, uterus was cut into 4–5mm segments, each with one embryo implantation site, were place near one another to maximize incidence of embryo capture on each section. Uterine segments were imbedded parallel to each other in paraffin. All uterine segments with implanted embryos were sectioned on microtome transversely into 5μm sections and transferred onto positively charged glass slides (Superfrost plus, FisherScientific, Cat# 22–037-246). Paraffin sections were deparaffinized, dehydrated, and subjected to 15 minutes of heat-induced antigen retrieval in a pressure cooker using antigen retrieval solution (10mM Sodium Citrate buffered to pH=6). Slides were blocked for 3 hours with PBS containing 5% BSA and 0.3% Triton X-100 and incubated with primary antibodies against SOX2 (1:200, Santa-Cruz, SC-365823), WNT5A (1:200, Santa-Cruz SC-365370). Indirect Immunofluorescence was performed using Alexa Fluor 488 Goat-anti-Rabbit IgG (1:400, Thermo A110034) and Alexa Fluor 594 Goat-anti-Mouse IgG (1:400, Thermo A11005). After applying secondary antibody, autofluorescence of red blood cells were reduced by incubating processed slides for 10 minutes at room temperature in quenching buffer (10mM CuSO4, 50mM NH4Cl). Finally, embryos were stained with (4′,6-diamidino-2-phenylindole) (DAPI at 300 nM in PBS, Sigma, D9564). Negative controls without primary antibody were processed at the same time.
BrdU incorporation in preimplantation embryos
Morulae and blastocysts were processed for BrdU incorporation analysis as previously described (Stuckey et al., 2011). Briefly, embryos were cultured for 1 hour in 20 μl droplet of KSOM + BSA media supplemented with 25 μM BrdU (BD Pharmingen, 51–2420KC) under mineral oil. Embryos were then washed three times in wash buffer (PBS containing 0.1% BSA, Sigma, A3311), then fixed in 4% paraformaldehyde for 10 min at room temperature. Embryos were washed again three times in wash buffer. Embryo permeabilization and DNA denaturation was performed simultaneously by incubating the embryos in 2M HCl/0.5% Triton-X100 in PBS for 20min (Triton X-100, Sigma, X100, HCl, Macron 2062–46). Embryos were washed again three times and placed in blocking solution (PBS containing 10% goat serum and 0.1% BSA) for 1 hour at room temperature. Embryos were incubated overnight at 4°C with anti-BrdU antibody in blocking buffer (1:100, Thermo, 17–5071-41), then processed for confocal imaging, as described in the previous section.
Phenotypical analyses of embryo implantation
For both Cdk2ap1ΔMT2B2/ΔMT2B2 and Cdk2ap1ΔCAN/ΔCAN deletion strains, uteri from littermate WTxWT and heterozygous crosses (Cdk2ap1ΔMT2B2/+ x Cdk2ap1ΔMT2B2/+), and littermate WTxWT and heterozygous (Cdk2ap1ΔCAN/+ x Cdk2ap1ΔCAN/+) were collected from female mice at specific developmental stages for embryo implantation analyses. Implantation was considered abnormal if sites were spaced either shorter or further than the expected normalized inter-embryo distance. Collected uterus was cleared of attached fat tissue and photographed next to ruler for scaling and measurement purposes. Embryos were then surgically removed, small tail segment collected, washed twice in PBS and collected for PCR based genotyping analysis, as previously described above.
Co-immunoprecipitation
Transfection of HEK293T cells with MSCV retroviral vectors (PGK driven Puro IRES GFP C-Terminal HA Tag as control vector, pMSCV-Cdk2ap1ΔN (MT2B2)-HA or pMSCV-Cdk2ap1CAN-HA) was performed by standard polyethylenimine (PEI) transfection (Polysciences, 23966–1). 10 μg of DNA were used for each 10cm dishes of HEK293T cells, where the ratio of DNA to PEI is 1:20. Transfected cells were collected at 48 hours, washed with ice-cold PBS and lysed in plate by adding 1ml ice cold lysis buffer (10mM Tris/HCl PH=7.5, 150mM NaCl, 0.5mM EDTA, 0.5% NP40,1uM PMSF). Cell lysate was transferred to individual tubes and homogenized on ice by passing through a 21-Gauge needle 10 times. Cleared cell lysate was transferred to a new tube after centrifugation at 10,000rpm at 4°C for 10min. An aliquot of 50 μl was set aside as “input”. The remaining lysate was incubated with 20 μl of anti-HA Affinity Gel (Sigma, EZview Red Anti-HA Affinity Gel, E6779) with rotation for 1 hour to overnight at 4°C. Samples were centrifuged at 10,000rpm at 4°C for 1min, and 50 μl of the supernatant was collected as a control sample for “depleted supernatant”. The pulled down pellet was washed with 750uL of lysis buffer for 3 times. Finally, loading buffer (2x Laemmli: 4% SDS, 20% glycerol, 120mM Tris-HCl, pH=6.8, 0.02% w/v bromophenol blue) was added to all samples, heated to 95°C for 10 minutes, and the pull-down samples was flash cooled on ice before western analyses. For each experiment, 0.5% of input (mentioned above), 0.5% of depleted supernatant, and 20% pulldown samples from the Co-IP experiment were loaded into 15% SDS-polyacrylamide gel and transferred onto a 0.45 μm nitrocellulose membrane (GE, 10600016). Blots were incubated with either rabbit-anti-Flag antibody (1:10,000, Cell Signaling Technologies, 2368S) or rabbit-anti-HA antibody (1:10,000, Cell Signaling Technologies, 3724), for 1 hour at room temperature, and then in HRP conjugated goat-anti-rabbit antibody (1:5,000, Santa Cruz, SC-2004), and immune detection was performed using Millipore chemiluminescent HRP substrate (Millipore, #WBKLS0100). Imaging was performed using XRS+ ChemiDoc imaging system (BioRad, 1708265).
Purification of Recombinant Cdk2ap1 Protein
To disrupt binding to CDK2, the previously described Cdk2 binding TER motif was mutated, Thr108Ala, Glu109Ala, Arg110Ala (Referred to as “MutTER” from here on). ORFs of mouse Cdk2ap1CAN, Cdk2ap1CAN-MutTER, Cdk2ap1ΔN (MT2B2) and Cdk2ap1ΔN (MT2B2)-MutTER were each cloned into the pET28a bacterial expression vector (EMD Biosciences, 69864). The vector backbone was modified so that the cloned ORF would be downstream of an N-terminal cassette, consisting of a His-Tag (6x), a Maltose Binding Protein (MBP), a short linker and a TEV cleavage site. Proteins were purified as previously described (Werner et al., 2018). Briefly, plasmids were transformed into E.coli LOBSTR expression cells (Kerafast, EC1002). Starter culture of 200 mL of LB liquid broth was grown at 30°C in the presence of Ampicillin (Vector resistance) and Chloramphenicol (LOBSTR Cell resistance) overnight. The following day, the culture was added to a pre-warmed (37°C) glassware containing 1.3L of LB growth media. When an optical density of 0.5 at 600nm was reached, the bacteria culture was chilled to 16°C. Expression of protein was induced by adding IPTG to 250 μM (GoldBio, 12481C5) and cultured overnight at 16°C. Cells were spun down and lysed in 20 mL of lysis buffer A (50 mM HEPES pH=7.5, 50 mM NaCl, 1 mM PMSF, 1 mM EDTA, 5 mg/mL Lysozyme, 30% glycerol) per 1.5 L of culture. Sample was incubated at Room Temperature (25°C) for 15 minutes while rocking, then 10 mL of lysis buffer B (50 mM HEPES pH=7.5, 300 mM NaCl, 1.5 mM PMSF, 15 mM β-mercaptoethanol, 30 mM imidazole, 20% Glycerol) per 1.5 L culture was added. Sample were then sonicated (On-pulse 10s, off-pulse 50s, amplitude 60%) on ice until proper viscosity was reached. Remove cell debris by centrifugation: 30,000 xg (19k rpm) in a F21S-8×50y rotor (Thermo Scientific), 60min, 4°C (tubes need to be balanced to within 0.1grams). His-tagged proteins were isolated using NI-NTA agarose (Qiagen, 30210), washed three times with wash buffer (50 mM HEPES pH=7.5, 150 mM NaCl, 1 mM PMSF, 5 mM β-mercaptoethanol, 20 mM imidazole, 20% glycerol), eluted with 2.5 mL elution buffer (50 mM HEPES pH=7.5,150 mM NaCl,5 mM β-mercaptoethanol, 250 mM imidazole, 20% glycerol). Samples were dialyzed overnight at 4°C (Fisher, 6–4033) in dialysis buffer (40 mM HEPES pH 7.5, 150 mM NaCl, 5 mM β-mercaptoethanol, 10% glycerol). Samples were further purified by size separation column purification via AKTA Chromatography through a Superdex 200 column (Millipore, G117–5175-01) in degassed purification buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM β-mercaptoethanol, 10% glycerol), followed by concentration of protein-containing fractions by Amicon Ultra centrifugal filter units (MWCO 3 kDa, Millipore, Z647993). Protein concentration was determined using Nanodrop 2000 with a measurement at A280nm. Concentrated proteins were aliquoted, flash-frozen in liquid nitrogen, and stored at −80°C.
CDK2 Kinase Assay
The CDK2 kinase assay was performed according to manufacturer’s instructions (Promega, CDK2/CyclinE1 Kinase Assay, V4489). Briefly, we combined 2 μl enzyme mix (4ng CDK2/CyclinE1) and 2 μl substrate mix (0.1μg/μL Histone H1 and 150μM ATP) with various previously diluted 1 μl concentrations of recombinant mouse Cdk2ap1CAN, Cdk2ap1CAN Mut-TER, Cdk2ap1ΔN (MT2B2) or, Cdk2ap1ΔN (MT2B2) Mut-TER proteins in 5 μl reactions and incubated at room temperature for 60 minutes. After incubation, 5μl of ADP-Glo luminescent reagent was added, followed by a 40 min incubation at room temperature. Then 10 μl of Kinase Luminescence Detection Reagent was added and incubated at room temperature for 30 minutes. Sample luminescence were individually measured using Promega GloMax 20/20 Luminometer.
Quantification and statistical analysis
Quantification and statistical analysis for experimental data
For experimental data, statistical analysis was performed in GraphPad Prism 9. All statistical details for each experiment were described in the figure legends. For embryo related experiment, “n” represents individual embryos. For all mouse experiments, “n” represents individual animals. No data were excluded from analysis. An unpaired Student’s T-test was used to compare two groups for most experiments. A P-value ≤ 0.05 was considered statistically significant.
Quantification of genes and retrotransposons
To obtain genomic coordinates of protein-coding genes and non-coding transcripts, we processed the gene annotation files provided by Refseq (Table S1). To obtain genomic coordinates of retrotransposon subfamilies, we downloaded the Repeatmasker output from UCSC and NCBI and selected for elements that belong to LINE, SINE and LTR. We used two quantitation methods to determine the expression profiles of retrotransposon subfamilies and protein-coding genes in preimplantation development: 1) We analyze both uniquely and multiply mapped RNA-seq reads using TEtranscripts (Jin et al., 2015) (v. 2.2.1, default parameters). 2) We analyzed only uniquely mapped RNA-seq reads with featureCounts (Liao et al., 2014) (v. 1.6.3, options -O -B -p --fracOverlap 0.1 -M --fraction -T 5 -Q 255 for paired end RNA-seq samples, -O --fracOverlap 0.1 -M --fraction -T 5 -Q 255 for single end RNA-seq samples). To avoid confounding between gene and retrotransposon expression, we excluded all the retrotransposons that overlap with Refseq annotated gene exons from our retrotransposon quantitation. The number of reads mapped to all the members of a retrotransposon subfamily were then combined to obtain retrotransposon subfamily-level expression.
Differential expression analysis on genes and retrotransposons
We combined the expression value of protein-coding genes and retrotransposon subfamilies into a single matrix and retained only those with at least one CPM (counts per million) in at least one sample. For datasets with more than 2 samples per developmental stage, we used edgeR (v.3.12.0)(Robinson et al., 2009) to test for differential expression during preimplantation development (negative binomial likelihood ratio test after full-quartile normalization (Risso et al., 2011) and RUVr normalization (Risso et al., 2014)). Genes or retrotransposon subfamilies with a false discovery rate less than 0.05 were defined as differentially expressed. For datasets with only one sample per developmental stage, we inferred the degree of differential expression by calculating the standard deviation per gene using its expression values across all developmental stages. All expressed protein coding genes or retrotransposon subfamilies were then ranked by averaged expression signal during the peak developmental stage (Table S1 and S2). To obtain the topmost highly and dynamically expressed candidates that were analyzed in Figure 1A, 1B, S1B, S1D, S1F, we first selected differentially expressed candidates with false discovery rate smaller than 0.05. We then ranked these differentially expressed candidates by their averaged expression level during the peak preimplantation developmental stage and selected the top candidates for subsequent analyses.
To illustrate the dynamic expression of protein-coding genes and retrotransposons, we generated heatmaps using z-scores of the most highly and differentially expressed protein-coding genes or retrotransposon subfamilies. Z-score was defined as the standard deviations by which the expression value of a gene or a retrotransposon subfamily is above or below its mean expression across all the preimplantation stages. Hierarchical clustering was then performed to group genes or retrotransposon subfamilies with similar expression patterns. Extreme z-scores (below 0.01 quantile or above 0.99 quantile) were capped for display purposes. To highlight the comparison among species, only a subset of preimplantation stages was shown in heatmaps. All the developmental stages that are available in the original studies were included in line plots.
Differential expression analysis on retrotransposon:gene junctions
For datasets with more than 2 samples per developmental stage, we first performed differential expression analysis with edgeR (v.3.12.0) (Robinson et al., 2009) to test for differential expression of retrotransposon:gene junction reads during preimplantation development. Negative binomial likelihood ratio test was performed after full-quartile normalization (Risso et al., 2011) and RUVr normalization (Risso et al., 2014). Junctions with a false discovery rate less than 0.05 were defined as differentially expressed. For datasets with only one sample per developmental stage, we inferred the degree of differential expression by calculating the standard deviation per gene using its expression values across all developmental stages. To obtain the topmost highly and dynamically expressed candidates that were analyzed in Figure 1E–G, S1G, S1I–L, we first selected differentially expressed candidates with false discovery rate smaller than 0.05. We then ranked these differentially expressed candidates by their averaged expression level during the peak preimplantation developmental stage and selected the top candidates for subsequent analyses.
Differential expression of Cdk2ap1 isoforms
We performed Cdk2ap1 isoform expression analyses using the following procedures. We first combined Cdk2ap1 RefSeq annotations with our transcript assembly results to obtain a comprehensive catalog of all Cdk2ap1 gene isoforms in each species. Interestingly, multiple Cdk2ap1 isoforms often exist for a given species, yet these isoforms encode either the canonical Cdk2ap1 protein or N-terminally truncated Cdk2ap1ΔN protein. To infer Cdk2ap1 isoform-level expression, we first computed Cdk2ap1 expression signal quantification at the gene level (sigGENE) by summing all RNA-seq reads mapped to Cdk2ap1 using featureCounts (Liao et al., 2014). We then counted the number of spliced reads across splicing junctions that are unique to each Cdk2ap1 canonical or N-terminally truncated isoforms (juncCAN and juncΔN ) using scripts derived from LeafCutter (Li et al., 2018). If more than one isoform were identified for canonical Cdk2ap1 protein or N-terminally truncated Cdk2ap1ΔN protein, we aggregated spliced reads across splicing junctions that are unique to all the canonical or all the N-terminally truncated isoforms. Cdk2ap1 isoform-level expression was then calculated by redistributing the gene-level expression based on the number of spliced reads across isoform specific junctions (sigCAN = (sigGENE x juncCAN) / (juncCAN + juncΔN), sigΔN = (sigGENE x juncΔN) / (juncCAN + juncΔN)). Inferred Cdk2ap1 isoform-kevel expression value in counts per million can be found in Table S6.
Additional Resources
Resources related to bioinformatic analyses, including information on raw and processed data, detailed documentation on the pipeline used for identifying retrotransposon:gene splicing junctions, as well as integrative browser sessions supported by the WashU epigenome browser (Li et al., 2019).
Resource available at: https://epigenome.wustl.edu/TE_Transcript_Assembly/index.html.
Supplementary Material
Figure S1. Retrotransposons mediate gene regulation in mammalian preimplantation development, related to Figure 1. A. Comparison of retrotransposon quantitation using uniquely mapped reads or using both uniquely and multiply mapped reads. Top, RNA-seq datasets from 8 mammalian species were processed with TEtranscripts (using uniquely and multiply mapped reads) or featureCounts (using only unique reads). A higher fraction of retrotransposon reads were observed in TEtranscripts analyses, particularly for mouse. Bottom, retrotransposon profiles of the top 100 most highly and differentially expressed retrotransposon subfamilies are shown as heatmaps for each mammalian species examined; and the percentage of transcriptome derived from retrotransposons are shown as line graphs. B. TEtranscripts analyses reveal similar expression patterns of protein-coding genes and retrotransposons in mouse and human preimplantation embryos. The top 100 most highly and differentially expressed protein-coding genes or retrotransposon subfamilies are shown as heatmaps; the percentage of reads that originate from protein-coding genes or retrotransposons are shown as line graphs. C. Percentage of uniquely mapped reads were quantified for intergenic retrotransposons (blue), intronic retrotransposons within protein-coding genes (green), and intronic retrotransposons within non-coding transcripts (orange). A large fraction of retrotransposon reads originate from intergenic retrotransposons and intronic retrotransposons within protein-coding genes. D. Intronic retrotransposon reads do not confound global retrotransposon expression profiles. We characterized protein-coding gene and retrotransposon subfamily expression profiles using TEtranscripts, including both uniquely and multiply mapped RNA-seq reads. Intergenic retrotransposons were profiled using uniquely mapped RNA-seq reads. Heatmaps showing the similar expression profiles of the top 100 most highly and differentially expressed protein-coding genes (left, TEtranscripts analyses), retrotransposon subfamilies (middle, TEtranscripts analyses) and intergenic retrotransposon subfamilies (right, featureCounts analyses, using uniquely mapped reads). E. Single embryo real time PCR analyses confirm the dynamic expression of multiple retrotransposon subfamilies. Error bars, ± s.e.m., P values were calculated using unpaired, two-tailed Student’s t test. ORR1A1, PN vs 2C, **P = 0.001, t=3.5, df=35; IAPez-int, Oo vs PN, ****P < 0.0001, t=5.7, df=48; RLTR45-LTR, 4C vs 8C, **P = 0.001 t=4.4, df=10; ERVB4-_2-I_Mm, 4C vs 8C, ***P = 0.0005, t=4.5, df=14. F. A subset of retrotransposon subfamilies exhibit similar expression patterns in mouse preimplantation embryos. The top 100 most highly and dynamically expressed retrotransposon subfamilies from Xue et al. 2013 mouse data were clustered with TCseq (Mengjun W and Lei Gu, 2017), and retrotransposon subfamilies with cluster membership bigger than 0.5 were plotted. Some retrotransposon subfamilies with the same expression pattern share similarities in sequences and retrotransposon classification (Table S1). G. Retrotransposon-dependent gene isoforms were dynamically expressed in mouse preimplantation embryos. A heatmap shows the dynamic expression pattern of the top 250 most highly and differentially expressed splicing junctions between a retrotransposon and a proximal gene exon with a normalized retrotransposon-gene junction read counts ≥ 30 for at least one sample in a preimplantation stage. H. Retrotransposon:gene splicing events predominantly impact host protein-coding genes in mouse preimplantation development. The percentage of protein-coding genes or non-coding transcripts in retrotransposon:gene isoforms were compared to those in GENCODE annotations (left), or those in expressed GENCODE annotated genes (right). Expressed genes are defined as those with >= 30 reads for at least one sample in a preimplantation stage. P-values (on the top of each bar) were calculated using Fisher’s exact test to compared the enrichment of protein-coding genes in retrotransposon:gene isoforms, versus that in GENCODE annotated gene (left) or in expressed GENCODE annotated genes (right). I. The top 250 most highly and dynamically expressed retrotransposon:gene isoforms in mouse preimplantation embryos were broken down to three categories, including those that contain retrotransposon derived promoter/5’UTR, internal exon and terminator. J. Retrotransposon:gene isoforms frequently alter canonical ORFs. Manual curation prediction of ORFs from the top 100 most highly and dynamically expressed retrotransposon:gene isoforms in mouse preimplantation embryos. (Left) A bar plot shows the number of retrotransposon:gene isoforms with predicted ORFs that are either intact or altered. N.D., not determined. (Right) Those with alternative ORFs were further categorized as intact ORFs, deletion, replacement, insertion, exaption, N-Del / N-Rep (retrotransposon-derived exons are predicted to either cause N-terminal deletions or N-terminal replacements of canonical ORF, due to uncertainty in ATG prediction), insertion or N.D. (not determined). K. Retrotransposon promoters are mostly species-specific. In the binary heatmap, rows represent datasets from 8 mammalian species examined, columns represent the top 100 highly and dynamically expressed retrotransposon subfamilies in each dataset, and tile colors indicate the usage of retrotransposon subfamilies as promoters. Datasets from the same species are more closely related than datasets from different species. L. Retrotransposons provide alternative promoters, internal exons and terminators for gene isoforms in mammalian preimplantation embryos. The top 200 most dynamically expressed retrotransposon:gene isoforms in each mammalian species are analyzed for the LTR, SINE and LINE contribution to retrotransposon derived promoters, internal exons and terminators. All retrotransposon derived promoters contain predicted transcription start sites. A, B, D, G. White, LTR; grey, LINE; black, SINE; black triangle, ZGA; Z-score, the number of standard deviations from the expression mean of a protein-coding gene or a retrotransposon subfamily. B, D. A subset of preimplantation stages are shown in heatmaps across all species to highlight the comparison among species. All the developmental stages that are available in the original datasets were included in line plots. F, G, H, I, J. Mouse RNA-seq data from Xue et al. 2013 were employed for the analyses. A, B, C, D, K, L. RNA-seq data were obtained from published datasets, documented in Tables S1. B, D, E, H, G. Oo, oocyte; PN, pronucleus; 2C, two cell embryo; e2C, early two cell embryo; m2C, mid two cell embryo; l2C, late 2C embryo; 4C, four cell embryo; 8C, eight cell embryo; M, morula; BL, blastocysts; eBL, early blastocyst; mBL, mid blastocyst; lBL, late blastocyst; ZGA, zygotic genome activation.
Figure S2. Cdk2ap1ΔN(MT2B2) and Cdk2ap1CAN exhibit distinct expression patterns, related to Figure 2. A. Cdk2ap1ΔN(MT2B2) and Cdk2ap1CAN exhibit distinct 5’ gene structure. The Cdk2ap1ΔN (MT2B2) isoform initiates transcription within the MT2B2 element, generates a transcript that skips the exon1 and splices directly into the exon2. The Cdk2ap1ΔN (MT2B2) isoform is predicted to be 1056nt in length, containing MT2B2 derived sequences in its 5’UTR (red) and utilizing a downstream ATG start codon within the exon 2. The Cdk2ap1CAN isoform is predicted to be 1271nt in length, which initiates transcription from the canonical exon1 (blue), utilizing an ATG within the exon 1 for translation. B. The Cdk2ap1ΔN(MT2B2) isoform produces a truncated Cdk2ap1 protein. Overexpression of Cdk2ap1CAN -Flag and Cdk2ap1ΔN(MT2B2)-Flag in HEK293T cells produce proteins of the expected sizes, demonstrating that the predicted ATG start codon in the Cdk2ap1ΔN(MT2B2) isoform is functional for translation. B. Real time PCR confirms the peak of Cdk2ap1CAN expression at 10.5 dpc embryos. C. Cdk2ap1ΔN(MT2B2) expression is enriched in the TE of blastocysts. Absolute real time PCR quantitation of Cdk2ap1CAN, Cdk2ap1ΔN(MT2B2) and Cdx2 was performed in isolated single cells of 4.0 dpc blastocysts. The expression of Cdx2 was used to distinguish TE cells from ICM cells (Not shown). Error bars, s.e.m.; Cdk2ap1CAN vs. Cdk2ap1ΔN(MT2B2) in TE, * P =0.047, t=2.3, df=10. P values are calculated using unpaired, two-tailed Student’s t-test. D. Preimplantation specific Cdk2ap1 expression is mostly derived from the Cdk2ap1ΔN(MT2B2) isoform. Using CRISPR-EZ, a V5 tag was engineered immediately after the canonical ATG for detection of Cdk2ap1CAN (Cdk2ap1CAN-Ex1-V5). Additionally, a V5 tag was engineered immediately after the alternative ATG in exon 2 for detection of the Cdk2ap1ΔN(MT2B2) isoform, and a stop codon (red hexagon) was introduced 12bp upstream of the Cdk2ap1ΔN(MT2B2) ATG to prevent Cdk2ap1CAN production (Cdk2ap1 –Ex2-V5). Finally, a V5 tag was engineered immediately before the stop codon shared by all Cdk2ap1 isoforms for detection of total Cdk2ap1 (Cdk2ap1-Ex4-V5). Using immunostaining of V5, we detected no expression of Cdk2ap1CAN-V5 in engineered blastocyst embryos, but a strong expression of Cdk2ap1ΔN(MT2B2) and total Cdk2ap1 in TE. The V5 expression patterns of Cdk2ap1 –Ex2-V5 and Cdk2ap1-Ex4-V5 blastocysts are nearly identical, confirming that Cdk2ap1ΔN(MT2B2) yields the majority of Cdk2ap1 proteins in preimplantation embryos. Two independent experiments were performed for each CRISPR-EZ editing experiment, with at least 5 embryos per condition. Scale bars, 20 μm. E. Diagrams illustrate the PCR genotyping strategies of CRISPR edited Cdk2ap1ΔMT2B2/ ΔMT2B2 (top) and Cdk2ap1ΔCAN/ΔCAN (bottom) embryos. Representative genotyping results are shown as electrophoresis images. F. Deletion of Cdk2ap1ΔN (MT2B2) causes delayed embryo development. Representative images of littermate controlled, 10.5dpc wildtype and Cdk2ap1ΔN (MT2B2)/ΔN (MT2B2) embryos (n=16 uteri collected from Cdk2ap1ΔN (MT2B2)/+× Cdk2ap1ΔN (MT2B2)/+ crosses). Embryo developmental delay is evident in 42.9% (9/21) of Cdk2ap1ΔMT2B2/ΔMT2B2 embryos compared to 7.7% (2/26) of WT embryos from the same uterus. Scale bars, 0.5 mm.
Figure S3. An MT2B2 retrotransposon promoter drives an N-terminally truncated Cdk2ap1ΔN (MT2B2) isoform to promote cell proliferation, related to Figure 3. A, B. Cdk2ap1ΔMT2B2/ΔMT2B2 embryos exhibited reduced cell number at 3.5 and 4.5dpc. A. A total of 29 embryos from littermate-controlled wildtype (n=11), Cdk2ap1ΔMT2B2/+ (n= 12) and Cdk2ap1ΔMT2B2/ΔMT2B2 (n=6) embryos were collected at 3.5 dpc from 6 Cdk2ap1ΔMT2B2/+ to Cdk2ap1ΔMT2B2/+ mating. Littermate controlled blastocysts were subjected to immunostaining of Nanog and Cdx2 for the quantitation of ICM cells and TE cells, respectively Representative confocal images are shown. Scale bar, 25 μm; error bars, s.d. Wildtype vs. Cdk2ap1ΔMT2B2/ΔMT2B2 embryos: ICM, ** P = 0.0011, t=3.7, df=15; TE, * P = 0.02, t=2.2, df=15. B. A total of 14 embryos from littermate-controlled wildtype (n=4), Cdk2ap1ΔMT2B2/+ (n= 5) and Cdk2ap1ΔMT2B2/ΔMT2B2 (n=5) embryos were collected at 4.5 dpc from 3 Cdk2ap1ΔMT2B2/+ to Cdk2ap1ΔMT2B2/+ mating. Littermate controlled blastocysts were subjected to immunostaining of Nanog and Cdx2 for the quantitation of ICM cells and TE cells, respectively Representative confocal images are shown. Scale bar, 25 μm; error bars, s.d.. Wildtype vs. Cdk2ap1ΔMT2B2/ΔMT2B2 embryos: ICM, **** P < 0.0001, t=8.2, df=7; TE, *** P = 0.001, t=737, df=7. C. ICM and TE cell fate specification is impaired in Cdk2ap1ΔMT2B2/ΔMT2B2, but not Cdk2ap1ΔCAN/ΔCAN blastocysts. Wildtype (n=5), Cdk2ap1ΔCAN/ΔCAN (n=5) and Cdk2ap1ΔMT2B2/ΔMT2B2 (n=17) blastocysts were collected at 4.0 dpc and subjected to immunostaining of Nanog and Cdx2. The presence of all Nanog and Cdx2 double positive cells in WT, Cdk2ap1ΔCAN/ΔCAN, and Cdk2ap1ΔMT2B2/ΔMT2B2 blastocysts were documented and compartment locations identified. Low levels of double positive cells in WT and Cdk2ap1ΔCAN/ΔCAN embryos suggests an impaired cell fate specification occurs specifically as a result of the MT2B2 deletion. D. Deletion of MT2B2 alters timing of uterine response during implantation. Uteri were collected, fixed and processing for immune-histochemistry after timed mating of wildtype × wildtype (n=5 Uteri, 27 implantations) and Cdk2ap1ΔMT2B2/ΔMT2B2 × Cdk2ap1ΔMT2B2/ΔMT2B2 (n=3 uteri, 9 implantations). Timepoints of 4.75dpc (left) and 5.0dpc (right) were chosen to investigate early implantation embryo and uterine responses. Embryo status and orientation was determined by Sox2 immunostaining. Blastocyst competency was monitored by Wnt5a immunostaining. In comparison, 91.3% (21/23) of WT embryos displayed expected signal, while only 25% (2/8) Cdk2ap1ΔMT2B2/ΔMT2B2 showed the appropriate signal. Remaining embryos served as experimental controls to determined background fluorescence. Boundary between embryo and uterus is shown as dotted line. Scale bar, 50μm.
Figure S4. Cdk2ap1ΔN (MT2B2) and Cdk2ap1CAN have opposite functions in cell proliferation, related to Figure 4. A. Acid Tyrode’s treatment optimization to increase mRNA delivery efficiency into zygotes by electroporation. H2b-Gfp mRNA was electroporated into acid Tyrode’s treated zygotes to determine the acid Tyrode’s treatment duration for optimal zona pellucida thinning. Efficiency of mRNA delivery was measured by the percentage of GFP positive embryos 6 hours after electroporation. Representative GFP images (left) and quantification of GFP positive embryos (right) are shown. Two independent experiments were performed. B and C. Cdk2ap1CAN and Cdk2ap1ΔN (MT2B2) have opposite effect on cell proliferation in wildtype preimplantation embryos. B. Ectopic expression of Cdk2ap1ΔN (MT2B2) increases S-Phase entry and cell proliferation in wildtype embryos at 3.0 dpc. H2b-Gfp, Cdk2ap1CAN or Cdk2ap1ΔN (MT2B2) mRNAs were each electroporated into wildtype pronuclear embryos, and resulted embryos were compared for BrdU incorporation in morula at 3.0 dpc. Representative immunostaining images (left) and quantitation of BrdU positive and total cell number (right) are shown. Violin plots are shown with median (red), as well as lower (25%) and upper (75%) quartiles (black lines). Scale bars, 20 μm. H2b-Gfp vs Cdk2ap1CAN in wildtype embryos: BrdU, **** P < 0.0001, t=4.9, df=119; total cell number, **** P < 0.0001, t=4.1, df=119. H2b-Gfp vs Cdk2ap1ΔN (MT2B2) in wildtype embryos: BrdU+, **** P < 0.0001, t=4.7, df=124; total cell number, *** P = 0.0005, t=3.6, df=124. C. Wildtype embryos overexpressing Cdk2ap1CAN or Cdk2ap1ΔN (MT2B2) were analyzed for ICM (Nanog+) and TE (Cdx2+) cell counts at 4.0 dpc using immunostaining (left). Quantitation is shown as violin plots with median (red line), lower (25%) and upper (75%) quartiles (black lines). H2b-Gfp vs Cdk2ap1CAN: ICM, * P = 0.03, t=2.2, df=49; TE, *P = 0.01, t=2.7, df=49. H2b-Gfp vs Cdk2ap1ΔN (MT2B2), ICM: n.s; TE, * P =0.03, t=2.2, df=40. D. Ectopic expression of Cdk2ap1ΔN (MT2B2) restores S-Phase entry and cell proliferation in Cdk2ap1ΔMT2B2/ΔMT2B2 embryos at 4.0 dpc. H2b-Gfp, Cdk2ap1CAN or Cdk2ap1ΔN (MT2B2) mRNAs were each electroporated into wildtype or Cdk2ap1ΔMT2B2/ΔMT2B2 pronuclear embryos, and resulted embryos were compared for BrdU incorporation in blastocysts at 4.0 dpc. Representative immunostaining images (left) and quantitation of BrdU positive and total cell number (right) are shown. Violin plots are shown with median (red), as well as lower (25%) and upper (75%) quartiles (black lines). Scale bars, 20 μm. H2b-Gfp in WT embryos vs Cdk2ap1ΔN (MT2B2) in Cdk2ap1ΔMT2B2/ΔMT2B2 embryos: BrdU, * P =0.02, t=2.4, df=21; total cell number, ns. H2b-Gfp vs Cdk2ap1CAN in Cdk2ap1ΔMT2B2/ΔMT2B2 embryos: BrdU, ** P =0.001, t=3.9, df=15; total cell number, *** P = 0.0002, t=4.9, df=15. H2b-Gfp vs Cdk2ap1ΔN (MT2B2) in Cdk2ap1ΔMT2B2/ΔMT2B2 embryos: BrdU, * P =0.02, t=2.6, df=13; total cell number, ** P =0.002, t=3.8, df=13. E. Cdk2ap1CAN and Cdk2ap1ΔN (MT2B2) both bind endogenous mouse Cdk2. In HEK293T cells transfected with either C-Terminal HA-Tagged Cdk2ap1CAN (Cdk2ap1CAN–HA) or C-Terminal HA-Tagged Cdk2ap1ΔN (MT2B2) (Cdk2ap1ΔN (MT2B2)-HA), immunoprecipitation of HA pull down endogenous Cdk2. F. Cdk2ap1CAN and Cdk2ap1ΔN (MT2B2) have opposite effects on Cdk2 kinase activity. HEK293T cells overexpressing Cdk2ap1CAN-HA, Cdk2ap1CAN-MutTER-HA, Cdk2ap1ΔN (MT2B2)-HA or Cdk2ap1ΔN (MT2B2)-MutTER-HA were subjected to immunoprecipitation with anti-HA antibodies. Immunoprecipitated lysates were each incubated with recombinant CDK2, CYCLIN E, HISTONE H1 and ATP in vitro. Their effects on CDK2 activity were analyzed in a kinase assay. Error bars are means ± s.e.m. Cdk2ap1CAN -HA vs Cdk2ap1CAN -MutTER-HA, **P = 0.009, t=4.7, df=4; Cdk2ap1ΔN (MT2B2) vs Cdk2ap1ΔN (MT2B2)-MutTER-HA, **P = 0.008, t=4.8, df=4. Three independent experiments were performed. All P values were calculated based on the unpaired two-tailed Student’s t test. n.s., not significant.
Figure S5. Multiple mouse retrotransposon promoters yield N-terminally altered gene isoforms that are conserved in human, related to Figure 5. Gene structure, protein motif analyses and sequence alignment are shown for Pemt (A), Gan14 (B) and Mkln1 (C). For Pemt, the transcriptional start site, as well as the splicing between retrotransposon and gene exon, were experimentally validated by 5’ RACE and RT-PCR.
Figure S6. CDK2AP1ΔN is expressed in human embryonic stem cells, related to Figure 6. A. A promoter from the L2a/Charlie4z region drives a strong expression of an N-terminally truncated CDK2AP1 isoform in human ES cells. Five human CDK2AP1 isoforms were quantified by RNA-seq for expression in H1 embryonic stem cells from ENCODE project (Encode Consortium, 2012). B. A diagram illustrates the approximate durations of preimplantation development in different mammalian species. Arrow, approximate timing of implantation.
Table S1.
RNA-seq analyses of retrotransposon expression in mouse, primate, cow, pig, goat and opossum preimplantation embryos, related to Figure 1 and S1.
Table S2.
RNA-seq analyses of protein-coding gene expression in mouse, primate, cow, pig, goat and opossum preimplantation embryos, related to Figure 1 and S1.
Table S3.
RNA-seq analyses of retrotransposon:gene junction reads in mouse, primate, cow, pig, goat and opossum preimplantation embryos, related to Figure 1 and S1.
Table S4.
Experimental validation of selected retrotransposon:gene isoforms, related to Figure 1, 2, and S5.
Table S5.
ORF and Conservation analyses on retrotransposon:gene isoforms in preimplantation embryos by manual curation, related to Figures 1, 5, S1, and S5
Table S6.
RNA-seq analyses of Cdk2ap1 gene isoforms in mouse, primate, cow, pig, goat and opossum preimplantation embryos, related to Figure 6.
Table S8.
Primer/oligo design for genotyping, CRISPR editing, real time PCR and RACE, related to STAR Methods.
Highlights:
Numerous retrotransposons act as preimplantation-specific, gene regulatory elements
An MT2B2 retrotransposon promoter is essential for mouse preimplantation development
MT2B2-driven Cdk2ap1ΔN and canonical Cdk2ap1 exhibit isoform-specific functions
Retrotransposon promoters can yield conserved gene isoforms with unique regulation
Acknowledgments
We thank M. Rape, A. Manford, F. Rodriquez, J. Cox, Y. Zhou, H. Huang, C. DiBiaggio, A. Herr, P Lishko, D. Yi and M. Kinisu for insightful advice, technical assistance and reagents, S. Dey, R Rogers, M Slatkin, M. Nachman and S Banker for stimulating discussion, M. Kinisu and S. Chen for manuscript revision, and members of the He lab for their support. A.J.M is supported by NIH (K99HD096108) and the Siebel Stem Cell Institute, T.P.S. is supported by a National Health and Medical Research Council Australia Fellowship. Z.X. is supported by NIH (R01NS096068), W.S., A.A. and T.W. are supported by NIH (R01HG007175, U24ES026699, U01CA200060, U01HG009391 and U41HG010972). D.R. is supported by “Programma per Giovani Ricercatori Rita Levi Montalcini” granted by the Italian Ministry of University and Research, and NIH-NCI (2U24CA180996). L.H. is a Thomas and Stacey Siebel Distinguished Chair Professor, supported by a HHMI Faculty Scholar award, a Bakar Fellow award, and NIH grants (1R01GM114414, R01CA139067, 1R21OD027053, GRANT12095758, R01NS120287).
Footnotes
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity statement
We worked to ensure gender balance in the authors who contributed to this paper.
Tables S1, S2 and S3 exceed size limit and can be found on third-party host (https://www.dropbox.com/sh/auo40kyfgu5jaiz/AABGEs2pR_EkHl3Pkgwldv4ya?dl=0)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alsayegh KN, Sheridan SD, Iyer S, and Rao RR (2018). Knockdown of CDK2AP1 in human embryonic stem cells reduces the threshold of differentiation. PLoS One 13, e0196817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardeljan D, Taylor MS, Ting DT, and Burns KH (2017). The Human Long Interspersed Element-1 Retrotransposon: An Emerging Biomarker of Neoplasia. Clin. Chem 63, 816–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayarpadikannan S, and Kim H-S (2014). The Impact of Transposable Elements in Genome Evolution and Genetic Instability and Their Implications in Various Diseases. Genomics Inform. 12, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batut P, Dobin A, Plessy C, Carninci P, and Gingeras TR (2013). High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res. 23, 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Garcia-Perez JL, Badge RM, and Moran JV (2011). LINE-1 Elements in Structural Variation and Disease. Annu. Rev. Genomics Hum. Genet 12, 187–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak T, Stirparo GG, Dietmann S, Hernando-Herraez I, Mohammed H, Reik W, Smith A, Sasaki E, Nichols J, and Bertone P (2018). Single cell transcriptome analysis of human, marmoset and mouse embryos reveals common and divergent features of preimplantation development. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo JI, Nozownik S, Danthi PS, and Benayoun BA (2020). Transposable elements, circular RNAs and mitochondrial transcription in age-related genomic regulation. Development 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Burke TW, and Kadonaga JT (1997). The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 11, 3020–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KH (2017). Transposable elements in cancer. Nat. Rev. Cancer 17, 415–424. [DOI] [PubMed] [Google Scholar]
- De Cecco M, Criscione SW, Peterson AL, Neretti N, Sedivy JM, and Kreiling JA (2013). Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging (Albany. NY). 5, 867–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Lee B, Lee AY-F, Modzelewski AJ, and He L (2016). Highly Efficient Mouse Genome Editing by CRISPR Ribonucleoprotein Electroporation of Zygotes. J. Biol. Chem. 291, 14457–14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Sun S, Moonen D, Lee C, Lee AY-F, Schaffer DV, and He L (2019). CRISPR-READI: Efficient Generation of Knockin Mice by CRISPR RNP Electroporation and AAV Donor Infection. Cell Rep. 27, 3780–3789.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Lin C-P, Risso D, Chen S, Kim TA, Tan MH, Li JB, Wu Y, Chen C, Xuan Z, et al. (2017). Deficiency of {microRNA} \textit{{miR}−34a} expands cell fate potential in pluripotent stem cells. Science (80-. ). 355, eaag1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary MNK, Friedman RZ, Wang JT, Jang HS, Zhuo X, and Wang T (2020). Co-opted transposons help perpetuate conserved higher-order chromosomal structures. Genome Biol. 21, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung N, Jonaid GM, Quinton S, Ross A, Sexton CE, Alberto A, Clymer C, Churchill D, Navarro Leija O., and Han MV (2019). Transcriptome analyses of tumor-adjacent somatic tissues reveal genes co-expressed with transposable elements. Mob. DNA 10, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Rumi M. a K., Soares MJ, and Baker JC (2013). Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat. Genet. 45, 325–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, and Feschotte C (2016). Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351, 1083–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosby RL, Chang N-C, and Feschotte C (2019). Host-transposon interactions: conflict, cooperation, and cooption. Genes Dev. 33, 1098–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, Hilton JA, Jain K, Baymuradov UK, Narayanan AK, et al. (2018). The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 46, D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Berrocal A, Morita T, Longden KD, and Stern DL (2016). Natural courtship song variation caused by an intronic retroelement in an ion channel gene. Nature 536, 329–332. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Vernochet C, Bawa O, Harper F, Pierron G, Opolon P, and Heidmann T (2009). Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc. Natl. Acad. Sci. U. S. A. 106, 12127–12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ED P, CE D, RB K, M K-S, AV T, J M, N Y, DM B, S E, DR M, et al. (2018). The Neuronal Gene Arc Encodes a Repurposed Retrotransposon Gag Protein That Mediates Intercellular RNA Transfer. Cell 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, and Lash AE (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium Encode (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. (2011). Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo ML, Dayan S, Kim Y, McBride J, Kupper TS, and Wong DTW (2006). Expression of cell-cycle regulator CDK2-associating protein 1 (p12CDK2AP1) in transgenic mice induces testicular and ovarian atrophy in vivo. Mol. Reprod. Dev. 73, 987–997. [DOI] [PubMed] [Google Scholar]
- Flemr M, Malik R, Franke V, Nejepinska J, Sedlacek R, Vlahovicek K, and Svoboda P (2013). A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell 155, 807–816. [DOI] [PubMed] [Google Scholar]
- Frankish A, Diekhans M, Ferreira A-M, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, et al. (2019). GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnier L, Belancio VP, and Mager DL (2019). Mouse germ line mutations due to retrotransposon insertions. Mob. DNA 10, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Perez JL, Widmann TJ, and Adams IR (2016). The impact of transposable elements on mammalian development. DEVELOPMENT 143, 4101–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes P, Richardson SR, Mager DL, and Faulkner GJ (2016). Transposable elements in the mammalian embryo: pioneers surviving through stealth and service. Genome Biol. 17, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford WD, Pfaff SL, and Macfarlan TS (2013). Transposable elements as genetic regulatory substrates in early development. Trends Cell Biol. 23, 218–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göke J, and Ng HH (2016). CTRL+INSERT: retrotransposons and their contribution to regulation and innovation of the transcriptome. EMBO Rep. 17, 1131–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL (2016). Restricting retrotransposons: a review. Mob. DNA 7, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi N, and Tramontano E (2018). Human Endogenous Retroviruses Are Ancient Acquired Elements Still Shaping Innate Immune Responses. Front. Immunol 9, 2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Kobayashi T, Dietmann S, and Surani MA (2017). Activation of Lineage Regulators and Transposable Elements across a Pluripotent Spectrum. Stem Cell Reports 8, 1645–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, and Kazazian HH (2016). Roles for retrotransposon insertions in human disease. Mob. DNA 7, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu MG, Hu G-F, Kim Y, Tsuji T, McBride J, Hinds P, and Wong DTW (2004). Role of p12(CDK2-AP1) in transforming growth factor-beta1-mediated growth suppression. Cancer Res. 64, 490–499. [DOI] [PubMed] [Google Scholar]
- Imbeault M, Helleboid P-Y, and Trono D (2017). KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 543, 550–554. [DOI] [PubMed] [Google Scholar]
- Inoue K, Ichiyanagi K, Fukuda K, Glinka M, and Sasaki H (2017). Switching of dominant retrotransposon silencing strategies from posttranscriptional to transcriptional mechanisms during male germ-cell development in mice. PLOS Genet. 13, e1006926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang HS, Shah NM, Du AY, Dailey ZZ, Pehrsson EC, Godoy PM, Zhang D, Li D, Xing X, Kim S, et al. (2019). Transposable elements drive widespread expression of oncogenes in human cancers. Nat. Genet 51, 611–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Tam OH, Paniagua E, and Hammell M (2015). TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics 31, 3593–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH, Wong C, Youssoufian H, Scott AF, Phillips DG, and Antonarakis SE (1988). Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332, 164–166. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D (2002). The human genome browser at UCSC. Genome Res. 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-J, Lee J, and Han K (2012). Transposable Elements: No More “Junk DNA”. Genomics Inform. 10, 226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Ohyama H, Patel V, Figueiredo M, and Wong DT (2005). Mutation of Cys105 inhibits dimerization of p12CDK2-AP1 and its growth suppressor effect. J. Biol. Chem. 280, 23273–23279. [DOI] [PubMed] [Google Scholar]
- Kim Y, McBride J, Kimlin L, Pae E-K, Deshpande A, and Wong DT (2009). Targeted Inactivation of p12Cdk2ap1, CDK2 Associating Protein 1, Leads to Early Embryonic Lethality. PLoS One 4, e4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Yang S, Gong F, Lu C, Zhang S, Lu G, and Lin G (2016). The Relationship between Cell Number, Division Behavior and Developmental Potential of Cleavage Stage Human Embryos: A Time-Lapse Study. PLoS One 11, e0153697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Rose CM, Cass AA, Williams AG, Darwish M, Lianoglou S, Haverty PM, Tong A-J, Blanchette C, Albert ML, et al. (2019). Transposable element expression in tumors is associated with immune infiltration and increased antigenicity. Nat. Commun. 10, 5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaka S, Zimin AV, Pertea GM, Razaghi R, Salzberg SL, and Pertea M (2019). Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, and Hedges SB (2017). TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 34, 1812–1819. [DOI] [PubMed] [Google Scholar]
- Lanciano S, and Cristofari G (2020). Measuring and interpreting transposable element expression. Nat. Rev. Genet. 21, 721–736. [DOI] [PubMed] [Google Scholar]
- Levis RW, Ganesan R, Houtchens K, Tolar LA, and Sheen F (1993). Transposons in place of telomeric repeats at a Drosophila telomere. Cell 75, 1083–1093. [DOI] [PubMed] [Google Scholar]
- Li D, Hsu S, Purushotham D, Sears RL, and Wang T (2019). WashU Epigenome Browser update 2019. Nucleic Acids Res. 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YI, Knowles DA, Humphrey J, Barbeira AN, Dickinson SP, Im HK, and Pritchard JK (2018). Annotation-free quantification of RNA splicing using LeafCutter. Nat. Genet. 50, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lo K, and Smale ST (1996). Generality of a functional initiator consensus sequence. Gene 182, 13–22. [DOI] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, and Pfaff SL (2012). Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). TECHNICAL NOTES.
- Maxwell PH, Burhans WC, and Curcio MJ (2011). Retrotransposition is associated with genome instability during chronological aging. Proc. Natl. Acad. Sci. U. S. A. 108, 20376–20381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei S, Qin Q, Wu Q, Sun H, Zheng R, Zang C, Zhu M, Wu J, Shi X, Taing L, et al. (2017). Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 45, D658–D662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengjun W, and Gu Lei (2017). TCseq: time course sequencing data analysis. Available Online.
- Miao B, Fu S, Lyu C, Gontarz P, Wang T, and Zhang B (2020). Tissue-specific usage of transposable element-derived promoters in mouse development. Genome Biol. 21, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modzelewski AJ, Chen S, Willis BJ, Lloyd KCK, Wood JA, and He L (2018). Efficient mouse genome engineering by CRISPR-EZ technology. Nat. Publ. Gr. 13, 1253–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molaro A, Falciatori I, Hodges E, Aravin AA, Marran K, Rafii S, McCombie WR, Smith AD, and Hannon GJ (2014). Two waves of de novo methylation during mouse germ cell development. Genes Dev. 28, 1544–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono R, Nakamura K, Inoue K, Naruse M, Usami T, Wakisaka-Saito N, Hino T, Suzuki-Migishima R, Ogonuki N, Miki H, et al. (2006). Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet. 38, 101–106. [DOI] [PubMed] [Google Scholar]
- Pasquesi GIM, Perry BW, Vandewege MW, Ruggiero RP, Schield DR, and Castoe TA (2020). Vertebrate Lineages Exhibit Diverse Patterns of Transposable Element Regulation and Expression across Tissues. Genome Biol. Evol. 12, 506–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, and Knowles BB (2004). Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell 7, 597–606. [DOI] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, and Mager DL (2012). Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu. Rev. Genet. 46, 21–42. [DOI] [PubMed] [Google Scholar]
- Risso D, Schwartz K, Sherlock G, and Dudoit S (2011). GC-Content Normalization for RNA-Seq Data. BMC Bioinformatics 12, 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D, Ngai J, Speed TP, and Dudoit S (2014). Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 32, 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2009). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Macia A, and Muotri AR (2019). Transposable Elements, Inflammation, and Neurological Disease. Front. Neurol 10, 894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekita Y, Wagatsuma H, Nakamura K, Ono R, Kagami M, Wakisaka N, Hino T, Suzuki-Migishima R, Kohda T, Ogura A, et al. (2008). Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat. Genet. 40, 243–248. [DOI] [PubMed] [Google Scholar]
- Shintani S, Ohyama H, Zhang X, McBride J, Matsuo K, Tsuji T, Hu MG, Hu G, Kohno Y, Lerman M, et al. (2000). p12(DOC-1) is a novel cyclin-dependent kinase 2-associated protein. Mol. Cell. Biol. 20, 6300–6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Patel RK, Palmer N, Grenier JK, Paduch D, Kaldis P, Grimson A, and Schimenti JC (2019). CDK2 kinase activity is a regulator of male germ cell fate. Development 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee B-K, Sheffield NC, Gräf S, Huss M, Keefe D, et al. (2011). Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 21, 1757–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt CG, Bartels SJJ, Brinkman AB, Tjeertes JV, Poser I, Stunnenberg HG, and Vermeulen M (2010). CDK2AP1/DOC-1 is a bona fide subunit of the Mi-2/NuRD complex. Mol. Biosyst. 6, 1700. [DOI] [PubMed] [Google Scholar]
- Stuckey DW, Clements M, Di-Gregorio A, Senner CE, Le Tissier P, Srinivas S, and Rodriguez TA (2011). Coordination of cell proliferation and anterior-posterior axis establishment in the mouse embryo. Development 138, 1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm Á, Ivics Z, and Vellai T (2015). The mechanism of ageing: primary role of transposable elements in genome disintegration. Cell. Mol. Life Sci. 72, 1839–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, Snyder MP, and Wang T (2014). Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 24, 1963–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram V, Choudhary MNK, Pehrsson E, Xing X, Fiore C, Pandey M, Maricque B, Udawatta M, Ngo D, Chen Y, et al. (2017). Functional cis-regulatory modules encoded by mouse-specific endogenous retrovirus. Nat. Commun. 8, 14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WWC, Dietmann S, Irie N, Leitch HG, Floros VI, Bradshaw CR, Hackett JA, Chinnery PF, and Surani MA (2015). A Unique Gene Regulatory Network Resets the Human Germline Epigenome for Development. Cell 161, 1453–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeneman BA, Shukla S, Dhanasekaran SM, Chinnaiyan AM, and Nesvizhskii AI (2015). Two-pass alignment improves novel splice junction quantification. Bioinformatics 32, btv642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, Burgess SM, Brachmann RK, and Haussler D (2007). Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc. Natl. Acad. Sci. U. S. A. 104, 18613–18618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JN, and Feschotte C (2020). A Field Guide to Eukaryotic Transposable Elements. Annu. Rev. Genet. 54, 539–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A, Baur R, Teerikorpi N, Kaya DU, and Rape M (2018). Multisite dependency of an E3 ligase controls monoubiquitylation-dependent cell fate decisions. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DTW, Kim JJ, Khalid O, Sun HH, and Kim Y (2012). Double edge: CDK2AP1 in cell-cycle regulation and epigenetic regulation. J. Dent. Res. 91, 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Hong C, Zhang B, Lowdon RF, Xing X, Li D, Zhou X, Lee HJ, Maire CL, Ligon KL, et al. (2013). DNA hypomethylation within specific transposable element families associates with tissue-specific enhancer landscape. Nat. Genet. 45, 836–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Huang K, Cai C, Cai L, Jiang C, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE, et al. (2013). Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500, 593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Retrotransposons mediate gene regulation in mammalian preimplantation development, related to Figure 1. A. Comparison of retrotransposon quantitation using uniquely mapped reads or using both uniquely and multiply mapped reads. Top, RNA-seq datasets from 8 mammalian species were processed with TEtranscripts (using uniquely and multiply mapped reads) or featureCounts (using only unique reads). A higher fraction of retrotransposon reads were observed in TEtranscripts analyses, particularly for mouse. Bottom, retrotransposon profiles of the top 100 most highly and differentially expressed retrotransposon subfamilies are shown as heatmaps for each mammalian species examined; and the percentage of transcriptome derived from retrotransposons are shown as line graphs. B. TEtranscripts analyses reveal similar expression patterns of protein-coding genes and retrotransposons in mouse and human preimplantation embryos. The top 100 most highly and differentially expressed protein-coding genes or retrotransposon subfamilies are shown as heatmaps; the percentage of reads that originate from protein-coding genes or retrotransposons are shown as line graphs. C. Percentage of uniquely mapped reads were quantified for intergenic retrotransposons (blue), intronic retrotransposons within protein-coding genes (green), and intronic retrotransposons within non-coding transcripts (orange). A large fraction of retrotransposon reads originate from intergenic retrotransposons and intronic retrotransposons within protein-coding genes. D. Intronic retrotransposon reads do not confound global retrotransposon expression profiles. We characterized protein-coding gene and retrotransposon subfamily expression profiles using TEtranscripts, including both uniquely and multiply mapped RNA-seq reads. Intergenic retrotransposons were profiled using uniquely mapped RNA-seq reads. Heatmaps showing the similar expression profiles of the top 100 most highly and differentially expressed protein-coding genes (left, TEtranscripts analyses), retrotransposon subfamilies (middle, TEtranscripts analyses) and intergenic retrotransposon subfamilies (right, featureCounts analyses, using uniquely mapped reads). E. Single embryo real time PCR analyses confirm the dynamic expression of multiple retrotransposon subfamilies. Error bars, ± s.e.m., P values were calculated using unpaired, two-tailed Student’s t test. ORR1A1, PN vs 2C, **P = 0.001, t=3.5, df=35; IAPez-int, Oo vs PN, ****P < 0.0001, t=5.7, df=48; RLTR45-LTR, 4C vs 8C, **P = 0.001 t=4.4, df=10; ERVB4-_2-I_Mm, 4C vs 8C, ***P = 0.0005, t=4.5, df=14. F. A subset of retrotransposon subfamilies exhibit similar expression patterns in mouse preimplantation embryos. The top 100 most highly and dynamically expressed retrotransposon subfamilies from Xue et al. 2013 mouse data were clustered with TCseq (Mengjun W and Lei Gu, 2017), and retrotransposon subfamilies with cluster membership bigger than 0.5 were plotted. Some retrotransposon subfamilies with the same expression pattern share similarities in sequences and retrotransposon classification (Table S1). G. Retrotransposon-dependent gene isoforms were dynamically expressed in mouse preimplantation embryos. A heatmap shows the dynamic expression pattern of the top 250 most highly and differentially expressed splicing junctions between a retrotransposon and a proximal gene exon with a normalized retrotransposon-gene junction read counts ≥ 30 for at least one sample in a preimplantation stage. H. Retrotransposon:gene splicing events predominantly impact host protein-coding genes in mouse preimplantation development. The percentage of protein-coding genes or non-coding transcripts in retrotransposon:gene isoforms were compared to those in GENCODE annotations (left), or those in expressed GENCODE annotated genes (right). Expressed genes are defined as those with >= 30 reads for at least one sample in a preimplantation stage. P-values (on the top of each bar) were calculated using Fisher’s exact test to compared the enrichment of protein-coding genes in retrotransposon:gene isoforms, versus that in GENCODE annotated gene (left) or in expressed GENCODE annotated genes (right). I. The top 250 most highly and dynamically expressed retrotransposon:gene isoforms in mouse preimplantation embryos were broken down to three categories, including those that contain retrotransposon derived promoter/5’UTR, internal exon and terminator. J. Retrotransposon:gene isoforms frequently alter canonical ORFs. Manual curation prediction of ORFs from the top 100 most highly and dynamically expressed retrotransposon:gene isoforms in mouse preimplantation embryos. (Left) A bar plot shows the number of retrotransposon:gene isoforms with predicted ORFs that are either intact or altered. N.D., not determined. (Right) Those with alternative ORFs were further categorized as intact ORFs, deletion, replacement, insertion, exaption, N-Del / N-Rep (retrotransposon-derived exons are predicted to either cause N-terminal deletions or N-terminal replacements of canonical ORF, due to uncertainty in ATG prediction), insertion or N.D. (not determined). K. Retrotransposon promoters are mostly species-specific. In the binary heatmap, rows represent datasets from 8 mammalian species examined, columns represent the top 100 highly and dynamically expressed retrotransposon subfamilies in each dataset, and tile colors indicate the usage of retrotransposon subfamilies as promoters. Datasets from the same species are more closely related than datasets from different species. L. Retrotransposons provide alternative promoters, internal exons and terminators for gene isoforms in mammalian preimplantation embryos. The top 200 most dynamically expressed retrotransposon:gene isoforms in each mammalian species are analyzed for the LTR, SINE and LINE contribution to retrotransposon derived promoters, internal exons and terminators. All retrotransposon derived promoters contain predicted transcription start sites. A, B, D, G. White, LTR; grey, LINE; black, SINE; black triangle, ZGA; Z-score, the number of standard deviations from the expression mean of a protein-coding gene or a retrotransposon subfamily. B, D. A subset of preimplantation stages are shown in heatmaps across all species to highlight the comparison among species. All the developmental stages that are available in the original datasets were included in line plots. F, G, H, I, J. Mouse RNA-seq data from Xue et al. 2013 were employed for the analyses. A, B, C, D, K, L. RNA-seq data were obtained from published datasets, documented in Tables S1. B, D, E, H, G. Oo, oocyte; PN, pronucleus; 2C, two cell embryo; e2C, early two cell embryo; m2C, mid two cell embryo; l2C, late 2C embryo; 4C, four cell embryo; 8C, eight cell embryo; M, morula; BL, blastocysts; eBL, early blastocyst; mBL, mid blastocyst; lBL, late blastocyst; ZGA, zygotic genome activation.
Figure S2. Cdk2ap1ΔN(MT2B2) and Cdk2ap1CAN exhibit distinct expression patterns, related to Figure 2. A. Cdk2ap1ΔN(MT2B2) and Cdk2ap1CAN exhibit distinct 5’ gene structure. The Cdk2ap1ΔN (MT2B2) isoform initiates transcription within the MT2B2 element, generates a transcript that skips the exon1 and splices directly into the exon2. The Cdk2ap1ΔN (MT2B2) isoform is predicted to be 1056nt in length, containing MT2B2 derived sequences in its 5’UTR (red) and utilizing a downstream ATG start codon within the exon 2. The Cdk2ap1CAN isoform is predicted to be 1271nt in length, which initiates transcription from the canonical exon1 (blue), utilizing an ATG within the exon 1 for translation. B. The Cdk2ap1ΔN(MT2B2) isoform produces a truncated Cdk2ap1 protein. Overexpression of Cdk2ap1CAN -Flag and Cdk2ap1ΔN(MT2B2)-Flag in HEK293T cells produce proteins of the expected sizes, demonstrating that the predicted ATG start codon in the Cdk2ap1ΔN(MT2B2) isoform is functional for translation. B. Real time PCR confirms the peak of Cdk2ap1CAN expression at 10.5 dpc embryos. C. Cdk2ap1ΔN(MT2B2) expression is enriched in the TE of blastocysts. Absolute real time PCR quantitation of Cdk2ap1CAN, Cdk2ap1ΔN(MT2B2) and Cdx2 was performed in isolated single cells of 4.0 dpc blastocysts. The expression of Cdx2 was used to distinguish TE cells from ICM cells (Not shown). Error bars, s.e.m.; Cdk2ap1CAN vs. Cdk2ap1ΔN(MT2B2) in TE, * P =0.047, t=2.3, df=10. P values are calculated using unpaired, two-tailed Student’s t-test. D. Preimplantation specific Cdk2ap1 expression is mostly derived from the Cdk2ap1ΔN(MT2B2) isoform. Using CRISPR-EZ, a V5 tag was engineered immediately after the canonical ATG for detection of Cdk2ap1CAN (Cdk2ap1CAN-Ex1-V5). Additionally, a V5 tag was engineered immediately after the alternative ATG in exon 2 for detection of the Cdk2ap1ΔN(MT2B2) isoform, and a stop codon (red hexagon) was introduced 12bp upstream of the Cdk2ap1ΔN(MT2B2) ATG to prevent Cdk2ap1CAN production (Cdk2ap1 –Ex2-V5). Finally, a V5 tag was engineered immediately before the stop codon shared by all Cdk2ap1 isoforms for detection of total Cdk2ap1 (Cdk2ap1-Ex4-V5). Using immunostaining of V5, we detected no expression of Cdk2ap1CAN-V5 in engineered blastocyst embryos, but a strong expression of Cdk2ap1ΔN(MT2B2) and total Cdk2ap1 in TE. The V5 expression patterns of Cdk2ap1 –Ex2-V5 and Cdk2ap1-Ex4-V5 blastocysts are nearly identical, confirming that Cdk2ap1ΔN(MT2B2) yields the majority of Cdk2ap1 proteins in preimplantation embryos. Two independent experiments were performed for each CRISPR-EZ editing experiment, with at least 5 embryos per condition. Scale bars, 20 μm. E. Diagrams illustrate the PCR genotyping strategies of CRISPR edited Cdk2ap1ΔMT2B2/ ΔMT2B2 (top) and Cdk2ap1ΔCAN/ΔCAN (bottom) embryos. Representative genotyping results are shown as electrophoresis images. F. Deletion of Cdk2ap1ΔN (MT2B2) causes delayed embryo development. Representative images of littermate controlled, 10.5dpc wildtype and Cdk2ap1ΔN (MT2B2)/ΔN (MT2B2) embryos (n=16 uteri collected from Cdk2ap1ΔN (MT2B2)/+× Cdk2ap1ΔN (MT2B2)/+ crosses). Embryo developmental delay is evident in 42.9% (9/21) of Cdk2ap1ΔMT2B2/ΔMT2B2 embryos compared to 7.7% (2/26) of WT embryos from the same uterus. Scale bars, 0.5 mm.
Figure S3. An MT2B2 retrotransposon promoter drives an N-terminally truncated Cdk2ap1ΔN (MT2B2) isoform to promote cell proliferation, related to Figure 3. A, B. Cdk2ap1ΔMT2B2/ΔMT2B2 embryos exhibited reduced cell number at 3.5 and 4.5dpc. A. A total of 29 embryos from littermate-controlled wildtype (n=11), Cdk2ap1ΔMT2B2/+ (n= 12) and Cdk2ap1ΔMT2B2/ΔMT2B2 (n=6) embryos were collected at 3.5 dpc from 6 Cdk2ap1ΔMT2B2/+ to Cdk2ap1ΔMT2B2/+ mating. Littermate controlled blastocysts were subjected to immunostaining of Nanog and Cdx2 for the quantitation of ICM cells and TE cells, respectively Representative confocal images are shown. Scale bar, 25 μm; error bars, s.d. Wildtype vs. Cdk2ap1ΔMT2B2/ΔMT2B2 embryos: ICM, ** P = 0.0011, t=3.7, df=15; TE, * P = 0.02, t=2.2, df=15. B. A total of 14 embryos from littermate-controlled wildtype (n=4), Cdk2ap1ΔMT2B2/+ (n= 5) and Cdk2ap1ΔMT2B2/ΔMT2B2 (n=5) embryos were collected at 4.5 dpc from 3 Cdk2ap1ΔMT2B2/+ to Cdk2ap1ΔMT2B2/+ mating. Littermate controlled blastocysts were subjected to immunostaining of Nanog and Cdx2 for the quantitation of ICM cells and TE cells, respectively Representative confocal images are shown. Scale bar, 25 μm; error bars, s.d.. Wildtype vs. Cdk2ap1ΔMT2B2/ΔMT2B2 embryos: ICM, **** P < 0.0001, t=8.2, df=7; TE, *** P = 0.001, t=737, df=7. C. ICM and TE cell fate specification is impaired in Cdk2ap1ΔMT2B2/ΔMT2B2, but not Cdk2ap1ΔCAN/ΔCAN blastocysts. Wildtype (n=5), Cdk2ap1ΔCAN/ΔCAN (n=5) and Cdk2ap1ΔMT2B2/ΔMT2B2 (n=17) blastocysts were collected at 4.0 dpc and subjected to immunostaining of Nanog and Cdx2. The presence of all Nanog and Cdx2 double positive cells in WT, Cdk2ap1ΔCAN/ΔCAN, and Cdk2ap1ΔMT2B2/ΔMT2B2 blastocysts were documented and compartment locations identified. Low levels of double positive cells in WT and Cdk2ap1ΔCAN/ΔCAN embryos suggests an impaired cell fate specification occurs specifically as a result of the MT2B2 deletion. D. Deletion of MT2B2 alters timing of uterine response during implantation. Uteri were collected, fixed and processing for immune-histochemistry after timed mating of wildtype × wildtype (n=5 Uteri, 27 implantations) and Cdk2ap1ΔMT2B2/ΔMT2B2 × Cdk2ap1ΔMT2B2/ΔMT2B2 (n=3 uteri, 9 implantations). Timepoints of 4.75dpc (left) and 5.0dpc (right) were chosen to investigate early implantation embryo and uterine responses. Embryo status and orientation was determined by Sox2 immunostaining. Blastocyst competency was monitored by Wnt5a immunostaining. In comparison, 91.3% (21/23) of WT embryos displayed expected signal, while only 25% (2/8) Cdk2ap1ΔMT2B2/ΔMT2B2 showed the appropriate signal. Remaining embryos served as experimental controls to determined background fluorescence. Boundary between embryo and uterus is shown as dotted line. Scale bar, 50μm.
Figure S4. Cdk2ap1ΔN (MT2B2) and Cdk2ap1CAN have opposite functions in cell proliferation, related to Figure 4. A. Acid Tyrode’s treatment optimization to increase mRNA delivery efficiency into zygotes by electroporation. H2b-Gfp mRNA was electroporated into acid Tyrode’s treated zygotes to determine the acid Tyrode’s treatment duration for optimal zona pellucida thinning. Efficiency of mRNA delivery was measured by the percentage of GFP positive embryos 6 hours after electroporation. Representative GFP images (left) and quantification of GFP positive embryos (right) are shown. Two independent experiments were performed. B and C. Cdk2ap1CAN and Cdk2ap1ΔN (MT2B2) have opposite effect on cell proliferation in wildtype preimplantation embryos. B. Ectopic expression of Cdk2ap1ΔN (MT2B2) increases S-Phase entry and cell proliferation in wildtype embryos at 3.0 dpc. H2b-Gfp, Cdk2ap1CAN or Cdk2ap1ΔN (MT2B2) mRNAs were each electroporated into wildtype pronuclear embryos, and resulted embryos were compared for BrdU incorporation in morula at 3.0 dpc. Representative immunostaining images (left) and quantitation of BrdU positive and total cell number (right) are shown. Violin plots are shown with median (red), as well as lower (25%) and upper (75%) quartiles (black lines). Scale bars, 20 μm. H2b-Gfp vs Cdk2ap1CAN in wildtype embryos: BrdU, **** P < 0.0001, t=4.9, df=119; total cell number, **** P < 0.0001, t=4.1, df=119. H2b-Gfp vs Cdk2ap1ΔN (MT2B2) in wildtype embryos: BrdU+, **** P < 0.0001, t=4.7, df=124; total cell number, *** P = 0.0005, t=3.6, df=124. C. Wildtype embryos overexpressing Cdk2ap1CAN or Cdk2ap1ΔN (MT2B2) were analyzed for ICM (Nanog+) and TE (Cdx2+) cell counts at 4.0 dpc using immunostaining (left). Quantitation is shown as violin plots with median (red line), lower (25%) and upper (75%) quartiles (black lines). H2b-Gfp vs Cdk2ap1CAN: ICM, * P = 0.03, t=2.2, df=49; TE, *P = 0.01, t=2.7, df=49. H2b-Gfp vs Cdk2ap1ΔN (MT2B2), ICM: n.s; TE, * P =0.03, t=2.2, df=40. D. Ectopic expression of Cdk2ap1ΔN (MT2B2) restores S-Phase entry and cell proliferation in Cdk2ap1ΔMT2B2/ΔMT2B2 embryos at 4.0 dpc. H2b-Gfp, Cdk2ap1CAN or Cdk2ap1ΔN (MT2B2) mRNAs were each electroporated into wildtype or Cdk2ap1ΔMT2B2/ΔMT2B2 pronuclear embryos, and resulted embryos were compared for BrdU incorporation in blastocysts at 4.0 dpc. Representative immunostaining images (left) and quantitation of BrdU positive and total cell number (right) are shown. Violin plots are shown with median (red), as well as lower (25%) and upper (75%) quartiles (black lines). Scale bars, 20 μm. H2b-Gfp in WT embryos vs Cdk2ap1ΔN (MT2B2) in Cdk2ap1ΔMT2B2/ΔMT2B2 embryos: BrdU, * P =0.02, t=2.4, df=21; total cell number, ns. H2b-Gfp vs Cdk2ap1CAN in Cdk2ap1ΔMT2B2/ΔMT2B2 embryos: BrdU, ** P =0.001, t=3.9, df=15; total cell number, *** P = 0.0002, t=4.9, df=15. H2b-Gfp vs Cdk2ap1ΔN (MT2B2) in Cdk2ap1ΔMT2B2/ΔMT2B2 embryos: BrdU, * P =0.02, t=2.6, df=13; total cell number, ** P =0.002, t=3.8, df=13. E. Cdk2ap1CAN and Cdk2ap1ΔN (MT2B2) both bind endogenous mouse Cdk2. In HEK293T cells transfected with either C-Terminal HA-Tagged Cdk2ap1CAN (Cdk2ap1CAN–HA) or C-Terminal HA-Tagged Cdk2ap1ΔN (MT2B2) (Cdk2ap1ΔN (MT2B2)-HA), immunoprecipitation of HA pull down endogenous Cdk2. F. Cdk2ap1CAN and Cdk2ap1ΔN (MT2B2) have opposite effects on Cdk2 kinase activity. HEK293T cells overexpressing Cdk2ap1CAN-HA, Cdk2ap1CAN-MutTER-HA, Cdk2ap1ΔN (MT2B2)-HA or Cdk2ap1ΔN (MT2B2)-MutTER-HA were subjected to immunoprecipitation with anti-HA antibodies. Immunoprecipitated lysates were each incubated with recombinant CDK2, CYCLIN E, HISTONE H1 and ATP in vitro. Their effects on CDK2 activity were analyzed in a kinase assay. Error bars are means ± s.e.m. Cdk2ap1CAN -HA vs Cdk2ap1CAN -MutTER-HA, **P = 0.009, t=4.7, df=4; Cdk2ap1ΔN (MT2B2) vs Cdk2ap1ΔN (MT2B2)-MutTER-HA, **P = 0.008, t=4.8, df=4. Three independent experiments were performed. All P values were calculated based on the unpaired two-tailed Student’s t test. n.s., not significant.
Figure S5. Multiple mouse retrotransposon promoters yield N-terminally altered gene isoforms that are conserved in human, related to Figure 5. Gene structure, protein motif analyses and sequence alignment are shown for Pemt (A), Gan14 (B) and Mkln1 (C). For Pemt, the transcriptional start site, as well as the splicing between retrotransposon and gene exon, were experimentally validated by 5’ RACE and RT-PCR.
Figure S6. CDK2AP1ΔN is expressed in human embryonic stem cells, related to Figure 6. A. A promoter from the L2a/Charlie4z region drives a strong expression of an N-terminally truncated CDK2AP1 isoform in human ES cells. Five human CDK2AP1 isoforms were quantified by RNA-seq for expression in H1 embryonic stem cells from ENCODE project (Encode Consortium, 2012). B. A diagram illustrates the approximate durations of preimplantation development in different mammalian species. Arrow, approximate timing of implantation.
Table S1.
RNA-seq analyses of retrotransposon expression in mouse, primate, cow, pig, goat and opossum preimplantation embryos, related to Figure 1 and S1.
Table S2.
RNA-seq analyses of protein-coding gene expression in mouse, primate, cow, pig, goat and opossum preimplantation embryos, related to Figure 1 and S1.
Table S3.
RNA-seq analyses of retrotransposon:gene junction reads in mouse, primate, cow, pig, goat and opossum preimplantation embryos, related to Figure 1 and S1.
Table S4.
Experimental validation of selected retrotransposon:gene isoforms, related to Figure 1, 2, and S5.
Table S5.
ORF and Conservation analyses on retrotransposon:gene isoforms in preimplantation embryos by manual curation, related to Figures 1, 5, S1, and S5
Table S6.
RNA-seq analyses of Cdk2ap1 gene isoforms in mouse, primate, cow, pig, goat and opossum preimplantation embryos, related to Figure 6.
Table S8.
Primer/oligo design for genotyping, CRISPR editing, real time PCR and RACE, related to STAR Methods.
Data Availability Statement
Our studies have employed published RNA-seq data, which are available from the Gene Expression Omnibus, ArrayExpress, or Short Read Archive, at accessions GSE44183, GSE45719, GSE36552, E-MTAB-7078, GSE86938, GSE143850, GSE25415, GSE129742, SRA076823, GSE139512, E-MTAB-7515, GSM733657, GSM646336, GSM748532, GSE23316.
Customized scripts used for quantifying retrotransposon:gene junction reads is publicly available as https://epigenome.wustl.edu/TE_Transcript_Assembly/tool.html.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


