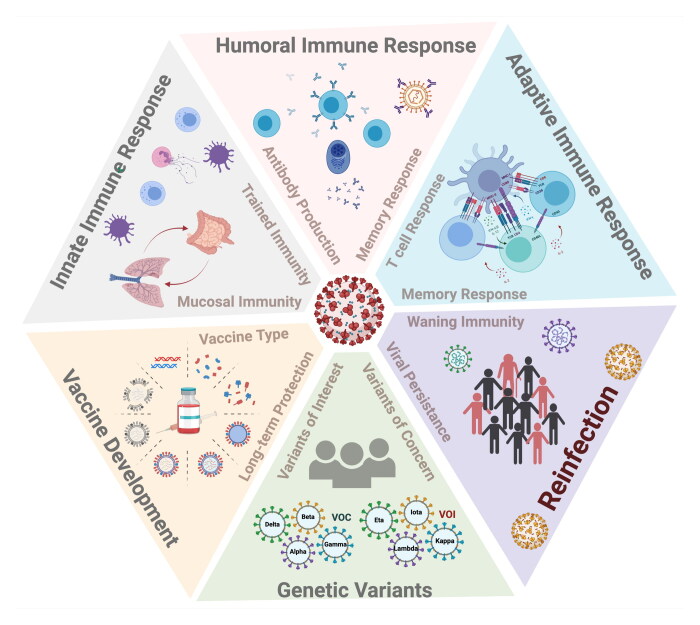
Abstract
Reinfection with SARS-CoV-2 is not frequent yet the incidence rate of it is increasing globally owing to the slow emergence of drift variants that pose a perpetual threat to vaccination strategies and have a greater propensity for disease reoccurrence. Long-term protection against SARS-CoV-2 reinfection relies on the induction of the innate as well as the adaptive immune response endowed with immune memory. However, a multitude of factors including the selection pressure, the waning immunity against SARS-CoV-2 over the first year after infection possibly favors evolution of more infectious immune escape variants, amplifying the risk of reinfection. Additionally, the correlates of immune protection, the novel SARS-CoV-2 variants of concern (VOC), the durability of the adaptive and mucosal immunity remain major challenges for the development of therapeutic and prophylactic interventions. Interestingly, a recent body of evidence indicated that the gastrointestinal (GI) tract is another important target organ for SARS-CoV-2 besides the respiratory system, potentially increasing the likelihood of reinfection by impacting the microbiome and the immune response via the gut-lung axis. In this review, we summarized the latest development in SARS-CoV-2 reinfection, and explored the untapped potential of trained immunity. We also highlighted the immune memory kinetics of the humoral and cell-mediated immune response, genetic drift of the emerging viral variants, and discussed the current challenges in vaccine development. Understanding the dynamics and the quality of immune response by unlocking the power of the innate, humoral and cell-mediated immunity during SARS-CoV-2 reinfection would open newer avenues for drug discovery and vaccine designs.
Keywords: COVID-19, genetic variants, reinfection, SARS-CoV-2, vaccines
Graphical Abstract
Introduction
The novel coronavirus, SARS-CoV-2, which came into the limelight during December 2019 have resulted over 236 million cases worldwide and around 4.8 million deaths as of 8th October 2021, according to the COVID-19 Map of the Johns Hopkins Coronavirus Resource Center. The landscape of immunological responses against SARS-CoV-2 infection is rapidly emerging in the context of disease pathogenesis, immunological response, virus evolution and vaccine development [1, 2] however an ongoing conundrum for clinicians and research scientists across the globe is the strength and durability of the immune response (Innate versus Cellular and Humoral Immunity). This issue of long term COVID-19 immunity gets further mystified by reinfection cases raising fresh concerns over the currently administered vaccines and their efficacy. With two major outbreaks by Coronaviruses-SARS-CoV in 2002–2004, and Middle East respiratory syndrome (MERS-CoV) in 2012 [3], it was reported that cross-protective immunity might exist against SARS-CoV-2 [4, 5]. However, this belief of preexisting cross-reactive immunity has been a matter of debate since the start of the pandemic [6, 7]. In general, immune memory is the primary source of long-term protective immunity [8, 9] and its repertoire/strength would help in determining protection against reinfection, disease risk, and vaccine efficacy. The immune response to viral infections can generally be classified into two categories with regard to the duration of the response – infections associated with (1) short-term and (2) long-term immunity. Sterilizing immunity develops against viruses such as measles and mumps wherein the infection induces a robust and long-lasting immunity while infections with flu and cold viruses result in short-term protection, and repeat infections, both host immune defense as well as the virulence attributes of the virus playing pivotal roles.
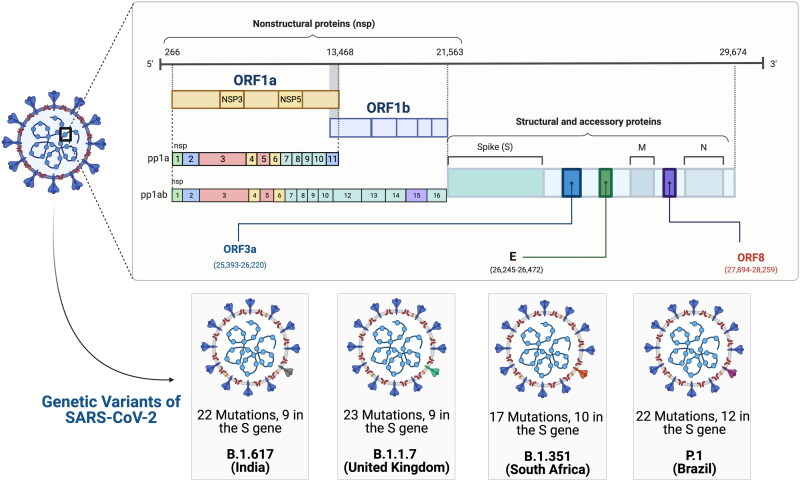
Infection caused by seasonal coronaviruses confers short term immunity [10] indicating that the novel SARS-CoV-2 falls in the first category of viral infections as investigations have revealed that COVID-19 infections induce immunity only for a short duration of 6–8 weeks. SARS-CoV-2 reinfections were a rare phenomenon a few months ago, pointing to the fact that SARS-CoV-2 infection may provide some level of protection against reinfection however, this scenario is now slowly changing as increasing evidence of greater number of reinfections emerge all over the world. Earlier studies on human and non-human primate models documented elevated levels of neutralizing antibodies against SARS-CoV-2 spike protein that offer considerable protection against reinfection [11–14]. Natural immunity developed after SARS-CoV-2 infection could be as effective as those with vaccination as primary infection declined risk of reinfection by 84% in 7 months in a large multicentric SARS-CoV-2 Immunity and Reinfection Evaluation study in the United Kingdom [15]. The rate of suspected reinfection was reported to be 1.9% in the UK SIREN study and 4.5% in Indian ICMR study [15, 16]. However, the possibilities of an exponential increase in reinfection rates may not be ruled out with waning immunity and evolving genetic mutants of SARS-CoV-2. The recent emergence of SARS-CoV-2 variants, within a span of a few months, especially in the UK (20I/501Y.V1, VOC 202012/01, or B.1.1.7) [17], the South Africa (20H/501Y.V2 or B.1.351) [18],and the Brazil (P.1) [19] variants strongly points toward rapid emergence of quasi species. It is tantalizing to speculate, albeit with a strong certainty, that the error prone RNA dependent RNA polymerase introduces mutations in genes targeted by the host immune response, a point of evidence is failure of the Astra Zeneca/Oxford vaccine to protect against the South African variant [20, 21]. Further evidence came from a Brazilian study that documented for the first time, a reinfection case where the patient recovered from the first SARS-CoV-2 infection got reinfected with a new viral variant harboring E484K spike mutation, well known for its infectivity and immune escape from neutralizing antibodies [22]. Additionally, a sudden surge in the novel SARS-CoV-2 strains harboring mutations (N440K, E484K, and V911I) in India and their rapid circulation in different states across the Indian continentis a cause of worry [23] (Figure 1). This raises serious concerns over reinfection as multiple mutations in the spike protein may lead to immune evasion. As the number of reinfection cases continue to rise, it is crucial to analyze the genetic, immunological and clinical profile of the reinfected individuals along with characterization of the viral genomes to understand the role of host and viral factors.
Figure 1.
Schematic representation of the genetics and natural evolution of SARS-CoV-2: SARS-CoV-2 virion is covered by three different proteins (S, M, E) and the genome RNA comprises of the 5′UTR consisting of frameshift ORF1a and ORF1b that forms 16 NSPs and the 3′-end encodes for 4 structural proteins (S, N, M, E) and 9 accessory proteins (Orf3a, Orf3b, Orf6, Orf7a, Orf7b, Orf8, Orf9b, Orf9c, Orf10). Four genetic variants of SARS-CoV-2 called as VOCs which includes the B.1.671.2 (Delta variant, first report from India), B.1.1.7 (alpha variant, first report from Britain), B.1.351 (beta variant first report from South Africa) and P.1 (gamma variant first report from Brazil) have evolved due to mutations in the 5′ UTR.
Abbreviations: Spike glycoprotein (S); Membrane (M); envelope (E); N (nucleocapsid protein); Open reading frame (ORF); Untranslated Region (UTR); Nonstructural Proteins (NSPs); Variant of Concern (VOC)
The dynamic behavior of SARS-CoV-2 infection, assessment of reinfection rates, impact of genetic variants on immune memory kinetics and its implication on global vaccination drive are some of the key areas highlighted in this review. This review will also address the potential challenges in immune memory development againstSARS-CoV-2 infection, and present perspective for the development of novel COVID-19 vaccine strategies in light of the increased SARS-CoV-2 reinfection cases.
Reinfection: a natural phenomenon
Reinfection is a natural phenomenon since ancient times. Across the vertebrate host species, evolution of the adaptive immune system, both humoral as well as cell mediated immunity, had surely been fine-tuned by reinvading microbes. The presence of innumerable antigen specific B and T cells, both resting as well as circulating immune memory cells capable of enhancing huge repertoire, is a clear proof of these events. In general, reinfection is defined as second time exposure of a person with the same or a different variant of the pathogen/virus after a period of recovery from illness [24] as shown in Figure 2. However, considering the over 236 million recorded SARS-CoV-2 cases globally as of now, the total number of documented SARS-CoV-2 re-infection and breakthrough infection cases worldwide is not that high. The expanding COVID-19 vaccine coverage may further reduce the chances of SARS-CoV-2 re-infection and breakthrough infection.
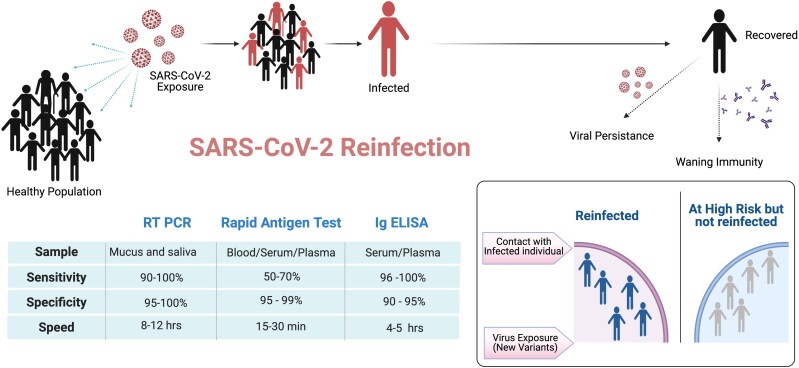
Figure 2.
Potential risk factors for SARS-CoV-2 reinfection. In a healthy population exposed to SARS-CoV-2, many individuals develop COVID-19 symptoms with different levels of disease severity. Three primary testing strategies namely RT-PCR, Rapid Antigen Testing and Immunoglobulin (Ig) ELISA differing in specificity, sensitivity and speed of testing are the preferred methods for diagnosing SARS-CoV-2 infection. Infected individuals with mild severity usually recover within 7-10 days but viral persistence may last for a longer duration. Waning immunity post infection, evolution of novel genetic variants and old age are some of the major risk factors for SARS-CoV-2 reinfection.
The reinfection has been defined in different ways by different agencies and authors:
The Center for Disease Control and Prevention [25] uses the following investigative criteria to define reinfection with SARS-CoV-2:
Detection of SARS-CoV-2 RNA (with Ct values <33 if detected by RT-PCR) >90 days after the first detection of viral RNA whether or not symptoms were present and paired respiratory specimens from each episode that belong to different clades of virus or have genomes with >2 nucleotide difference per month.
Cases in which detection of SARS-CoV-2 RNA is present 45–89 days apart are considered reinfections if the second symptomatic episode had no obvious alternate explanation for the COVID-19-like symptoms or a close contact with a person known to have laboratory-confirmed COVID-19 and paired respiratory specimens are available with the Ct values < 33 and sequence diversity noted above.
The European CDC has proposed whole-genome sequencing and phylogenetic analysis to confirm diagnosis of SARS-CoV-2 reinfection however, the access to these expensive resources might not be the possible in developing countries [25]. Likewise, efforts were also made by Yahav et al. who proposed three different criteria (laboratory, clinical and epidemiological) to redefine SARS-CoV-2 reinfection as the definition might differ based on different situations [24].
True reinfection in case of SARS-CoV-2 needs more clarity and a clear distinction should be made from prolonged viral shedding and reactivation. Viral shedding is very common for respiratory viruses such as influenza wherein the virus replicates inside patient’s body and is released into the environment for a long period of time. Recently, Li et al. reported prolonged shedding of SARS-CoV-2 RNA with a median duration of 53 days and a maximum of 83 days in 36 patients [26]. Additionally, Yuan et al. showed that 14.5% (25/172) of discharged COVID-19 patients had positive RT PCR test after a negative RT-PCR test [192]. SARS-CoV-2 RNA shedding studies are still in their infancy as the virus has recently transgressed the host species barrier. Silent spread of SARS-CoV-2 infection by asymptomatic patients for an extended period of time has been supported by many studies [27–29]. Moreover, there is a strong possibility that the reverse-transcription PCR (RT-PCR) might give false negative results due to different sampling source, kit’s sensitivity/specificity and the sampling procedure [30, 31] (Figure 2). It has also been demonstrated that stool samples were found positive in many cases after the respiratory samples tested negative for RT-PCR, indicating prolonged viral shedding and potential transmission via fecal-oral route [26, 32]. In fact, gastrointestinal tract has been reported to act as a reservoir for SARS-CoV-2 when the nasal and throat swab test negative for SARS-CoV-2 [33–36]. To define true reinfection cases, isolation of the complete virus, phylogenetic analysis of the isolated strains from different episodes, proof of virus infectivity in both episodes by virus culture from multiple specimens, time elapsed between two episodes and immunological characteristics at both time points will enable a clear vision of reinfection pathology [25, 37]. The odds of reinfection cases to be viral reactivation is also a possibility. Latency is commonly reported for herpes viruses like cytomegalovirus (CMV) and Epstein Barr virus (EBV)and also in Human Immunodeficiency Virus [38, 39]. Coronaviruses, although not known for latency, its genome consisting of many unknown and yet to be explored functional genes, have exhibited strange traits in human pathogenesis. Ye et al. reported the first possible case of viral reactivation in 9.1% discharged patients previously diagnosed with COVID-19 [40]. Likewise, Yadav et al. in a recent finding showed reactivation of SARS-CoV-2 in a child with neuroblastoma after recovering fully from the first infection. Genetic analysis showed that the virus isolated from the two episodes were not distinctly different, a possible case of virus reactivation during the course of chemotherapy [41]. Interestingly, Siqueira et al. added another dimension to the prevailing dilemma and reported existence of multiple viral variants in dynamic frequencies during the first clinical episode, with variant selection over time in SARS-CoV-2 infections [42]. Yet, it is still hard to clearly distinguish reactivation from reinfection and many different criteria’s/parameters should be taken into account while diagnosing SARS-CoV-2 reinfection cases.
In most of the SARS-CoV-2 reinfection cases, reinfection was caused by different viral clades suggesting that the immunity generated against one variant did not provide full protection against another viral variant. However, the reinfection case reported from the State of Nevada, US) [43], infection was caused by the viral strain responsible for both episodes, and there could be many other cases not yet published where the reinfecting strain might be similar to the first variant. Genome sequencing is a pre-requisite to define reinfection cases however, only about 5% of samples are sequenced as sequencing is a big expensive task. In most of the reported cases of SARS-CoV-2 reinfection cases, the antibody titers in infected patients declined over a period and offered only partial protection, most patients with repeat infections, recovered after the second exposure, immunocompetent young patients with no comorbidity had a milder reinfection episode while elderly immunocompromised individuals with comorbidities were more vulnerable to develop severe disease pathology. Further evidence in support of this came from a recently published population-level observational study from Denmark where Hansen et al. used the national PCR-test data from 2020 to estimate protection toward repeat infection with SARS-CoV-2. They reported that the first SARS-CoV-2 infection confers approximately 80–83% protection in people younger than 65 for about six months while the level of protection is lower for people ≥65 at approximately 47% [44]. The findings are in concordance with other cohort studies from the UK, Qatar and the USA that reported reinfection to be a rare phenomenon [45–47]. An interesting observation about the reinfected individuals was that they did not have detectable antibodies even after the onset of symptoms, implying the absence of robust neutralizing antibody response in the first exposure that could have resulted from down play of the host immune system or from the presence of a strong virus induced immune regulation [48, 49]. There could be combination of host’s innate and adaptive immunological as well as viral factors that may drive severity of symptoms following re-infection. It includes the extent of long-term transcriptional, epigenetic and functional reprogramming to reduce innate inflammatory response during re-infection; sufficient reservoirs of specific memory like NK cells, memory T and B cells and their rapid expansion following reinfection; the presence of neutralizing antibodies in sufficient titers in blood and respiratory tract during re-infection; the specificity of neutralizing antibodies that may cross-reactive with variants; non-healed respiratory tract integrity and germinal centers; Immune-senescence and others. These factors will be discussed in detail within the upcoming chapters of this review.
It is also important to mention that besides these documented reinfection cases, undocumented mild or asymptomatic cases may also exist. SARS-CoV-2 RNA Testing on nasopharyngeal specimen by RT-PCR is done for screening asymptomatic reinfection, similar to that for screening SARS-CoV-2 infection. The duration of negative time period between two positive tests should differ by 45–89 days as per CDC guideline. Population-based survey in Qatar estimated that only 9.3%(95% CI: 7.9–11.0%) had a documented PCR-confirmed infection prior to antibody testing, suggesting reinfection (undocumented infections) could possibly be ten-fold higher than documented [50]. Hence, it is important to comprehend the patho-physiology of Covid-19 reinfection to better understand the protective immune response. Studies must be designed to delineate the factors responsible for host immune protection as well as a clear understanding of how the virus establishes a reinfection in the partially protected individuals.
Host virus interaction (the host immune response)
Innate immune response
Response to viral infection
The innate immune system acts as a frontline warrior and offers triple-layer protection (physical barrier, cells, molecular factors) against invading pathogens. Mucous secretion and production of anti-microbial compounds by epithelial cells lining the upper respiratory tract [51] help in preventing infection. However, if the pathogen is able to breach this physical barrier, then to counteract viral replication and limit the viral spread, the second layer of protection comes into action. It is made up of innate immune cells such as dendritic cells that express pattern recognition receptors (PRRs) and recognize the pathogen-associated molecular patterns (PAMPS) [52–54]. Toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), and nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), are important PRRs [55, 56]. This event triggers a cascade of downstream signaling pathways leading to induction of nuclear transcription factors, in particular nuclear factor κB (NF-κB), interferon response factor 3 (IRF3), and IRF7 [57]. They eventually stimulate the expression of type I and III interferons (IFNs) and other proinflammatory cytokines (IL-1β, IL-6, IL-8, IL-12, and tumor necrosis factor [TNF] as shown in Figure 3 [53, 58, 59]. Type I IFNs further increase expression levels of endosomal TLRs, including TLR3, TLR7, TLR8, and TLR9. IFNs mediate the recruitment of other immune cells such as neutrophils, natural killer cells and also help in activating the adaptive immune system [60].
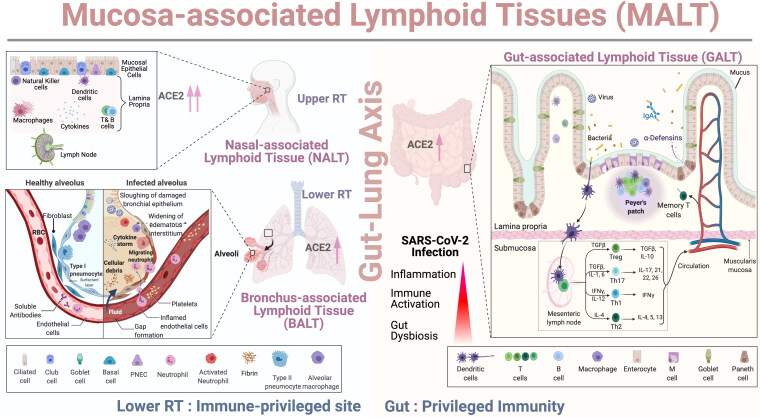
Figure 3.
Role of Mucosal and Gut Immunity in SARS-CoV-2 Infection: As shown in the schematic representation, the upper respiratory tract (URT) have specialized lymphoid structure called Nasal Associated Lymphoid Tissue (NALT). The mucosal epithelial cells covering NALT express high levels of ACE2 receptors which facilitates binding of spike protein of SARS-CoV-2. The lamina propria consisted of a mixed population of T cells, B cells, NK cells, macrophages and dendritic cells while the lower respiratory tract (LRT) is possibly an immune-privileged site as a single layer of pneumocytes forms the healthy alveolus and has very few immune cells in the vicinity. Type 1 and type II pneumocytes express ACE2 receptors and SARS-CoV-2 infection damages the respiratory epithelium, widening the interstitium followed by accumulation of fluid in the alveoli along with cellular debris. Immune cells such as neutrophils, macrophages migrate from blood vessels to infected alveolus and leads to hyperinflammation/cytokine storm, thrombosis along with disruption of the “Gut-lung axis.” The gut associated lymphoid tissue (GALT) consists of multi-follicular Peyer’s patches, plasma cells, T cells present in the lamina propria, and mesenteric lymph nodes. Dendritic cells capture microbial antigen and carry it through lamina propria, submucosa to draining mesenteric lymph node where they interact with helper T cells (Th cells). Th cells differentiate into Th1, Th2, regulatory T cells (Tregs), Th17 cells and memory T cell pool which then migrate to the gut and respond against SARS-CoV-2 infection.
SARS-CoV-2 infection: Spread from upper to lower respiratory tract
SARS-CoV-2 infection begins when the spike glycoprotein trimer [2] of the virus binds to the angiotensin-converting enzyme 2 (ACE2) highly expressed by the nasal epithelia lining the URT and decreased levels in the lower respiratory tract (LRT) as depicted in Figure 3 [61–63]. Nasal-associated lymphoid tissue (NALT) in the URT serve as an inductive site for the initiation of the mucosal immune response (Figure 3) and for active virus replication during the early days of infection [64]. The existence of a well-organized mucosa-associated lymphoid tissue (MALT) like tonsils, adenoids, Waldeyer’s ring in the URT inhibits the majority of the respiratory invaders however, a few that are able to enter the sacrosanct space induce severe inflammation resulting in acute red hepatization of the lungs as seen in S. pneumoniae or H. influenza pneumonia infections [65, 66]. On the other side, the lower respiratory tract (LRT) is (almost) an immune-privileged site, and the alveoli have evolved into a highly specialized organ for blood gas exchange. The walls of alveoli contain an extensive network of blood vessels and are made of a single layer of cells protected by a few immune cells circulating through the interalveolar space (pulmonary capillaries). Lack of an organized lymphoid structure makes it one of the most susceptible binding sites for SARS-CoV-2 where the virus is capable of causing substantial damage. Goblet cells that lubricate lungs by mucous production, and ciliated cells that clear the debris from the lungs are the preferred sites for virus binding and replication as they display the highest expression levels of the ACE2 gene in the URT (Figure 3) [62, 67–69]. Many other receptors including Neuropilin-1 have also been suggested as entry points for SARS-CoV-2, however, their role in COVID outcome or immune dysregulation is still under investigation [70, 71]. ACE2 receptors are not exclusive to the lungs, they are also expressed by other tissues as well, including the heart, kidney, pancreas, brain, and gastrointestinal (GI) tract [69, 72].
SARS-CoV-2 infection: Impaired innate immune response
Upon binding, the virus enters inside the host cell by receptor-mediated endocytosis as shown in Figure 4 followed by RNA release into the host cell cytoplasm, production of viral proteins, and formation of replication/transcription complexes [72, 73]. The important PRR involved in recognizing SARS-CoV-2 is yet to be determined, previous findings on SARS-CoV points to endosomal TLR3 and TLR7 as well as RIG-I to be the potential candidates [74, 75]. The innate immune system gets activated through these PRRs which then turns on NF-kb signaling cascade in monocytes, plasmacytoid dendritic cells (DCs), and macrophages. An in-silico study predicted that TLR4 possesses a strong binding affinity to S protein and is also one of the most efficient innate immune receptors for induction of inflammatory responses [76]. The pathogenic role of TLR4 in inducing excessive inflammation in COVID-19 patients has also been further confirmed by other groups [72, 77, 78]. Similarly, pro-inflammatory cytokines such as IL-2, IL-6, TNF-a, GM-CSF, and IFN-γ are produced in higher amounts in severe COVID-19 patients [79, 80], and IL-6 correlated positively with disease severity [81, 82]. Once activated, many different innate immune players come into action, for instance, enhanced peripheral blood neutrophil/lymphocyte ratio is regarded as a strong predictor of mortality [83–85]. Neutrophil infiltration in pulmonary capillaries leads to acute capillaritis, extravasation into the alveolar space, and immunothrombosis/capillary leak syndrome in other organs [86] (Figure 3). Other innate immune cells including macrophages, monocytes, and NK cells have been found to be equally affected by SARS-CoV-2 infection [80] although a connecting link between these events has not been found yet. This innate-immune mediated damage results in the accumulation of protein-rich edema fluid within the pulmonary interstitium and alveoli eventually leading to hypoxia [87, 88] (Figure 3).
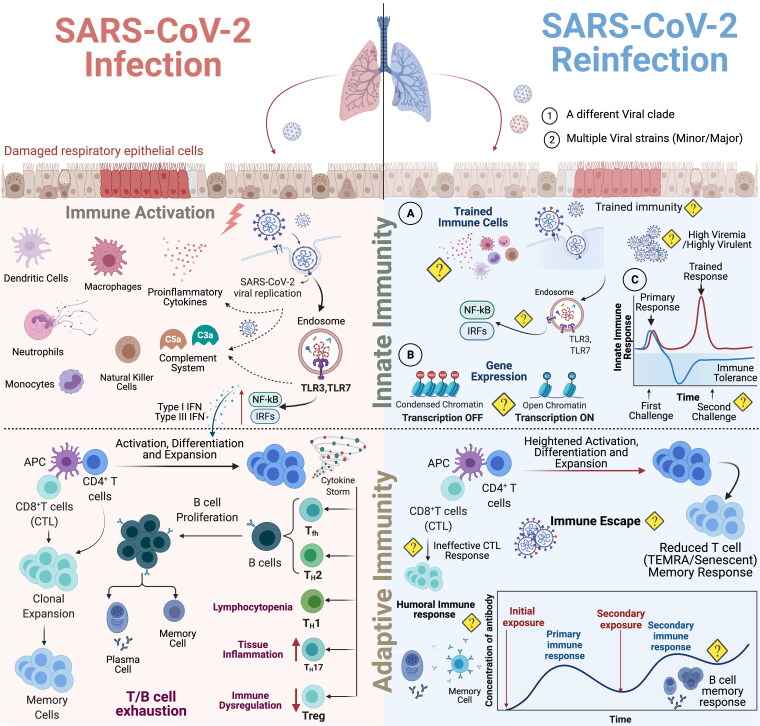
Figure 4.
Comparative immunological response (Innate vs adaptive) in SARS-CoV-2 infection and reinfection. SARS-CoV-2 attach to ACE2 receptor highly expressed by the respiratory epithelium that eventually results in epithelial cell damage as the virus replicates and these damaged cells in turn activates different part of the immune system. Following activation, immune cells differentiate into dendritic cells and macrophages. SARS-CoV-2 antigens acts on endosomal TLR3 and TLR7 which then turns on NF-κB signaling cascade in monocytes, plasmacytoid dendritic cells (DCs), and macrophages to produce Type 1 interferon for activation, differentiation and expansion of CD4+ T cells. There is also production of pro-inflammatory cytokines such as IL-6 and activation of complement cascade with release of C3a and C5a. Neutrophils via extracellular traps and reactive oxygen species cause mucositis, capillaritis, thrombosis and capillary leak syndrome. Antigen presenting cells carrying viral antigen interact with T cells and activate adaptive immunity. CD8 + T cells perform cytotoxic function while CD4+ T cells differentiate into Tfh, Th2, Th1, Th17 and Tregs. Tfh and Th2 facilitates B cell proliferation which further differentiate into antibodies producing plasma cells. CD4+/CD8+ T and B cells undergo extensive exhaustion leading to lymphocytopenia during an active SARS-CoV-2 infection, however, few cells still undergo clonal expansion to form memory cell pool [63]. During SARS-CoV-2 reinfection thathappens possibility by different viral clade or a virulent strain, the innate immune cells might have trained immunity to limit viral replication. However, there are many unanswered immunological questions during SARS-CoV-2 reinfection [1] Is there an induction of trained immunity during SARS-CoV-2 reinfection? If yes, then how would the trained immune cells behave during high viremia and in the presence of highly virulent SARS-CoV-2 strain? [63] How is the NF-κB signaling cascade and production of type 1 interferons regulated during the second encounter? [3] What are the epigenetic changes and how they impact the immune response during reinfection? [4] What component determines disease severity: immune tolerance or trained immunity? SARS-CoV-2 reinfection might result in heightened activation, differentiation and expansion of the T and B cell memory pool triggering adaptive immune response. The magnitude and breadth of the secondary humoral immune response necessary to contain SARS-CoV-2 reinfection needs further investigation.
SARS-CoV-2 infection: Innate immune evasion
To promote its own replication and transmissibility, SARS-CoV-2 deploy several tactics to evade host antiviral defense. One way is by shutting down the host cellular machinery and the evidence for this came from an in-vitro and in-vivo study where SARS-CoV-2 evaded the immune response using its nonstructural protein 1 (Nsp1) that blocked retinoic acid-inducible gene I–dependent innate immune responses and inhibited host cellular translational machinery [89]. Besides that, many different SARS-CoV-2 genes by inhibiting the translocation of IRF-3 and/or NFkB to the nucleus act as strong suppressors of the innate immune response. Furthermore, modulation of dendritic cell function, dysregulation of IFN-γ production, exhaustion of NK cell-mediated cytotoxicity [90] and antigenic variation [91] are some other ways to escape the innate immune defense. Likewise, hyperactivation of the complement cascade by SARS-CoV-2 triggers a vicious cycle of “hypercytokinaemia” [92, 93] that aids in elevating levels of pro-inflammatory modulators, such as C3a and C5a [94–96] (Figure 4) and increases the susceptibility to develop the severe acute respiratory distress syndrome [97]. Therefore, this modulation of the innate antiviral response induced by both host and viral factors gives a good head start to the viral replication in the URT and hyper-inflammation in the lungs, resulting in conditions that lead to COVID-19 severity. Additionally, hyper-inflammation in the respiratory bronchioles will increase the probability of airway collapse and opening, further increasing virus-laden aerosol generation [98]. As supported by recent investigation though lacking perfect experimental data, it is highly likely that the droplets and aerosols exhaled from the alveoli are more infectious than the droplets from the other airway sites and these droplets/aerosols possibly help in the airborne route of SARS-Cov-2 transmission [99–101].
With the emergence of the new SARS-CoV-2 variants, it is essential to better understand the innate immune response not only localized in the respiratory tract but also at other locations such as the GI tract. In a recent ex-vivo investigation, Chu et al. demonstrated robust activation of the innate immune response against SARS-CoV-2 infection in the human intestinal tissue including the activation of both type I and type III interferons [102]. However, this response has been found to be largely attenuated in human lung cell lines [103, 104], lung organoids [105], animals [104], and patient lungs [104].
SARS-CoV-2 reinfection: Are we missing the “trained immunity”?
Immune memory, a classical feature of adaptive immunity now also envelopes innate immunity as emerging evidence supports the notion that innate immunity can maintain memory, termed as “trained immunity or innate immune memory” and plays a key role in improving host protection against reinfection [106, 107]. Prior exposure to vaccines such as Bacille Calmette–Guérin (BCG) or pathogenic components have been shown to trigger trained innate immunity conferring cross-protection against many different infections [108, 109]. An integrated network of epigenetic rewiring and metabolic reprogramming allow innate immune cells such as myeloid and natural killer (NK) cells to remain in a “trained” state by allowing increased accessibility to proinflammatory genes, facilitating a heightened immune activation and long-term protection against reinfection (Figure 4) [51, 107, 110–112]. Investigations deciphering the clinical and immunological profile of SARS-CoV-2 reinfection cases are still in their early phases as most of the research efforts are primarily directed toward unlocking the SARS-CoV-2 immune paradox. During an asymptomatic/mild SARS-CoV-2 reinfection as is the case in most of the re-infected patients, the innate immune response is expected to respond to the viral antigen in a very similar fashion as the first challenge (Figure 4) however, the critical factors that would determine the outcome of the response definitely rely on the viral load, the virulence of the viral strain, and the strength of the innate immune memory. In a cohort of patients re-infected with SARS-CoV-2, Fintelman-Rodrigues et al. reported enhanced innate immune response during the first episode but, insufficient to provide protection against reinfection. From this preliminary investigation, it is clear that reinfection is a natural phenomenon that occurs frequently than previously thought and re-challenge may be necessary to achieve full protection against the virus though more conclusive studies are required to prove it [113].
In the case of children, it has been observed that they show a good level of resistance against SARS-CoV-2 infection globally possibly due to a highly efficient trained immunity developed through heavy vaccination against viral infections such as measles, mumps, rubella, and influenza [114, 115]. Along the same line, Yao et al. reported induction of long-lasting trained immunity in alveolar macrophages following acute respiratory viral exposure in mice models [116] pointing to the fact that trained immunity is very much active even in the lungs. However, in the case of SARS-CoV-2 reinfection, dysregulated activation of trained immunity possibility leads to either hyper inflammation or immune tolerance, depending on whether trained immunity is amplified or dampened as shown in Figure 4. It is not clear to what extent this trained immunity will affect susceptibility to reinfections. In the context of SARS-Cov-2 reinfection, the most important questions that need to be addressed are: what spectrum of innate immune cells develop trained immunity and what is the duration, kinetics, and robustness of this immune response after the first challenge? Does the trained immunity differ significantly at different anatomical sites such as the gut or lungs? Precise mechanisms that allow trained cells to undergo epigenetic and metabolic reprogramming to induce an effective trained immunity should be evaluated during SARS-CoV-2 infection and reinfection (Figure 4). Clearly, more studies are needed to investigate the impact of trained immunity on SARS-CoV-2 reinfection as they would lay a strong foundation for better vaccination strategies and effective ways to trigger long-lasting trained immunity against SARS-CoV-2.
Adaptive immune response
Response to viral infection
T cell mediated cellular immune responses and B cell-mediated humoral responses form an integral component of the adaptive immunity. Innate immune players such as DCs and macrophages, also referred to as antigen-presenting cells (APC) help in inducing this response. Secretory immunoglobulin A (s-IgA), IgM and IgG are major neutralizing antibodies present at the mucosal sites lining the nasal cavity (NALT) and URT during primary viral infections. However, these mucosal antibodies persist for a much shorter period of time than serum antibodies, which might explain in part the reason for shorter immunity duration to mucosal pathogens in particular, SARS-CoV-2 than systemic viral infections [117, 118]. Following antigen activation in the draining secondary lymphoid tissues, primed B and T cells migrate from NALT to lungs (BALT) where they further differentiate into antigen-specific, and memory cells. Within the B cell compartment, some activated B cells become short-lived plasma cells, while others become long-lived memory cells [119, 120]. Virus specific CD8+ T cells kill infected/abnormal cells through cytotoxic effector functions, and CD4+ T cells migrate to inflammatory sites to offer helper/effector functions (Th1/Th2/Th17), regulate inflammatory response (T-reg) and provide support to B cells (Tfh cells) (Figure 4) [121]. These long-lived antigen specific memory B/T cells can initiate strong recall responses with an accelerated kinetics during reinfection [122, 123].
SARS-CoV-2 infection: Impaired adaptive immune response
SARS-CoV-2 infection leads to a high level of heterogeneity in the breadth and magnitude of the adaptive immune response [9, 124]. Reduction in both CD4+ and CD8+ T cells (lymphopenia) along with functional impairment and elevated levels of activation/and or exhaustion markers are some of the key features of COVID-19 disease severity, which resolves when patients recover [79, 125–129]. SARS-CoV-2-specific CD4+ and CD8+ T cells memory responses have been demonstrated in the majority of patients recovered from COVID-19 even in the absence of detectable circulating antibodies [130–132]. Additionally, majority of SARS-CoV-2 specific CD4+ T cells exhibited a central memory phenotype while CD8+ T cells were skewed toward a more effector phenotype [1, 133, 134]. Notably, the frequency of regulatory T cells (T-regs) were reduced in severe COVID-19 patients with an exponential increase Th-17 population. Together, this dysregulation in the regulatory, the effector and the memory arm of the immune system shifts the subtle balance leading to severe inflammatory conditions, and perhaps also contributes to SARS-CoV-2 reinfection [135, 136]. To date, it remains largely unclear how well the SARS-CoV-2 T cell memory is established and how the memory T cells respond upon re-exposure to viral antigens. The magnitude and quality of secondary T cell immune response during reinfection will however depend on wide variety of factors like severity of primary infection [137], viral load [1], fraction of T cells converted into memory phenotype, time duration between primary infection and reinfection [43, 48, 138], status of co-morbidities [139, 140] and other yet to be discovered factors. Furthermore, old age could also contribute to greatly compromise the generation of naive T cells and TCR diversity, in both CD4+ T cells and CD8+ T cell populations [141].
Like the T cell compartment, the humoral component has also been shown to be significantly affected by the SARS-CoV-2 infection [142, 143] and is characterized by a marked expansion of the plasmablasts and depletion of IgM memory B cells [144] in extremely severe cases. Studies have confirmed that detectable levels of IgM, IgG and IgA could be found in the blood circulation up to 8 months after the first exposure, and neutralizing antibodies though decline over time but remain detectable post 6 months of infection [9, 145]. Interestingly, Gaebler et al. showed dynamic evolution of the memory B cell response during the first six months post SARS-CoV-2 infection. Persistence of antigen in the tissues resulted in antibody evolution via somatic mutations with increased neutralization potency. This evolution of the memory B cells would lead to a highly robust and effective immune response upon reinfection [146]. Breton et al. highlighted maintenance of polyfunctional SARS-CoV-2 specific memory immune cells and antibodies following natural infection [147, 148]. The stable maintenance memory B cell pool could provide an alternative way to help mitigate subsequent infections, although the kinetics of reinfection and protective potential of memory B cells in SARS-CoV-2 immunity are yet to be determined.
SARS-CoV-2 reinfection: “puzzling” immune memory
Despite an effective, multi-faceted immune memory generated by SARS-CoV-2 infection as mentioned above, there have now been more than 70 cases, along with 36000 suspected cases of SARS-CoV-2 reinfection worldwide. Reinfection cases reported from India and their characteristics are summarized in Table 1. A significant under-reporting of SARS-CoV-2 reinfections has also been observed as the stringent criteria set by the scientific journals for reporting of reinfection cases cannot be applied in every health setting owing to the lack of resources to sequence viral genome in all reported infections [149]. Moreover, the current belief that reinfection is a rare phenomenon is possibly an understatement as individuals with asymptomatic or mild reinfection may not come for testing. A recent study from Brazil estimated a high risk of reinfection, approximately 7% in health-care workers [150], and the rate of reinfection was reported to be 4.5% in SARS-CoV-2 infected individuals by another epidemiological study from India [16]. However, there is a complete paucity of immunological data on SARS-CoV-2 reinfection, hence, there is a need for more studies to investigate which responses (humoral versus cellular) are better correlates of immune protection against SARS-CoV-2 reinfection.
Table 1.
SARS-CoV-2 reinfections cases in india and their clinical characteristics.
| S.No. | References | (Age/Sex) | Comorbidity | Clinical symptoms of first infection | Clinical symptoms of reinfection | Confirmed reinfection (PCR/Antibody) | Intervening period (Days) | Sequencing Data (Genetic Variants) |
|---|---|---|---|---|---|---|---|---|
| 1 | (193) | 52/M | None | Asymptomatic | low-grade fever and body ache | RT PCR | 73 | clade from the first episode = 19A clade from the second episode = 20A |
| 2 | (39) | 47/M | None | Asymptomatic (civil official) |
Mild (fever, cough, and malaise) | RT PCR | 39 | 15 and 17 genetic variants in the genomes from the two episodes, respectively, of which 14variants were common between the two episodes. 22882T > G (Spike: N440K) variant in the genomes isolated from both episodes of infection |
| 3 | Patient 1, (178) | 27/M | None | Sore throat, rhinitis (2 days) | Cough, fever, myalgia (1 week) | RT PCR | 65 | clade from the first episode = 20A clade from the second episode = 19A |
| 4 | Patient 2, (178) | 31/M | None | asymptomatic | Myalgia, malaise (2 days) | RT PCR | 64 | clade from the first episode = 20B clade from the second episode = 20B |
| 5 | Patient 3, (178) | 24/F | None | Sore throat, rhinitis, Myalgia (5 days) |
Sore throat, rhinitis, Myalgia, Cough, fever (3 weeks) | RT PCR | 54 | clade from the first episode = 19A clade from the second episode = 20B |
| 6 | Patient 4, (178) | 51/F | Controlled hypertension | Fever, myalgia, dry cough (1 week) | Fever, myalgia, dry cough, nausea, abdominal pain, breathlessness (6 weeks) | RT PCR | 136 | clade from the first episode = 19A clade from the second episode = 20B |
| 7 | Patient 1, (195) | 61/M | None | Asymptomatic | Weakness, cough (2 days) | RT PCR | 75 | 10 unique variations between the viral genomes of both episodes (no variation in spike protein) |
| 8 | Patient 2, (195) | 38/M | Steroid usage (TB meningitis) | Hospitalized with headache and fever (1 day) | Fever (5 days) | RT PCR | 18 | 3 unique variations between both episodes ( one synonymous mutation in spike protein) |
| 9 | Patient 1 (194) | 25/M | None | Asymptomatic (Health care worker) | Asymptomatic (Health care worker) |
RT PCR | 108 | 9 unique variant differences between first and second episode of infection. |
| 10 | Patient 2, (194) | 28/F | None | Asymptomatic (Health care worker) |
Asymptomatic (Health care worker) |
RT PCR | 111 | 10 unique variant differences between first and second episode of infection. Genetic variation 22882T > G (S:N440K) within the receptor binding domain found in reinfection. |
| 11 | Patient 1,(196) | 18/F | Had PTB 5yrs ago | Fever, Cough (5 days) |
High grade fever, cough, dyspnea (2 days) | RT PCR | 5 | Not sequenced |
| 12 | Patient 2, (196) | 78/M | Coronary Artery disease | Fever, Cough (5 days) |
Fever, Cough Dyspnea (1 day) | RT PCR | 43 | Not sequenced |
| 13 | Patient 3, (196) | 42/M | None | Fever, Cough Dyspnea (4 days) | High grade fever and worsened dyspnea | RT PCR | 4 | Not sequenced |
| 14 | Patient 4, (196) | 25/F | None | Asymptomatic | Fever, Cough (2 days) |
RT PCR | 6 | Not sequenced |
| 15 | Patient 5, (196) | 64/M | None | Fever, Cough (2 days) |
Fever, Cough Dyspnea (3 days) | RT PCR | 17 | Not sequenced |
Abbreviations: M, Male; F, Female; RT, Real-time (RT-PCR).
The reinfection case from Hong-Kong [48] reported that the levels of IgG rise rapidly within 5–7 days during the second encounter [48] and the neutralizing antibody response observed in the reinfection case from Belgium [138] possibly downgraded the severity of reinfection. Interestingly, reinfection with all four seasonal coronaviruses has been shown to induce a short-lasting immunity for as little as 6–12 months measured in terms of a rapid decline in the antibody titters [10] therefore, it is important to investigate whether SARS-CoV-2 would also follow the same antibody dynamics like other coronaviruses or not. Another key point to be highlighted is that the SARS-CoV-2 strains in almost all reinfection cases were different from the primary strain that might have allowed the virus to evade the humoral as well as the cell mediated immunity. For instance, the spike protein mutants of SARS-CoV-2 (e.g., emerging Brazil and South Africa variants) have been shown to escape efficiently form the majority of neutralizing antibodies produced by long-lasting memory B cells [151]. Furthermore, memory B cell response wane over time, much before memory T cell response, favoring reinfection [152]. This clearly points to the fact that both waning immunity and novel emerging SARS-CoV-2 variants are the root cause of the SARS-CoV-2 reinfection. Therefore, based on the published literature, there could be two possible scenarios following natural SARS-CoV-2 infection; 1) a high level of protection may result in the prevention of subsequent infection; 2) a dysregulated immune response, virulent viral strain and a strong viral replication, would potentially lead to reinfection with a severe clinical outcome (Figure 4). During an asymptomatic or mild episode of SARS-CoV-2 reinfection, the immune system may respond in a very robust manner by activating different compartments of the adaptive immune system. Neutralizing antibodies might respond immediately by secreting high levels of IgA or IgG at the mucosal sites. Activated memory B and T cells present in the tissues or lungs proliferate rapidly and amplify the cascade of the immune cells by increasing the levels of neutralizing antibodies as well as cytokines such as IFN-γ thereby reducing the disease severity and viral replication. Preliminary studies in macaques reinfected with the identical SARS-CoV-2 strain showed that a primary infection with SARS-CoV-2 protects against subsequent reinfection as the response was found to be driven by the memory cell component with an enhanced neutralizing antibody response [153, 154]. However, in severe cases of SARS-CoV-2 reinfection, reported from the US, the Netherlands, Ecuador and India, a whole host of reasons might have contributed to disease severity. For example, antibody-dependent enhancement which occur due to preexisting non-neutralizing antibodies that interact with complement components/or virions could enhance subsequent infection thereby increasing the COVID-19 disease severity, however its clinical role in COVID-19 pathology is yet to be established [155, 156]
Immune system disruption is another potential way by which the virus can counter-attack the immune barrier. In that context, Kaneko et al. reported complete absence of germinal centers in the post mortem thoracic lymph nodes and spleens of severely ill COVID-19 patients suggesting that the long-term development of memory/protection may be difficult as the patients were unable to generate highly effective, long-lasting antibodies that would fight the virus for years [157]. It is therefore possible that many of the SARS-CoV-2-infected population with low immune memory would be vulnerable to reinfection. Thus, it is crucial to understand the kinetics and interrelationship of the four major players of the protective immunity: memory CD4+ T cells, and/or memory CD8+ T cells, memory B cells and antibodies to identify the durability, specificity and kinetics of memory cell response against SARS-CoV-2 infection. On a last note, there is also an urgent need for studies focused on the mucosal immune response besides humoral and cell-mediated immunity as mucosal surfaces in the upper (NALT) and lower respiratory tract (BALT) are the prime targets of SARS-CoV-2, memory response at these sites could prevent viral entry, and systemic viral transmission.
SARS-CoV-2 genetic variants: the viral storm
Viruses are evolving since the dawn of life as the process follows the same Darwinian principles of evolution for life, involving natural selection, genetic variation and survival of the fittest [158]. SARS-CoV-2 is also following its natural trajectory just like any other virus and it does not have any special animosity toward human beings. The virus first crossed the species barrier possibly by jumping from bats to human [159] and it is now continuously evolving to adapt and have better transmissibility. SARS-CoV-2 is a 30 kb size single stranded positive sense RNA virus as shown in Figure 1 having 88% homology with bat-SL-CoVZC45 and bat-SL-CoVZXC21, 79.5% homology with SARS-CoV and around 50% with MERS-CoV [160–162]. SARS-CoV-2 genome encodes multiple genes including structural: S (spike), E (envelope), M (membrane), N (nucleocapsid); nonstructural: ORF1a, ORF1b; and accessory genes:ORF3a, ORF6, ORF7a, ORF7b, ORF8 and ORF10 [163] (Figure 1). World’s largest database of novel coronavirus genome sequences, The Global Initiative on Sharing All influenza Data (GISAID) has shown that the mutation rate of this novel virus is comparatively slower than other viruses such as HIV yet it has resulted in several major strains over time [164]. Some mutations in the receptor-binding domain of the virus’s spike protein have allowed better binding capacity to host cells [164, 165].
During the early COVID-19 pandemic, D614G substitution in the spike protein was reported at multiple geographical locations [164]. This eventually impacted the SARS-CoV-2 infectivity and D614G variant then became a dominant mutation globally replacing initial Wuhan strain by June 2020 [164]. Although more infectious, D614G variant (now associated with B.1 lineage; https://cov-lineages.org/lineages/lineage_B.1.html) alone was not more virulent than initial Wuhan strain nor affected vaccine efficiency [164, 166]. CDC has devised and regularly updates a federal classification system for SARS-CoV-2 variants that identify and defines four classes of variants: variants being monitored, variants of interest, variants of concern (VOC), and variants of high consequence based on disease severity, transmissibility, impact on diagnosis, treatment strategies/vaccines [167]. As of Oct 4 2021, there are no variants of high consequence. B.1.526 (New York), B.1.525 (New York), P.2 (Brazil) were previously designated as variants of interest but now there are no designated variant of interest. Variant of concern occupied the center stage because of increased transmissibility, more severe disease and reduced neutralization by antibodies. Designated VOC include delta variants (B.1.617.2 and AY lineages) which were first identified in India. B.1.1.7 (Alpha, United Kingdom), P.1 (Gamma, Japan/Brazil), B.1.351 (Beta, South Africa), B.1.427 (Epsilon, California) and B.1.429 (Epsilon, California) which were previously VOC are now classified as variants being monitored along with others like B.1.617.1(kappa), B.1.617.3, P.2(zeta), B.1.621(mu), B.1.621.1 (mu), B.1.525 (eta) and B.1.526 (lota). However, WHO has a slightly different designation for these variants as shown in Table 2. B.1.1.7 UK were shown to be more contagious (>50%) and lethal than the wild form. B.1.351 South African strain and B.1.1.28 Brazil strain had E484K mutation that possibly conferred escape immunity against previous natural infection or vaccine induced immunity [168, 169]. India witnessed a huge surge in daily cases in early May 2021 (>4,00,000 cases/day) possibly due to delta variant B.1.617.2. The delta variant became globally predominant during July-Sept. 2021. Itwas among the variants with highest spike mutations and it was first declared as a variant of concern by Public Health England on 7th May 2021 [170]. The list of major genetic variants of SARS-CoV-2 and their impact on virulence as well as on vaccine induced immunity is shown in Table 3.
Table 2.
Currently designated Variants of concern, Variants of interest and Variants under Monitoring by World Health Organization (as on 4th Oct. 2021).
| Designation | WHO label |
Pango
lineage |
GISAID clade |
Nextstrain
clade |
Country of origin | Date of designation |
|---|---|---|---|---|---|---|
| Variants of concern (VOC) | Alpha | B.1.1.7 | GRY | 20I (V1) | United Kingdom | 18th Dec. 2020 |
| Beta | B.1.351 | GH/501Y.V2 | 20H (V2) | South Africa | 18th Dec. 2020 | |
| Gamma | P.1 | GR/501Y.V3 | 20J (V3) | Brazil |
11th Jan. 2021 | |
| Delta | B.1.617.2 | G/478K.V1 | 21A | India | 11th May 2021 | |
| Variants of Interest | Lambda | C.37 | GR/452Q.V1 | 21G | Peru | 14th Jun. 2021 |
| Mu | B.1.621 | GH | 21H | Colombia | 30th Aug. 2021 | |
| Variants Under Monitoring | R.1 | GR | – | Multiple countries |
7th Apr. 2021 | |
| B.1.466.2 | GH | – | Indonesia |
28th Apr. 2021 | ||
| B.1.1.318 | GR | – | Multiple countries |
02th Jun. 2021 | ||
| B.1.1.519 | GR | 20B/S.732A | Multiple countries |
02th Jun. 2021 | ||
| C.36.3 | GR | – | Multiple countries |
16th Jun. 2021 | ||
| B.1.214.2 | G | – | Multiple countries | 30th Jun. 2021 | ||
| Epsilon | B.1.427 B.1.429 |
GH/452R.V1 | 21C | United States of America | 6th Jul. 2021 |
|
| B.1.1.523 | GR | – | Multiple countries | 14th Jul. 2021 | ||
| B.1.619 | G | 20A/S.126A | Multiple countries | 14th Jul. 2021 | ||
| B.1.620 | G | – | Multiple countries | 14th Jul. 2021 | ||
| C.1.2 | GR | – | South Africa | 1st Sep. 2021 | ||
| Kappa | B.1.617.1 | G/452R.V3 | 21B | India | 20th Sep. 2021 | |
| Lota | B.1.526 | GH/253G.V1 | 21F | United States of America | 20th Sep. 2021 | |
| Eta | B.1.525 | G/484K.V3 | 21D | Multiple countries |
20th Sep. 2021 |
Table 3.
List of emerging COVID-19 variants and their potential impact on virulence and vaccine induced immunity.
|
Emerging COVID-19 variants |
Origin | Gene/Mutation | Important amino acid replacement | Impact on virulence and vaccine |
|---|---|---|---|---|
| B.1 | Unknown | A23403G | D614Gin spike protein | Increased infectivity but no effect on severity of disease or vaccine efficiency |
| 20A.EU1, B.1.177 | Spain | C22227T, C28932T, G29645T | A222V and D614G in spike protein | Increased infectivity; Slightly less effective neutralization by antibodies. |
| 20A.EU2 | France | C4543T, G5629T, G22992A | S477N in spike protein | modest increase in infectivity and ACE binding |
| Cluster 5 variant/B.1.1.298 | Denmark | del69_70 | Y453F, I692V, M1229I in spike proteins | Convalescent sera showed reduced neutralization activity; enhanced ACE-2 affinity; Human cases rare |
|
B.1.1.7 |
UK | 14 non-synonymous; 6 synonymous and 3 deletions | E484K, N501Y, D614G, P681H, (7 mutations and 2 deletion in spike protein) | Enhanced ACE-2 binding PLUS 50% increased infectivity PLUS modestdecline in neutralization activity of sera from previous SARS-CoV-2 exposure/post vaccination |
| B.1.351 | South Africa | 12 non-synonymous mutations and two deletion |
E484K, N501Y, K417N, D614G, A701V (9 mutations and 1 deletion in spike protein) |
Enhanced ACE-2 binding PLUS 50% increased infectivity PLUS significant decline in neutralization activity of sera from previous SARS-CoV-2 exposure/post vaccination |
|
B.1.1.28/P.1 |
Brazil | 17 non-synonymous mutations; 1 deletion; 4 synonymous mutations |
E484K, K417N/T, N501Y, D614G (12 mutations in spike protein) |
Enhanced ACE-2 binding PLUS increased infectivity PLUS significant decline in neutralization activity of sera from previous SARS-CoV-2 exposure/post vaccination |
| P.2 | Brazil | 10 mutations | E484K, D614G | Decline in neutralization activity of sera from previous SARS-CoV-2 exposure/post vaccination |
| B.1.526 | New York, USA | 15 mutations and 1 deletion | E484K, D614G, S477N, A701V | Decline in neutralization activity of sera from previous SARS-CoV-2 exposure/post vaccination |
| B.1.525 | New York, USA | 12 mutations and 5 deletions | E484K, D614G, Q677H, F888L | significant decline in neutralization activity of sera from previous SARS-CoV-2 exposure/post vaccination |
| B.1.427 | California, USA | 11 mutations | L452R, D614G, S13I, W152C, | 20% higher infectivity; declined neutralization activity of sera from post |
| B.1.429 | California, USA | 10 mutations | L452R, D614G, S13I, W152C, | 20% higher infectivity declined neutralization activity of sera from post vaccination |
| B.1.617.2 | India | 25 mutations | T19R, G142D, E156G, del157/158, L452R, T478K, D614G, P681R, D950N | Highly transmissible to become the predominant variant globally. Potentially declined neutralization activity of sera from post vaccination |
Note: D614G enhanced infectivity; N501Y enhanced ACE-2 affinity and E484K enhanced escape from antibody neutralization.
Emerging genetic variants could be associated with reinfections and could cause an elevation in COVID-19 cases globally. Manaus city of Brazil achieved 76% COVID-19 sero-prevalence in October 2020, but it experienced second COVID-19 wave in Dec 2020–Jan 2021 [19]. 42% of COVID-19 patients in Manaus and 51% of COVID-19 patients in Amazonas state had E484K P.1 lineage in mid Dec 2020 [171, 172] E484K P.1 lineage drastically increased to 91% in Jan 2021 in Amazonas state, and E484K spike mutation has been found in at least 3 Brazilian reinfection cases [22, 173, 174]. It may not necessarily mean that only genetic variants of SARS-CoV-2 would cause reinfection as coronaviruses, in general, are known to cause reinfection multiple times in a year [175]. These synchronic, monophyletic set of lineage-representatives have been defined as clades [176]. Although SARS-CoV-2 reinfection clade has been shown to be different in most of the reported reinfection cases but in few exceptional cases, for instance, the reinfection case from Nevada case, same clade was found in both the first and second episode of infection with a variation of 6 single nucleotides and 1 multi nucleotide in its non-spike genes [43]. Likewise, reinfection with the same SARS-CoV-2 clade has also been reported from India [177].
The evolving SARS-CoV-2 variants are raising serious concerns, although these variants may not affect the RT-PCR based diagnosis as most PCR test kits detect multiple non-spike targets (i.e., genes for ORF1ab, RNA-dependent RNA polymerase, N protein, E protein). Yet, it is imperative to continuously evaluate and monitor routine PCR tests as new variants emerge to prevent the risk of false negative COVID-19 diagnosis.
COVID-19 vaccines
Although the process of vaccine development needs years of research and testing before being marketed, yet the COVID-19 epidemic in 2020 embarked the scientists around the globe to race against the time. At present, more than 70 vaccines have entered clinical trials on humans and nearly 20 have touched the final testing stage. A dozen vaccines now have been authorized around the globe; many more are still in their development phase. The messenger RNA (mRNA) based coronavirus vaccines “Comiranaty” and “Moderna” targeting the spike protein announced by Pfizer-BioNTech and Moderna-National Institute of Health (NIH) have an efficacy rate of 90% and 94.5% respectively. Both the vaccines got their emergency use authorization in the United States in December 2020. Another DNA-based vaccine approach was adopted independently by Gamealeya Research Institute (Russia), the University of Oxford in collaboration with AstraZeneca, Johnson & Johnson, and CanSino Biologics in partnership with the Academy of Military Medical Sciences, China respectively. Herein “Sputnik V,” “AZD1222,” (also known as “Covishield”), “Ad26.COV2.S” and “Convidecia” were developed by genetically modifying adenoviruses that carry genes encoding the spike protein against SARS-CoV-2. A vaccine based on the inactivated form of coronavirus was followed independently by the Beijing Institute of Biological Products (“BBIBP-CorV”), Sinovac Biotech (“CoronaVac”), and Bharat Biotech in collaboration with the Indian Council of Medical Research and National Institute of Virology, India (“Covaxin”). These vaccines have been reported to have an efficacy rate of 72.51%, 50.38%, and 81% respectively [178]. In February 2021, the World Health Organization (WHO) announced the emergency use of the AstraZeneca vaccine in adults and the vaccine is now being administered in many countries across the globe.
As the development and distribution of vaccines accelerated globally, the virus also continues to accrue mutations in its genome. In a recent yet-to-be peer-reviewed study, Wibmer et al. showed that SARS-CoV-2 501Y.V2, another novel lineage of the coronavirus harbored mutations in nine parts of its spike protein and this lineage completely escaped from neutralizing antibodies in COVID-19 convalescent plasma. This novel strain that emerged rapidly during the second half of 2020 has been reported to be extremely resistant to neutralizing antibodies elicited by previous circulating viral strains and highlighted the prospect of reinfection with antigenically distinct variants. They have also been predicted to have reduced efficacy against spike-protein-based vaccine-induced antibodies [179]. Some of these mutations provide fitness advantage and rapid transmission of the particular viral clade [164], such as the viral variant with an Asp614Gly spike substitution and the recently dispersed variant of concern from the United Kingdom (B.1.1.7) [180]. Results from a recent pre-print study indicates that the Oxford-AstraZeneca vaccine efficacy is preserved against the B.1.1.7 variant of SARS-CoV-2 [181]. However, another pre-print study from South Africa refuted the previously made claims by reporting the efficacy of the same vaccine (AZD1222) to be only 21.9% against the original strain causing mild or moderate COVID-19, and 10.4% against the viral B.1.351 variant [20]. Despite these controversial reports, the vaccine makers are still claiming the effectiveness of their vaccines against the emerging variants, however, efforts should be made in redesigning vaccines that are more effective against the variants. These variants pose the highest risk of reinfection as reports indicate immune escape in documented reinfection cases with a viral variant at rates similar to uninfected people (Figure 3) [180]. As the virus continues to evolve, reinfection cases are also going to occur more frequently as the variants become more prevalent and adopt mechanisms in escaping the natural immunity generated against the previous infection [182]. Although, neutralizing antibodies against SARS-CoV-2 provides some level of protection against reinfection, however, a reduction in the neutralizing potential against viral variants over time may indicate a reduced capacity to protect against repeat infections [183]. More specifically, there is no data available on how these novel viral variants and their mutations will impact vaccination and the rate of SARS-CoV-2 reinfection. The pressing question whether reinfection occurs due to the inefficacy of the immune system or from the enhanced viral virulence acquired via SARS-CoV-2 genomic alterations needs to be addressed by the scientific community. Hence, it is crucial to monitor virus evolution through genetic surveillance programs highlighting transmissibility, immune evasion, severity, and vaccine escape. It is also necessary to test the neutralizing ability of antibodies against virus variants generated in response to vaccination and investigate the dynamic immune response in individuals infected with viral variants. This will allow better determination of the magnitude and breadth of immune response during reinfection and its protective effect on transmission. The efficacy of mRNA and adenoviral vector-based vaccines targeting the spike glycoprotein could be challenged by emerging variants and could impact the long-term immunity generated by natural infection and vaccination. Therefore, with new variants on the horizons, vaccine manufacturers have already begun developing booster shots/or second-generation vaccines, and the likelihood of having a bivalent or multivalent vaccine harboring both the original strain as well as the escape viral variants are far greater in the future. The current vaccines approved in various countries or in pipeline are tabulated in Table 4. The landscape of vaccination in India is shown in Figure 5.
Table 4.
Current COVID-19 vaccines under use and in pipeline.
| Vaccine Name | Manufacturer | Vaccine basis | Vaccine approval |
|---|---|---|---|
| Comirnaty (also known as tozinameran or BNT162b2) |
Pfizer-BioNTech | mRNA |
Approved in U.S |
| mRNA-1273 or Spikevax | Moderna | mRNA | Approved in Switzerland. |
| ZyCoV-D | Zydus | DNA | Emergency use in India |
| Sputnik V (also known as Gam-Covid-Vac) |
Gamaleya Research Institute | Ad5 and Ad26 | Emergency use in Russia |
| Vaxzevria (also known as AZD1222, or Covishield in India) |
University of Oxford-AstraZeneca | ChAdOx1 | Approved for use in Brazil. Emergency use in other countries |
| Convidecia (also known as Ad5-nCoV) |
CanSino Biologics- Academy of Military Medical Sciences. |
Ad5 | Approved for use in China |
| Ad26.COV2.S | Johnson & Johnson | Ad26 | Emergency use in Andorra, Australia, Bahamas, other countries |
| EpiVacCorona, Aurora-CoV | Vector Institute | peptides | Approved for use in Turkmenistan. |
| ZF2001, Zifivax | Anhui Zhifei Longcom-Institute of Medical Biology at the Chinese Academy of Medical Sciences | Adjuvant-RBD | Emergency use in China, Uzbekistan. |
| Soberana 2, or PastoCoVac (in Iran) | Finlay Vaccine Institute | spike protein fused to a standard tetanus vaccine | Emergency use in Cuba, Iran, Nicaragua |
| Abdala | Center for Genetic Engineering and Biotechnology of Cuba | RBD | Emergency use in Cuba, Nicaragua, Venezuela, Vietnam. |
| BBIBP-CorV | Beijing Institute of Biological Products-Sinopharm | inactivated | Approved for use in Bahrain, China, United Arab Emirates. |
| CoronaVac (formerly PiCoVacc) | Sinovac | Inactivated | Approved for use in China. |
| Wuhan Vaccine | Sinopharm-Wuhan | Inactivated | Approved for use in China. |
| Covaxin (also known as BBV152 A, B, C) | Bharat Biotech- Indian Council of Medical Research-National Institute of Virology |
inactivated | Emergecy use in India |
| QazVac | Research Institute for Biological Safety Problems | inactivated | Early use in Kazakhstan, Kyrgyzstan |
| COVIran Barekat | Shafa Pharmed Pars | inactivated | Emergency use in Iran. |
| CoviVac | Chumakov Center | inactivated | Early use in Russia. |
| Vaccines in pipeline | |||
| AG0302-COVID19 | AnGes-Osaka University-Takara Bio | DNA | |
| HGC019 | Gennova Biopharmaceuticals-HDT Bio | self-amplifying RNA | |
| COVID-eVax | Takis Biotech-Rottapharm Biotech, | DNA | |
| DS-5670 | Daiichi Sankyo-University of Tokyo | mRNA | |
| EXG-5003 | Elixirgen Therapeutics | RNA | |
| EG-COVID | Eyegene | mRNA | |
| ChulaCov19 | Chulalongkorn University | mRNA | |
| Covigenix VAX-001 | Entos Pharmaceuticals | DNA | |
| CORVax12 | OncoSec Immunotherapies | loop of DNA that encodes both the spike protein and IL-12 | |
| COVIGEN | BioNet-Asia-Technovalia | DNA | |
| SCOV1, SCOV2 | Scancell | DNA | |
| AdCLD-CoV19 | Cellid | Ad5 and Ad35 | |
| BCD-250 | BIOCAD | adenovirus-associated virus AAV-5 | |
| – | ImmunityBio | Ad5 | |
| Meissa Vaccines | RSV | ||
| CVXGA1 | University of Georgia-University of Iowa | canine parainfluenza virus | |
| COVID-19-EDV | EnGeneIC | ||
| Soberana 1 | Finlay Vaccine Institute | RBD | |
| EuCorVac-19 | EuBiologics | protein | |
| VBI-2902a | VBI Vaccines | virus-like protein shells | |
| AKS-452 | Akston Biosciences | RBD | |
| IVX-411 | Icosavax-Seqirus | RBD | |
| QazCoVac-P | Research Institute for Biological Safety Problems | protein | |
| St. Petersburg Scientific Research Institute of Vaccines and Sera | protein subunit | ||
| Mambisa | Center for Genetic Engineering and Biotechnology of Cuba | RBD | |
Source: The New York Times.
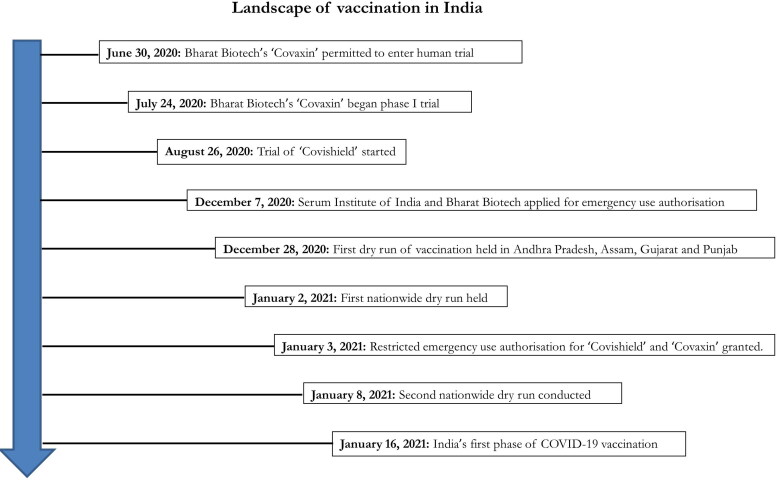
Figure 5.
Landscape of vaccination in India.
Taken together, although the evolving virus is posing a challenge, it doesn’t necessarily imply the failure of scientists all over the world to produce an effective vaccine. The present vaccines although may prove less effective to the evolving virus, yet these are capable of neutralizing the virus to a certain extent. Additionally, these vaccines boost the immune system: cell mediated as well as humoral immune response leading to reduction in severity of disease during reinfection.
The need to vaccinate children below 18 years
The question of COVID-19 vaccination in children below 18 years of age has vexed the scientists and health professionals all over the world since the COVID-19 vaccination program were initiated. One the one hand, relatively less severity, morbidity and mortality reported in children and young adults all over the world demands that the older people and most vulnerable population should get vaccinated first. On the other hand, children and young adults who although suffered from mild to moderate COVID-19 disease during the pandemic could act as carrier to cause disease in older people. Since the results of studies in adults cannot be extrapolated in children, and the safety data of vaccination in children is yet to be made available, this population has to wait till the safety and efficacy of COVID-19 vaccination could be established. The issue that urges to vaccinate children is education. Since the start of the epidemic, the educational institutions have been closed for the safety. However, the risk of transmission of COVID-19 in educational institutions can be minimized by vaccinating the teachers and other staff working in the school.
Breakthrough infection and reinfection: the ground report
As of now, there are >1.07 lakh suspected cases and 362 confirmed cases of SARS-CoV-2 re-infection (COVID-19 reinfection tracker; BNO news). The breakthrough infection following COVID-19 vaccines varies from country to country depending upon the type of vaccine administered. As in United States till 27th September 2021, 22,115 breakthrough cases were reported after vaccinating >183 million people against COVID-19 (CDC, COVID-19 breakthrough cases investigations and reporting). Out of 22,115 breakthrough cases, 5,226 (23.6%) were fatal. In India, 0.26 million people tested COVID-19 positive after administration of 531.4 million vaccine doses as on 3rd August 2021. Among them, 0.17 million got breakthrough infection in just one vaccine dose while 87,049 breakthrough infections were reported after full vaccination (NCDC, Delhi).
The viral load is substantially reduced for breakthrough infection occurring 12–37 days after the first dose of BNT162b2 mRNA vaccine as demonstrated in the analysis of the real-world data set [184] and thereby suppress the onward transmission. The quantification of viral load and comparison with first infection is not available in majority of reinfection cases.
The age of the patient could be critically important in deaths during breakthrough infection where CDC reported that 86% deaths were in people aged 65 or more during breakthrough infection (CDC, COVID-19 breakthrough cases investigations and reporting). This is contrary to SARS-CoV-2 infection and reinfection where deaths could be seen in a wide range of age. Unlike other age groups, people aged 65 or more may have substantially reduced generation of T cells which impairs the cellular immune response during the breakthrough infection.
The virulence of viral strain (same or different) plays an important role in deciding the outcome of breakthrough infection. The delta variant is associated with high viral load, prolonged PCR positivity and low levels of vaccine-induced neutralizing antibodies even if the symptoms are mild or asymptomatic during breakthrough infection following Oxford-AstraZeneca vaccination [185]. It suggests that virulent strain may still cause onward transmission even after full vaccination. UK public health England has already warned of increased risk of COVID-19 reinfection with the delta variant.
The strength of host immunity is overall potent in breakthrough infection, so moderate or severe symptoms are rarely seen, and majority of studies have mostly reported asymptomatic and mild symptoms. The severity of the disease during possible reinfection declined or remained unchanged in 97.3% in a meta-analysis of 1128 patients [186]. However, severe symptoms were observed when the interval between infection and reinfection was shorter (less than 2 months) in another analysis of 123 patients of repeat SARS-CoV-2 [187].
There is scarcity of information on the breakthrough infection in those recovered and having received at least one dose of vaccination. Such cases could be rare due to precautions taken to ensure suitable time interval of 2–3 months between COVID-19 infection and first vaccine dose. In such cases, even the first dose of vaccine can rapidly activate the memory response. Further, among previously infected individuals, the non-vaccinated ones are associated with 2.34 times the odds of reinfection in comparison to the fully vaccinated ones [188]. This emphasis the need to vaccinate all adults irrespective of their past exposure.
Conclusion
As stated by Vardhana and Wolchok, COVID-19 is a case of innate immune hyper activation and adaptive immune dysregulation [189]. The road to a robust protective immune response against Covid-19, apparently, is fraught with potholes of reinfections. Enhancement of repertoire of the adaptive immune response is solely based on repeated exposure to the same or similar pathogenic microorganisms fulfilling Darwin’s hypothesis of survival of the fittest. The recent increase in SARS-CoV-2 reinfections, even in fully vaccinated individuals, is a cause for major concern. Rapid antigenic drift in emerging variants combined with the decline of post-covid-19 antibodies within a short span of 6–8 months may confuse immunological memory and the existence of comorbidities like chronic obstructive airways disease with reduced ciliary function, age-related senescence, uncontrolled diabetes may pause major risk to reinfection also. Failure of the respiratory immune system in neutralizing the virus resulting in escape into the alveoli, a highly specialized organ with reduced immune components, will always remain a challenge. The fact that ACE2 receptors, the binding ligand of the virus, are expressed in the alveolar epithelial cells is another complicating matter since some viral particles of SARS-CoV-2 may continuously escape to alveoli even when most of them are trapped in URT. Innovative ways that may prime mucosal immune response associated with the respiratory tract, both innate as well as the adaptive immune system, is the need of the hour to prevent SARS-CoV-2 reinfections. Therefore, it is crucial to have worldwide surveillance programs for emerging variants that are likely to cause immune evasion and delineate the viral attributes responsible for modulating the protective immune response in the host. Involvement of the gastrointestinal tract in the transmission of SARS-CoV-2 infection has garnered a lot of attention from the scientific community in the recent days. Besides fever and cold that are the most commonly reported symptoms of COVID-19, an increasing number of individuals had been presenting with nausea, vomiting and diarrhea suggesting that SARS-CoV-2 is slowly evolving with enhanced tropism for the gastrointestinal tract [190, 191]. The intestinal mucosal with biggest surface epithelium and its accompanying larger volume lymphoid tissues (both organized and diffuse) offer a level playing field for the virus to compromise the mucosal immunity as well as amplify their spillover effect on the systemic immune response which could predispose individuals to reinfections many times over. From the diagnostic point of view, while the limited number of respiratory tract epithelial cells and their lymphoid tissue, in comparison to intestinal mucosa, may control the viral replication leading to a negative RT-PCR test, the war of the virus may still raze on in the intestinal tract shedding large number in the stool, favoring fecal-oral transmission. In addition, dysbiosis in gut microbiota by SARS-CoV-2 infection significantly impacts the lung immunity by modulating the innate and adaptive immune responses, and it also influences the disease severity. In recovered COVID-19 patients, this dysregulated gut-lung axis can play a prominent role in persistent inflammation increasing the likelihood of reinfection. The development of many highly efficacious vaccines over the last one hundred years has not fully strengthened our understanding of the immunological correlates of protection, baring a few e.g., pneumococcal vaccine (opsonophagocytic assay), Hepatitis B vaccine (Neutralizing antibodies). The emergence of a large number of variants across the globe, some more virulent with higher transmission rates, may derail the current vaccination strategies, and these new viral variants with their distinct capacity to evade polyclonal antibody responses could potentially increase the rate of SARS-CoV-2 reinfections. Therefore, search for a utopic broadly neutralizing antibody, as documented in certain long-term survivors of pediatric HIV-infected children is highly enticing. Detailed analysis of the complete genome of the SARS-CoV-2 virus may delineate epitopes (both B and T cells) that could offer cross-protection against variants and prevent reinfections; hence a vaccine composed of a cocktail of antigens may offer a ray of hope. Likewise, oral vaccination approach for instance the nasal spray along with other oral vaccines (virus-like particles (VLPs)/attenuated virus with modified genome) might boost the mucosal immune response with local memory B cells and tissue-resident memory T cells and may reduce the chances of repeat infections. Studies analyzing the clinical and immunological profile of SARS-CoV-2 reinfection cases should be integrated with vaccination follow-up investigations to thoroughly evaluate correlates of immune protection against SARS-CoV-2.
The light at the end of the tunnel, we ardently hope, is not far away with the arrival of more and more information regarding the host-viral relationship which may help to develop better strategies to prevent reinfection with this deadly virus. Patients who have recovered from documented COVID-19 are mostly protected but reinfection is still a possibility therefore, recovered people should comply with all the control measures as the fight against COVID-19 is a marathon and not a sprint. Till then, wearing a mask, sanitization of the hands, social distancing, and following COVID-appropriate behaviors is the best way to keep the virus at bay.
Disclaimer
It was not possible in this review to cite all original research studies relevant to the topic. We apologize to those authors whose published work related to COVID-19 and reinfection could not be referenced.
Funding Statement
The authors are thankful to Indian Council of Medical Research, New Delhi for financial support (8/3/2019-ECD-2 and 5/8-I(10)/2020-ECD-2).
Declaration of Interest
Authors declared no potential conflicts of interest.
References
- 1.Chen Z, John Wherry E.. T cell responses in patients with COVID-19. Nat Rev Immunol 2020;20(9):529–536. doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taefehshokr N, Taefehshokr S, Heit B.. Mechanisms of dysregulated humoral and cellular immunity by SARS-CoV-2. Pathogens 2020;9(12):1027. doi: 10.3390/pathogens9121027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Wit E, van Doremalen N, Falzarano D, Munster VJ.. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aran D, Beachler DC, Lanes S, Overhage JM.. Prior presumed coronavirus infection reduces COVID-19 risk: A cohort study. J Infect 2020;81(6):923–930. doi: 10.1016/j.jinf.2020.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng OW, Chia A, Tan AT, Jadi RS, et al. . Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34(17):2008–2014. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sette A, Crotty S.. Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat Rev Immunol. 2020;20(8):457–458. doi: 10.1038/s41577-020-0389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doshi P. Covid-19: Do many people have pre-existing immunity? BMJ. 2020;370:m3563. doi: 10.1136/bmj.m3563. [DOI] [PubMed] [Google Scholar]
- 8.Stephens DS, McElrath MJ.. COVID-19 and the path to immunity. JAMA. 2020;324(13):1279–1281. doi: 10.1001/jama.2020.16656. [DOI] [PubMed] [Google Scholar]
- 9.Dan JM, Mateus J, Kato Y, et al. . Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371(6529):eabf4063.doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edridge AWD, Kaczorowska J, Hoste ACR, et al. . Seasonal coronavirus protective immunity is short-lasting. Nat Med. 2020;26(11):1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- 11.Muecksch F, Wise H, Batchelor B, et al. . Longitudinal serological analysis and neutralizing antibody levels in coronavirus disease 2019 convalescent patients. J Infect Dis. 2021;223(3):389–398. doi: 10.1093/infdis/jiaa659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Bi Y, Xiao H, et al. . A novel DNA and protein combination COVID-19 vaccine formulation provides full protection against SARS-CoV-2 in rhesus macaques. Emerg Microbes Infect 2021;10(1):342–355. doi: 10.1080/22221751.2021.1887767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu J, Tostanoski LH, Peter L, et al. . DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369(6505):806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang AT, Garcia-Carreras B, Hitchings MDT, et al. A systematic review of antibody mediated immunity to coronaviruses: antibody kinetics, correlates of protection, and association of antibody responses with severity of disease. medRxiv: the preprint server for health sciences. 2020. [DOI] [PMC free article] [PubMed]
- 15.Hall VJ, Foulkes S, Charlett A, SIREN Study Group, et al.. SARS-CoV-2 infection rates of antibody-positive compared with antibody-negative health-care workers in England: a large, multicentre, prospective cohort study (SIREN). Lancet. 2021;397(10283):1459–1469. doi: 10.1016/S0140-6736(21)00675-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukherjee A, Anand T, Agarwal A, et al. . SARS-CoV-2 re-infection: development of an epidemiological definition from India. Epidemiol Infect. 2021;149:e82. doi: 10.1017/S0950268821000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Volz E, Mishra S, Chand M, COVID-19 Genomics UK (COG-UK) Consortium, et al.. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593(7858):266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- 18.Tegally H, Wilkinson E, Giovanetti M, et al. . Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592(7854):438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 19.Sabino EC, Buss LF, Carvalho MPS, et al. . Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397(10273):452–455. doi: 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madhi SA, Baillie V, Cutland CL, Wits-VIDA COVID Group, et al.. Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N Engl J Med. 2021;384(20):1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irfan N, Chagla Z.. In South Africa, a 2-dose Oxford/AZ vaccine did not prevent mild to moderate COVID-19 (cases mainly B.1.351 variant). Ann Intern Med. 2021;174(5):JC50. doi: 10.7326/ACPJ202105180-050. [DOI] [PubMed] [Google Scholar]
- 22.Nonaka CKV, Franco MM, Graf T, et al. . Genomic evidence of SARS-CoV-2 reinfection involving E484K spike mutation, Brazil. Emerg Infect Dis. 2021;27(5):1522–1524. doi: 10.3201/eid2705.210191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srivastava S, Banu S, Singh P, Sowpati DT, Mishra RK.. SARS-CoV-2 genomics: An Indian perspective on sequencing viral variants. J Biosci. 2021;46(1):22. doi: 10.1007/s12038-021-00145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yahav D, Yelin D, Eckerle I, et al. . Definitions for coronavirus disease 2019 reinfection, relapse and PCR re-positivity. Clin Microbiol Infect. 2021;27(3):315–318. doi: 10.1016/j.cmi.2020.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ECDC . Reinfection with SARS-CoV: considerations for public health response. 2020.
- 26.Hoang VT, Dao TL, Gautret P.. Recurrence of positive SARS-CoV-2 in patients recovered from COVID-19. J Med Virol. 2020;92(11):2366–2367. doi: 10.1002/jmv.26056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oran DP, Topol EJ. Prevalence of asymptomatic SARS-CoV-2 infection: a narrative review. Ann Intern Med. 2020;173(5):362–367. doi: 10.7326/M20-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Kang H, Liu X, Tong Z.. Asymptomatic cases with SARS-CoV-2 infection. J Med Virol. 2020;92(9):1401–1403. doi: 10.1002/jmv.25990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothe C, Schunk M, Sothmann P, et al. . Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382(10):970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tahamtan A, Ardebili A.. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev Mol Diagn. 2020;20(5):453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arevalo-Rodriguez I, Buitrago-Garcia D, Simancas-Racines D, et al. . False-negative results of initial RT-PCR assays for COVID-19: A systematic review. PloS One. 2020;15(12):e0242958. doi: 10.1371/journal.pone.0242958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang X, Luo M, Zou Z, Wang X, Chen C, Qiu J.. Asymptomatic SARS-CoV-2 infected case with viral detection positive in stool but negative in nasopharyngeal samples lasts for 42 days. J Med Virol. 2020;92(10):1807–1809. doi: 10.1002/jmv.25941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prakash S, Shukla S, Mishra H, et al. . SARS-CoV-2-RNA persists longer in faecal sample as compared to nasal and throat swab samples of COVID-19 patients’; an observational study. Indian J Med Microbiol. 2021;39(1):122–124. doi: 10.1016/j.ijmmb.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ge R, Chen Z, Liu X, Zhang Q, Zhu G, Xiao Q.. Positive stool test results suggest that the discharge standard for COVID-19 needs improvement. Jpn J Infect Dis. 2021;74(1):76–78. doi: 10.7883/yoken.JJID.2020.265. [DOI] [PubMed] [Google Scholar]
- 35.Bwire GM, Majigo MV, Njiro BJ, Mawazo A.. Detection profile of SARS-CoV-2 using RT-PCR in different types of clinical specimens: A systematic review and meta-analysis. J Med Virol. 2021;93(2):719–725. doi: 10.1002/jmv.26349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Y, Guo C, Tang L, et al. . Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5(5):434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Falahi S, Kenarkoohi A.. COVID-19 reinfection: prolonged shedding or true reinfection? New Microbes New Infect. 2020;38:100812. doi: 10.1016/j.nmni.2020.100812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rani PR, Imran M, Lakshmi JV, et al. . Symptomatic reinfection of SARS-CoV-2 with spike protein variant N440K associated with immune escape. J Med Virol. 2021;93(7):4163–4165. doi: 10.1002/jmv.26997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kapoor R, Nair RK, Nayan N, Bhalla S, Singh J.. Reinfection or reactivation of coronavirus-19 in patients with hematologic malignancies: case report series. SN Compr Clin Med. 2021;3(2):670–675. doi: 10.1007/s42399-021-00790-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye G, Pan Z, Pan Y, et al. . Clinical characteristics of severe acute respiratory syndrome coronavirus 2 reactivation. J Infect. 2020;80(5):e14–e7. doi: 10.1016/j.jinf.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yadav SP, Thakkar D, Bhoyar RC, et al. . Asymptomatic reactivation of SARS-CoV-2 in a child with neuroblastoma characterised by whole genome sequencing. IDCases. 2021;23:e01018. doi: 10.1016/j.idcr.2020.e01018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siqueira JD, Goes LR, Alves BM, et al. . Distinguishing SARS-CoV-2 bonafide re-infection from pre-existing minor variant reactivation. Infect Genet Evol. 2021;90:104772. doi: 10.1016/j.meegid.2021.104772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tillett RL, Sevinsky JR, Hartley PD, et al. . Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis. 2021;21(1):52–58. doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hansen CH, Michlmayr D, Gubbels SM, Molbak K, Ethelberg S.. Assessment of protection against reinfection with SARS-CoV-2 among 4 million PCR-tested individuals in Denmark in 2020: a population-level observational study. Lancet. 2021;397(10280):1204–1212. doi: 10.1016/S0140-6736(21)00575-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lumley SF, O’Donnell D, Stoesser NE, Oxford University Hospitals Staff Testing Group, et al.. Antibody status and incidence of SARS-CoV-2 infection in health care workers. N Engl J Med. 2021;384(6):533–540. doi: 10.1056/NEJMoa2034545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harvey RA, Rassen JA, Kabelac CA, et al. . Association of SARS-CoV-2 seropositive antibody test with risk of future infection. JAMA Intern Med. 2021;181(5):672–679. doi: 10.1001/jamainternmed.2021.0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abu-Raddad LJ, Chemaitelly H, Malek JA, Ahmed AA, et al. Assessment of the risk of SARS-CoV-2 reinfection in an intense re-exposure setting. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2020. [DOI] [PMC free article] [PubMed]
- 48.To KK, Hung IF, Ip JD, et al. . COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis. 2021;73(9):e2946–e2951. doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torres DA, Ribeiro L, Riello A, Horovitz DDG, Pinto LFR, Croda J.. Reinfection of COVID-19 after 3 months with a distinct and more aggressive clinical presentation: case report. J Med Virol. 2021;93(4):1857–1859. doi: 10.1002/jmv.26637. [DOI] [PubMed] [Google Scholar]
- 50.Al-Thani MH, Farag E, Bertollini R, et al. Seroprevalence of SARS-CoV-2 infection in the craft and manual worker population of Qatar. medRxiv: the preprint server for health sciences. 2020. 2020.11.24.20237719.
- 51.Netea MG, Giamarellos-Bourboulis EJ, Dominguez-Andres J, et al. . Trained immunity: a tool for reducing susceptibility to and the severity of SARS-CoV-2 infection. Cell. 2020;181(5):969–977. doi: 10.1016/j.cell.2020.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawai T, Akira S.. Innate immune recognition of viral infection. Nat Immunol. 2006;7(2):131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 53.Streicher F, Jouvenet N.. Stimulation of innate immunity by host and viral RNAs. Trends Immunol. 2019;40(12):1134–1148. doi: 10.1016/j.it.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 54.Rokni M, Ghasemi V, Tavakoli Z.. Immune responses and pathogenesis of SARS-CoV-2 during an outbreak in Iran: comparison with SARS and MERS. Rev Med Virol. 2020;30(3):e2107. doi: 10.1002/rmv.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoo JK, Kim TS, Hufford MM, Braciale TJ.. Viral infection of the lung: host response and sequelae. J Allergy Clin Immunol. 2013;132(6):1263–1276; quiz 77. doi: 10.1016/j.jaci.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li G, Fan Y, Lai Y, et al. . Coronavirus infections and immune responses. J Med Virol. 2020;92(4):424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carty M, Guy C, Bowie AG.. Detection of viral infections by innate immunity. Biochem Pharmacol. 2021;183:114316. doi: 10.1016/j.bcp.2020.114316. [DOI] [PubMed] [Google Scholar]
- 58.Kikkert M. Innate immune evasion by human respiratory RNA viruses. J Innate Immun. 2020;12(1):4–20. doi: 10.1159/000503030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park A, Iwasaki A.. Type I and Type III interferons – induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe. 2020;27(6):870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chau AS, Weber AG, Maria NI, et al. . The longitudinal immune response to coronavirus disease 2019: chasing the cytokine storm. Arthritis Rheumatol. 2021;73(1):23–35. doi: 10.1002/art.41526. [DOI] [PubMed] [Google Scholar]
- 61.Hou YJ, Okuda K, Edwards CE, et al. . SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182(2):429–446 e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ortiz ME, Thurman A, Pezzulo AA, et al. . Heterogeneous expression of the SARS-Coronavirus-2 receptor ACE2 in the human respiratory tract. EBioMedicine. 2020;60:102976. doi: 10.1016/j.ebiom.2020.102976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ziegler CGK, Allon SJ, Nyquist SK, HCA Lung Biological Network, et al.. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181(5):1016–1035 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodriguez-Argente F, Alba-Dominguez M, Ortiz-Munoz E, Ortega-Gonzalez A.. Oromucosal immunomodulation as clinical spectrum mitigating factor in SARS-CoV-2 infection. Scand J Immunol. 2021;93(1):e12972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mo P, Xing Y, Xiao Y, et al. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2020. [DOI] [PMC free article] [PubMed]
- 66.Zhan N, Guo Y, Tian S, et al. . Clinical characteristics of COVID-19 complicated with pleural effusion. BMC Infect Dis. 2021;21(1):176. doi: 10.1186/s12879-021-05856-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sungnak W, Huang N, Becavin C, HCA Lung Biological Network, et al.. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26(5):681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miyazawa M. Immunopathogenesis of SARS-CoV-2-induced pneumonia: lessons from influenza virus infection. Inflamm Regen. 2020;40:39. doi: 10.1186/s41232-020-00148-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H.. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mayi BS, Leibowitz JA, Woods AT, Ammon KA, Liu AE, Raja A.. The role of Neuropilin-1 in COVID-19. PLoS Pathog. 2021;17(1):e1009153. doi: 10.1371/journal.ppat.1009153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ulrich H, Pillat MM.. CD147 as a target for COVID-19 treatment: suggested effects of azithromycin and stem cell engagement. Stem Cell Rev Rep. 2020;16(3):434–440. doi: 10.1007/s12015-020-09976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khanmohammadi S, Rezaei N.. Role of toll-like receptors in the pathogenesis of COVID-19. J Med Virol. 2021;93(5):2735–2739. doi: 10.1002/jmv.26826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Angelopoulou A, Alexandris N, Konstantinou E, et al. . Imiquimod – a toll like receptor 7 agonist – is an ideal option for management of COVID 19. Environ Res. 2020;188:109858. doi: 10.1016/j.envres.2020.109858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schultze JL, Aschenbrenner AC.. COVID-19 and the human innate immune system. Cell. 2021;184(7):1671–1692. doi: 10.1016/j.cell.2021.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mazaleuskaya L, Veltrop R, Ikpeze N, Martin-Garcia J, Navas-Martin S.. Protective role of toll-like receptor 3-induced type I interferon in murine coronavirus infection of macrophages. Viruses. 2012;4(5):901–923. doi: 10.3390/v4050901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Choudhury A, Mukherjee S.. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J Med Virol. 2020;92(10):2105–2113. doi: 10.1002/jmv.25987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sohn KM, Lee SG, Kim HJ, et al. . COVID-19 patients upregulate toll-like receptor 4-mediated inflammatory signaling that mimics bacterial sepsis. J Korean Med Sci. 2020;35(38):e343. doi: 10.3346/jkms.2020.35.e343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Khadke S, Ahmed N, Ahmed N, et al. . Harnessing the immune system to overcome cytokine storm and reduce viral load in COVID-19: a review of the phases of illness and therapeutic agents. Virol J. 2020;17(1):154. doi: 10.1186/s12985-020-01415-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang C, Wang Y, Li X, et al. . Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kang YW, Park S, Lee KJ, Moon D, Kim YM, Lee SW.. Understanding the host innate immune responses against SARS-CoV-2 infection and COVID-19 pathogenesis. Immune Netw. 2021;21(1):e1. doi: 10.4110/in.2021.21.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gong J, Dong H, Xia QS, et al. . Correlation analysis between disease severity and inflammation-related parameters in patients with COVID-19: a retrospective study. BMC Infect Dis. 2020;20(1):963. doi: 10.1186/s12879-020-05681-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen X, Zhao B, Qu Y, et al. . Detectable serum severe acute respiratory syndrome coronavirus 2 viral load (RNAemia) is closely correlated with drastically elevated interleukin 6 level in critically ill patients with coronavirus disease 2019. Clin Infect Dis. 2020;71(8):1937–1942. doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Qin C, Zhou L, Hu Z, et al. . Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fu J, Kong J, Wang W, et al. . The clinical implication of dynamic neutrophil to lymphocyte ratio and D-dimer in COVID-19: a retrospective study in Suzhou China. Thrombosis Research. 2020;192:3–8. doi: 10.1016/j.thromres.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu J, Liu Y, Xiang P, et al. . Neutrophil-to-lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage. J Transl Med. 2020;18(1):206. doi: 10.1186/s12967-020-02374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Barnes BJ, Adrover JM, Baxter-Stoltzfus A, et al. . Targeting potential drivers of COVID-19: Neutrophil extracellular traps. J Experim Med. 2020;217(6):e20200652. doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ashraf UM, Abokor AA, Edwards JM, et al. . SARS-CoV-2, ACE2 expression, and systemic organ invasion. Physiol Genom. 2021;53(2):51–60. doi: 10.1152/physiolgenomics.00087.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thompson BT, Chambers RC, Liu KD.. Acute respiratory distress syndrome. N Engl J Med. 2017;377(19):1904–1905. doi: 10.1056/NEJMra1608077. [DOI] [PubMed] [Google Scholar]
- 89.Thoms M, Buschauer R, Ameismeier M, et al. . Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369(6508):1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bouayad A. Innate immune evasion by SARS-CoV-2: comparison with SARS-CoV. Rev Med Virol. 2020;30(6):1–9. doi: 10.1002/rmv.2135. [DOI] [PubMed] [Google Scholar]
- 91.Richter AM, Waterfield E, Jain AK, Canaan AJ, Allison BA, Levy JG.. Liposomal delivery of a photosensitizer, benzoporphyrin derivative monoacid ring A (BPD), to tumor tissue in a mouse tumor model. Photochem Photobiol. 1993;57(6):1000–1006. doi: 10.1111/j.1751-1097.1993.tb02962.x. [DOI] [PubMed] [Google Scholar]
- 92.Scala S, Pacelli R.. Fighting the host reaction to SARS-COv-2 in critically ill patients: the possible contribution of off-label drugs. Front Immunol. 2020;11:1201. doi: 10.3389/fimmu.2020.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Das A, Rana S.. The role of human C5a as a non-genomic target in corticosteroid therapy for management of severe COVID19. Comput Biol Chem. 2021;92:107482. doi: 10.1016/j.compbiolchem.2021.107482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mahmudpour M, Roozbeh J, Keshavarz M, Farrokhi S, Nabipour I.. COVID-19 cytokine storm: the anger of inflammation. Cytokine. 2020;133:155151. doi: 10.1016/j.cyto.2020.155151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Carvelli J, Demaria O, Vely F, Explore COVID-19 Marseille Immunopole group, et al.. Association of COVID-19 inflammation with activation of the C5a-C5aR1 axis. Nature. 2020;588(7836):146–150. doi: 10.1038/s41586-020-2600-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Perico L, Benigni A, Casiraghi F, Ng LFP, Renia L, Remuzzi G.. Immunity, endothelial injury and complement-induced coagulopathy in COVID-19. Nat Rev Nephrol. 2021;17(1):46–64. doi: 10.1038/s41581-020-00357-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Holter JC, Pischke SE, de Boer E, et al. . Systemic complement activation is associated with respiratory failure in COVID-19 hospitalized patients. Proc Natl Acad Sci U S A. 2020;117(40):25018–25025. doi: 10.1073/pnas.2010540117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.April Si X, Talaat M, Xi J.. SARS COV-2 virus-laden droplets coughed from deep lungs: Numerical quantification in a single-path whole respiratory tract geometry. Phys Fluids (1994). 2021;33(2):023306. doi: 10.1063/5.0040914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Greenhalgh T, Jimenez JL, Prather KA, Tufekci Z, Fisman D, Schooley R.. Ten scientific reasons in support of airborne transmission of SARS-CoV-2. Lancet. 2021;397(10285):1603–1605. doi: 10.1016/S0140-6736(21)00869-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.The Lancet Respiratory M . COVID-19 transmission-up in the air. Lancet Respirator Med. 2020;8(12):1159. doi: 10.1016/S2213-2600(20)30514-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Klompas M, Baker MA, Rhee C.. Airborne transmission of SARS-CoV-2: theoretical considerations and available evidence. Jama. 2020;324(5):441–442. doi: 10.1001/jama.2020.12458. [DOI] [PubMed] [Google Scholar]
- 102.Chu H, Chan JF, Wang Y, et al. SARS-CoV-2 induces a more robust innate immune response and replicates less efficiently than SARS-CoV in the human intestines: an ex vivo study with implications on pathogenesis of COVID-19. Cell Mol Gastroenterol Hepatol. 2021;11(3):771–781. doi: 10.1016/j.jcmgh.2020.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shuai H, Chu H, Hou Y, et al. Differential immune activation profile of SARS-CoV-2 and SARS-CoV infection in human lung and intestinal cells: Implications for treatment with IFN-β and IFN inducer. J Infect. 2020;81(4):e1–e10. doi: 10.1016/j.jinf.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Blanco-Melo D, Nilsson-Payant BE, Liu WC, et al. . Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181(5):1036–1045 e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Han Y, Yang L, Duan X, et al. Identification of Candidate COVID-19 Therapeutics using hPSC-derived Lung Organoids. bioRxiv: the preprint server for biology. 2020.
- 106.Netea MG, van der Meer JW.. Trained immunity: an ancient way of remembering. Cell Host Microbe. 2017;21(3):297–300. doi: 10.1016/j.chom.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 107.Netea MG, Dominguez-Andres J, Barreiro LB, et al. . Defining trained immunity and its role in health and disease. Nat Rev Immunol. 2020;20(6):375–388. doi: 10.1038/s41577-020-0285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.van der Poel J, Sukhai RN.. [Initial symptoms in post-streptococcal glomerulonephritis]. Ned Tijdschr Geneeskd. 1989;133(9):425–427. [PubMed] [Google Scholar]
- 109.Mantovani A, Netea MG.. Trained innate immunity, epigenetics, and Covid-19. N Engl J Med. 2020;383(11):1078–1080. doi: 10.1056/NEJMcibr2011679. [DOI] [PubMed] [Google Scholar]
- 110.Escobar LE, Molina-Cruz A, Barillas-Mury C.. BCG vaccine protection from severe coronavirus disease 2019 (COVID-19). Proc Natl Acad Sci U S A. 2020;117(30):17720–17726. doi: 10.1073/pnas.2008410117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.de Laval B, Maurizio J, Kandalla PK, et al. C/EBPβ-dependent epigenetic memory induces trained immunity in hematopoietic stem cells. Cell Stem Cell. 2020;26(5):657–674 e8. doi: 10.1016/j.stem.2020.01.017. [DOI] [PubMed] [Google Scholar]
- 112.Netea MG, Joosten LA, Latz E, et al. . Trained immunity: a program of innate immune memory in health and disease. Science. 2016;352(6284):aaf1098. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fintelman-Rodrigues N, da Silva APD, Dos Santos MC, et al. . Genetic evidence and host immune response in persons reinfected with SARS-CoV-2, Brazil. Emerg Infect Dis. 2021;27(5):1446–1453. doi: 10.3201/eid2705.204912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dhochak N, Singhal T, Kabra SK, Lodha R.. Pathophysiology of COVID-19: why children fare better than adults? Indian J Pediatr. 2020;87(7):537–546. doi: 10.1007/s12098-020-03322-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mohapatra PR, Mishra B, Behera B.. BCG vaccination induced protection from COVID-19. Indian J Tuberc. 2021;68(1):119–124. doi: 10.1016/j.ijtb.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yao Y, Jeyanathan M, Haddadi S, et al. . Induction of autonomous memory alveolar macrophages requires T cell help and is critical to trained immunity. Cell. 2018;175(6):1634–1650 e17. doi: 10.1016/j.cell.2018.09.042. [DOI] [PubMed] [Google Scholar]
- 117.Chen X, Liu S, Goraya MU, Maarouf M, Huang S, Chen JL.. Host immune response to influenza A virus infection. Front Immunol. 2018;9:320. doi: 10.3389/fimmu.2018.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zuercher AW, Coffin SE, Thurnheer MC, Fundova P, Cebra JJ.. Nasal-associated lymphoid tissue is a mucosal inductive site for virus-specific humoral and cellular immune responses. J Immunol. 2002;168(4):1796–1803. doi: 10.4049/jimmunol.168.4.1796. [DOI] [PubMed] [Google Scholar]
- 119.Akkaya M, Kwak K, Pierce SK.. B cell memory: building two walls of protection against pathogens. Nat Rev Immunol. 2020;20(4):229–238. doi: 10.1038/s41577-019-0244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ionescu L, Urschel S.. Memory B cells and long-lived plasma cells. Transplantation. 2019;103(5):890–898. doi: 10.1097/TP.0000000000002594. [DOI] [PubMed] [Google Scholar]
- 121.Doherty PC, Topham DJ, Tripp RA, Cardin RD, Brooks JW, Stevenson PG.. Effector CD4+ and CD8+ T-cell mechanisms in the control of respiratory virus infections. Immunol Rev. 1997;159:105–117. doi: 10.1111/j.1600-065x.1997.tb01010.x. [DOI] [PubMed] [Google Scholar]
- 122.Ando M, Ito M, Srirat T, Kondo T, Yoshimura A.. Memory T cell, exhaustion, and tumor immunity. Immunol Med. 2020;43(1):1–9. doi: 10.1080/25785826.2019.1698261. [DOI] [PubMed] [Google Scholar]
- 123.Jameson SC, Masopust D.. Understanding subset diversity in T cell memory. Immunity. 2018;48(2):214–226. doi: 10.1016/j.immuni.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Snyder TM, Gittelman RM, Klinger M, et al. Magnitude and dynamics of the T-cell response to SARS-CoV-2 Infection at both individual and population levels. medRxiv: the preprint server for health sciences. 2020.
- 125.Liu J, Li S, Liu J, et al. . Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55:102763. doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen G, Wu D, Guo W, et al. . Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kuri-Cervantes L, Pampena MB, Meng W, et al. . Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci Immunol. 2020;5(49):eabd7114.doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mazzoni A, Salvati L, Maggi L, et al. . Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J Clin Invest. 2020;130(9):4694–4703. doi: 10.1172/JCI138554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zheng HY, Zhang M, Yang CX, et al. . Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol. 2020;17(5):541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sekine T, Perez-Potti A, Rivera-Ballesteros O, Karolinska COVID-19 Study Group, et al.. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183(1):158–168 e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Le Bert N, Tan AT, Kunasegaran K, et al. . SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584(7821):457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 132.Grifoni A, Weiskopf D, Ramirez SI, et al. . Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501 e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.DiPiazza AT, Graham BS, Ruckwardt TJ.. T cell immunity to SARS-CoV-2 following natural infection and vaccination. Biochem Biophys Res Commun. 2021;538:211–217. doi: 10.1016/j.bbrc.2020.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Weiskopf D, Schmitz KS, Raadsen MP, et al. . Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol. 2020;5(48):eabd2071.doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rahimzadeh M, Naderi N.. Toward an understanding of regulatory T cells in COVID-19: A systematic review. J Med Virol. 2021;93(7):4167–4181. doi: 10.1002/jmv.26891. [DOI] [PubMed] [Google Scholar]
- 136.De Biasi S, Meschiari M, Gibellini L, et al. . Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat Commun. 2020;11(1):3434. doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zuo J, Dowell AC, Pearce H, et al. . Robust SARS-CoV-2-specific T cell immunity is maintained at 6 months following primary infection. Nat Immunol. 2021;22(5):620–626. doi: 10.1038/s41590-021-00902-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Selhorst P, Van Ierssel S, Michiels J, et al. Symptomatic SARS-CoV-2 reinfection of a health care worker in a Belgian nosocomial outbreak despite primary neutralizing antibody response. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2020. [DOI] [PMC free article] [PubMed]
- 139.Griffin DE. Are T cells helpful for COVID-19: the relationship between response and risk. J Clin Invest. 2020;130(12):6222–6224. doi: 10.1172/JCI142081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sattler A, Angermair S, Stockmann H, et al. . SARS-CoV-2-specific T cell responses and correlations with COVID-19 patient predisposition. J Clin Invest. 2020;130(12):6477–6489. doi: 10.1172/JCI140965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Naylor K, Li G, Vallejo AN, et al. . The influence of age on T cell generation and TCR diversity. J Immunol. 2005;174(11):7446–7452. doi: 10.4049/jimmunol.174.11.7446. [DOI] [PubMed] [Google Scholar]
- 142.Liu B, Han J, Cheng X, et al. . Reduced numbers of T cells and B cells correlates with persistent SARS-CoV-2 presence in non-severe COVID-19 patients. Sci Rep. 2020;10(1):17718. doi: 10.1038/s41598-020-73955-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xu B, Fan CY, Wang AL, et al. . Suppressed T cell-mediated immunity in patients with COVID-19: A clinical retrospective study in Wuhan, China. The Journal of Infection. 2020;81(1):e51–e60. doi: 10.1016/j.jinf.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lenti MV, Aronico N, Pellegrino I, et al. . Depletion of circulating IgM memory B cells predicts unfavourable outcome in COVID-19. Sci Rep. 2020;10(1):20836. doi: 10.1038/s41598-020-77945-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rodda LB, Netland J, Shehata L, et al. . Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell. 2021;184(1):169–183 e17. doi: 10.1016/j.cell.2020.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gaebler C, Wang Z, Lorenzi JCC, et al. . Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591(7851):639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Poon MML, Farber DL.. Lasting memories of SARS-CoV-2 infection. The Journal of Experimental Medicine. 2021;218(4):e20210210.doi: 10.1084/jem.20210210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Breton G, Mendoza P, Hagglof T, et al. . Persistent cellular immunity to SARS-CoV-2 infection. The Journal of Experimental Medicine. 2021;218(4):e20202515.doi: 10.1084/jem.20202515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cromer D, Juno JA, Khoury D, Reynaldi A, et al. . Prospects for durable immune control of SARS-CoV-2 and prevention of reinfection. Nat Rev Immunol. 2021;21(6):395–404. doi: 10.1038/s41577-021-00550-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Izzard A, Wilders S, Smith C, et al. . Improved treatment completion for tuberculosis patients: The case for a dedicated social care team. J Infect. 2021;82(3):e1–e3. doi: 10.1016/j.jinf.2020.12.019. [DOI] [PubMed] [Google Scholar]
- 151.Sakharkar M, Rappazzo CG, Wieland-Alter WF, et al. . Prolonged evolution of the human B cell response to SARS-CoV-2 infection. Sci Immunol. 2021;6(56):eabg6916.doi: 10.1126/sciimmunol.abg6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tang F, Quan Y, Xin ZT, et al. . Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J Immunol. 2011;186(12):7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- 153.Deng W, Bao L, Liu J, et al. . Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science. 2020;369(6505):818–823. doi: 10.1126/science.abc5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chandrashekar A, Liu J, Martinot AJ, et al. . SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369(6505):812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lee WS, Wheatley AK, Kent SJ, DeKosky BJ.. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat Microbiol. 2020;5(10):1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PubMed] [Google Scholar]
- 156.Karthik K, Senthilkumar TMA, Udhayavel S, Raj GD.. Role of antibody-dependent enhancement (ADE) in the virulence of SARS-CoV-2 and its mitigation strategies for the development of vaccines and immunotherapies to counter COVID-19. Hum Vaccin Immunother. 2020;16(12):3055–3060. doi: 10.1080/21645515.2020.1796425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kaneko N, Kuo HH, Boucau J, et al. The Loss of Bcl-6 Expressing T Follicular Helper Cells and the Absence of Germinal Centers in COVID-19. SSRN. 2020. 3652322. [DOI] [PMC free article] [PubMed]
- 158.Vl P. Evolution of viruses. Encyclopedia Virol. 2008:174–184. doi: 10.1016/B978-012374410-4.00706-8. [DOI] [Google Scholar]
- 159.MacLean OA, Lytras S, Weaver S, et al. . Natural selection in the evolution of SARS-CoV-2 in bats created a generalist virus and highly capable human pathogen. PLoS Biol. 2021;19(3):e3001115. doi: 10.1371/journal.pbio.3001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wu F, Zhao S, Yu B, et al. . A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhou P, Yang XL, Wang XG, et al. . A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lu R, Zhao X, Li J, et al. . Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wu A, Peng Y, Huang B, et al. . Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Korber B, Fischer WM, Gnanakaran S, Sheffield COVID-19 Genomics Group, et al.. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827 e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Isabel S, Grana-Miraglia L, Gutierrez JM, et al. . Evolutionary and structural analyses of SARS-CoV-2 D614G spike protein mutation now documented worldwide. Sci Rep. 2020;10(1):14031. doi: 10.1038/s41598-020-70827-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zou J, Xie X, Fontes-Garfias CR, et al. . The effect of SARS-CoV-2 D614G mutation on BNT162b2 vaccine-elicited neutralization. NPJ Vaccines. 2021;6(1):44. doi: 10.1038/s41541-021-00313-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.CDC . SARS-CoV-2 Variant classifications and definitions. 2020.
- 168.Cele S, Gazy I, Jackson L, COMMIT-KZN Team, et al.. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021;593(7857):142–146. doi: 10.1038/s41586-021-03471-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Planas D, Bruel T, Grzelak L, et al. . Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med. 2021;27(5):917–924. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- 170.England PH. Confirmed cases of COVID-19 variants identified in UK. 2021.
- 171.Imc NRF, Candido D, Franco LAM, et al. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. https://virologicalorg/. 2020.
- 172.Resende P. Update of the SARS-CoV-2 genomic surveillance in the Amazonas state, Brazil. https://virologicalorg/. 2021.
- 173.MSd S, Demoliner M, Hansen AW, et al. Early detection of SARS-CoV-2 P.1 variant in Southern Brazil and reinfection of the same patient by P.2. Research Square; 2021. [DOI] [PMC free article] [PubMed]
- 174.Jfb PCR, Vasconcelo R. H T d, Arantes I, et al. Spike E484K mutation in the first SARS-CoV-2 reinfection case confirmed in Brazil. 2021.
- 175.Galanti M, Shaman J.. Direct observation of repeated infections with endemic coronaviruses. J Infect Dis. 2021;223(3):409–415. doi: 10.1093/infdis/jiaa392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mishler B, Wilkins J.. The hunting of the SNaRC: a snarky solution to the species problem. Philosophy, Theory, and Practice in Biology. 2018;10(20210712) doi: 10.3998/ptpbio.16039257.0010.001. [DOI] [Google Scholar]
- 177.Shastri J, Parikh S, Agrawal S, et al. . Clinical, serological, whole genome sequence analyses to confirm SARS-CoV-2 reinfection in patients from Mumbai, India. Front Med (Lausanne). 2021;8:631769. doi: 10.3389/fmed.2021.631769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Jc CZ, Wee S-L. Coronavirus vaccine tracker. The New York Times. 2020, p. 21.
- 179.Wibmer CK, Ayres F, Hermanus T, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. bioRxiv: the preprint server for biology. 2021. [DOI] [PubMed]
- 180.England PH. Investigation of novel SARS-CoV-2 variant. 2020.
- 181.Emary K, Golubchik T, Aley P, et al. . Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): an exploratory analysis of a randomised controlled trial. The Lancet. 2021;397(10282):1351-1362. doi: 10.2139/ssrn.3779160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Callaway E. Fast-spreading COVID variant can elude immune responses. Nature. 2021;589(7843):500–501. doi: 10.1038/d41586-021-00121-z. [DOI] [PubMed] [Google Scholar]
- 183.Prevost J, Finzi A.. The great escape? SARS-CoV-2 variants evading neutralizing responses. Cell Host Microbe. 2021;29(3):322–324. doi: 10.1016/j.chom.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Levine-Tiefenbrun M, Yelin I, Katz R, et al. . Initial report of decreased SARS-CoV-2 viral load after inoculation with the BNT162b2 vaccine. Nat Med. 2021;27(5):790–792. doi: 10.1038/s41591-021-01316-7. [DOI] [PubMed] [Google Scholar]
- 185.Chau NVV, Ngoc NM, Nguyet LA, et al. . An observational study of breakthrough SARS-CoV-2 Delta variant infections among vaccinated healthcare workers in Vietnam. EClinicalMed. 2021;41:101143. doi: 10.1016/j.eclinm.2021.101143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Piri SM, Edalatfar M, Shool S, Jalalian MN, Tavakolpour S.. A systematic review on the recurrence of SARS-CoV-2 virus: frequency, risk factors, and possible explanations. Infect Dis (Lond). 2021;53(5):315–324. doi: 10.1080/23744235.2020.1871066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Vancsa S, Dembrovszky F, Farkas N, et al. . Repeated SARS-CoV-2 Positivity: Analysis of 123 Cases. Viruses. 2021;13(3):512. doi: 10.3390/v13030512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Cavanaugh AM, Spicer KB, Thoroughman D, Glick C, Winter K.. Reduced Risk of Reinfection with SARS-CoV-2 After COVID-19 Vaccination – Kentucky, May-June 2021. MMWR Morb Mortal Wkly Rep. 2021;70(32):1081–1083. doi: 10.15585/mmwr.mm7032e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Vardhana SA, Wolchok JD.. The many faces of the anti-COVID immune response. J Experim Med. 2020;589(7843):500-501.doi: 10.1084/jem.20200678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Guan WJ, Ni ZY, Hu Y, China Medical Treatment Expert Group for Covid-19, et al.. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lin L, Jiang X, Zhang Z, et al. . Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69(6):997–1001. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- 192.Yuan J, Kou S, Liang Y, Zeng J, Pan Y, Liu L. Polymerase Chain Reaction Assays Reverted to Positive in 25 Discharged Patients With COVID-19. Clinical Infectious Diseases. 2020;71(16):2230-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]