Abstract
Species-specific bacterial identification of clinical specimens is often limited to a few species due to the difficulty of performing multiplex reactions. In addition, discrimination of amplicons is time-consuming and laborious, consisting of gel electrophoresis, probe hybridization, or sequencing technology. In order to simplify the process of bacterial identification, we combined anchored in situ amplification on a microelectronic chip array with discrimination and detection on the same platform. Here, we describe the simultaneous amplification and discrimination of six gene sequences which are representative of different bacterial identification assays: Escherichia coli gyrA, Salmonella gyrA, Campylobacter gyrA, E. coli parC, Staphylococcus mecA, and Chlamydia cryptic plasmid. The assay can detect both plasmid and transposon genes and can also discriminate strains carrying antibiotic resistance single-nucleotide polymorphism mutations. Finally, the assay is similarly capable of discriminating between bacterial species through reporter-specific discrimination and allele-specific amplification. Anchored strand displacement amplification allows multiplex amplification and complex genotype discrimination on the same platform. This assay simplifies the bacterial identification process greatly, allowing molecular biology techniques to be performed with minimal processing of samples and practical experience.
Genotype identification has been widely recognized as an effective tool in the identification and characterization of infectious disease organisms (7, 12, 16). Molecular biology-based bacterial identification assays have the potential of decreasing the time necessary for and increasing the specificity of bacterial determinations, making them efficacious alternatives to traditional biochemical and microbiological culture techniques. For example, Chlamydia trachomatis and Mycobacterium tuberculosis are traditionally very difficult organisms to culture. Molecular biology techniques circumvent the need for long culturing protocols by using amplification of either DNA (PCR, ligase chain reaction [LCR], strand displacement amplification [SDA], and nucleic acid sequence-based amplification [NASBA]) or RNA (reverse transcription-PCR, NASBA, reverse transcription-SDA, and transcription-mediated amplification [TMA]) targets. However, molecular biology techniques are also advantageous because of the amount of information that can be obtained from a single assay in a short period of time. As a result, the 1999 National Committee for Clinical Laboratory Standards (13a) guidelines have for the first time mandated the use of molecular biology methods in clinical laboratories that perform bacterial identification assays.
There are many examples of molecular biology-based assays used in the laboratory. Genotypic identification of bacterial samples is used to discriminate and identify bacteria at either the genus, species, or strain level (1, 9). Genotypic identification of antimicrobial resistance is also used as an aid in the treatment of infectious diseases (15, 19, 20, 24). Conventional antimicrobial susceptibility testing provides only phenotypic profiling of a potential pathogen. Low-level antimicrobial activity and heterogeneous populations of antimicrobial agent-resistant pathogens are difficult to detect with these techniques. Molecular analysis of pathogens provides a more definite means of obtaining the antimicrobial status of microorganisms by identifying organisms that possess the genetic material necessary for resistance.
As with all amplification assays, multiplex target or signal amplification is difficult. There have been numerous reports of multiplex PCR assays developed for many bacterial identification assays (15, 20). However, multiplex PCR assays, as well as all other target and signal amplification assays, are still limited in the amounts of templates that can be amplified simultaneously because of the difficulty of optimizing primer and reagent conditions. In addition, discrimination of closely matched amplicon sequences is time-consuming and laborious, consisting of either gel electrophoresis, probe hybridization, or sequencing technology.
Microchip arrays are capable of analyzing hundreds to thousands of different loci simultaneously in a relatively short period of time (3, 13). However, most microchip array systems require large amounts of template DNA, or targets from multiplex amplification, in order to detect DNA or RNA at fairly low levels (2, 13). We recently described an assay on an electronic microchip array which was capable of multiplex amplification of 10 targets simultaneously, with little decrease in amplification efficiency (22). Anchored SDA on microelectronic chips encompasses amplification on the surface of the array, requiring very small amounts of input DNA. The flexibility of the microelectronic chip array has been demonstrated repeatedly in many types of genetic discrimination assays, including single-nucleotide polymorphism (SNP) (8, 18) and short tandem repeat (17) discrimination assays. Here, we describe an in situ amplification assay using complex genomic DNA samples on a microelectronic chip array. We have also combined amplification with genetic discrimination assays for the identification of bacterial species and antimicrobial resistance and for SNP discrimination of fluoroquinolone resistance in Campylobacter isolates. This technology greatly simplifies molecular genotyping assays by requiring substantially less sample processing and technical expertise than most molecular biology-based assays.
MATERIALS AND METHODS
Strains, culture conditions, and isolation of genomic DNA.
Bacterial strains were obtained from various sources, including the American Type Culture Collection (Manassas, Va.), the Centers for Disease Control and Prevention (Atlanta, Ga.), the National Cell and Tissue Collection, and BD Biosciences (Hunt Valley, Md.). All bacterial species were cultured as recommended by the American Type Culture Collection. Genomic DNA was isolated from liquid culture cells or plate scrapings using DNeasy genomic DNA isolation kits (Qiagen, Valencia, Calif.). All amplification systems were tested with at least three different strains.
SDA primers.
Oligonucleotides (Table 1) were synthesized at Integrated DNA Technologies (Coralville, Iowa). Oligonucleotides were coupled with either BODIPY Texas red (BTR), Cy3, or Cy5 fluorophore or biotin at the 5′ end.
TABLE 1.
Bacterial anchored SDA primersa
| Gene | Designation | Function | Sequence |
|---|---|---|---|
| Factor V | 1933 | Amplification | B-ACCGCATCGAATGCATGTCCTCGGGTCTCTGGGCTAATAGGA |
| 1934 | Amplification | B-ACGATTCAGCTCCAGACTTCTCGGGTCAGAATTTCTCAAAGG | |
| 975 | Bumper primer | ACTACAGTGACGTGGACATC | |
| 976 | Bumper primer | TGTTATCACACTGGTGCTAA | |
| 1935 | Reporter | BTR-ACTTCTAATCTGTAAGAGCAG | |
| Chlamydia | 1937 | Amplification | B-ACCGCATCGAATGCATGTCCTCGGGTACAACATCAACACCTG |
| 1938 | Amplification | B-CGATTCAGCTCCAGACTTCTCGGGTGAGACTGTTAAAGATA | |
| 1939 | Noncleavable | B-ACCGCATCGAATGCATGTCCTCCGGTACAACATCAACACCTG | |
| 1940 | Noncleavable | B-CGATTCAGCTCCAGACTTCTCCGGTGAGACTGTTAAAGATA | |
| BL1 | Bumper primer | CAGCAAATAATCCTTGG | |
| BL2 | Bumper primer | CATTGGTTGATGGATTATT | |
| 1941 | Reporter | BTR-GTCGCAGCCAAAATG | |
| 1942 | Reporter | BTR-TTCCATCAGAAGCTGT | |
| mecA | 3365 | Amplification | B-GCAAACTTAATTGGCAAATCCGCTCGGGCAGAACTCA |
| 3366 | Amplification | B-ACATCTTTAACATTAATAGCCATCCTCGGGTTTGGATTATC | |
| 3370 | Noncleavable | B-GCAAACTTAATTGGCAAATCCGCTCCGGCAGAACTCA | |
| 3371 | Noncleavable | B-ACATCTTTAACATTAATAGCCATCCTCCGGTTTGGATTATC | |
| 3367 | Bumper primer | TAAAGAAGATATTTATAGATC | |
| 3368 | Bumper primer | GGCATTGTAGCTAGCCATTCC | |
| 3369 | Reporter | Cy3-GGTTTATATCATATG | |
| Campylobacter | 3328 | Amplification | B-AAAGTAGAACAGATTTTGTCAAATCTCGGGGTATAGTGG |
| 3329 | Amplification | B-ACTTGGATATCTCATAGAAAAATCTCGGGCCATTCTAAC | |
| 3335 | Noncleavable | B-AAAGTAGAACAGATTTTGTCAAATCTCCGGGTATAGTGG | |
| 3336 | Noncleavable | B-ACTTGGATATCTCATAGAAAAATCTCCGGCCATTCTAAC | |
| 3332 | Bumper primer | TTTTATATGCTATGCAAAAT | |
| 3333 | Bumper primer | GATCCAAAGTTGCCTTGTCC | |
| 3334 | Stabilizer | GTCGTTATCACCCACATGGAGATA | |
| 3330 | Reporter | Cy3-CAGCAGTTTAT | |
| 3331 | Reporter | Cy3-TAGCAGTTTAT | |
| E. coli parC | 2764 | Amplification | B-TCGCATTAACCTCAATATGATCTCGGGGGATGGTCG |
| 2765 | Amplification | B-TCTCCAGACGATAGTTCAGTCGCTCGGGCACGGTATC | |
| 2768 | Noncleavable | B-TCGCATTAACCTCAATATGATCTCCGGGGATGGTCG | |
| 2769 | Noncleavable | B-TCTCCAGACGATAGTTCAGTCGCTCCGGCACGGTATC | |
| 2766 | Bumper primer | CACCGATCTGGAAAAGAGCTA | |
| 2767 | Bumper primer | CTTCGAGGATATGCAGGCGC | |
| 3321 | Reporter | Cy3-ATTGCCAGAGG | |
| gyrA | 3303 | Amplification | B-TGACTGGAACAAAGCCTATAAAAAATCTCGGGGTGTCGTT |
| 3304 | Amplification | B-CAGCGAGAATGGCTGCGCCATACGCTCGGGCGTGTCATA | |
| 3311 | Noncleavable | B-TGACTGGAACAAAGCCTATAAAAAATCTCCGGGTGTCGTT | |
| 3312 | Noncleavable | B-CAGCGAGAATGGCTGCGCCATACGCTCCGGCGTGTCATA | |
| 3306 | Bumper primer | TACTTTACGCCATGAACGTA | |
| 3307 | Bumper primer | GAACCGAAGTTACCCTGACC | |
| 3310 | Stabilizer | TGGGGATGGTATTTACCGATTACGT | |
| 3308 | Reporter | Cy3-CCGAGTCACCA | |
| 3309 | Reporter | Cy5-CGGAATCGCCG |
B, 5′ end labeled with biotin; BTR, 5′ end labeled with BTR; Cy3, 5′ end labeled with Cy3 fluorophore; Cy5, 5′ end labeled with Cy5 fluorophore.
Microelectronic chips, amplification primer deposition, and hybridization of templates.
Microelectronic chips and the permeation layer were prepared as described previously (5, 8). In brief, the APEX chip used for these experiments consists of a 5- by 5-array of 80-μm circular microelectrodes with 200-μm microelectrodes at each corner of the array. Chips were mounted on a micromanipulator stage, and the microelectrodes were activated by a power supply and appropriately controlled relay switches. All electronic manipulations on the microchip were done in 50 mM histidine buffer. Biotinylated, BTR-labeled T12 oligonucleotide [BTR-dT(12)] was used as a control for streptavidin integrity in the permeation layer. All SDA primers were coupled with biotin at the 5′ terminus. After hydration of the microchip in 50 mM histidine buffer, SDA primer sets were electronically addressed (0.8 μA/site, pulsed for 1 min) to specific array sites as indicated in each experiment. The chips were washed with 50 mM histidine buffer between each application of oligonucleotide sets. The SDA primers were then electronically hybridized with genomic DNA templates (0.8 μA/site, pulsed for 3.5 min). The chips were washed with water after hybridization with the templates, incubated in a 100-μg/ml bovine serum albumin solution for 10 to 30 min at room temperature, and washed again with water.
Anchored SDA.
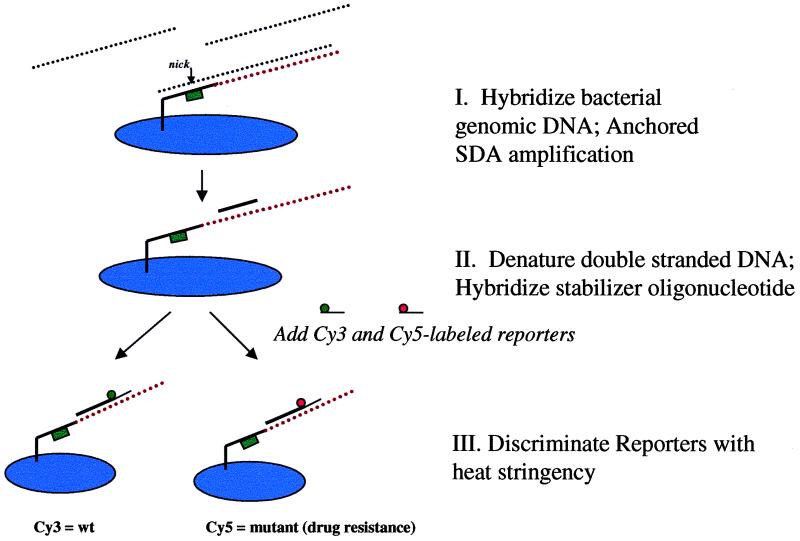
Figure 1 is a schematic representation of anchored SDA. For simplicity, only one strand is shown in the illustration. Biotinylated primers were first electronically addressed to individual locations on the microchip array (Fig. 1, panel I) as mentioned above. Templates were then electronically hybridized to the biotinylated primers, and the amplification reaction was initiated with the addition of bumper primers and enzymes (Fig. 1, panel II). This step was performed by preheating the microchips to 60°C for 5 min in a humidifying chamber. The water was removed and replaced with 10 μl of prewarmed SDA mixture (6 mM morpholinepropanesulfonic acid [MOPS] [pH 7.8]; 1.7 mM [each] dCTP [thiolated], dTTP, dATP, and dGTP; 85 mM KCl; 18 mM MgCl2; 23 mM NaCl; 3.5 mM Tris-HCl [pH 7.9]; 0.035 mM dithiothreitol; 1.5 U of BsoBI; 0.8 U of BstI polymerase; 25 nM each bumper primer) (21). The microchips were incubated for 30 min at 60°C in a humidifying chamber. The reaction was stopped by removing the chips and washing them approximately five times with 0.5× SSC (1× SSC is 0.075 M NaCl plus 0.0075 M sodium citrate [pH 7.2]) at room temperature.
FIG. 1.
Schematic representation of anchored SDA and bacterial discrimination. Biotinylated SDA amplification primer sets are first addressed to spatially distinct areas on the microchip array using electronic biasing. Templates are then electronically hybridized to the SDA primer sets, and the SDA reaction is performed in situ with the addition of bumper primers and reaction reagents to the microchip (panel I). The amplification primers contain a BsoBI restriction endonuclease site, essential for SDA. The bumper primers (not shown) are needed only to remove the initial primer extension product from the target template, allowing the primer-extended strand to bind to its complementary amplification primer. Primer extension of the complementary amplification primer and subsequent incorporation of a thiolated nucleotide (dCTP) over the BsoBI site induce nicking by BsoBI in the amplification primer region. The presence of a nick signals polymerase binding and simultaneous strand displacement-primer extension, resulting in exponential amplification of the target DNA. The SDA reaction is stopped by removal of the supernatant, double-stranded DNA products are denatured on the microchip, and internal reporters are hybridized to the amplicon products remaining on the chip (panel II). Discrimination is performed by increasing heat (thermal stringency) until only one reporter species (Cy3 or Cy5) remains (panel III). wt, wild type.
After amplification, the anchored amplicons were denatured with the addition of an alkali solution (0.5× SSC, pH 12.5, 4 min) and washed five times at room temperature (0.5× SSC, pH 7.2). The amplicons were then hybridized with 1.0 μM stabilizer oligonucleotides in 4× SSC (pH 7.2) at room temperature for 3 min. After extensive washing (5 to 10 times) with 0.5× SSC, a 0.5 μM concentration of reporter oligonucleotides was hybridized for 5 min in 4× SSC at room temperature (Fig. 1, panel III). The reporters were coupled at the 5′ end with either Cy3, Cy5, or BTR fluorophore. Extensive washing (5 to 10 times) with 0.5× SSC (pH 7.2), followed by 0.2× SSC–1.0% sodium dodecyl sulfate and 0.2× SSC washes at room temperature, removed most of the unbound reporter oligonucleotides. Temperature stringency in 50 mM NaPO4 (pH 7.7) was applied to discriminate SNPs and other closely matched reporters. Temperature was ramped up in 3°C increments and maintained for at least 3 min at each step. Images were taken at every step, and the microchips were washed with 50 mM NaPO4 (pH 7.7) at every temperature interval. The entire assay from template hybridization to reporter discrimination took approximately 70 to 90 min, depending on the number of templates to be hybridized on the microchip array.
Reporter discrimination using base-stacking energy transfer.
One of the advantages of anchored SDA lies in its ability to amplify and detect target DNA on the same platform. Achieving amplification of and discrimination between very similar DNA targets is difficult to accomplish with a single-platform assay. We took advantage of base-stacking energy transfer techniques to assist in reporter discrimination of SNPs (14, 17, 23). Amplification primers were designed surrounding the Thr86Ile mutation in the gyrase A region of type II DNA topoisomerase in Campylobacter jejuni; this mutation was previously shown to confer fluoroquinolone resistance in bacteria carrying it (24). Base-stacking reporters were used such that the 5′ end of the labeled reporter would base stack alongside a longer stabilizer oligonucleotide (14, 17, 23). If the amplified target presented a perfect match to the labeled reporter, then the base-stacking energies between the stabilizer oligonucleotide and the labeled reporter would be favorable, allowing the shorter reporter to remain hybridized to the amplified target at elevated temperatures. However, if the amplified target presented a mismatch to the labeled reporter, then the base-stacking energy of the stabilizer oligonucleotide would be absent, allowing removal of the labeled reporter at elevated temperatures. In order to distinguish the presence of the Thr86Ile mutation, two reporters were designed, with either a Cy3 or a Cy5 fluorophore label coupled at the 5′ end. The wild-type reporter, with the wild-type C at the 5′ end, was end labeled with Cy3, whereas the mutant reporter, with a T at the 5′ end, was end labeled with Cy5. After temperature stringency steps were applied to the amplified product on the microelectronic chip array, the genotype could be determined by noting the fluorophore type remaining on the microchip.
RESULTS
Chlamydia identification using a plasmid-dependent gene.
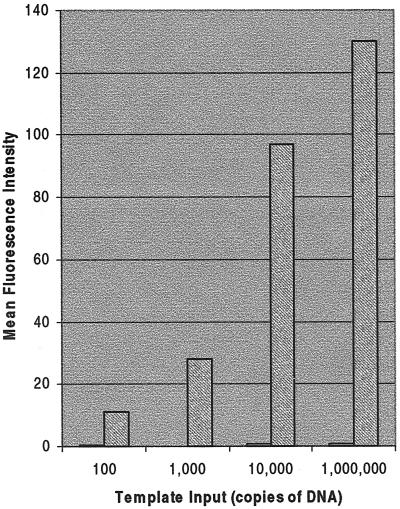
C. trachomatis was amplified by anchored SDA using primers to the cryptic plasmid as the target sequence (10). Figure 2 shows the results of a titration experiment with various amounts of Chlamydia template input. Noncleavable primers, which have a single base mutation in the recognition site for BsoBI, were used as a control for background binding of hybridized template and to ensure that amplification was occurring on the microchip surface (22). Template input levels of 1,000 copies of DNA could be routinely detected using anchored SDA in multiple experiments. Template input levels of 100 copies gave variable results (data not shown), indicating the threshold of amplification to be between 100 and 1,000 copies of plasmid DNA. No amplification could be detected when other bacterial sources were hybridized to the Chlamydia SDA primers (Table 1), indicating a high specificity of amplification. The latter is due to both the specificity of the SDA primers for amplification and the specificity of the reporter oligonucleotide for hybridization to the correct amplicon.
FIG. 2.
Amplification of C. trachomatis cryptic plasmid by anchored SDA. Anchored SDA was performed with increasing amounts of template DNA input to determine the sensitivity of the system. Representative fluorescent images of each titration reaction are made after reporter hybridization and washing. Gray bars show SDA primers; white bars show mutated SDA primers for control of template background binding (noncleavable; bars are barely visible above the x axis).
Antimicrobial resistance discrimination using anchored SDA.
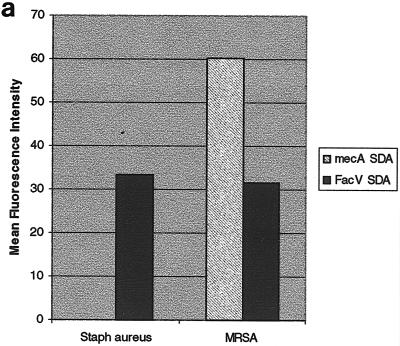
Staphylococcus aureus methicillin resistance was used as a model system for antimicrobial resistance determination with a novel gene insertion mechanism. In this experiment, genomic DNA from cultured methicillin-sensitive and methicillin-resistant S. aureus was purified and hybridized to the microelectronic chip array. As shown in Fig. 3a, only S. aureus samples that have been characterized as methicillin resistant showed any positive signal. An anchored SDA system from the human factor V gene (22) was used as a control to show that amplification was functional in the methicillin-sensitive amplification reaction. These results demonstrate the specificity and accuracy of anchored SDA.
FIG. 3.
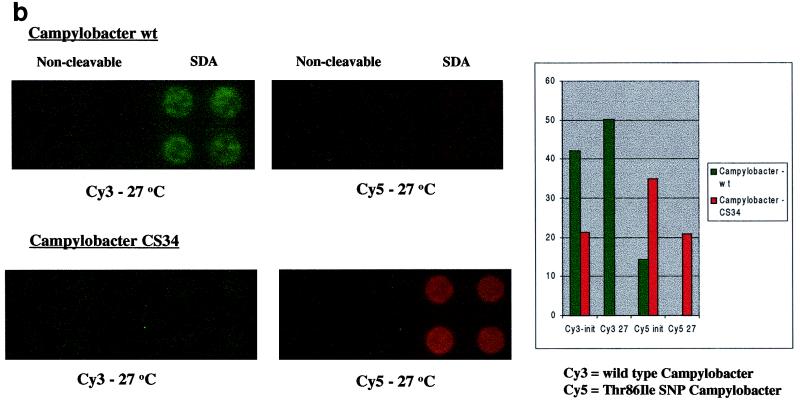
(a) Detection of the mecA gene in S. aureus by anchored SDA. Anchored SDA was performed with either methicillin-susceptible (Staph aureus) or methicillin-resistant (MRSA) S. aureus genomic DNA. Factor V-anchored SDA was performed on the same microchip array as a control for the efficiency of the anchored SDA reaction. (b) Discrimination of fluoroquinolone-resistant C. jejuni using anchored SDA. Anchored SDA was performed with 50 ng of C. jejuni genomic DNA samples expressing a wild-type phenotype (wt; top fluorescent image) or containing a fluoroquinolone-resistant point mutation (CS34; bottom fluorescent image). Wild-type reporters (Cy3) and fluoroquinolone-resistant mutant reporters (Cy5) were hybridized to the microchip array, and thermal stringency was applied to discriminate the amplification reaction. Reporter signals remaining after thermal stringency were quantified to determine the genotype of the hybridized genomic DNA sample (graph). init, initial temperature; 27, 27°C.
Targeting of critical proteins used by antimicrobial agents to access bacterial cells can also confer antibiotic resistance. We tested our system by using an SNP, recently found in C. jejuni samples (24), which confers resistance against fluoroquinolone antibiotics by interfering with the putative active site of gyrase A (4). Figure 3b shows the results of amplification with either wild-type genomic DNA or genomic DNA from clinical isolate samples with a confirmed fluoroquinolone resistance mutation at the C position of codon 86 (24). Amplification with wild-type genomic DNA resulted in only the wild-type Cy3 reporter remaining on targeted microchip sites, indicating the correct discrimination pattern. However, if genomic DNA from, in this example, CS34 was used for target amplification, only the mutant Cy5 reporter remained on the microchip electrode sites, again indicating the correct discrimination pattern. Two other clinical isolates with a confirmed SNP at the same site, CS5 and CS50 (24), were also used as target sources, with identical results (data not shown). These results demonstrate the ability to perform both amplification and complex genotyping discrimination on the same platform.
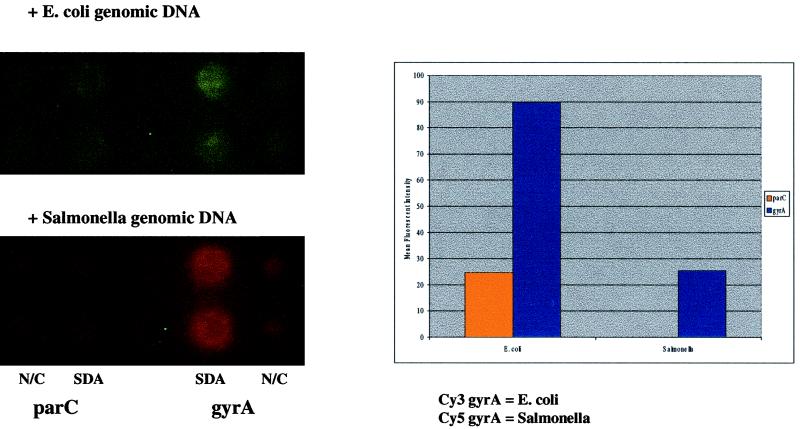
E. coli and Salmonella discrimination using anchored SDA.
Bacterial gyrase A genes have been extensively studied as a model system for molecular phylogenetic reconstruction due to the presence of highly conserved motifs interspersed with regions of divergent sequences (11). This relatively conserved sequence organization allows for the design of common amplification primers for amplification of bacterial sequences, followed by the use of specific reporters for bacterial identification discrimination assays. We used sequences from the gyrA region of type II DNA topoisomerase to amplify and discriminate E. coli and Salmonella genomic DNAs in our assay (Fig. 4). The SDA primers were designed on the basis of a region that is conserved between E. coli and Salmonella, allowing both organisms to be amplified if present in a sample. Campylobacter, Chlamydia, and S. aureus genomic DNAs were not amplified using this set of SDA primers (Table 1). After amplification of the samples, microchips were hybridized with base-stacking reporters specific for either E. coli (Cy3) or Salmonella (Cy5). As shown in Fig. 4, the correct signal was dependent on the input DNA origin. Only a Cy3 signal could be seen when E. coli was used as the DNA template for amplification. In contrast, only a Cy5 signal could be seen when Salmonella was used as the DNA template. The assay was repeated with six other Salmonella samples and seven other E. coli samples, with identical results (data not shown).
FIG. 4.
E. coli and Salmonella bacterial identification using reporter-specific and allele-specific SDA primers. A duplex anchored SDA was performed to identify E. coli and Salmonella genomic DNA samples after amplification. Amplification primers for parC and gyrA amplifications were addressed to separate locations on the microchip array. Approximately 100 pg of E. coli and Salmonella genomic DNA was addressed to all primer sets and hybridized with reporters for both E. coli gyrA and parC and Salmonella gyrA amplicons. Reporter signals remaining after application of thermal stringency were quantified to determine the genotype of the hybridized genomic DNA sample (graph). N/C, noncleavable.
Reporter discrimination is a relatively straightforward method for bacterial identification assays. However, discrimination can also be designed into amplification primers, allowing amplification of one set of sequences but not others. Allele-specific amplification has been demonstrated previously using SDA (6). We combined both reporter discrimination and allele-specific amplification in the same assay to provide a more flexible and secure means of bacterial identification. In addition to reporter discrimination of E. coli and Salmonella, Fig. 4 also demonstrates the use of allele-specific amplification of E. coli and Salmonella genomic DNA samples with amplification primers from the parC region of type II DNA topoisomerase. The parC amplification primers were designed such that E. coli but not Salmonella samples would be amplified. As shown in Fig. 4 (graph), only E. coli samples amplified a product when hybridized to parC amplification primers. No product could be detected for Salmonella genomic DNA samples, either on the microchip array or in solution SDA assays using polyacrylamide gels for visualizing amplicon products (data not shown).
Assay sensitivity and cross-reactivity with other bacterial genomic DNAs.
Table 2 lists the sensitivities of the amplification systems as well as the cross-reactivity of other bacterial genomic DNAs with each primer set. Since these primer sets were not optimized, the levels of sensitivity differed for the systems. Campylobacter gyrA and E. coli or Salmonella gyrA amplification primers were the most sensitive, with detection occurring at 100 copies of template input. Chlamydia cryptic plasmid and Staphylococci mecA amplification primers were next, needing approximately 1,000 copies for successful amplification. The least sensitive system was for the E. coli parC gene, needing at least 10,000 copies for successful amplification. Genomic DNAs from 10 other bacterial sources were tested for cross-reactivity. None of the primer sets exhibited any level of amplification with these genomic DNA sources.
TABLE 2.
Sensitivity of anchored SDA and cross-reactivity with other bacterial genomic DNAsa
| SDA primer set | No. of copies needed for amplification of DNA from:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli | Typhi | C. jejuni | MRSA | C. trachomatis | P. hemo | Enterococcus faecalis | Bacteroides fragilis | Kebsiella oxytoca | Yersinia enterocolitica | Shigella sonnei | Plesiomonas shigelloides | Pseudomonas aeruginosa | S. aureus | Vibrio parahaemolyticus | |
| E. coli or Salmonella gyrA | 100 | 100 | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Campylobacter gyrA | — | — | 100 | — | — | — | — | — | — | — | — | — | — | — | — |
| E. coli parC | 10,000 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| mecA | — | — | — | 1,000 | — | — | — | — | — | — | — | — | — | — | — |
| Chlamydia cryptic plasmid | — | — | — | — | 1,000 | — | — | — | — | — | — | — | — | — | — |
Titration experiments were performed for each anchored SDA system, and images were quantified to determine the lower limit of amplification sensitivity. The number of copies necessary for consistent amplification of template DNA is indicated. Genomic DNA from other bacterial species (100 ng) was also tested with every system to determine the degree of cross-reactivity. Typhi, Salmonella enterica, serovar Typhi; MRSA, methicillin-resistant S. aureus. —, no amplification.
Multiplex amplification.
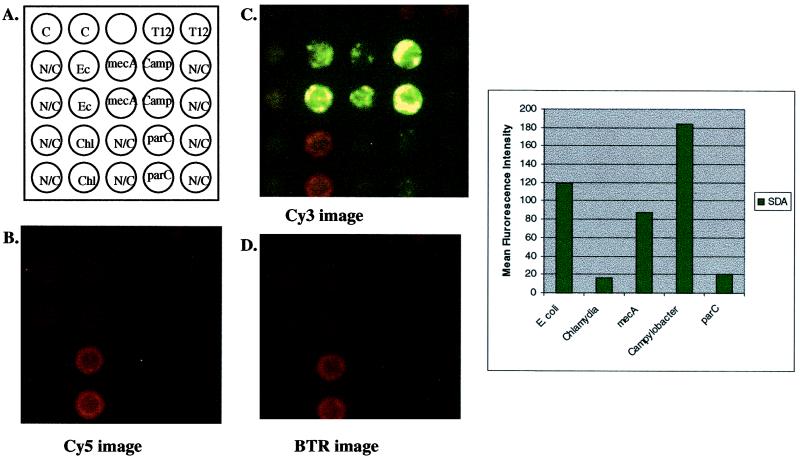
Although antimicrobial resistance and strain discrimination are important in molecular diagnostic assays, it would be ideal to have a high-throughput assay that can screen a large number of genes simultaneously. In order to demonstrate this concept, the maximum numbers of amplification systems for a 25-site microchip array, with the proper controls, were amplified simultaneously. E. coli, C. jejuni, methicillin-resistant S. aureus, and Chlamydia cryptic plasmid genomic DNAs were hybridized to the microchip array and amplified simultaneously. As shown in Fig. 5, all systems amplified simultaneously, giving correct discrimination patterns for the template DNA inputs used. Quantification of the signals (Fig. 5, graph) showed that the least sensitive system (parC) (Table 1) also amplified poorly in the multiplex amplification situation, whereas the most sensitive system (E. coli or Salmonella gyrA and Campylobacter gyrA) (Table 1) amplified target DNA best. Quantification of the Chlamydia cryptic plasmid amplification results could not be compared to that of the other systems, since a different fluorophore (BTR) was used as the reporter moiety. The results indicated that, independent of the numbers of amplification systems present, the different primer sets did not influence the amplification efficiency of other, neighboring primer sets. These findings confirm the previous findings (22) that anchored SDA primer sets act as discrete amplification units on the microelectronic chip array.
FIG. 5.
Multiplex amplification of E. coli gyrA, Staphylococcus mecA, Campylobacter gyrA, C. trachomatis cryptic plasmid, and E. coli parC. (A) Schematic representation of a multiplex (n = 5) amplification on the microelectronic chip array. Noncleavable (N/C) primers were included for each primer set to control for background hybridization of template DNA. Approximately 104 copies each of E. coli, methicillin-resistant S. aureus, fluoroquinolone-sensitive Campylobacter, and C. trachomatis cryptic plasmid genomic DNAs were combined and hybridized to each amplification primer set on the microelectronic chip array. Anchored SDA was initiated by the addition of enzymes and bumper primers for all systems. After amplification, reporters from all systems (wild-type and mutant reporters) were added and thermal stringency was applied at 29°C. Images were quantified for genotype identification of hybridized genomic DNA. (B) Cy5 fluorescent image after thermal stringency. Note that there is some bleed-through of the BTR signal seen in this image, due to overlapping emission of the BTR fluorophore at this wavelength. (C) Cy3 fluorescent image after thermal stringency. Note again the bleed-through of the BTR signal due to overlapping emission of the BTR fluorophore at this wavelength. (D) BTR image after thermal stringency. Quantification of signals after thermal stringency is shown in the graph.
DISCUSSION
Anchored SDA of bacterial gene targets is an efficient method for multiplex amplification and discrimination of antimicrobial agent-resistant strains as well as for bacterial identification. The assay provides a specific and reliable means for culture confirmation and can be sensitive to an input level of approximately 100 copies of DNA. It is important to note that the primer sets described here were not optimized for either primer-primer interactions or primer amplification efficiency. Anchored SDA allows multiplex amplification to occur because, as has been shown previously (22), the amplification primer sets act as discrete units on the microelectronic chip array. In anchored SDA, amplification primer sets are spatially separated, creating distinct zones of amplification that share only enzymes and reagents. Electronic addressing of amplification primers to distinct regions on the microchip allows a reduction in primer-primer interactions while maintaining a completely open format that simplifies the amplification procedure greatly. In addition to electronic addressing, the electronic microchip format also allows electronic hybridization, which has been shown previously to be essential for anchored SDA of genomic DNA samples (22). Electronic hybridization may increase the efficiency of the reaction by both facilitating strand separation of target DNA in a low-ionic-strength environment and concentrating targets onto the array site (5). Electronic hybridization also confers advantages in time, allowing hybridization reactions to be completed in minutes instead of hours (5, 8). These two aspects, combined with the flexibility of the assay in addressing any oligonucleotide or DNA sequence to any site on the microchip array, make the microelectronic chip a very attractive platform for molecular biology applications.
The flexibility of anchored SDA for bacterial identification makes it an ideal candidate for a task- or group-specific assay design. In this work, we have shown that five different genotypic assays (plasmid, transposon, SNP analysis, allele-specific amplification, and reporter-specific discrimination) can be accomplished simultaneously using anchored SDA and discrimination on the microelectronic chip platform. In the future, microchip arrays could include amplification primer sets, for example, for all known food-borne or respiratory pathogens on the same microchip, enabling amplification and discrimination of a whole class of bacterial pathogens on a single microchip array. In addition, anchored SDA could further discriminate, again on the same platform, possible antimicrobial resistance markers or other genetic markers that may be present in the same sample. Anchored SDA allows many different types of assays to be accommodated on the same platform, including RNA amplification (22), without adjusting for special conditions such as matching hybridization temperatures or altering stringency conditions, as in other amplification or microchip assays. This flexibility allows multiplex amplification and discrimination on the same platform, potentially streamlining the development of any nucleic-acid-based bacterial determination assay.
As demonstrated here, anchored SDA can readily discriminate SNPs. This result suggests the immediate application of this assay for culture confirmation in clinical laboratories. That 100 copies of DNA are sufficient for antimicrobial resistance determination suggests that other sample sources, including blood culture bottles, could be used in conjunction with anchored SDA. However, increases in sensitivity are necessary in order to analyze samples that have limited target possibilities, including blood, sputum, or urine samples. Studies are under way to increase the sensitivity of the assay through optimization of amplification primer design and reagent conditions, as well as increased amplicon retention and reporter sensitivity.
Anchored SDA incorporates amplification and detection on the same platform. With the development of automated protocols for anchored SDA, the process of bacterial identification could be simplified greatly by minimizing the sample processing and transfer of amplicons needed for other molecular biology-based assays. These features make anchored SDA an ideal candidate for multiplex point-of-care applications. Anchored SDA can enhance the ability of miniaturized on-site instrumentation by decreasing the complexity of the necessary molecular biology reactions and manipulations while simultaneously allowing efficient multiplex amplification reactions and discrimination. Through integration of anchored SDA into an on-site instrument, the level of technical expertise necessary can be minimized, allowing many current clinical laboratories access to powerful molecular biology-based assays.
ACKNOWLEDGMENTS
We gratefully acknowledge the contributions of Beth Mather, Michael Heller, Richard Anderson, Bruce Wallace, and Douglas Malinowski. We thank Michael Moore and Harry J. Leonhardt for critical reading of the manuscript. We are especially grateful to Halleh Ahadian for technical assistance.
REFERENCES
- 1.Cebula T A, Payne W L, Feng P. Simultaneous identification of strains of Escherichia coli serotype O157:H7 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J Clin Microbiol. 1995;33:248–250. doi: 10.1128/jcm.33.1.248-250.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chee M, Yang R, Hubbell E, Berno A, Huang X C, Stern D, Winkler J, Lockhart D J, Morris M S, Fodor S P. Accessing genetic information with high-density DNA arrays. Science. 1996;25:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- 3.Debouck C, Goodfellow P N. DNA microarrays in drug discovery and development. Nat Genet. 1999;21:48–50. doi: 10.1038/4475. [DOI] [PubMed] [Google Scholar]
- 4.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edman C F, Raymond D E, Wu D J, Tu E, Sosnowski R G, Butler W F, Nerenberg M, Heller M J. Electric field directed nucleic acid hybridization on microchips. Nucleic Acids Res. 1997;25:4907–4914. doi: 10.1093/nar/25.24.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edman C F, Mehta P, Press R, Spargo C A, Walker G T, Nerenberg M. Pathogen analysis and genetic predisposition testing using microelectronic arrays and isothermal amplification. J Investig Med. 2000;48:93–101. [PubMed] [Google Scholar]
- 7.Fredericks D N, Relman D A. Application of polymerase chain reaction to the diagnosis of infectious disease. Clin Infect Dis. 1999;29:475–488. doi: 10.1086/598618. [DOI] [PubMed] [Google Scholar]
- 8.Gilles P N, Wu D J, Foster C B, Dillon P J, Chanock S J. Single nucleotide polymorphic discrimination by an electronic dot blot assay on semiconductor microchips. Nat Biotechnol. 1999;17:365–370. doi: 10.1038/7921. [DOI] [PubMed] [Google Scholar]
- 9.Gunzberg S T, Tornieporth N G, Riley L W. Identification of enteropathogenic Escherichia coli by PCR-based detection of the bundle-forming pilus gene. J Clin Microbiol. 1995;33:1375–1377. doi: 10.1128/jcm.33.5.1375-1377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatt C, Ward M E, Clarke I N. Analysis of the entire nucleotide sequence of the cryptic plasmid of Chlamydia trachomatis serovar L1. Evidence for involvement in DNA replication. Nucleic Acids Res. 1998;11:4053–4067. doi: 10.1093/nar/16.9.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang W M. Bacterial diversity based on type II DNA topoisomerase genes. Annu Rev Genet. 1996;30:79–107. doi: 10.1146/annurev.genet.30.1.79. [DOI] [PubMed] [Google Scholar]
- 12.Lisby G. Application of nucleic acid amplification in clinical microbiology. Mol Biotechnol. 1999;12:75–99. doi: 10.1385/MB:12:1:75. [DOI] [PubMed] [Google Scholar]
- 13.Lockhart D J, Dong H, Byrne M C, Follettie M, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, Brown E L. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 13a.National Committee for Clinical Laboratory Standards. Molecular diagnostic methods for infectious diseases. Approved guideline MM3-A. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 14.Parinov S, Barsky V, Yershov G, Kirillov E, Timofeev E, Belgovskiy A, Mirzabekov A. DNA sequencing by hybridization to microchip octa- and decanucleotides extended by stacked pentanucleotides. Nucleic Acids Res. 1996;24:2998–3004. doi: 10.1093/nar/24.15.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel R, Uhl J R, Kohner P, Hopkins M K, Cockerill F R., III Multiplex PCR detection of vanA, vanB, vanC-1, and vanC-2/3 genes in enterococci. J Clin Microbiol. 1997;35:703–707. doi: 10.1128/jcm.35.3.703-707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfaller A M, Herwaldt L A. The clinical microbiology laboratory and infection control: emerging pathogens, antimicrobial resistance, and new technology. Clin Infect Dis. 1997;25:858–870. doi: 10.1086/515557. [DOI] [PubMed] [Google Scholar]
- 17.Radtkey R, Feng L, Muralhidar M, Duhon M, Canter D, DiPierro D, Fallon S, Tu E, McElfresh K, Nerenberg M, Sosnowski R. Rapid, high fidelity analysis of simple sequence repeats on an electronically active DNA microchip. Nucleic Acids Res. 2000;28:E17. doi: 10.1093/nar/28.7.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sosnowski R G, Tu E, Butler W F, O'Connell J P, Heller M J. Rapid determination of single-base mismatch mutations in DNA hybrids by direct electric field control. Proc Natl Acad Sci USA. 1997;94:1119–1123. doi: 10.1073/pnas.94.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ubukata K, Nakagami S, Nitta A, Yamane A, Kawakami S, Sugiura M, Konno M. Rapid detection of the mecA gene in methicillin-resistant staphylococci by enzymatic detection of polymerase chain reaction products. J Clin Microbiol. 1992;30:1728–1733. doi: 10.1128/jcm.30.7.1728-1733.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vannuffel P, Gigi J, Ezzedine H, Vandercam B, Delmee M, Wauters G, Gala J-L. Specific detection of methicillin-resistant Staphylococcus species by multiplex PCR. J Clin Microbiol. 1995;33:2864–2867. doi: 10.1128/jcm.33.11.2864-2867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker G T, Little M C, Nadeau J G, Shank D D. Isothermal, in vitro amplification of DNA by a restriction enzyme/DNA polymerase system. Proc Natl Acad Sci USA. 1992;89:392–396. doi: 10.1073/pnas.89.1.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westin L, Xu X, Miller C, Wang L, Edman C F, Nerenberg M. Anchored multiplex amplification on a microelectronic chip array. Nat Biotechnol. 2000;18:199–204. doi: 10.1038/72658. [DOI] [PubMed] [Google Scholar]
- 23.Yershov G, Barsky V, Belgovskiy A, Kirillov E, Kreindlin E, Ivanov I, Parinov S, Guschin D, Drobishev A, Dubiley S, Mirzabekov A. DNA analysis and diagnostics on oligonucleotide microchips. Proc Natl Acad Sci USA. 1996;93:4913–4918. doi: 10.1073/pnas.93.10.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zirnstein G, Li Y, Swaminathan B, Angulo F. Ciprofloxacin resistance in Campylobacter jejuni isolates: detection of gyrA resistance mutations by mismatch amplification mutation assay PCR and DNA sequence analysis. J Clin Microbiol. 1999;37:3276–3280. doi: 10.1128/jcm.37.10.3276-3280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]