Abstract
An outbreak of shigellosis occurred in a township of Nantou Conuty in central Taiwan from August to October in 1996. The infections extended to two neighboring townships and continued to the end of 1996. Forty cases were confirmed during the period, in contrast to only one confirmed case in Nantou County in 1996 before the outbreak. All of these 41 cases in 1996 were identified as infections with Shigella flexneri serotype 2a. In order to trace the source of the infections, the 41 isolates recovered were analyzed by plasmid profile and pulsed-field gel electrophoresis (PFGE). There was no correlation between the plasmid profile results and the PFGE results, and the latter were used for subtyping of the 41 isolates. Twenty-two isolates (53%) had the same NotI and XbaI PFGE patterns, and 4 isolates (10%) had an additional unstable plasmid band in their NotI patterns but otherwise had the same NotI and XbaI patterns as the 22 isolates. These 26 isolates were designated the outbreak strain, and of these, 24 appeared in eight villages in one township and 2 appeared in a neighboring township. Fourteen of the remaining 15 isolates, including the isolate recovered 7 months before the outbreak, had both NotI and XbaI PFGE patterns closely related to those of the outbreak strain, indicating that Shigella infections were endemic in the area. By tracing the first isolation dates of the outbreak strain in individual villages and the neighboring township, it was found that the strain spread along the major arterial road and its branch road as time passed. Our molecular typing results and epidemiological data demonstrated the endemic nature of the outbreak strain as well as a person-to-person mode of transmission for the widespread infections the strain caused.
Shigellosis is an acute gastroenteritis caused by Shigella species, including Shigella dysenteriae, Shigella flexneri, Shigella boydii, and Shigella sonnei. It is one of the most common causes of morbidity and mortality in children with diarrhea in developing countries. Worldwide, about 1,100,000 deaths are caused by the disease per year, and two-thirds of the patients are children under 5 years of age (5). The disease is highly contagious due to its low infection dose (3). Epidemics usually occur in areas with crowding and poor sanitary conditions, where transmission from person to person is common, or when food or water is contaminated by the organism (5, 9, 15, 17). In Taiwan, about 250 to 550 cases of shigellosis were identified annually during the years from 1995 to 1999, with an average annual incidence rate of 1 to 3 cases per 100,000 persons. The infections are caused mostly by S. flexneri and S. sonnei, whereas infections caused by S. dysenteriae and S. boydii are rare and seen only in cases of imported disease (13).
The Central Mountain Range in the middle of Taiwan geographically separate the east of the island from the west. Sporadic cases of shigellosis, caused mostly by S. flexneri, appear in some mountainous townships of central and eastern Taiwan, while point-source outbreaks caused by S. flexneri and S. sonnei occur occasionally at schools, institutions, and communities in industrial western Taiwan. During August through October in 1996, an outbreak of shigellosis occurred in a mountainous township of Nantou County in central Taiwan and the infections continued to the end of the year. Cases, all identified as infections with S. flexneri serotype 2a, were widely distributed in villages of the township as well as two neighboring townships. In this study, isolates recovered from Nantou County in 1996 were collected and analyzed by plasmid profile and pulsed-field gel electrophoresis (PFGE). The PFGE patterns rather than the plasmid profiles were used for subtyping of the isolates such that an outbreak strain and its mode of transmission were defined.
MATERIALS AND METHODS
Bacterial isolates.
During 1995 through 1997, stool specimens from patients with either diarrhea or dysentery (bloody diarrhea) and their asymptomatic contacts in Nantou county were collected and screened for Shigella species by conventional biochemical methods (10) in two local hospitals (Puli Christian Hospital and Puli Veteran Hospital) and C.-S. Chiou's laboratory. The contacts included family members, close relatives, close friends, neighbors, and classmates of the patients. Shigella species recovered were serotyped in C.-S. Chiou's laboratory by a slide agglutination test with commercial polyclonal antiserum (Denka Seiken Co. LTD, Tokyo, Japan). The 41 isolates recovered in 1996, all identified as S. flexneri serotype 2a, and 4 geographically unrelated S. flexneri 2a strains were chosen for further PFGE and plasmid profile analyses. The four geographically unrelated strains were recovered separately from Hwalien County (one strain) and Taitung County (two strains) during June and July in 1996. These were obtained from the Sixth Branch Office of the Center for Disease Control in Taiwan. A fourth strain was purchased from the American Type Culture Center (ATCC; Manassas, Va.). Sources and characteristics of these 45 isolates are described in Table 1. Bacterial isolates were cultured in Luria broth (LB) and stored in 15% glycerol at −70°C.
TABLE 1.
Sources, plasmid profiles, and PFGE patternsa of the 45 Shigella isolates
| Isolateb | Isolation date | Source of isolation
|
Plasmid profile | PFGE patternc with enzyme:
|
||
|---|---|---|---|---|---|---|
| County/township | Village | NotI | XbaI | |||
| Related isolates | ||||||
| SH2182d | August 12 | Nantou/Renai | Tsunyang | I | N1 | X1 |
| SH2214d | August 19 | Nantou/Renai | Tsunyang | I | N1 | X1 |
| SH2302 | August 27 | Nantou/Renai | Tsunyang | I | N1 | X1 |
| SH2308e, g | September 4 | Nantou/Renai | Tsunyang | I | N1 | X1 |
| SH2557e | September 4 | Nantou/Renai | Tsunyang | I | N1(1) (A) | X1 |
| (SH2343)d | September 5 | Nantou/Renai | Tsunyang | I | N1(1) (A) | X1 |
| SH3160 | September 12 | Nantou/Renai | Tsunyang | I | N1 | X1 |
| SH3896 | August 18 | Nantou/Renai | Chingying | I | N1 | X1 |
| SH2229 | August 20 | Nantou/Renai | Chingying | I | N1 | X1 |
| SH2291 | August 31 | Nantou/Renai | Chingying | I | N1 | X1 |
| (SH3094)g | September 14 | Nantou/Renai | Chingying | I | N1 | X1 |
| SH2286 | August 29 | Nantou/Renai | Fahsiang | I | N1 | X1 |
| SH4347f | October 18 | Nantou/Renai | Fahsiang | I | N1 | X1 |
| SH4798f | October 31 | Nantou/Renai | Fahsiang | I | N1(1) (A) | X1 |
| SH2372d, f | September 5 | Nantou/Renai | Chinai | I | N1 | X1 |
| (SH2371)d | September 5 | Nantou/Renai | Chinai | I | N1 | X1 |
| (SH2374)f | September 5 | Nantou/Renai | Chinai | I | N1 | X1 |
| SH4377e | October 23 | Nantou/Renai | Chinai | I | N1 | X1 |
| (SH2585)e | September 10 | Nantou/Renai | Nanfeng | I | N1 | X1 |
| SH4029 | September 25 | Nantou/Renai | Wanfeng | I | N1 | X1 |
| SH4232 | October 12 | Nantou/Renai | Lihsing | I | N1 | X1 |
| SH4217 | October 14 | Nantou/Renai | Lihsing | I | N1 | X1 |
| SH4332 | October 24 | Nantou/Renai | Lihsing | I | N1 | X1 |
| SH4418e | October 31 | Nantou/Renai | Huchu | I | N1 | X1 |
| SH4520 | November 14 | Nantou/Puli | I | N1 | X1 | |
| SH4785 | December 31 | Nantou/Puli | I | N1(1) (A) | X1 | |
| SH4799 | December 30 | Nantou/Renai | Fahsiang | I | N1 | X13 |
| SH1105 | January 3 | Nantou/Puli | IV | N13 | X1 | |
| SH2558 | September 7 | Nantou/Puli | I | N12 (B) | X1 | |
| SH2594 | September 9 | Nantou/Renai | Hotso | I | N12 (B) | X1 |
| SH3364 | September 19 | Nantou/Hsini | I | N12 (B) | X1 | |
| SH2590e | September 4 | Nantou/Renai | Tsunyang | I | N11 | X11 |
| SH2683 | September 13 | Nantou/Renai | Chinai | V | N13 | X12 (C) |
| SH2953 | September 14 | Nantou/Renai | Nanfeng | I | N13 | X12 (C) |
| SH2576 | September 5 | Nantou/Renai | Tsunyang | I | N14 (D) | X12 (E) |
| SH3010 | September 13 | Nantou/Renai | Tsunyang | I | N14 (D) | X12 (E) |
| SH3006e | September 13 | Nantou/Renai | Tsunyang | I | N14 (D) | X12 (E) |
| SH2955e | September 14 | Nantou/Renai | Tsunyang | I | N14 (D) | X12 (E) |
| SH3162 | September 13 | Nantou/Renai | Nanfeng | I | N14 | X12 |
| SH3151 | September 12 | Nantou/Hsini | I | N14 | X13 | |
| Unrelated isolates | ||||||
| SH2276 | August 23 | Nantou/Renai | Tatung | I | N2 | X2 |
| SH46993 | July 29 | Taitung | I | N23 | X3 | |
| SH46949 | June 22 | Taitung | III | N3 | X4 | |
| SH46959 | July 9 | Hualian | II | N4 | X5 | |
| ATCC 29903 | II | N5 | X6 | |||
Each NotI and XbaI PFGE main pattern was assigned a letter and number designation (N1 to N5 and X1 to X6). Isolates having patterns differing from these main patterns (main types) by one to three bands were considered closely related and assigned into subtypes of the main type, designated by subscripts (A11, A12, A23, etc.). Subscript 1 (without parentheses) represents addition of one band to the pattern, whereas subscript 1 with parentheses represents addition of one band to the pattern but the added band is relatively weak. Subscript 2 represents deletion of one band from the pattern, subscript 3 represents a two-band difference, and subscript 4 represents a three-band difference from the main pattern (7).
Isolates without parentheses were recovered from patients with symptoms of diarrhea or dysentery, and isolates with parentheses were recovered from asymptomatic contacts.
Patterns with the same letters beside them in parentheses represent identical PFGE patterns.
Isolates were recovered from members of two separate families: (i) SH2182, SH2214, and SH2343 and (ii) SH2372 and SH2371.
Isolates were recovered from close relatives and friends, which included three separate groups: (i) SH2308, SH2557, SH2585, and SH2590; (ii) SH4377 and SH4418; and (iii) SH3006 and SH2955.
Isolates were recovered from separate next-door neighbors living at two different locations: (i) SH4347 and SH4798 and (ii) SH2372 and SH2374.
Isolates were recovered from classmates.
Plasmid profile analysis.
Lysates of Shigella isolates were prepared by the rapid alkaline lysis procedure described by Kado and Liu (4). Plasmids were fractionated in 0.7% agarose gel, and supercoiled DNA ladders (Life Technologies, GIBCO BRL, Gaithersburg, Md.) were used as size standards. DNA bands were visualized by ethidium bromide staining and with UV and photographed.
PFGE of total genomic DNA.
Genomic DNA for PFGE was prepared in agarose plugs using the method described by Soldati and Piffaretti (16). Slices of agarose plugs were digested with 20 U of NotI or XbaI for 20 h, and electrophoresis was carried out in 0.8% agarose gel with a Rotaphor type V apparatus (Biometra, Göttingen, Germany) in 0.5× TBE buffer (0.045 M Tris-borate, 0.1 mM EDTA [pH 8.3]) at 13°C. The NotI-digested DNA fragments were separated by ramped switches of pulse time logarithmically from 60 to 10 s, of angle linearly from 120 to 110°, and of pulse logarithmically from 200 to 150 V (program 8) for 18 h. This was followed by ramped switches of pulse time logarithmically from 100 to 10 s, of angle linearly from 120 to 110°, and of pulse logarithmically from 200 to 150 V (program 9) for 10 h. The XbaI-digested DNA fragments were separated by ramped switches of pulse time logarithmically from 12 to 2 s, of angle linearly from 120 to 110°, and of pulse logarithmically from 180 to 120 V (program 5) for 21 h. This was followed by ramped switches of pulse time logarithmically from 30 to 5 s, of angle linearly from 120 to 110°, and of pulse logarithmically from 180 to 150 V (program 6) for 23 h. Yeast chromosomal DNA and ladders of phage λ DNA concatemers (New England BioLabs, Inc., Beverly, Mass.) were used as size standards. DNA bands were visualized by ethidium bromide staining and with UV and photographed.
PFGE pattern analysis.
PFGE patterns were analyzed by visual inspection of the photographs of the stained gels, and an index isolate was included in each photograph for comparison of isolates in separate photographs. NotI digests produced PFGE patterns of 15 to 19 DNA bands between 1,150 and 50 kb, while XbaI digests produced PFGE patterns with 20 to 24 DNA bands between 350 and 40 kb. PFGE patterns were classified according to the work of Luna et al. (7). When four or more bands in the PFGE patterns were different from each other, they were designated main types and given a name with the first letter of the enzyme and an assigned number (N1 to N5 for NotI and X1 to X6 for XbaI). When three or fewer bands in the PFGE patterns were different from those of the main type, they were designated subtypes and given a subscripted number (N11, N12, X11, etc.). Subtypes are considered to be related to the main types, as the band difference could be explained by one genetic event (18). PFGE patterns that had four or more different bands were considered unrelated. Isolates that had identical or related (with three or fewer different bands) NotI and XbaI PFGE patterns were considered to be genetically related, whereas isolates that had unrelated (with four or more different bands) NotI, XbaI, or both NotI and XbaI patterns were considered to be genetically unrelated. This classification is more stringent than the criteria suggested by Tenover et al. (18) and has been used in subtyping of Streptococcus pneumoniae and Neisseria gonorrhoeae (7, 14, 19).
Collection of the first to the seventh subcultures of the isolates.
Fresh colonies of Shigella isolates were obtained by streaking the −70°C stock onto LB plates. One fresh colony of an isolate was inoculated into a tube of 5 ml of LB and grown at 37°C overnight, which was designated the first subculture. One-one hundredth volume of the first subculture was inoculated into another tube of 5 ml of LB and grown at 37°C overnight, which was designated the second subculture. This process was repeated five times, and the third to the seventh subcultures were obtained.
Epidemiological data.
The epidemiological data of the patients were obtained from standardized case report forms filled in by the county public health authorities. The reports included basic information of the patients such as date of onset, sex, age, residency, symptoms, medical treatment, and travel history. In some cases, patients were interviewed to find out connections with other patients and contacts.
Estimation of similarity among isolates and construction of a dendrogram.
Genetic similarities between pairs of isolates were calculated using Nei and Li's F statistic (12) (also known as the Dice coefficient or coefficient of similarity) by the equation F = 2Nij/(Ni + Nj), where Ni is the total number of DNA bands from isolate i, and Nj is the total number of DNA bands for isolate j, and Nij is the number of fragments identical in the two isolates. A matrix of F values for all pairs of isolates was constructed and used for construction of a dendrogram by the unweighted pair group method using arithmetic averages program of NTSYS-PC software (Numerical Taxonomy and Multivariate Analysis System, version 1.50) from Applied Biostatistics Inc. (Setauket, N.Y.).
RESULTS
Cases of shigellosis in 1995, 1996, and 1997.
Bacteria of Shigella spp. were recovered from stool specimens of patients and their contacts in Nantou County from 1995 through 1997 for confirmation of shigellosis. As shown in Table 2, the numbers of confirmed cases in 1995, 1996, and 1997 were 8, 41, and 59, respectively. Except for one case in 1997 that appeared in Nantou City, all other cases appeared in the Renai, Puli, Hsini, and Yuchr townships. The Hsini, Yuchr, and Puli townships are located adjacent to the Renai township. Figure 1 indicates the geographic relationship of the four townships. Of the eight cases in 1995, four appeared in the Renai, three appeared in the Hsini, and 1 appeared in the Yuchr township. These eight cases occurred between April and December and were identified as S. flexneri infections with serotypes 2a, 3a, and y. Of the 41 cases in 1996, 35, 4, and 2 cases appeared in the Renai, Puli, and Hsini townships, respectively, and all were identified as S. flexneri 2a infections. Thirty-four of the 35 cases in Renai Township appeared in August to October, indicating that an outbreak occurred in that township during this period in 1996. The infections continued into 1997, with 38, 14, 4, and 2 cases, respectively, in the Renai, Puli, Hsini, and Yuchr townships and 1 case in Nantou City. Eight of the 14 cases in Puli Township, 2 of the 4 cases in Hsini Township, and each of the 2 cases in Yuchr Township appeared in the first 3 months of 1997 and were also identified as S. flexeria 2a infections.
TABLE 2.
Numbers and serotypes of Shigella species recovered monthly from townships of Nantou County in 1995, 1996, and 1997a
| Month | No. of Shigella isolates recovered in indicated township or city in:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1995
|
1996
|
1997
|
|||||||||
| Renai | Hsini | Yuchr | Renai | Puli | Hsini | Renai | Puli | Hsini | Yuchr | Nantou | |
| January | 1 | 2 | 3 | ||||||||
| February | 1 | 2 | 1 | 1 | |||||||
| March | 2 (1 was 3b) | 3 | 1 | ||||||||
| April | 1c | 2 (1 was 3b) | |||||||||
| May | 6 | 2 (Ss) | |||||||||
| June | 1 (y) | 6 | 1 | ||||||||
| July | 4 | 1 (Sb) | |||||||||
| August | 2 | 1 | 8 | 2 | 1 (Ss) | ||||||
| September | 1 (3a) | 19 | 1 | 2 | 6 (2 were Ss) | 2 (1 was Ss) | |||||
| October | 7 | 3 | |||||||||
| November | 1 | 5 | 1 | ||||||||
| December | 1 (y)d | 1 | 1 | 1 | 1 (Ss) | ||||||
| Total | 4 | 3 | 1 | 35 | 4 | 2 | 38 | 14 | 4 | 2 | 1 |
S. flexneri isolates with serotypes other than 2a are indicated with “3a”, “3b”, or “y” in parentheses; species of S. boydii and S. sonnei are also indicated in parentheses with “Sb” and “Ss,” respectively. The isolate of S. boydii was recovered from a patient infected with an imported isolate. Eight isolates were recovered in 1995, 41 isolates were recovered in 1996, and 59 isolates were recovered in 1997.
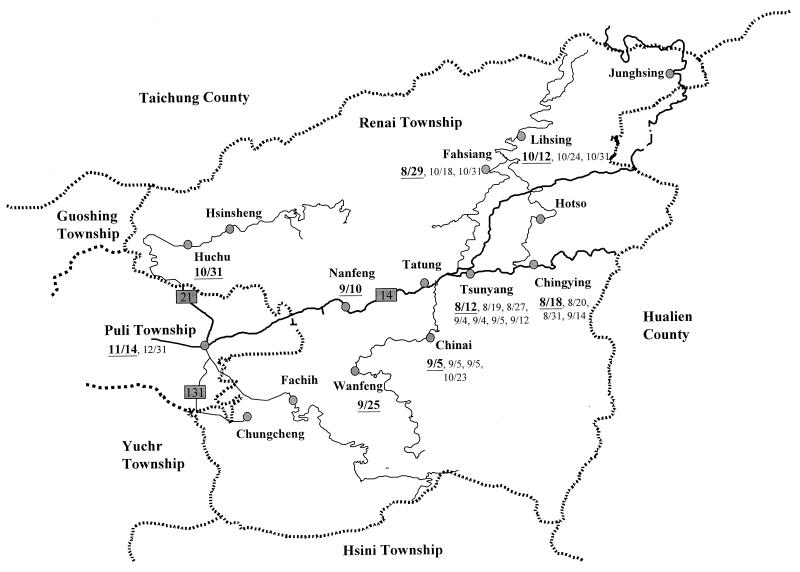
FIG. 1.
Map of 14 villages in Renai Township and the neighboring townships depicting the distribution of the S. flexneri 2a outbreak strain. The dates when an isolate of the outbreak strain was recovered from a specific village or township are indicated next to the villages or township, with the first date being in boldface and underlined. Dates are shown as month/day. Continuous lines represent the road systems, thick lines represent the main road, and thin lines represent branch roads.
Epidemiological data of the 41 cases in 1996.
The 41 cases in 1996 were selected for further investigation. The subjects included 27 females and 14 males who were either without any symptoms (contacts) or with symptoms of watery diarrhea or bloody diarrhea (dysentery). Table 3 shows the age distribution of the 41 cases. It was found that 51% (21 of 41 cases) occurred in children under the age of 12 years, with 34% (14 of 41 cases) occurring in children of preschool age (<7 years). The five contacts that did not develop any symptoms were aged 2, 8, 11, 11, and 39 years (data not shown). As shown in Table 1, 19 (46%) of the 41 cases had epidemiological links. Among them, three, four, two, two, two, two, two, and two cases occurred separately in family members, close relatives, family members, neighbors, classmates, neighbors, close friends, and close relatives, respectively.
TABLE 3.
Age distribution of the 41 patients of Nantou County in 1996a
| Age group (yr) | No. of males infected | No. of females infected | Total |
|---|---|---|---|
| 0–6 | 6 (5) | 8 (7) | 14 (12) |
| 7–12 | 2 | 5 (4) | 7 (4) |
| 13–24 | 1 | 2 (1) | 3 (1) |
| 25–36 | 0 | 1 | 1 |
| 37–48 | 0 | 2 (2) | 2 (2) |
| 49–60 | 0 | 4 (2) | 4 (2) |
| 61–72 | 1 (1) | 5 (3) | 6 (4) |
| 73–80 | 3 | 0 | 3 |
| 81 | 1 (1) | 0 | 1 (1) |
| Total | 14 (7) | 27 (19) | 41 (26) |
Numbers in parentheses are numbers of patients of the indicated age groups infected with the outbreak strain.
Plasmid profiles.
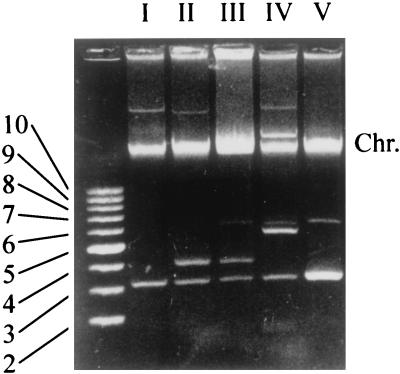
Plasmid profile analysis was carried out to subtype the 41 S. flexneri 2a isolates recovered from Nantou County in 1996. Four geographically unrelated flexneri 2a strains, i.e., three eastern Taiwan strains and one ATCC strain, were analyzed together for comparison. Because large plasmids tend to be lost during cell storage and subculturing or plasmid extractions, only plasmid bands below the chromosomal DNA band were taken into account for profile assignment. The results are shown in Table 1 and Fig. 2. The 45 isolates each contained one to three small plasmids ranging from 3.2 to 7 kb with a 3.2-kb plasmid in common, and a total of five plasmid profiles were identified. Of the 41 isolates recovered from Nantou County, 39 had plasmid profile I, one had profile IV, and the other one had profile V. The three eastern Taiwan strains separately had profiles I, II, and III, and the ATCC strain (ATCC 29903) had profile II.
FIG. 2.
Plasmid profiles of the 45 S. flexneri 2a isolates. Only plasmid profiles of representative isolates are shown, and lanes are labeled I to V, corresponding to the plasmid profile. The leftmost lane contains supercoiled DNA size standards, and numbers at left indicate the sizes of the corresponding plasmids in kilobases. Chr., chromosomal DNA.
PFGE analysis of the 45 isolates.
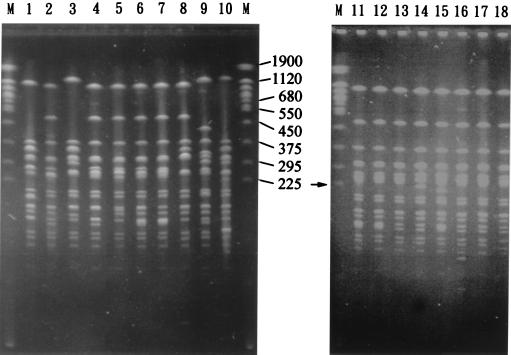
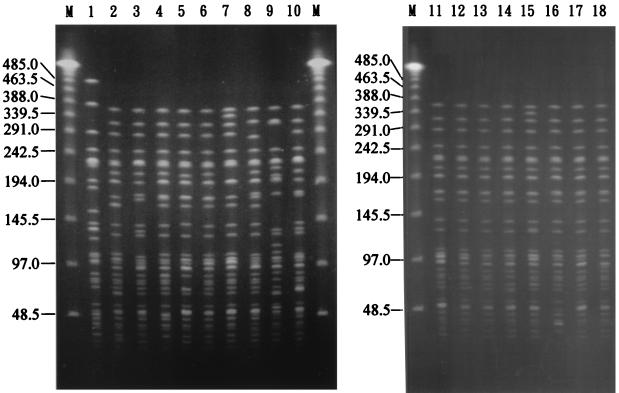
Genomic DNAs of the 45 S. flexneri 2a isolates were restricted by NotI or XbaI and analyzed by PFGE. As shown in Fig. 3 and 4 and Table 1, five NotI PFGE types (N1 to N5), six XbaI PFGE types (X1 to X6), and subtypes of N1, N2, and X1 were identified. Twenty-two isolates of Nantou County had identical NotI and XbaI PFGE patterns (the N1X1 pattern). Compared with these 22 isolates with the N1X1 pattern, 4 isolates (SH2343, SH2557, SH4785, and SH4798) had an additional relatively weak 225-kb band in their NotI patterns and an identical XbaI pattern (the N1(1)X1 pattern), 1 isolate (SH4799) had an identical NotI pattern and a difference of two bands in its XbaI pattern (the N1X1(3) pattern), and 13 isolates had differences of three or fewer bands in both their NotI and XbaI patterns. All of these 40 isolates were recovered from Nantou County and are considered to be closely related genetically. The one remaining isolate (SH2276) from Nantou County, the three eastern Taiwan isolates, and the ATCC isolate each had a difference of more than three bands among their NotI and/or XbaI patterns, and each also had a difference of more than three bands from the N1 and X1 patterns. They each represented a NotI-XbaI PFGE type and were considered genetically unrelated to each other and to the 40 isolates from Nantou County.
FIG. 3.
PFGE patterns of NotI-digested genomic DNAs of the 45 S. flexneri 2a isolates. Only NotI patterns from representative isolates are shown. Lane 1, ATCC 29903 (the N5 pattern); lane 2, SH1105 (the N13 pattern); lane 3, SH2276 (the N2 pattern); lane 4, SH2683 (the N13 pattern); lane 5, SH3151 (the N14 pattern); lane 6, SH4232 (the N1 pattern); lane 7, SH4799 (the N1 pattern); lane 8, SH46949 (the N3 pattern); lane 9, SH46959 (the N4 pattern); lane 10, SH46993 (the N23 pattern); lane 11, SH2343 (the N1(1) pattern); lane 12, SH2182 (the N1 pattern); lane 13, SH2558 (the N12 pattern); lane 14, SH2576 (the N14 pattern); lane 15, SH2590 (the N11 pattern); lane 16, SH2953 (the N13 pattern); lane 17, SH2594 (the N12 pattern); lane 18, SH3162 (the N14 pattern). Lanes M contain yeast chromosomes that were used as molecular size standards, with sizes of selected chromosomes indicated in kilobases. The arrow indicates the 225-kb band in lane 11.
FIG. 4.
PFGE patterns of XbaI-digested genomic DNAs of the 45 S. flexneri 2a isolates. Patterns from the representative isolates with their NotI patterns shown in Fig. 3 are shown in the same order here. Lane 1, ATCC 29903 (the X6 pattern); lane 2, SH1105 (the X1 pattern); lane 3, SH2276 (the X2 pattern); lane 4, SH2683 (the X12 pattern); lane 5, SH3151 (the X13 pattern); lane 6, SH4232 (the X1 pattern); lane 7, SH4799 (the X13 pattern); lane 8, SH46949 (the X4 pattern); lane 9, SH46959 (the X5 pattern); lane 10, SH46993 (the X3 pattern); lane 11, SH2343 (the X1 pattern); lane 12, SH2182 (the X1 pattern); lane 13, SH2558 (the X1 pattern); lane 14, SH2576 (the X12 pattern); lane 15, SH2590 (the X11 pattern); lane 16, SH2953 (the X12 pattern); lane 17, SH2594 (the X1 pattern); lane 18, SH3162 (the X12 pattern). Lanes M contain concatemers of bacteriophage λ DNA that were used as molecular size standards, with sizes of selected λ DNA concatemers indicated in kilobases.
Designation of the outbreak strain.
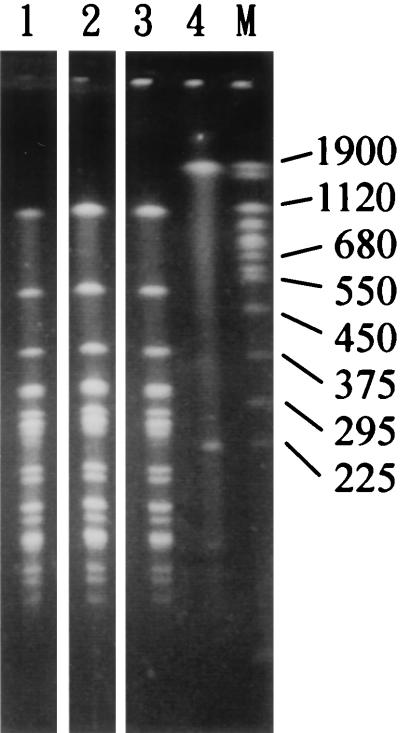
For the four isolates of the N1(1)X1 pattern (SH2343, SH2557, SH4785, and SH4798), it was found that the intensities of the 225-kb band in their NotI patterns were not only relatively weak but also variable (data not shown). It was hypothesized that the 225-kb band might represent an endogenous unstable plasmid. To investigate this, PFGE was carried out with both undigested and NotI-digested genomic DNAs of an isolate of the N1(1)X1 pattern (SH4798), and the result is shown in Fig. 5. Two bands were observed with the undigested DNA of SH4798, a 1,900-kb band and a 225-kb band, and only a 1,900-kb band was observed with undigested DNA of an isolate of the N1X1 pattern (SH2308). While the 1,900-kb band might represent the chromosomal DNA, the 225-kb band probably represents an endogenous plasmid.
FIG. 5.
PFGE of NotI-digested DNAs of isolates of the N1 and the N1(1) patterns and undigested DNA of an isolate of the N1(1) pattern. Lane 1, NotI-digested DNA of SH4798 (the N1(1) pattern); lane 2, NotI-digested DNA of SH2308 (the N1 pattern); lane 3, NotI-digested DNA of SH2343 (the N1(1) pattern); lane 4, undigested DNA of SH2343 (the N1(1) pattern). Lane M contains yeast chromosomes that were used as molecular size standards, with sizes of selected chromosomes indicated in kilobases.
Two isolates of the N1(1)X1 pattern (SH2343 and SH4798) and one isolate of the N1X1 pattern (SH2308) were subcultured for seven consecutive days and examined by PFGE for the presence of the 225-kb band. In addition, the seventh subculture from each of the three isolates was streaked on an LB plate and a single colony was picked for inoculation into a tube of LB. For each isolate, PFGE was performed with the undigested genomic DNAs of the first, third, and seventh subcultures and the overnight culture of a single colony derived from the seventh subculture. The results indicated that the 225-kb plasmid band was seen only with the first and the third subcultures of SH2343 and SH4798, with the intensities of the third subcultures being much lower than that of the first subcultures. All of the subcultures of SH2308, the seventh subcultures of SH2343 and SH4798, and the overnight cultures of the single colonies derived from the seventh subcultures of the three isolates did not show the 225-kb plasmid band by PFGE (data not shown). Thus, isolates of the N1(1)X1 pattern could transform into isolates of the N1X1 pattern through fewer than seven subcultures. Isolates of both patterns were considered clones of the same strain. Since this strain accounted for 63% (26 of 41 isolates) of the isolates recovered from Nantou County and was distributed widely into eight villages and two townships (Table 1), it was designated the outbreak strain according to the criteria of Tenover et al. (18).
Genetic similarity.
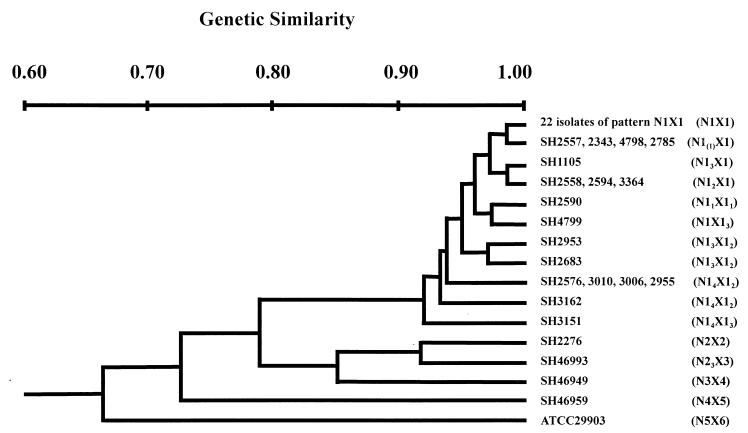
The genetic similarities of the 45 isolates based on their PFGE patterns were calculated and are represented by the dendrogram shown in Fig. 6. Five clusters were generated at an F value of <0.9, corresponding to a genetic distance of less than 90% similarity between clusters and greater than 90% within clusters. All of the 41 isolates, except SH2276, from Nantou County were within the largest cluster. Isolate SH2276 and an isolate from Taitung County in eastern Taiwan (SH46993) formed another cluster. The other two eastern Taiwan isolates, one from Taitung County and the other from Hualian County, and the ATCC strain each formed a distinct cluster.
FIG. 6.
Dendrogram of the 45 S. flexneri 2a isolates based on their NotI and XbaI PFGE patterns. The NotI and XbaI patterns of the isolates are indicated in parentheses.
Geographical distribution of the 41 isolates.
The 41 isolates of Nantou County appeared in 11 of the 14 villages in Renai Township and the neighboring Puli and Hsini townships (Table 1). Of these, 26 isolates belonged to the outbreak strain and appeared in eight villages (24 isolates) and Puli Township (2 isolates). The remaining 15 isolates, 14 of which were closely related to the outbreak strain, appeared in seven villages and the Puli and Hsini townships. Of these, four isolates (SH2576, SH3010, SH3006, and SH2955) and three isolates (SH2558, SH2594, and SH3364) separately had identical PFGE patterns, but the former four isolates appeared in the same village (Tsunyang Village) and the latter three isolates appeared in three separate locations (Hotso Village and the Puli and Hsini townships).
The isolation dates of the 26 isolates of the outbreak strain as well as the locations where they appeared are depicted on a map of the Renai Township, which is shown in Fig. 1. The outbreak strain first appeared in Tsunyang Village on August 12 and then appeared in Chingying Village on August 18. Tsunyang Village had the most cases of infections (n = 7) with the outbreak strain. It is also the place through which residents of Chingying Village have to pass on their way to Puli Township. Subsequently, the outbreak strain sequentially appeared in the Fahsiang, Chinai, Nanfeng, Wanfeng, Lihsing, and Huchu villages (on 29 August, 5 September, 10 September, 25 September, 12 October, and 31 October, respectively) and finally in Puli Township on 14 November (Fig. 1). Except for Huchu, all of these villages are situated on Road 14 and its branch road. Huchu Village is located on Road 21. The isolate that appeared in Huchu Village (SH4418) was recovered from a patient who became infected while taking care of her grandson in Chinai Village. An isolate of the outbreak strain was also recovered from her grandson on 23 October (SH4377) (Table 1).
DISCUSSION
Renai Township occupies 1,273 km2 and has 15,000 people residing in 14 villages (Fig. 1). While only 8% of the households use tap water, 92% of the households use water piped directly from mountain springs. Almost all of the households have toilet facilities. Before August 1996, sporadic cases of shigellosis were identified in the township. From August through October in 1996, an outbreak of shigellosis occurred in the township and infections continued to the end of the year. Ten villages of the township and two neighboring townships were affected, and S. flexneri serotype 2a was recovered in all confirmed cases (Table 1).
For the 41 confirmed cases in Renai Township and the two neighboring townships in 1996, over half (52%) of the infections occurred in children under the age of 12 years and 34% of the infections occurred in children of preschool age (<7 years). Similar results were reported from a survey of Shigella infections in the United States from 1974 to 1980, in which the age group with the highest rate of infection comprised children under 5 years of age (2). To find out whether a single strain or multiple strains were responsible for the infections and to trace their sources, the 41 isolates recovered were analyzed by two typing procedures, as with the analyses of others (1, 6, 16, 20). Three eastern Taiwan strains and one ATCC strain were analyzed for comparison. Plasmid typing with the 45 isolates identified five plasmid profiles (profiles I to V) with limited heterogeneity (Table 1 and Fig. 2). Since 95% (39 of 41 isolates) of the isolates from Nantou County had the same plasmid profile (profile I) and since the same plasmid profile (profile II) was found for the isolate from Hualian County as for the ATCC strain, plasmid typing appeared to be inappropriate for subtyping these isolates for epidemiological studies.
PFGE has a high discriminatory power and has been suggested as a preferred typing technique for Shigella spp. (1, 16). Our results of PFGE typing showed five NotI and six XbaI PFGE types and subtypes of two NotI types and one XbaI type for the 45 isolates. Unlike the plasmid profile results, the PFGE patterns correlated well with the geographical locations of the isolates (Table 1 and Fig. 4). It was found that 40 isolates of plasmid profile I belonged to two NotI types and 2 isolates of plasmid profile II belonged to two XbaI PFGE types. On the other hand, isolates of the same NotI and XbaI PFGE type could have different plasmid profiles (e.g., SH1105, SH2683, and isolates of the N1X1 type) (Table 1). It is concluded that there is no correlation between PFGE type and plasmid profile for these S. flexneri isolates. Similar results were obtained in another study of S. flexneri isolates by Navia et al. (11) and a study of Legionella pneumophila isolates by Marrie et al. (8).
Tenover et al. (18) defined isolates with differences of three or fewer bands in their PFGE patterns with one restriction enzyme to be closely related, while they considered those with differences of four to five bands to be possibly related. A more stringent definition for closely related isolates has been used by other investigators (7, 14, 19) and by us in our study, where closely related isolates have three or fewer bands in their PFGE patterns with each of two restriction enzymes or with at least two of the three restriction enzymes. Arbeit (1) defined genetically related isolates as those with a similarity of ≥90% in their restriction fragment length polymorphism (PFGE) patterns. Clustering of the 45 isolates by both the NotI PFGE and XbaI PFGE patterns revealed five clusters at a genetic distance of ≥90% within clusters. Forty isolates were placed in one large cluster. We found that these were the same 40 isolates that could also be defined to be closely related by either NotI PFGE or XbaI PFGE alone or by both NotI and XbaI PFGE (Table 1 and Fig. 4). However, an inconclusive result was obtained with one Nantou County isolate (SH2276) and an eastern Taiwan isolate (SH46993). Both isolates were defined to be closely related by either clustering or by a difference of three bands in their NotI patterns. However, they could be defined to be possibly related by a difference of four bands in their XbaI patterns and yet unrelated by their NotI and XbaI patterns, as indicated in Table 1. The patient with the SH2276 infection had no history of traveling outside of Nantou County for at least 1 year before and after the infection. Epidemiological linkage of SH2276 with the eastern Taiwan isolate is questionable. At this point, we consider the two isolates to be unrelated.
One strain (SH1105) isolated 7 months before the outbreak was closely related to the outbreak strain (Table 1). Therefore, except for one isolate (SH2276), 39 of the 40 isolates recovered during and after the outbreak were indigenous and circulated in the area where S. flexneri infection is endemic. The outbreak strain may be among the most adaptive strains and/or the most virulent strain in its tendency to spread (Table 1).
The outbreak strain caused 26 (63%) of the 41 infections identified, which extended to eight villages of Renai Township and Puli Township. Apart from the 26 infections, another four and three infections were also likely to have been caused by the same strains, as they were found to have identical NotI and XbaI PFGE patterns. Fifteen infections caused by the outbreak strain had epidemiological links, as did one infection caused by the outbreak strain, one infection caused by a closely related isolate, and two infections caused by two closely related isolates (Table 1). These isolates thus were closely related not only genetically but also epidemiologically. The infections mostly occurred from August to October 1996. During July and August 1996, two typhoons hit central Taiwan in succession and destroyed many water supply facilities in Renai Township. It took 2 months to repair the facilities. We suggest that the Shigella strains had been dormant in the community and that the deterioration of sanitary and hygiene conditions favored the spread of the organism, resulting in the outbreak.
Our results indicate that the outbreak strain spread among villages of Renai Township and the neighboring Puli Township along Road 14 and its branch road as time passed. Road 14 is the only road system that connects most of the villages and Puli Township which residents of Renai Township frequently visit. It is likely that the outbreak strain spread via a person-to-person mode of transmission. Our study demonstrates that personal contact was responsible not only for small-scale transmission in households or communities, but also for transmission over a much wider geographic area.
ACKNOWLEDGMENTS
This work was supported by research grant DOH-86-DC-023 from the Department of Health.
We thank staff from the Renai Township Health Station for assistance in the epidemiological investigation, from Puli Christian Hospital and Puli Veteran Hospital for providing Shigella isolates, and from the Sixth Branch Office of the Center for Disease Control for providing three eastern Taiwan Shigella isolates as reference strains.
REFERENCES
- 1.Arbeit R D. Laboratory procedures for the epidemiologic analysis of microorganisms. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: American Society for Microbiology; 1999. pp. 116–137. [Google Scholar]
- 2.Blaser M J, Pollard R A, Feldman R A. Shigella infections in the United States, 1974–1980. J Infect Dis. 1983;147:771–775. doi: 10.1093/infdis/147.4.771. [DOI] [PubMed] [Google Scholar]
- 3.DuPont H L, Levine M M, Hornick R B, Formal S B. Inoculum size in shigellosis and implications for expected mode of transmission. J Infect Dis. 1989;159:1126–1128. doi: 10.1093/infdis/159.6.1126. [DOI] [PubMed] [Google Scholar]
- 4.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotloff K L, Winickoff J P, Ivanoff B, Clemens J D, Swerdlow D L, Sansonetti P J, Adak G K, Levine M M. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull W H O. 1999;77:651–666. [PMC free article] [PubMed] [Google Scholar]
- 6.Litwin C M, Leonard R B, Carroll K C, Drummond W K, Pavia A T. Characterization of endemic strains of Shigella sonnei by use of plasmid DNA analysis and pulsed-field gel electrophoresis to detect patterns of transmission. J Infect Dis. 1997;175:864–870. doi: 10.1086/513983. [DOI] [PubMed] [Google Scholar]
- 7.Luna V A, Jernigan D B, Tice A, Kellner J D, Roberts M C. A novel multiresistant Streptococcus pneumoniae serogroup 19 clone from Washington state identified by pulsed-field gel electrophoresis and restriction fragment length patterns. J Clin Microbiol. 2000;38:1575–1580. doi: 10.1128/jcm.38.4.1575-1580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marrie T J, Tyler S, Bezanson G, Dendy C, Johnson W. Analysis of Legionella pneumophila serogroup 1 isolates by pulsed-field gel electrophoresis. J Clin Microbiol. 1999;37:251–254. doi: 10.1128/jcm.37.1.251-254.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin D L, Gustafson T L, Pelosi J W, Suarez L, Pierce G V. Contaminated produce, a common source for two outbreaks of Shigella gastroenteritis. Am J Epidemiol. 1986;124:299–305. doi: 10.1093/oxfordjournals.aje.a114388. [DOI] [PubMed] [Google Scholar]
- 10.Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken F C, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: American Society for Microbiology; 1995. [Google Scholar]
- 11.Navia M M, Capitano L, Ruiz J, Vargas M, Urassa H, Schellemberg D, Gascon J, Vila J. Typing and characterization of mechanisms of resistance of Shigella spp. isolated from feces of children under 5 years of age from Ifakara, Tanzania. J Clin Microbiol. 1999;37:3113–3117. doi: 10.1128/jcm.37.10.3113-3117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nei M, Li W H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan T M. Bacillary dysentery in Taiwan, 1995–1996. Epidemiol Bull. 1997;13:151–161. [Google Scholar]
- 14.Rudolph K M, Parkinson A J, Roberts M C. Molecular analysis by pulsed-field gel electrophoresis and antibiogram of Streptococcus pneumoniae serotype 6B isolates from selected areas within the United States. J Clin Microbiol. 1998;36:2703–2707. doi: 10.1128/jcm.36.9.2703-2707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samonis G, Elting L, Skoulika E, Maraki S, Tselentis Y. An outbreak of diarrhoeal disease attributed to Shigella sonnei. Epidemiol Infect. 1994;112:235–245. doi: 10.1017/s0950268800057642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soldati L, Piffaretti J C. Molecular typing of Shigella strains using pulsed field gel electrophoresis and genome hybridization with insertion sequences. Res Microbiol. 1991;142:489–498. doi: 10.1016/0923-2508(91)90182-a. [DOI] [PubMed] [Google Scholar]
- 17.Swaddiwudhipong W, Karintraratana S, Kavinum S. A common-source outbreak of shigellosis involving a piped public water supply in northern Thai communities. J Trop Med Hyg. 1995;98:145–150. [PubMed] [Google Scholar]
- 18.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia M, Whittington W L, Holmes K K, Plummer F A, Roberts M C. Pulsed-field gel electrophoresis for genomic analysis of Neisseria gonorrhoeae. J Infect Dis. 1995;171:455–458. doi: 10.1093/infdis/171.2.455. [DOI] [PubMed] [Google Scholar]
- 20.Yagupsky P, Loeffelholz M, Bell K, Menegus M A. Use of multiple markers for investigation of an epidemic of Shigella sonnei infections in Monroe County, New York. J Clin Microbiol. 1991;29:2850–2855. doi: 10.1128/jcm.29.12.2850-2855.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]