Abstract
Background
Mutations in DYRK1A are a cause of microcephaly, autism spectrum disorder (ASD), and intellectual disability (ID); however, the underlying cellular and molecular mechanisms are not well understood.
Methods
We generated a conditional mouse model using Emx1-cre, including conditional heterozygous and homozygous knockouts, to investigate the necessity of Dyrk1a in the cortex during development. We employed unbiased, high throughput phospho-proteomics to identify dysregulated signaling mechanisms in the developing Dyrk1a mutant cortex as well as classic genetic modifier approaches and pharmacological therapeutic intervention to rescue microcephaly and neuronal undergrowth caused by Dyrk1a mutations.
Results
We found that cortical deletion of Dyrk1a in mice causes decreased brain mass and neuronal size, structural hypoconnectivity, and autism-relevant behaviors. Using phospho-proteomic screening, we identified growth-associated signaling cascades dysregulated upon Dyrk1a deletion, including TrkB/BDNF, an important regulator of ERK/MAPK and mTOR signaling. Genetic suppression of Pten or pharmacological treatment with IGF-1, both of which impinge on these signaling cascades, rescued microcephaly and neuronal undergrowth in neonatal mutants.
Conclusions
Altogether, these findings identify a previously unknown mechanism through which Dyrk1a mutations disrupt growth factor signaling in the developing brain, thus influencing neuronal growth and connectivity. Our results place Dyrk1a as a critical regulator of a biological pathway known to be dysregulated in humans with autism spectrum disorder and intellectual disability. Additionally, these data position Dyrk1a within a larger group of ASD/ID risk genes that impinge on growth-associated signaling cascades to regulate brain size and connectivity, suggesting a point of convergence for multiple autism etiologies.
Keywords: Autism spectrum disorder, microcephaly, mTOR, neuronal growth, Dyrk1a, cerebral cortex
Introduction
Loss of function mutations in DYRK1A (OMIM #600855) are strongly associated with autism spectrum disorder (ASD) and intellectual disability (ID). A recent study found that 48% and 81% of individuals with DYRK1A mutations exhibited ASD and ID, respectively (1). Primary microcephaly, described as decreased head circumference at birth (2–4), occurs in as many as 90% of these patients (1). Other clinical findings in this population include developmental delay, anxiety, seizures, speech and motor difficulties, and vision abnormalities (5, 6).
Dyrk1a is a serine/threonine kinase (7) that targets proteins critical for proliferation and differentiation in the developing mammalian brain, including Gsk3β (8), CyclinD1 (9), Foxo1 (10), Notch Intracellular Domain (NICD) (11), and P53 (12). Mutations in Dyrk1a impact cell cycle and proliferation through these phosphorylation targets. Germline Dyrk1a+/− mice exhibit decreased brain and body mass in adulthood as well as behavioral deficits (13–15) and altered proliferation (9, 12, 13). Dyrk1a mutations also cause decreased complexity of cultured neurons (16). However, the molecular mechanisms governing these phenotypes are currently unknown.
Multiple signaling cascades have been implicated in the pathogenesis of ASD, including Wnt/GSK3β, mTOR, and ERK/MAPK signaling (17–21). These molecular cascades are critical for regulating growth and proliferation in the developing mammalian brain (22–24) and have been proposed as signaling “hubs” for various etiologies of ASD (21, 25, 26). Convergence between Wnt/GSK3β, ERK/MAPK and PI3K/Akt/mTOR pathways in the developing brain (27–29) illustrate the abundant crosstalk between signaling cascades implicated in ASD pathobiology. Multiple studies indicate that Dyrk1a may interact with these ASD-associated signaling cascades. For example, Dyrk1a overexpression causes increased MAPK signaling in PC12 cells (30) and increased ERK/Akt activation in mice (31). Dyrk1a is a priming kinase for GSK3β (32), and PI3K/Akt/mTOR signaling is hyperactivated in human brain tissue from patients with Down syndrome, caused by trisomy of chromosome 21 including DYRK1A (33). While these findings are suggestive, the role of these cascades in ASD/ID-relevant brain and behavioral phenotypes caused by Dyrk1a mutations is unexplored.
Methods and Materials
Please see Supplementary Methods and Materials for more details.
Mice:
All mice used in the study were obtained from the Jackson Laboratory or MMRC and have been previously described, including C57BL/6-Dyrk1atm1Jdc/J (Dyrk1aloxP/loxP, stock #027801), Gt (ROSA)26Sortm14 (CAG-tdTomato)Hze/J (Ai14 or tdTomato, stock #007914), Ptentm1Hwu (PtenloxP/loxP, stock #006440), Emx1tm1 (cre)Krj (Emx1-cre+/−, stock #005628), and B6.FVB (Cg)-Tg (Rbp4-cre)KL100Gsat/Mmucd (Rbp4-cre+/−, stock #037128-UCD). WT and Emx1-cre−; Dyrk1aloxP/+ mice were used as controls. To avoid germline recombination in males, Emx1-cre−;Dyrk1aloxP/loxP males were bred with Emx1-cre+;Dyrk1aloxP/+ females. The same breeding strategy was used for experiments involving Rbp4-cre, mTORloxP/loxP, and PtenloxP/+. Genomic DNA isolated from ear samples was used for PCR to confirm genotypes. All animal experiments were conducted in accordance with National Institutes of Health and Association for Assessment and Accreditation of Laboratory Animal Care guidelines and were approved by The Scripps Research Institute’s Institutional Animal Care and Use Committee. Mixed sexes were used for all cellular and molecular assays. Male mice were used for behavioral experiments. Mixed sexes of even numbers were used for proteomic analysis, and females were used for the phospho-enrichment and analysis.
Results
Conditional mutations in Dyrk1a cause microcephaly and ASD-relevant behaviors
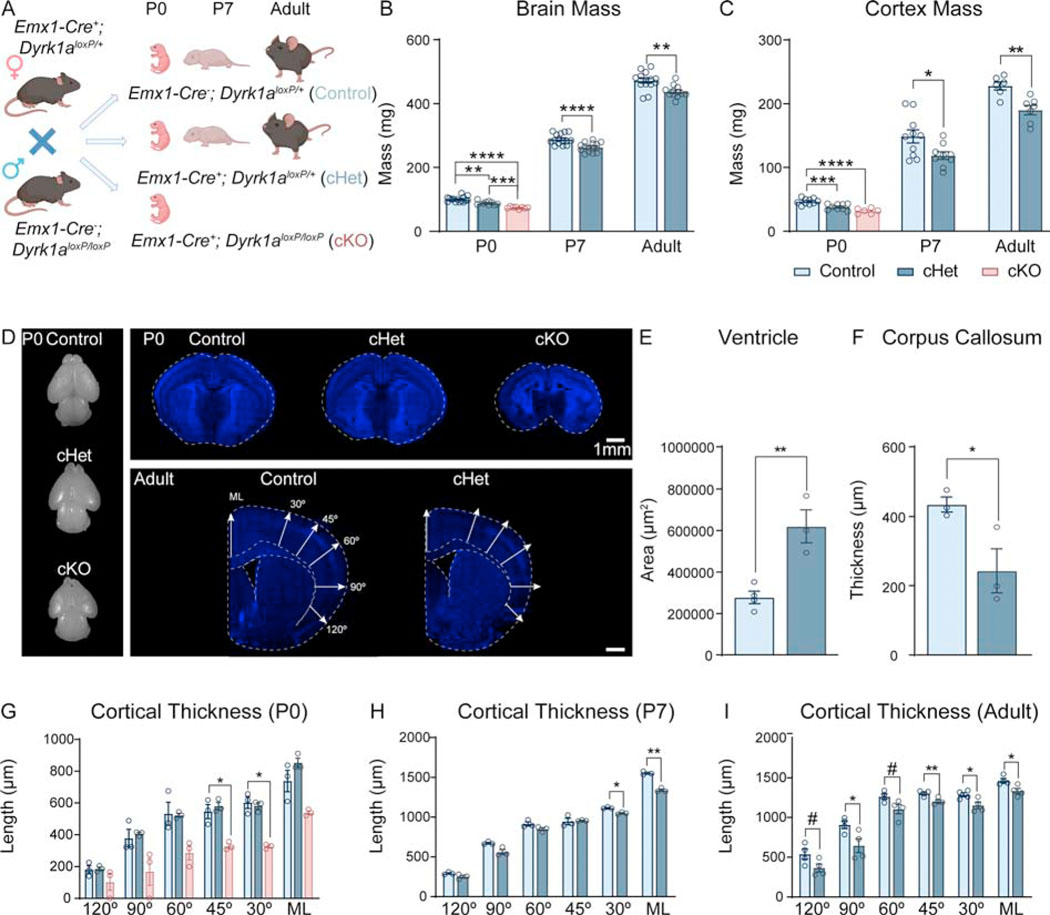
To circumvent pleiotropic effects outside the brain and isolate the effects of Dyrk1a mutation in the cerebral cortex, which is heavily impacted in microcephaly, we employed a conditional approach. Using an Emx1-cre driver, which is expressed in ~88% of neurons in the neocortex and hippocampus as well as a subset of cells in the astrocyte and oligodendrocyte lineages (34), we generated heterozygous (Emx1-cre+; Dyrk1aloxP/+, “cHet”) and homozygous (Emx1-cre+; Dyrk1aloxP/loxP, “cKO”) mutants (Figure 1A, S1). cKOs survive to birth but die by the end of postnatal day 0 (P0). Thus, cKO data was only collected at P0 for all assays. We found that cHets and cKOs exhibit decreased brain and cortex mass at birth (P0), throughout development (P7), and in adulthood (>P56) (Figure 1B,C) indicating that cortical mutations in Dyrk1a cause microcephaly. cKOs exhibit morphological abnormalities including increased ventricle size, lack of a corpus callosum, and an absent lateral cortex (Figure 1D, top panel). Adult cHets also exhibit significantly enlarged ventricles and a thinner corpus callosum (Figure 1E,F). We measured cortical thickness as the distance from the ventricle to pial surface at several positions on the mediolateral axis (Figure 1D (bottom panel)) and found that the cHet cortex is not altered at birth. However, the cKO cortex is markedly thinner (Figure 1G). P7 and adult cHets exhibit decreased cortical thickness at multiple measurement points (Figure 1H,I).
Figure 1: Cortical mutations in Dyrk1a model microcephaly and ASD-relevant behaviors.
(A) Breeding scheme used to generate cHets and cKOs. cKOs do not survive past birth and are analyzed only at P0 as reflected in the data points.
(B-C) Cortical deletion of Dyrk1a causes decreased brain (B) and cortex (C) mass at birth, throughout development, and in adulthood. P0 analyzed by one-way ANOVA with Tukey’s post hoc multiple comparisons tests showed significant effects of genotype on brain (F2,29=41.15, P<.0001) and cortex mass (F2,21=22.14, P<.0001). P7 and adult timepoints were analyzed by independent sample t-test (P7 brain: t28=4.472, P=.00186; P7 cortex: t17=2.437, P=.026081; adult brain: t25=3.602, P=.001366 adult cortex: t11=3.811, P=.002889). N=8–15/genotype per timepoint.
(D) Dorsal view of dissected whole brains at birth from each genotype (left panel). Representative images of coronal brain sections showcasing the morphological abnormalities in the cKO brain at birth (top panel). Representative coronal brain images of adult control cHet brains at Bregma 0.26mm (bottom right panel) with schematic showing measurements used in (G-I) to determine cortical thickness (ML=midline).
(E) The ventricles in adult cHet brains are significantly larger compared to controls (t5=4.551, P=.0061).
(F) The corpus callosum is significantly thinner in adult cHets (t4=2.833, P=.0472). Ventricle and corpus callosum measurements were analyzed by independent sample t-tests. N=3–4/genotype.
(G) The thickness of the cortex in the cKO brain is significantly reduced at multiple measurement points with no gross alterations in the cHets at birth. Analysis by repeated measures two-way ANOVA (F10,36=1.244, P=.2977) shows no significant interaction of angle measured X genotype. Despite absence of significant interactions between layer and genotype, planned comparisons between genotypes at each layer (with Tukey’s post hoc multiple comparisons tests) showed significant differences (one-way ANOVAs; 120°: F2,6=1.893, P=.2305; 90°: F2,6=4.871, P=.0554; 60°: F2,6=7.852, P=.0211; 45°: F2,6=19.99, P=.0022; 30°: F2,6=38.20, P=.0004; ML°: F2,6=13.89, P=.0056).
(H) At P7, the cortex is thinner at multiple angles in cHets. Repeated measure two-way ANOVA with Sidak’s post hoc multiple comparisons tests showed a significant interaction between genotype and measurement angle (F5,20=12.52, P<.0001).
(I) Adult cHet cortex is thinner at multiple measurement angles as analyzed by repeated measures two-way ANOVA (F5,30=.8433, P=.5299). While there were no significant interactions between layer and genotype, planned comparisons between genotypes at each layer showed significant differences (independent sample t-tests; 120°: t6=2.14646, P=.075474; 90°: t6 =2.67184, P=.036936; 60°: t6=2.34475, P=.057470; 45°: t6=3.75555, P=.009448; 30°: t6=2.74530, P=.033499; ML°: t6=2.96656, P=.025069).
N=3–4/genotype.
Results from post hoc and t-tests indicated on graphs. Error bars represent mean +/−SEM. *P<.05, **P<.01, ***P<.001, ****P<.0001.
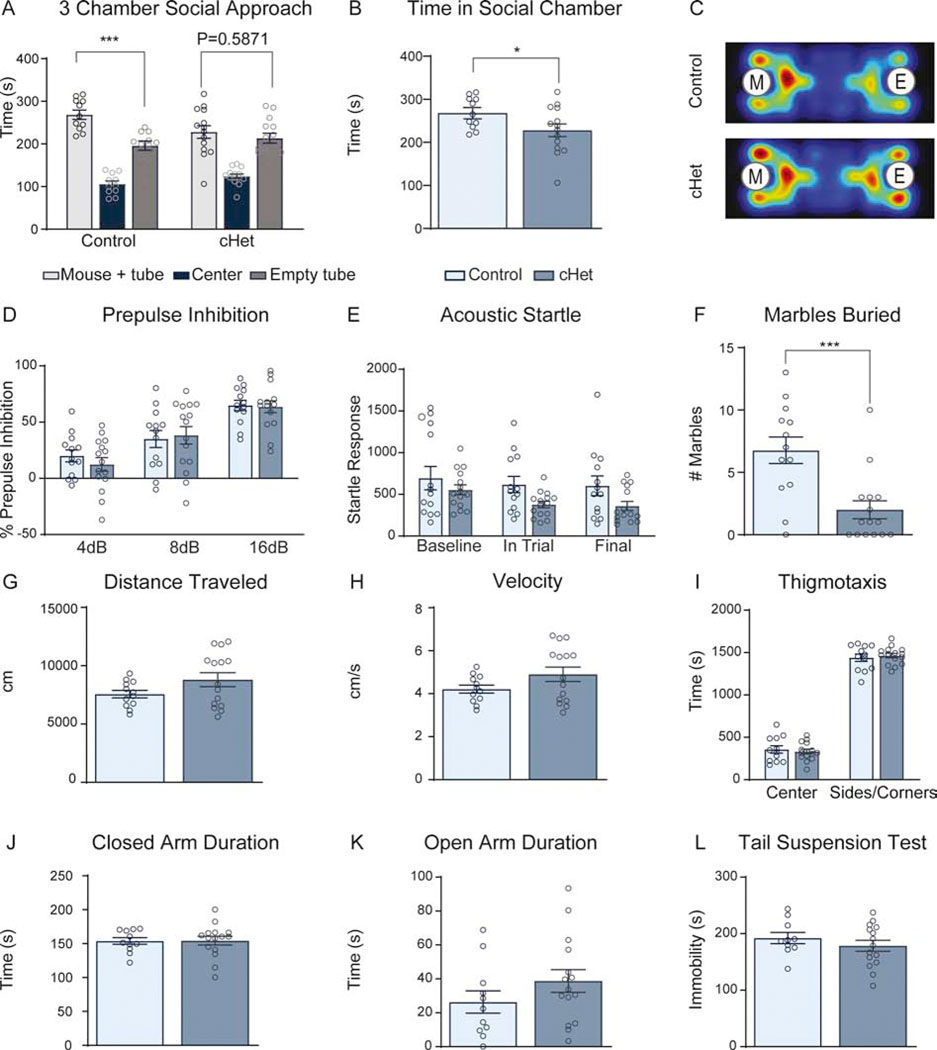
To determine whether cHets model phenotypes relevant to patients with DYRK1A mutations, we investigated ASD-relevant behaviors. In the three-chamber social approach assay, control mice exhibit a significant preference for interaction with a stimulus mouse, while cHets spend equal amounts of time in the chamber containing a stimulus mouse and the chamber with an empty tube (Figure 2A). cHets spend significantly less time in the social chamber, suggesting a social deficit (Figure 2B,C). Further, cHets bury significantly fewer marbles thereby displaying an altered repetitive behavior (Figure 2F). cHets do not exhibit alterations in sensorimotor gating (Figure 2D,E), locomotion (Figure 2G,H), anxiety-like behavior (Figure 2I–K), or depression-like behavior (Figure 2L). These data show that a cortical Dyrk1a mutation is sufficient to cause microcephaly and ASD-relevant behavioral deficits.
Figure 2: cHets exhibit autism-relevant behaviors but no alterations in locomotor activity, anxiety-like behavior, or depressive-like behavior.
(A) Analysis of 3-chamber social approach assay shows that controls exhibit a significant preference for the chamber with the stimulus mouse compared to the chamber with the empty tube (paired sample t-test within control group, t10=3.576, P=.0050). cHets do not exhibit this preference and spend equal time in the social and nonsocial chambers (paired sample t-test within cHet group, t13=.5569, P=.5871).
(B) cHets spend significantly less time with the social stimulus compared to controls. Analyzed by independent sample t-test (t23=2.082, P=.0487).
(C) Averaged heat map from controls and cHets during 3-chamber social approach assay where “M” represents the chamber with the stimulus mouse and “E” represents the chamber with the empty tube. N=11–14 males/genotype.
(D) In the pre-pulse inhibition assay, there is no difference in percent inhibition between controls and cHets. Repeated measures two-way ANOVA shows no significant interaction between genotype and volume of pre-pulse (F2,52=.9034, P=.4114); however, there is a significant effect of pre-pulse volume (F2,52=.70.39, P<.0001).
(E) Acoustic startle measured within the pre-pulse inhibition assay shows no differences between controls and cHets. While there is no significant interaction between timing and genotype shown by repeated measures two-way ANOVA, (F2,52 =.6061, P=.5493), there is a trend towards an effect of genotype (F1,26 =3.638, P=.0676) and significant effect of timing of startle (F1.408,36.62 =4.339, P=.0319).
(F) cHets bury significantly fewer marbles in 30 minutes compared to controls (t26=3.806, P=.0008).
(G-I) In the open field test, cHets do not exhibit any alterations in distance traveled (G; t25=1.705, P=.1005), velocity (H; t25=1.698, P=.1018), or thigmotaxis (I; Center; t25=.498910, P=.622205; Sides/corners; t25=.511655, P=.613383).
(J-K) In the elevated plus maze, cHets do not exhibit alterations in closed arm duration (J; t24=.04611, P=.9636) or open arm duration (K; t24=1.283, P=.2117).
(L) In the tail suspension test, cHets do not exhibit any alterations in immobility (t23=.9501, P=.3519). For all behavioral experiments, N=11–14 males/genotype. Analyzed by independent sample t-test unless otherwise specified.
Results from post hoc and t-tests indicated on graphs. Error bars represent mean +/− SEM. *P<.05, **P<.01, ***P<.001, ****P<.0001.
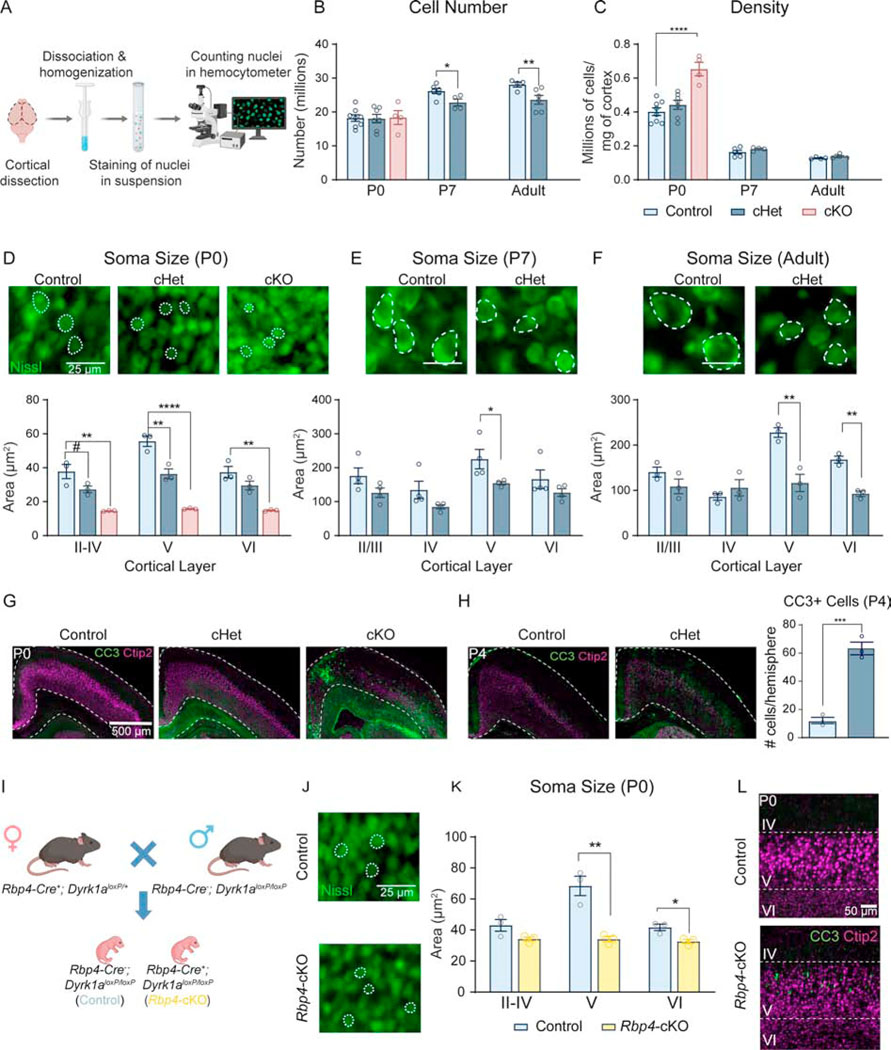
Microcephaly in Dyrk1a conditional mutants corresponds to reduced cell size at birth and fewer cells during postnatal development
Since germline Dyrk1a+/− mice exhibit altered neurogenesis and cell cycle exit (9), we predicted that decreased cell number would correspond with microcephaly at birth. We conducted isotropic fractionator to obtain the absolute number of cells (Figure 3A) (35). Despite decreased cortical mass and thickness upon Dyrk1a deletion, cell number is unexpectedly not altered in cHets or cKOs at birth. The density of cells in the cKO cortex is significantly increased, suggesting a decrease in cell size or spacing. P7 cHets exhibit decreased cell number relative to both controls and P0 cHets, but cell density is not altered. We observed the same effect in adult cHets, with the mutant cortex containing significantly fewer cells and unchanged cellular density (Figure 3B,C). These findings suggest that decreased cell number is not driving microcephaly in conditional mutants at birth, but rather there is a decrease in cell number in the mutants between P0 and P7.
Figure 3: Microcephaly in conditional Dyrk1a mutants is driven by decreased cell size at birth and decreased cell number after birth.
(A) Schematic of isotropic fractionator protocol.
(B) At birth, P0 cHets and cKOs do not exhibit alterations in cell number in the cortex. At P7 and adulthood, cell number is significantly reduced relative to controls. P0 analyzed by one-way ANOVA with Tukey’s post hoc multiple comparisons tests (shown on graph; F2,16=.01390, P=.9862). P7 and adult timepoints analyzed by independent sample t-test (P7: t8=2.663, P=.028690; adult: t8=2.579, P=.032651).
(C) At P0, cortical cell density is significantly increased in cKOs and is normalized by P7 and through adulthood. P0 analyzed by one-way ANOVA with Tukey’s post hoc multiple comparisons tests (shown on graph; F2,16=18.31, P<.0001). P7 and adult timepoints analyzed by independent sample t-tests (P7: t8=1.345, P=.215528; adult: t8=1.942, P=.0881). For all ages, N=4–8/genotype.
(D) Representative images of Nissl-stained cell somas in layer V of somatosensory cortex at birth with quantification showing significant decreases in multiple layers of P0 cHet and cKO cortices. Analyzed by one-way ANOVA [I-IV (F2,6=18.83, P=.0026), V (F2,6=65.83, P<.0001), VI (F2,6=21.62 P=.0018)] with Tukey’s post hoc multiple comparisons tests.
(E) Representative Nissl-stained cell somas at P7 with quantification showing a significant decrease in layer V of somatosensory cortex in cHets [independent sample t-tests: II/III (t6=1.842, P=.114989), IV (t6=1.897, P=.106656), V (t6=2.462, P=.048976), VI (t6=1.328, P=.23571)].
(F) Representative images of Nissl-stained cell somas in layer V of somatosensory cortex in adult animals with quantification showing that somas in layers V and VI are significantly smaller in cHets [independent sample t-tests II/III (t4=1.612, P=.182235), IV (t4=1.045, P=.355062), V (t4=5.107, P=.006961), VI (t4=7.862, P=.001414)]. For all ages, N=3–4/genotype.
(G) Representative images of CC3 staining in the cortex at birth.
(H) Representative images of increased CC3 in the cortex at P4 with quantification of CC3+ cells per cortical hemisphere using immunohistochemistry. Analyzed by independent sample t-test (t4=10.21, P=.0005). N=3/genotype.
(I) Breeding scheme used to generate Rbp4-cre+; Dyrk1aloxP/loxP pups.
(J) Representative images of Nissl-stained cell somas in layer V of somatosensory cortex at birth in Rbp4-cKOs
(K) Quantification of soma size using Nissl-stained sections shows that Rbp4-cKOs exhibit decreased soma size in layer V of somatosensory cortex compared to controls [independent sample t-tests: II-IV (t4 =4.00343, P=.091760), V (t4=6.48343, P=.006136), VI (t4=2.46921, P=.021545)]. N=3/genotype.
(L) Representative images of CC3 staining in the cortex at birth shows that Rbp4-cKOs exhibit increased CC3+ apoptotic cells in layer V.
Results from post hoc and t-tests indicated on graphs. Error bars represent mean +/− SEM. *P<.05, **P<.01, ***P<.001, ****P<.0001.
Because Emx1 is expressed in both neurons and glia, we investigated the specific cellular population driving the decreased cell number by flow cytometry of dissociated nuclei stained with neuronal marker Neu-N. At P0, in which no difference in cHet total cortical cell number is observed, the proportions (percentage of total nuclei) of Neu-N+ and Neu-N– cells are unchanged in cHets and cKOs, consistent with unchanged total numbers of both neuronal and non-neuronal populations. At P7, the decrease in cHet total cortical cell number corresponds with a significant decrease in the proportion of Neu-N+ cells and a significant increase in the proportion of Neu-N- cells, consistent with a decrease in the total number of neurons and no change in the total number of non-neuronal cells. In adults, the decrease in cHet total cortical cell number corresponds with no change in the proportion of Neu-N+ or Neu-N– cells, consistent with a decreased total number of both neuronal and non-neuronal cells. Altogether, the data show that at P0, the populations of putative neurons and putative glia are unchanged. At P7, the observed decrease in cell number may be driven by a decrease in the neuronal population, which persists into adulthood with the addition of a decrease in the putative glial population (Figure S2).
To determine the source of increased cell density in cKOs at birth, we employed fluorescent Nissl staining to measure cell soma size in the somatosensory cortex (S1). We observed a decrease in cell soma size across all layers in cKOs at birth, aligning with our finding of increased cell density (Figure 3D). At P0, cHets exhibit decreased cell soma size in layers II-IV and V. At P7, cell somas in layer V are significantly smaller, and this decrease persists into adulthood in addition to a significant decrease in cell soma size in layer VI (Figure 3E,F). This decrease of cortical cell number in cHets at P7 but not at birth prompted us to investigate cell death as a potential mechanism. We observed increased apoptosis, as measured by apoptotic marker cleaved caspase 3 (CC3), throughout the cortex at P4 (Figure 3H). Staining of the P0 cKO cortex revealed a marked induction of apoptosis, particularly near the midline (Figure 3G).
Because layer V soma size is impacted in Emx1-cre cHets and cKOs across ages, we tested whether the putative layer V (Ctip2+) population is altered in density in cHets and cKOs. We quantified the number of Ctip2+ cells normalized to DAPI+ cells using 10-bin analysis and found that at P0, the density of Ctip2+ cells is comparable in cHets but altered in cKOs (Figure S3A). At P7 and adulthood, cHets exhibit decreased density of Ctip2+ neurons in the bins corresponding to layer V (at P7) and layers V and VI (in adults) (Figure S3B,C). These data indicate that conditional Dyrk1a mutations cause decreased neuronal cell number by P7 and provide layer V as a potential neuronal population driving this decrease.
Dyrk1a (RNA (36) and protein (37)) is enriched in, but is not exclusive to, layer V pyramidal neurons. This, together with decreased layer V soma size and Ctip2+ cell density at multiple timepoints in cHets led us to investigate the sufficiency of layer V-specific deletion of Dyrk1a to drive these cellular phenotypes. We generated conditional knockouts using Rbp4-Cre, which depletes Dyrk1a in layer V from development (Rbp4-cre+; Dyrk1aloxP/loxP) (Figure 3I). At P0, Rbp4-cKOs exhibit decreased soma size in layer V and a subtle yet significant decrease in layer VI soma size (Figure 3J,K). We also observed a robust induction of apoptosis within layer V of Rbp4-cKOs (Figure 3L). Altogether, microcephaly in Dyrk1a mutants corresponds to smaller cells at birth and fewer cells after birth, which may be driven by induction of apoptosis. Additionally, deletion of Dyrk1a in layer V neurons is sufficient to drive both the reduction in cell soma size and increased apoptosis.
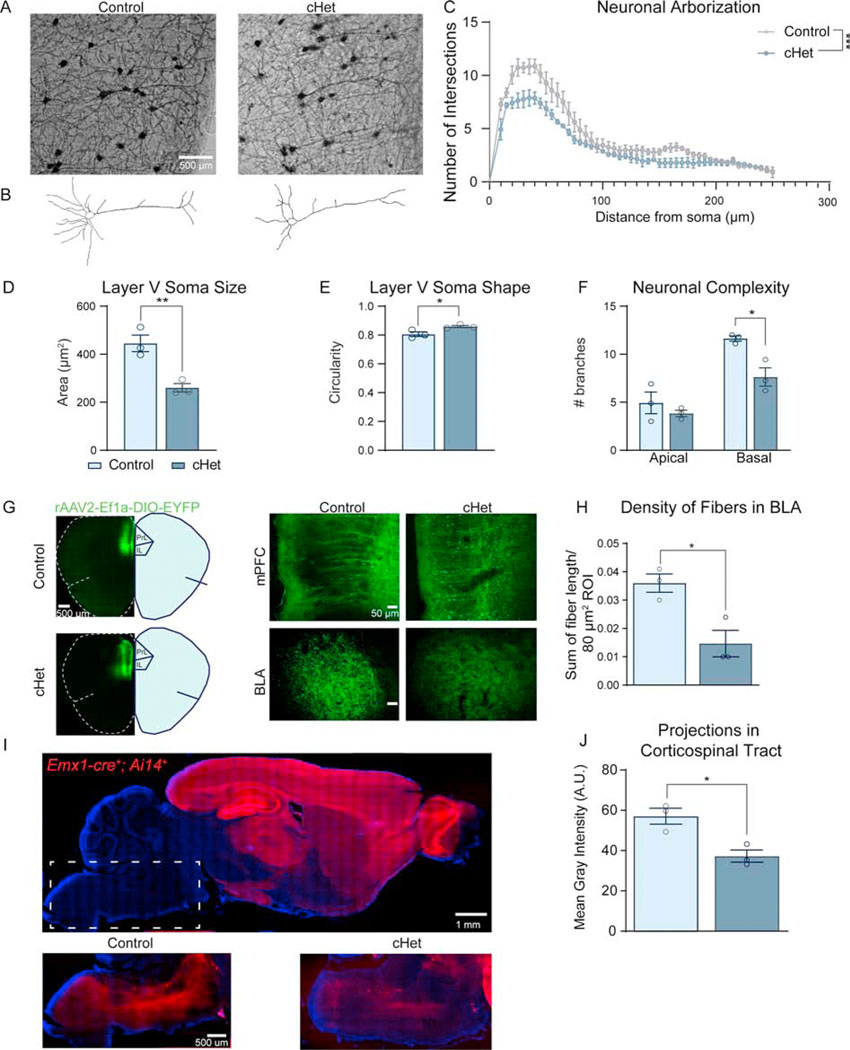
Cortical pyramidal neurons display altered morphology and axonal projections upon Dyrk1a mutation
To investigate a potential cellular mechanism driving altered social behavior, we investigated whether cHets display altered neuronal complexity in vivo by measuring Golgi-stained layer V neurons in the mPFC. These neurons send projections to subcortical regions (i.e., BLA, VTA) and are responsible for encoding social behavior and other ASD-relevant behaviors (38). We found that cHets exhibit significantly less complex layer V neurons, particularly close to the soma (Figure 4A–C). This significant decrease in soma size of cHet neurons (Figure 4D) validates our findings from fluorescent Nissl staining (Figure 3F). Additionally, basal dendritic branching is significantly decreased, while apical dendrites are unaffected (Figure 4F). After observing that somas of layer V neurons in mutant brains were more circular than controls, we quantified circularity [4π (area/perimeter2)] where 1.0 indicates a perfect circle. We found that cHet somas are significantly more circular than controls, exhibiting a mean value closer to 1.0 (Figure 4E). A similar decrease in complexity and soma size as well as an increase in circularity of layer II/III pyramidal neurons suggests that cortical mutations in Dyrk1a have a widespread impact on neuronal morphology and axonal projections (Figure S4).
Figure 4: Dyrk1a cHets exhibit altered neuronal morphology of layer V pyramidal neurons and decreased cortico-subcortical projection density.
(A) Representative Golgi-stained layer V area of the mPFC from adult control and cHet brains.
(B) Reconstructed neurons from the representative images in (A) using Neuromantic software.
(C) Sholl analysis of neuronal arborization on individual neurons shows that cHets exhibit significantly decreased neuronal arborization (independent sample t-test of area under the curve generated for each genotype: t4=8.818, P=.0009). N=10–15 neurons/mouse, 3 mice/genotype across two sections.
(D) Adult cHets exhibit decreased soma size in layer V pyramidal neurons in the mPFC (t4=4.775, P=.0088).
(E) cHet pyramidal neurons exhibit more circular somas measured by the circularity plugin in ImageJ (t4=2.991, P=.0403).
(F) cHet layer V pyramidal neurons contain less complex basal branches, measured by counting the endpoints on the reconstructed neurons (apical: t4=.9301, P=.404961; basal: t4=4.026, P=.015785).
(G) Overview of injection site in the mPFC where 50nL AAV-DIO-EYFP was injected in adult mice (Bregma 1.98mm). Zoomed inset shows comparable density of virus in the mPFC. Inset of the BLA shows decreased EYFP+ projections in the BLA in cHets (Bregma −1.58mm).
(H) cHets exhibit decreased projection density in the BLA (t4=2.933, P=.0427). Measured by fiber reconstruction in ImageJ relative to size of the ROI. N=3/genotype.
(I) Control sagittal brain section showing tdTomato expression under the Emx1-cre promoter. Dotted rectangle delineates the area in which the image was taken with enhancement of the red fluorophore expressed in the fibers of the corticospinal tract (CST). When comparing matched sections from a control and cHet, the qualitative difference is striking.
(J) Quantification of the fiber density from the cortex to the corticospinal tract shows cHets have decreased intensity of the red fluorophore expressed in these fibers (t4=3.977, P=.0164). Analyzed by averaging the mean gray intensity in the CST in 3 matched sections per animal normalized to the size of the CST. N=3/genotype. Results analyzed by independent sample t-tests unless otherwise specified.
Results from post hoc and t-tests indicated on graphs. Error bars represent mean +/− SEM. *P<.05, **P<.01, ***P<.001, ****P<.0001.
We next investigated a circuit that encodes social behavior and is dependent on layer V neurons. Using an anterograde Cre-dependent virus (rAAV2-Ef1a-DIO-EYFP), we traced projections from the mPFC to the BLA in adult mice to measure axonal projections in vivo. Reconstruction of fibers in the BLA showed decreased density in cHets with comparable injection site density (Figure 4G,H), providing a substrate of altered axonal projections that may cause social deficits in cHets. To determine whether there are more widespread effects on layer V projecting circuits caused by Dyrk1a mutation, we investigated the corticospinal tract (CST), which contains the longest projections in the mammalian brain (39) and thus may be sensitive to alterations in growth. Using Cre-dependent reporter tdTomato in sagittal sections, we found that cHets exhibit significantly decreased density of CST projections, suggesting that multiple circuits dependent on layer V pyramidal neurons are altered in cHets (Figure 4I,J). Both decreased arborization of pyramidal neurons and decreased axonal projections in cortico-subcortical circuits provide a potential cellular link between alterations in brain growth and autism-relevant behavior.
Growth signaling pathways are altered in conditional Dyrk1a mutants
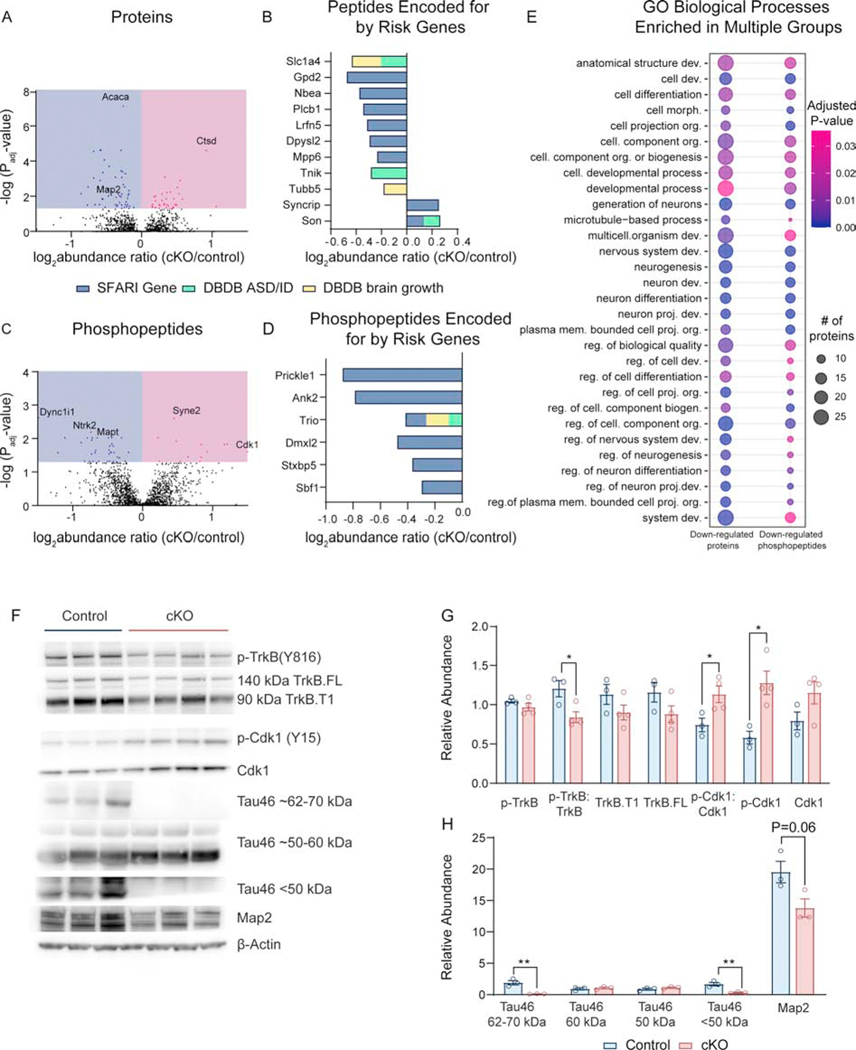
To investigate altered signaling cascades through which Dyrk1a mutations cause microcephaly, decreased neuronal growth, and ASD-relevant behaviors, we conducted a phospho-proteomic and proteomic screen via high-resolution tandem mass spectrometry coupled to liquid chromatography (LC-MS/MS). Our tandem mass tag (TMT) approach and phosphopeptide enrichment in P0 control and cKO cortices identified 89 significantly altered proteins with abnormal abundance in the cKO cortex at birth (56 decreased and 33 increased) (Figure 5A, Supplemental Table 1). Ingenuity Pathway Analysis (IPA) identified axonal guidance signaling, RhoA signaling, and semaphorin signaling in neurons as significantly altered canonical pathways. IPA also identified BDNF, mTOR, and IGF-1 as predicted upstream regulators of the altered proteome. We then surveyed the altered proteome for proteins encoded by risk genes for ASD, ID, and brain growth abnormalities using SFARI gene and the Developmental Brain Database (DBDB), specifically using the categories “autism”, “intellectual disability”, “microcephaly”, “megalencephaly”, and “hemimegalencephaly”. We found that several proteins in the dataset are encoded for by ASD/ID risk genes or genes that impact brain growth (Figure 5B).
Figure 5: Dyrk1a targets trkB-BDNF signaling cascades in the developing cortex.
(A) Volcano plot showing number of significantly altered proteins after FDR correction. A false discovery rate (FDR) of 0.01 was used and only peptides that had an adjusted P value of <0.05 after correction were analyzed. Additionally, only phosphopeptides with a probability value > 75% were included. Individual proteins were used to compare cKOs to controls using the ANOVA option in Proteome Discoverer, and adjusted P values were generated. N=6–7 mice/genotype, males and females.
(B) Log2 abundance ratio of proteins (identified in proteome data set) that are encoded for by ASD/ID risk genes or genes involved in regulating brain size as identified in DBDB. For SFARI gene, only genes with confidence score 1–3 were considered.
(C) Volcano plot showing number of significantly altered peptides after FDR correction. N=3–4 female mice/genotype.
(D) Log2 abundance ratio of proteins (identified in phospho-proteome dataset) that are encoded for by ASD/ID risk genes or genes involved in regulating brain size as identified in DBDB. For SFARI gene, only genes with confidence score 1–3 were considered.
(E) Dot plot of GO terms enriched in both down-regulated proteins and down-regulated phosphopeptides. Abbreviations: development (dev.), cellular (cell.), morphogenesis (morph.), projection (proj.), membrane (mem.), and regulation (reg.). Dot size corresponds to the number of proteins in the dataset identified under the GO term. Color corresponds to FDR-corrected P value. The size of the dot is relative to the dataset, such that the smallest dot represents 6 proteins.
(F) Western blot of P0 control and cKO cortical lysate using antibodies against phospho-proteins and total proteins identified by mass spectrometry.
(G) Quantification of western blot where phospho-proteins are normalized to total protein and to a loading control (β-actin). Analyzed by multiple t-tests. N=3–4/genotype.
Results from post hoc and t-tests indicated on graphs. Error bars represent mean +/− SEM. *P<.05, **P<.01, ***P<.001, ****P<.0001.
After phosphopeptide enrichment, 51 significantly altered phosphopeptide groups (35 phospho-proteins) were discovered (24 decreased and 11 increased) (Figure 5C, Supplemental Table 2). IPA identified synaptogenesis signaling, axonal guidance, and ERK/MAPK as altered pathways. BDNF, ERK, and MAPK were found to be predicted upstream regulators. Using the same analysis against SFARI gene and DBDB, we identified multiple proteins in the altered phospho-proteome encoded for by ASD/ID/brain growth genes (Figure 5D).
Gene ontology (GO) enrichment analysis revealed significant overlap shared between the down-regulated protein and down-regulated phosphopeptide groups, including GO terms “anatomical structure development” and “neuron projection development” (Figure 5E). No biological processes were significantly enriched after correction in either the up-regulated phosphopeptide group or up-regulated protein group.
Noting BDNF as a predicted upstream regulator of both the altered proteome and phospho-proteome, we are interested to report significantly decreased phosphorylation of TrkB (Y816) in Dyrk1a cKOs. To validate this and other significantly altered targets in the phospho-proteome, we performed western blots on P0 controls and cKOs (Figure 5F,G). Indeed, p-TrkB (Y816) is decreased in Dyrk1a cKOs when normalized to total TrkB. We also validated increased inhibitory phosphorylation on Cdk1 relative to total Cdk1 and β-actin. Phosphorylation of MAPT/Tau was also identified as significantly decreased in the cKO dataset. Dyrk1a is known to phosphorylate Tau at Thr212, Ser202, and Ser404(40); however, the peptide identified by mass spectrometry in this experiment is phosphorylated at Ser704, which does not contain the Dyrk1a consensus sequence and thus may not be a direct target of Dyrk1a. We found that multiple isoforms of Tau are depleted in Dyrk1a cKOs. Map2 was found to be significantly decreased in the proteome, and indeed was found to be decreased in cKO cortical lysate (Figure 5F,G). These data implicate signaling cascades that were previously not known to be altered by Dyrk1a mutations (Figure S5).
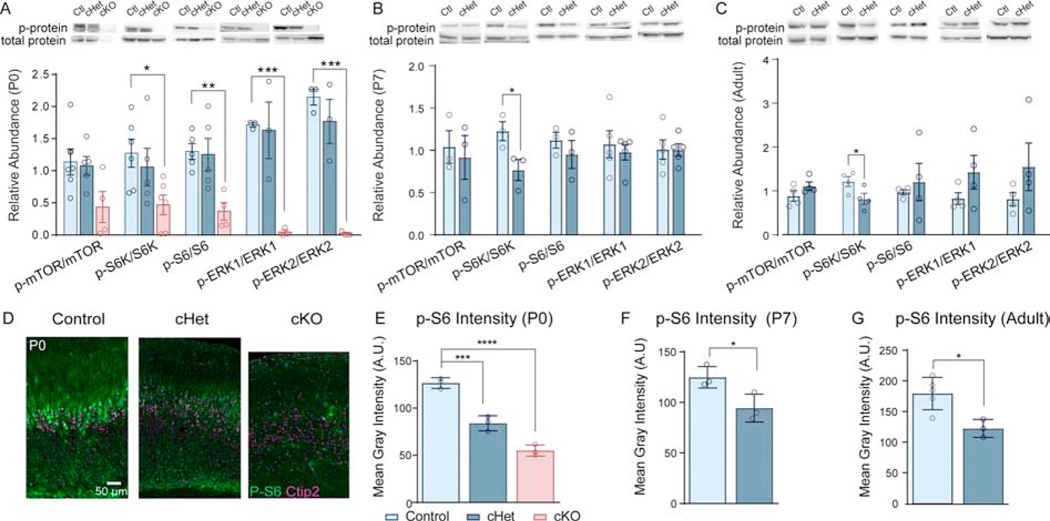
As mTOR was identified by IPA in Dyrk1a cKOs, and because mTOR is regulated by BDNF/TrkB signaling (41, 42), we next investigated whether activation of the mTOR signaling cascade is altered in cortical tissue. We performed western blots on cortical lysate at P0, P7, and adult timepoints. At P0, cHets showed no difference in the activation of mTOR signaling components; however, cKOs exhibit a severe reduction in phosphorylation of S6, S6K1, and ERK1/2 (Figure 6A,S6). This remarkably low activation of ERK1/2 aligns with the prediction by IPA that ERK/MAPK signaling is significantly disrupted in Dyrk1a cKOs and corroborates down-regulation of BDNF-TrkB signaling. Both P7 and adult cHets exhibit a significant decrease in phosphorylation of S6K1 (Figure 6B,C,S6). To determine whether there is a cell-selective effect of decreased mTOR signaling in Dyrk1a mutants, we used phosphorylation of S6 (Ser235/246), which is enriched in layer V pyramidal neurons (20), as a readout of mTORC1 activity. We found decreased p-S6 per putative layer V neuron (Ctip2+) in cHets at P0, P7, and adulthood (Figure 6D–G). In cKOs, p-S6 is virtually ablated in layer V neurons. We did note bright, fluorescent bodies that did not colocalize with DAPI or Ctip2. These have been noted in other mouse models of mTOR ablation (data not shown) and warrant further investigation (Figure 6D).
Figure 6: Activation of the mTOR pathway is decreased in layer V and throughout the cortex of conditional Dyrk1a mutants.
(A) P0 cKO cortices exhibit decreased activation of mTOR signaling molecules. Western blot of cortical lysate from P0 pups with quantification measured as relative abundance in ImageJ. Phospho-protein was normalized to total protein. Analyzed by one-way ANOVA with Sidak’s post hoc multiple comparisons tests (p-mTOR/mTOR: F2,12=3.406, P=.0674; p-S6K/S6K: F2,14=3.757, P=.0494; p-S6/S6: F2,11=7.371, P=.0093; p-ERK1/ERK1: F2,6=13.55, P=.006; p-ERK2/ERK2: F2,6=28.9, P=.0008).
(B) P7 cHets exhibit decreased activation of S6K in the cortex (t4=2.85833, P=.046007). Quantified using relative abundance in ImageJ.
(C) Adult cHets exhibit decreased activation of S6K in the cortex (t6=2.49104, P=.047096). Quantified using relative abundance in ImageJ. For all ages, N=3–5/genotype.
(D) Representative images from coronal sections in S1 showing decreased p-S6 intensity in P0 cHets and cKOs.
(E) p-S6 intensity quantified by measuring mean gray intensity of the green fluorophore (Alexafluor 488) per Ctip2+ layer V neuron. 25–30 neurons measured per section in 2–3 plane-matched sections per animal. Analyzed by one-way ANOVA with Sidak’s post hoc multiple comparisons tests (F2,6=88.93 P<.0001).
(F) p-S6 per layer V neuron is decreased at P7 (t4=3.013, P=.0394).
(G) p-S6 per layer V neuron is decreased in adults (t6=3.374, P=.0150). For all ages, N=3/genotype. Results analyzed by independent sample t-tests unless otherwise specified.
Results from post hoc and t-tests indicated on graphs. Error bars represent mean +/− 940 SEM. *P<.05, **P<.01, ***P<.001, ****P<.0001.
To test the necessity of mTOR for regulating cell soma size, we generated conditional mTOR cKOs (Emx1-cre+; mTORloxP/loxP) (Figure S7C,D). At P0, mTOR cKOs exhibit decreased soma size, comparable to Dyrk1a cKOs (Figure S7E,F). This phenocopy of Dyrk1a mutants by mTOR deletion suggests that decreased mTOR signaling may drive decreased cell size in Dyrk1a mutants. To determine whether decreased mTOR activation can be driven by deletion of Dyrk1a in layer V neurons, we measured p-S6 (Ser235/236) in P0 Rbp4-cKOs. Similar to Emx1-cKOs, P0 Rbp4-cKOs exhibit markedly decreased p-S6 intensity in layer V neurons (Figure S7A,B). These data provide a novel molecular mechanism by which Dyrk1a impacts growth signaling cascades, including mTOR and BDNF-TrkB, to regulate growth and connectivity.
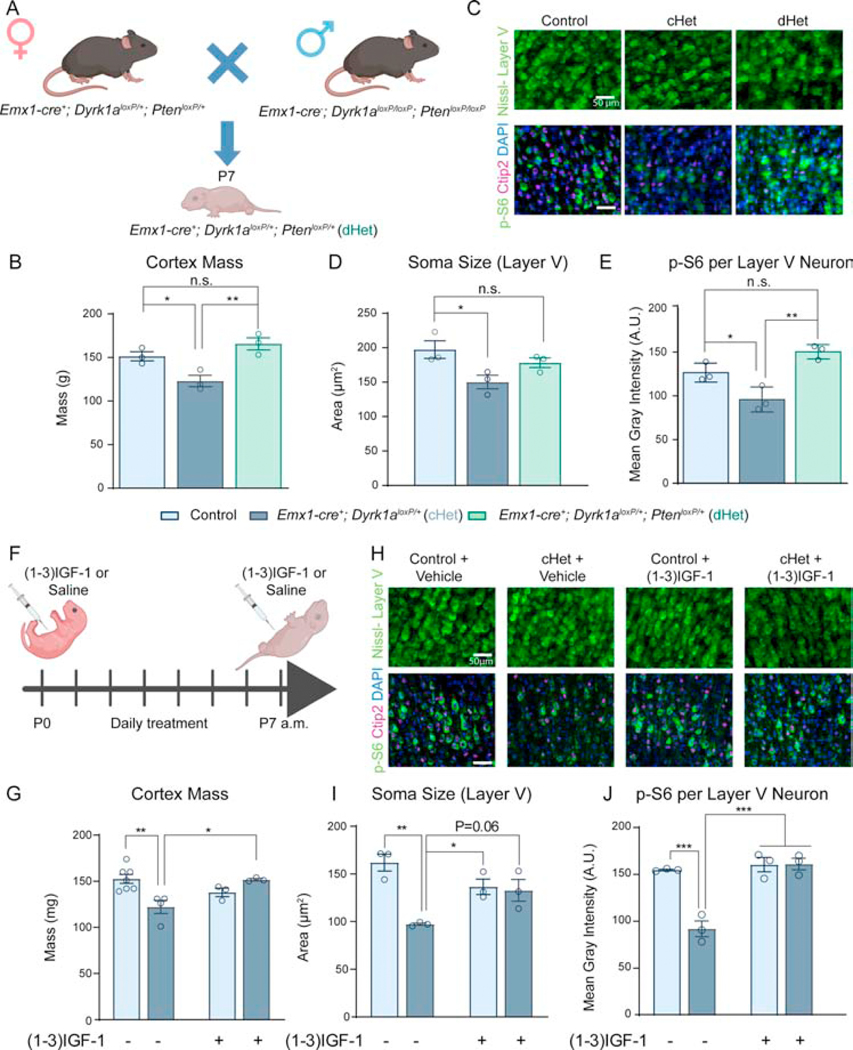
Genetic suppression of Pten or pharmacological treatment with IGF-1 rescues microcephaly and neuronal undergrowth in cHets
Given the decreased activation of BDNF-TrkB and mTOR signaling and the decrease in overall cortex mass and cellular size, we hypothesized that increasing mTOR signaling would rescue the observed deficits. Since Pten is a negative regulator of the PI3K/Akt/mTOR pathway (43), decreasing Pten dosage in cHets would remove the “brake” on mTOR signaling and be expected to rescue the observed microcephaly and decreased neuronal size. We generated conditional double heterozygous mutants (Emx1-cre+; Dyrk1aloxP/+; PtenloxP/+, “dHets”) to genetically suppress Pten in Dyrk1a cHets (Figure 7A). At P7, the cortex mass of dHets is similar to controls (Figure 7B). dHets also exhibit rescued soma size of layer V neurons, and p-S6 per layer V neuron is significantly higher than cHets (Figure 7C–E). Thus, genetic suppression of Pten in Dyrk1a mutants rescues the decreased cortical mass and cellular size through increased mTOR signaling. Moreover, the data suggest epistatic regulation between Pten and Dyrk1a whereby Pten must be present at normal levels to observe the effects of Dyrk1a mutations.
Figure 7: Genetic suppression of Pten and pharmacological treatment with (1–3)IGF-1 rescues microcephaly and cellular undergrowth in neonatal cHets.
(A) Schematic of a genetic rescue approach. dHet (Emx1-cre; Dyrk1aloxP/+; PtenloxP/+) pups were collected at P7 for analysis.
(B) Genetic suppression of Pten in Dyrk1a cHets rescues cortex mass (F2,6=11.76, P=.0084). N=3/genotype.
(C) Representative images of Nissl-stained layer V cell somas (top) and p-S6/Ctip2 stained cells (bottom).
(D) Quantification of cell soma size in layer V shows a partial rescue of cell size in dHets (F2,6=6.724, P=.0294). N=3/genotype.
(E) Quantification of p-S6 fluorescence per layer V neuron as identified by positive staining for Ctip2. dHets rescued the decreased p-S6 in cHets (F2,6=17.71, P=.003). N=3/genotype.
(F) Schematic of neonatal (1–3)IGF-1 injection paradigm. Pups were injected with 10μg/g (1–3)IGF-1 in saline in the morning from P1 daily until P7. They were sacrificed 8 hours post-injection on P7.
(G) (1–3)IGF-1 injection rescues the decreased cortex mass observed in cHets treated with vehicle. Two-way ANOVA with Tukey’s post hoc multiple comparisons tests showed a significant interaction of drug and genotype (F1,13=13.70, P=.0027). N=3–7/genotype.
(H) Representative images of Nissl-stained layer V cell somas (top) and p-S6/Ctip2 stained cells (bottom).
(I) Neonatal (1–3)IGF-1 injection rescues the decreased soma size observed in cHets treated with vehicle. Two-way ANOVA with Tukey’s post hoc multiple comparisons tests showed a significant interaction of drug and genotype (F1,8=13.4, P=.0064). N=3/genotype.
(J) Neonatal (1–3)IGF-1 injection rescues the decreased p-S6 in layer V neurons observed in vehicle-treated cHets. Two-way ANOVA with Tukey’s post hoc multiple comparisons tests showed a significant interaction of drug and genotype (F1,8=24.19, P=.0012). N=3/genotype. Results analyzed by one-way ANOVA with Tukey’s post hoc multiple comparisons tests unless otherwise specified.
Results from post hoc and t-tests indicated on graphs. Error bars represent mean +/−SEM. *P<.05, **P<.01, ***P<.001, ****P<.0001.
We next aimed to increase mTOR signaling via pharmacological intervention. Because activation of TrkB receptor is decreased in cKOs, treating mice with exogenous BDNF may not be effective. To circumvent this issue while stimulating the same signaling cascades, we injected neonatal pups with (1–3)IGF-1 (GPE), a tripeptide cleaved from the N-terminus of IGF-1 (44). Pups were injected daily from P0 to P7 with 10μg/g (1–3)IGF-1 or vehicle based on a previously described treatment paradigm (45). Eight hours post-injection on P7, pups were sacrificed, and brains were fixed (Figure 7F). Dyrk1a cHets treated with (1–3)IGF-1 exhibit cortex mass similar to vehicle-treated controls, and (1–3)IGF-1 treatment had no effect on cortex mass of controls (Figure 7G). (1–3)IGF-1 treated Dyrk1a cHets also display soma size comparable to controls as well as normal levels of p-S6 per layer V neuron (Figure 7H–J). Taken together, these data show that neonatal (1–3)IGF-1 treatment in Dyrk1a cHets rescues cortical and neuronal undergrowth corresponding with increased p-S6 levels.
Lastly, to test whether (1–3)IGF-1 treatment rescues the decreased complexity of Dyrk1a cHet neurons, we cultured control and cHet primary cortical neurons. Neurons analyzed were restricted to Ctip2+ neurons, as we are focused on the impact of Dyrk1a mutations on the growth and connectivity of layer V pyramidal neurons. On DIV15, vehicle-treated cHet neurons displayed significantly decreased complexity compared to controls. This was rescued by treatment with 100ng/mL (1–3)IGF-1 daily from DIV13 to DIV15 while (1–3)IGF-1 did not affect control neurons (Figure S8). These data show that (1–3)IGF-1 treatment rescues the decreased neuronal complexity in vitro in Dyrk1a cHets.
Discussion
DYRK1A haploinsufficiency causes ASD, ID, and microcephaly in humans (4, 5). Trisomy of chromosome 21 (including DYRK1A) causes Down syndrome and microcephaly (OMIM #190685). These patient genetic findings confirm a critical role of Dyrk1a in regulating brain growth and the development of behavior and cognition. Utilizing both heterozygous and homozygous conditional mutants has enabled us to study a clinically relevant model with similar construct validity to patients and a loss-of-function model to investigate the role of Dyrk1a, respectively. The investigation of cKOs is particularly interesting because germline Dyrk1a knockouts (Dyrk1a−/−) die by embryonic day 13.5 (13). Here we describe the altered trajectory of postnatal brain growth and found that cortical Dyrk1a mutations cause decreased brain and cortex mass at birth, during development, and in adulthood. cHets also exhibit cortical thinning and enlarged ventricles, which may be explained by the excessive cell death observed. Interestingly, MRI reveals patients with DYRK1A mutations also display enlarged ventricles and a hypoplastic corpus callosum (4). In addition to morphological aberrations, mouse models of Dyrk1a mutations exhibit behavioral abnormalities, including altered social behavior (14, 15, 46, 47), and our finding of altered social interest in cHets illustrates the necessity of Dyrk1a in the cortex for encoding this behavior. Thus, we have shown that the conditional Dyrk1a mutant accurately models phenotypes observed in humans with DYRK1A mutations in both brain size/morphology and ASD-relevant behavioral deficits.
Dyrk1a plays an evolutionarily conserved role in regulating proliferation in neural progenitor cells in both vertebrates and invertebrates (48–50). Thus, we were surprised that cortical cell number was not changed in cHets or cKOs as compared to controls at birth. This may differ from findings in germline Dyrk1a+/− mutants due to the timing of Dyrk1a deletion using Emx1-Cre. However, germline Dyrk1a+/− mutants exhibit increased density in the cortex, which suggests the presence of smaller cells (13). This supports our finding of decreased cell size at all ages in the cHet and cKO cortex. Layer V pyramidal neurons are one of the largest neuronal types in the brain (51), send subcortical projections that are critical for social behavior and other ASD-relevant behaviors (52, 53), and their size is correlated with the amount of mTOR activity in the neuron (20). The decreased complexity of layer V neurons in the mPFC together with the decrease in neuronal number and layer V (Ctip2+) neuronal density suggest potential cellular mechanisms responsible for altered connectivity and behavior in Dyrk1a mutants. A proposed cellular mechanism driving the altered postnatal growth trajectory in cHets is summarized in Figure S9.
Using high-throughput proteomics and phospho-proteomics, we identified signatures of altered growth, development, and microtubule dynamics in cKOs at birth. The role of Dyrk1a in regulating microtubule dynamics has been described (54), but implication of both BDNF-TrkB signaling and mTOR signaling is novel. The nature of the interaction between Dyrk1a and BDNF remains unclear; however, our findings provide a mechanism by which Dyrk1a regulates survival and growth through BDNF-TrkB signaling. It is important to note that mutations in TrkB cause decreased cellular size and neuronal complexity as well as a compressed cortex (55). TrkB is a critical receptor that activates multiple growth signaling cascades, suggesting changes in activation of TrkB by Dyrk1a will result in alterations in plasticity through CAMK2 and PKC, transcription and translation through AKT/mTOR, and growth and differentiation through ERK (56). Additionally, Xu et al. noted “rounder” cell somas in cortical pyramidal neurons of TrkB mutants, supporting our finding that Golgi-stained pyramidal neurons in cHets exhibit more circular cell somas (55).
Concordant with the predicted involvement of mTOR signaling in Dyrk1a mutants by IPA, p-S6 levels are decreased in layer V neurons at all ages in the conditional Dyrk1a mutant, and mTOR cKO phenocopies the pyramidal neuronal undergrowth observed in Dyrk1a cKOs. While these data underscore a novel role of mTOR dysregulation in Dyrk1a mutants, our findings cannot rule out other contributing mechanisms. Signaling through the mTOR pathway is critical for regulating neuronal growth via protein synthesis and controlling proliferation, differentiation, migration, and autophagy in diverse neuronal cell types (28, 57). Deviations from optimal levels of protein synthesis—either increased or decreased—are detrimental for synaptic connectivity and cognitive function (58). Genetic suppression of Pten in cHets not only rescued the observed microcephaly and decreased neuronal growth but also provided evidence of a novel genetic interaction between two highly penetrant ASD/ID risk genes that impact brain growth. We also utilized (1–3)IGF-1 treatment, which has been used in Shank3 and Mecp2 mutant mouse models of ASD to rescue growth and behavioral deficits (45, 59, 60). To our knowledge, this is the first investigation of the efficacy of an FDA-approved drug (Increlex, FDA reference ID 3517143) to rescue neurodevelopmental deficits caused by Dyrk1a haploinsufficiency. Neonatal treatment with (1–3)IGF-1 rescued brain and neuronal undergrowth in neonatal cHets, corresponding with an increase in p-S6 levels. Additionally, (1–3)IGF-1 treatment rescued neuronal complexity in primary neurons in vitro, suggesting the potential to rescue altered connectivity and behavior in vivo.
Genetic suppression of mTOR signaling components mTOR, IGF-1, and Erk1/2 causes microcephaly, decreased neuronal size, and social deficits (23, 24, 61), and dysregulated mTOR signaling has been identified in models of ASD/ID and abnormal patterns of brain growth, e.g., Pten and Mecp2 mutants (20, 62). Our findings add Dyrk1a to an increasing list of genes that impinge on growth-associated signaling cascades, including mTOR, to cause altered connectivity and/or growth, including Mecp2, Fmr1, Tsc, Pten, Nf1, and Ube3a (58, 63). Future work will investigate whether increasing mTOR signaling during development is sufficient to rescue altered connectivity and behavior in adult cHet mice. Overall, we have identified a novel molecular mechanism through which mutations in Dyrk1a cause microcephaly and decreased neuronal size via decreased growth factor signaling and tested a therapeutically relevant avenue to rescue the observed deficits.
Supplementary Material
Acknowledgements and Disclosures:
We thank all members of the Page lab for insightful discussions and Dr. Ori Cohen for assistance in in vitro primary neuronal culture experiments. Schematic images were made using Biorender.com. We are grateful for funding provided by gift funds from Ms. Nancy Lurie Marks, National Institute of Health (NIH) grant RO1MH108519, and the RJ Foundation.
Footnotes
The authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stessman HA, Xiong B, Coe BP, Wang T, Hoekzema K, Fenckova M, Kvarnung M, Gerdts J, Trinh S, Cosemans N, Vives L, Lin J, Turner TN, Santen G, Ruivenkamp C, Kriek M, van Haeringen A, Aten E, Friend K, Liebelt J, Barnett C, Haan E, Shaw M, Gecz J, Anderlid BM, Nordgren A, Lindstrand A, Schwartz C, Kooy RF, Vandeweyer G, Helsmoortel C, Romano C, Alberti A, Vinci M, Avola E, Giusto S, Courchesne E, Pramparo T, Pierce K, Nalabolu S, Amaral DG, Scheffer IE, Delatycki MB, Lockhart PJ, Hormozdiari F, Harich B, Castells-Nobau A, Xia K, Peeters H, Nordenskjold M, Schenck A, Bernier RA, Eichler EE. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat Genet. 2017;49(4):515–26. Epub 2017/02/14. doi: 10.1038/ng.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Bon BW, Hoischen A, Hehir-Kwa J, de Brouwer AP, Ruivenkamp C, Gijsbers AC, Marcelis CL, de Leeuw N, Veltman JA, Brunner HG, de Vries BB. Intragenic deletion in DYRK1A leads to mental retardation and primary microcephaly. Clin Genet. 2011;79(3):296–9. Epub 2011/02/08. doi: 10.1111/j.1399-0004.2010.01544.x. [DOI] [PubMed] [Google Scholar]
- 3.Moller RS, Kubart S, Hoeltzenbein M, Heye B, Vogel I, Hansen CP, Menzel C, Ullmann R, Tommerup N, Ropers HH, Tumer Z, Kalscheuer VM. Truncation of the Down syndrome candidate gene DYRK1A in two unrelated patients with microcephaly. Am J Hum Genet. 2008;82(5):1165–70. Epub 2008/04/15. doi: 10.1016/j.ajhg.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luco SM, Pohl D, Sell E, Wagner JD, Dyment DA, Daoud H. Case report of novel DYRK1A mutations in 2 individuals with syndromic intellectual disability and a review of the literature. BMC Med Genet. 2016;17:15. Epub 2016/02/29. doi: 10.1186/s12881-016-0276-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ji J, Lee H, Argiropoulos B, Dorrani N, Mann J, Martinez-Agosto JA, Gomez-Ospina N, Gallant N, Bernstein JA, Hudgins L, Slattery L, Isidor B, Le Caignec C, David A, Obersztyn E, Wisniowiecka-Kowalnik B, Fox M, Deignan JL, Vilain E, Hendricks E, Horton Harr M, Noon SE, Jackson JR, Wilkens A, Mirzaa G, Salamon N, Abramson J, Zackai EH, Krantz I, Innes AM, Nelson SF, Grody WW, Quintero-Rivera F. DYRK1A haploinsufficiency causes a new recognizable syndrome with microcephaly, intellectual disability, speech impairment, and distinct facies. Eur J Hum Genet. 2015;23(11):1473–81. Epub 2015/05/07. doi: 10.1038/ejhg.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Earl RK, Turner TN, Mefford HC, Hudac CM, Gerdts J, Eichler EE, Bernier RA. Clinical phenotype of ASD-associated DYRK1A haploinsufficiency. Mol Autism. 2017;8:54. Epub 2017/10/17. doi: 10.1186/s13229-017-0173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker W, Sippl W. Activation, regulation, and inhibition of DYRK1A. FEBS J. 2011;278(2):246–56. Epub 2010/12/04. doi: 10.1111/j.1742-4658.2010.07956.x. [DOI] [PubMed] [Google Scholar]
- 8.Song WJ, Song EA, Jung MS, Choi SH, Baik HH, Jin BK, Kim JH, Chung SH. Phosphorylation and inactivation of glycogen synthase kinase 3beta (GSK3beta) by dual-specificity tyrosine phosphorylation-regulated kinase 1A (Dyrk1A). J Biol Chem. 2015;290(4):2321–33. Epub 2014/12/06. doi: 10.1074/jbc.M114.594952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Najas S, Arranz J, Lochhead PA, Ashford AL, Oxley D, Delabar JM, Cook SJ, Barallobre MJ, Arbones ML. DYRK1A-mediated Cyclin D1 Degradation in Neural Stem Cells Contributes to the Neurogenic Cortical Defects in Down Syndrome. EBioMedicine. 2015;2(2):120–34. Epub 2015/07/03. doi: 10.1016/j.ebiom.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woods YL, Rena G, Morrice N, Barthel A, Becker W, Guo S, Unterman TG, Cohen P. The kinase DYRK1A phosphorylates the transcription factor FKHR at Ser329 in vitro, a novel in vivo phosphorylation site. Biochem J. 2001;355(Pt 3):597–607. Epub 2001/04/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammerle B, Ulin E, Guimera J, Becker W, Guillemot F, Tejedor FJ. Transient expression of Mnb/Dyrk1a couples cell cycle exit and differentiation of neuronal precursors by inducing p27KIP1 expression and suppressing NOTCH signaling. Development. 2011;138(12):2543–54. Epub 2011/05/26. doi: 10.1242/dev.066167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park J, Oh Y, Yoo L, Jung MS, Song WJ, Lee SH, Seo H, Chung KC. Dyrk1A phosphorylates p53 and inhibits proliferation of embryonic neuronal cells. J Biol Chem. 2010;285(41):31895–906. Epub 2010/08/11. doi: 10.1074/jbc.M110.147520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fotaki V, Dierssen M, Alcantara S, Martinez S, Marti E, Casas C, Visa J, Soriano E, Estivill X, Arbones ML. Dyrk1A haploinsufficiency affects viability and causes developmental delay and abnormal brain morphology in mice. Mol Cell Biol. 2002;22(18):6636–47. Epub 2002/08/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arque G, de Lagran MM, Arbones ML, Dierssen M. Age-associated motor and visuo-spatial learning phenotype in Dyrk1A heterozygous mutant mice. Neurobiol Dis. 2009;36(2):312–9. Epub 2009/08/08. doi: 10.1016/j.nbd.2009.07.027. [DOI] [PubMed] [Google Scholar]
- 15.Arque G, Fotaki V, Fernandez D, Martinez de Lagran M, Arbones ML, Dierssen M. Impaired spatial learning strategies and novel object recognition in mice haploinsufficient for the dual specificity tyrosine-regulated kinase-1A (Dyrk1A). PLoS One. 2008;3(7):e2575. Epub 2008/07/24. doi: 10.1371/journal.pone.0002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dang T, Duan WY, Yu B, Tong DL, Cheng C, Zhang YF, Wu W, Ye K, Zhang WX, Wu M, Wu BB, An Y, Qiu ZL, Wu BL. Autism-associated Dyrk1a truncation mutants impair neuronal dendritic and spine growth and interfere with postnatal cortical development. Mol Psychiatry. 2018;23(3):747–58. Epub 2017/02/09. doi: 10.1038/mp.2016.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franklin AV, King MK, Palomo V, Martinez A, McMahon LL, Jope RS. Glycogen synthase kinase-3 inhibitors reverse deficits in long-term potentiation and cognition in fragile X mice. Biol Psychiatry. 2014;75(3):198–206. Epub 2013/09/18. doi: 10.1016/j.biopsych.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mines MA, Yuskaitis CJ, King MK, Beurel E, Jope RS. GSK3 influences social preference and anxiety-related behaviors during social interaction in a mouse model of fragile X syndrome and autism. PLoS One. 2010;5(3):e9706. Epub 2010/03/20. doi: 10.1371/journal.pone.0009706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Huang WC, Sejourne J, Clipperton-Allen AE, Page DT. Pten Mutations Alter Brain Growth Trajectory and Allocation of Cell Types through Elevated beta-Catenin Signaling. J Neurosci. 2015;35(28):10252–67. Epub 2015/07/17. doi: 10.1523/JNEUROSCI.5272-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang WC, Chen Y, Page DT. Hyperconnectivity of prefrontal cortex to amygdala projections in a mouse model of macrocephaly/autism syndrome. Nat Commun. 2016;7:13421. Epub 2016/11/16. doi: 10.1038/ncomms13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vithayathil J, Pucilowska J, Landreth GE. ERK/MAPK signaling and autism spectrum disorders. Prog Brain Res. 2018;241:63–112. Epub 2018/11/19. doi: 10.1016/bs.pbr.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 22.Kim WY, Wang X, Wu Y, Doble BW, Patel S, Woodgett JR, Snider WD. GSK-3 is a master regulator of neural progenitor homeostasis. Nat Neurosci. 2009;12(11):1390–7. Epub 2009/10/06. doi: 10.1038/nn.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cloetta D, Thomanetz V, Baranek C, Lustenberger RM, Lin S, Oliveri F, Atanasoski S, Ruegg MA. Inactivation of mTORC1 in the developing brain causes microcephaly and affects gliogenesis. J Neurosci. 2013;33(18):7799–810. Epub 2013/05/03. doi: 10.1523/JNEUROSCI.3294-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pucilowska J, Puzerey PA, Karlo JC, Galan RF, Landreth GE. Disrupted ERK signaling during cortical development leads to abnormal progenitor proliferation, neuronal and network excitability and behavior, modeling human neuro-cardio-facial-cutaneous and related syndromes. J Neurosci. 2012;32(25):8663–77. Epub 2012/06/23. doi: 10.1523/JNEUROSCI.1107-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caracci MO, Avila ME, De Ferrari GV. Synaptic Wnt/GSK3beta Signaling Hub in Autism. Neural Plast. 2016;2016:9603751. Epub 2016/02/18. doi: 10.1155/2016/9603751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winden KD, Ebrahimi-Fakhari D, Sahin M. Abnormal mTOR Activation in Autism. Annu Rev Neurosci. 2018;41:1–23. Epub 2018/03/01. doi: 10.1146/annurev-neuro-080317-061747. [DOI] [PubMed] [Google Scholar]
- 27.Ka M, Condorelli G, Woodgett JR, Kim WY. mTOR regulates brain morphogenesis by mediating GSK3 signaling. Development. 2014;141(21):4076–86. Epub 2014/10/03. doi: 10.1242/dev.108282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lipton JO, Sahin M. The neurology of mTOR. Neuron. 2014;84(2):275–91. Epub 2014/11/07. doi: 10.1016/j.neuron.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gazestani VH, Pramparo T, Nalabolu S, Kellman BP, Murray S, Lopez L, Pierce K, Courchesne E, Lewis NE. A perturbed gene network containing PI3K-AKT, RAS-ERK and WNT-beta-catenin pathways in leukocytes is linked to ASD genetics and symptom severity. Nat Neurosci. 2019;22(10):1624–34. Epub 2019/09/26. doi: 10.1038/s41593-019-0489-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelly PA, Rahmani Z. DYRK1A enhances the mitogen-activated protein kinase cascade in PC12 cells by forming a complex with Ras, B-Raf, and MEK1. Mol Biol Cell. 2005;16(8):3562–73. Epub 2005/05/27. doi: 10.1091/mbc.e04-12-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abekhoukh S, Planque C, Ripoll C, Urbaniak P, Paul JL, Delabar JM, Janel N. Dyrk1A, a serine/threonine kinase, is involved in ERK and Akt activation in the brain of hyperhomocysteinemic mice. Mol Neurobiol. 2013;47(1):105–16. Epub 2012/08/28. doi: 10.1007/s12035-012-8326-1. [DOI] [PubMed] [Google Scholar]
- 32.Woods YL, Cohen P, Becker W, Jakes R, Goedert M, Wang X, Proud CG. The kinase DYRK phosphorylates protein-synthesis initiation factor eIF2Bepsilon at Ser539 and the microtubule-associated protein tau at Thr212: potential role for DYRK as a glycogen synthase kinase 3-priming kinase. Biochem J. 2001;355(Pt 3):609–15. Epub 2001/04/20. doi: 10.1042/bj3550609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perluigi M, Pupo G, Tramutola A, Cini C, Coccia R, Barone E, Head E, Butterfield DA, Di Domenico F. Neuropathological role of PI3K/Akt/mTOR axis in Down syndrome brain. Biochim Biophys Acta. 2014;1842(7):1144–53. Epub 2014/04/17. doi: 10.1016/j.bbadis.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22(15):6309–14. Epub 2002/08/02. doi: 20026564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herculano-Houzel S, von Bartheld CS, Miller DJ, Kaas JH. How to count cells: the advantages and disadvantages of the isotropic fractionator compared with stereology. Cell Tissue Res. 2015;360(1):2942. Epub 2015/03/06. doi: 10.1007/s00441-015-2127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, Goeva A, Nemesh J, Kamitaki N, Brumbaugh S, Kulp D, McCarroll SA. Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell. 2018;174(4):1015–30 e16. Epub 2018/08/11. doi: 10.1016/j.cell.2018.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hammerle B, Elizalde C, Tejedor FJ. The spatio-temporal and subcellular expression of the candidate Down syndrome gene Mnb/Dyrk1A in the developing mouse brain suggests distinct sequential roles in neuronal development. Eur J Neurosci. 2008;27(5):1061–74. Epub 2008/03/28. doi: 10.1111/j.1460-9568.2008.06092.x. [DOI] [PubMed] [Google Scholar]
- 38.Ko J. Neuroanatomical Substrates of Rodent Social Behavior: The Medial Prefrontal Cortex and Its Projection Patterns. Front Neural Circuits. 2017;11:41. Epub 2017/07/01. doi: 10.3389/fncir.2017.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Canty AJ, Murphy M. Molecular mechanisms of axon guidance in the developing corticospinal tract. Prog Neurobiol. 2008;85(2):214–35. Epub 2008/04/02. doi: 10.1016/j.pneurobio.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 40.Ryoo SR, Jeong HK, Radnaabazar C, Yoo JJ, Cho HJ, Lee HW, Kim IS, Cheon YH, Ahn YS, Chung SH, Song WJ. DYRK1A-mediated hyperphosphorylation of Tau. A functional link between Down syndrome and Alzheimer disease. J Biol Chem. 2007;282(48):34850–7. Epub 2007/10/02. doi: 10.1074/jbc.M707358200. [DOI] [PubMed] [Google Scholar]
- 41.Huang EJ, Reichardt LF. Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem. 2003;72:609–42. Epub 2003/04/05. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- 42.Slipczuk L, Bekinschtein P, Katche C, Cammarota M, Izquierdo I, Medina JH. BDNF activates mTOR to regulate GluR1 expression required for memory formation. PLoS One. 2009;4(6):e6007. Epub 2009/06/24. doi: 10.1371/journal.pone.0006007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carracedo A, Pandolfi PP. The PTEN-PI3K pathway: of feedbacks and cross-talks. Oncogene. 2008;27(41):5527–41. Epub 2008/09/17. doi: 10.1038/onc.2008.247. [DOI] [PubMed] [Google Scholar]
- 44.Sara VR, Carlsson-Skwirut C, Bergman T, Jornvall H, Roberts PJ, Crawford M, Hakansson LN, Civalero I, Nordberg A. Identification of Gly-Pro-Glu (GPE), the aminoterminal tripeptide of insulin-like growth factor 1 which is truncated in brain, as a novel neuroactive peptide. Biochem Biophys Res Commun. 1989;165(2):766–71. Epub 1989/12/15. doi: 10.1016/s0006-291x(89)80032-4. [DOI] [PubMed] [Google Scholar]
- 45.Tropea D, Giacometti E, Wilson NR, Beard C, McCurry C, Fu DD, Flannery R, Jaenisch R, Sur M. Partial reversal of Rett Syndrome-like symptoms in MeCP2 mutant mice. Proc Natl Acad Sci U S A. 2009;106(6):2029–34. Epub 2009/02/12. doi: 10.1073/pnas.0812394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arranz J, Balducci E, Arato K, Sanchez-Elexpuru G, Najas S, Parras A, Rebollo E, Pijuan I, Erb I, Verde G, Sahun I, Barallobre MJ, Lucas JJ, Sanchez MP, de la Luna S, Arbones ML. Impaired development of neocortical circuits contributes to the neurological alterations in DYRK1A haploinsufficiency syndrome. Neurobiol Dis. 2019;127:210–22. Epub 2019/03/05. doi: 10.1016/j.nbd.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Souchet B, Guedj F, Sahun I, Duchon A, Daubigney F, Badel A, Yanagawa Y, Barallobre MJ, Dierssen M, Yu E, Herault Y, Arbones M, Janel N, Creau N, Delabar JM. Excitation/inhibition balance and learning are modified by Dyrk1a gene dosage. Neurobiol Dis. 2014;69:65–75. Epub 2014/05/08. doi: 10.1016/j.nbd.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 48.Tejedor F, Zhu XR, Kaltenbach E, Ackermann A, Baumann A, Canal I, Heisenberg M, Fischbach KF, Pongs O. minibrain: a new protein kinase family involved in postembryonic neurogenesis in Drosophila. Neuron. 1995;14(2):287–301. Epub 1995/02/01. doi: 10.1016/0896-6273(95)90286-4. [DOI] [PubMed] [Google Scholar]
- 49.Hammerle B, Vera-Samper E, Speicher S, Arencibia R, Martinez S, Tejedor FJ. Mnb/Dyrk1A is transiently expressed and asymmetrically segregated in neural progenitor cells at the transition to neurogenic divisions. Dev Biol. 2002;246(2):259–73. Epub 2002/06/08. doi: 10.1006/dbio.2002.0675. [DOI] [PubMed] [Google Scholar]
- 50.Yabut O, Domogauer J, D’Arcangelo G. Dyrk1A overexpression inhibits proliferation and induces premature neuronal differentiation of neural progenitor cells. J Neurosci. 2010;30(11):4004–14. Epub 2010/03/20. doi: 10.1523/JNEUROSCI.4711-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spruston N. Pyramidal neurons: dendritic structure and synaptic integration. Nat Rev Neurosci. 2008;9(3):206–21. Epub 2008/02/14. doi: 10.1038/nrn2286. [DOI] [PubMed] [Google Scholar]
- 52.Cheriyan J, Kaushik MK, Ferreira AN, Sheets PL. Specific Targeting of the Basolateral Amygdala to Projectionally Defined Pyramidal Neurons in Prelimbic and Infralimbic Cortex. eNeuro. 2016;3(2). Epub 2016/03/30. doi: 10.1523/ENEURO.0002-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adolphs R. Cognitive neuroscience of human social behaviour. Nat Rev Neurosci. 2003;4(3):165–78. Epub 2003/03/04. doi: 10.1038/nrn1056. [DOI] [PubMed] [Google Scholar]
- 54.Ori-McKenney KM, McKenney RJ, Huang HH, Li T, Meltzer S, Jan LY, Vale RD, Wiita AP, Jan YN. Phosphorylation of beta-Tubulin by the Down Syndrome Kinase, Minibrain/DYRK1a, Regulates Microtubule Dynamics and Dendrite Morphogenesis. Neuron. 2016;90(3):551–63. Epub 2016/04/27. doi: 10.1016/j.neuron.2016.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu B, Zang K, Ruff NL, Zhang YA, McConnell SK, Stryker MP, Reichardt LF. Cortical degeneration in the absence of neurotrophin signaling: dendritic retraction and neuronal loss after removal of the receptor TrkB. Neuron. 2000;26(1):233–45. Epub 2000/05/08. doi: 10.1016/s0896-6273(00)81153-8. [DOI] [PubMed] [Google Scholar]
- 56.Minichiello L. TrkB signalling pathways in LTP and learning. Nat Rev Neurosci. 2009;10(12):850–60. Epub 2009/11/21. doi: 10.1038/nrn2738. [DOI] [PubMed] [Google Scholar]
- 57.Takei N, Nawa H. mTOR signaling and its roles in normal and abnormal brain development. Front Mol Neurosci. 2014;7:28. Epub 2014/05/06. doi: 10.3389/fnmol.2014.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kelleher RJ 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135(3):401–6. Epub 2008/11/06. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 59.Bozdagi O, Tavassoli T, Buxbaum JD. Insulin-like growth factor-1 rescues synaptic and motor deficits in a mouse model of autism and developmental delay. Mol Autism. 2013;4(1):9. Epub 2013/04/30. doi: 10.1186/2040-2392-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Castro J, Garcia RI, Kwok S, Banerjee A, Petravicz J, Woodson J, Mellios N, Tropea D, Sur M. Functional recovery with recombinant human IGF1 treatment in a mouse model of Rett Syndrome. Proc Natl Acad Sci U S A. 2014;111(27):9941–6. Epub 2014/06/25. doi: 10.1073/pnas.1311685111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beck KD, Powell-Braxton L, Widmer HR, Valverde J, Hefti F. Igf1 gene disruption results in reduced brain size, CNS hypomyelination, and loss of hippocampal granule and striatal parvalbumin-containing neurons. Neuron. 1995;14(4):717–30. Epub 1995/04/01. doi: 10.1016/0896-6273(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 62.Rangasamy S, Olfers S, Gerald B, Hilbert A, Svejda S, Narayanan V. Reduced neuronal size and mTOR pathway activity in the Mecp2 A140V Rett syndrome mouse model. F1000Res. 2016;5:2269. Epub 2016/10/27. doi: 10.12688/f1000research.8156.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Switon K, Kotulska K, Janusz-Kaminska A, Zmorzynska J, Jaworski J. Molecular neurobiology of mTOR. Neuroscience. 2017;341:112–53. Epub 2016/11/28. doi: 10.1016/j.neuroscience.2016.11.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.