ABSTRACT
The rapid emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has created a global health emergency. While most human disease is mild to moderate, some infections lead to a severe disease characterized by acute respiratory distress, hypoxia, anosmia, ageusia, and, in some instances, neurological involvement. Small-animal models reproducing severe disease, including neurological sequela, are needed to characterize the pathophysiological mechanism(s) of disease and to identify medical countermeasures. Transgenic mice expressing the human angiotensin-converting enzyme 2 (hACE2) viral receptor under the control of the K18 promoter develop severe and lethal respiratory disease subsequent to SARS-CoV-2 intranasal challenge when high viral doses are used. Here, we report on SARS-CoV-2 infection of hamsters engineered to express the hACE2 receptor under the control of the K18 promoter. K18-hACE2 hamsters infected with a relatively low dose of 100 or 1,000 PFU of SARS-CoV-2 developed a severe and lethal disease, with most animals succumbing by day 5 postinfection. Hamsters developed severe lesions and inflammation within the upper and lower respiratory system, including infection of the nasal cavities causing marked destruction of the olfactory epithelium as well as severe bronchopneumonia that extended deep into the alveoli. Additionally, SARS-CoV-2 infection spread to the central nervous system (CNS), including the brain stem and spinal cord. Wild-type (WT) hamsters naturally support SARS-CoV-2 infection, with the primary lesions present in the respiratory tract and nasal cavity. Overall, infection in the K18-hACE2 hamsters is more extensive than that in WT hamsters, with more CNS involvement and a lethal outcome. These findings demonstrate the K18-hACE2 hamster model will be valuable for studying SARS-CoV-2.
KEYWORDS: SARS-CoV-2, transgenic hamsters, angiotensin-converting enzyme 2, K18-hACE2, nasal cavity, cardiac lesions, neuropathology, olfactory bulb
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a betacoronavirus and the causative agent of coronavirus disease 2019 (COVID-19), a febrile respiratory human disease that emerged in China in late 2019 and subsequently spread throughout the world (1, 2). SARS-CoV-2 infection in humans results in a spectrum of respiratory disease severity ranging from mild transient respiratory illness to an acute respiratory distress syndrome (ARDS) (3, 4). More severe aspects of disease are predominantly observed in the older population (>50 years) and those with underlying health conditions, such as hypertension or cardiovascular disorders (5, 6). SARS-CoV-2 human infections can also cause vascular damage and coagulopathies that lead to infarction. Acute disease often presents with elevated levels of inflammatory cytokines (e.g., interleukin 6 [IL-6]) that likely contribute to the pathogenic process (7, 8). Additionally, neurological disease, such as headache, anosmia (loss of smell), ataxia, meningitis, seizures, and impaired consciousness, has been reported in more severe cases and may contribute to mortality (9–11). As of 1 December 2021, SARS-CoV-2 has infected over 200 million people and resulted in over 5 million deaths worldwide.
Similar to SARS-CoV (12), SARS-CoV-2 binds to target cells via an interaction between the 13- kDa viral spike protein and the host angiotensin-converting enzyme 2 (ACE2) protein (13–16). ACE2 interaction with the spike protein is essential for host tropism, and reduced affinity between these two molecules greatly impacts host susceptibility to infection. For example, SARS-CoV-2 has low affinity for murine ACE2 (17), and accordingly, mice are naturally refractory to infection by ACE2-utilizing human coronaviruses (18–21). In contrast, wild-type (WT) golden Syrian hamster ACE2 has higher affinity for the SARS-CoV-2 spike protein (22), and these animals are naturally susceptible to infection. Disease in hamsters is generally localized to the respiratory system, where intranasal exposure results in mild to moderate pathological lesions and high levels of viral replication (22, 23). SARS-CoV-2 also targets the nasal turbinates and has been shown to infect the olfactory neuroepithelium, including olfactory sensory neurons and support cells (24, 25). Lower infectious doses and more prolonged disease can ensue when hamsters are immunosuppressed pharmacologically (cyclophosphamide) (26). Disease in immunocompetent hamsters is generally limited, and the animals usually start to gain weight after day 6 postinfection and recover. Hamsters have been the primary small-animal model used to evaluate medical countermeasures and to study viral pathogenesis of SARS-CoV-2.
Animal models reproducing the most severe aspects of COVID-19, including central nervous system (CNS) involvement, are needed to more clearly understand the pathogenic mechanisms driving acute disease. Mice ectopically expressing the human ACE2 (hACE2) receptor under the control of the keratin 18 promoter are highly susceptible to SARS-CoV and SARS-CoV-2 (20, 27–29). These mice develop a lethal disease that is characterized by respiratory damage, in addition to infection of the nasal cavity and brain. Because hamsters, unlike mice, are naturally permissive to SARS-CoV-2, here we investigated if ectopic expression of the hACE2 receptor would produce a severe disease model resembling human COVID-19, including more extreme disease manifestations, such as damage to the olfactory system and injury to the central nervous system. We demonstrate that intranasal challenge with SARS-CoV-2 in K18-hACE2 hamsters causes severe and lethal disease that affects both upper and lower respiratory systems and the central nervous system, causing extensive damage to the tissues of nasal cavity, lung, and neurons of the brain.
RESULTS
K18-hACE2 hamsters develop severe, lethal disease upon intranasal exposure to SARS-CoV-2.
Two strains of hACE2 transgenic hamster, K18-hACE2-M41 and K18-hACE2-M51 (9 animals per group), were infected with two doses of SARS-CoV-2 strain WA1 (100 and 1,000 PFU) and monitored for survival and weight loss (Fig. 1A). Three in each dose group were euthanized for tissue collection on day 3, and six per group were followed for up to 15 days. On day 2, infected animals in all groups began to lose weight. All K18-hACE2-M41 hamsters succumbed to infection by day 5. Most K18-hACE2-M51 hamsters died before the end of the study, with a single survivor in the 100 PFU-infected group and two survivors in the 1,000 PFU group. There was no statistical survival difference between any group (log rank; P > 0.05). The viral load in the lungs was determined by plaque assay from tissues collected at a designated time point (day 3; 3/group), at time of death (day 5; 4 to 6/group), or at the study endpoint (day 15; n = 3) (Fig. 1A). Lung titers were similar for all groups, with 1-log-higher titers on day 3 (∼107 PFU/g) versus day 5 (∼106 PFU/g) (Fig. 1B). No viable virus was detected on day 15 in any surviving animal (n = 3). These findings indicated that the K18-hACE2 hamsters are sensitive to SARS-CoV-2 and develop severe and lethal disease after intranasal exposure. Uninfected control hamsters (2 per hamster strain; 4 total) were also examined, and one tested positive for virus on day 3 with a titer of 8,330 PFU/g (Fig. 1B).
FIG 1.
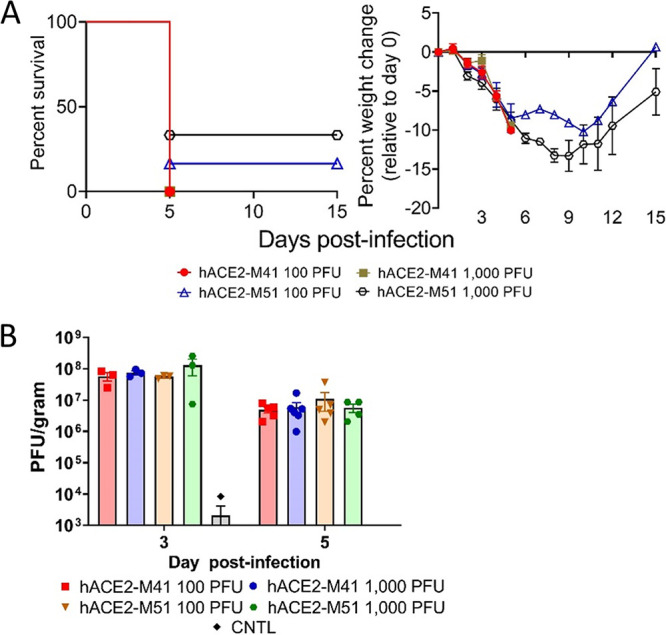
SARS-CoV-2 infection in K18-hACE2 Syrian hamsters. (A) Male and female K18-hACE2 transgenic hamster strains M41 and M51 (days 0 to 3, n = 9/group; days 3+, n = 6/group) were infected with 100 PFU (5 males and 4 females per hamster strain) or 1,000 PFU (4 males and 5 females per hamster strain) of SARS-CoV-2 by the intranasal route. Survival and weight loss (with standard errors of the means [SEM]) were monitored and plotted using Prism software. (B) Titers in lungs were examined by plaque assay on day 3 (n = 3) or 5 (n = 4 to 6). Means and SEM were graphed. Each dot represents an individual hamster.
SARS-CoV-2-infected hamsters develop mild lung lesions.
Lung lesions were observed in tissue collected from both the upper and lower airways of animals on days 3 and 5 (Fig. 2A and Fig. S1) but were resolved in bronchi and bronchioles in day 15 surviving hamsters; however, day 15 hamsters still had various degrees of residual inflammation in alveoli at this late time point. Lesions from hamsters on both days 3 and 5 were characterized by degeneration and necrosis of bronchial epithelium and multifocal areas of alveolar inflammation of minimal to moderate severity, which was associated with hemorrhage, edema, and fibrin exudation. Inflammation was also present within the lumen of bronchi and bronchioles and often extended through the mucosa and into the outer layers of affected airways. On day 5, regeneration of the bronchiole epithelium was detected. Lesions were generally more consistent and severe in bronchi and bronchioles than in alveoli on both days 3 and 5, with a subset of hamsters having minimal or absent inflammation in alveoli. Inflammation corresponded to the presence of viral RNA (vRNA) labeled by in situ hybridization (ISH) (Fig. 2B). More vRNA labeling was detected in the bronchioles and bronchi than the alveoli. Increases in the presence of myeloperoxidase-positive (MPO+) cells, neutrophil granulocytes, and ionized calcium binding adaptor molecule 1-positive (Iba1+) macrophages were observed on day 5 by immunofluorescence assay (IFA) (Fig. 2C). SARS-CoV-2 antigen was detected in both E-cadherin+ and Iba-1+ cells, suggesting that both epithelial cells and macrophages were infected (Fig. 2D and E). Coinciding with viral replication and lung lesions was the increase presence of IL-1β and CXCL10 RNA, a proinflammatory cytokine and chemokine, respectively (Fig. 2F). Overall, these data indicate that SARS-CoV-2 infects lungs of K18-hACE2 hamsters with lesions ranging in severity from minimal to moderate and predominantly affects bronchi and bronchioles at days 3 to 5.
FIG 2.
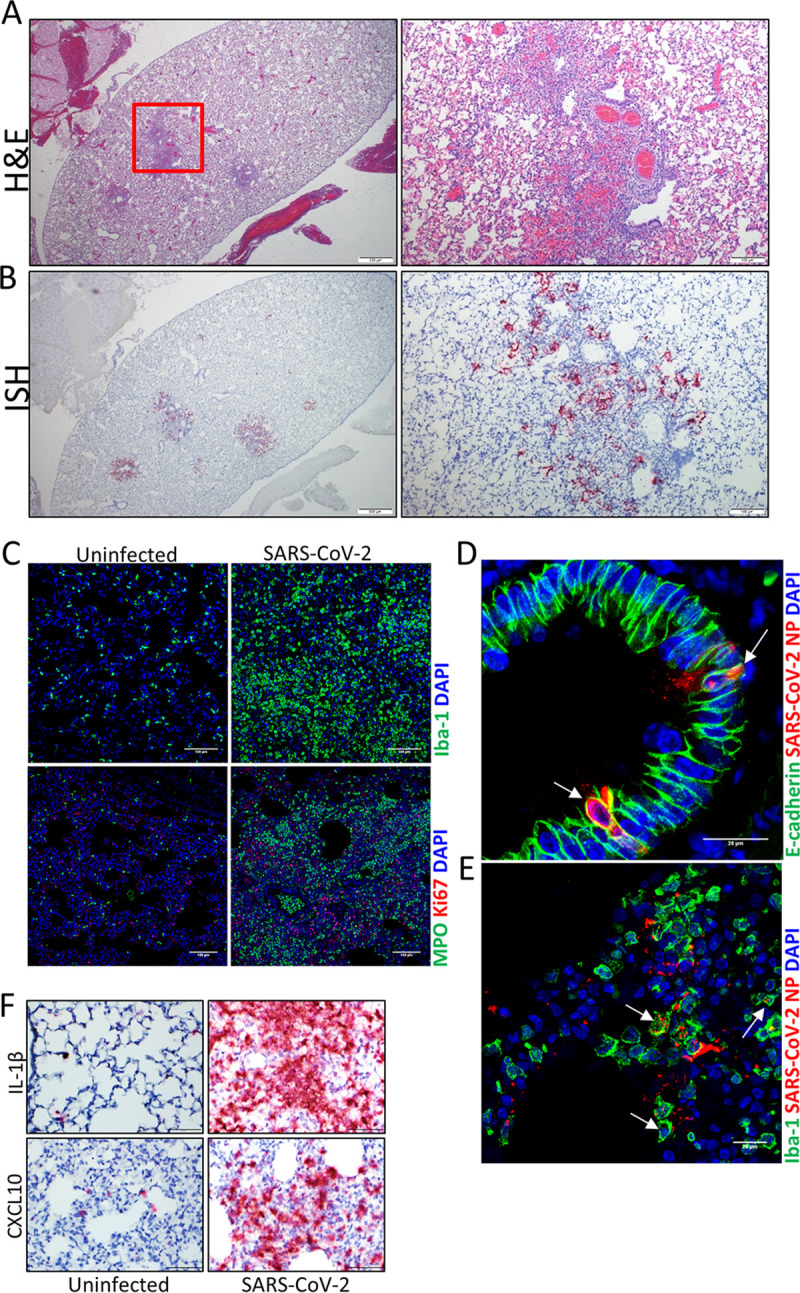
SARS-CoV-2 respiratory infection in K18-hACE2 hamsters. (A) Representative H&E stain showing multifocal areas of alveolar inflammation with minimal overall severity. (Right) Expansion of the red box showing mononuclear inflammation in alveoli with hemorrhage and edema adjacent to bronchioles, and perivascular inflammation. (B) Representative ISH images (corresponding to the area in panel A) showing the presence of SARS-CoV-2 RNA (red) in the lungs in areas of inflammation identified in the H&E panels. Cells were counterstained with hematoxylin (blue). (C) IFA showing the present of Iba-1+ (macrophages) or MPO+ (granulocytes) cells in the lungs of uninfected and infected hamsters. Nuclei are stained with DAPI (blue). (D) IFA costaining for viral nucleoprotein (NP) protein (red) and E-cadherin (green) in infected lung tissues. Arrows point to double-positive cells. Nuclei are stained with DAPI (blue). (E) IFA showing costaining of NP and the macrophage marker Iba-1 (green) in infected lungs. Arrows indicate double-positive cells. Nuclei are stained with DAPI (blue). (F) ISH staining for IL-1β and CXCL10 mRNA expression (red) in the lungs of uninfected and infected hamsters. Cells were counterstained with hematoxylin (blue).
Lung pathology in day 5 animals. (A) H&E staining showing multifocal to coalescing areas of alveolar inflammation with moderate severity. The right panels are an expansion of boxed areas, showing mononuclear inflammation in alveoli (septa and lumen) with hemorrhage, edema, and fibrin exudation. There was regeneration of bronchiole epithelium (bottom left, arrows). Lower panels show ISH staining for SARS-CoV-2 genome in same region seen in the H&E panels. Note viral labeling (red) in areas of inflammation and rarely in bronchiole epithelium (expanded view of the boxed area in the right panel). (B) Lung from a different day 5 animal shows degeneration, necrosis, and loss of bronchiole epithelium and peribronchiole inflammation. (Middle) ISH staining of the same panel as H&E. (Right) Magnified H&E image of the black box in the left panel, highlighting degeneration, necrosis, and sloughing of bronchiole epithelium as well as mild mononuclear inflammation in the bronchiole adventitial layer. Download FIG S1, PDF file, 0.6 MB (648.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Nasal cavities and olfactory bulbs of K18-hACE2 hamsters infected by SARS-CoV-2 show signs of acute pathology.
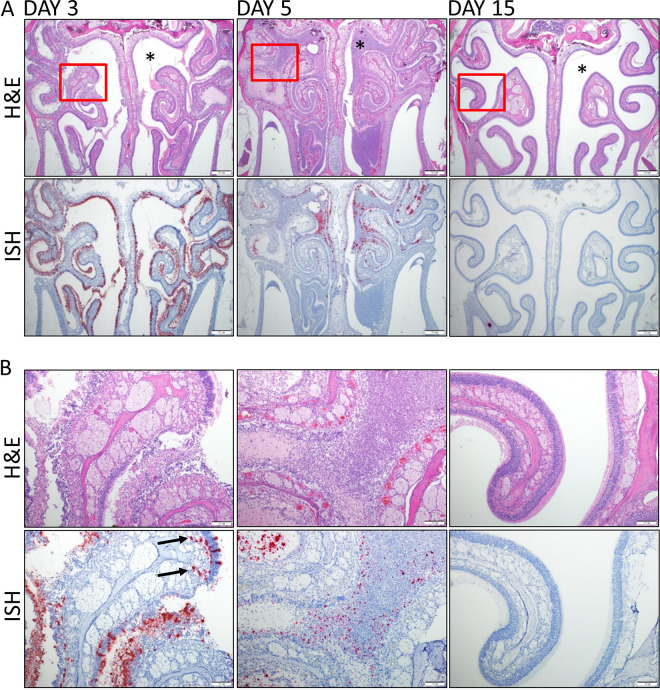
SARS-CoV-2 infection led to severe damage within the nasal turbinates (Fig. 3). Damage included heterophilic and necrotic inflammatory exudate filling the nasal cavity, which was most pronounced on day 5. Other lesions included inflammation of the epithelium and underlying lamina propria along with ulceration and regeneration of the olfactory, respiratory and transitional epithelium. Sloughing and loss of olfactory, respiratory, and transitional epithelium was detected in the majority of animals; however, the olfactory epithelium was the most severely impacted, with widespread degeneration and necrosis present on both days 3 and 5. Respiratory epithelium also showed signs of atrophy, degeneration, and necrosis, but to a lesser degree than the olfactory epithelium. Markedly less damage occurred in the transitional epithelium, which was not generally ulcerated or necrotic. Lesions were consistent between the two strains of hamsters and between the two doses. vRNA was detected by ISH throughout the nasal cavity in both the respiratory and olfactory epithelium but was generally less abundant in the respiratory epithelium. Sloughed cellular debris (exudate) within the nasal cavity was positive for vRNA by ISH. vRNA was also prominently detected in the vomeronasal organ and to a lesser degree in Steno’s gland ducts. In the olfactory bulb in a subset of animals, vRNA was present either focally in a single location or multifocally in all layers of the olfactory bulb, with a minimal to mild severity (Fig. S2). Additionally, viral particles were detected by electron microscopy within the nasal cavity (Fig. S3). vRNA decreased from day 3 to day 5, while there was an increase in tissue lesions. These data revealed that the nasal cavity is a target of SARS-CoV-2 in the K18-hACE2 hamsters and infection results in acute tissue injury.
FIG 3.
SARS-CoV-2 infection of the nasal cavity. (A) Representative H&E and ISH staining of hamsters’ nasal cavities on days 3, 5, and 15. Corresponding ISH images are shown below the H&E stains, with viral labeling in red and with marked staining of the nasal and olfactory epithelium on day 3. Viral genomic RNA (red) was detected on day 3, decreased on day 5, and was absent on day 15. ISH panels were counterstained with hematoxylin (blue). Nasal cavity exudate was limited on day 3 and absent on day 15 but partially occluded the cavity on day 5 (asterisks). (B) Higher magnification of the area in the box in the top image. The day 3 ISH image shows viral labeling in the deeper lamina propria and olfactory nerve layer below the olfactory epithelium (arrows).
Viral RNA in the olfactory bulb. In situ hybridization for SARS-CoV-2 RNA shows prominent labeling in the all layers of the olfactory bulb in a day 5 animal, including (from outside in) the olfactory nerve layer, glomerular layer, external plexiform layer, mitral cell layer, internal plexiform layer, and granule cell layer. Labeling is absent in an uninfected control. Download FIG S2, PDF file, 0.4 MB (376.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Electron micrograph of the nasal cavity. (A and B) Representative images of macrophages showing the cytoplasmic presence of SARS-CoV-2. Right panels show higher magnifications of the boxed areas in the left panels. (C) SARS-CoV-2-infected epithelial cell exhibiting potential virus budding from the plasma membrane (arrows) and in the apical lumen (arrowhead). Download FIG S3, PDF file, 0.6 MB (592KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
SARS-CoV-2 infiltrates the central nervous system of infected hamsters and produces severe lesions.
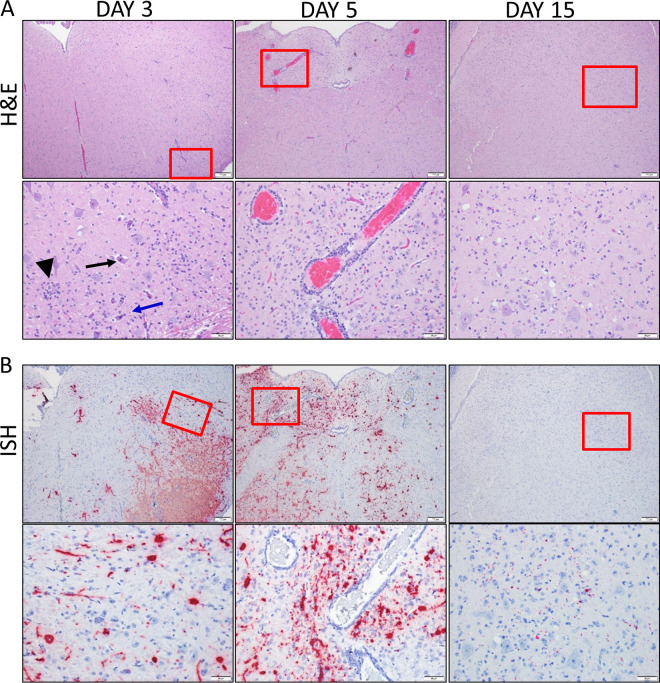
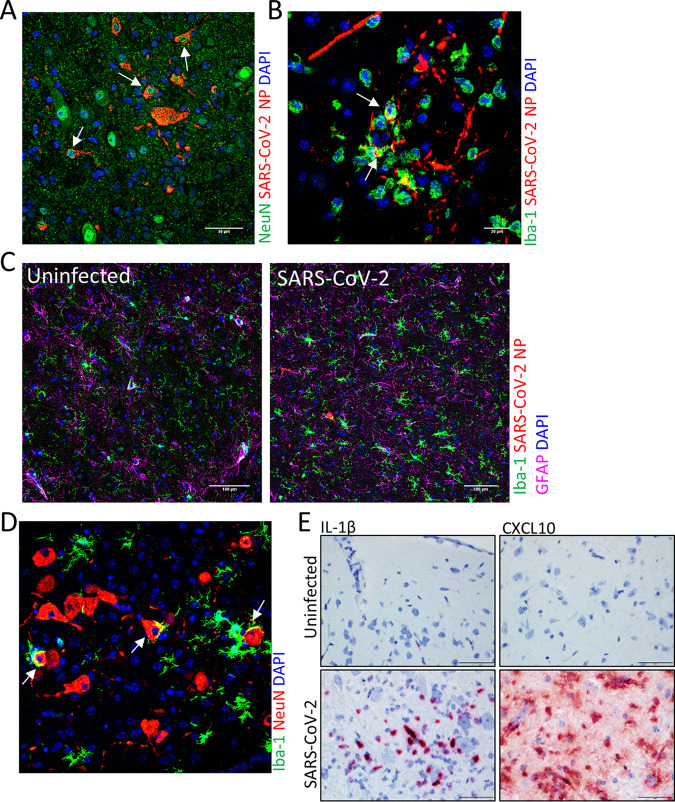
SARS-CoV-2 infection caused extensive damage to the CNS in the K18-hACE2 hamsters (Fig. 4). The major pathological findings consisted of multifocal gliosis throughout the gray matter, meningitis, perivascular inflammation, and neuronal necrosis (Fig. 4A). In several animals, these findings were predominantly in the brain stem (medulla). On day 3, gliosis was present only in the brain stem. In some animals, neuronal necrosis was associated with neurophagia and satellitosis. vRNA was detected by ISH in most animals and generally increased in intensity on day 5 but was only minimally detected by day 15 (Fig. 4B). vRNA was present in striatum, pallidum, hypothalamus, thalamus, amygdala, midbrain, and brain stem (pons and medulla) but was infrequent in the cerebral cortex and cerebellum. IFA revealed that viral antigen was present in both neurons (NeuN+ cells) and microglial cells (Iba-1+ cells) (Fig. 5A and B). Furthermore, gliosis was confirmed by IFA staining of microglial cells (Iba-1+) (Fig. 5C). Neurophagia was also further confirmed by IFA, with the presence of Iba-1+ cells engulfing NeuN+ neurons (Fig. 5D). IL-1β and CXCL10 RNA were also detected in the brain of infected, but not uninfected, animals (Fig. 5E).
FIG 4.
SARS-CoV-2 infiltration of the brain. (A) Representative H&E staining of the brain stem (medulla) from days 3, 5 and 15. The boxes in the top images indicate the areas shown at higher magnification in the lower panels. Areas of gliosis, glial nodules (black arrowhead), neuronal necrosis (black arrow), and neurophagia (blue arrow) were observed. Perivascular cuffing and gliosis surrounding affected vessels was prominent in day 5. (B) Corresponding ISH images shown with viral RNA labeling in red. The red box in the top images indicates areas shown at higher magnification in the lower panels. Viral RNA labeling was present in neuron processes, glia, and neurons at days 3 and 5. ISH signal was reduced on day 15 and localized predominantly in the glia and neuropil. ISH panels were counterstained with hematoxylin (blue).
FIG 5.
IFA staining of SARS-CoV-2 infected hamster brain. (A) IFA for the neuron marker NeuN (green) and SARS-CoV-2 NP (red). NP was detected in NeuN+ neurons (arrows). Nuclei were stained with DAPI (blue). (B) IFA for the microglial cell marker Iba-1 (green) and SARS-CoV-2 NP (red). NP costaining in Iba-1+ cells is shown by arrows. Nuclei were stained with DAPI (blue). (C) Iba-1 (green) and GFAP (pink) markers for microgliosis and astrogliosis, respectively, in uninfected and infected brain sections and viral NP (red) using IFA. Nuclei were stained with DAPI (blue). (D) Costaining for the microglial cell marker Iba-1 (green) and the neuron marker NeuN (red). Neurophagia is indicated by the arrows. Nuclei were stained with DAPI (blue). (E) ISH staining for IL-1β and CXCL10 expression (red) in the lungs of uninfected or infected hamsters. Cells were counterstained with hematoxylin (blue).
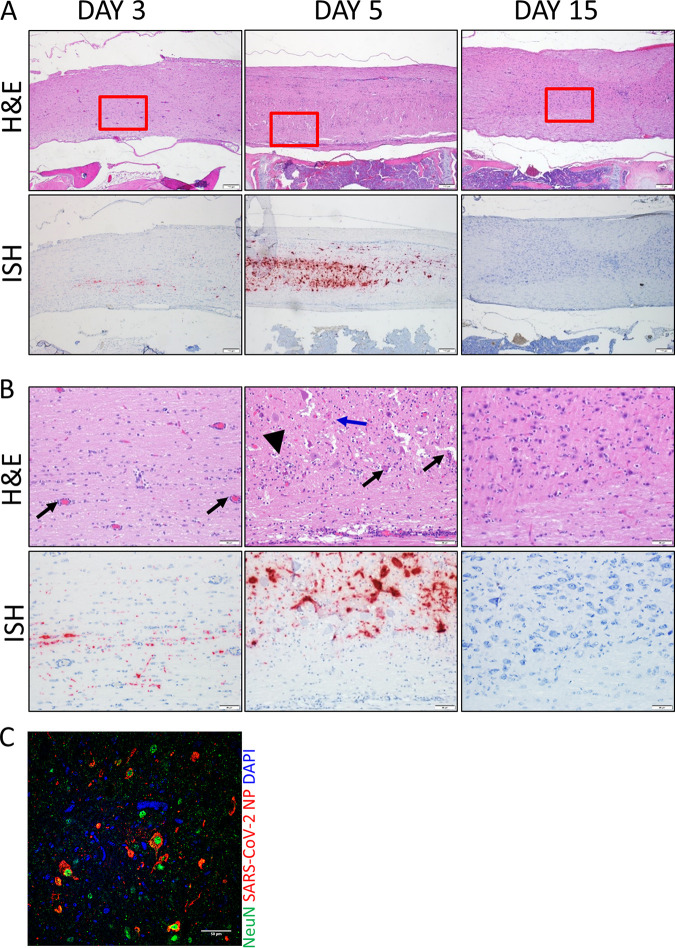
In addition to the brain, SARS-CoV-2 genomic RNA was also detected in the spinal cord on day 3, increasing on day 5, but was absent on day 15 (Fig. 6A and B). Lesions were predominantly found in spinal cord gray matter, and, similar to the brain, infection was associated with tissue injury, including gliosis, meningitis, perivascular inflammation, neuronal necrosis, and neurophagia (Fig. 6B). In contrast to the brain, viral antigen was detected only in spinal cord neurons (NeuN+ cells) and not microglial cells (Fig. 6C). These findings demonstrate that the CNS is an important target of SARS-CoV-2 in K18-hACE2 hamsters.
FIG 6.
SARS-CoV-2 in the spinal cord of infected hamsters. (A and B) Spinal cord H&E and ISH images from days 3, 5, and 15 (animals 21, 3, and 28, respectively). The boxes in the top images indicate the areas shown at higher magnification in panel B. Perivascular inflammation was present on day 3 (black arrows), but on day 5, there were gliosis, neuronal necrosis (blue arrow), neuronophagia (arrowhead), and increased perivascular inflammation. Cells were counterstained with hematoxylin (blue). (C) IFA for the neuron marker NeuN (green) and SARS-CoV-2 NP (red). NP was detected in NeuN+ neurons (arrows) of spinal cords. Nuclei were stained with DAPI (blue).
SARS-CoV-2 infection results in cardiac injury.
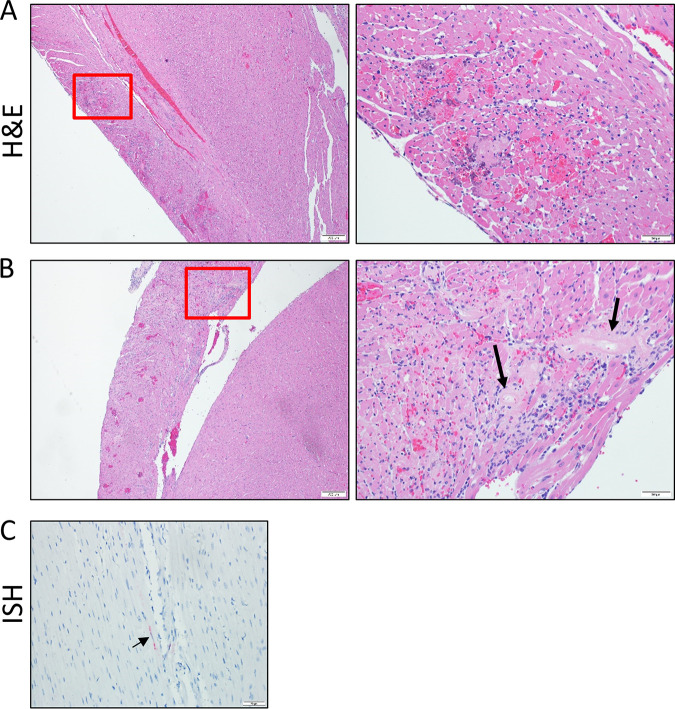
Hearts from 4 of 6 (67%) K18-hACE2 hamsters examined on day 5 showed evidence of myocardial injury (Fig. 7A and B). The predominant lesions included myocardial inflammation, characterized by infiltration of macrophages and heterophils around degenerating and necrotic myofibers, often associated with small hemorrhages, and necrosis. Necrosis was characterized by shrunken, irregularly shaped myofibers with pyknotic nuclei, loss of cross striations, myofibril fragmentation, and hypereosinophilic sarcoplasm. Areas of necrosis were small and mostly centered around small-caliber vessels. Most of the heart lesions were localized to the right ventricular free wall. Cardiac injury was generally not associated with the presence of virus, and only a single hamster had a small area of vRNA staining by ISH within the myocardium (Fig. 7C). Therefore, the precise relationship between viral infection and the cardiac lesions is unclear. No lesions were present on day 3 or 15 in infected hamsters, and none were identified in uninfected control K18-hACE2 hamsters. These findings indicated that SARS-CoV-2 infection may impact the heart in the K18-hACE2 hamster model through a bystander effect or means other than viral infection.
FIG 7.
Heart lesions in SARS-CoV-2-infected hamsters. (A and B) Representative H&E stain of heart tissue from two infected hamsters. The right panels are higher magnifications of the boxed areas in the left panels. Both hearts show areas of cardiac myocyte necrosis and mononuclear inflammation with hemorrhage in the right ventricle. Prominent hyalinized vessels with loss of cellular detail are indicated by arrows. (C) Animal from panel A showing a small area of viral RNA by ISH (red), indicated by the arrow.
Infection of a single control animal.
This study had four uninfected control K18-hACE2 animals in barrier caging located in proximity to infected animals. Tissues from uninfected control animals were taken at the first time point (day 3). Three animals had no virus or lesions in the nasal cavity; however, in a fourth animal, we detected vRNA by ISH (Fig. S4). No lesions were detected in this animal histopathologically and no other tissues were positive. The low level of virus noted in this animal was also detected in lung homogenates by plaque assay (Fig. 2B). These serendipitous data indicated that K18-hACE2 hamsters are highly sensitive to infection and are suitable for the study of natural viral transmission.
Infection of control animals. Representative ISH and H&E staining of the nasal cavity of negative-control hamsters. A single control animal was incidentally exposed to the virus, as indicated by ISH (red). during the challenge period. These tissues were harvested on day 3. Animals were in different locations in the same room in barrier cages and purposely not manipulated (weights, etc.). Download FIG S4, PDF file, 0.4 MB (400.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DISCUSSION
K18-hACE2 hamsters are hypersensitive to SARS-CoV-2.
Our findings reveal ectopic expression of hACE2 in Syrian hamsters produces an animal model that is highly susceptible to SARS-CoV-2 resulting in severe and lethal disease. Infection also resulted in increases in proinflammatory cytokine (IL-1β) and chemokine (CXCL10) transcripts in lung, nasal tissue, and brain that is consistent with the cytokine storm produced during severe human infections. CXCL10 is a chemokine known to be increased in some COVID-19 cases that require intensive care unit admission (30). In contrast to nontransgenic wild-type (WT) hamsters (22, 23), damage to the lungs in K18-hACE2 hamsters was minimal to moderate and generally more common and severe in the bronchioles than the alveoli. The infection was primarily contained within the upper respiratory tract with only moderate changes in the lower respiratory tract. While both macrophages (Iba-1+ cells) and epithelial cells had viral antigen, more work would be required to determine if macrophages were supporting viral replication. The principal tissue targeted by SARS-CoV-2 in this model was the nasal cavity, which resulted in acute tissue damage, including loss of olfactory epithelium. Even in WT hamsters, the nasal cavity is an important target, and at least one study found that viral replication was higher in this region than in the lungs (22). In our model, SARS-CoV-2 replication in the nasal epithelium, detected by ISH, is extensive on day 3 but decreases by day 5. However, tissue damage continues to increase and is severe by day 5, with the olfactory epithelium showing the most extensive destruction and loss. The presence of virus in Steno’s glands and prominently in the vomeronasal organ in the Syrian hamster model have also been described elsewhere (25). Severe infection in the nasal cavity causing an exudate, sloughing of tissues, and debris accumulation likely contributed to severe morbidity and perhaps even lethality, as hamsters are obligately nasal breathers.
Neuropathology in the K18-hACE2 hamster system.
In humans, evidence suggests that SARS-CoV-2 can impact the central nervous system directly and indirectly (9, 31–35). SARS-CoV-2 RNA has been detected in cerebrospinal fluid (CSF) in a patient with encephalitis, suggesting that the virus infiltrates the CNS (36, 37). SARS-CoV-2 infection has also been shown to result in acute transverse myelitis (38). The pathophysiological mechanisms of human SARS-CoV-2 CNS injury are poorly understood. Previous studies in nontransgenic (non-Tg) hamsters identified SARS-CoV-2 in the olfactory bulb and in other regions of the brain (24). In several K18-hACE2 hamsters, virus was present in all layers of the olfactory bulb, but in others, it was present only in specific layers, such as the glomerular layer, nerve fiber layer, or granular layer. Virus was also found in several other regions of the brains of K18-hACE2 hamsters. The primary targeted cell type was neurons; however, viral antigen was also found in microglial cells. More studies will be required to determine if these cell types are productively infected by SARS-CoV-2 or if these immune cells engulf virus-infected cell debris; at least one study suggested that neurons support productive infection (39). Additionally, virus was found within the spinal cord, which has not been previously reported in the hamster system.
At least one study has suggested that lethal human cases may involve infection of the respiratory centers of the brain (40, 41). This area was infected in the K18-hACE2 hamsters, possibly indicating that this model system would be useful in investigating the role infection of respiratory centers has in morbidity. Infection through the olfactory bulb may be the major route by which SARS-CoV-2 enters the brain, as has been speculated with regard to infection in humans (42). Many regions of the brain testing positive for vRNA have primary or secondary (via the olfactory cortex) connections to the olfactory bulb. However, the degree of infection, shown by viral labeling, and inflammation in the olfactory bulb was much lower in the olfactory cortex than in other regions of the brain, suggesting that another route of infection may be involved (i.e., directly hematogenous or via leukocyte trafficking). Additionally, the presence of virus in the spinal cord suggests that virus may spread via a route other than the olfactory pathways. Infection by way of the cerebral spinal fluid or ventricular system is possible but also less likely, given that infection of the choroid plexus was of minimal severity compared to the infection in the adjacent nervous tissue and there was no significant damage or inflammation observed in the ependymal cells of the choroid plexus. Further studies will be required to more clearly understand how SARS-CoV-2 infects tissues of the central nervous system of K18-hACE2 hamsters. Given that human CNS involvement may be more common than previously appreciated (9, 31–35), the K18-hACE2 hamster system may provide a unique platform to better understand neuropathology of SARS-CoV-2.
Cardiac injury in K18-hACE2 hamsters.
Lesions in the heart have been described in human SARS-CoV-2 infections (43–45) and were present in several K18-hACE2 hamsters that succumbed to infection. However, myocarditis with associated cardiomyocyte necrosis similar to what was seen in our study appears to be rare in humans (44). Individual cardiac myocyte necrosis has also been described elsewhere in human cases of COVID-19 with absence of significant inflammation (43). However, a direct role of virus in human heart lesions is unclear. In our study, only a single animal (of 14 with heart lesions) had vRNA identified in the heart, and only in a very small focal area that lacked inflammation or necrosis. Myocardial degeneration and interstitial edema were observed in another study involving non-Tg Syrian hamsters with SARS-CoV-2 infection, but viral protein was not detected in the heart (22). These findings suggest that while cardiac lesions may develop in the hamster models of SARS-CoV-2 infection, and potentially in humans, the exact pathogenesis is unclear. Lesions may develop secondary to factors related to viral infection more generally (and not SARS-CoV-2 infection specifically), such as a result of the hyperactive inflammatory response or ischemia caused by thrombosis. While high levels of inflammatory transcripts were found in infected tissues, no fibrin thrombi were observed in the heart or other tissues in this study. Heart lesions in the K18-hACE2 animals may be a result of demand ischemia caused by the occlusion of the nasal cavity resulting in hypoxia or of stressed induced catecholamine release. Demand ischemia resulting in heart injury in humans has been proposed (45). It is not clear why the majority of lesions were isolated in the right ventricular free wall, with fewer lesions in the right atrium, left ventricle, and interventricular septum. Nevertheless, the K18-hACE2 hamster system may be beneficial in the study of cardiomyopathy observed in human cases of COVID-19.
K18-hACE2 hamsters versus K18-hACE2 mice as lethal infection models for SARS-CoV-2.
We and others have reported that mice expressing hACE2 under the control of the K18 promoter develop lethal disease upon exposure to SARS-CoV-2 (20, 28, 29). Infection in the mice and hamsters exhibited several differences. In the mice, the lungs underwent extensive injury, with the lesions and virus localized to the alveoli. Mice also developed vasculitis in pulmonary vessels. This contrasts sharply with the K18-hACE2 hamsters, where lung lesions were mild, sporadic, and localized predominantly to bronchioles. Nasal cavity lesions in the hamsters were more severe than in mice and were characterized by degeneration and necrosis of epithelium with inflammation. Histologic lesions in the olfactory bulb were generally similar to those in the K18-hACE2 mouse model; however, the degree of vRNA labeling was reduced in the K18-hACE2 hamster model. The regions of the brain demonstrating infection in this model have both similarities and differences compared to what was seen in the K18-hACE2 mouse model. In this K18-hACE2 hamster model, there was more involvement of the brain stem, with both vRNA labeling and histological changes such as gliosis and neuronal necrosis. Furthermore, the infectious dose required for severe disease and lethality in the K18-hACE2 mouse is multifold higher than in hamsters (≥20,000 PFU versus 100 PFU). This could be due to the native hamster ACE2 having high affinity for the SARS-CoV-2 spike protein, whereas the murine ACE2 has poor affinity (17, 22), but it is also possible that this resulted from the intrinsic differences between the immune systems in the two species.
Wild-type hamsters versus K18-hACE2 hamsters.
Wild-type hamsters are naturally susceptible to SARS-CoV-2 and develop a transient respiratory disease (22–25). Accordingly, the WT hamster model has been used extensively to evaluate SARS-CoV-2 countermeasures and disease features. In general, the K18-hACE2 hamster model is a more severe disease model than the WT hamster model even when lower virus doses are used, but there are some specific differences. Our group has infected both model systems with the WA01-2020 strain of SARS-CoV-2 from the same production lot, enabling a critical model comparison (WT versus K18-hACE2 hamsters). Consistent with published findings (24, 25), we found that SARS-CoV-2 infects the nasal cavity of WT (Fig. S5) hamsters, causing transient pathological disturbances; however, in the K18-hACE2 model, pathology in the nasal cavity is more extensive. In WT hamsters, SARS-CoV-2 caused more severe lung lesions and there was increased viral labeling (Fig. S6) compared to the day 3 lung lesions from transgenic hamsters. In contrast, on day 7, infected WT lungs showed evidence of healing (Fig. S7), whereas ongoing damage was prevalent in the K18-hACE2 animals that died on day 5. In the K18-hACE2 hamsters, the CNS is also more greatly impacted than in WT hamsters. The dose required for observable weight loss in the WT hamsters is orders of magnitude higher than that for the K18-hACE-2 hamsters. Thus, the K18-hACE2 hamster model seems to better mimic natural human infection, which is likely also caused by a low infection dose.
Nasal turbinate histopathology in wild-type Syrian hamsters. H&E and ISH staining in nasal turbinates of wild-type hamsters on days 3 and 7 from a separate study, presented here for comparison. Note that the level of inflammation and damage to the olfactory and respiratory epithelium, as well as the viral labeling, are roughly similar to what was seen in the K18-ACE2 hamsters on day 3. On day 7, the levels of inflammation and labeling were significantly decreased and there was regeneration of epithelium. This contrasts with the day 5 transgenic animals in this study, where the histologic lesions (epithelial necrosis, inflammation, exudate) increased in severity on day 5 compared to day 3. Red boxes in upper images correspond to higher-magnification images below. Download FIG S5, PDF file, 0.5 MB (515.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Lung pathology from day 3 SARS-CoV-2 infected wild-type Syrian hamsters. H&E and ISH staining in SARS-CoV-2-exposed wild-type hamsters harvested on day 3. Note the presence of areas of alveolar inflammation, bronchiole epithelial degeneration/regeneration, and perivascular inflammation affecting roughly half the section, which corresponds to areas of strong viral labeling. Lung lesions and viral labeling were increased in severity compared to the lung lesions from day 3 transgenic hamsters. Red boxes in upper images correspond to higher-magnification images below. Download FIG S6, PDF file, 0.7 MB (685.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Lung pathology from day 7 SARS-CoV-2-infected wild-type Syrian hamsters. H&E and ISH staining in SARS-CoV-2-exposed wild-type hamsters harvested on day 7. Note the presence of patchy areas of increased cell density characterized by prominent type II pneumocyte hyperplasia, residual alveolar inflammation, and perivascular inflammation; there is only rare viral labeling seen in few bronchiole epithelial cells (arrows) and inflammatory cells. These findings contrast with the day 5 transgenic animals in this study, where there was ongoing damage to bronchiole epithelium, much less type II pneumocyte hyperplasia, and more prominent viral labeling. Red boxes in upper images correspond to higher-magnification images below. Download FIG S7, PDF file, 0.6 MB (585KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Study limitations.
This study has some notable limitations. Because of limited animal numbers, we did not thoroughly investigate physiological (e.g., hypoxia, body temperature, and blood chemistry) or behavioral (e.g., activity) effects observed in severe human COVID-19 in the K18-hACE2 mouse model. As with most rodent models, sample volumes result in inherent limitation on the range of analysis that can be performed, and in the case of hamsters, there are fewer reagents available than there are for mice or nonhuman primates.
Hamster models for the study of SARS-CoV-2.
Here, we showed that the K18-hACE2 hamster model is a low-virus-dose and lethal-disease model that has features overlapping those of severe COVID-19 in humans. Previously we developed an immunosuppressed WT hamster model using cyclophosphamide (CyP) treatment that uses a much lower challenge dose of virus, comparable to the K18-hACE2 hamster system (26). Thus, there are at least three hamster systems available for the study of SARS-CoV-2 disease progression and for the development of medical countermeasures. Because hamster ACE2 facilitates SARS-CoV-2 infection, WT hamsters may be the preferred overall model for the study of SARS-CoV-2 pathogenesis. CyP-treated WT hamsters develop more severe pathology, including lower airway injury, and have more prolonged weight loss than untreated WT hamsters. The additional of CyP to WT hamster infection studies may also allow a deeper study of more severe respiratory disease in the hamster system and also allow the use of a more natural viral challenge dose. Ectopic expression of hACE2 enhances disease but promotes a more severe infection in the nasal cavity and the CNS. Thus, the K18-hACE2 model may be useful for studies aimed at understanding SARS-CoV-2 interaction with the CNS and the olfactory epithelium. The K18-hACE2 hamster system can also be used to evaluate immunotherapeutics (46). Because this is a lethal model, it may allow more stringent evaluation of medical countermeasures and avoid the need for more detailed analysis, such as pathology, as is required in the WT hamster system. Our data indicate that K18-hACE2 hamsters, like WT hamsters, are highly sensitive to infection by SARS-CoV-2, because an uninfected control animal in close proximity to the infected animals became unexpectedly infected by the nasal route. This was despite barrier precautions and handling protocols used to prevent exposure of the uninfected animals. Unlike the nonlethal WT hamster model, the readout using the K18-hACE2 hamsters would be lethal disease. Accordingly, K18-hACE2 hamsters could be used to evaluate natural SARS-CoV-2 transmission and to develop physical and pharmacological countermeasures to thwart spread to naive hosts and release from infected hosts. However, as no rodent model is a complete transcript of human disease, a careful understanding of the questions being addressed or countermeasure products being evaluated should be considered before choosing a particular system.
MATERIALS AND METHODS
Viruses and cells.
A third-passage SARS-CoV-2 strain USA-WA1/2020 viral stock was obtained from the CDC and was originally isolated from a human nonfatal case in January 2020 (47). A master stock of virus (passage 5) was propagated as described previously (20). Virus was quantified by plaque assay and determined to be endotoxin free. All virus work was handled in biosafety level 3 (BSL-3) containment at USAMRIID.
Hamster virus challenge.
Development of the K18-hACE2 transgenic hamster will be reported in detail elsewhere. Briefly, the K18-hACE2 transgenic hamster lines were generated with a piggyBac vector, pmhyGENIE-3 carrying the K18-hACE2 cassette transferred from a pK18-hACE2 plasmid. The piggyBac-K18-hACE2 vector was then used to produce K18-hACE2 transgenic hamsters via pronuclear injection. Two transgenic hamster strains were developed and used in this study, K18-hACE2-M41 and K18-hACE2-M51, each carrying a single copy of the K18-hACAE2 transgenic cassette but at different genomic loci. Hamsters were infected intranasally with 100 or 1,000 PFU of SARS-CoV-2 strain WA-1/2020 diluted in a total volume of 50 μl of phosphate-buffered saline (PBS) (25 μl per naris). Animals were uniquely identified with numerical ear tags.
Ethics statement.
All animal studies were conducted at an AAALAC-accredited facility in compliance with federal statutes and regulations relating to animals and experiments involving animals and adheres to principles in the Guide for the Care and Use of Laboratory Animals (48). Animals meeting pre-established criteria were humanly euthanized after consultation with veterinary staff.
Plaque assay.
Plaque titrations for SARS-CoV-2 were conducted as previously described (26). Briefly, 10-fold dilutions of clarified tissue homogenate, or stock virus, starting at a 1:10 dilution were adsorbed for 1 h on Vero 76 (ATCC CRL-1587) cell monolayers in 6-well tissue culture plates and then overlaid with 0.6% agarose. After 2 days (day 2) in a 37°C, 5% CO2 incubator, an overlay containing 5% neutral red was added. On day 3, plaques were counted and the number of PFU per milliliter was calculated. The number of PFU per gram of lung was calculated as the PFU per milliliter of lung homogenate divided by the mass of lung tissue per volume of homogenate (in grams per milliliter).
Histology.
Tissues were immersed in 10% neutral buffered formalin for 14 days. Tissues were then trimmed and processed according to standard protocols (49), then cut at 5 to 6 μm on a rotary microtome, mounted onto glass slides and stained with hematoxylin and eosin (H&E). Examination of the tissue was performed by a board-certified veterinary pathologist.
In situ hybridization.
To detect SARS-CoV-2 genomic RNA or host RNA for L-1β and CXCL10 in formalin-fixed paraffin-embedded tissues, in situ hybridization (ISH) was performed using the RNAscope 2.5 HD RED kit (Advanced Cell Diagnostics, Newark, CA, USA) as described previously (20, 26, 50). Briefly, 40 ZZ ISH probes targeting SARS-CoV-2 genomic RNA fragment from position 21571 to 25392 (GenBank no. LC528233.1; catalog no. 854841), 20 ZZ ISH probes targeting IL-1β (GenBank no. XM_005068610.3; catalog no. 1062331-C1), and 20 ZZ ISH probes targeting CXCL10 (GenBank no. NM_001281344.1; catalog no. 1062371-C1) were designed and synthesized by Advanced Cell Diagnostics. Tissue sections were deparaffinized with xylene, underwent a series of ethanol washes and peroxidase blocking, and were then heated in kit-provided antigen retrieval buffer and digested by kit-provided proteinase. Sections were exposed to ISH target probe pairs and incubated at 40°C in a hybridization oven for 2 h. After rinsing, ISH signal was amplified using kit-provided preamplifier and amplifier conjugated to alkaline phosphatase and incubated with a Fast Red substrate solution for 10 min at room temperature. Sections were then stained with hematoxylin, air dried, and coverslipped.
Immunofluorescence assay method.
Formalin-fixed paraffin-embedded (FFPE) tissue sections were deparaffinized using xylene and a series of ethanol washes. The sections were heated in Tris-EDTA buffer (10 mM Tris base, 1 mM EDTA solution, 0.05% Tween 20; pH 9.0) for 15 min to reverse formaldehyde cross-links. After rinses with PBS (pH 7.4), the sections were blocked with PBT (PBS plus 0.1% Tween 20) containing 5% normal goat serum overnight at 4°C. Then the sections were incubated with primary antibodies: rabbit polyclonal anti-myeloperoxidase (MPO) at a dilution of 1:200 (A039829-2; Dako Agilent Pathology Solutions, Carpinteria, CA, USA), chicken polyclonal anti-glial fibrillary acidic protein (GFAP) antibody at a dilution of 1:1,000 (ab4674; Abcam, Cambridge, MA, USA), mouse monoclonal anti-NeuN antibody at a dilution of 1:200 (MAB377; Millipore Sigma, Burlington, MA, USA), rabbit monoclonal anti-Iba 1 antibody at a dilution of 1:500 (ab178846; Abcam), rabbit monoclonal anti-SARS-CoV-2 nucleocapsid protein (no. 40143-R001; Sino Biological, Chesterbrook, PA, USA), mouse monoclonal anti-E-cadherin antibody at a dilution of 1:100 (33-4000; Thermo Fisher Scientific, Waltham, MA, USA), and/or mouse monoclonal anti-Ki67 at a dilution of 1:200 (clone B56; BD Biosciences, San Jose, CA, USA) for 2 h at room temperature. After rinses with PBS, the sections were incubated with secondary goat anti-rabbit antibody conjugated with Alexa Fluor 488 at a dilution of 1:500 (Thermo Fisher Scientific, Waltham, MA, USA), goat anti-mouse antibody conjugated with Alexa Fluor 568 at a dilution of 1:500, and/or goat anti-chicken antibody conjugated with Alexa Fluor 633 (Thermo Fisher Scientific) at a dilution of 1:500 for 1 h at room temperature. Sections were coverslipped using Vectashield mounting medium with DAPI (4′,6-diamidino-2-phenylindole; Vector Laboratories, Burlingame, CA, USA). Images were captured on a Zeiss LSM 880 confocal system (Zeiss, Oberkochen, Germany) and processed using ImageJ software (National Institutes of Health, Bethesda, MD).
Electron microscopy.
FFPE tissue sections were deparaffinized using xylene and a series of ethanol washes. Samples were fixed with 4% paraformaldehyde, 1% glutaraldehyde, 0.1 M sodium cacodylate buffer for 1 h and then rinsed with buffer. Samples were then postfixed with 2% osmium tetroxide for 1 h, rinsed with buffer, and then dehydrated through an increasing ethanol series, followed by propylene oxide dehydrations for 10-min incubations. Samples were then infiltrated with propylene oxide and Embed812 epoxy resin. Samples were further infiltrated with pure epoxy resin for 24 h, embedded, and allowed to polymerize at 60°C overnight. Sections were cut at 70 to 80 nm and further contrast stained with 1% uranyl acetate and Reynolds’ lead citrate prior to imaging on a JEOL 1011 transmission electron microscope.
ACKNOWLEDGMENTS
Funding was provided through the Defense Health Agency and awarded to J.W.G. and J.W.H. The hACE2 transgenic hamster lines were established through NIH contract /75N93020F00001/A38 (NIH/NIAID). The piggyBac vector, pmhyGENIE-3, was a gift from Stefan Moisyadi at University of Hawaii. The pK18-hACE2 plasmid was a gift from Paul McCray and Stanley Perlman at University of Iowa.
We thank the USAMRIID histology and molecular pathology lab for their technical assistance. Opinions, interpretations, conclusions, and recommendations are ours and are not necessarily endorsed by the U.S. Army or the Department of Defense.
We have no conflicts of interest.
Contributor Information
Joseph W. Golden, Email: joseph.w.golden.civ@mail.mil.
Jay W. Hooper, Email: jay.w.hooper.civ@mail.mil.
Himanshu Batra, The Catholic University of America.
Marthandan Mahalingam, Catholic University of America.
REFERENCES
- 1.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. 2020. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, Wu Y, Zhang L, Yu Z, Fang M, Yu T, Wang Y, Pan S, Zou X, Yuan S, Shang Y. 2020. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med 8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes BJ, Adrover JM, Baxter-Stoltzfus A, et al. 2020. Targeting potential drivers of COVID-19: neutrophil extracellular traps. J Exp Med 217:e20200652. doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salimi S, Hamlyn JM. 2020. COVID-19 and crosstalk with the hallmarks of aging. J Gerontol A Biol Sci Med Sci 75:e34–e41. doi: 10.1093/gerona/glaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang D, Comish P, Kang R. 2020. The hallmarks of COVID-19 disease. PLoS Pathog 16:e1008536. doi: 10.1371/journal.ppat.1008536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye Q, Wang B, Mao J. 2020. The pathogenesis and treatment of the “cytokine storm” in COVID-19. J Infect 80:607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vardhana SA, Wolchok JD. 2020. The many faces of the anti-COVID immune response. J Exp Med 217:e20200678. doi: 10.1084/jem.20200678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DosSantos MF, Devalle S, Aran V, Capra D, Roque NR, Coelho-Aguiar JDM, de Sampaio e Spohr TCL, Subilhaga JG, Pereira CM, D'Andrea Meira I, Niemeyer Soares Filho P, Moura-Neto V. 2020. Neuromechanisms of SARS-CoV-2: a review. Front Neuroanat 14:37. doi: 10.3389/fnana.2020.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hopkins C, Surda P, Whitehead E, Kumar BN. 2020. Early recovery following new onset anosmia during the COVID-19 pandemic—an observational cohort study. J Otolaryngol Head Neck Surg 49:26. doi: 10.1186/s40463-020-00423-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Natoli S, Oliveira V, Calabresi P, Maia LF, Pisani A. 2020. Does SARS-CoV-2 invade the brain? Translational lessons from animal models. Eur J Neurol 27:1764–1773. doi: 10.1111/ene.14277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. 2020. Structural basis of receptor recognition by SARS-CoV-2. Nature 581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280.E8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, Graham BS, McLellan JS. 2020. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. 2020. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 17.Wan Y, Shang J, Graham R, Baric RS, Li F. 2020. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol 94:e00127-20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gretebeck LM, Subbarao K. 2015. Animal models for SARS and MERS coronaviruses. Curr Opin Virol 13:123–129. doi: 10.1016/j.coviro.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subbarao K, McAuliffe J, Vogel L, Fahle G, Fischer S, Tatti K, Packard M, Shieh W-J, Zaki S, Murphy B. 2004. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J Virol 78:3572–3577. doi: 10.1128/jvi.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golden JW, Cline CR, Zeng X, Garrison AR, Carey BD, Mucker EM, White LE, Shamblin JD, Brocato RL, Liu J, Babka AM, Rauch HB, Smith JM, Hollidge BS, Fitzpatrick C, Badger CV, Hooper JW. 2020. Human angiotensin-converting enzyme 2 transgenic mice infected with SARS-CoV-2 develop severe and fatal respiratory disease. JCI Insight 5:e142032. doi: 10.1172/jci.insight.142032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golden JW, Zeng X, Cline CR, Garrison AR, White LE, Fitzpatrick CJ, Kwilas SA, Bowling PA, Fiallos JO, Moore JL, Sifford WB, Ricks KM, Mucker EM, Smith JM, Hooper JW. 2021. Human convalescent plasma protects K18-hACE2 mice against severe respiratory disease. J Gen Virol 102:001599. doi: 10.1099/jgv.0.001599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan JFW, Zhang AJ, Yuan S, Poon VKM, Chan CCS, Lee ACY, Chan WM, Fan Z, Tsoi HW, Wen L, Liang R, Cao J, Chen Y, Tang K, Luo C, Cai JP, Kok KH, Chu H, Chan KH, Sridhar S, Chen Z, Chen H, To KKW, Yuen KY. 2020. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis 71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imai M, Iwatsuki-Horimoto K, Hatta M, Loeber S, Halfmann PJ, Nakajima N, Watanabe T, Ujie M, Takahashi K, Ito M, Yamada S, Fan S, Chiba S, Kuroda M, Guan L, Takada K, Armbrust T, Balogh A, Furusawa Y, Okuda M, Ueki H, Yasuhara A, Sakai-Tagawa Y, Lopes TJS, Kiso M, Yamayoshi S, Kinoshita N, Ohmagari N, Hattori SI, Takeda M, Mitsuya H, Krammer F, Suzuki T, Kawaoka Y. 2020. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci U S A 117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Melo GD, Lazarini F, Levallois S, Hautefort C, Michel V, Larrous F, Verillaud B, Apricio C, Wagner S, Gheusi G, Kergoat L, Kornobis E, Donati F, Cokelaer T, Hervochon R, Madec Y, Roze E, Salmon D, Bourhy H, Lecuit M, Lledo PM. 2021. COVID-19-related anosmia is associated with viral persistence and inflammation in human olfactory epithelium and brain infection in hamsters. Sci Transl Med 13:eabf8396. doi: 10.1126/scitranslmed.abf8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bryche B, St Albin A, Murri S, Lacôte S, Pulido C, Ar Gouilh M, Lesellier S, Servat A, Wasniewski M, Picard-Meyer E, Monchatre-Leroy E, Volmer R, Rampin O, Le Goffic R, Marianneau P, Meunier N. 2020. Massive transient damage of the olfactory epithelium associated with infection of sustentacular cells by SARS-CoV-2 in golden Syrian hamsters. Brain Behav Immun 89:579–586. doi: 10.1016/j.bbi.2020.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brocato RL, Principe LM, Kim RK, Zeng X, Williams JA, Liu Y, Li R, Smith JM, Golden JW, Gangemi D, Youssef S, Wang Z, Glanville J, Hooper JW. 2020. Disruption of adaptive immunity enhances disease in SARS-CoV-2-infected Syrian hamsters. J Virol 94:e01683-20. doi: 10.1128/JVI.01683-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCray PB, Pewe L, Wohlford-Lenane C, Hickey M, Manzel L, Shi L, Netland J, Jia HP, Halabi C, Sigmund CD, Meyerholz DK, Kirby P, Look DC, Perlman S. 2007. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol 81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng J, Wong LR, Li K, Verma AK, Ortiz ME, Wohlford-Lenane C, Leidinger MR, Knudson CM, Meyerholz DK, McCray PB, Jr., Perlman S. 2021. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature 589:603–607. doi: 10.1038/s41586-020-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winkler ES, Bailey AL, Kafai NM, Nair S, McCune BT, Yu J, Fox JM, Chen RE, Earnest JT, Keeler SP, Ritter JH, Kang L-I, Dort S, Robichaud A, Head R, Holtzman MJ, Diamond MS. 2020. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol 21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coperchini F, Chiovato L, Croce L, Magri F, Rotondi M. 2020. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev 53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song E, Zhang C, Israelow B, Lu-Culligan A, Prado AV, Skriabine S, Lu P, Weizman OE, Liu F, Dai Y, Szigeti-Buck K, Yasumoto Y, Wang G, Castaldi C, Heltke J, Ng E, Wheeler J, Alfajaro MM, Levavasseur E, Fontes B, Ravindra NG, Van Dijk D, Mane S, Gunel M, Ring A, Kazmi SAJ, Zhang K, Wilen CB, Horvath TL, Plu I, Haik S, Thomas JL, Louvi A, Farhadian SF, Huttner A, Seilhean D, Renier N, Bilguvar K, Iwasaki A. 2021. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J Exp Med 218. doi: 10.1084/jem.20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellul MA, Benjamin L, Singh B, Lant S, Michael BD, Easton A, Kneen R, Defres S, Sejvar J, Solomon T. 2020. Neurological associations of COVID-19. Lancet Neurol 19:767–783. doi: 10.1016/S1474-4422(20)30221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frontera JA, Sabadia S, Lalchan R, Fang T, Flusty B, Millar-Vernetti P, Snyder T, Berger S, Yang D, Granger A, Morgan N, Patel P, Gutman J, Melmed K, Agarwal S, Bokhari M, Andino A, Valdes E, Omari M, Kvernland A, Lillemoe K, Chou SH-Y, McNett M, Helbok R, Mainali S, Fink EL, Robertson C, Schober M, Suarez JI, Ziai W, Menon D, Friedman D, Friedman D, Holmes M, Huang J, Thawani S, Howard J, Abou-Fayssal N, Krieger P, Lewis A, Lord AS, Zhou T, Kahn DE, Czeisler BM, Torres J, Yaghi S, Ishida K, Scher E, de Havenon A, Placantonakis D, Liu M, Wisniewski T, Troxel AB, Balcer L, Galetta S. 2021. A prospective study of neurologic disorders in hospitalized patients with COVID-19 in New York City. Neurology 96:e575–e586. doi: 10.1212/WNL.0000000000010979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanafi R, Roger PA, Perin B, Kuchcinski G, Deleval N, Dallery F, Michel D, Hacein-Bey L, Pruvo JP, Outteryck O, Constans JM. 2020. COVID-19 neurologic complication with CNS vasculitis-like pattern. AJNR Am J Neuroradiol 41:1384–1387. doi: 10.3174/ajnr.A6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reichard RR, Kashani KB, Boire NA, Constantopoulos E, Guo Y, Lucchinetti CF. 2020. Neuropathology of COVID-19: a spectrum of vascular and acute disseminated encephalomyelitis (ADEM)-like pathology. Acta Neuropathol 140:1–6. doi: 10.1007/s00401-020-02166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Virhammar J, Kumlien E, Fällmar D, Frithiof R, Jackmann S, Sköld MK, Kadir M, Frick J, Lindeberg J, Olivero-Reinius H, Ryttlefors M, Cunningham JL, Wikström J, Grabowska A, Bondeson K, Bergquist J, Zetterberg H, Rostami E. 2020. Acute necrotizing encephalopathy with SARS-CoV-2 RNA confirmed in cerebrospinal fluid. Neurology 95:445–449. doi: 10.1212/WNL.0000000000010250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis A, Jain R, Frontera J, Placantonakis DG, Galetta S, Balcer L, Melmed KR. 2021. COVID-19 associated brain/spinal cord lesions and leptomeningeal enhancement: a meta-analysis of the relationship to CSF SARS-CoV-2. J Neuroimaging 31:826–848. doi: 10.1111/jon.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palahuta HV, Fartushna OY, Yevtushenko SK, Hnepa YY. 2021. Acute transverse myelitis as a neurological complication of Covid-19: a case report. Wiad Lek 74:1045–1049. doi: 10.36740/WLek202104144. [DOI] [PubMed] [Google Scholar]
- 39.Chu H, Chan JF-W, Yuen TT-T, Shuai H, Yuan S, Wang Y, Hu B, Yip CC-Y, Tsang JO-L, Huang X, Chai Y, Yang D, Hou Y, Chik KK-H, Zhang X, Fung AY-F, Tsoi H-W, Cai J-P, Chan W-M, Ip JD, Chu AW-H, Zhou J, Lung DC, Kok K-H, To KK-W, Tsang OT-Y, Chan K-H, Yuen K-Y. 2020. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe 1:e14–e23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bocci T, Bulfamante G, Campiglio L, Coppola S, Falleni M, Chiumello D, Priori A. 2021. Brainstem clinical and neurophysiological involvement in COVID-19. J Neurol 268:3598–3600. doi: 10.1007/s00415-021-10474-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meinhardt J, Radke J, Dittmayer C, Franz J, Thomas C, Mothes R, Laue M, Schneider J, Brünink S, Greuel S, Lehmann M, Hassan O, Aschman T, Schumann E, Chua RL, Conrad C, Eils R, Stenzel W, Windgassen M, Rößler L, Goebel H-H, Gelderblom HR, Martin H, Nitsche A, Schulz-Schaeffer WJ, Hakroush S, Winkler MS, Tampe B, Scheibe F, Körtvélyessy P, Reinhold D, Siegmund B, Kühl AA, Elezkurtaj S, Horst D, Oesterhelweg L, Tsokos M, Ingold-Heppner B, Stadelmann C, Drosten C, Corman VM, Radbruch H, Heppner FL. 2021. Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat Neurosci 24:168–175. doi: 10.1038/s41593-020-00758-5. [DOI] [PubMed] [Google Scholar]
- 42.Burks SM, Rosas-Hernandez H, Alejandro Ramirez-Lee M, Cuevas E, Talpos JC. 2021. Can SARS-CoV-2 infect the central nervous system via the olfactory bulb or the blood-brain barrier? Brain Behav Immun 95:7–14. doi: 10.1016/j.bbi.2020.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fox SE, Akmatbekov A, Harbert JL, Li G, Quincy Brown J, Vander Heide RS. 2020. Pulmonary and cardiac pathology in African American patients with COVID-19: an autopsy series from New Orleans. Lancet Respir Med 8:681–686. doi: 10.1016/S2213-2600(20)30243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haslbauer JD, Tzankov A, Mertz KD, Schwab N, Nienhold R, Twerenbold R, Leibundgut G, Stalder AK, Matter M, Glatz K. 2021. Characterisation of cardiac pathology in 23 autopsies of lethal COVID-19. J Pathol Clin Res 7:326–337. doi: 10.1002/cjp2.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cajanding RJM. 2021. Comprehensive review of cardiovascular involvement in COVID-19. AACN Adv Crit Care 32:169–187. doi: 10.4037/aacnacc2021302. [DOI] [PubMed] [Google Scholar]
- 46.Gilliland T, Liu Y, Li R, Dunn M, Cottle E, Terada Y, Ryckman Z, Alcorn M, Vasilatos S, Lundy J, Larson D, Wu H, Luke T, Bausch C, Egland K, Sullivan E, Wang Z, Klimstra WB. 2021. Protection of human ACE2 transgenic Syrian hamsters from SARS CoV-2 variants by human polyclonal IgG from hyper-immunized transchromosomic bovines. bioRxiv. doi: 10.1101/2021.07.26.453840. [DOI]
- 47.Harcourt J, Tamin A, Lu X, Kamili S, Sakthivel SK, Murray J, Queen K, Tao Y, Paden CR, Zhang J, Li Y, Uehara A, Wang H, Goldsmith C, Bullock HA, Wang L, Whitaker B, Lynch B, Gautam R, Schindewolf C, Lokugamage KG, Scharton D, Plante JA, Mirchandani D, Widen SG, Narayanan K, Makino S, Ksiazek TG, Plante KS, Weaver SC, Lindstrom S, Tong S, Menachery VD, Thornburg NJ. 2020. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg Infect Dis 26:1266–1273. doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]
- 49.Prophet EB, Mills B, Arrington JB, Sobin LH. 1992. Laboratory methods for histotechnology. Armed Forces Institute of Pathology, Washington, DC. [Google Scholar]
- 50.Liu J, Babka AM, Kearney BJ, Radoshitzky SR, Kuhn JH, Zeng X. 2020. Molecular detection of SARS-CoV-2 in formalin fixed paraffin embedded specimens. JCI Insight 5:e139042. doi: 10.1172/jci.insight.139042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lung pathology in day 5 animals. (A) H&E staining showing multifocal to coalescing areas of alveolar inflammation with moderate severity. The right panels are an expansion of boxed areas, showing mononuclear inflammation in alveoli (septa and lumen) with hemorrhage, edema, and fibrin exudation. There was regeneration of bronchiole epithelium (bottom left, arrows). Lower panels show ISH staining for SARS-CoV-2 genome in same region seen in the H&E panels. Note viral labeling (red) in areas of inflammation and rarely in bronchiole epithelium (expanded view of the boxed area in the right panel). (B) Lung from a different day 5 animal shows degeneration, necrosis, and loss of bronchiole epithelium and peribronchiole inflammation. (Middle) ISH staining of the same panel as H&E. (Right) Magnified H&E image of the black box in the left panel, highlighting degeneration, necrosis, and sloughing of bronchiole epithelium as well as mild mononuclear inflammation in the bronchiole adventitial layer. Download FIG S1, PDF file, 0.6 MB (648.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Viral RNA in the olfactory bulb. In situ hybridization for SARS-CoV-2 RNA shows prominent labeling in the all layers of the olfactory bulb in a day 5 animal, including (from outside in) the olfactory nerve layer, glomerular layer, external plexiform layer, mitral cell layer, internal plexiform layer, and granule cell layer. Labeling is absent in an uninfected control. Download FIG S2, PDF file, 0.4 MB (376.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Electron micrograph of the nasal cavity. (A and B) Representative images of macrophages showing the cytoplasmic presence of SARS-CoV-2. Right panels show higher magnifications of the boxed areas in the left panels. (C) SARS-CoV-2-infected epithelial cell exhibiting potential virus budding from the plasma membrane (arrows) and in the apical lumen (arrowhead). Download FIG S3, PDF file, 0.6 MB (592KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Infection of control animals. Representative ISH and H&E staining of the nasal cavity of negative-control hamsters. A single control animal was incidentally exposed to the virus, as indicated by ISH (red). during the challenge period. These tissues were harvested on day 3. Animals were in different locations in the same room in barrier cages and purposely not manipulated (weights, etc.). Download FIG S4, PDF file, 0.4 MB (400.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Nasal turbinate histopathology in wild-type Syrian hamsters. H&E and ISH staining in nasal turbinates of wild-type hamsters on days 3 and 7 from a separate study, presented here for comparison. Note that the level of inflammation and damage to the olfactory and respiratory epithelium, as well as the viral labeling, are roughly similar to what was seen in the K18-ACE2 hamsters on day 3. On day 7, the levels of inflammation and labeling were significantly decreased and there was regeneration of epithelium. This contrasts with the day 5 transgenic animals in this study, where the histologic lesions (epithelial necrosis, inflammation, exudate) increased in severity on day 5 compared to day 3. Red boxes in upper images correspond to higher-magnification images below. Download FIG S5, PDF file, 0.5 MB (515.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Lung pathology from day 3 SARS-CoV-2 infected wild-type Syrian hamsters. H&E and ISH staining in SARS-CoV-2-exposed wild-type hamsters harvested on day 3. Note the presence of areas of alveolar inflammation, bronchiole epithelial degeneration/regeneration, and perivascular inflammation affecting roughly half the section, which corresponds to areas of strong viral labeling. Lung lesions and viral labeling were increased in severity compared to the lung lesions from day 3 transgenic hamsters. Red boxes in upper images correspond to higher-magnification images below. Download FIG S6, PDF file, 0.7 MB (685.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Lung pathology from day 7 SARS-CoV-2-infected wild-type Syrian hamsters. H&E and ISH staining in SARS-CoV-2-exposed wild-type hamsters harvested on day 7. Note the presence of patchy areas of increased cell density characterized by prominent type II pneumocyte hyperplasia, residual alveolar inflammation, and perivascular inflammation; there is only rare viral labeling seen in few bronchiole epithelial cells (arrows) and inflammatory cells. These findings contrast with the day 5 transgenic animals in this study, where there was ongoing damage to bronchiole epithelium, much less type II pneumocyte hyperplasia, and more prominent viral labeling. Red boxes in upper images correspond to higher-magnification images below. Download FIG S7, PDF file, 0.6 MB (585KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.