ABSTRACT
Acinetobacter baumannii infection poses a major health threat, with recurrent treatment failure due to antibiotic resistance, notably to carbapenems. While genomic analyses of clinical strains indicate that homologous recombination plays a major role in the acquisition of antibiotic resistance genes, the underlying mechanisms of horizontal gene transfer often remain speculative. Our understanding of the acquisition of antibiotic resistance is hampered by the lack of experimental systems able to reproduce genomic observations. We here report the detection of recombination events occurring spontaneously in mixed bacterial populations and which can result in the acquisition of resistance to carbapenems. We show that natural transformation is the main driver of intrastrain but also interstrain recombination events between A. baumannii clinical isolates and pathogenic species of Acinetobacter. We observed that interbacterial natural transformation in mixed populations is more efficient at promoting the acquisition of large resistance islands (AbaR4 and AbaR1) than when the same bacteria are supplied with large amounts of purified genomic DNA. Importantly, analysis of the genomes of the recombinant progeny revealed large recombination tracts (from 13 to 123 kb) similar to those observed in the genomes of clinical isolates. Moreover, we highlight that transforming DNA availability is a key determinant of the rate of recombinants and results from both spontaneous release and interbacterial predatory behavior. In the light of our results, natural transformation should be considered a leading mechanism of genome recombination and horizontal gene transfer of antibiotic resistance genes in Acinetobacter baumannii.
KEYWORDS: AbaR, Acinetobacter baumannii, OXA23, type VI secretion system, antibiotic resistance, carbapenem resistance, horizontal gene transfer, natural transformation, resistance island
INTRODUCTION
Acinetobacter baumannii is a Gram-negative bacterium responsible for a wide range of infections in both humans and animals (1, 2). This multidrug-resistant (MDR) agent poses a health threat, particularly in intensive care units, where it can lead to bacteremia and ventilator-associated pneumonia. Consequently, secondary infections with multidrug-resistant A. baumannii have been reported during the COVID-19 pandemic (3, 4). A. baumannii infections are frequently resistant to multiple antibiotics, including to carbapenems. For Europe only, a combined resistance to fluoroquinolones, aminoglycosides, and carbapenems is observed for nearly 30% of invasive Acinetobacter sp. isolates (5). In A. baumannii, carbapenem resistance is mainly associated with the expression of OXA23 carbapenemase, encoded by the blaOXA-23 gene (6). The composite transposon Tn2006, formed by two ISAba1 insertion sequences framing the blaOXA-23 gene, is the main genetic context for the blaOXA-23 gene and can be found in different locations, including plasmids, but it is most often found inserted in the large resistance island AbaR4 (6, 7). Such large and diverse A. baumannii resistance islands (Ab-RI) are potential contributors to the multidrug resistance phenotype observed in A. baumannii. The first description of an Ab-RI was reported in 2006 in the epidemic strain AYE with the AbaR1 island consisting in an 86-kb-long genomic structure (8). Since then, analysis of more than 3,000 A. baumannii genomes revealed that Ab-RI are present in nearly 65% of them (9). These genomic islands present a great diversity in gene content, with often multiple putative antibiotic resistance genes (10, 11). Ab-RI were presumed to be initially acquired through plasmid conjugation followed by chromosomal insertion and to evolve through multiple insertions and rearrangements of insertion sequences (12, 13). However, genome analyses also support their horizontal transfer between distantly related isolates, as exemplified by the acquisition of the 21-kb-long AbGRI3 resistance island (14) and the 35-kb-long AbGRI5 (15). Both acquisitions involved large recombination events (up to 34 kb-long) at the sequence flanking the island. Indeed, high rates of genome recombination are a hallmark of A. baumannii genomes (16, 17). Recombination events may have led to the acquisition by MDR strains of parC or gyrA alleles conferring resistance to fluoroquinolones and of ISAba1 upstream of the ampC gene, leading to resistance to 3rd-generation cephalosporins (14, 17, 18). Genome analysis of isolates from a longitudinal study also offered a glimpse of the gene transfer and recombination going on in the hospital setting, with the acquisition of the blaOXA-72 variant of the blaOXA24/40-type carbapenemase gene and recombination affecting the blaOXA-51 locus (19).
Despite their importance in the evolution of A. baumannii into an MDR pathogen, the mechanisms leading to genome recombination and recombination-dependent acquisition of resistance genes and Ab-RI remain elusive. Indeed, few studies have experimentally investigated the horizontal transfer of chromosomal antibiotic resistance genes in A. baumannii. The chromosomal transfer of an 11-kb-long Tn215 harboring blaNDM-1 during mating of isolate R2090 to the reference strain CIP 70.10 was experimentally observed and involved the acquisition of a 65-kb region through homologous recombination (20). While transduction by prophages was suggested, the mechanism of transfer was not elucidated. Phage particles, present in prepared fractions from culture supernatant of the clinical isolate NU-60, were found to mediate chromosomal transfer to the reference strain ATCC 17978 (21). If generalized transduction by strain-specific prophages is a potential mechanism of resistance gene transmission, natural transformation is another potential and highly conserved route. Natural transformation allows bacteria to actively import exogenous DNA, and provided there is sufficient sequence identity, it integrates the recipient cell’s genome by homologous recombination (22). Most A. baumannii isolates and also closely related Acinetobacter nosocomialis were found to be capable of natural transformation when presented with purified DNA (23–27). Yet we currently have a limited understanding of the role of natural transformation in genome dynamics and the spread of antibiotic resistance in A. baumannii populations.
In the current study, we explored the intraspecies and intragenus transfer of antibiotic resistance, notably to carbapenems, which is spontaneously occurring in mixed populations of pathogenic Acinetobacter spp. We provide evidence that natural transformation is the main transfer route and fosters recombination events and the acquisition of multiple resistance genes carried by large genomic island. Our results obtained experimentally replicate the large recombination events observed in A. baumannii chromosomes and suggest a major role played by natural transformation in the dynamics of A. baumannii genomes.
RESULTS
Intra- and interspecies recombinants are produced in mixed populations of pathogenic Acinetobacter.
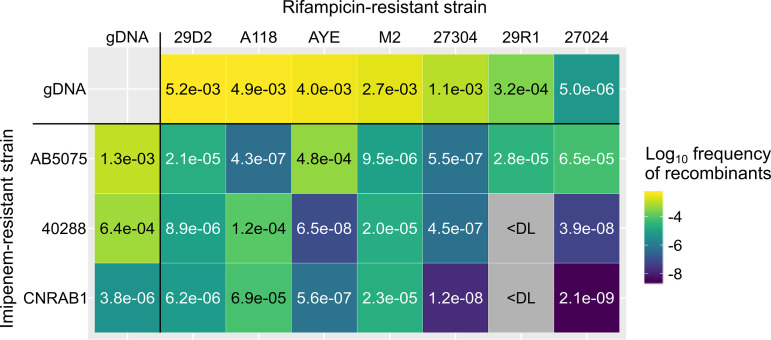
To test the possible transfer of antibiotic resistance in bacterial populations, we first assessed if recombinants could arise in mixed cultures of isolates harboring distinct resistance determinants. Three imipenem-resistant (Imir) A. baumannii clinical isolates (AB5075, 40288, and CNRAB1) and seven imipenem-sensitive (Imis) isolates of either A. baumannii (29D2, A118, AYE, 27304, 29R1, and 27024) or A. nosocomialis (M2) were selected. Rifampicin-resistant (Rifr) mutants of the Imis isolates were obtained. Each of these imipenem-sensitive and Rifr isolates was grown for 24 h in mixed culture with each Imir isolate. The frequency of Rifr Imir recombinants in the mixed cultures was then determined. Rifr Imir recombinants were detected in 19 of the 21 tested combinations, with highly variable frequencies ranging from 2.10 × 10−9 to 4.82 × 10−4 (Fig. 1). Importantly, an imipenem-sensitive isolate did not become Imir in the absence of an Imir isolate. And similarly, in the absence of Rifr isolates, Imir isolates spontaneously develop resistance to rifampicin at frequencies below the detection limit of this assay (10−9). This strongly suggests that Rifr Imir recombinants result from horizontal gene transfer (HGT) between the mixed isolates. All tested isolates were capable of natural transformation under the tested growth conditions when presented with purified genomic DNA (gDNA) extracted from their own Rifr derivative (Fig. 1). We could not correlate transformation efficiency with nucleotide relatedness, as most pairs had comparable nucleotide identities (around 98% [see Fig. S1 in the supplemental material]), with the exception of the pairing with A. nosocomialis strain M2, which had the lowest nucleotide identities (around 92%) but high recombinant frequencies. Most isolates showed high transformability (transformation frequencies > 1 × 10−3), while two isolates (CNRAB1 and 27024) presented lower transformation frequencies (1 × 10−6). Pairing these two isolates generated only a few recombinants, while pairing with a more transformable isolate generated up to 10,000 times more recombinants, suggesting that at least one of the two isolates must be transformable to obtain recombinants. Yet some combinations of highly transformable isolates were poorly productive in recombinants (40288 × AYE), suggesting that factors other than the intrinsic transformability of isolates play a role in the production of recombinant progeny.
FIG 1.
Recombinants are produced in mixed cultures of A. baumannii and A. nosocomialis. Bacterial suspensions of rifampicin-resistant (Rifr) isolates of carbapenem-sensitive strains (29D2, A118, AYE, M2, 27304, 29R1, and 27024) were mixed with the carbapenem-resistant (Imir) clinical isolates AB5075, 40288, and CNRAB1 using cultures adjusted to an OD600 of 0.01. The mixture (2.5 μL) was deposited on the surface of tryptone-NaCl medium and incubated overnight at 37°C. Rifr and Imir recombinants were determined after 24 h of mixed culture. Recombinant frequencies represent the ratio of Rifr and Imir CFU over the total CFU counts. Frequency of recombinants is presented as a heat map with indicated average frequencies from two or three biological replicates with three technical replicates each. <DL, below detection limit (∼10−9). Top row and outermost left column show the transformation frequency displayed under the same condition by the unmixed isolates, tested by incubating the original isolates with genomic DNA (gDNA) extracted from their Rifr derivatives and select with rifampicin. The transformation frequencies (ratio of Rifr CFU by total CFU counts) are represented by a color gradient from blue to yellow. Data shown are the means of natural transformation assays done in two or three biological replicates with three technical replicates each.
Recombinant frequency as a function of genomic relatedness between paired strains. Analysis of recombinant frequencies for paired cotransformants as presented in Fig. 1 in the main text depending on the pairwise average nucleotide identity (ANI). For each pairing, the imipenem-resistant strain is highlighted in panel A and the rifampicin-resistant one in panel B. Download FIG S1, TIF file, 0.4 MB (400.5KB, tif) .
Copyright © 2022 Godeux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Pathogenic Acinetobacter rapidly acquire carbapenem resistance by natural transformation in mixed culture.
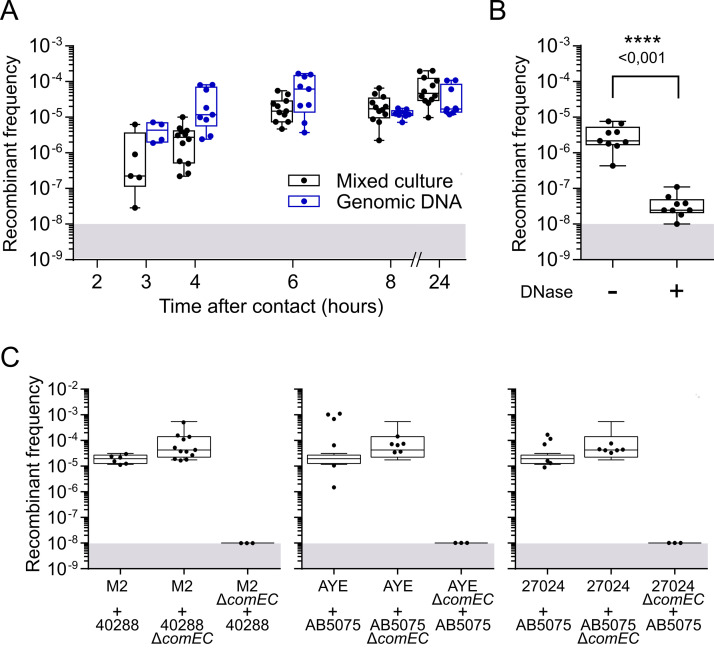
The conditions under which we observed the rise of recombinants are permissive to natural transformation, yet we could not exclude other mechanisms of horizontal gene transfer. Natural transformation was previously reported to occur during exponential growth of A. baumannii (27, 28). Therefore, we sought to determine whether the kinetics of increasing recombinant numbers under mixed culture are consistent with a role of natural transformation. Focusing on the M2 × 40288 combination of two pathogenic Acinetobacter strains, we could detect recombinants as early as 3 h after mixing the two isolates (Fig. 2A). The frequency of recombinants reached a plateau at 6 h. The kinetics of recombinant emergence were strikingly similar when we provided the M2 isolate with genomic DNA of 40288, further indicating that recombinants emerge by natural transformation (Fig. 2A). Moreover, in a 4-h-long mixed-culture experiment, addition of DNase I reduced the size of the recombinant population by 2 orders of magnitude, ruling out the role of phages or outer membrane vesicles in recombinant production (Fig. 2B). To further validate the role of natural transformation, we tested the emergence of recombinants with ΔcomEC::aacC4 derivatives, deficient in DNA import. Combination of the M2 strain with the 40288 ΔcomEC mutant did not alter the level of recombinants (Fig. 2C). However, inactivation of comEC in the M2 strain completely abolished recombinants production indicating that the recombinants are formed from M2 bacteria that have acquired carbapenem resistance by natural transformation. Consistent with the role of natural transformation, acquisition of carbapenem resistance in mixed culture requires functional type IV pili and recombination machinery, as M2 ΔpilA::kan and ΔrecA::aacC4 derivatives did not produce any recombinants (Fig. S2). Similarly, the results demonstrate that the A. baumannii strains AYE and 27024 use natural transformation to acquire carbapenem resistance from AB5075 (Fig. 2C). Interestingly, carbapenem-resistant transformants of 27024 are produced at higher rates than the transformation rate of 27024 when tested with its own gDNA, conferring resistance to rifampicin (Fig. 1). This suggests that acquisition of carbapenem resistance is more effective than the acquisition of the mutation in the rpoB gene, conferring resistance to rifampicin. The mutations in this housekeeping gene are probably highly costly in terms of fitness (29, 30), which may explain the absence of recombinants resulting from the acquisition of the rifampicin resistance by transformable carbapenem-resistant isolates. In conclusion, in mixed populations of Acinetobacter, carbapenem resistance can rapidly and efficiently spread to susceptible isolates by natural transformation.
FIG 2.
Acquisition of carbapenem resistance by natural transformation in a mixed culture. (A) Kinetics of emergence of carbapenem- and rifampicin-resistant recombinants in a mixed culture of M2 Rifr and 40288 (Imir). Bacterial suspensions of M2 Rifr at an OD600 of 0.01 were mixed with either an equal volume of a suspension of 40288 at an OD600 of 0.01 or gDNA extracted from 40288 (at 200 ng/μL). The mixture (2.5 μL) was deposited on the surface of tryptone-NaCl medium and incubated overnight at 37°C. Recombinant frequencies represent the ratio of Rifr and Imir CFU over the total. The limit of detection (10−8) is indicated by the gray area. (B) Sensitivity of recombinant frequencies to DNase I treatment under the same conditions as for panel A, with a mixed culture of 6 h. (C) Recombinants emerge by the ComEC-dependent acquisition of carbapenem resistance by susceptible isolates. Rifr isolates of M2, AYE, 27024, or their comEC mutants were mixed with the carbapenem-resistant (Imir) clinical isolates 40288 and AB5075 or their comEC mutants. Rifr and Imir recombinants were determined after 24 h of mixed culture. The boxplots represent the distributions of Rifr and Imir recombinants divided by the total CFU count (recombinant frequencies). A horizontal line represents the median. The limit of detection (10−8) is indicated by the gray area. In panel B, statistical analysis was conducted using the nonparametric Mann-Whitney-Wilcoxon test (two tailed). P values are indicated.
Recombinants emerge in mixed culture in a PilA- and RecA-dependent manner. Rifampicin-resistant (Rifr) isolates of M2 or its pilA::kan or recA::aacC4 mutant were mixed with the carbapenem-resistant (Imir) clinical isolate 40288 ΔcomEC mutant. Rifr and Imir recombinants were determined after 24 h of mixed culture. The boxplots represent the distributions of Rifr and Imir recombinants divided by the total CFU count (recombinant frequencies). A horizontal line represents the median. The limit of detection (10−8) is indicated by the gray area. Download FIG S2, TIF file, 0.1 MB (129.5KB, tif) .
Copyright © 2022 Godeux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Chromosomal DNA transfer is stimulated by T6SS-killing activity but primarily relies on contact-independent spontaneous release of DNA.
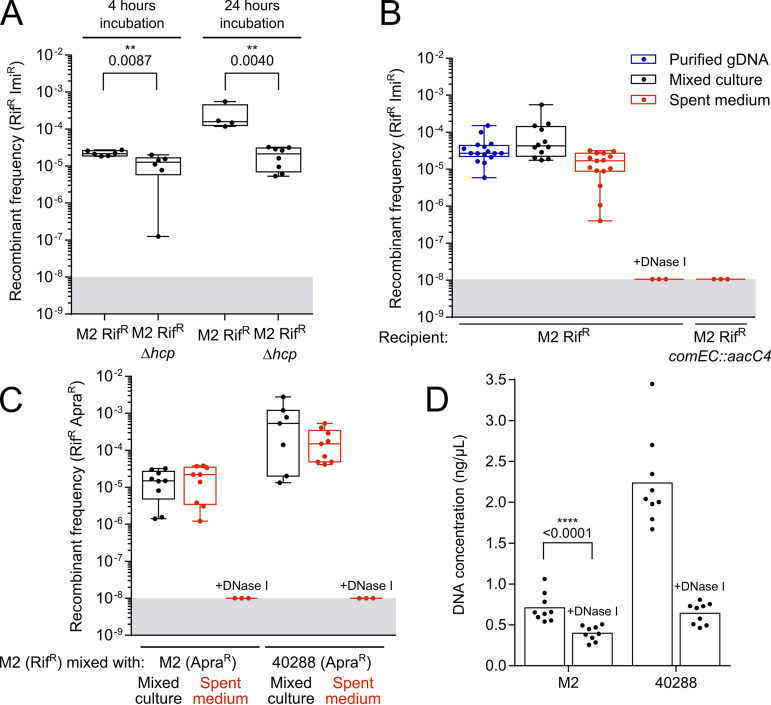
The essential role of natural transformation in the acquisition of carbapenem resistance suggests that chromosomal DNA of the carbapenem-resistant isolate is released during mixed culture. In Vibrio cholerae, the type VI secretion system (T6SS) mediates the killing of nonkin cells, promoting the release of DNA from prey cells, which is then imported by natural transformation by the predatory bacteria (31). Acinetobacter baylyi also produces a T6SS to kill prey, allowing the uptake of their DNA (32). In order to determine the contribution of T6SS-dependent killing to the chromosomal DNA transfer in the Acinetobacter mixed culture, we examined the combination of strains 40288 and M2, the latter being known for producing an active T6SS in LB medium (33). Under these conditions, we confirmed that M2 efficiently kills the 40288 strain, with a reduction of the population of 40288 by 4 orders of magnitude (Fig. S3A). A Δhcp mutant of the M2 strain could not kill 40288, demonstrating that killing is indeed T6SS dependent (Fig. S3A). However, under the condition in which we observed transformation-dependent chromosomal transfer, M2 T6SS is presumably poorly active, with comparable viable 40288 cells titers when mixed for 4 h with either M2 or its Δhcp derivative (Fig. S3B). Yet inactivation of M2 T6SS slightly lowered the rate of recombinants at 4 h, and reduced it by 1 order of magnitude at 24 h (Fig. 3A). This indicates that residual T6SS activity contributes to DNA release and chromosomal DNA acquisition by the M2 strain. Yet even in the absence of a functional T6SS, recombinant frequency remained high (10−5). We thus hypothesized that DNA is also released independently of contact-dependent killing mediated by the T6SS. To test this hypothesis, we collected culture supernatant of 40288 at 4 h in the absence of M2. The M2 strain presented with this spent medium could acquire carbapenem resistance almost as efficiently as when grown with 40288 or when presented with purified genomic DNA from 40288 (Fig. 3B). No carbapenem-resistant recombinant was observed when the spent medium was treated with DNase I or if the M2 strain was inactivated for DNA uptake (ΔcomEC). This demonstrated that contact-independent DNA release is sufficient to promote chromosomal DNA acquisition by nonkin cells.
FIG 3.
Contribution of contact-dependent T6SS-killing activity and contact-independent DNA release to the acquisition of the carbapenem resistance in mixed cultures. (A) Contribution of the T6SS to the frequency of recombinants generated in a mixed culture of 40288 and M2 Rifr. 40288 was mixed with the M2 Rifr strain or the M2 Rifr Δhcp mutant, defective for T6SS activity. Imipenem-resistant transformants of the M2 strain were determined after 4 h or 24 h of mixed culture. Transformation frequencies represent the ratio of M2 Rifr Imir CFU over the total count of M2 Rifr CFU. (B) Contribution of contact-independent DNA release by 40288 to the acquisition of the imipenem resistance by M2 in mixed culture. Bacterial suspensions of M2 Rifr at an OD600 of 0.01 were mixed with an equal volume of either genomic DNA extracted from 40288 (at 200 ng/μL), a suspension of 40288 at an OD600 of 0.01, or filtered spent medium of a 4-h culture in tryptone-NaCl medium of 40288. The mixture (2.5 μL) was deposited on the surface of tryptone-NaCl medium and incubated for 6 h at 37°C. Recombinant frequencies represent the ratio of Rifr and Imir CFU over the total. (C) Contact-independent DNA release promotes both intra- and interstrain recombination. The frequency of intrastrain recombinants from M2 Rifr and M2 Aprar strains was determined in mixed culture or when M2 Rifr was exposed to the spent medium of the M2 Aprar strain. To test for interstrain recombination, M2 Rifr was mixed with 40288 Aprar or with the spent medium from that strain. Recombinant frequencies represent the ratio of Rifr and Aprar CFU over the total. (D) Fluorescence-based quantification of DNA present in the spent medium of strains M2 and 40288. Samples treated with DNase I represent the detection limit of the assay (∼0.4 ng). When displayed, P values of statistical analysis were obtained using the nonparametric Mann-Whitney-Wilcoxon test (two tailed). The limit of detection (10−8) is indicated by the gray area.
T6SS-dependent killing activity of the M2 strain on the 40288 isolate in LB medium and under conditions of mixed culture. Recovery of the A. baumannii strain 40288 incubated either with the wild-type M2 strain (WT) or with its hcp gene mutant derivative (Δhcp) is shown. (A) Cocultivation of 40288 with the M2 strain during in LB medium as described previously (1) resulted in a 4-log decrease of the 40288 population, while inactivating the T6SS of M2 (Δhcp) abolished the killing activity. (B) In the mixed-culture setup used to observe the rise of recombinants, mixed culture of 40288 with M2 for 4 h did not significantly reduce the population size of 40288. Download FIG S3, TIF file, 0.4 MB (392.5KB, tif) .
Copyright © 2022 Godeux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Given that DNA release is independent of the presence of another strain, DNA release and transformation may also occur between individuals of the same strain. We tested this possibility by examining the ability of M2 Rifr individuals to acquire the apramycin resistance (Aprar) marker from an M2 ΔcomEC mutant. In this mixed-culture setup, Rifr Aprar recombinants occurred at a frequency of 10−5 (Fig. 3C). The same frequency was observed when using spent medium of the M2 Aprar ΔcomEC mutant to transform M2 Rifr. Together, these findings show that M2 cells spontaneously release DNA that can serve to transform other cells of the population. Yet in a mixed-culture setup, M2 acquired the apramycin resistance marker from 40288 ΔcomEC at higher rates than from itself (M2 ΔcomEC) (Fig. 3C). The same result was obtained with the spent medium, raising the possibility that the DNA released by 40288 was more potent in producing transformants. As M2 can be transformed with the same efficiency with either M2 or 40288 gDNA (Fig. S4), we hypothesized that 40288 simply releases more DNA than the M2 strain. Indeed, quantification of DNA in spent medium from 4-h cultures showed that 40288 released at least 3 times more DNA than M2 (Fig. 3D). Yet surprisingly, quantities of DNA in the spent media remained very low, with concentrations of around 2 ng/μL for 40288 strain and 0.7 ng/μL for M2 strain, almost as low as the detection limit (0.4 ng/μL, obtained with DNase I-treated samples). However, this small amount collected at 4 h of growth was sufficient to generate transformants at frequencies comparable to those obtained with about 100-times-larger amounts of purified gDNA in a 6-h window (Fig. 3B). Overall, our results indicate that the amount of spontaneously released DNA is the major determinant of the rate of recombinants generated from individuals of the same or different strains. In conclusion, the T6SS-dependent and contact-independent release of small amounts of genomic DNA by pathogenic Acinetobacter fuels horizontal gene transfer and the acquisition of antibiotic resistance genes.
Strain M2 of A. nosocomialis is equally transformed with DNA from itself and from the distantly related strain 40288 of A. baumannii. Strain M2 was incubated under transformation conditions (>16 h) with genomic DNA extracted either from an M2 ΔcomEC::aacC4 (Aprar) or from a 40288 ΔcomEC::aacC4 (Aprar) strain. Transformation frequency represents the CFU count on an apramycin plate divided by the total CFU count on nonselective plates. Download FIG S4, TIF file, 0.1 MB (131.3KB, tif) .
Copyright © 2022 Godeux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Acquisition of resistance can result from the horizontal transfer of large genomic islands.
To characterize the chromosomal transfer of the imipenem resistance at a genetic level, we sought to identify the imipenem resistance determinant in the 40288 donor strain and in transformants. 40288 is a carbapenem-resistant strain isolated from a diseased animal and carries the blaOXA-23 carbapenem resistance gene (34). An initial assembly of the 40288 genome using Illumina reads resulted in 54 contigs, and the blaOXA-23 carbapenem resistance gene was found in a 2.4-kb contig whose exact location in the genome remained unresolved. A hybrid assembly combining the Illumina reads with long reads (Oxford Nanopore technology) resulted in the complete genome of 40288 showing a single circular contig of 4,084 kb. The genome revealed the presence of the blaOXA-23 gene as part of the 4.8-kb Tn2006 transposon inserted at two distinct locations. One Tn2006 transposon is within the large AbaR4 island of 16 kb inserted into the comM gene. We previously demonstrated that in A. baumannii the presence of a resistance island in this gene reduces the transformation level (35). Consistently, acquisition of AbaR4 into the comM gene subsequently lowers the transformation level of the recombinant strains 10-fold (Fig. S5A). The other copy of Tn2006 is inserted into the beginning of the vgrG gene, encoding a T6SS component. This gene, in a putative operon with a LysM domain effector, is hereafter referred as vgrG3 (36). Insertion of Tn2006 into the vgrG3 gene did not impair T6SS-mediated killing, as introduction of this insertion in the vgrG3 gene of the M2 strain did not alter its T6SS-medated killing of Escherichia coli and A. baumannii, in contrast to the case with a Δhcp mutant (Fig. S5B and C).
Phenotypic outputs of acquisition of AbaR4 and Tn2006 from strain 40288. (A) Acquisition of AbaR4 in the comM gene lowers the transformation levels of the recipient strain. Bacterial suspensions of M2 Rifr or two of its derivatives that acquired AbaR4 in the comM gene in previous mixed-culture experiments were mixed with an equal volume of 40288 ΔcomEC::aacC4, all strains at an OD600 of 0.01. The mixture (2.5 μL) was deposited on the surface of transformation medium. Recombinant frequency represents the ratio of M2 Rifr Aprar CFU over the total count of Rifr CFU. P values of statistical analysis were obtained using the nonparametric Mann-Whitney-Wilcoxon test (two tailed). The limit of detection (10−8) is indicated by the gray area. (B) T6SS-mediated killing assay of a VgrG3 impaired mutant of A. nosocomialis strain M2 in LB medium. Killing assays were performed as described for Fig. S1A. Escherichia coli strain JW2912 (Kanr) and A. baumannii strain 40288 were both used as prey and were incubated either with the wild-type M2 strain, its hcp gene mutant derivative (Δhcp), or an imipenem-resistant transformant carrying an insertion into the vgrG3 gene (vgrG3::Tn2006; two transformants tested). Download FIG S5, TIF file, 0.6 MB (596.1KB, tif) .
Copyright © 2022 Godeux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
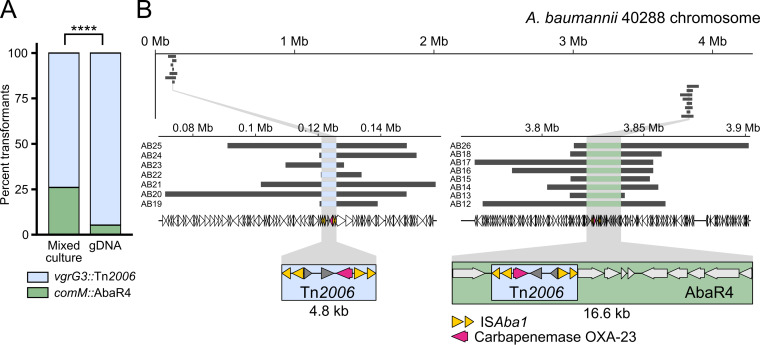
Importantly, we found that either copy of blaOXA-23 confers the same level of resistance to imipenem on the M2 strain (Table S2). However, the rate of their horizontal transfer may be affected by their genetic context. Acquisition of the blaOXA-23 gene from vgrG3::Tn2006 requires incorporation of 4.8 kb of heterologous sequence, while acquisition of the other copy would involve acquisition of the whole AbaR4 island of 16.6 kb. When genomic DNA from 40288 is provided to the M2 strain, the vast majority of imipenem-resistant transformants results from the acquisition of vgrG3::Tn2006, with only 5% of transformants acquiring the 16-kb-long AbaR4 island in comM (Fig. 4A). In contrast, under the mixed-culture condition, this percentage increased 5-fold (Fig. 4A), suggesting that the mixed culture allowed the acquisition of large DNA fragments. To further determine the length of imported DNA molecules under these conditions, we characterized by whole-genome sequencing the extent of DNA recombination which had occurred in the M2 transformants. To this end, 15 imipenem-resistant transformants of M2 obtained from mixed cultures with 40288 were analyzed: 7 resulting from the acquisition of the vgrG3::Tn2006 insertion and 8 from the acquisition of comM::AbaR4. We used the nearly 300,000 single nucleotide polymorphisms (SNPs) between the two strains to identify the parts of the 40288 genome which had been incorporated into the M2 genome (Fig. S6). The succession of acquired SNPs revealed discontinuous recombination tracts on each side of the acquired heterologous sequence (Fig. S6). The discontinuous tracts most likely reflect multiple strand invasion events from the same imported DNA molecule (37). We thus used the position of the outermost acquired SNP to estimate the length of the DNA molecule of 40288 which was imported by the M2 recipients (Fig. 4B). We estimated that recombinants which have acquired Tn2006 in vgrG3 had imported DNA fragments ranging from 13 to 77 kb, while those acquiring AbaR4 had imported DNA fragments ranging from 27 to 89 kb (Table S3). Thus, transformants resulted from the recombination of DNA molecules much larger than the acquired heterologous sequences. This suggests that the mixed culture is prone to allow the acquisition of large heterologous sequences such as those of genomic islands.
FIG 4.
Genomic analysis of the acquisition of imipenem resistance. (A) PCR-based analysis of the chromosomal location of the Tn2006 transposon and associated blaOXA-23 gene, conferring resistance to imipenem. One hundred Imir and blaOXA-23 PCR-positive CFU from 2 independent transformation assays were analyzed by PCR probing insertion in the comM gene. Statistical significance was calculated using Pearson’s chi-squared test returning a P value of <0.0001 (****). (B) Graphical representation of the chromosome of the 40288 strain (thin black line) and location of the acquired DNA fragments (light gray thick lines) by transformants of the A. nosocomialis strain M2 during mixed culture with 40288. Bottom left and right diagrams represent close-ups of the regions in which the blaOXA-23 gene is acquired as part of the Tn2006 element (blue box) in the vgrG3 gene and as part of the Tn2006 element within the AbaR island (green box) inserted in the comM gene. Acquired regions were determined by sequencing the genomes of imipenem-resistant recombinants of M2 which had acquired blaOXA-23 on a NovaSeq instrument. Variant calling and identification of converted markers (SNPs of 40288 acquired by M2) were used to delineate the acquired regions (see Materials and Methods).
Genomic analysis of the acquisition of imipenem resistance. Graphical representation of the chromosome of the 40288 strain (boxed area) and location of the acquired DNA fragments (light gray thick lines) by transformants of A. nosocomialis strain M2 during mixed culture with 40288. Top and bottom panels represent close-ups of the regions in which the blaOXA-23 gene is acquired as part of the Tn2006 element (top box) in the vgrG3 gene and as part of the Tn2006 element within the AbaR island (bottom box) inserted in the comM gene. Acquired regions were determined by sequencing the genomes of imipenem-resistant recombinants of M2 which had acquired blaOXA-23 on a NovaSeq instrument. Variant calling and identification of converted markers (SNPs of 40288 acquired by M2) were used to delineate the acquired regions (see Materials and Methods). Thin green lines represent coverage, and the outermost right scale indicates the results of read screening corresponding to the AbaR sequence in recombinants. Orange triangles represent individual SNPs used to determine recombination tracts. Download FIG S6, TIF file, 0.5 MB (545.2KB, tif) .
Copyright © 2022 Godeux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Antimicrobial susceptibility profiles of mixed-culture transformants. Download Table S2, PDF file, 0.03 MB (32.5KB, pdf) .
Copyright © 2022 Godeux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Estimated length of the imported DNA molecules leading to the detected recombination tracts. REFERENCE 1. Fournier P-E, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, Richet H, Robert C, Mangenot S, Abergel C, Nordmann P, Weissenbach J, Raoult D, Claverie J-M. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet 2:e7. https://doi.org/10.1371/journal.pgen.0020007. Download Table S3, PDF file, 0.03 MB (32.1KB, pdf) .
Copyright © 2022 Godeux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
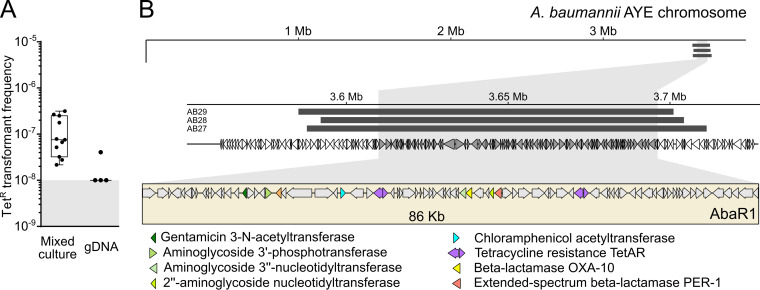
The AbaR4 island is one of the many representatives of resistance islands that can be found in A. baumannii clinical isolates (9). The largest resistance island described so far, the 86-kb-long AbaR1, is found in the AYE strain (8). We thus tested whether this island could be acquired by the M2 strain. In a previous work, we showed that the AbaR1 island conferred resistance to aminoglycosides, tetracycline, and some beta-lactams (35). We thus used tetracycline resistance as a marker for the transfer of AbaR1 from A. baumannii strain AYE (donor strain) to A. nosocomialis strain M2 (recipient strain). To test the acquisition of AbaR1 by M2, we used the rifampicin-resistant derivative of strain M2 and a derivative of the strain AYE impaired for natural transformation (ΔcomEC::aacC4). The mixed-culture condition resulted in recombinants emerging at a frequency of about 10−7 (Fig. 5A). It is noteworthy that only few transformants were detected when using purified gDNA, confirming that the mixed culture brought together conditions favorable to the transfer of the large resistance island (Fig. 5A). The resulting transformants displayed the resistance profile expected from the known resistance associated with AbaR1 (Table S2), indicating that the entire 86-kb-long island had indeed been acquired. Genome sequencing of three tetracycline-resistant transformants confirmed that the entire AbaR1 island (86 kb) was inserted into the genome of the recipient cells (Fig. 5B and Fig. S7). The recombination tracts flanking the comM locus indicated that the imported DNA molecules ranged from 112 to 123 kb (Fig. 5B and Fig. S7). Taken together, these results demonstrate that recombinants can emerge from the encounter of Acinetobacter isolates, even if their genomes display relatively low sequence identity (90%). The interbacterial transfer can result in the efficient recombination of homologous regions and the acquisition of large heterologous segments up to 86 kb, such as AbaR islands conferring resistance to multiple antibiotics, including carbapenems.
FIG 5.
Genomic analysis of the acquisition of the AbaR1 island. (A) Transformation frequencies of the acquisition of tetracycline resistance carried by the AbaR1 island of the AYE strain, in a mixed-culture setup between the AYE strain impaired for natural transformation (comEC::aacC4) and the M2 Rifr strain and by supplying M2 Rifr with genomic DNA extracted from AYE. (B) Graphical representation of the chromosome of the AYE strain (thin black line) and location of the acquired DNA fragments (light gray thick lines) by transformants of the A. nosocomialis strain M2 during mixed culture with AYE. Bottom diagram, close-up view of the acquired fragments and the AbaR1 island. The genomes of tetracycline-resistant recombinants of M2 were sequenced. Variant calling and identification of converted markers (SNPs of AYE acquired by M2) were used to delineate the acquired regions (see Materials and Methods).
Genomic analysis of the acquisition of the AbaR1 island. Graphical representation of the acquired fragments and the AbaR1 island in the M2 transformants. Variant calling and identification of converted markers (SNPs of AYE acquired by M2) were used to delineate the acquired regions (see Materials and Methods). Thin green lines represent coverage, and the outermost right scale indicates the results of read screening corresponding to the AbaR sequence in recombinants. Orange triangles represent individual SNPs used to determine recombination tracts. Download FIG S7, TIF file, 0.2 MB (195.6KB, tif) .
Copyright © 2022 Godeux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
To gain insight into the mechanism leading to emergence of carbapenem and more broadly multidrug resistance in pathogenic Acinetobacter spp., we devised an experimental system to monitor emergence of recombinant progeny and diffusion of antibiotic resistance between multiple combinations of pathogenic Acinetobacter strains. We observed that resistance is efficiently acquired within mixed populations of A. baumannii strains and even between A. baumannii and A. nosocomialis. Investigating in more detail a specific mixed population, we demonstrated that rapid gene transfer occurs through natural transformation in communities of sessile cells, leading to antibiotic resistance. Resistance genes were either on a composite transposon (Tn2006 carrying blaOXA-23) or on large resistance islands (AbaR4 and AbaR1). So far, demonstrations of gene acquisition by natural transformation in A. baumannii and A. nosocomialis were obtained using purified DNA (23–28). Our results indicate that this form of DNA is a poor substrate for transformation in comparison to the DNA released in the extracellular milieu during growth or upon bacterial interaction. DNA purification is known to lead to some DNA degradation, with nicks which will terminate the import of the single-stranded DNA (ssDNA) (38). Mixed culture allowed a transfer of up to 123 kb from strain A. baumannii AYE to A. nosocomialis, corresponding to over 3% of the recipient strain’s genome. Our results corroborate seminal observations in Streptococcus pneumoniae, in which mixed sessile populations were found to support larger recombination events (up to 29 kb) (39). Similarly, interaction of V. cholerae strains also promotes transfer of a large genomic region (up to 168 kb) (40). Importantly, our experiments recapitulate the observed recombination events associated with the acquisition by A. baumannii strains of AbGRI3 and AbGRI5 from other strains of the global clone 2 (GC2) lineage (14, 15). Acquisition by natural transformation would therefore explain the acquisition of AbaRs described for GC2 strains (41) but also for other Acinetobacter species from the A. baumannii-A. calcoaceticus complex, namely, A. nosocomialis, A. haemolyticus, A. pittii, and A. seifertii (9, 42, 43). Our experimental observation leads us to reconsider the role of natural transformation in the acquisition of large islands of resistance by A. baumannii previously attributed to generalized transduction based on presumed limited size of acquisition enabled by natural transformation (20). Transfer of a very large resistance island such as AbaR1 is less frequent, as the odds of importing an intact DNA are likely lower for long DNA fragments. Moreover, acquisition of new genetic material has a potential cost to bacterial replication (44). In the specific case of AbaR1, importing numerous antibiotic and heavy metal resistance genes may be detrimental for the recipient strain, with a temporary fitness cost that would affect the apparent transformation frequency (45, 46). Another consequence of acquisition by natural transformation of these genomic islands is that their horizontal transfer would be conservative of their insertion site. Genomics studies suggests that Ab-RI were initially introduced into A. baumannii through plasmid conjugation, with subsequent insertion in the chromosome (12, 13). Ab-RI retain the features of ancestral Tn7 elements (47), and although transposition activity tested with AbaR1 was not detectable (48), Ab-RI are found inserted in over 50 chromosomal insertion sites (9). Yet the vast majority of Ab-RI appear to be inserted in a specific locus, the comM gene (9, 11, 12), which encodes a putative helicase involved in natural transformation (35, 49, 50). Supported by genomic evidence (14, 15), our results suggest that HGT of Ab-RI can occur through homologous recombination produced by natural transformation. This mechanism of HGT would propagate the inactivation of comM and the apparent fixation of some Ab-RI at this specific locus.
We investigated the role of the T6SS in the transfer of resistance upon bacterial interaction. The competition mechanism was an evident culprit to trigger the release of DNA from prey cells, as described for A. baylyi and V. cholerae (31, 32, 40). Although we found that the T6SS is partially involved in acquisition of resistance genes, we found that T6SS-mediated killing is potentially impaired under conditions compatible with natural transformation. Our results corroborate previous findings that T6SS development is dependent on growth conditions, with the observation of a reduced T6SS activity in minimal medium for A. nosocomialis M2 as determined by Hcp secretion (51). Contribution of the T6SS to DNA release and diffusion of resistance determinants may, however, depend on strain combination, as T6SS activity was shown to be highly dependent on the strains in competitions (36, 52, 53). In addition, our results show that the direct negative interaction between cells is not the sole mechanism promoting release of DNA and subsequent acquisition of the resistance determinant. We found that the contact-independent release of DNA during growth is sufficient to mediate high rates of acquisition of resistance through natural transformation. In addition to passive release by autolysis upon cell death, several mechanisms of active DNA release in the extracellular compartment had been described for other transformable species. In Neisseria gonorrhoeae, genomic DNA is actively secreted through a type IV secretion system by the donor strain, delivering a more potent substrate of transformation than DNA released upon autolysis (54, 55). In Pseudomonas aeruginosa, DNA release is dependent on a cryptic prophage endolysin that is required for formation of biofilm, where transformation can also occur (56–58). Indeed, extracellular DNA is also a critical component of the biofilm matrix (59), and treatment with DNase leads to severe biofilm alteration for several pathogenic bacteria, including A. baumannii (60). However, as suggested by our results, it is conceivable that the level of DNA release may vary between strains of A. baumannii and thereby influence biofilm formation, the diffusion of resistance genes, and their acquisition by neighboring cells.
Altogether, this work highlights the importance of one of the major HGT mechanisms in bacteria, natural transformation, in the acquisition by A. baumannii of resistance to clinically relevant antibiotics. By naturally releasing high-quality DNA in their environment, some strains of A. baumannii may be highly potent donors for the uptake and recombination of large DNA fragments by natural transformation. This mechanism may enable a rapid and direct acquisition of multiple antibiotic resistance genes by A. baumannii and explain the evolution by recombination observed at the genomic scale.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains or strains used in this study are listed in Table S1. Acinetobacter baumannii isolates and strains were grown in lysogeny broth (LB; Lennox) or tryptone-NaCl medium (5 g/L of tryptone, 2.5 g/L NaCl). All experiments were performed at 37°C. Antibiotic concentrations were 15 μg/mL for tetracycline, 50 μg/mL for kanamycin, 1.6 μg/mL for imipenem (always spread extemporaneously on an LB agar plate), and 100 μg/mL for rifampicin.
Bacterial strains, plasmids, and primers used in this study. Download Table S1, PDF file, 0.1 MB (78.1KB, pdf) .
Copyright © 2022 Godeux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Construction of bacterial strains.
All the oligonucleotides used in this study for genetic modification are listed in Table S1. Gene disruptions were performed using a scarless genome editing strategy described previously (35). The oligonucleotides used for strain construction are listed in Table S1. Rifampicin-resistant derivatives were obtained by selecting spontaneous mutants from an overnight culture in liquid LB on LB agar supplemented with rifampicin.
Detection of recombinants in mixed cultures.
Isolates to be tested for the production of recombinants were grown overnight on LB agar plates and then inoculated and grown in 2 mL of LB until the cultures reach an optical density at 600 nm (OD600) of 1. The bacterial broths were then diluted to an OD600 of 0.01 in phosphate-buffered saline (PBS). Then equal volumes of bacterial suspensions were mixed by pairs, each consisting of an imipenem-resistant (Imir) and rifampicin-resistant (Rifr) isolate. The mixture (2.5 μL) was deposited on the surface of 1 mL of tryptone-NaCl medium solidified with 2% agarose D3 (Euromedex) poured into 2-mL microtubes or in wells of 24-well plates and incubated overnight at 37°C. Bacteria were then resuspended in PBS and were plated onto LB agar plates without antibiotics and LB agar plates containing rifampicin (100 μg/mL) and imipenem (1.6 μg/mL) with either beads or easySpiral Pro (Interscience). Recombinant frequencies were determined through calculation of the ratio of the number of CFU on rifampicin and imipenem plates to the total number of CFU on plates without antibiotics.
Transformation assay.
Transformation assays were performed as previously described (35) except that then bacterial suspensions of the recipient strain were mixed with an equal volume of either genomic DNA extracted from the donor strain (concentration, 200 ng/μL), a bacterial suspension of the donor strain at an OD600 of 0.01 for mixed culture, or a filtered spent medium. DNase I treatment was performed at a final concentration of 0.3 U/μL (Sigma). All the transformation assays were performed on at least two separate occasions with three independent transformation reactions (three different bacterial cultures). All the independent data points are plotted. For kinetics of transformation, bacteria were recovered at indicated times points with bacteria harvested from an independent sample and plated as previously described.
DNA measurement in spent media.
Supernatants were obtained by centrifugation (3,000 × g, 3 min) of a culture of the donor strain for 4 h in 37°C tryptone-NaCl liquid medium with agitation at 200 rpm. The supernatant was then filtered (0.22-μm-size pore filter). DNA quantification was performed by fluorescent labeling with Quant-iT reagent (Invitrogen Qubit dsDNA HS assay kit; Thermo Fisher Scientific) measured with Infinite M200 PRO (TECAN) and Magellan 7.1 SP1 (TECAN).
Genome sequencing, assembly, and annotations.
The genome of 40288 was sequenced using Illumina technology (paired-end, read length 100 nt, HiSeq instrument) and Oxford Nanopore technology (rapid barcoding kit, MinION instrument) with a read coverage of 100×. Short (Illumina) and long (Oxford Nanopore) reads were used to assemble the genome with Unicycler (61). This generated a complete genome consisting of a circular chromosome of 4,084,922 bp and circular plasmid of 145,711 bp. Genes were annotated with Prokka (62), using the A. baumannii AB5075-UW genome as a reference (63). The genome of the human clinical isolate CNRAB1 was sequenced using similar procedures.
Variant calling and detection of recombination events.
Genomes were sequenced on a NovaSeq instrument (Illumina, 2 × 150-bp paired end). Reads were subsampled to 50× coverage of the donor genome using seqtk version 1.3-r106 (https://github.com/lh3/seqtk). Reads of recombinants were mapped to their corresponding donor genome (M2, 40288, or AYE) using bwa mem version 0.7.17-r1888 (64) and sorted using samtools version 1.10.3 (65). Donor (40288 or AYE) and recipient (M2) genome short reads were simulated using wgsim version 1.10 (https://github.com/lh3/wgsim) and mapped to the donor genome (40288 or AYE) using the same parameters as for transformant sequencing reads. Reads were then tallied on the donor genome using bcftools version 1.10, to keep only sites with read coverage above 25×, where haplotype differs between donor and recipient and match donor haplotype. The variant calling and identification of converted markers were performed using twgt version 0.1.0.5 (https://gitlab.com/bacterial-gbgc/twgt/-/tree/master). The resulting VCF file was then parsed using VariantAnnotation (66) using R version 3.6.6. With Tn2006, AbaR4, and AbaR1 being absent from the recipient genome, their insertions in the transformant genome were confirmed via read screening: their sequence together with 1 kb of flanking sequence was sketched using mash sketch and the presence of this sketch in sequencing reads was confirmed using mash screen version 2.2.2 (67).
Data availability.
Genome sequences are available within BioProject PRJNA741866. Raw sequencing reads are available in Sequence Read Archive (SRA) within PRJNA741866.
ACKNOWLEDGMENTS
We warmly thank Agnese Lupo and Marisa Haenni (ANSES, Lyon, France) for sharing the clinical isolate 40288 and sequencing and antimicrobial susceptibility data.
This work was supported by the LABEX ECOFECT (ANR-11-LABX-0048) of Université de Lyon, within the program “Investissements d’Avenir” (ANR-11-IDEX-0007) operated by the French National Research Agency (ANR) and by the RESPOND program of the Université de Lyon (UDL). A.-S.G. was also supported by VetAgro Sup (Ph.D. funding).
Footnotes
[This article was published on 25 January 2022 with Elin Svedholm's surname misspelled. This error was corrected in the current version, posted on 25 February 2022.]
Contributor Information
Xavier Charpentier, Email: xavier.charpentier@univ-lyon1.fr.
Maria-Halima Laaberki, Email: maria-halima.laaberki@vetagro-sup.fr.
Robert A. Bonomo, Louis Stokes Veterans Affairs Medical Center
REFERENCES
- 1.Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol 5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 2.van der Kolk JH, Endimiani A, Graubner C, Gerber V, Perreten V. 2019. Acinetobacter in veterinary medicine, with an emphasis on Acinetobacter baumannii. J Glob Antimicrob Resist 16:59–71. doi: 10.1016/j.jgar.2018.08.011. [DOI] [PubMed] [Google Scholar]
- 3.Patel A, Emerick M, Cabunoc MK, Williams MH, Preas MA, Schrank G, Rabinowitz R, Luethy P, Johnson JK, Leekha S. 2021. Rapid spread and control of multidrug-resistant Gram-negative bacteria in COVID-19 patient care units. Emerg Infect Dis 27:1234–1237. doi: 10.3201/eid2704.204036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perez S, Innes GK, Walters MS, Mehr J, Arias J, Greeley R, Chew D. 2020. Increase in hospital-acquired carbapenem-resistant Acinetobacter baumannii infection and colonization in an acute care hospital during a surge in COVID-19 admissions—New Jersey, February–July 2020. MMWR Morb Mortal Wkly Rep 69:1827–1831. doi: 10.15585/mmwr.mm6948e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.European Centre for Disease Prevention and Control. 2020. Antimicrobial resistance in the EU/EEA (EARS-Net)—annual epidemiological report 2019. ECDC, Stockholm, Sweden. [Google Scholar]
- 6.Hamidian M, Nigro SJ. 2019. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microbial Genomics 5:e000306. doi: 10.1099/mgen.0.000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nigro S, Hall RM. 2015. Distribution of the blaOXA-23-containing transposons Tn2006 and Tn2008 in Australian carbapenem-resistant Acinetobacter baumannii isolates. J Antimicrob Chemother 70:2409–2411. doi: 10.1093/jac/dkv102. [DOI] [PubMed] [Google Scholar]
- 8.Fournier P-E, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, Richet H, Robert C, Mangenot S, Abergel C, Nordmann P, Weissenbach J, Raoult D, Claverie J-M. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet 2:e7. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bi D, Xie R, Zheng J, Yang H, Zhu X, Ou H-Y, Wei Q. 2019. Large-scale identification of AbaR-type genomic islands in Acinetobacter baumannii reveals diverse insertion sites and clonal lineage-specific antimicrobial resistance gene profiles. Antimicrob Agents Chemother 63:e02526-18. doi: 10.1128/AAC.02526-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonnin RA, Poirel L, Nordmann P. 2012. AbaR-type transposon structures in Acinetobacter baumannii. J Antimicrob Chemother 67:234–236. doi: 10.1093/jac/dkr413. [DOI] [PubMed] [Google Scholar]
- 11.Hamidian M, Hall RM. 2018. The AbaR antibiotic resistance islands found in Acinetobacter baumannii global clone 1—structure, origin and evolution. Drug Resist Updat 41:26–39. doi: 10.1016/j.drup.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Hamidian M, Hall RM. 2017. Origin of the AbGRI1 antibiotic resistance island found in the comM gene of Acinetobacter baumannii GC2 isolates. J Antimicrob Chemother 72:2944–2947. doi: 10.1093/jac/dkx206. [DOI] [PubMed] [Google Scholar]
- 13.Hamidian M, Kenyon JJ, Holt KE, Pickard D, Hall RM. 2014. A conjugative plasmid carrying the carbapenem resistance gene blaOXA-23 in AbaR4 in an extensively resistant GC1 Acinetobacter baumannii isolate. J Antimicrob Chemother 69:2625–2628. doi: 10.1093/jac/dku188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamidian M, Hawkey J, Wick R, Holt KE, Hall RM. 2019. Evolution of a clade of Acinetobacter baumannii global clone 1, lineage 1 via acquisition of carbapenem- and aminoglycoside-resistance genes and dispersion of ISAba1. Microb Genom 5:e000242. doi: 10.1099/mgen.0.000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hua X, Moran RA, Xu Q, He J, Fang Y, Zhang L, van Schaik W, Yu Y. 2021. Acquisition of a genomic resistance island (AbGRI5) from global clone 2 through homologous recombination in a clinical Acinetobacter baumannii isolate. J Antimicrob Chemother 76:65–69. doi: 10.1093/jac/dkaa389. [DOI] [PubMed] [Google Scholar]
- 16.Snitkin ES, Zelazny AM, Montero CI, Stock F, Mijares L, Murray PR, Segre JA, NISC Comparative Sequence Program. 2011. Genome-wide recombination drives diversification of epidemic strains of Acinetobacter baumannii. Proc Natl Acad Sci USA 108:13758–13763. doi: 10.1073/pnas.1104404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holt K, Kenyon JJ, Hamidian M, Schultz MB, Pickard DJ, Dougan G, Hall R. 2016. Five decades of genome evolution in the globally distributed, extensively antibiotic-resistant Acinetobacter baumannii global clone 1. Microb Genom 2:e000052. doi: 10.1099/mgen.0.000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Douraghi M, Kenyon JJ, Aris P, Asadian M, Ghourchian S, Hamidian M. 2020. Accumulation of antibiotic resistance genes in carbapenem-resistant Acinetobacter baumannii isolates belonging to lineage 2, global clone 1, from outbreaks in 2012–2013 at a Tehran burns hospital. mSphere 5:e00164-20. doi: 10.1128/mSphere.00164-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright MS, Iovleva A, Jacobs MR, Bonomo RA, Adams MD. 2016. Genome dynamics of multidrug-resistant Acinetobacter baumannii during infection and treatment. Genome Med 8:26. doi: 10.1186/s13073-016-0279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krahn T, Wibberg D, Maus I, Winkler A, Bontron S, Sczyrba A, Nordmann P, Pühler A, Poirel L, Schlüter A. 2016. Intraspecies transfer of the chromosomal Acinetobacter baumannii bla NDM-1 carbapenemase gene. Antimicrob Agents Chemother 60:3032–3040. doi: 10.1128/AAC.00124-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wachino J-I, Jin W, Kimura K, Arakawa Y. 2019. Intercellular transfer of chromosomal antimicrobial resistance genes between Acinetobacter baumannii strains mediated by prophages. Antimicrob Agents Chemother 63:e00334-19. doi: 10.1128/AAC.00334-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston C, Martin B, Fichant G, Polard P, Claverys J-P. 2014. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol 12:181–196. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 23.Godeux A-S, Lupo A, Haenni M, Guette-Marquet S, Wilharm G, Laaberki M-H, Charpentier X. 2018. Fluorescence-based detection of natural transformation in drug-resistant Acinetobacter baumannii. J Bacteriol 200:e00181-18. doi: 10.1128/JB.00181-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harding CM, Tracy EN, Carruthers MD, Rather PN, Actis LA, Munson RS. 2013. Acinetobacter baumannii strain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. mBio 4:e00360-13. doi: 10.1128/mBio.00360-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Traglia GM, Chua K, Centrón D, Tolmasky ME, Ramírez MS. 2014. Whole-genome sequence analysis of the naturally competent Acinetobacter baumannii clinical isolate A118. Genome Biol Evol 6:2235–2239. doi: 10.1093/gbe/evu176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilharm G, Piesker J, Laue M, Skiebe E. 2013. DNA uptake by the nosocomial pathogen Acinetobacter baumannii occurs during movement along wet surfaces. J Bacteriol 195:4146–4153. doi: 10.1128/JB.00754-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vesel N, Blokesch M. 2021. Pilus production in Acinetobacter baumannii is growth phase dependent and essential for natural transformation. J Bacteriol 203:e00034-21. doi: 10.1128/JB.00034-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Y, He L, Tao X, Meng F, Zhang J. 2019. High DNA uptake capacity of international clone II Acinetobacter baumannii detected by a novel planktonic natural transformation assay. Front Microbiol 10:2165. doi: 10.3389/fmicb.2019.02165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall AR, Iles JC, MacLean RC. 2011. The fitness cost of rifampicin resistance in Pseudomonas aeruginosa depends on demand for RNA polymerase. Genetics 187:817–822. doi: 10.1534/genetics.110.124628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brandis G, Pietsch F, Alemayehu R, Hughes D. 2015. Comprehensive phenotypic characterization of rifampicin resistance mutations in Salmonella provides insight into the evolution of resistance in Mycobacterium tuberculosis. J Antimicrob Chemother 70:680–685. doi: 10.1093/jac/dku434. [DOI] [PubMed] [Google Scholar]
- 31.Borgeaud S, Metzger LC, Scrignari T, Blokesch M. 2015. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347:63–67. doi: 10.1126/science.1260064. [DOI] [PubMed] [Google Scholar]
- 32.Ringel PD, Hu D, Basler M. 2017. The role of type VI secretion system effectors in target cell lysis and subsequent horizontal gene transfer. Cell Rep 21:3927–3940. doi: 10.1016/j.celrep.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 33.Carruthers MD, Nicholson PA, Tracy EN, Munson RS, Jr.. 2013. Acinetobacter baumannii utilizes a type VI secretion system for bacterial competition. PLoS One 8:e59388. doi: 10.1371/journal.pone.0059388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lupo A, Châtre P, Ponsin C, Saras E, Boulouis H-J, Keck N, Haenni M, Madec J-Y. 2017. Clonal spread of Acinetobacter baumannii sequence type 25 carrying blaOXA-23 in companion animals in France. Antimicrob Agents Chemother 61:e01881-16. doi: 10.1128/AAC.01881-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Godeux A-S, Svedholm E, Lupo A, Haenni M, Venner S, Laaberki M-H, Charpentier X. 2020. Scarless removal of large resistance island AbaR results in antibiotic susceptibility and increased natural transformability in Acinetobacter baumannii. Antimicrob Agents Chemother 64:e00951-20. doi: 10.1128/AAC.00951-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fitzsimons TC, Lewis JM, Wright A, Kleifeld O, Schittenhelm RB, Powell D, Harper M, Boyce JD. 2018. Identification of novel Acinetobacter baumannii type VI secretion system antibacterial effector and immunity pairs. Infect Immun 86:e00297-18. doi: 10.1128/IAI.00297-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alfsnes K, Frye SA, Eriksson J, Eldholm V, Brynildsrud OB, Bohlin J, Harrison OB, Hood DW, Maiden MCJ, Tønjum T, Ambur OH. 2018. A genomic view of experimental intraspecies and interspecies transformation of a rifampicin-resistance allele into Neisseria meningitidis. Microb Genom 4:e000222. doi: 10.1099/mgen.0.000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dubnau D, Blokesch M. 2019. Mechanisms of DNA uptake by naturally competent bacteria. Annu Rev Genet 53:217–237. doi: 10.1146/annurev-genet-112618-043641. [DOI] [PubMed] [Google Scholar]
- 39.Cowley LA, Petersen FC, Junges R, Jimenez MJD, Morrison DA, Hanage WP. 2018. Evolution via recombination: cell-to-cell contact facilitates larger recombination events in Streptococcus pneumoniae. PLoS Genet 14:e1007410. doi: 10.1371/journal.pgen.1007410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matthey N, Stutzmann S, Stoudmann C, Guex N, Iseli C, Blokesch M. 2019. Neighbor predation linked to natural competence fosters the transfer of large genomic regions in Vibrio cholerae. Elife 8:e48212. doi: 10.7554/eLife.48212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iacono M, Villa L, Fortini D, Bordoni R, Imperi F, Bonnal RJP, Sicheritz-Ponten T, De Bellis G, Visca P, Cassone A, Carattoli A. 2008. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob Agents Chemother 52:2616–2625. doi: 10.1128/AAC.01643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim DH, Choi J-Y, Kim HW, Kim SH, Chung DR, Peck KR, Thamlikitkul V, So TM-K, Yasin RMD, Hsueh P-R, Carlos CC, Hsu LY, Buntaran L, Lalitha MK, Song J-H, Ko KS. 2013. Spread of carbapenem-resistant Acinetobacter baumannii global clone 2 in Asia and AbaR-type resistance islands. Antimicrob Agents Chemother 57:5239–5246. doi: 10.1128/AAC.00633-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim DH, Ko KS. 2015. AbaR-type genomic islands in non-baumannii Acinetobacter species isolates from South Korea. Antimicrob Agents Chemother 59:5824–5826. doi: 10.1128/AAC.01175-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baltrus DA. 2013. Exploring the costs of horizontal gene transfer. Trends Ecol Evol 28:489–495. doi: 10.1016/j.tree.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 45.Gullberg E, Albrecht LM, Karlsson C, Sandegren L, Andersson DI. 2014. Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. mBio 5:e01918-14. doi: 10.1128/mBio.01918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vogwill T, MacLean RC. 2015. The genetic basis of the fitness costs of antimicrobial resistance: a meta-analysis approach. Evol Appl 8:284–295. doi: 10.1111/eva.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bi D, Zheng J, Xie R, Zhu Y, Wei R, Ou H-Y, Wei Q, Qin H. 2020. Comparative analysis of AbaR-type genomic islands reveals distinct patterns of genetic features in elements with different backbones. mSphere 5:e00349-20. doi: 10.1128/mSphere.00349-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rose A. 2010. TnAbaR1: a novel Tn7-related transposon in Acinetobacter baumannii that contributes to the accumulation and dissemination of large repertoires of resistance genes. Bioscience Horizons 3:40–48. doi: 10.1093/biohorizons/hzq006. [DOI] [Google Scholar]
- 49.Nero TM, Dalia TN, Wang JC-Y, Kysela DT, Bochman ML, Dalia AB. 2018. ComM is a hexameric helicase that promotes branch migration during natural transformation in diverse Gram-negative species. Nucleic Acids Res 46:6099–6111. doi: 10.1093/nar/gky343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sinha S, Mell JC, Redfield RJ. 2012. Seventeen Sxy-dependent cyclic AMP receptor protein site-regulated genes are needed for natural transformation in Haemophilus influenzae. J Bacteriol 194:5245–5254. doi: 10.1128/JB.00671-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harding CM, Pulido MR, Venanzio GD, Kinsella RL, Webb AI, Scott NE, Pachón J, Feldman MF. 2017. Pathogenic Acinetobacter species have a functional type I secretion system and contact-dependent inhibition systems. J Biol Chem 292:9075–9087. doi: 10.1074/jbc.M117.781575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Repizo GD, Gagné S, Foucault-Grunenwald M-L, Borges V, Charpentier X, Limansky AS, Gomes JP, Viale AM, Salcedo SP. 2015. Differential role of the T6SS in Acinetobacter baumannii virulence. PLoS One 10:e0138265. doi: 10.1371/journal.pone.0138265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weber BS, Miyata ST, Iwashkiw JA, Mortensen BL, Skaar EP, Pukatzki S, Feldman MF. 2013. Genomic and functional analysis of the type VI secretion system in Acinetobacter. PLoS One 8:e55142. doi: 10.1371/journal.pone.0055142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dillard JP, Seifert HS. 2001. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol Microbiol 41:263–277. doi: 10.1046/j.1365-2958.2001.02520.x. [DOI] [PubMed] [Google Scholar]
- 55.Hamilton HL, Domínguez NM, Schwartz KJ, Hackett KT, Dillard JP. 2005. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol Microbiol 55:1704–1721. doi: 10.1111/j.1365-2958.2005.04521.x. [DOI] [PubMed] [Google Scholar]
- 56.Turnbull L, Toyofuku M, Hynen AL, Kurosawa M, Pessi G, Petty NK, Osvath SR, Cárcamo-Oyarce G, Gloag ES, Shimoni R, Omasits U, Ito S, Yap X, Monahan LG, Cavaliere R, Ahrens CH, Charles IG, Nomura N, Eberl L, Whitchurch CB. 2016. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat Commun 7:11220. doi: 10.1038/ncomms11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hynen AL, Lazenby JJ, Savva GM, McCaughey LC, Turnbull L, Nolan LM, Whitchurch CB. 2021. Multiple holins contribute to extracellular DNA release in Pseudomonas aeruginosa biofilms. Microbiology (Reading) 167:000990. doi: 10.1099/mic.0.000990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nolan LM, Turnbull L, Katrib M, Osvath SR, Losa D, Lazenby JJ, Whitchurch CBY. 2020. 2020. Pseudomonas aeruginosa is capable of natural transformation in biofilms. Microbiology (Reading) 166:995–1003. doi: 10.1099/mic.0.000956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Okshevsky M, Meyer RL. 2015. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit Rev Microbiol 41:341–352. doi: 10.3109/1040841X.2013.841639. [DOI] [PubMed] [Google Scholar]
- 60.Tetz GV, Artemenko NK, Tetz VV. 2009. Effect of DNase and antibiotics on biofilm characteristics. Antimicrob Agents Chemother 53:1204–1209. doi: 10.1128/AAC.00471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 63.Jacobs AC, Thompson MG, Black CC, Kessler JL, Clark LP, McQueary CN, Gancz HY, Corey BW, Moon JK, Si Y, Owen MT, Hallock JD, Kwak YI, Summers A, Li CZ, Rasko DA, Penwell WF, Honnold CL, Wise MC, Waterman PE, Lesho EP, Stewart RL, Actis LA, Palys TJ, Craft DW, Zurawski DV. 2014. AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. mBio 5:e01076-14. doi: 10.1128/mBio.01076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Obenchain V, Lawrence M, Carey V, Gogarten S, Shannon P, Morgan M. 2014. VariantAnnotation: a Bioconductor package for exploration and annotation of genetic variants. Bioinformatics 30:2076–2078. doi: 10.1093/bioinformatics/btu168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2016. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Recombinant frequency as a function of genomic relatedness between paired strains. Analysis of recombinant frequencies for paired cotransformants as presented in Fig. 1 in the main text depending on the pairwise average nucleotide identity (ANI). For each pairing, the imipenem-resistant strain is highlighted in panel A and the rifampicin-resistant one in panel B. Download FIG S1, TIF file, 0.4 MB (400.5KB, tif) .
Copyright © 2022 Godeux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Recombinants emerge in mixed culture in a PilA- and RecA-dependent manner. Rifampicin-resistant (Rifr) isolates of M2 or its pilA::kan or recA::aacC4 mutant were mixed with the carbapenem-resistant (Imir) clinical isolate 40288 ΔcomEC mutant. Rifr and Imir recombinants were determined after 24 h of mixed culture. The boxplots represent the distributions of Rifr and Imir recombinants divided by the total CFU count (recombinant frequencies). A horizontal line represents the median. The limit of detection (10−8) is indicated by the gray area. Download FIG S2, TIF file, 0.1 MB (129.5KB, tif) .
Copyright © 2022 Godeux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
T6SS-dependent killing activity of the M2 strain on the 40288 isolate in LB medium and under conditions of mixed culture. Recovery of the A. baumannii strain 40288 incubated either with the wild-type M2 strain (WT) or with its hcp gene mutant derivative (Δhcp) is shown. (A) Cocultivation of 40288 with the M2 strain during in LB medium as described previously (1) resulted in a 4-log decrease of the 40288 population, while inactivating the T6SS of M2 (Δhcp) abolished the killing activity. (B) In the mixed-culture setup used to observe the rise of recombinants, mixed culture of 40288 with M2 for 4 h did not significantly reduce the population size of 40288. Download FIG S3, TIF file, 0.4 MB (392.5KB, tif) .
Copyright © 2022 Godeux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strain M2 of A. nosocomialis is equally transformed with DNA from itself and from the distantly related strain 40288 of A. baumannii. Strain M2 was incubated under transformation conditions (>16 h) with genomic DNA extracted either from an M2 ΔcomEC::aacC4 (Aprar) or from a 40288 ΔcomEC::aacC4 (Aprar) strain. Transformation frequency represents the CFU count on an apramycin plate divided by the total CFU count on nonselective plates. Download FIG S4, TIF file, 0.1 MB (131.3KB, tif) .
Copyright © 2022 Godeux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phenotypic outputs of acquisition of AbaR4 and Tn2006 from strain 40288. (A) Acquisition of AbaR4 in the comM gene lowers the transformation levels of the recipient strain. Bacterial suspensions of M2 Rifr or two of its derivatives that acquired AbaR4 in the comM gene in previous mixed-culture experiments were mixed with an equal volume of 40288 ΔcomEC::aacC4, all strains at an OD600 of 0.01. The mixture (2.5 μL) was deposited on the surface of transformation medium. Recombinant frequency represents the ratio of M2 Rifr Aprar CFU over the total count of Rifr CFU. P values of statistical analysis were obtained using the nonparametric Mann-Whitney-Wilcoxon test (two tailed). The limit of detection (10−8) is indicated by the gray area. (B) T6SS-mediated killing assay of a VgrG3 impaired mutant of A. nosocomialis strain M2 in LB medium. Killing assays were performed as described for Fig. S1A. Escherichia coli strain JW2912 (Kanr) and A. baumannii strain 40288 were both used as prey and were incubated either with the wild-type M2 strain, its hcp gene mutant derivative (Δhcp), or an imipenem-resistant transformant carrying an insertion into the vgrG3 gene (vgrG3::Tn2006; two transformants tested). Download FIG S5, TIF file, 0.6 MB (596.1KB, tif) .
Copyright © 2022 Godeux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genomic analysis of the acquisition of imipenem resistance. Graphical representation of the chromosome of the 40288 strain (boxed area) and location of the acquired DNA fragments (light gray thick lines) by transformants of A. nosocomialis strain M2 during mixed culture with 40288. Top and bottom panels represent close-ups of the regions in which the blaOXA-23 gene is acquired as part of the Tn2006 element (top box) in the vgrG3 gene and as part of the Tn2006 element within the AbaR island (bottom box) inserted in the comM gene. Acquired regions were determined by sequencing the genomes of imipenem-resistant recombinants of M2 which had acquired blaOXA-23 on a NovaSeq instrument. Variant calling and identification of converted markers (SNPs of 40288 acquired by M2) were used to delineate the acquired regions (see Materials and Methods). Thin green lines represent coverage, and the outermost right scale indicates the results of read screening corresponding to the AbaR sequence in recombinants. Orange triangles represent individual SNPs used to determine recombination tracts. Download FIG S6, TIF file, 0.5 MB (545.2KB, tif) .
Copyright © 2022 Godeux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Antimicrobial susceptibility profiles of mixed-culture transformants. Download Table S2, PDF file, 0.03 MB (32.5KB, pdf) .
Copyright © 2022 Godeux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Estimated length of the imported DNA molecules leading to the detected recombination tracts. REFERENCE 1. Fournier P-E, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, Richet H, Robert C, Mangenot S, Abergel C, Nordmann P, Weissenbach J, Raoult D, Claverie J-M. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet 2:e7. https://doi.org/10.1371/journal.pgen.0020007. Download Table S3, PDF file, 0.03 MB (32.1KB, pdf) .
Copyright © 2022 Godeux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genomic analysis of the acquisition of the AbaR1 island. Graphical representation of the acquired fragments and the AbaR1 island in the M2 transformants. Variant calling and identification of converted markers (SNPs of AYE acquired by M2) were used to delineate the acquired regions (see Materials and Methods). Thin green lines represent coverage, and the outermost right scale indicates the results of read screening corresponding to the AbaR sequence in recombinants. Orange triangles represent individual SNPs used to determine recombination tracts. Download FIG S7, TIF file, 0.2 MB (195.6KB, tif) .
Copyright © 2022 Godeux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial strains, plasmids, and primers used in this study. Download Table S1, PDF file, 0.1 MB (78.1KB, pdf) .
Copyright © 2022 Godeux et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Genome sequences are available within BioProject PRJNA741866. Raw sequencing reads are available in Sequence Read Archive (SRA) within PRJNA741866.