Abstract
Rationale
Both genetic variants and chronic obstructive pulmonary disease (COPD) contribute to the risk of incident severe coronavirus disease (COVID-19). Whether genetic risk of incident severe COVID-19 is the same regardless of preexisting COPD is unknown.
Objectives
In this study, we aimed to investigate the potential interaction between genetic risk and COPD in relation to severe COVID-19.
Methods
We constructed a polygenic risk score for severe COVID-19 by using 112 single-nucleotide polymorphisms in 430,582 participants from the UK Biobank study. We examined the associations of genetic risk and COPD with severe COVID-19 by using logistic regression models.
Results
Of 430,582 participants, 712 developed severe COVID-19 as of February 22, 2021, of whom 19.8% had preexisting COPD. Compared with participants at low genetic risk, those at intermediate genetic risk (odds ratio [OR], 1.34; 95% confidence interval [CI], 1.09–1.66) and high genetic risk (OR, 1.50; 95% CI, 1.18–1.92) had higher risk of severe COVID-19 (P for trend = 0.001), and the association was independent of COPD (P for interaction = 0.76). COPD was associated with a higher risk of incident severe COVID-19 (OR, 1.37; 95% CI, 1.12–1.67; P = 0.002). Participants at high genetic risk and with COPD had a higher risk of severe COVID-19 (OR, 2.05; 95% CI, 1.35–3.04; P < 0.001) than those at low genetic risk and without COPD.
Conclusions
The polygenic risk score, which combines multiple risk alleles, can be effectively used in screening for high-risk populations of severe COVID-19. High genetic risk correlates with a higher risk of severe COVID-19, regardless of preexisting COPD.
Keywords: severe COVID-19, genetic risk, COPD
The ongoing pandemic due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has affected more than 149 million people worldwide as of April 29, 2021 (1), with the median infection fatality rate of 0.00–1.63% (2). The severity of coronavirus disease (COVID-19) ranged from mild to severe illness that might readily result in death (3). A surge of severe cases requiring intensive care unit (ICU) admission has been observed (4). There has been a compelling need to identify the risk factors of severe COVID-19, which would enable the risk stratification, triage, and optimization of resource allocation.
Using a case–control design, recent genome-wide association studies (GWASs) have identified several DNA polymorphisms that might substantially influence the risk of developing severe COVID-19, including rs11385942 on LZTFL1 (leucine zipper transcription factor-like 1), rs657152 on ABO (ABO blood group), rs429358 on APOE (apolipoprotein E), rs142984500 on angiotensin-converting enzyme 2, and rs12329760 on TMPRSS2 (transmembrane serine protease 2) (5–9). However, the effect size of each of these GWAS variants is generally small. For example, some people who test negative for risk gene mutations may be at high genetic risk because of other unmeasured genetic factors. Although each individual variant only explains a small proportion of severe COVID-19 risk, the combination of many genetic variants into a single polygenic risk score (PRS) explains a greater proportion of the risk. Therefore, aggregating multiple single-nucleotide polymorphisms (SNPs) to generate a composite PRS might help explain the genetic risk of polygenic diseases and provide a quantitative measure of the genetic risk (10, 11). A previous study has revealed a statistically significant association of PRS based on six DNA polymorphisms and risk of incident severe COVID-19 among athletes (12). However, the previous study was limited by the relatively small sample size and the relatively few genetic polymorphisms in the PRS assessment. PRSs based on larger GWASs and larger sample size, and including more variants, tend to exhibit higher predictive performance (13).
According to the previous studies, both angiotensin-converting enzyme 2 and TMPRSS2 were found to be significantly upregulated in patients with chronic obstructive pulmonary disease (COPD) compared with healthy subjects, and the ABO locus influences blood biomarker measurements in COPD (14–16). Accumulating evidence indicated that COPD might be a risk factor for severe COVID-19 (17–21). For instance, a meta-analysis focusing on comorbidities in 1,558 patients documented a high odds ratio (OR) for severe COVID-19 in patients with preexisting COPD (OR, 5.97), as compared with the relatively lower OR of other common comorbidities such as hypertension (OR, 2.29), diabetes (OR, 2.47), and cardiovascular diseases (OR, 3.89) (18). However, the relevance of composite PRS and COPD in individual patients with severe COVID-19 remains uncertain, and the interplay between genetic risk and COPD in the context of severe COVID-19 is largely unknown.
Therefore, based on the UK Biobank study, we investigated the association between genetic risk and incident severe COVID-19 in different subgroups with or without preexisting COPD. We also sought to investigate the potential interactions between genetic risk and COPD.
Methods
Study Population
The study population and design of the UK Biobank study have been described previously (22–24). Briefly, the UK Biobank study is a prospective, population-based cohort study with more than 500,000 participants aged 40–69 years who attended one of the 22 participating sites across the UK (England, Scotland, and Wales) between 2006 and 2010. The study collects extensive data from questionnaires, interviews, health records, physical measures, biological samples, and radiology. Blood samples are also obtained for genotyping. For all participants, retrospective and prospective linkage to electronic health data are available, including Hospital Episode Statistics (HES). The HES provides detailed information for participants admitted to the hospital and includes coded data on the diagnoses. Data on mortality are updated from National Health Service Digital for participants in England and Wales and from the National Health Service Central Register for participants in Scotland. The UK Biobank study received approval from the North West Multicenter Research Ethics Committee (11/NW/0382), and all participants provided informed consent.
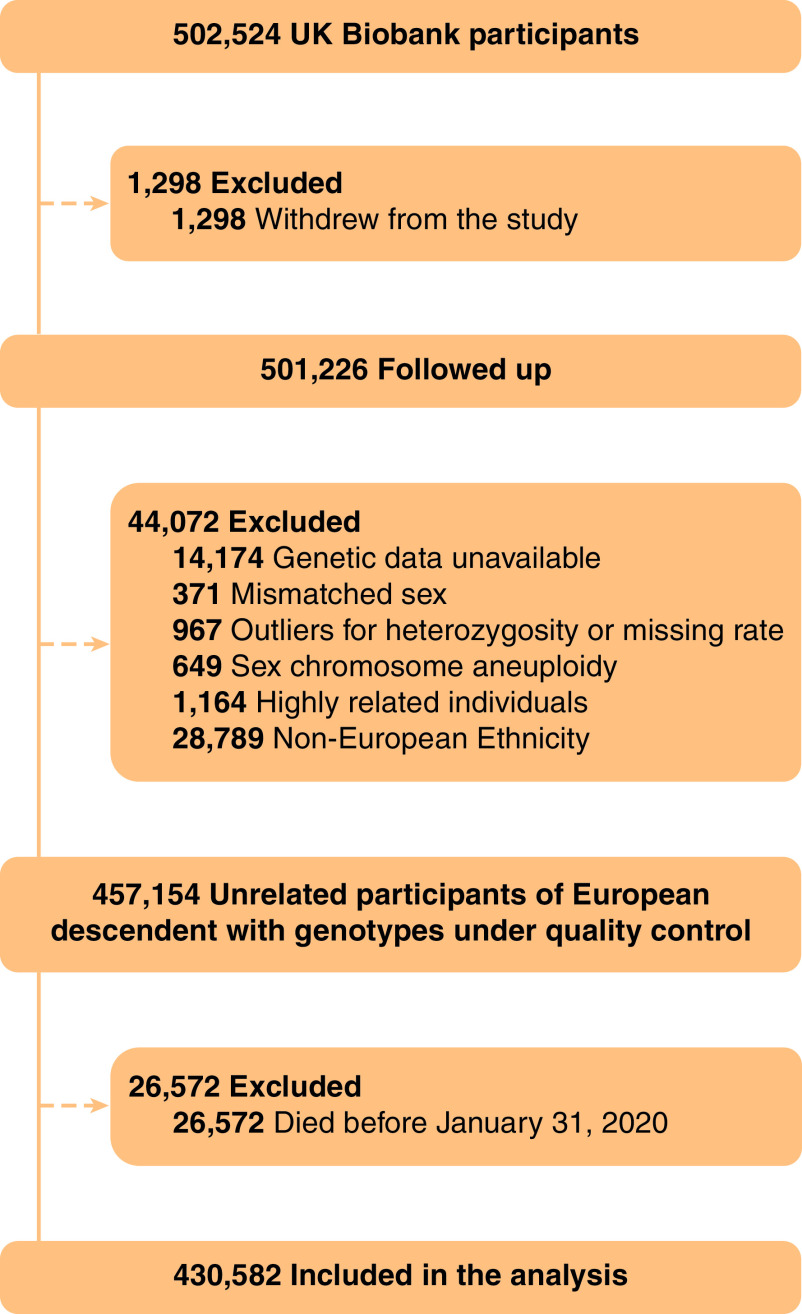
Participants were excluded if they withdrew from the study (n = 1,298), their genotype data did not meet the quality control conditions, they were highly related individuals (second degree or greater: kinship coefficient ⩾ 0.0884), or they were non-European ancestry (n = 44,072). In addition, participants who died before January 31, 2020, that is, the start of the COVID-19 outbreak in the UK, were also excluded (n = 26,572). In total, our analysis included 430,582 participants (Figure 1).
Figure 1.
Flow of participants through the study.
Genotyping and PRS
The GWAS data were derived from participants of predominantly European ancestry included in 14 cohort studies, which collectively consisted of 4,933 cases and 1,398,672 controls, released on October 20, 2020 (Table E1 in the online supplement). Details regarding the cohort-specific genotyping platform and risk scores are available at www.covid19hg.org (6). The PRS was calculated by using an additive model for the common genetic variants associated with severe COVID-19. Briefly, the number of risk alleles (0, 1, or 2) for individual participants was summed by multiplication with the effect size (25–27). The SNPs and effect sizes were obtained and pruned by P value < 1 × 10−6, linkage disequilibrium R2 < 0.1, and minor allele frequency > 1% from the previously reported GWAS studies (6, 25–28). A total of 112 SNPs was taken into account (Table E2). The PRS was then z-standardized based on the values for all individual participants and categorized into three risk categories: low (the lowest quintile), intermediate (quintiles 2–4), and high (the highest quintile) risk, as described previously (29, 30).
Ascertainment of COPD
In the present study, the spirometry, self-reported diagnoses, and diagnostic codes, covering the period 2006–2010 (i.e., during study enrollment) to January 2020, were used to identify individuals with diagnosed COPD. Details of the spirometry protocol in the UK Biobank have been previously reported (22, 31). Trained medical technicians and nurses at the UK Biobank Assessment Centre performed spirometric measurements on participants using the Vitalograph Pneumotrac 6800 in accordance with American Thoracic Society and European Respiratory Society guidelines. Each participant was given three attempts to obtain two reproducible maneuvers (32). The reproducibility of the first two blows was compared using the spirometer software, and if the differences between both forced vital capacity (FVC) and forced expiratory volume in 1 second (FEV1) of the first two blows were more than 5%, a third blow was required. No post-bronchodilator measures were performed. Acceptability of spirometry data was assessed by quality appraisal of a sample of maneuvers, as previously described (33). COPD outcomes were defined by the Global Lung Function Initiative 2012 reference values for the lower limit of normal (34), which were computed using the Global Lung Function Initiative R macro (35). Individuals with an FEV1/FVC ratio below the lower limit of normal were classified as having COPD (36). In addition, to avoid misdiagnosis of COPD, COPD was supplementally defined based on the codes from fields 20002 (self-reported diagnoses) and the International Classification of Diseases, Ninth and Tenth Revision codes (ICD-9 and ICD-10). Detailed information on the diagnosis codes is provided in Table E3. Of 7,901 participants who were excluded from consideration of COPD diagnosis with spirometry because of smoking tobacco or using inhalers 1 hour before testing, 793 participants had a diagnosis of COPD based on self-reported diagnoses, ICD-9, and ICD-10.
Ascertainment of Severe COVID-19
In the present study, severe COVID-19 was defined as an admission to an ICU according to data from the HES, death captured from 1 day before to up to 14 days after the SARS-CoV-2 test date, or death with COVID-19 (ICD-10 codes U0.71) as the underlying cause according to data from the death registers (last updated on February 22, 2021).
Ascertainment of Covariates
The covariates of the present study are as follows: age; sex; education; socioeconomic status (household income and Townsend deprivation index [TDI]); physical activity; smoking status; passive smoking; alcohol consumption; body mass index (BMI); diet; comorbidities, including cardiovascular disease (CVD), hypertension, diabetes, chronic respiratory infection, and asthma; the relatedness of individuals in the sample; and first 10 principal components of ancestry. The TDI is a composite measure of poverty based on unemployment, non–car ownership, non–home ownership, and household crowding, with negative values indicating higher socioeconomic status (37). BMI (kg/m2) was calculated for all participants by UK Biobank based on their measured weight and height. Duration of physical activity was ascertained with the touchscreen questionnaires based on the validated International Physical Activity Questionnaire (38). A healthy diet was calculated based on the Dietary Approaches to Stop Hypertension recommendation, which has been shown to be associated with multiple cancer types (39, 40). Passive smoking exposure refers to exposure more than 1 hour per week to other people’s tobacco smoke at home or outside. The information pertaining to the comorbidities was derived from the electronic medical records. Details of these measurements can be found on the website of the UK Biobank (www.ukbiobank.ac.uk).
Statistical Analysis
The baseline characteristics of the included participants were summarized by the severity of COVID-19 as numbers (percentages) for categorical variables, as means (standard deviations) for normal continuous variables, and medians (interquartile ranges) for skewed variables.
Logistic regression models were used to assess the associations of genetic risk, COPD, and their combination with severe COVID-19. The ORs together with the 95% confidence intervals (CIs) were calculated. Two sets of models were used. Model 1 was adjusted for age (years), sex (male or female), education (lower or high qualification), household income (<£18,000, £18,000–£30,999, £31,000–£51,999, £52,000–£100,000, or >£100,000), TDI, physical activity (some or no), smoking status (never, former, or current), passive smoking (no or yes), alcohol consumption (never, 1–2, 3–4, ⩾5 times per week), BMI (kg/m2), Dietary Approaches to Stop Hypertension score, CVD (no or yes), hypertension (no or yes), diabetes (no or yes), chronic respiratory infection (no or yes), asthma (no or yes), relatedness of individuals in the sample, and first 10 principal components of ancestry. In model 2, additional variables were controlled for preexisting COPD in the analysis of the association between genetic risk and severe COVID-19 and PRS in the analysis of the association between COPD and severe COVID-19. In addition, we included an interaction term in the regression model to test for any statistical interaction between the genetic risk categories and COPD.
We performed subgroup analyses stratified by age (<60 or ⩾60 yr), sex (male or female), and chronic respiratory infection (yes or no). For sensitivity analysis, we restricted analysis to participants with SARS-CoV-2 test, restricted analysis to participants with positive SARS-CoV-2 test result, excluded participants based on shared relatedness (third degree or greater: kinship coefficient ⩾0.0442), restricted COPD with spirometric definition, and excluded participants who died of other causes after February 1, 2020, because they were not followed-up for a sufficiently long period for further ascertainment of the acquisition of COVID-19. Furthermore, to verify the predictive performance of PRS for severe COVID-19, we estimated the area under the curve using receiver operating characteristic (ROC).
All statistical analyses were performed in R software, version 3.6 (R Project for Statistical Computing).
Results
Participant Characteristics
Table 1 shows the participants’ characteristics. Of the 430,582 participants, 237,533 (55.2%) were female, with a mean age of 56.5 years. Overall, 45,648 (10.6%) of the 430,582 participants had preexisting COPD. As of February 22, 2021, 712 severe COVID-19 events occurred. Compared with the general population, participants with severe COVID-19 seem to be older, be male, be current smokers, and have higher TDI, lower household income, and lower educational attainment. In addition, they were more likely to have a higher prevalence of preexisting cardiovascular disease, hypertension, diabetes, and COPD (19.8%) and had higher genetic risks.
Table 1.
Characteristics of the participants
| Characteristics | Overall (N = 430,582) | General Population (n = 429,870) | Incident Severe COVID-19 (n = 712) |
|---|---|---|---|
| Age, yr, mean (SD) | 56.5 (8.0) | 56.5 (8.0) | 62.1 (6.6) |
| Sex | |||
| Female | 237,533 (55.2) | 237,305 (55.2) | 228 (32.0) |
| Male | 193,049 (44.8) | 192,565 (44.8) | 484 (68.0) |
| BMI, kg/m2, mean (SD) | 27.3 (4.7) | 27.3 (4.7) | 29.8 (5.5) |
| DASH score, mean (SD) | 22.2 (4.0) | 22.2 (4.0) | 21.9 (4.2) |
| Physical activity, min/wk | |||
| Regular physical activity | 251,946 (58.5) | 251,546 (58.5) | 400 (56.2) |
| Some physical activity | 130,825 (30.4) | 130,626 (30.4) | 199 (27.9) |
| No regular physical activity | 47,811 (11.1) | 47,698 (11.1) | 113 (15.9) |
| Smoking status | |||
| Never | 236,689 (55.0) | 236,437 (55.0) | 252 (35.4) |
| Former | 151,344 (35.1) | 150,986 (35.1) | 358 (50.3) |
| Current | 42,549 (9.9) | 42,447 (9.9) | 102 (14.3) |
| Passive smoking | |||
| No | 341,713 (79.4) | 341,185 (79.4) | 528 (74.2) |
| Yes | 88,869 (20.6) | 88,685 (20.6) | 184 (25.8) |
| Alcohol consumption, times per wk | |||
| Never | 122,261 (28.4) | 122,000 (28.4) | 261 (36.7) |
| 1–2 | 114,078 (26.5) | 113,886 (26.5) | 192 (27.0) |
| 3–4 | 104,045 (24.2) | 103,918 (24.2) | 127 (17.8) |
| ⩾5 | 90,198 (20.9) | 90,066 (21.0) | 132 (18.5) |
| TDI, median (interquartile range) | −2.3 (−3.7 to 0.2) | −2.3 (−3.7 to 0.2) | −1.2 (−3.1 to 1.9) |
| Household income, £ | |||
| <18,000 | 94,566 (22.0) | 94,248 (21.9) | 318 (44.7) |
| 18,000–30,999 | 110,169 (25.6) | 109,985 (25.6) | 184 (25.8) |
| 31,000–51,999 | 113,996 (26.5) | 113,878 (26.5) | 118 (16.6) |
| 52,000–100,000 | 88,515 (20.6) | 88,434 (20.6) | 81 (11.4) |
| >100,000 | 23,336 (5.4) | 23,325 (5.4) | 11 (1.5) |
| Education | |||
| Lower qualification | 221,944 (51.5) | 221,472 (51.5) | 472 (66.3) |
| Higher qualification | 208,638 (48.5) | 208,398 (48.5) | 240 (33.7) |
| Comorbidities | |||
| CVD | 22,953 (5.3) | 22,818 (5.3) | 135 (19.0) |
| Hypertension | 100,818 (23.4) | 100,461 (23.4) | 357 (50.1) |
| Diabetes | 28,398 (6.6) | 28,231 (6.6) | 167 (23.5) |
| Chronic respiratory infection | 8,602 (2.0) | 8,585 (2.0) | 17 (2.4) |
| Asthma | 56,204 (13.1) | 56,078 (13.0) | 126 (17.7) |
| COPD | 45,648 (10.6) | 45,507 (10.6) | 141 (19.8) |
| Genetic risk category | |||
| Low | 86,099 (20.0) | 85,990 (20.0) | 109 (15.3) |
| Intermediate | 258,363 (60.0) | 257,925 (60.0) | 438 (61.5) |
| High | 86,120 (20.0) | 85,955 (20.0) | 165 (23.2) |
Definition of abbreviations: BMI = body mass index; COPD = chronic obstructive pulmonary disease; COVID-19 = coronavirus disease; CVD = cardiovascular disease; DASH = Dietary Approaches to Stop Hypertension; SD = standard deviation; TDI = Townsend deprivation index.
Data are presented as n (%) unless otherwise indicated.
Association between Genetic Risk and Incident Severe COVID-19
The calculated PRS was normally distributed (Figure E1). Table 2 presents the ORs of participants at intermediate and high genetic risk compared with those at low genetic risk. Higher genetic risk was associated with a higher risk of incident severe COVID-19 (P for trend = 0.001). Participants at high genetic risk had a significantly higher risk of incident severe COVID-19 than those at low genetic risk, with an adjusted odds ratio of 1.50 (95% CI, 1.18–1.92; P = 0.001). Results were broadly similar when the PRS was divided into quintiles (Table E4). Further adjustment for COPD (model 2) did not materially alter this finding (Table 2). The association between genetic risk and the risk of incident severe COVID-19 was similar in subgroups that were stratified by age, sex, and chronic respiratory infection (P for interaction > 0.05; Tables E5 and E6). In the sensitivity analysis, these results did not markedly change after restricting analyses to participants with the SARS-CoV-2 test (Table E7) and participants with positive SARS-CoV-2 test results (Table E8).
Table 2.
Risk of severe COVID-19 according to genetic risk
| Genetic Risk | Total No. of Participants | No. of Severe COVID-19 Cases (%) | Model 1* |
Model 2† |
||||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P Value | P for Trend | OR (95% CI) | P Value | P for Trend | |||
| Low | 86,099 | 109 (0.13) | 1 (reference) | 0.001 | 1 (reference) | 0.001 | ||
| Intermediate | 258,363 | 438 (0.17) | 1.34 (1.09–1.66) | 0.006 | 1.34 (1.09–1.66) | 0.006 | ||
| High | 86,120 | 165 (0.19) | 1.50 (1.18–1.92) | 0.001 | 1.50 (1.18–1.92) | 0.001 | ||
Definition of abbreviations: CI = confidence interval; COVID-19 = coronavirus disease; OR = odds ratio.
Model 1: Logistic regression adjusted for age, sex, assessment centers, Townsend deprivation index, income, education, body mass index, diet, physical activity, smoking, alcohol consumption, passive smoking, cardiovascular disease, hypertension, diabetes, chronic respiratory infections, asthma, relatedness of individuals in the sample, and first 10 principal components of ancestry; P value for trend calculated treating the polygenic risk score as a continuous variable.
Model 2: Logistic regression adjusted for model 1 and preexisting chronic obstructive pulmonary disease; P value for trend calculated treating the polygenic risk score as a continuous variable.
ROC curves were constructed and area under the curve was calculated for the following combinations: model 1: age, sex, and comorbidities (CVD, hypertension, diabetes, chronic respiratory infections, asthma, and COPD); model 2: age, sex, comorbidities, and PRS (Figure E2). For the base model, model 1, area under the curve was 0.789 (95% CI, 0.772–0.806), whereas that for model 2 was 0.794 (95% CI, 0.777–0.810; P = 0.002). Areas under the ROC are nearly identical, although the difference is statistically significant because of the large sample size.
Association between COPD and Incident Severe COVID-19
Table 3 presents the ORs of participants with COPD compared with those without. Participants with COPD had a higher risk of incident severe COVID-19 than those without, with an adjusted OR of 1.37 (95% CI, 1.12–1.67; P = 0.002). Further adjustment for the PRS (model 2) did not materially alter this result (Table 3). We observed a significant interaction effect between COPD and sex on the risks of severe COVID-19 (P for interaction = 0.006; Table E9). However, the association between COPD and the risk of severe COVID-19 was similar in subgroups that were stratified by age and chronic respiratory infection (P for interaction > 0.05; Tables E9 and E10).
Table 3.
Risk of severe COVID-19 according to COPD
| Preexisting COPD | Total No. of Participants | No. of Severe COVID-19 Cases (%) | Model 1* |
Model 2† |
||
|---|---|---|---|---|---|---|
| OR (95% CI) | P Value | OR (95% CI) | P Value | |||
| No | 384,934 | 571 (0.15) | 1 (reference) | 1 (reference) | ||
| Yes | 45,648 | 141 (0.31) | 1.37 (1.12–1.67) | 0.002 | 1.37 (1.12–1.67) | 0.002 |
Definition of abbreviations: CI = confidence interval; COPD = chronic obstructive pulmonary disease; COVID-19 = coronavirus disease; OR = odds ratio.
Model 1: Logistic regression adjusted for age, sex, assessment centers, Townsend deprivation index, income, education, body mass index, diet, physical activity, smoking, alcohol consumption, passive smoking, cardiovascular disease, hypertension, diabetes, chronic respiratory infections, asthma, relatedness of individuals in the sample, and first 10 principal components of ancestry.
Model 2: Logistic regression adjusted for model 1 and polygenic risk score.
Association of Genetic Risk and COPD with Incident Severe COVID-19
Figure 2 shows the risk of incident severe COVID-19 for the combined genetic risk and COPD. An additive effect on the risk of severe COVID-19 was found for the genetic risk and COPD. The highest risk of incident severe COVID-19 was observed in participants at high genetic risk and with preexisting COPD (OR, 2.05; 95% CI, 1.35–3.04; P < 0.001). The test for statistical interaction between the PRS and COPD in relation to the incident severe COVID-19 was not significant (P = 0.76). Furthermore, there is no statistically significant association between the PRS and COPD among the control population (P for trend = 0.75; Table E11). In the sensitivity analysis, these results did not markedly change after excluding participants based on shared relatedness (Table E12), excluding participants who died for other causes after February 1, 2020 (Table E13), and restricting COPD with spirometric definition (Table E14).
Figure 2.
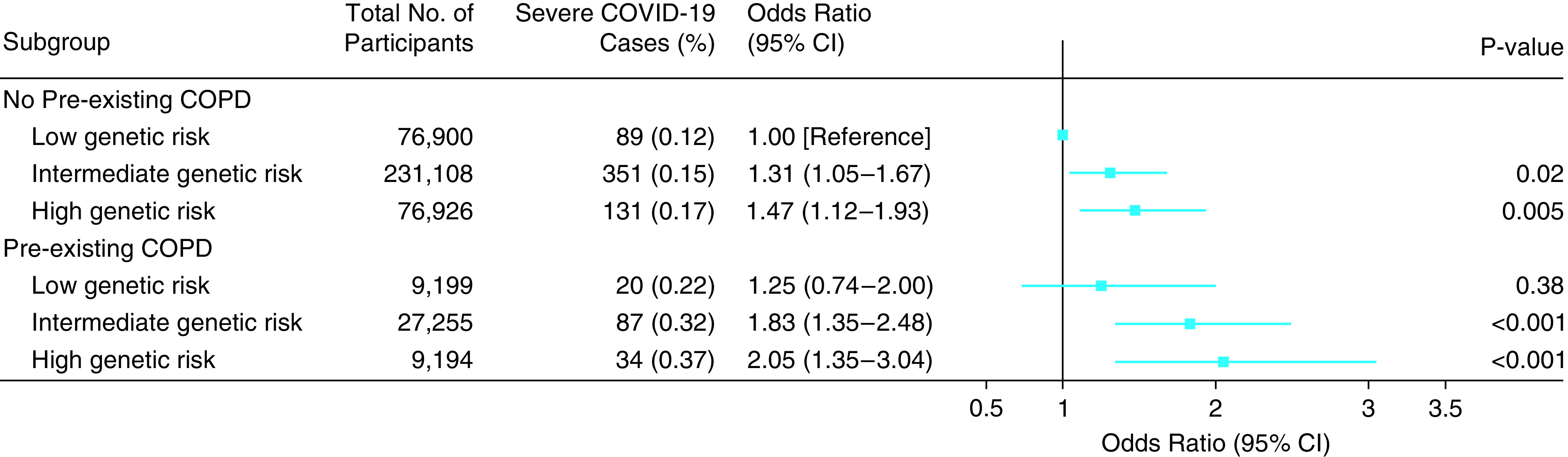
Risk of severe COVID-19 according to genetic risk and COPD. The results were obtained after adjusting for age, sex, assessment centers, Townsend deprivation index, income, education, body mass index, diet, physical activity, smoking, alcohol consumption, passive smoking, cardiovascular disease, hypertension, diabetes, chronic respiratory infections, asthma, relatedness of individuals in the sample, and first 10 principal components of ancestry; P value for trend calculated treating the polygenic risk score as a continuous variable. CI = confidence interval; COPD = chronic obstructive pulmonary disease; COVID-19 = coronavirus disease.
Discussion
In this population-based cohort study of more than 430,000 participants, both high genetic risk and COPD were independently associated with an increased risk of incident severe COVID-19. The risk of incident severe COVID-19 at high genetic risk combined with preexisting COPD was 2.05-fold higher than that at low genetic risk and without preexisting COPD.
There are common variants that were previously identified to be associated with the high risk of severe COVID-19, and their combined impact can be indicated by the PRS (5, 8). Our results demonstrated that the PRS was associated with the risk of developing severe COVID-19. Although the previous study has evaluated the predictive performance of PRSs based on six DNA polymorphisms in severe COVID-19 among sports players to guide screening (12), the limited sample size with case–control design in previous studies often led to discouraging results. In the present study, we found that our PRS was significantly associated with the risk of severe COVID-19 in a prospective cohort study. Our study has included a larger sample size and a significantly more comprehensive indicator of the genetic risk to increase the power for risk estimation. We have also adjusted for multiple potential confounding factors (economic and social background, lifestyle factors, comorbidities, etc.). Several genetic variants used in the present study associated with severe COVID-19 may affect the immune response and transmission of SARS-CoV-2 (8, 41, 42). Furthermore, our results demonstrated the lack of interactions between genetic risk and preexisting COPD, which suggests that it provides information regarding severe COVID-19 that is independent of preexisting COPD. These findings in the present study provided additional insights into the genetic basis of severe COVID-19, which would further advance our understanding of severe COVID-19 susceptibility.
From a clinical perspective, methodologically similar to assessing other diseases by using PRS, our PRS may help to identify individuals at the greatest risk for severe COVID-19 (30, 43). The risk of incident severe COVID-19 at high genetic risk was 1.50-fold higher than that at low genetic risk, suggesting that our PRS potentially was suitable to the risk stratification among those with COVID-19. In addition, the availability of the PRS throughout the life course suggests that PRS might be helpful for predicting severe COVID-19 in scenarios in which up-to-date clinical information is not available (e.g., COPD). From a disease management perspective, the distinct incidence risk for populations within different categories of genetic risk as defined by the PRS provides support to carry out PRS-informed severe COVID-19 screening and individualized prevention, independent of clinical risk factors such as age, sex, and COPD exposure. Furthermore, individuals with higher genetic risk might be warned to be more likely to maintain social distancing and carry out life planning to reduce the risk of developing severe COVID-19. Therefore, one potential application is using the PRS to help predict susceptibility to severe COVID-19 in initially healthy people, so as to focus preventive interventions on those at the highest risk of severe COVID-19 (10).
Several studies have consistently reported an increased risk of developing severe COVID-19 in participants with preexisting COPD (17–20). However, these findings are limited by small sample sizes and incomplete data on comorbidities, and the relevance of composite genetic risk and COPD in predicting the risk of severe COVID-19 remains unclear. Based on the population-based cohort study, our results demonstrated that participants with preexisting COPD had increased risks of developing severe COVID-19 as compared with participants without preexisting COPD, regardless of the individuals’ genetic risk. Moreover, the real-life setting of our large-scale cohort study has provided further support that COPD is a risk factor for severe COVID-19, even in participants genetically not predisposed to developing severe COVID-19. Although the underlying mechanisms remain to be elucidated, it is suggested that angiotensin-converting enzyme 2 plays an important role in this association. Studies of cellular mechanisms suggest that angiotensin-converting enzyme 2 expression, which is the main receptor for the cellular entry of SARS-CoV-2, is significantly upregulated in participants with COPD (7, 44). The findings in the present study indicate that patients with COPD should follow basic infection-control measures to help prevent severe COVID-19, and medical workers should pay more attention to patients with COPD who are infected with COVID-19.
There are several limitations that should be taken into account. First, we have limited information about the exposure to SARS-CoV-2 among participants without COVID-19 in the UK Biobank, which might have resulted in the selection bias, as in previous studies (45, 46). However, this concern is mitigated because these results did not markedly change after restricting analyses to participants with SARS-CoV-2 test and participants with positive SARS-CoV-2 test results. Second, additional variants or genetic patterns associated with severe COVID-19 are likely to be identified in the future, which may prove useful for refining estimates of genetic risk. Third, because the PRS was calculated based on the GWAS of European ancestry, we must cautiously generalize the inferences to other populations. Individuals of Black, Asian, and other minority race or ethnicity groups may be at increased risk for SARS-CoV-2 infection and potentially worse clinical outcomes, including ICU admission and mortality, when compared with White individuals (47). Race refers to self-identifying groups based on beliefs concerning shared culture, ancestry, and history (48). And ancestry is determined biographically or genetically. Although race may be a social construct, differences in genetic ancestry that happen to correlate to many of today’s racial constructs are real (49). As such, the findings in our study highlight that future GWASs and large-scale epidemiologic research should consider predicting the risk of incident severe COVID-19 among non-European ancestry groups. Fourth, the measures did not include information about the epidemiology of COVID-19 and medical care practices to treat COVID-19 infections, such as type of virus and use of drug, possibly leading to imprecise measurements. Finally, there are very few severe COVID-19 cases among participants in the UK Biobank, which might reduce the power in these analyses. Thus, further research involving more patients with severe COVID-19 is needed in the future.
Conclusions
PRSs combining multiple risk alleles are predictive of incident severe COVID-19. Meanwhile, high genetic risk was associated with a higher risk of incident severe COVID-19, regardless of preexisting COPD. These findings could have important implications for understanding the mechanisms underlying severe COVID-19 and provide future opportunities for risk stratification and early intervention.
Acknowledgments
Acknowledgment
The authors thank all the investigators and participants in the UK Biobank.
Footnotes
Supported by the Guangdong Province Higher Vocational Colleges and Schools Pearl River Scholar Funded Scheme (2019), the Zhejiang University special scientific research fund for COVID-19 prevention and control (2020XGZX041), and Zhongnanshan Medical Foundation of Guangdong Province. This research has been conducted using the UK Biobank resource under application number 43795.
Data availability: All available data are published in the current manuscript.
Author Contributions: Conception and study design: Q.-M.H., P.-D.Z., S.-G.Y., W.-J.G., and C.M. Acquisition, analysis, or interpretation of data: Q.-M.H., P.-D.Z., Z.-H.L., J.-M.Z., D.L., X.-R.Z., W.-F.Z., Y.-J.Z., D.S., F.L., and W.-Q.S. Manuscript preparation and critical revision: Q.-M.H., P.-D.Z., S.-G.Y., W.-J.G., and C.M. Obtained funding: C.M.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.COVID-19 coronavirus pandemic. https://www.worldometers.info/coronavirus
- 2. Ioannidis JPA. Infection fatality rate of COVID-19 inferred from seroprevalence data. Bull World Health Organ . 2021;99:19–33F. doi: 10.2471/BLT.20.265892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vetter P, Vu DL, L'Huillier AG, Schibler M, Kaiser L, Jacquerioz F. Clinical features of covid-19. BMJ . 2020;369:m1470. doi: 10.1136/bmj.m1470. [DOI] [PubMed] [Google Scholar]
- 4. Phua J, Weng L, Ling L, Egi M, Lim CM, Divatia JV, et al. Asian Critical Care Clinical Trials Group Intensive care management of coronavirus disease 2019 (COVID-19): challenges and recommendations. Lancet Respir Med . 2020;8:506–517. doi: 10.1016/S2213-2600(20)30161-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P, et al. Severe Covid-19 GWAS Group Genomewide association study of severe Covid-19 with respiratory failure. N Engl J Med . 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. COVID-19 Host Genetics Initiative. The COVID-19 Host Genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur J Hum Genet . 2020;28:715–718. doi: 10.1038/s41431-020-0636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature . 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kuo CL, Pilling LC, Atkins JL, Masoli JAH, Delgado J, Kuchel GA, et al. APOE e4 genotype predicts severe COVID-19 in the UK Biobank community cohort. J Gerontol A Biol Sci Med Sci . 2020;75:2231–2232. doi: 10.1093/gerona/glaa131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guo X, Chen Z, Xia Y, Lin W, Li H. Investigation of the genetic variation in ACE2 on the structural recognition by the novel coronavirus (SARS-CoV-2) J Transl Med . 2020;18:321. doi: 10.1186/s12967-020-02486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet . 2018;19:581–590. doi: 10.1038/s41576-018-0018-x. [DOI] [PubMed] [Google Scholar]
- 11. Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, et al. International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature . 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ahmetov II, Borisov OV, Semenova EA, Andryushchenko ON, Andryushchenko LB, Generozov EV, et al. Team sport, power, and combat athletes are at high genetic risk for coronavirus disease-2019 severity. J Sport Health Sci . 2020;9:430–431. doi: 10.1016/j.jshs.2020.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shrine N, Guyatt AL, Erzurumluoglu AM, Jackson VE, Hobbs BD, Melbourne CA, et al. Understanding Society Scientific Group New genetic signals for lung function highlight pathways and chronic obstructive pulmonary disease associations across multiple ancestries. Nat Genet . 2019;51:481–493. doi: 10.1038/s41588-018-0321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.biobankuk release BNsUBrt. 2018 [updated 2018 Aug 1; accessed 2020 Apr 12]. Available from: http://www.nealelab.is/uk-biobank/.
- 15. Saheb Sharif-Askari N, Saheb Sharif-Askari F, Alabed M, Temsah MH, Al Heialy S, Hamid Q, et al. Airways expression of SARS-CoV-2 receptor, ACE2, and TMPRSS2 is lower in children than adults and increases with smoking and COPD. Mol Ther Methods Clin Dev . 2020;18:1–6. doi: 10.1016/j.omtm.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun W, Kechris K, Jacobson S, Drummond MB, Hawkins GA, Yang J, et al. SPIROMICS Research Group COPDGene Investigators. Common genetic polymorphisms influence blood biomarker measurements in COPD. PLoS Genet . 2016;12:e1006011. doi: 10.1371/journal.pgen.1006011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lippi G, Henry BM. Chronic obstructive pulmonary disease is associated with severe coronavirus disease 2019 (COVID-19) Respir Med . 2020;167:105941. doi: 10.1016/j.rmed.2020.105941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang B, Li R, Lu Z, Huang Y. Does comorbidity increase the risk of patients with COVID-19: evidence from meta-analysis. Aging (Albany NY) . 2020;12:6049–6057. doi: 10.18632/aging.103000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao Q, Meng M, Kumar R, Wu Y, Huang J, Lian N, et al. The impact of COPD and smoking history on the severity of COVID-19: A systemic review and meta-analysis. J Med Virol . 2020;92:1915–1921. doi: 10.1002/jmv.25889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang L, He W, Yu X, Hu D, Bao M, Liu H, et al. Coronavirus disease 2019 in elderly patients: Characteristics and prognostic factors based on 4-week follow-up. J Infect . 2020;80:639–645. doi: 10.1016/j.jinf.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ovsyannikova IG, Haralambieva IH, Crooke SN, Poland GA, Kennedy RB. The role of host genetics in the immune response to SARS-CoV-2 and COVID-19 susceptibility and severity. Immunol Rev . 2020;296:205–219. doi: 10.1111/imr.12897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med . 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Palmer LJUK. UK Biobank: bank on it. Lancet . 2007;369:1980–1982. doi: 10.1016/S0140-6736(07)60924-6. [DOI] [PubMed] [Google Scholar]
- 24. Li ZH, Zhong WF, Liu S, Kraus VB, Zhang YJ, Gao X, et al. Associations of habitual fish oil supplementation with cardiovascular outcomes and all cause mortality: evidence from a large population based cohort study. BMJ . 2020;368:m456. doi: 10.1136/bmj.m456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McKay JD, Hung RJ, Han Y, Zong X, Carreras-Torres R, Christiani DC, et al. SpiroMeta Consortium Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat Genet . 2017;49:1126–1132. doi: 10.1038/ng.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dai J, Lv J, Zhu M, Wang Y, Qin N, Ma H, et al. Identification of risk loci and a polygenic risk score for lung cancer: a large-scale prospective cohort study in Chinese populations. Lancet Respir Med . 2019;7:881–891. doi: 10.1016/S2213-2600(19)30144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Broderick P, Wang Y, Vijayakrishnan J, Matakidou A, Spitz MR, Eisen T, et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res . 2009;69:6633–6641. doi: 10.1158/0008-5472.CAN-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li Y, Sheu CC, Ye Y, de Andrade M, Wang L, Chang SC, et al. Genetic variants and risk of lung cancer in never smokers: a genome-wide association study. Lancet Oncol . 2010;11:321–330. doi: 10.1016/S1470-2045(10)70042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lourida I, Hannon E, Littlejohns TJ, Langa KM, Hyppönen E, Kuzma E, et al. Association of lifestyle and genetic risk with incidence of dementia. JAMA . 2019;322:430–437. doi: 10.1001/jama.2019.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Khera AV, Emdin CA, Drake I, Natarajan P, Bick AG, Cook NR, et al. Genetic risk, adherence to a healthy lifestyle, and coronary disease. N Engl J Med . 2016;375:2349–2358. doi: 10.1056/NEJMoa1605086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Doiron D, de Hoogh K, Probst-Hensch N, Fortier I, Cai Y, De Matteis S, et al. Air pollution, lung function and COPD: results from the population-based UK Biobank study. Eur Respir J . 2019;54:1802140. doi: 10.1183/13993003.02140-2018. [DOI] [PubMed] [Google Scholar]
- 32. Miller MR, Hankinson J, Brusasco V, Burgos F, Casaburi R, Coates A, et al. ATS/ERS Task Force Standardisation of spirometry. Eur Respir J . 2005;26:319–338. doi: 10.1183/09031936.05.00034805. [DOI] [PubMed] [Google Scholar]
- 33. De Matteis S, Jarvis D, Hutchings S, Darnton A, Fishwick D, Sadhra S, et al. Occupations associated with COPD risk in the large population-based UK Biobank cohort study. Occup Environ Med . 2016;73:378–384. doi: 10.1136/oemed-2015-103406. [DOI] [PubMed] [Google Scholar]
- 34. Quanjer PH, Stanojevic S, Cole TJ, Baur X, Hall GL, Culver BH, et al. ERS Global Lung Function Initiative Multi-ethnic reference values for spirometry for the 3-95-yr age range: the global lung function 2012 equations. Eur Respir J . 2012;40:1324–1343. doi: 10.1183/09031936.00080312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Society ER.2017http://www.ers-education.org/guidelines/global-lung-function-initiative/spirometry-tools/r-macro.aspx.
- 36.Global Initiative for Chronic Obstructive Lung Disease (GOLD) http://goldcopd.org
- 37. Tyrrell J, Jones SE, Beaumont R, Astley CM, Lovell R, Yaghootkar H, et al. Height, body mass index, and socioeconomic status: mendelian randomisation study in UK Biobank. BMJ . 2016;352:i582. doi: 10.1136/bmj.i582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bassett DR., Jr International physical activity questionnaire: 12-country reliability and validity. Med Sci Sports Exerc . 2003;35:1396. doi: 10.1249/01.MSS.0000078923.96621.1D. [DOI] [PubMed] [Google Scholar]
- 39. Ali Mohsenpour M, Fallah-Moshkani R, Ghiasvand R, Khosravi-Boroujeni H, Mehdi Ahmadi S, Brauer P, et al. Adherence to Dietary Approaches to Stop Hypertension (DASH)-style diet and the risk of cancer: a systematic review and meta-analysis of cohort studies. J Am Coll Nutr . 2019;38:513–525. doi: 10.1080/07315724.2018.1554460. [DOI] [PubMed] [Google Scholar]
- 40. Appel LJ, Moore TJ, Obarzanek E, Vollmer WM, Svetkey LP, Sacks FM, et al. DASH Collaborative Research Group A clinical trial of the effects of dietary patterns on blood pressure. N Engl J Med . 1997;336:1117–1124. doi: 10.1056/NEJM199704173361601. [DOI] [PubMed] [Google Scholar]
- 41. Schreiber G. The molecular basis for differential type I interferon signaling. J Biol Chem . 2017;292:7285–7294. doi: 10.1074/jbc.R116.774562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mendonca P, Soliman KFA. Flavonoids activation of the transcription factor Nrf2 as a hypothesis approach for the prevention and modulation of SARS-CoV-2 infection severity. Antioxidants. 2020;9:659. doi: 10.3390/antiox9080659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mavaddat N, Pharoah PD, Michailidou K, Tyrer J, Brook MN, Bolla MK, et al. Prediction of breast cancer risk based on profiling with common genetic variants. J Natl Cancer Inst . 2015;107:djv036. doi: 10.1093/jnci/djv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leung JM, Yang CX, Tam A, Shaipanich T, Hackett TL, Singhera GK, et al. ACE-2 expression in the small airway epithelia of smokers and COPD patients: implications for COVID-19. Eur Respir J. 2020;55:2000688. doi: 10.1183/13993003.00688-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhu Z, Hasegawa K, Ma B, Fujiogi M, Camargo CA, Jr, Liang L. Association of asthma and its genetic predisposition with the risk of severe COVID-19. J Allergy Clin Immunol . 2020;146:327–329.e4. doi: 10.1016/j.jaci.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhu Z, Hasegawa K, Ma B, Fujiogi M, Camargo CA, Jr, Liang L. Association of obesity and its genetic predisposition with the risk of severe COVID-19: Analysis of population-based cohort data. Metabolism . 2020;112:154345. doi: 10.1016/j.metabol.2020.154345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pan D, Sze S, Minhas JS, Bangash MN, Pareek N, Divall P, et al. The impact of ethnicity on clinical outcomes in COVID-19: a systematic review. EClinicalMedicine . 2020;23:100404. doi: 10.1016/j.eclinm.2020.100404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Little W, McGivern R, Keirns N, Strayer E, Griffiths H, Cody-Rydzewski S, et al. 2014. https://opentextbc.ca/introductiontosociology/chapter/chapter11-race-and-ethnicity/
- 49.Reich D.2018https://www.nytimes.com/2018/03/23/opinion/sunday/genetics-race.html.